Abstract
Translational readthrough-promoting drugs enhance the incorporation of amino acids at stop codons and can thus bypass premature termination during protein synthesis. The polymerase (Pol) proteins of Moloney murine leukemia virus (MoMLV) are synthesized as a large Gag–Pol fusion protein, formed by the readthrough of a stop codon at the end of the gag ORF. The downstream pol ORF lacks its own start codon, and Pol protein synthesis is wholly dependent on translation of the upstream gag gene and the readthrough event for expression. Here, we explored the effects of readthrough-promoting drugs – aminoglycoside antibiotics and the small molecule ataluren – on the efficiency of readthrough of the stop codon in the context of the MoMLV genome. We showed that these compounds increased readthrough of the stop codon at the MoMLV gag–pol junction in vivo above the already high basal level and that the resulting elevated gag–pol readthrough had deleterious effects on virus replication. We also showed that readthrough efficiency could be driven to even higher levels in vitro, and that the combination of the small molecules and the RNA structure at the MoMLV stop codon could achieve extremely high readthrough efficiencies.
Introduction
Retroviruses rely on recoding – alternative interpretations of mRNA during translation – to overcome termination at a stop codon separating two ORFs and thereby control production of viral proteins. The retroviral pol gene, encoding enzymes for virus replication, lies directly downstream of the gag gene, encoding viral structural proteins. The gag ORF ends with a stop codon, and during conventional reading of the viral mRNA by the translational machinery, only the Gag polyprotein precursor is produced. The Pol proteins are synthesized only when the stop codon is misread and translation continues into the pol reading frame to produce a large Gag–Pol fusion protein (Murphy & Arlinghaus, 1978; Murphy et al., 1978; Oppermann et al., 1977). The recoding event can be mediated by one of two mechanisms in different viruses. Many retroviruses, such as avian retroviruses (Jacks & Varmus, 1985), human immunodeficiency virus type 1 (HIV-1) (Wilson et al., 1988), human T-cell leukemia virus type 1 (HTLV-1) (Nam et al., 1993) and HTLV-2 (Honigman et al., 1995) and mouse mammary tumor virus (Moore et al., 1987), and also some coronaviruses (e.g. severe acute respiratory syndrome coronavirus), utilize ribosomal frameshifting (reviewed by Plant & Dinman, 2008). A frameshift event readjusts the position of the ribosome on the mRNA to an alternative reading frame. The stop codon is thereby bypassed in the new frame, and translation continues into the downstream gene. In contrast, murine and feline leukaemia viruses use a readthrough event to synthesize the Gag–Pol precursor (Yoshinaka et al., 1985a, b); the translating ribosome remains in the same reading frame, but the UAG gag stop codon is misread as a sense codon, and the amino acid glutamine is inserted at the point of normal termination (reviewed by Levin, 1993; Rein & Levin, 1992). Recoding in retroviruses, and in many other viruses, requires the presence of a secondary structure in the viral mRNA that forms downstream of the site of recoding (Du et al., 1997; Panganiban, 1988; Parkin et al., 1992; Wills et al., 1991). This structure is often a two-stem, two-loop pseudoknot that varies in sequence and length among viruses and is utilized to induce a frameshift in some retroviruses and a readthrough in others (Chamorro et al., 1992; Chen et al., 1995; Le et al., 1991). The Moloney murine leukemia virus (MoMLV) pseudoknot has been studied extensively by mutational and biochemical means (Alam et al., 1999; Felsenstein & Goff, 1992; Feng et al., 1989a, 1992; Wills et al., 1991). Recently, RNA structures of this pseudoknot have been determined by nuclear magnetic resonance, revealing alternative conformations that control the level of the structure's activity as a cis-acting readthrough-promoting factor (Houck-Loomis et al., 2011).
Readthrough can also be induced on eukaryotic cellular mRNAs by aminoglycoside antibiotics and readthrough-promoting drugs (Burke & Mogg, 1985; Fan-Minogue & Bedwell, 2008; Manuvakhova et al., 2000; Murphy et al., 2006; Welch et al., 2007). Aminoglycosides are naturally occurring amino-modified sugars, traditionally used as antibiotics, that interfere with translation by promoting miscoding during the elongation process through interactions with the rRNA of the decoding centre (Carter et al., 2000; Fourmy et al., 1996; Ogle et al., 2001). The interaction mimics the rRNA conformations that allow incorporation of near and non-cognate amino acids, and can result in insertion of an amino acid at a stop codon (reviewed by Zaher & Green, 2009). Most aminoglycosides work preferentially on prokaryotic ribosomes due to subtle differences in rRNAs between kingdoms and to better uptake by prokaryotic cells (Hermann, 2005; Xie et al., 2010), but at higher concentrations, aminoglycosides can promote miscoding in mammalian cells. This effect is exploited in treatment of diseases caused by alleles with premature termination codons such as Duchenne's muscular dystrophy, cystic fibrosis and cancer with truncated p53 expression (reviewed by Rowe & Clancy, 2009). Treatment with aminoglycosides induces readthrough at a frequency that allows enough translation of the full-length protein to restore normal function. At high enough doses, aminoglycosides cause gross defects in eukaryotic translation, allowing their use in selection for drug resistance in tissue culture (Southern & Berg, 1982). Nephrotoxicity and non-reversible ototoxicity are serious side effects from the administration of aminoglycosides, which limit their clinical use (reviewed by Karasawa & Steyger, 2011). Screens for small molecules able to promote readthrough with reduced toxicity, however, have revealed potential alternatives (Du et al., 2008, 2009; Nudelman et al., 2009; Welch et al., 2007). One such compound, ataluren (formerly PTC124), has been shown to increase readthrough and has demonstrated therapeutic effects in clinical trials for treatment of muscular dystrophy and cystic fibrosis (Kerem et al., 2008; Sermet-Gaudelus et al., 2010; Welch et al., 2007; Wilschanski et al., 2011).
A specific ratio of Gag:Gag–Pol products is needed for proper virus replication, and thus the recoding event at the gag–pol junction of the murine leukaemia viruses is tightly regulated to occur at a frequency of about 5 % during translation. An abnormally high production of Gag–Pol protein has been shown to have negative effects on assembly and maturation of infectious particles (Felsenstein & Goff, 1988; Shehu-Xhilaga et al., 2001). Manipulating the Gag : Gag–Pol ratio downward by mutation has been shown to be strongly deleterious to virus replication, while increasing the ratio by separate expression of these proteins, by silencing expression of the termination factors eRF1 or -3, or with aminoglycosides, was well tolerated in one study (Csibra et al., 2014).
Here, we examined the effects of readthrough-promoting drugs on eukaryotic recoding in the context of retroviral readthrough sequences. We investigated the ability of aminoglycosides and the small molecule ataluren to increase readthrough of the MoMLV gag–pol junction beyond the normal levels of recoding induced by the pseudoknot. We showed that pseudoknot-mediated readthrough can be increased by certain drugs both in vivo and in vitro, thereby demonstrating that the normal efficiency of readthrough induced by the pseudoknot is not a maximal limiting level. We also showed that an abnormal increase in readthrough can modestly inhibit virus replication.
Results
Aminoglycosides increase readthrough of the stop codon at the MoMLV gag–pol junction
We tested the effect of several aminoglycosides and ataluren on readthrough of the stop codon at the MoMLV gag–pol junction in human 293A cells. We transfected cells with a dual-luciferase reporter DNA (MoMLV-PK) in which the MoMLV gag–pol junction region containing the gag stop codon (UAG), pseudoknot and relevant flanking nucleotides was placed downstream from the coding sequences for Renilla luciferase and upstream of the coding sequences for firefly luciferase (Green et al., 2012). The efficiency of readthrough was then determined by measuring the ratio of firefly luciferase to Renilla luciferase activity. To normalize levels and to control for general effects on translation not related to readthrough, we performed parallel transfections of a control plasmid in which the UAG stop codon had been changed to CAG, directing maximal readthrough. The percentage readthrough was calculated as the ratio of firefly : Renilla luciferase in the MoMLV WT (containing the stop codon) context divided by the ratio of firefly : Renilla luciferase expressed by the no-stop control plasmid, multiplied by 100. Finally, the fold increase in these values upon addition of drug was determined in comparison with the value of the no-drug control, which was set to unity. We note that this readout of the efficiency of stop codon readthrough yields experimental values with a very tight distribution and very small errors across repeat assays (Houck-Loomis et al., 2011; Green et al., 2012).
We initially tested G418 at several concentrations in 293A cells and found that readthrough was consistently and significantly increased in a dose-dependent manner (Fig. 1a, b). We tested several other aminoglycosides: paromomycin (humetin), which is used to treat protozoan infections such as leishmaniasis (Banerjee et al., 2011; Musa et al., 2010) and cryptosporidiosis (reviewed by Cabada & White, 2010); gentamicin, a less toxic derivative of G418 that is widely used therapeutically; amikacin (amikin), used in the treatment of enterobacteria (Al-Tawfiq et al., 2009) and some tuberculosis infections (Caminero et al., 2010; Filippini et al., 2010); and the non-aminoglycoside ataluren. G418 had the greatest effect on readthrough, with changes of more than threefold at the highest concentrations used (Fig. 1a). Ataluren and paromomycin increased the occurrence of readthrough over twofold. Gentamicin had a slight effect on increasing readthrough in some experiments, but the effects were not clearly dose dependent or consistently observed in all experiments. Amikacin treatment also resulted in only a slight increase in readthrough.
Fig. 1.
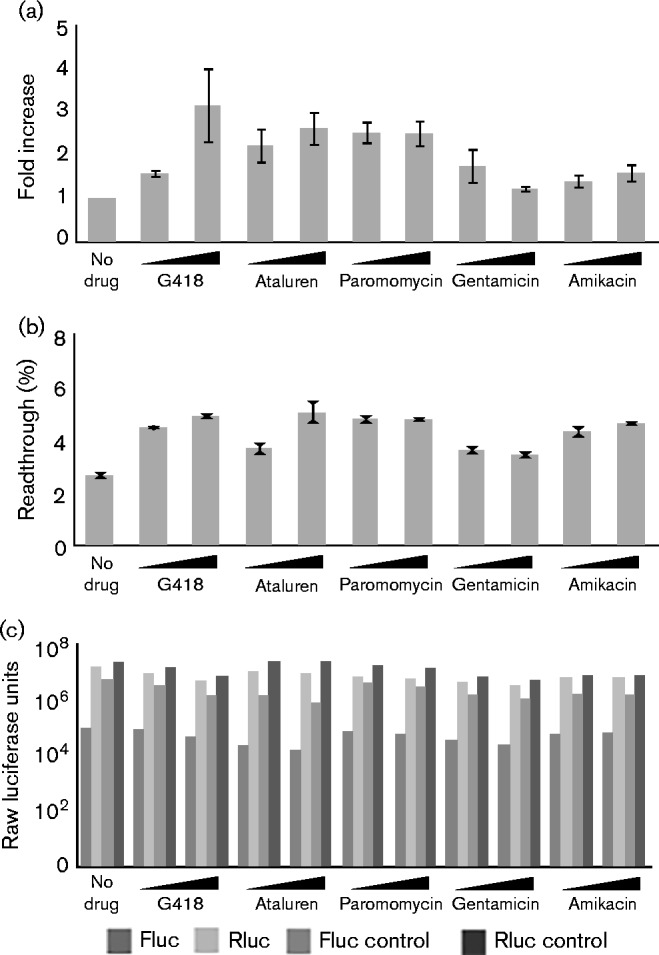
Effects of aminoglycosides and ataluren on readthrough at the MoMLV gag–pol junction. (a) Fold increase in readthrough at the MoMLV gag–pol junction in 293A cells induced by various small molecules, relative to the no-drug control. 293A cells were transfected with MoMLV-PK dual-luciferase reporter, and treated with increasing concentrations of aminoglycosides or ataluren as described in Methods. Readthrough efficiency was determined from the ratio of luciferase activities relative to the ratio of a control construct lacking the stop codon, and then expressed relative to the no-drug control, which was set to unity. The graph shows results from three trials per condition. Error bars represent sem of mean fold changes in the various trials (n = 3). Drug concentrations were: G418, 0.25 and 0.5 mg ml− 1; ataluren, 0.5 and 1 μg ml− 1; paromomycin, 0.5 and 1 μg ml− 1; gentamicin, 25 and 50 μg ml− 1; amikacin, 1.5 and 3 mg ml− 1. (b) Absolute levels of readthrough efficiency in drug-treated 293A cells relative to the control construct lacking a stop codon. The graph shows a representative single experiment of 293A cells transfected with MoMLV-PK dual-luciferase reporter and treated with drugs. Error bars represent sd among triplicate luciferase readings in this experiment (n = 3). (c) Raw luciferase values in drug-treated 293A cells for Fluc and Rluc constructs with the stop codon, and for control constructs without the stop codon, as indicated, used to generate the data in (a) and (b). The graph shows the mean luciferase values of triplicate readings of each condition (n = 3).
Ataluren has been reported to stabilize firefly luciferase and prevent its degradation, and so might cause higher signals by increasing the luciferase half-life rather than by effects on translation (Auld et al., 2009, 2010). We observed no increase in firefly luciferase levels expressed by the control plasmid in ataluren-treated versus the no-drug control in our trials (Fig. 1c) and therefore concluded that increases in firefly luciferase signal in the presence of ataluren in our experimental system could be attributed to increased translation and not stabilization of the reporter protein.
G418 at higher concentrations had some global impact on translation, as all luciferase signals decreased slightly (Fig. 1c; compare Rluc and Rluc-control levels for G418 with the corresponding no-drug values). Gentamicin, although considered less toxic than G418, also decreased overall luciferase levels, with little effect on readthrough. Similarly, amikacin, which is well tolerated as a therapeutic antibiotic, also decreased overall translation. These decreases in luciferase signals may be due to a general inhibition of translation, or to an increase in misreading and production of non-functional luciferase proteins. Global effects on cell metabolism or viability caused by drug toxicity may also be a factor, particularly in the case of G418.
To verify that the increase in readthrough was directly attributable to the presence of the active forms of the readthrough-promoting drugs, we repeated the assays in neomycin (neo)-resistant 293T cells (Fig. 2). 293T cells contain the bacterial resistance gene neo, which encodes an aminoglycoside 3′-phosphotransferase, an enzyme that modifies the antibiotics and renders them inactive. All of the readthrough-promoting activities of the aminoglycosides were reduced in these cells. G418 demonstrated a significantly reduced ability to increase readthrough of the MoMLV gag–pol junction as expected, and paromomycin, gentamicin and amikacin did not show any increase in readthrough in these cells. However, ataluren, which has a molecular structure unrelated to that of aminoglycosides and for which the mechanism of action is not precisely known, retained its readthrough-promoting activity. This result suggested that the increase in readthrough by the aminoglycosides is sensitive to neo-induced phosphorylation and that the mechanism of action of ataluren is distinct from that of the aminoglycosides.
Fig. 2.
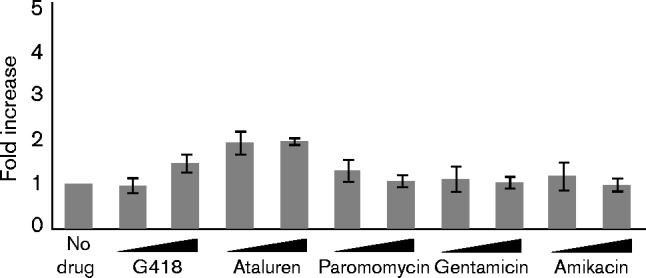
Effects of aminoglycosides and ataluren on readthrough at the MoMLV gag–pol junction in neo-resistant 293T cells. 293T cells were transfected with the MoMLV-PK dual-luciferase reporter and treated with aminoglycosides or ataluren at two concentrations, as detailed in Fig. 1. The graph shows a representative experiment showing the fold increase of readthrough at the MoMLV gag–pol junction in neo-resistant 293T cells. Error bars represent sem of average fold changes among triplicate readings in this experiment (n = 3).
Aminoglycosides and ataluren decrease virus replication
We next examined the effect of the drug-induced increases in readthrough on the production of infectious virus particles. To measure virus production, we transfected 293A cells with a mixture of three DNAs: a plasmid expressing an MoMLV vector genome expressing the firefly luciferase gene, a plasmid expressing the vesicular stomatitis virus envelope glycoprotein (VSV-G) and a plasmid containing the complete MoMLV gag–pol genes. The cells were then treated with readthrough-promoting drugs to alter the Gag : Gag–Pol ratio. To test for effects of the drugs on transfection efficiency or on the health or viability of the cells, a fourth DNA, encoding Renilla luciferase, was included with the transfecting DNAs, and levels of Renilla luciferase were determined. Culture medium from the virus-producing cells was collected 48 h after transfection and serial dilutions of these virus harvests were used to infect naïve 293A cells. Expression of firefly luciferase in the infected-cell lysates was measured to gauge the amount of infectious virus produced by the drug-treated cells (Fig. 3a). We note that, although cells infected with reporter virus are exposed to low levels of residual drug in the medium, there is no recoding signal in the reporter vector, and firefly luciferase expression is independent of readthrough.
Fig. 3.
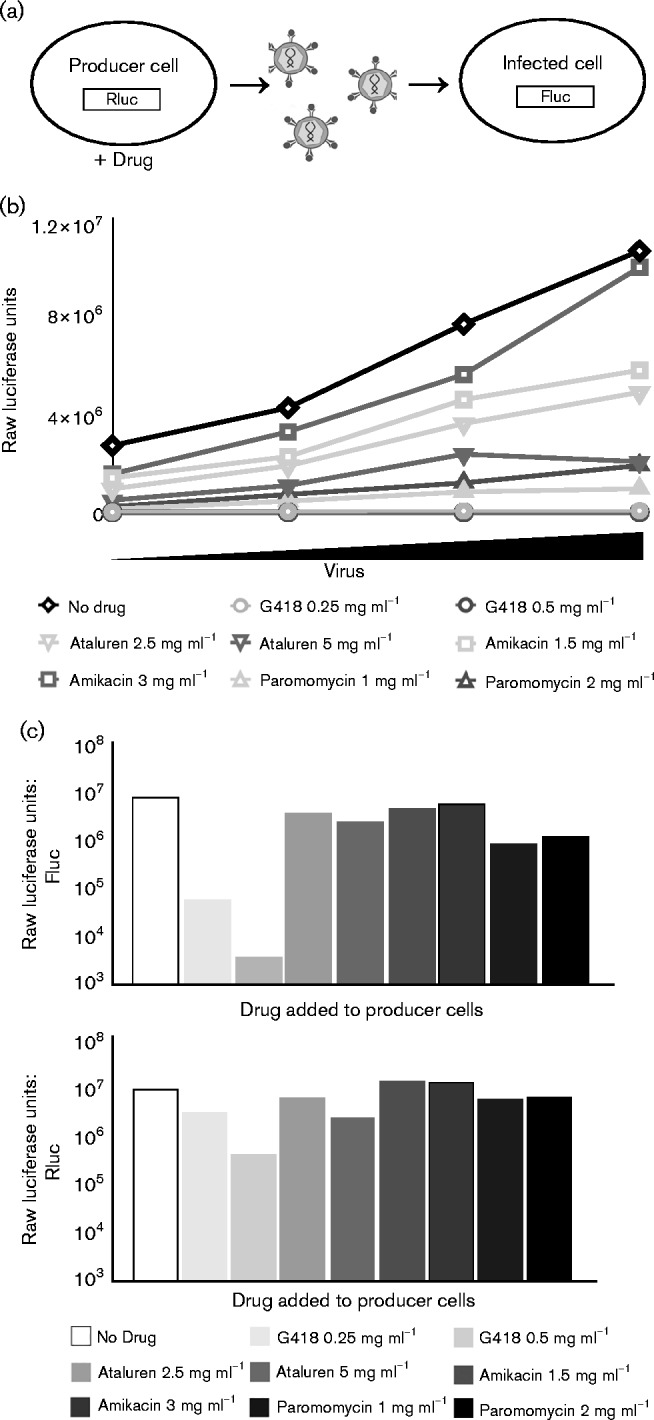
Effects of aminoglycosides and ataluren on virus replication. (a) Schematic of the transducing virus assay. Producer cells were generated by transfection with plasmids expressing MoMLV gag–pol and VSV-G env, a viral vector carrying firefly luciferase as a reporter gene, and a DNA encoding Renilla luciferase. Producer cells were treated with readthrough-promoting drugs at the indicated concentrations, and virus was harvested from producer cells and used to infect naïve cells. Firefly luciferase activity was measured in infected cells as an indicator of production of virus. Renilla luciferase activity was measured in producer cells as an indicator of overall translational competence. (b) Firefly luciferase activity in infected cells, after infection with virus collected from producer cells treated with the indicated drugs. Cells were infected with increasing concentrations of virus collected from drug-treated cells (serial dilutions of 1 : 16, 1 : 8, 1 : 4, 1 : 2, titres increasing from left to right). Values are the means of duplicate assays (n = 2). (c) Example of infections with the 1 : 8 virus dilution used in (b). Upper panel: virus production as measured by expression of firefly luciferase in infected cells. Lower panel: translational competence of producer cells treated with drugs as measured by Renilla luciferase activity.
Four compounds – G418, ataluren, amikacin and paromomycin – were tested for their effects on virus production. Cells treated with G418 produced very little virus (Fig. 3b), with a reduction of over 4 logs in transduced levels of firefly luciferase compared with virus produced in untreated cells (Fig. 3c). However, Renilla luciferase expression in the transfected producer cells was also profoundly decreased, suggesting that the effect on virus was probably attributable to general drug toxicity on the transfected cells. Paromomycin treatment had the next most significant effect on production of virus, with transduction of firefly luciferase into infected cells reduced by approximately 10-fold. Renilla luciferase expression in the drug-treated producer cells was only slightly reduced from the no-drug control, suggesting very limited toxicity. Ataluren treatment caused only a very small decrease in virus production and transduction of firefly luciferase activity in infected cells, and minimal effects on Renilla levels in the transfected cells. Interestingly, despite amikacin's modest but significant ability to increase readthrough of the MoMLV gag–pol junction dual-luciferase reporter, the drug showed a negligible effect on virus production and no change, or even a modest increase, in Renilla signal in the producer cells.
Aminoglycosides increase MoMLV readthrough in vitro
To assess the effects of the readthrough-promoting drugs without the limitations of drug uptake and toxicity in live cells, we performed coupled in vitro transcription and translation (IVT) reactions that expressed the MoMLV gag–pol junction dual-luciferase reporter. We prepared IVT reactions with reporter constructs and increasing concentrations of aminoglycosides, and measured luciferase levels generated during the reactions as before. We tested G418, paromomycin and ataluren because of their enhancement of readthrough in vivo. G418 caused a dramatic enhancement of MoMLV readthrough in a dose-dependent manner (Fig. 4a, b). G418 had little effect on readthrough efficiency at the lowest concentrations tested (5 and 20 ng ml− 1), and a small increase in readthrough with no global effect on translation, compared with the no-drug control, at moderate concentrations. Readthrough was increased twofold at 300 ng ml− 1 but with some reduction in control reporter luciferase levels (Fig. 4c). This trend of increased readthrough with decreased control luciferase expression continued with consecutively higher G418 levels. The highest final concentration of 20 μg ml− 1 induced very high levels of readthrough, but the overall translation efficiency was profoundly impaired at this drug level. We concluded that, although G418 increases readthrough in a dose-dependent manner in vitro, it also negatively affects translation of all mRNAs.
Fig. 4.
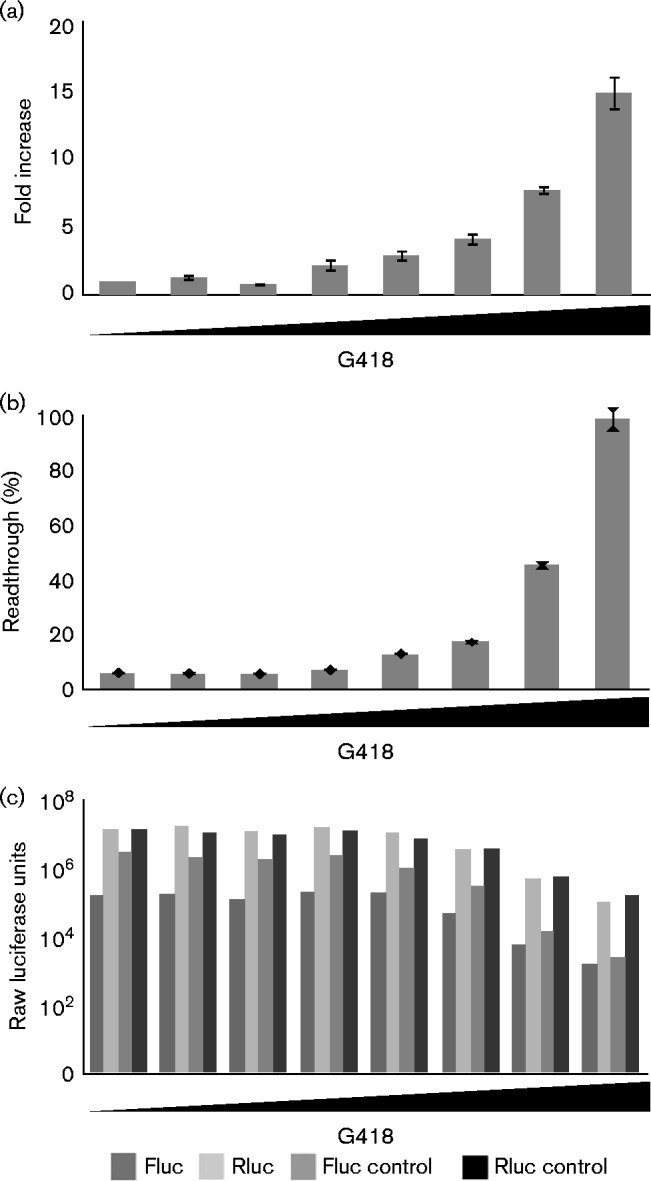
Effects of G418 on readthrough of the MoMLV gag–pol junction in vitro. (a) Mean fold increase above the no-drug control of readthrough of the MoMLV gag–pol junction during in vitro translation reaction with increasing concentrations of G418 [final concentrations shown are 0 (no drug), 5 ng ml− 1, 20 ng ml− 1, 80 ng ml− 1, 300 ng ml− 1, 1.25 μg ml− 1, 5 μg ml− 1 and 20 μg ml− 1], with means calculated from three trials at each concentration (n = 3). Error bars represent sem of average fold changes in the various trials. (b) Readthrough levels of the MoMLV gag–pol junction during in vitro translation reaction with increasing concentrations of G418; values are means of triplicate experiments (n = 3). Error bars represent SEM of average fold changes in the various trials. (c) Raw Fluc and Rluc luciferase values in IVT reactions performed with construct with a stop codon, and for control construct without astop codon, as indicated, used to generate the data in panels (a) and (b). The graph shows mean luciferase values of triplicate readings of each condition (n = 3).
Paromomycin also caused a dose-dependent increase in readthrough of the MoMLV gag–pol reporter in vitro. As with G418, the readthrough efficiencies attained in vitro were considerably higher than the maximum measured in vivo in 293A cells, with the highest levels of paromomycin inducing a sixfold increase (Fig. 5a), which corresponded to over 40 % readthrough efficiency (Fig. 5b). In contrast to the findings with G418, the raw levels of luciferase remained relatively stable, even at high concentrations of paromomycin, indicating that general translation was not impaired (Fig. 5c).
Fig. 5.
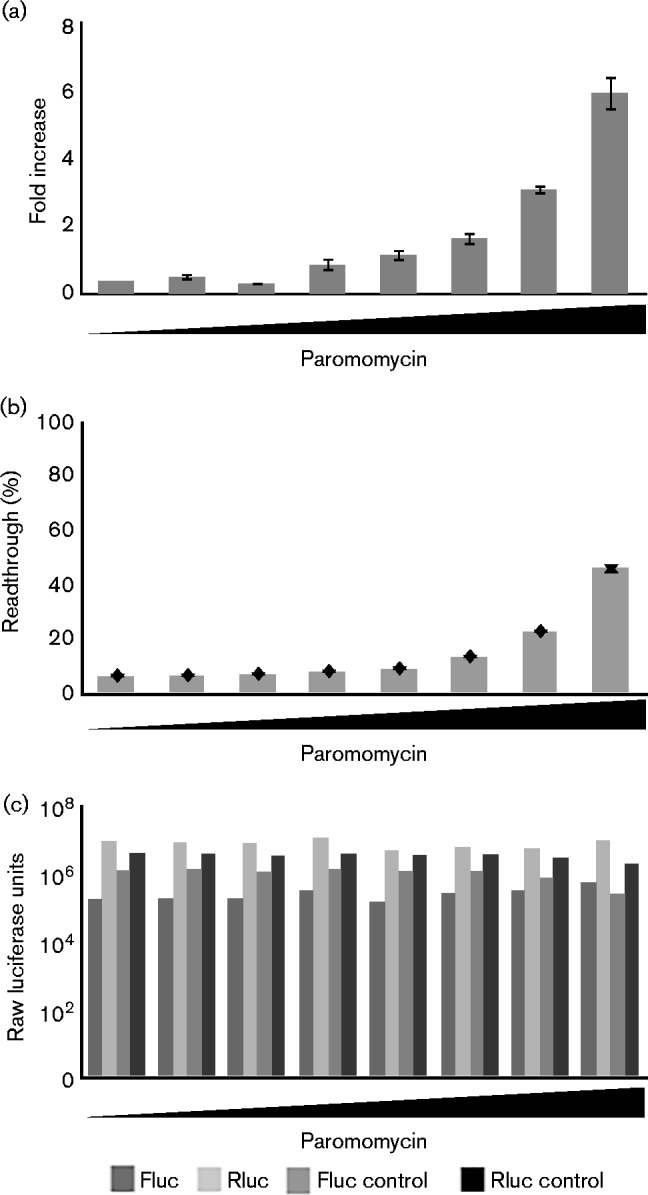
Effects of paromomycin on readthrough of the MoMLV gag–pol junction in vitro. (a) Mean fold increase in readthrough of the MoMLV gag–pol junction during in vitro translation reactions with increasing concentrations of paromomycin [final concentrations: 0 (no drug), 5 ng ml− 1, 20 ng ml− 1, 80 ng ml− 1, 300 ng ml− 1, 1.25 μg ml− 1, 5 μg ml− 1 and 20 μg ml− 1]; means were calculated from at least three trials per concentration. Error bars represent standard error of average fold changes in the various trials. (b) Readthrough levels of MoMLV gag–pol junction during in vitro translation reactions with increasing concentrations of paromomycin; representative trial shown; values are means of triplicate experiments (n = 3). Error bars represent sem of average fold changes in the various trials. (c) Raw Fluc and Rluc luciferase values in IVT reactions performed with construct with a stop codon, and for control construct without a stop codon, as indicated, used to generate the data in (a) and (b). The graph shows mean luciferase values of triplicate readings of each condition (n = 3).
We also tested ataluren for effects on readthrough in vitro but were unable to observe any significant increases in readthrough (data not shown). These results suggested that ataluren may act indirectly in vivo or require cellular components that are missing from the in vitro reactions for its activity.
Aminoglycosides increase readthrough of all three stop codons, of a hypoactive pseudoknot mutant and without viral pseudoknot sequences
We next examined the effects of G418 and paromomycin on a number of sequence variants of the MoMLV gag–pol junction. We tested luciferase constructs in which the WT UAG terminator was replaced by UGA and UAA stop codons. Viruses carrying these alternative stop codons are known to express Gag–Pol, with the insertion of distinct amino acids in the readthrough protein (Feng et al., 1989b, 1990), and are at least partially replication competent (Jones et al., 1989; Odawara et al., 1991). G418 and paromomycin both increased readthrough with both alternative stop codons (Fig. 6a, b). Readthrough of UGA had a high basal level without drug, and was increased by the drugs to a very high absolute level, with a maximal increase of about fivefold. UGA has been observed to have a higher incidence of basal readthrough in non-viral mRNAs and appears to be more sensitive to drug-induced enhancement of readthrough than UAA. Readthrough of UAA, which is resistant to readthrough under conditions of normal translation (Manuvakhova et al., 2000), had a low basal level and increased with both drugs, with a high fold increase of about 20-fold. These results indicated that, in the context of the MoMLV gag–pol junction, the same stop codon drug effects were observed as in studies of non-pseudoknot drug-mediated readthrough events.
Fig. 6.
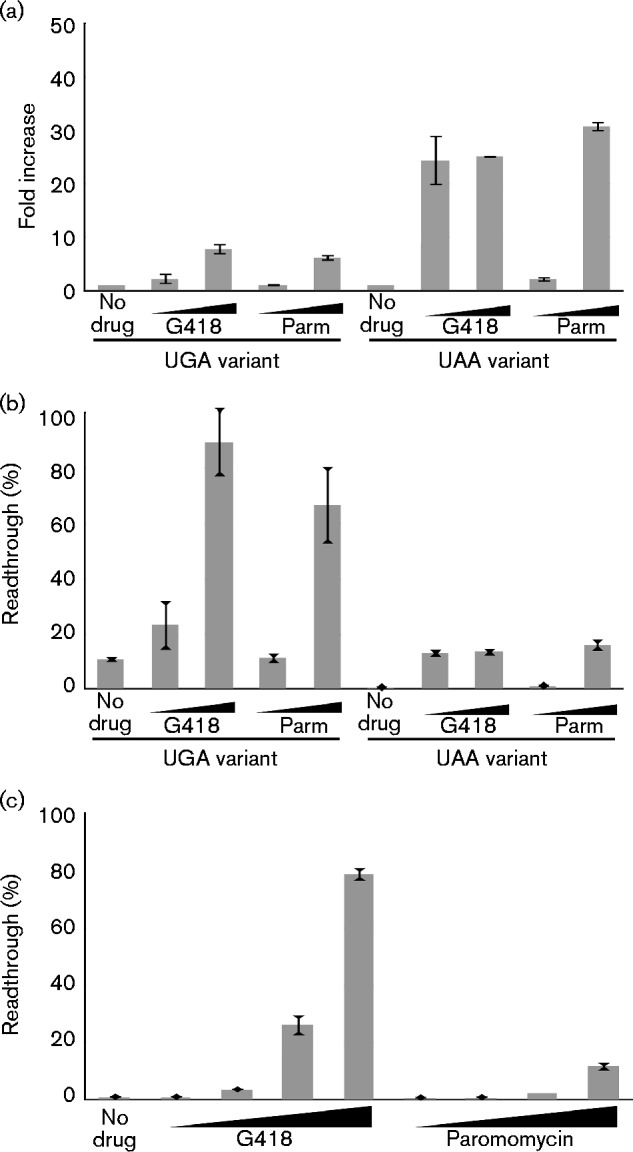
Effects of G418 and paromomycin on MoMLV gag–pol junction mutants. (a) Mean fold increase in readthrough frequencies of MoMLV gag–pol junction variants with UGA (left) and UAA (right) stop codons replacing the WT UAG stop codon during in vitro translation reactions containing two concentrations of G418 (80 and 300 ng ml− 1) or paromomycin (Parm; 1.5 and 5 μg ml− 1), expressed relative to the no-drug control, which was set to unity. Error bars represent sem of duplicate luciferase readings (n = 2). (b) Readthrough frequencies used in generating graph shown in (a). Error bars represent sem of duplicate luciferase readings (n = 2). (c) Readthrough frequencies of MoMLV gag–pol hypoactive pseudoknot mutant G11C during in vitro translation reactions with increasing concentrations of G418 or paromomycin (5 ng ml− 1, 80 ng ml− 1, 1.25 μg ml− 1 and 20 μg ml− 1). Error bars represent sem of triplicate luciferase readings (n = 3).
We additionally tested the effects of G418 and paromomycin on a pseudoknot mutant with impaired readthrough-promoting ability. The G11C pseudoknot mutant contains a change of the 11th nt after the gag stop codon, which disrupts base pairing in the first stem of the pseudoknot structure. Viruses with this mutation do not replicate (Felsenstein & Goff, 1992), probably due to impaired readthrough. The hypoactive G11C luciferase reporter showed enhanced readthrough in IVT reactions supplemented with increasing amounts of G418 (Fig. 6c). Paromomycin was also able to increase readthrough of this reporter, albeit with more modest fold changes. Notably, both drugs increased the readthrough of the hypoactive pseudoknot mutant to a lower level than they did with the WT pseudoknot. These results indicated that deficits in the readthrough-promoting activity of the G11C mutant pseudoknot are not fully overcome by the action of readthrough-promoting drugs.
To determine whether the paromomycin induced a specific or distinctive enhancement of readthrough in the presence of the pseudoknot structure, we directly compared the MoMLV pseudoknot and a no-pseudoknot reporter in the IVT system. The experimental premature termination codon (PTC) reporter contained the same UAG-G stop codon sequence as our MoMLV gag–pol junction dual-luciferase reporter, but the pseudoknot sequence was replaced with a portion of the collagen ORF, which is not known to contain any secondary structure; a matched control plasmid contained a CAG-G sequence to allow maximum expression of the luciferases to use as normalization. Both PTC and MoMLV reporters showed an increase in readthrough with paromomycin. The basal readthrough level of the PTC reporter was very low (0.4 %), and increased 16-fold to 6.8 % with the highest concentration of paromomycin (Fig. 7). This was a greater fold increase in readthrough for the PTC reporter than for the MoMLV gag–pol junction reporter, although the maximal readthrough frequency of the MoMLV reporter was much higher at almost 50 %. This result suggests that paromomycin is highly active even without the pseudoknot.
Fig. 7.
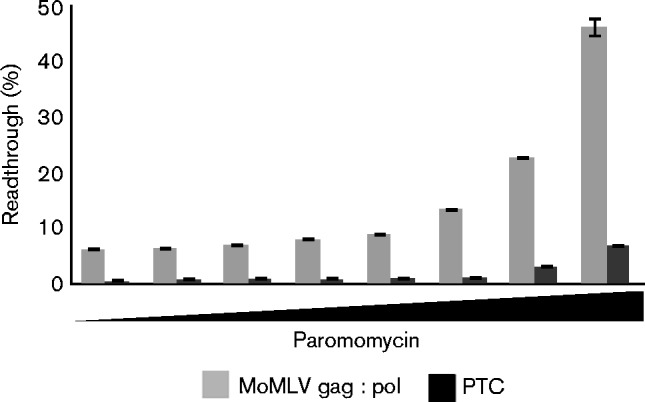
Effects of paromomycin on readthrough of a no-pseudoknot PTC and WT MoMLV gag–pol sequence. The readthrough frequency for a dual-luciferase reporter with a no-pseudoknot PTC and for the WT MoMLV reporter were determined in the presence of increasing concentrations of paromomycin (no drug; 5 ng ml− 1, 20 ng ml− 1, 80 ng ml− 1, 300 ng ml− 1, 1.25 μg ml− 1, 5 μg ml− 1 and 20 μg ml− 1). Error bars indicate sd of means from representative experiments with each reporter (n = 3).
Discussion
We examined the effect of aminoglycoside antibiotics and the small molecule ataluren on the readthrough efficiency of the MoMLV gag–pol junction. We showed that exposure of mammalian cells to specific aminoglycosides resulted in a dose-dependent increase in the already high basal readthrough levels at the MoMLV junction as measured by our dual-luciferase reporter system. The various drugs demonstrated distinctive behaviours. G418 and paromomycin exhibited the greatest effects on readthrough in vivo, while amikacin and gentamicin demonstrated little effect on MoMLV readthrough in this setting. Exposure of cells to ataluren also increased readthrough of the MoMLV gag–pol reporter. It has been reported that ataluren can interact reversibly with firefly luciferase to stabilize it and prevent its degradation; thus, high levels of readthrough can be mimicked by an accumulation of the reporter over the course of the assay (Auld et al., 2009, 2010), but this process did not seem to be occurring in our experimental conditions. We also showed that the aminoglycoside effects on readthrough were not observed in 293T ‘neo’-resistant cells. Therefore, unsurprisingly, inactivation of the drugs by modifying enzymes in drug-resistant cells prevented their ability to enhance readthrough.
The detailed mechanisms of action of the aminoglycosides on readthrough are not clear. Most likely, they involve interactions with the ribosomal A site of the elongating ribosome, subtly altering the function of the decoding machinery and affecting the ratio of binding of the termination factors versus tRNA at the time of recognition of the stop codon. The increase in readthrough probably does not involve interactions of the drug with secondary structures in the viral mRNA, such as the pseudoknot itself, although this possibility cannot be strictly ruled out. The aminoglycosides do interact specifically with at least one viral RNA, the RNA dimerization elements of the HIV-1 genome (Ennifar et al., 2003), and can prevent structural rearrangements required for the complete dimerization of the two copies of the HIV RNA genome during virion assembly (Bernacchi et al., 2007; Ennifar et al., 2006). This structure has no known role in recoding, however, and the MoMLV pseudoknot has no obvious structural similarity to the HIV-1 RNA dimerization region or to the ribosomal A site.
We showed that exposure of virus-producing cells to readthrough-promoting drugs can affect virus production. The titre of reporter virus was reduced in all cells infected with virus produced in the presence of drugs. Virus from G418-treated cells was reduced from 2 to 4 logs compared with the no-drug control. However, G418 was clearly toxic: Renilla luciferase levels in producer cells exposed to G418 were reduced in all trials, and a viability assay of G418-treated cells confirmed that viability was reduced as G418 concentration and readthrough efficiency increased (data not shown). Therefore, we were unable to determine whether it was the readthrough-promoting action of G418 or its general toxicity to the producer cells that was responsible for the decreased virus yield. Paromomycin had the greatest effect on virus replication with the least disruption of translation in producer cells (measured by Renilla luciferase expression), with up to a 10-fold reduction in virus yield, suggesting that this compound is the most promising lead for an effective antiviral drug. The structural differences responsible for the differential toxicity are not clear. Both G418 and paromomycin have an OH at the 6′ position of ring I, which may be responsible for facilitating an interaction with the 18S rRNA that favours readthrough. However, the structure of an additional sugar (ring III) is different between the two, and is attached at a different position on ring II. The combination of ring III structure and linkage may contribute to the toxicity of G418 and/or higher tolerance of cells for paromomycin.
We also examined the effects of G418 and paromomycin on readthrough in an in vitro translation system. We found that G418 promoted high levels of readthrough but that the increases were associated with overall translational inhibition, consistent with the toxicity in vivo. Paromomycin caused moderate increases in readthrough frequency in a dose-dependent manner with less negative impact on overall translation. G418 and paromomycin also stimulated readthrough of alternative stop codons in the MoMLV gag–pol context, and of a hypoactive pseudoknot mutant. We were surprised to observe that ataluren showed no effects in our in vitro reactions. Its activity may require components in cells but missing from the in vitro reactions, or present at concentrations too low to contribute. The readouts used here could in principle provide an assay for the identification of factors that might recreate the ataluren effects seen in vivo.
A major conclusion from these experiments is that readthrough promoted by the MoMLV pseudoknot can be pushed beyond normal levels upon exposure to certain aminoglycosides. Thus, the results showed that the mechanism by which the MoMLV pseudoknot stimulates readthrough does not normally operate at maximal efficiency. These two stimulators of readthrough can act together and probably act independently. The results suggest that there may be more than one point of regulation that can be altered to promote miscoding. Finally, we note that our limited survey of aminoglycoside drug effects on MoMLV readthrough suggests that it may well be possible to usefully alter the rate of miscoding to reduce virus replication without cytotoxicity. The effects of paromomycin and ataluren are encouraging, indicating that similar drugs might have useful antiviral activity. Although any such compounds that act through this mechanism are not likely to be as potent as direct inhibitors of the viral reverse transcriptase, protease or integrase, the distinct target of their action would suggest that they would retain activity towards the more common drug-resistant viral mutants.
Methods
Tissue culture
293A and 293T cells were grown in Dulbecco's Modified Eagle's Medium with 10 % FBS supplemented with l-glutamine plus 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were maintained in a 5 % CO2 atmosphere at 37 °C, and were passaged at approximately 70 % confluence.
Transfection and dosing of cell cultures
Cells were plated at densities of 250 000 cells per well in six-well plates the day before transfection. Cells were transfected with 1 μg experimental or control reporter DNA using Fugene 6 Transfection reagent (Roche) at a ratio of 3 μl Fugene : 1 μg DNA; all transfections were adjusted with empty vector to contain the same total amount of DNA per well per experiment. At 2 h post-transfection, the medium was replaced and supplemented with aminoglycosides or ataluren. Drugs were used at the following final concentrations: G418, 0.25 and 0.5 mg ml− 1; ataluren, 0.5 and 1 μg ml− 1; paromomycin, 0.5 and 1.0 μg ml− 1; amikacin, 1.5 and 3.0 mg ml− 1; gentamicin, 25 and 50 μg ml− 1. Cells were harvested for dual-luciferase analysis at 24 h post-transfection by lysis with 150 μl Promega Passive Lysis buffer per well.
Dual-luciferase assay
The dual-luciferase reporter system used here was essentially as originally described (Grentzmann et al., 1998), and made use of MoMLV-based reporter DNA and assays utilized previously (Green et al., 2012). Cell lysates from transfections (20 μl from each well) were transferred to opaque 96-well plates in triplicate. Dual-luciferase assays were performed using a Promega Dual Luciferase kit with buffers diluted 1 : 2 with dH2O. Assays were read on an Omega plate reader for a 24 s interval per well, with readings taken every half second for a total of 48 readings. One hundred microlitres of firefly luciferase reagent was dispensed for the first 12 s. Firefly signal was quenched by Renilla luciferase reagent addition at 12.5 s. Values from each interval of firefly (fluc) and Renilla (rluc) luciferase were summed in the Omega Mars Data Analysis program and exported to a spreadsheet program (Numbers) for analysis. The percentage recoding was calculated as [(fluc WT/rluc WT) × (rluc control/fluc control)] × 100, with WT representing signals from expression of plasmids containing the native recoding sequence and control representing signals from plasmids designed to eliminate recoding elements to allow maximal expression of both luciferases.
Transducing virus assay
293A cells were plated at 250 000 cells per well in six-well plates and transfected the next day with 0.5 μg of a mix of three plasmids encoding components of a recombinant luciferase reporter virus [MoMLV gag–pol (pCMV-intron), firefly luciferase with the MoMLV packaging signal (pFbluc) and VSV-G envelope (pMDG)], along with DNA encoding Renilla luciferase and the human thymidine kinase reporter (pHRLTK) as a measure of translational health in the producer cell. At 2 h post-transfection, the medium was replaced and supplemented with readthrough-promoting drugs. At 24 h post-transfection, virus-containing supernatants were collected from producer cells and used to infect naïve 293A cells at 1 : 2, 1 : 4, 1 : 8 and 1 : 16 dilutions. Producer cells were assayed for Renilla luciferase activity after virus collection. Infected cells were assayed for firefly luciferase activity at 24 h post-infection.
Assay for enhanced readthrough during translation in vitro
Drug effects on readthrough occurring during in vitro translation reactions were determined by assaying expression of dual-luciferase reporter plasmids using Promega T7 TnT (Transcription and Translation) reagents. Reactions were assembled on ice with the following components: 6.25 μl TnT lysate, 0.5 μl TnT reaction buffer, 0.25 μl T7 polymerase, 0.25 complete amino acid mix, 0.25 μl reporter plasmid at 1 μg μl− 1, 0.5 μl drug solution and 4.5 μl nuclease-free H2O. Reactions were incubated for 2 h at 30 °C. Reaction mixes were diluted with passive lysis buffer, and assayed for Renilla and firefly luciferase activity as described above for cell lysates.
Acknowledgements
This work was supported by the National Cancer Institute (grant no. R01 CA 30488). S. P. G. is an Investigator of the Howard Hughes Medical Institute. We thank Joseph Colacino of PTC Therapeutics for comments on the manuscript and for generously providing PTC124.
References
- Al-Tawfiq J.A., Antony A., Abed M.S. (2009). Antimicrobial resistance rates of Enterobacter spp.: a seven-year surveillance study Med Princ Pract 18100–104 10.1159/000189806. [DOI] [PubMed] [Google Scholar]
- Alam S.L., Wills N.M., Ingram J.A., Atkins J.F., Gesteland R.F. (1999). Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus J Mol Biol 288837–852 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- Auld D.S., Thorne N., Maguire W.F., Inglese J. (2009). Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression Proc Natl Acad Sci U S A 1063585–3590 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld D.S., Lovell S., Thorne N., Lea W.A., Maloney D.J., Shen M., Rai G., Battaile K.P., Thomas C.J., other authors (2010). Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124 Proc Natl Acad Sci U S A 1074878–4883 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., De M., Ali N. (2011). Combination therapy with paromomycin-associated stearylamine-bearing liposomes cures experimental visceral leishmaniasis through Th1-biased immunomodulation Antimicrob Agents Chemother 551661–1670 10.1128/AAC.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S., Freisz S., Maechling C., Spiess B., Marquet R., Dumas P., Ennifar E. (2007). Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion Nucleic Acids Res 357128–7139 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J.F., Mogg A.E. (1985). Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin Nucleic Acids Res 136265–6272 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabada M.M., White A.C., Jr (2010). Treatment of cryptosporidiosis: do we know what we think we know? Curr Opin Infect Dis 23494–499 10.1097/QCO.0b013e32833de052. [DOI] [PubMed] [Google Scholar]
- Caminero J.A., Sotgiu G., Zumla A., Migliori G.B. (2010). Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis Lancet Infect Dis 10621–629 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. (2000). Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics Nature 407340–348 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H.E. (1992). An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA Proc Natl Acad Sci U S A 89713–717 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr, Varmus H.E. (1995). Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting EMBO J 14842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibra E., Brierley I., Irigoyen N. (2014). Modulation of stop codon read-through efficiency and its effect on the replication of murine leukemia virus J Virol 8810364–10376 10.1128/JVI.00898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Holland J.A., Hansen M.R., Giedroc D.P., Hoffman D.W. (1997). Base-pairings within the RNA pseudoknot associated with the simian retrovirus-1 gag–pro frameshift site J Mol Biol 270464–470 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- Du M., Liu X., Welch E.M., Hirawat S., Peltz S.W., Bedwell D.M. (2008). PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model Proc Natl Acad Sci U S A 1052064–2069 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Damoiseaux R., Nahas S., Gao K., Hu H., Pollard J.M., Goldstine J., Jung M.E., Henning S.M., other authors (2009). Nonaminoglycoside compounds induce readthrough of nonsense mutations J Exp Med 2062285–2297 10.1084/jem.20081940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar E., Paillart J.C., Marquet R., Ehresmann B., Ehresmann C., Dumas P., Walter P. (2003). HIV-1 RNA dimerization initiation site is structurally similar to the ribosomal A site and binds aminoglycoside antibiotics J Biol Chem 2782723–2730 10.1074/jbc.M205726200. [DOI] [PubMed] [Google Scholar]
- Ennifar E., Paillart J.C., Bodlenner A., Walter P., Weibel J.M., Aubertin A.M., Pale P., Dumas P., Marquet R. (2006). Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell Nucleic Acids Res 342328–2339 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan-Minogue H., Bedwell D.M. (2008). Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity RNA 14148–157 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K.M., Goff S.P. (1988). Expression of the gag–pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing J Virol 622179–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K.M., Goff S.P. (1992). Mutational analysis of the gag–pol junction of Moloney murine leukemia virus: requirements for expression of the gag–pol fusion protein J Virol 666601–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Hatfield D.L., Rein A., Levin J.G. (1989a). Translational readthrough of the murine leukemia virus gag gene amber codon does not require virus-induced alteration of tRNA J Virol 632405–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Levin J.G., Hatfield D.L., Schaefer T.S., Gorelick R.J., Rein A. (1989b). Suppression of UAA and UGA termination codons in mutant murine leukemia viruses J Virol 632870–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Copeland T.D., Oroszlan S., Rein A., Levin J.G. (1990). Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag–pol junction of Moloney murine leukemia virus Proc Natl Acad Sci USA 878860–8863 10.1073/pnas.87.22.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Yuan H., Rein A., Levin J.G. (1992). Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot J Virol 665127–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini P., Iona E., Piccaro G., Peyron P., Neyrolles O., Fattorini L. (2010). Activity of drug combinations against dormant Mycobacterium tuberculosis Antimicrob Agents Chemother 542712–2715 10.1128/AAC.01736-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy D., Recht M.I., Blanchard S.C., Puglisi J.D. (1996). Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic Science 2741367–1371 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Green L., Houck-Loomis B., Yueh A., Goff S.P. (2012). Large ribosomal protein 4 increases efficiency of viral recoding sequences J Virol 868949–8958 10.1128/JVI.01053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. (1998). A dual-luciferase reporter system for studying recoding signals RNA 4479–486. [PMC free article] [PubMed] [Google Scholar]
- Hermann T. (2005). Drugs targeting the ribosome Curr Opin Struct Biol 15355–366 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Honigman A., Falk H., Mador N., Rosental T., Panet A. (1995). Translation efficiency of the human T-cell leukemia virus (HTLV-2) gag gene modulates the frequency of ribosomal frameshifting Virology 208312–318 10.1006/viro.1995.1154. [DOI] [PubMed] [Google Scholar]
- Houck-Loomis B., Durney M.A., Salguero C., Shankar N., Nagle J.M., Goff S.P., D'Souza V.M. (2011). An equilibrium-dependent retroviral mRNA switch regulates translational recoding Nature 480561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H.E. (1985). Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting Science 2301237–1242 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Jones D.S., Nemoto F., Kuchino Y., Masuda M., Yoshikura H., Nishimura S. (1989). The effect of specific mutations at and around the gag-pol gene junction of Moloney murine leukaemia virus Nucleic Acids Res 175933–5945 10.1093/nar/17.15.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa T., Steyger P.S. (2011). Intracellular mechanisms of aminoglycoside-induced cytotoxicity Integr Biol (Camb) 3879–886 10.1039/c1ib00034a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E., Hirawat S., Armoni S., Yaakov Y., Shoseyov D., Cohen M., Nissim-Rafinia M., Blau H., Rivlin J., other authors (2008). Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial Lancet 372719–727 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- Le S.Y., Shapiro B.A., Chen J.H., Nussinov R., Maizel J.V. (1991). RNA pseudoknots downstream of the frameshift sites of retroviruses Genet Anal Tech Appl 8191–205 10.1016/1050-3862(91)90013-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J.G., Hatfield D.L., Oroszlan S., Rein A. (1993). Mechanisms of Translational Suppression Used in the Biosynthesis of Reverse Transcriptaser, pp. 5-31. In Reverse Transcriptase. Edited by Skalka A.-M., Goff S. P.Cold Spring Harbor: Cold Spring Harbor Press. [Google Scholar]
- Manuvakhova M., Keeling K., Bedwell D.M. (2000). Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system RNA 61044–1055 10.1017/S1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. (1987). Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol J Virol 61480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E.C., Jr, Arlinghaus R.B. (1978). Cell-free synthesis of Rauscher murine leukemia virus “gag” and “gag-pol” precursor polyproteins from virion 35S RNA in a mRNA-dependent translation system derived from mouse tissue culture cells Virology 86329–343 10.1016/0042-6822(78)90074-0. [DOI] [PubMed] [Google Scholar]
- Murphy E.C., Jr, Kopchick J.J., Watson K.F., Arlinghaus R.B. (1978). Cell-free synthesis of a precursor polyprotein containing both gag and pol gene products by Rauscher murine leukemia virus 35S RNA Cell 13359–369 10.1016/0092-8674(78)90204-0. [DOI] [PubMed] [Google Scholar]
- Murphy G.J., Mostoslavsky G., Kotton D.N., Mulligan R.C. (2006). Exogenous control of mammalian gene expression via modulation of translational termination Nat Med 121093–1099 10.1038/nm1376. [DOI] [PubMed] [Google Scholar]
- Musa A.M., Younis B., Fadlalla A., Royce C., Balasegaram M., Wasunna M., Hailu A., Edwards T., Omollo R., other authors (2010). Paromomycin for the treatment of visceral leishmaniasis in Sudan: a randomized, open-label, dose-finding study PLoS Negl Trop Dis 4e855. 10.1371/journal.pntd.0000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S.H., Copeland T.D., Hatanaka M., Oroszlan S. (1993). Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I J Virol 67196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman I., Rebibo-Sabbah A., Cherniavsky M., Belakhov V., Hainrichson M., Chen F., Schacht J., Pilch D.S., Ben-Yosef T., Baasov T. (2009). Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations J Med Chem 522836–2845 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odawara T., Yoshikura H., Ohshima M., Tanaka T., Jones D.S., Nemoto F., Kuchino Y., Iwamoto A. (1991). Analysis of Moloney murine leukemia virus revertants mutated at the gag–pol junction J Virol 656376–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. (2001). Recognition of cognate transfer RNA by the 30S ribosomal subunit Science 292897–902 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Oppermann H., Bishop J.M., Varmus H.E., Levintow L. (1977). A joint produce of the genes gag and pol of avian sarcoma virus: a possible precursor of reverse transcriptase Cell 12993–1005 10.1016/0092-8674(77)90164-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A.T. (1988). Retroviral gag gene amber codon suppression is caused by an intrinsic cis-acting component of the viral mRNA J Virol 623574–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T., Chamorro M., Varmus H.E. (1992). Human immunodeficiency virus type 1 gag–pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo J Virol 665147–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Dinman J.D. (2008). The role of programmed-1 ribosomal frameshifting in coronavirus propagation Front Biosci 134873–4881 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Levin J.G. (1992). Readthrough suppression in the mammalian type C retroviruses and what it has taught us New Biol 4283–289. [PubMed] [Google Scholar]
- Rowe S.M., Clancy J.P. (2009). Pharmaceuticals targeting nonsense mutations in genetic diseases: progress in development BioDrugs 23165–174 10.2165/00063030-200923030-00003. [DOI] [PubMed] [Google Scholar]
- Sermet-Gaudelus I., Boeck K.D., Casimir G.J., Vermeulen F., Leal T., Mogenet A., Roussel D., Fritsch J., Hanssens L., other authors (2010). Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis Am J Respir Crit Care Med 1821262–1272 10.1164/rccm.201001-0137OC. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M., Crowe S.M., Mak J. (2001). Maintenance of the Gag/Gag–Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity J Virol 751834–1841 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P.J., Berg P. (1982). Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter J Mol Appl Genet 1327–341. [PubMed] [Google Scholar]
- Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., other authors (2007). PTC124 targets genetic disorders caused by nonsense mutations Nature 44787–91 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- Wills N.M., Gesteland R.F., Atkins J.F. (1991). Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon Proc Natl Acad Sci U S A 886991–6995 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilschanski M., Miller L.L., Shoseyov D., Blau H., Rivlin J., Aviram M., Cohen M., Armoni S., Yaakov Y., other authors (2011). Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis Eur Respir J 3859–69 10.1183/09031936.00120910. [DOI] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S.E., Rathjen P.D., Kingsman S.M., Kingsman A.J. (1988). HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems Cell 551159–1169 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Xie Y., Dix A.V., Tor Y. (2010). Antibiotic selectivity for prokaryotic vs. eukaryotic decoding sites Chem Commun (Camb) 465542–5544 10.1039/c0cc00423e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T.D., Oroszlan S. (1985a). Murine leukemia virus protease is encoded by the gag–pol gene and is synthesized through suppression of an amber termination codon Proc Natl Acad Sci U S A 821618–1622 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T.D., Oroszlan S. (1985b). Translational readthrough of an amber termination codon during synthesis of feline leukemia virus protease J Virol 55870–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H.S., Green R. (2009). Fidelity at the molecular level: lessons from protein synthesis Cell 136746–762 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]


