Abstract
The genus Orbivirus of the family Reoviridae comprises 22 virus species including the Changuinola virus (CGLV) serogroup. The complete genome sequences of 13 CGLV serotypes isolated between 1961 and 1988 from distinct geographical areas of the Brazilian Amazon region were obtained. All viral sequences were obtained from single-passaged CGLV strains grown in Vero cells. CGLVs are the only orbiviruses known to be transmitted by phlebotomine sandflies. Ultrastructure and molecular analysis by electron microscopy and gel electrophoresis, respectively, revealed viral particles with typical orbivirus size and morphology, as well as the presence of a segmented genome with 10 segments. Full-length nucleotide sequencing of each of the ten RNA segments of the 13 CGLV serotypes provided basic information regarding the genome organization, encoded proteins and genetic traits. Segment 2 (encoding VP2) of the CGLV is uncommonly larger in comparison to those found in other orbiviruses and shows varying sizes even among different CGLV serotypes. Phylogenetic analysis support previous serological findings, which indicate that CGLV constitutes a separate serogroup within the genus Orbivirus. In addition, six out of 13 analysed CGLV serotypes showed reassortment of their genome segments.
Introduction
The genus Orbivirus is one of the 15 distinct genera currently included in the family Reoviridae; the genus contains 22 distinct virus species recognized by the International Committee for the Taxonomy of Viruses (ICTV) (Attoui et al., 2012). Viruses included in the genus Orbivirus are transmitted by culicoid midges, ticks, phlebotomine sandflies, as well as anopheline and culicine mosquitoes. Orbiviruses are icosahedral, non-enveloped and have a genome consisting of 10 double-stranded RNA (dsRNA) segments encoding seven structural proteins (VP1–VP7), and at least three non-structural proteins (NS1–NS3) (Verwoerd et al., 1972).
Insect-borne orbiviruses that have been reported to infect humans belong to the species Changuinola virus (CGLV), Corriparta virus (CORV), Lebombo virus and Orungo virus (Attoui et al., 2005), while the human pathogenic tick-borne orbivirus species include Kemerovo virus (KEMV), Tribeč virus (TRBV) and Lipovnik virus (Dilcher et al., 2012).
CGLV (BT-436) is the prototype of the CGLV serogroup and was originally isolated from a pool of phlebotomine sandflies (Lutzomyia sp.) collected in Panama in 1960. A second antigenically related agent, Irituia virus (BE AN 28873), was recovered from the serum of a rice rat (Oryzomys goeldi) collected in 1961 in the Brazilian Amazon Basin in the municipality of Ipixuna, state of Pará, Brazil (3° S 49° W). Subsequently, many additional isolates of Changuinola serogroup viruses have been obtained from sandflies and sloths in Panama, Colombia and other regions of Brazil (Seymour et al., 1983; Tesh et al., 1974; Travassos da Rosa et al., 1984, 1998).
A single isolate of CGLV was also obtained from the blood of a febrile entomology field worker in Panama in 1966. To date, this is the only reported case of human illness associated with CGLV infection (Karabatsos, 1985; Peralta & Shelokov, 1966).
The Changuinola serogroup consists of a large number of antigenetically related viruses that have been isolated from phlebotomine sandflies, mosquitoes and various species of wild mammals. To date, these viruses have been found only in Panama and tropical regions of South America, and they are presumed to be arthropod-borne (Travassos da Rosa et al., 1984). By complement fixation (CF) test, the Changuinola group viruses are broadly cross-reactive and cannot be differentiated; consequently most of the available isolates have never been characterized beyond the serogroup level. Based on mouse neutralization tests, the Panamanian prototype BT-436 and 11 Brazilian isolates were shown to differ antigenetically, and 12 distinct CGLV serotypes have now been recognized by the ICTV. These include Changuinola, Almeirim, Altamira, Caninde, Gurupi, Irituia, Jamanxi, Jari, Monte Dourado, Ourem, Purus and Saraca viruses (Attoui et al., 2012). However, there are most likely more, since many of the existing isolates have never been examined by neutralization testing.
The first complete genome sequence of a CGLV serotype, Irituia virus (BE AN 28873), was published in 2013 (Silva et al., 2013). The full genome sequence of a second serotype, Xaraira virus (BE AR 490492), was published in 2014 (Jaafar et al., 2014). In the present study, we determined the full genome sequence of all 10 segments of Irituia, Caninde, Jari, Ourem, Saraca and Gurupi viruses and seven additional CGLV serotypes isolated from wild animals and phlebotomine sandflies in the Brazilian Amazon region between 1961 and 1988. Analysis of their complete genomes and phylogenetic relationships indicate that the Changuinola group represents a genetically diverse species with multiple serotypes, many of which show signs of reassortment.
Results
Electron microscopy and electrophoretic profile
Ultra structural analysis of ultrathin sections of Vero cells infected with Irituia virus (BE AN 28873) showed icosahedral particles in the cytoplasm, with a mean diameter of 75 nm. These particles were observed individually and aggregated (viroplasm) in the cytoplasm (Fig. 1). Analysis of the RNA electrophoretic profile of ten CGLVs, using agarose gel electrophoresis, revealed the presence of ten genome segments that were classified according to their molecular mass into three distinct groups: group 1, high molecular mass (S1–S3); group 2, intermediate molecular mass (S4–S6); and group 3, low molecular mass (S7–S10) (Fig. 2).
Fig. 1.
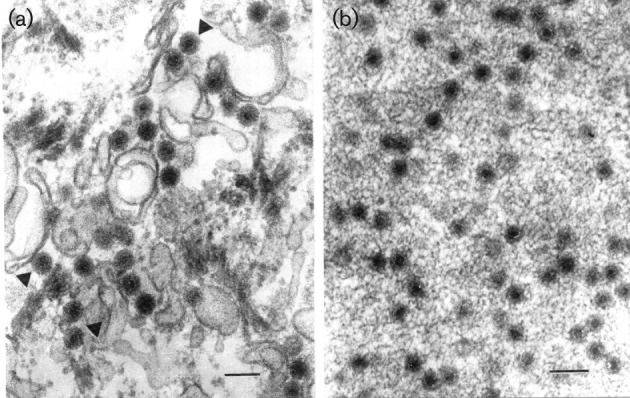
Electron micrographs of serotype Irituia virus (BE AR 28873). (a) Virions 75 nm in diameter in the cytoplasm of a Vero cell (arrowheads). (b) A portion of a fibrillar aggregate (viroplasm) in the cytoplasm of a Vero cell with forming viruses. Bars, 100 nm.
Fig. 2.
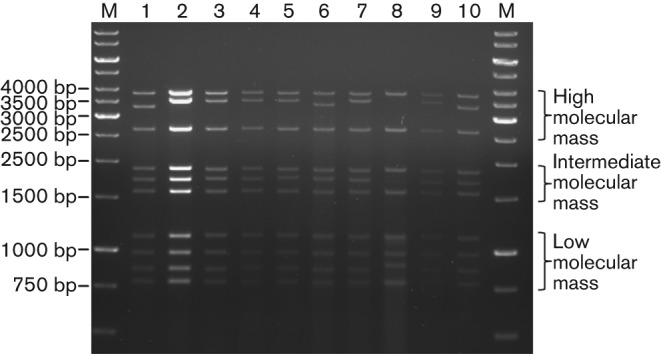
Agarose gel electrophoretic profiles of selected CGLV serotypes. Lane M: molecular mass marker (size is shown in bp). BE AR 35646 (lane 1); BE AN 28873 (lane 2); BE AR 425269 (lane 3); BE AR 434080 (lane 4); BE AR 478620 (lane 5); BE AR 440489 (lane 6); BE AR 440541 (lane 7); BE AR 385278 (lane 8; in this lane, segments 2 and 5 had very low concentrations); BE AR 397956 (lane 9); BE AR 397374 (lane 10).
Genetic characterization
Full-length sequences were obtained for all 13 viral isolates listed in Table 1 for each of the ten dsRNA segments. The overall coverage of the genomes and the number and percentage of virus-specific reads are summarized in Table S1 (available in the online Supplementary Material). The analysis of similarity and homology with nucleotide and amino acid sequences available in the GenBank database (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi) and in the InterProScan database (http://www.ebi.ac.uk/Tools/pfa/iprscan/) demonstrated that the nucleotide and amino acid sequences of the CGLVs match with other known orbivirus genomes and proteins. The nucleotide sequences (S1 to S10) and respective proteins (VP1 to VP7 and NS1 to NS3) were deposited in GenBank (Table 1).
Table 1. CGLV serotypes isolated from 1961 to 1988 in the Amazon region of Brazil.
| Prototype number | Serotype | GenBank accession numbers | Isolation information | ||
| Host | Date | Material (place*) | |||
| BE AR 35646 | Gurupi virus | KF680168–KF680177 | Phlebotominae spp. | 1961 | Pool (BR 14, km 94) |
| BE AN 28873 | Irituia virus | KF624614–KF624623 | Oryzomys goeldi | 1961 | Blood (BR 14, km 94) |
| BE AR 41067 | Ourem virus | KF680178–KF680187 | Phlebotominae spp. | 1962 | Pool (BR 14, km 94) |
| BE AR 54342 | Caninde virus | KF690591–KF690600 | Phlebotominae spp. | 1963 | Pool (BR 14, km 94) |
| BE AN 385199 | Jari virus | KF680188–KF680197 | Choloepus didactylus | 1980 | Viscera (Jari, Monte Dourado) |
| BE AR 385278 | Saraca virus | KF690601–KF690610 | Phlebotominae spp. | 1980 | Pool (Porto de Trombetas, Oriximiná) |
| BE AR 397956 | Tumucumaque virus | KF668547–KF668556 | Lutzomyia umbratilis | 1981 | Pool (Jari, Monte Dourado) |
| BE AR 397374 | Jutai virus | KF668527–KF668536 | Lutzomyia umbratilis | 1981 | Pool (Jari, Monte Dourado) |
| BE AR 425269 | Aracai virus | KF668557–KF668566 | Phlebotominae spp. | 1984 | Pool (Tucurui) |
| BE AR 434080 | Tapirope virus | KF690611–KF690620 | Phlebotominae spp. | 1984 | Pool (Serra Norte, Marabá) |
| BE AR 440489 | Jandia virus | KF668537–KF668546 | Phlebotominae spp. | 1985 | Pool (Tucurui) |
| BE AR 440541 | Timbozal virus | KF690581–KF690590 | Phlebotominae spp. | 1985 | Pool (Tucurui) |
| BE AR 478620 | Balbina virus | KF690621–KF690630 | Phlebotominae spp. | 1988 | Pool (Balbina) |
All virus isolates were obtained in the state of Pará, except for Balbina virus, which was recovered at the Balbina hydro-electric dam in Balbina, Amazonas state. Kilometre markers are indicated for locations on Brazil road BR 14.
The predicted proteins ranged from 1309 aa (segment 1; VP1) to 231 aa (segment 10; NS3) in length (Table S2). Among the different CGLV serotypes, the length of some genome segments and the length of the encoded proteins varied significantly, whereas for other genome segments the length was rather constant (Table S2). The length of segments S1, S3, S4, S6 and S7 varied only by about 1 bp among the different serotypes, leading to encoded proteins of identical length. In contrast, the length of segment 2 varied by about 245 nt, yielding VP2 proteins with size differences of about 78 aa. In addition, the length of segment 9 in BE AR 385278, with 940 nt, was significantly longer than that of segment 9 in the other CGLV isolates (see PAGE in Fig. 2), yielding a VP6 (Hel) protein that is 10 to 12 aa bigger than other CGLV VP6 (Hel) proteins. Size differences can also be seen in segment 5, where this segment is 52 nt shorter in BE AN 385199 than the longest segment 5 sequence. Regarding the non-coding regions (NCRs), the 5′ termini were composed of short sequences ranging from 7 bp for segment 4 (VP4) to 34 bp for segment 5 (NS1). The 3′ termini were longer, with NCRs ranging from 32 bp for segment 2 (VP2) to 108 bp for segment 5 (NS1).
Analysis of the 5′ and 3′ NCRs demonstrated that all of the segments share five conserved nucleotides at their 5′ and 3′ ends (5′-GUAAA-----CUUAC-3′). Segment 2, which encodes the outer capsid protein VP2, showed the highest genetic variability among the 13 studied CGLV serotypes, with only 19.2 % identity (712 nt). The amino acid alignment showed 17.1 % identity. The size of segment 2 is rather variable, ranging from 3450 to 3695 nt in the different CGLV serotypes. Furthermore, the G+C content of CGLV serotypes showed values ranging between 33.1 and 42.1 mol%.
All CGLV serotypes encode the helicase protein on segment 9 (VP6, ORF +2); however, an additional putative protein was also found to be encoded on the same segment but in a different reading frame (ORF +3). This putative protein ranged from 87 aa (ORF of 261 nt) to 89 aa (ORF of 267 nt) in length with a mean identity of 59.4 % (from 33.7 % to 97 %). The most divergent strain was the Saraca virus (BE AR 385278), showing a mean identity of 36.15 %.
Phylogenetic analysis
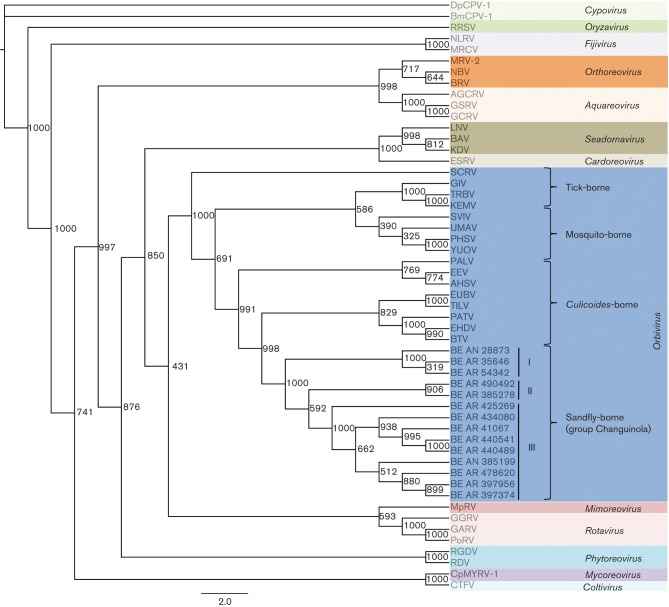
Phylogenetic reconstruction based on multiple alignments of VP1 amino acid sequences generated trees with similar topologies regardless of the method used. In this case, the maximum likelihood (ML) tree was selected as the best phylogenetic tree to represent the genetic relationship for CGLVs, as it was the one that best represented the known taxonomy. The analysis using the VP1 protein (segment 1, viral polymerase) confirmed that the CGLVs belong to the genus Orbivirus and are more closely related to Bluetongue virus (BTV), Epizootic hemorrhagic disease virus (EHDV), Pata virus (PATV), Eubenangee virus (EUBV) and Tilligerry virus (TILV), which are all Culicoides-borne orbiviruses (Fig. 3).
Fig. 3.
Phylogenetic analysis using the ML method of different genera within the family Reoviridae. Based on the complete amino acid sequences of the viral RNA polymerase (VP1). Different phylogenetic groups are labelled in different coloured boxes. The genus Orbivirus is represented as a separate group (dark blue) within the tree. Numbers at each main node of the tree correspond to bootstrap values (1000 replicates). Viral abbreviations and GenBank accession numbers are available in Table S3. The scale bar corresponds to 2 % of genetic divergence among amino acid sequences.
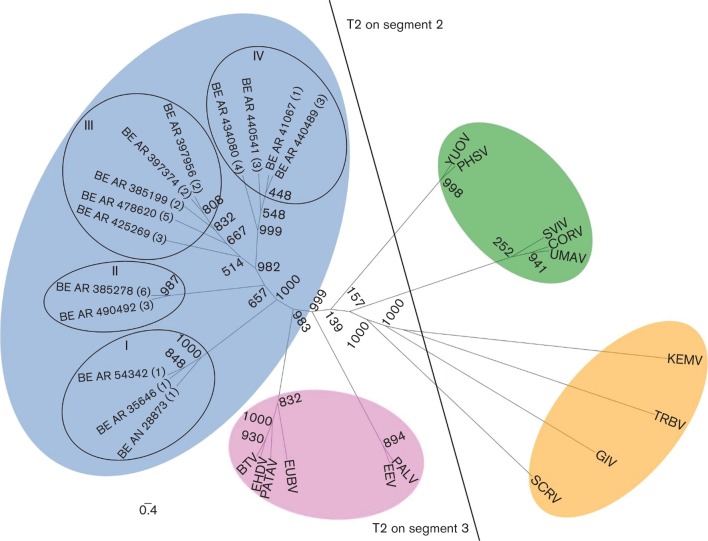
The phylogenetic relationship based on the amino acid sequences of orbivirus T2 proteins (segment 3 for Culicoides-borne viruses and segment 2 for mosquito-borne and tick-borne orbiviruses) demonstrated that the CGLV serotypes studied belong to a new group near the Culicoides-borne group (Fig. 4). Figs S1 to S8 summarize the genetic relationships for the proteins encoded by the other genome segments.
Fig. 4.
Relationships between complete amino acid sequences of T2 proteins of different orbiviruses. Maximum-likelihood unrooted tree showing distinct phylogenetic groups based on the T2 protein. Group ‘T2 on segment 3′ includes viruses in which the T2 protein [in this case VP3 (T2)] is encoded on segment 3: Culicoides-borne (pink) and sandfly-borne (blue) viruses composed of different serotypes of the Changuinola group. Group ‘T2 on segment 2’ includes viruses in which the T2 protein [in this case VP2 (T2) or VP3 (T2)] is encoded on segment 2: mosquito-borne (green) and tick-borne (orange). Bootstrap values are placed at each main node of the tree. Roman numerals (I to IV) represent the phylogenetic lineages. Arabic numerals (1 to 5) correspond to different geographical locations from where the viruses were isolated. (1) Belém–Brasilia highway km 94, Pará state; (2) Monte Dourado/Jari, Pará state; (3) base 4/Tucuruí, Pará state; (4) Serra Norte/Carajás, Pará state; (5) Balbina/Pte. Figueiredo, Amazonas state; (6) Porto Trombetas, Pará state. Full names of virus isolates and accession numbers of T2 protein sequences used for comparative analysis are listed in Table S4. The scale bar represents 0.4 % genetic divergence.
Genetic reassortment
The analysis of genetic reassortment demonstrated that six out of 13 CGLVs showed strong phylogenetic permutation signs (>90 %) involving at least one reassorted RNA segment: Ourem virus (BE AR 41067), Tapirope virus (BE AR 434080), Jandia virus (BE AR 440489), Tumucumaque virus (BE AR 397956), Jari virus (BE AN 385199) and Jutai virus (BE AR 397374) (Figs 5, S9 and S10). Reassortment events mainly involved the segments S2, S4, S5, S8, S9 and S10 (encoding VP2, VP4, NS1, NS2, VP6 and NS3, respectively). No reassortment event was observed for S1, S3, S6 or S7 (encoding VP1, VP3, VP5 and VP7 proteins, respectively).
Fig. 5.
Scheme of genetic reassortment between CGLV serotypes. Each hexagon represents a different CGLV serotype. The 10 internal lines represent all genome segments. The arrows indicate shared segments. The sample Tapirope virus (BE AR 434080) presents a reassortment event with at least two different CGLVs. Jutai virus (BE AR 397374) appears to harbour segments which originate from four different viruses.
Discussion
Between 1960 and 1990, a large number of arthropod-borne viruses were isolated in the Brazilian Amazon region and described as new members of the Changuinola serogroup (Travassos da Rosa et al., 1998). In the past, arbovirus classification was based on several criteria, which included virion morphology and size, the nature of the RNA (segmented, non-segmented, ssRNA or dsRNA) and antigenic relationships. More recently, the analysis of genetic relationships within a given group of viruses, using a specific signature gene (e.g. polymerase gene) has been used (Attoui et al., 2005).
Based on these criteria, 12 different serotypes are currently recognized within the Changuinola species complex (Attoui et al., 2012). This classification is supported by the icosahedral ultrastructure of the virions (Fig. 1), electrophoretic profiles demonstrating 10 dsRNA segments (Fig. 2), and their genetic relationship with other members of the genus Orbivirus on the basis of phylogenetic reconstructions using the amino acid sequences of the polymerase and T2 proteins (Figs 3 and 4). These molecular and structural data are in agreement with antigenic, biological, and chemical properties described previously for members of the Changuinola serogroup (Jaafar et al., 2014; Silva et al., 2013; Travassos da Rosa et al., 1984).
The VP2 protein is the most variable protein found in orbiviruses (Attoui et al., 2012). This outer shell protein is associated with virus antigen specificity, as well as with virus–host cell attachment, virulence, and virus entry into host cells (Hwang & Li, 1993; Kahlon et al., 1983). High genetic divergence and size heterogeneity have been observed among VP2 proteins from mosquito, Culicoides and tick-associated orbiviruses. In fact the VP2 proteins of tick-transmitted orbiviruses are half the size of those of mosquito- and Culicoides-associated orbiviruses (Schoehn et al., 1997).
In the case of the Brazilian CGLV isolates, the VP2 proteins are the largest VP2 proteins of all known orbiviruses. Considerable segment 2 size heterogeneity was even found among different CGLV serotypes (Table S2) reflected in proteins of different sizes. For BE AR 35646, BE AR 385278, BE AR 54342, BE AR 397374 and BE AR 440489 respective segment 2 sizes of 3450 nt, 3450 nt, 3503 nt, 3536 nt and 3548 nt are 113–245 nt smaller than in the segments 2 of the remaining Changuinola serotypes (3661–3695 nt). These differences could be related to viral protein selection and evolutionary adaptation to a given host. Although many conserved motifs were found among CGLV isolates, unique sequences or motifs were observed in the region of VP2 comprising amino acid positions 210–380 aa (Fig. S11). Furthermore, three isolates BE AR 35646 (Gurupi virus), BE AR 54342 (Caninde virus) and BE AR 385278 (Saraca virus) had a deletion of 70 aa in their VP2. It is important to assess how this deletion might affect VP2 structure and function.
Cysteine residues are unique among coding amino acids because they contain a reactive sulfhydryl group. Therefore, two cysteine residues may form a cystine (disulfide link) between various parts of the same protein or between two separate polypeptide chains (Miseta & Csutora, 2000). Several cysteine residues were found to be conserved among the CGLV VP2 proteins (Fig. S9). This suggests that the basic structure of the VP2 protein, and consequently its function, remains unchanged for the Brazilian CGLV serotypes. However, further studies are necessary to assess and validate the antigenic sites of the CGLV group members.
In addition to the viral helicase VP6 (reading frame +2), segment 9 of CGLV seems to code for an additional putative protein, since all CGLV strains contain a 261 to 267 nt long ORF (reading frame +3) overlapping VP6 at the same position. VP6 overlapping ORF′s in different reading frames have been discovered in several other orbivirus genomes (Belhouchet et al., 2010; Dilcher et al., 2012; Firth, 2008), For Culicoides-borne Bluetongue virus (BTV) and tick-borne Great Island virus (GIV) it has been shown that this VP6 overlapping ORF encodes a fourth non-structural protein, NS4, which can also be detected in infected mammalian cells, where it seems to counteract the antiviral response of the host. In addition it has been shown that it protects DNA from degradation by DNase, indicating an ability to bind dsDNA (Belhouchet et al., 2011; Ratinier et al., 2011).
The genetic relationship of the genus Orbivirus in the Reoviridae was previously analysed using basic information on both polymerase (VP1) and T2 proteins (Attoui et al., 2009). The viral polymerase protein has the highest degree of genetic conservation among the orbiviruses, and has been used for taxonomic classification (Kapoor et al., 2013; Vieira et al., 2009). The phylogenetic analysis of the VP1 proteins performed in this study, in comparison to other reoviruses, groups the Brazilian CGLV isolates as members of the genus Orbivirus. These results confirm previous serological grouping (Travassos da Rosa et al., 1984). Within the clade corresponding to orbiviruses, the closest relative group to the Brazilian CGLV subclade are the orbiviruses transmitted by Culicoides. Similar results were also observed by others, using partial CGLV VP1 nt sequences (132 nt) (Palacios et al., 2011). The T2 protein is largely used to assess the evolutionary relationship of orbiviruses and their natural hosts (Attoui et al., 2001, 2005, 2009; Belaganahalli et al., 2012; Dilcher et al., 2012; Jaafar et al., 2014; Kapoor et al., 2013; Vieira et al., 2009). The analysis of the T2 protein of the 13 Brazilian CGLV isolates also confirmed their closest genetic association with Culicoides orbiviruses (Fig. 4). These findings suggest a possible ecological, epidemiological and evolutionary relationship between the CGLV clade transmitted by phlebotomine sandflies and orbiviruses transmitted by Culicoides midges (Fig. 4).
The evolutionary process of segmented RNA viruses occurs basically by three mechanisms (mutation, genetic reassortment and genetic recombination) (Bishop & Beaty, 1988; Bolker et al., 2009; Talbi et al., 2010). Although several studies have been performed, the effective contribution of these three mechanisms for biogenesis of segmented viruses is not well understood. However, in the specific case of Brazilian orbiviruses, genetic reassortment appears to be directly associated with virus biodiversity since 6 out of 13 Brazilian CGLV serotypes demonstrated reassortment events (Fig. 5).
The results of our phylogenetic analysis, using the polymerase and T2 proteins, demonstrated that the Brazilian CGLVs isolated in distant geographical locations of the Brazilian Amazon (e.g. Pará and Amazonas states) were generally genetically diverse (Figs 3 and 4), suggesting that the geographical isolation of a group of viruses can lead to genetic modifications and in specific cases to a virus speciation over a period of time (Bolker et al., 2009; Talbi et al., 2010).
The combined analysis and the results using phylogenetic topologies, percentage of permutation trees in Simplot analysis, and geographical information, suggested that genetic reassortment and geographical location play an important role in viral biodiversity among Changuinola serogroup members. However, further studies including complete sequencing of other CGLV serotypes isolated in Brazil and in other geographical regions are needed for a better understanding of the factors involved in viral diversity.
Methods
Virus strains.
Thirteen CGLV serotypes recovered from distinct geographical areas of the Brazilian Amazon region and isolated between 1961 and 1988 were used in this study. The 13 CGLV serotypes were initially isolated by intracerebral inoculation of newborn mice; viruses were single-passaged in Vero cells to obtain viral RNA for sequencing. The studied strains were obtained from the World Health Organization/Pan American Health Organization Collaborating Center for Arbovirus Reference Research at the Department of Arbovirology and Hemorrhagic Fevers, Evandro Chagas Institute, Brazilian Ministry of Health, Ananindeua, Brazil. A summary of information about the 13 viral isolates (serotype, designation, geographical area, source and year of isolation) is presented in Table 1.
Transmission electron microscopy.
Vero cells infected with Irituia virus (BE AN 28873) were used for ultrastructural analysis. Ultrathin sections of infected cells were first fixed for at least 1 h in a mixture of 2.5 % formaldehyde prepared from paraformaldehyde powder, and 0.1 % glutaraldehyde in 0.05 M cacodylate buffer pH 7.3 to which 0.03 % picric acid and 0.03 % CaCl2 were added. The monolayers were washed in 0.1 M cacodylate buffer; cells were scraped off and processed further as a pellet. The pellets were post-fixed in 1 % OsO4 in 0.1 M cacodylate buffer pH 7.3 for 1 h, washed with distilled water and stained en bloc with 2 % aqueous uranyl acetate for 20 min at 60 °C. The pellets were dehydrated in ethanol, processed through propylene oxide and embedded in Poly/Bed 812 (Polysciences). Ultrathin sections were cut on a Leica EM UC7 ultramicrotome (Leica Microsystems), stained with lead citrate and examined in a Philips 201 transmission electron microscope at 60 kV.
Treatment to remove contaminant host RNA.
Approximately 20 ml of cell culture supernatant from infected Vero cells, showing 80–90 % cytopathic effect (CPE) was collected between 3 and 5 days post-infection; this material was first centrifuged for 10 min at 2000 r.p.m. (700 g) and then for 5 min at 4000 r.p.m. (2800 g) and passed through 0.2 µm filters, followed by PEG-precipitation to enrich for virus particles (Dilcher et al., 2012). Subsequently, the virus pellets were resuspended in 250 µl PBS, and 25 µl RNase A (final concentration, 2 mg ml−1) was added to digest cellular RNAs. The samples were kept at 37 °C for 30 min after which 225 µl PBS was added.
Nucleic acid isolation and gel electrophoresis.
The dsRNA was extracted from virus pellets using a guanidinium isothiocyanate (Trizol) procedure according to the manufacturer's instructions (peqGOLDTrifast FL; Peqlab). To improve the RNA precipitation, 2 µl glycogen (35 µg µl−1) was added together with one volume of 2-propanol per sample and kept overnight at −20 °C. Samples were then centrifuged for 15 min at 12 000 g and 4 °C and washed twice with 75 % ethanol. After drying, each RNA pellet was resuspended in 25 µl RNase-free diethyl pyrocarbonate (DEPC)-treated water and incubated for 5 min at 57 °C in a heating block. After DNA digestion via the Turbo-DNA-free kit (Ambion) and Pellet-Paint precipitation of the purified RNAs (Pellet Paint NF Co-Precipitant; Novagen), cellular ssRNAs that might still have been present were precipitated overnight in 2 M LiCl at 4 °C (Attoui et al., 2000). The viral dsRNA was precipitated from the supernatant by addition of 0.25 volume 7.5 M ammonium acetate and 1 volume (RNA+ammonium acetate) 2-propanol for 2 h at −20 °C. The RNA pellet was washed twice with 70 % ethanol and once with 100 % ethanol and resuspended in 18 µl DEPC-treated water after drying. The RNA concentration was measured using a Nanodrop 2000 device (Peqlab). Electrophoresis was carried out in long 1 % TAE agarose gels at 4 °C in a cold room. A total of 1–2 µg of purified dsRNA was used per lane and subsequently run for 2 h at 90 V, then for 4 h at 100–120 V (total 6 h) at 4 °C. The RNA segments were visualized via staining with ethidium bromide. The sizes of the RNA bands were estimated using the GeneRuler 1 kb DNA Ladder (Thermo Scientific).
Genome sequencing.
Complete genomes were obtained using the pyrosequencing approach (Shendure & Ji, 2008). In order to recover the 5′ and 3′ ends of the dsRNA segments, the same anchor primer sequence used in the FLAC-method (full-length amplification of cDNAs) was ligated to the 3′ ends of the viral RNAs prior to pyrosequencing as previously described (Maan et al., 2007; Potgieter et al., 2009). 500 ng of purified viral dsRNA was ligated to 500 ng of FLAC-anchor primer. The concentration of the adaptor-ligated dsRNA was determined using a NanoDrop 2000 (Peqlab). Then, 60 ng of adaptor-ligated viral RNA was amplified and converted to cDNA using a TransPlex Whole Transcriptome Amplification kit (WTA2; Sigma-Aldrich).
After purification with the QIAquick PCR Purification kit (Qiagen), an additional size exclusion step via Ampure-XP beads (Agencourt) in a ratio of 1 volume WTA2 product to 0.7 volume Ampure-XP beads was used to remove fragments shorter than 300 bp. Furthermore, 300 ng of the whole genome amplified cDNA were used for titanium shotgun library preparation and pyrosequencing as described in the FLX Titanium Rapid Library Preparation Protocol starting with the end-repair step (Roche Applied Sciences) (Margulies et al., 2005). In this case, the DNA fragmentation by nebulization step was skipped. In the RL adaptor ligation step, RL-MID adapters were used to allow pooling of several samples.
A pyrosequencing library was prepared and used for sequencing on a GS FLX pyrosequencer (Roche, 454 Life Sciences) at the Department of Virology, University Medical Center, Göttingen, Germany. In parallel, the same libraries were sequenced on a GS FLX pyrosequencer (Roche, 454 Life Sciences) in the Center for Technological Innovation at the Evandro Chagas Institute, Ministry of Health, Brazil.
Genome assembly and phylogenetic analysis.
The assembly of the obtained sequences was carried out using the GS De Novo Assembler program (Newbler v. 2.6). For further analysis, the Geneious v. 6.1.4 software and a set of programs (SeqMan Pro, SeqBuilder, Protean) available in the dnastar package (Lasergene) were used. The dataset used to reconstruct the phylogenetic trees consisted of amino acid sequences obtained for each genome segment.
The alignment was made using the Mafft v. 7 program (Katoh & Standley, 2013). The reconstruction of phylogenetic trees was made using the ML method (Myung, 2003), implemented in the PhyML v. 3.0 (https://github.com/stephaneguindon/phyml-downloads/releases/download/stable/phyml-20120412.tar.gz) computational program. For determination of the reliability of the tree topology, bootstrap analysis (Felsenstein, 1985) was carried out on 1000 replicates. Confidence values used as criteria for group inclusion or exclusion were calculated based on the mean of the amino acid sequence identities within and among selected members of the different genera of the family Reoviridae.
Evaluation of genetic reassortment.
In order to evaluate the possibility of natural genetic reassortment among the 13 CGLV serotypes analysed, concatenate sequences of the 10 dsRNA segments of each virus were used. Multiple sequence alignments were carried out using Mafft v. 7 (Katoh & Standley, 2013) and Simplot v. 3.5.1 (Lole et al., 1999) for evaluation of RNA segment shifting. Values of permutation trees were assigned as percentages. Genetic reassortment was considered when the percentage of permutation trees was more than 90 % across a given entire genomic segment.
Acknowledgements
Work in Brazil was supported by the Evandro Chagas Institute, Ministry of Health; Coordination of Improvement of Higher Education Personnel (CAPES) process no. 301641/2010-2 and National Counsel of Technological and Scientific Development (CNPq) and grant no. 302032/2011-8. Work at the University of Texas Medical Branch was supported by the National Institutes of Health contract HHSN272201000040I/HHSN27200004/DO4.
Supplementary Data
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the nucleotide sequences (S1 to S10) obtained for each of the 13 viral isolates are KF680168–KF680177, KF624614–KF624623, KF680178–KF680187, KF690591–KF690600, KF680188–KF680197, KF690601–KF690610, KF668547–KF668556, KF668527–KF668536, KF668557–KF668566, KF690611–KF690620, KF668537–KF668546, KF690581–KF690590 and KF690621–KF690630.
Four supplementary tables and 11 supplementary figures are available with the online version of this paper.
References
- Attoui H., Billoir F., Cantaloube J. F., Biagini P., de Micco P., de Lamballerie X. (2000). Strategies for the sequence determination of viral dsRNA genomes. J Virol Methods 89, 147–158. 10.1016/S0166-0934(00)00212-3 [DOI] [PubMed] [Google Scholar]
- Attoui H., Stirling J. M., Munderloh U. G., Billoir F., Brookes S. M., Burroughs J. N., de Micco P., Mertens P. P., de Lamballerie X. (2001). Complete sequence characterization of the genome of the St Croix River virus, a new orbivirus isolated from cells of Ixodes scapularis. J Gen Virol 82, 795–804. [DOI] [PubMed] [Google Scholar]
- Attoui H., Mohd Jaafar F., Belhouchet M., Aldrovandi N., Tao S., Chen B., Liang G., Tesh R. B., de Micco P., de Lamballerie X. (2005). Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. J Gen Virol 86, 3409–3417. 10.1099/vir.0.81258-0 [DOI] [PubMed] [Google Scholar]
- Attoui H., Mendez-Lopez M. R., Rao S., Hurtado-Alendes A., Lizaraso-Caparo F., Jaafar F. M., Samuel A. R., Belhouchet M., Pritchard L. I. & other authors (2009). Peruvian horse sickness virus and Yunnan orbivirus, isolated from vertebrates and mosquitoes in Peru and Australia. Virology 394, 298–310. 10.1016/j.virol.2009.08.032 [DOI] [PubMed] [Google Scholar]
- Attoui H., Mertens P. P. C., Becnel J., Belaganahalli S., Bergoin M., Brussaard C. P., Chappell J. D., Ciarlet M., del Vas M. & other authors (2012). Family Reoviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 541–637. Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. Amsterdam: Elsevier Academic Press. [Google Scholar]
- Belaganahalli M. N., Maan S., Maan N. S., Nomikou K., Pritchard I., Lunt R., Kirkland P. D., Attoui H., Brownlie J., Mertens P. P. C. (2012). Full genome sequencing and genetic characterization of Eubenangee viruses identify Pata virus as a distinct species within the genus Orbivirus. PLoS ONE 7, e31911. 10.1371/journal.pone.0031911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhouchet M., Mohd Jaafar F., Tesh R., Grimes J., Maan S., Mertens P. P. C., Attoui H. (2010). Complete sequence of Great Island virus and comparison with the T2 and outer-capsid proteins of Kemerovo, Lipovnik and Tribec viruses (genus Orbivirus, family Reoviridae). J Gen Virol 91, 2985–2993. 10.1099/vir.0.024760-0 [DOI] [PubMed] [Google Scholar]
- Belhouchet M., Mohd Jaafar F., Firth A. E., Grimes J. M., Mertens P. P. C., Attoui H. (2011). Detection of a fourth orbivirus non-structural protein. PLoS ONE 6, e25697. 10.1371/journal.pone.0025697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Beaty B. J. (1988). Molecular and biochemical studies of the evolution, infection and transmission of insect bunyaviruses. Philos Trans R Soc Lond B Biol Sci 321, 463–483. 10.1098/rstb.1988.0103 [DOI] [PubMed] [Google Scholar]
- Bolker B. M., Brooks M. E., Clark C. J., Geange S. W., Poulsen J. R., Stevens M. H. H., White J.-S. S. (2009). Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol 24, 127–135. 10.1016/j.tree.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Dilcher M., Hasib L., Lechner M., Wieseke N., Middendorf M., Marz M., Koch A., Spiegel M., Dobler G. & other authors (2012). Genetic characterization of Tribeč virus and Kemerovo virus, two tick-transmitted human-pathogenic Orbiviruses. Virology 423, 68–76. 10.1016/j.virol.2011.11.020 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Firth A. E. (2008). Bioinformatic analysis suggests that the Orbivirus VP6 cistron encodes an overlapping gene. Virol J 5, 48. 10.1186/1743-422X-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G. Y., Li J. K. K. (1993). Identification and localization of a serotypic neutralization determinant on the VP2 protein of bluetongue virus 13. Virology 195, 859–862. 10.1006/viro.1993.1445 [DOI] [PubMed] [Google Scholar]
- Jaafar F. M., Belhouchet M., Belaganahalli M., Tesh R. B., Mertens P. P. C., Attoui H. (2014). Full-genome characterisation of Orungo, Lebombo and Changuinola viruses provides evidence for co-evolution of orbiviruses with their arthropod vectors. PLoS ONE 9, e86392. 10.1371/journal.pone.0086392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlon J., Sugiyama K., Roy P. (1983). Molecular basis of bluetongue virus neutralization. J Virol 48, 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Tesh R. B., Duraisamy R., Popov V. L., Travassos da Rosa A. P. A., Lipkin W. I. (2013). A novel mosquito-borne Orbivirus species found in South-east Asia. J Gen Virol 94, 1051–1057. 10.1099/vir.0.046748-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabatsos N. (1985). International Catalogue of Arboviruses, 3rd edn San Antonio: American Society of Tropical Medicine and Hygiene. [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., Ingersoll R., Sheppard H. W., Ray S. C. (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan S., Rao S., Maan N. S., Anthony S. J., Attoui H., Samuel A. R., Mertens P. P. C. (2007). Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods 143, 132–139. 10.1016/j.jviromet.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y.-J. & other authors (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miseta A., Csutora P. (2000). Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol 17, 1232–1239. 10.1093/oxfordjournals.molbev.a026406 [DOI] [PubMed] [Google Scholar]
- Myung I. J. (2003). Tutorial on maximum likelihood estimation. J Math Psychol 47, 90–100. 10.1016/S0022-2496(02)00028-7 [DOI] [Google Scholar]
- Palacios G., Cowled C., Bussetti A. V., Savji N., Weir R., Wick I., Travassos da Rosa A., Calisher C. H., Tesh R. B. & other authors (2011). Rapid molecular strategy for orbivirus detection and characterization. J Clin Microbiol 49, 2314–2317. 10.1128/JCM.00337-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta P. H., Shelokov A. (1966). Isolation and characterization of arboviruses from Almirante, Republic of Panama. Am J Trop Med Hyg 15, 369–378. [DOI] [PubMed] [Google Scholar]
- Potgieter A. C., Page N. A., Liebenberg J., Wright I. M., Landt O., van Dijk A. A. (2009). Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90, 1423–1432. 10.1099/vir.0.009381-0 [DOI] [PubMed] [Google Scholar]
- Ratinier M., Caporale M., Golder M., Franzoni G., Allan K., Nunes S. F., Armezzani A., Bayoumy A., Rixon F. & other authors (2011). Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog 7, e1002477. 10.1371/journal.ppat.1002477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoehn G., Moss S. R., Nuttall P. A., Hewat E. A. (1997). Structure of Broadhaven virus by cryoelectron microscopy: correlation of structural and antigenic properties of Broadhaven virus and bluetongue virus outer capsid proteins. Virology 235, 191–200. 10.1006/viro.1997.8685 [DOI] [PubMed] [Google Scholar]
- Seymour C., Peralta P. H., Montgomery G. G. (1983). Viruses isolated from Panamanian sloths. Am J Trop Med Hyg 32, 1435–1444. [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H. (2008). Next-generation DNA sequencing. Nat Biotechnol 26, 1135–1145. 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- Silva S. P., Dilcher M., Weidmann M., Carvalho V. L., Casseb A. R., Silva E. V. P., Nunes K. N. B., Chiang J. O., Martins L. C. & other authors (2013). Changuinola virus serogroup, new genomes within the genus Orbivirus (family Reoviridae) isolated in the Brazilian Amazon region. Genome Announc 1, e00940–13. 10.1128/genomeA.00940-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbi C., Lemey P., Suchard M. A., Abdelatif E., Elharrak M., Nourlil J., Faouzi A., Echevarría J. E., Vazquez Morón S. & other authors (2010). Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog 6, e1001166. 10.1371/journal.ppat.1001166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Chaniotis B. N., Peralta P. H., Johnson K. M. (1974). Ecology of viruses isolated from Panamanian phlebotomine sandflies. Am J Trop Med Hyg 23, 258–269. [DOI] [PubMed] [Google Scholar]
- Travassos da Rosa A. P. A., Tesh R. B., Pinheiro F. P., Travassos da Rosa J. F. S., Peralta P. H., Knudson D. L. (1984). Characterization of the Changuinola serogroup viruses (Reoviridae: Orbivirus). Intervirology 21, 38–49. 10.1159/000149501 [DOI] [PubMed] [Google Scholar]
- Travassos da Rosa J. F. S., Travassos da Rosa A. P. A., Pinheiro F. P., Vasconcelos P. F. C., Rodrigues S. G. (1998). An overview of arbovirology in Brazil and neighboring countries.
- Verwoerd D. W., Els H. J., De Villiers E. M., Huismans H. (1972). Structure of the bluetongue virus capsid. J Virol 10, 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C. M., Nunes M. R. T., da Silva E. V. P., Carvalho V. L., Nunes Neto J. P., Cruz A. C. R., Casseb S. M. M., Vasconcelos H. B., Quaresma J. A. S., Vasconcelos P. F. D. C. (2009). Full-length sequencing and genetic characterization of Breu Branco virus (Reoviridae, Orbivirus) and two related strains isolated from Anopheles mosquitoes. J Gen Virol 90, 2183–2190. 10.1099/vir.0.010165-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.