Abstract
We investigated whether the abundance of bitter receptor mRNA expression from human taste papillae is related to an individual’s perceptual ratings of bitter intensity and habitual intake of bitter drinks. Ratings of the bitterness of caffeine and quinine and three other bitter stimuli (urea, propylthiouracil, and denatonium benzoate) were compared with relative taste papilla mRNA abundance of bitter receptors that respond to the corresponding bitter stimuli in cell-based assays (TAS2R4, TAS2R10, TAS2R38, TAS2R43, and TAS2R46). We calculated caffeine and quinine intake from a food frequency questionnaire. The bitterness of caffeine was related to the abundance of the combined mRNA expression of these known receptors, r = 0.47, p = .05, and self-reported daily caffeine intake, t(18) = 2.78, p = .012. The results of linear modeling indicated that 47% of the variance among subjects in the rating of caffeine bitterness was accounted for by these two factors (habitual caffeine intake and taste receptor mRNA abundance). We observed no such relationships for quinine but consumption of its primary dietary form (tonic water) was uncommon. Overall, diet and TAS2R gene expression in taste papillae are related to individual differences in caffeine perception.
Keywords: bitter taste, psychophysics, human, mRNA expression, TAS2R, copy number variation
Introduction
Of the primary taste qualities, bitter perception varies most among people (Knaapila et al., 2012). Bitterness arises when chemicals contact bitter receptors on taste cells. Investigators discovered the TAS2R gene family of receptors that detect bitter compounds more than a decade ago (Adler et al., 2000; Chandrashekar et al., 2000; Matsunami, Montmayeur, & Buck, 2000). This family contains about 25 G protein-coupled receptors, but the exact number varies due to copy number variation (Nozawa, Kawahara, & Nei, 2007; Pronin et al., 2007; Roudnitzky, Bufe, et al., 2011; Wong et al., 2007). The conscious experience of bitterness differs among people in part due to genetic variation in these sensory receptors. Investigators demonstrated this principle for particular receptor-ligand pairs, most notably the receptor T2R38 and the ligands phenylthiocarbamide and propylthiouracil (PROP; Bufe et al., 2005; Kim et al., 2003). The TAS2R38 gene has two main haplotypes, PAV (P49, A262, and V296) and AVI (A49, V262, and I296), that determine individual differences in the perceived bitterness of these receptor agonists (Behrens, Gunn, Ramos, Meyerhof, & Wooding, 2013; Campbell et al., 2012; Duffy et al., 2004; Genick et al., 2011; Mennella, Pepino, Duke, & Reed, 2010; Mennella, Pepino, & Reed, 2005; Prodi et al., 2004; Reed et al., 2010; Timpson et al., 2007).
Even within a TAS2R38 haplotype group, there can be wide variation in sensitivity to phenylthiocarbamide and PROP. We recently investigated whether these person-to-person differences in bitter perception were related to the expression of this receptor’s mRNA (Lipchock, Mennella, Spielman, & Reed, 2013). Variation in mRNA abundance in taste cells may reflect the number of receptors present in the taste papillae, which in turn may explain some of the wide variation in bitter taste perception among those sharing the same genotype (Mennella et al., 2010; Newcomb, Xia, Reed, 2012). We previously focused on the TAS2R38 gene and individuals who were heterozygotes (PAV or AVI) for its two common taster (PAV) and nontaster (AVI) alleles. Subjects with more mRNA expression of the taster allele reported more bitterness from the ligand (Lipchock et al., 2013).
There are large differences among people in their perception of the bitterness of common compounds such as caffeine which are due (in part) to inborn genetic variation (Ledda et al., 2014). Likewise, the perception of quinine differs among people by genotype (Reed et al., 2010). However, we do not know the extent to which gene expression may also account for these differences, as they do for TAS2R38 mRNA and its ligands. We wanted to understand whether the positive relationship between mRNA bitter receptor expression and bitter taste perception is a general observation or is unique to the T2R38 receptor and its ligands. Thus, in the present study, we examined this relationship for caffeine and quinine and the TAS2R mRNA for some receptors stimulated by these agonists (Meyerhof et al., 2010).
Subjects and Methods
Overview
Subjects rated the bitterness intensity of several compounds using the psychophysical measures explained later and immediately afterward provided a sample of tongue taste tissue. They also completed a food frequency questionnaire about their habitual intake of specific food items, for example, coffee.
Subjects
The study population (N = 20) was 60% female, 19 to 40 years of age (31 ± 1 years), and represented the ethnic diversity of Philadelphia (35% of African ancestry, 55% of European ancestry, and 10% other or mixed race). All were healthy, normotensive nonsmokers. Subjects were selected based on TAS2R38 genotype in human taste tissue; 18 were heterozygotes, one was homozygous PAV/PAV (taster) and one was homozygous AVI/AVI (nontaster); data on allele-specific variation in TAS2R38 expression levels and bitter taste perception were previously published (Lipchock et al., 2013). The Office of Regulatory Affairs at the University of Pennsylvania approved all procedures for the study, and each subject gave informed consent. We registered this trial at clinicaltrials.gov (NCT01399944).
Psychophysics
After being trained to use the general Labeled Magnitude Scale (gLMS), subjects tasted a variety of bitter compounds (see later or Table 1) and rated them on a computerized scale using Compusense five Plus software (Compusense Inc., Guelph, ON, Canada). We tested subjects individually between 8:30 a.m. and 1:30 p.m. in a closed room designed for sensory (psychophysics) studies. They were instructed to eat their typical breakfast or lunch and to arrive at the testing location having not eaten for at least 1 hr.
Table 1.
Bitter Receptors and Taste Compounds Tested.
| Bitter receptor gene | Liganda |
|---|---|
| TAS2R4 | Denatonium benzoate, quinine |
| TAS2R10 | Denatonium benzoate, caffeine, quinine |
| TAS2R38 | PROP |
| TAS2R43 | Denatonium benzoate, caffeine, quinine |
| TAS2R46 | Denatonium benzoate, caffeine, quinine |
Note. PROP = propylthiouracil.
Recognized by the protein product of the bitter receptor gene. No known bitter receptors are reported to respond to urea.
Subjects tasted 5mL of each stimulus, presented in random order. The stimuli comprised five bitter compounds (560 μM PROP, 500mM urea, 8mM caffeine, 492nM denatonium benzoate, 119 μM quinine hydrochloride) and a control stimulus for general taste sensitivity (100mM sodium chloride). Concentrations were selected based on a previous study of individual differences in perception (Delwiche, Buletic, & Breslin, 2001). Subjects swished the stimulus solution in their mouths for 5 s, expectorated, and rated the bitterness and saltiness using the gLMS. (Subjects also made other ratings but did not present them here for brevity.) A 1-min interval, in which subjects rinsed twice with deionized water, separated the tasting of each stimulus. To ensure that differences in perception were specific to taste sensations and not to scale use, subjects were also asked to use the gLMS to rate the heaviness of six opaque, sand-filled jars ranging from 235 to 955 g; heaviness ratings were used to normalize taste intensity ratings (Delwiche et al., 2001).
Taste Tissue Collection
We collected tissue in a surgical suite between 8:30 a.m. and 1:30 p.m. on the same day as taste testing, immediately after the psychophysical testing. Before tissue collection, a dentist interviewed subjects about medical and dental history and inspected the tongue to confirm that each subject had no oral disease. We measured heart rate and blood pressure to ensure they were in the normal range. Under sterile conditions, six to eight fungiform papillae were removed from the dorsal surface of the anterior tongue using curved spring microscissors (McPherson-Vannas type, Roboz; for detailed description of methods, see Spielman, Pepino, Feldman, & Brand, 2010). Each papilla was immediately transferred with forceps into Ringer buffer (130mM NaCl, 5mM KCl, 1mM CaCl2, 1mM MgCl2, 1mM Na-pyruvate, 20mM HEPES, pH 7.2), and after six to eight papillae (7.2 ± 0.2) were removed, combined, and transferred to RNA later (Invitrogen, Life Technologies Corp. Carlsbad CA) and stored at −80°C.
RNA Extraction, cDNA Preparation, and Gene Expression Analyses
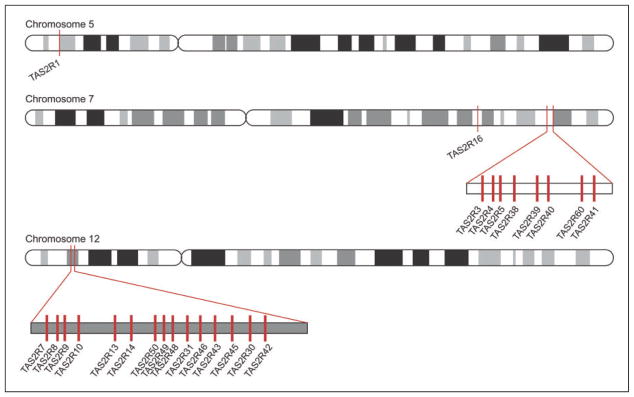
Within 3 days of papilla collection, the tissue was thawed on ice and homogenized using Kontes disposable pellet pestles and 1.5mL microtubes (Kimble Chase, Vineland, NJ). RNA was extracted with the ZR-Duet DNA/RNA MiniPrep kit (Zymo Research Corporation, Irvine, CA), and cDNA was generated using the WT-Ovation RNA Amplification System (NuGEN Technologies, Inc., San Carlos, CA). Samples were treated with DNAase (Zymo Research, Irvine, CA) for 30 min to remove residual genomic DNA. TAS2R mRNA was measured for TAS2R4, TAS2R10, TAS2R38, TAS2R43, and TAS2R46 (Table 1). We chose TAS2Rs that were shown in cell-based assays to be activated by at least three of the five bitter compounds tested here. We show their chromosomal positions relative to the other bitter receptors in Figure 1. Information about their physical location is useful because genetic variants in adjacent receptors are often coinherited. We measured GNAT3 gene expression to ensure the papillae contained the appropriate cell type (Type II taste receptor cells); gene expression for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was measured as a general housekeeping gene. In all cases, we measured gene expression using TaqMan real-time polymerase chain reaction (PCR) gene expression assays (Invitrogen, 4326317E, Hs00249946_s1, Hs00256794_s1, Hs00604294_s1, Hs00820217_s1, and Hs00853124_s1, and Hs01385400_m1). We compared gene expression of GNAT3 and all TAS2Rs to GAPDH using the delta–delta Ct method (Livak & Schmittgen, 2001).
Figure 1.
Bitter receptor genes and their location on human chromosomes.
We repeated assays in triplicate, and the median was calculated. The log of gene expression relative to the GAPDH control gene expression was used in all analyses, with values of 0 changed to 0.001 before log transformation.
Follow-up Assessment of Copy Number Variation of TAS2R43
TAS2R43 expression was very low compared with the other genes analyzed, and 14 of the 20 subjects had no detectable expression. We hypothesized that this result might be due to a deletion of the gene and therefore measured TAS2R43 copy number for all participants. Copy number variation of the TAS2R43 gene was determined using genomic DNA that was extracted from saliva following the directions of the manufacturer (Epicenter, Madison, WI) and following previously published methods (Wooding et al., 2012). The presence of TAS2R43 in the genome was determined using the TaqMan real-time PCR gene expression assay for TAS2R43 (Hs00820217_s1). RNaseP was used to determine the number of copies of TAS2R43 using the equation copies of TAS2R43 per genome = (2((CtRNaseP−CtTAS2R43)−(AvgCtRNaseP−AvgCtTAS2R43))) × 2. In addition to copy number genotype, we also assayed two additional variants. Using the genomic DNA, we genotyped the samples for those variants previously reported to be associated with perception of caffeine and quinine (Supplemental Table 1).
Bitter agonists often stimulate more than one receptor, and we tested the hypothesis that perception might be more tightly related to bitter receptor mRNA expression when multiple receptors were considered additively. mRNA expression levels from subsets of receptors were summed using information about receptor-ligand interactions summarized in Table 1. For example, we summed mRNA expression for TAS2R10, TAS2R43, and TAS2R46 to evaluate the bitterness of caffeine because these three receptors have been demonstrated to interact with caffeine (Meyerhof et al., 2010).
Habitual Consumption of Caffeine and Tonic Water
Subjects completed a questionnaire to quantify their habitual use of caffeine (e.g., coffee) and quinine (i.e., in tonic water) using a modified version of the Harvard Service food-frequency check-list, a semiquantitative dietary assessment tool (Suitor, Gardner, & Willett, 1989). Reported intake (servings/month) for beverage items was determined by using the following scoring system: never = 0, less than once per month = 0.5, 1 to 3 times per month = 2, once per week = 4, 2 to 4 times per week = 12, 5 to 6 times per week = 22, and one or more times per day = 30 servings/month (Feskanich et al., 1993; Paul, Rhodes, Kramer, Baer, & Rumpler, 2005). In the case of caffeine, we considered aggregated reports of coffee, tea, and carbonated beverages into a single frequency rating (daily user, yes or no); in the case of quinine, we used reported intake of a single item, tonic water and divided people into two groups based on reported use (never vs. occasional).
Statistical Analyses
All analyses were conducted using either Statistica version 10 (StatSoft, Inc., Tulsa, OK) or GraphPad Prism 6 (version 6.05; GraphPad Software, La Jolla, CA). We calculated Pearson correlation coefficients between gene expression values and perceptual ratings except for TASR43 mRNA expression (which was bimodal owing to the deletion of this gene in some people) in which case we used a nonparametric method (Spearman correlation) to determine relationships between mRNA abundance and bitterness ratings. To probe for relationships between genetic variants and bitterness ratings, we used a one-way analysis of variance using genotype as a factor and bitterness ratings as the outcome measure. We analyzed differences between daily users of caffeine versus those that consumed caffeine less frequently for differences in bitterness perception or gene expression by t test. We analyzed the tonic water consumption groups (never vs. occasional) the same way. Finally, we used a general linear model to evaluate the contribution of each single variable that affected caffeine bitterness ratings. And p < .05 was used as a statistical criterion for significance.
Results
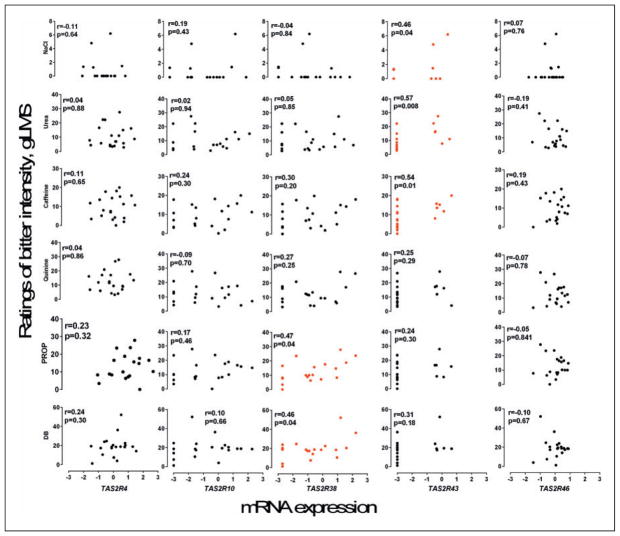
We show the distributions of relative gene expression and ratings of bitterness for each compound tested in Figure 2. Expression of TAS2R38 was correlated with bitterness ratings of PROP, r(18) = 0.51, p = .023. Similarly, people expression of TAS2R43 mRNA was correlated with the bitterness of urea, r(18) = 0.57, p = .008, and caffeine, r(18) = 0.54, p = .014. TAS2R43 expression was also related to the bitterness of sodium chloride, r(18) = 0.46, p = .04, but not its saltiness, r(18) = −0.02, p = .92. mRNA abundance of TAS2R4, TAS2R10, and TAS2R46 was not individually related to bitterness of any of the compounds tested.
Figure 2.
mRNA expression for each bitter receptor gene versus subject ratings of bitterness for each taste compound. Each dot represents data from one person (N = 20 subjects). Red dots highlight cases were ratings of bitterness were related to mRNA abundance, p < .05.
DB = denatonium benzoate; PROP = propylthiouracil.
Inborn copy number variants explained in part the bimodal distribution of TAS2R43 gene expression. Six subjects had two deleted copies of the TAS2R43 gene and had no measurable TAS2R43 mRNA expression. 8 of the 14 subjects with at least one copy of the TAS2R43 gene also showed no mRNA expression; for the remaining six subjects, expression of TAS2R43 mRNA was not related to genomic copy number, r = 0.08, p = .87. There was no significant relationship among people grouped by other genotypes (rs2708377 for caffeine or rs10772420 for quinine; Supplemental Table 1). This result is likely due to the small sample sizes for each genotype group and low statistical power.
Summed receptor mRNA expression, based on known responsiveness to ligands in cell-based assays, was significantly correlated with the perception of caffeine, r(18) = 0.47, p = .036, but not quinine, r(18) = 0.06, p = .79, or denatonium benzoate, r(18) = 0.27, p = .24 (Figure 3). This type of analysis could not be conducted for urea because the receptor(s) is unknown, or for PROP, because only one receptor responds to this ligand in most cell-based assays.
Figure 3.
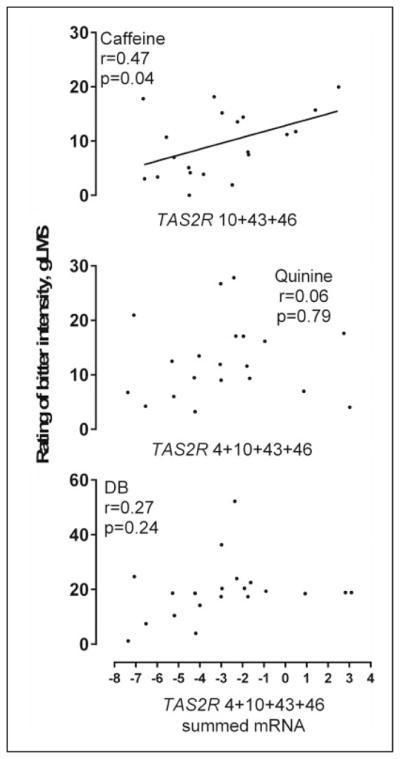
Summed mRNA expression for those bitter receptors tested that are known to respond to specific ligands tested.
DB = denatonium benzoate.
Regarding effects of regular caffeine intake on bitterness ratings, we observed that subjects fell into one of two groups. Either they were daily consumers of caffeine or not; therefore we classified subjects as daily consumers (N = 11; at least one serving per day, 30 servings a month) or infrequent consumers (N = 9; less than 30 servings a month; Supplemental Table 1). Tonic water was rarely consumed; thus we grouped the occasional consumers together (N = 5; 1–3 times per month or less than once per month) compared with those who reported never consuming tonic water (N = 15). Daily consumption of caffeine was related to reported bitterness (t(18) = 2.78, p = .012) but not to summed mRNA expression (t(18) = 0.50, p = .63). There was no relationship between tonic water consumption and ratings of quinine bitterness (t(18) = 0.94, p = .36) or summed mRNA abundance (t(18) = 0.54, p = .60; Figure 4).
Figure 4.
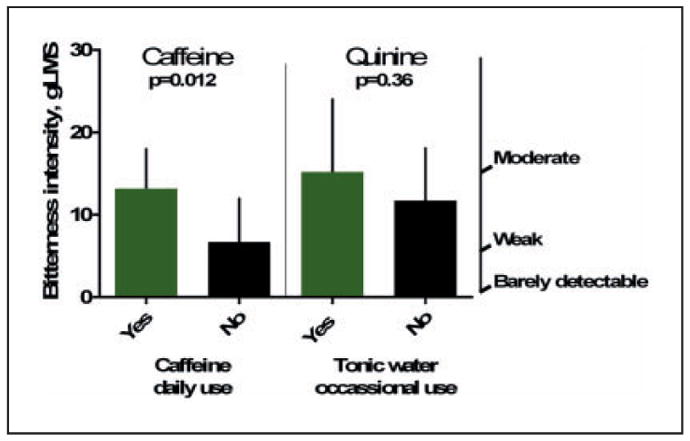
Ratings of perceived intensity of caffeine (left) and quinine (right) on the general Labeled Magnitude Scale scale that is anchored by descriptors shown on the right-hand x-axis. Subjects were grouped by daily caffeine use (yes or no) and by never or occasional use of tonic water, a beverage which contains quinine.
We probed for the relationship between diet and gene expression on caffeine bitterness ratings with a general linear model, using the two variables in the above analysis that were related to the intensity ratings of the bitterness of caffeine. Thus, the daily consumption of caffeine was a categorical variable (yes or no), and the summed gene expression was a continuous variable, that is, ranging from −6.6 to 2.6 (relative to the housekeeping gene). This model accounted for 47% of the total variance in caffeine bitterness ratings, with equal and mostly independent contributions for diet versus gene expression (Table 2).
Table 2.
Results of General Linear Model for Ratings of Caffeine Bitterness.
| Variable | df | F | p Value | η2 |
|---|---|---|---|---|
| Daily consumption | 1 | 5.4 | .038 | 0.32 |
| Summed mRNA | 1 | 7.9 | .012 | 0.24 |
| % Total variance (r2) | 0.47 |
Discussion
Approximately 25 human genes code for bitter taste receptors and specialized cells in human taste papillae express their mRNA. In this study, subjects who produced more relevant bitter receptor mRNA perceived more bitterness when they tasted caffeine. The idea that perception is tied to receptor abundance has roots in earlier observations that papilla number on the tongue predicts taste intensity (Miller & Reedy, 1990) and is further supported by the observation that individual differences in the perception of different bitters form clusters (Delwiche et al., 2001). These clusters follow a pattern that suggests they are determined in part by genetic variation in shared receptors (Roura et al., 2015). Thus, the results of this study are somewhat consistent with the broader hypothesis that when more receptors are activated, the signal to the brain is interpreted as more intense. We did not see these same relationship for quinine or denatonium benzoate, although this outcome may be due to experimental limitations, for example, we did not test all bitter receptor mRNA expression and thus may have missed critical receptors that have high ligand affinity. Thus, confirmation of this general hypothesis awaits direct quantification of the receptor density of the complete receptor repertoire and more complete knowledge of receptor-ligand interactions of all receptor haplotypes.
The TAS2R38 gene and its protein product are unusual in two respects: There are two equally common forms that differ markedly in function (Kim et al., 2003), and the receptor is an extreme specialist, responding only to a few structurally related ligands (Bufe et al., 2005; Meyerhof et al., 2010). We previously reported that the amount of mRNA of the taster form of the TAS2R38 bitter receptor was associated with the bitter perception of its ligand. Here, we examined whether this result would generalize to other receptors that are less specialized. While neither the five bitter compounds nor the receptors tested here are exhaustive, the relationship of mRNA to perception does appear to be a feature of human taste biology if only for caffeine. Caffeine is a ligand for TAS2R10, TAS2R43, and TAS2R46 (Meyerhof et al., 2010), and the amount of mRNA from these three genes together was associated with subjects’ ratings of caffeine bitterness.
The TAS2R43 gene is absent from the genome of many people (Pronin et al., 2007; Roudnitzky, Bufe, et al., 2011; Wooding et al., 2012), and previous reports of this structural polymorphism were confirmed in this study: 6 of 20 subjects (30%) had two deleted copies of the TAS2R43 gene, and 14 of 20 (70%) had no corresponding mRNA. The abundance of TAS2R43 mRNA was related to the perceptual ratings of the bitterness of caffeine, urea, and sodium chloride. However, this result does not mean that this particular receptor necessarily binds those particular ligands. The deletion polymorphism may be a proxy for genomic variants among other nearby receptors in chromosome 12 (linkage disequilibrium; Figure 1); Thus, this result points to an influential genomic region rather than to this particular gene. Genes that code for salivary proline-rich proteins are also in this genetic vicinity (Matsunami et al., 2000), and these proteins may play a supporting or even key role in individual differences in taste perception (Dsamou et al., 2012). While the association between urea perception (and the bitterness of sodium chloride) and TAS2R43 mRNA abundance is worth noting, it does not specifically identify these as a receptor-ligand pairs.
This study had at least one limitation: the lack of control over the underlying genotypes for most of the bitter receptors. Because of the design of the study, almost all subjects were heterozygous for the two common haplotypes of the TAS2R38 gene, so that we could distinguish the effects of TAS2R38 gene expression from genotype. We did not attempt to control the genotypes of the other bitter receptors, which also have known effects on perception (Ledda et al., 2014; Reed et al., 2010). Assessing the joint effects of gene expression and genotypes more broadly is a logical next step but not an easy one because of the extreme varieties of human haplotypes and their complex effects on taste perception (Roudnitzky, Behrens, et al., 2015; Roudnitzky, Bufe, et al., 2011). This approach will require larger sample sizes than those used here.
A salt solution was included as a control, but we learned that there is a bitter component to its perception. People with more TAS2R43 expression reported that 100mM sodium chloride was more bitter than did those with less expression. Recently, Oka et al. (2013) reported that high salt (> 300mM) but not low salt (< 100mM) solutions recruit aversive taste pathways by activating sour- and bitter-taste-sensing cells. Other data suggest that bitter transduction pathways may play a role even at lower salt concentration like that tested here (Tordoff et al., 2014). The preceding two studies were conducted in rodents but they nevertheless may be informative. They are consistent with our data that suggest that the bitter taste of salt concentrations may be due in part to stimulation of bitter receptors although we mention the caveat as to their nature and location above.
Other investigators have tested the idea that caffeine consumption may affect the perception of caffeine bitterness. They did so by asking subjects to ingest caffeine either acutely or chronically and measuring how these manipulation may change how low a concentration of caffeine the subject can detect (Mela, Mattes, Tanimura, & Garcia-Medina, 1992). The results of these studies were inconsistent and differed by study population, perhaps pointing to inborn genetic or cultural influences like habitual diet. Thus, regulators of bitter receptor transcription in taste cells are almost entirely unknown, but exposure to bitter compounds (especially early in life) may push expression up and sensitize the animal later in life (Cheled-Shoval, Behrens, Meyerhof, Niv, & Uni, 2014; Lipchock et al., 2013). Our next step is to identify the driving force behind individual transcriptional differences, which has implications for improving diet, designing better-accepted pediatric medicine formulations (Giacoia, Taylor-Zapata, & Zajicek, 2012; Mennella, Spector, Reed, & Coldwell, 2013), and understanding bitter gene regulation in pathways outside the tongue (Jeon, Zhu, Larson, & Osborne, 2008; Lee et al., 2012).
Supplementary Material
Acknowledgments
The authors acknowledge the expert technical assistance of Sara Castor, Laura Overton née Lukasewycz, Loma Inamdar, Kristi Roberts, Christina Furia, Anna Lysenko, Rebecca James, Charles J. Arayata, Hakan Ozdener, and Brad Fesi and the editorial assistance of Patricia Watson. Michael G Tordoff and Gary K. Beauchamp commented on the manuscript prior to publication. The authors’ responsibilities were as follows—S. V. L., J. A. M., and D. R. R.: designed the research, analyzed data, performed statistical analysis, and wrote the manuscript; S. V. L., L. D. H., and A. I. S.: conducted the research; and J. A. M. and D. R. R.: share primary responsibility for the final content. C. J. M. and J. E. D. performed statistical analysis and collected data. All authors read and approved the final manuscript.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health [T32DC000014 and F32DC011975 to S. V. L., P30DC011735 and R21DC013886 to D. R. R., and R01DC011287 to J. A. M.].
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary material for this paper can be found at http://journals.sagepub.com/doi/suppl/10.1177/0301006616686098.
Contributor Information
Sarah V. Lipchock, Monell Chemical Senses Center, Philadelphia, PA, USA
Andrew I. Spielman, Department of Basic Science and Craniofacial Biology, College of Dentistry, New York University, NY, USA
Julie A. Mennella, Monell Chemical Senses Center, Philadelphia, PA, USA
Corrine J. Mansfield, Monell Chemical Senses Center, Philadelphia, PA, USA
Liang-Dar Hwang, Monell Chemical Senses Center, Philadelphia, PA, USA.
Jennifer E. Douglas, Monell Chemical Senses Center, Philadelphia, PA, USA
Danielle R. Reed, Monell Chemical Senses Center, Philadelphia, PA, USA
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Behrens M, Gunn HC, Ramos PC, Meyerhof W, Wooding SP. Genetic, functional, and phenotypic diversity in TAS2R38-mediated bitter taste perception. Chemical Senses. 2013;38:475–484. doi: 10.1093/chemse/bjt016. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, … Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Current Biology. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Ranciaro A, Froment A, Hirbo J, Omar S, Bodo JM, … Tishkoff SA. Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa. Molecular Biology and Evolution. 2012;29:1141–1153. doi: 10.1093/molbev/msr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Cheled-Shoval SL, Behrens M, Meyerhof W, Niv MY, Uni Z. Perinatal administration of a bitter tastant influences gene expression in chicken palate and duodenum. Journal of Agricultural and Food Chemistry. 2014;62:12512–12520. doi: 10.1021/jf502219a. [DOI] [PubMed] [Google Scholar]
- Delwiche JF, Buletic Z, Breslin PA. Covariation in individuals’ sensitivities to bitter compounds: Evidence supporting multiple receptor/transduction mechanisms. Perception & Psychophysics. 2001;63:761–776. doi: 10.3758/bf03194436. [DOI] [PubMed] [Google Scholar]
- Dsamou M, Palicki O, Septier C, Chabanet C, Lucchi G, Ducoroy P, … Morzel M. Salivary protein profiles and sensitivity to the bitter taste of caffeine. Chemical Senses. 2012;37:87–95. doi: 10.1093/chemse/bjr070. [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, … Bartoshuk LM. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcoholism: Clinical and Experimental Research. 2004;28:1629–1637. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feskanich D, Rimm EB, Giovannucci EL, Colditz GA, Stampfer MJ, Litin LB, … Willett WC. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. Journal of the American Dietetic Association. 1993;93:790–796. doi: 10.1016/0002-8223(93)91754-e. [DOI] [PubMed] [Google Scholar]
- Genick UK, Kutalik Z, Ledda M, Souza Destito MC, Souza MM, Cirillo CA, … le Coutre J. Sensitivity of genome-wide-association signals to phenotyping strategy: The PROP-TAS2R38 taste association as a benchmark. PLoS One. 2011;6:e27745. doi: 10.1371/journal.pone.0027745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacoia GP, Taylor-Zapata P, Zajicek A. Eunice Kennedy Shriver National Institute of Child Health and Human Development pediatrics formulation initiative: Proceedings from the second workshop on pediatric formulations. Clinical Therapeutics. 2012;34:S1–S10. doi: 10.1016/j.clinthera.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Jeon TI, Zhu B, Larson JL, Osborne TF. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. The Journal of Clinical Investigation. 2008;118:3693–3700. doi: 10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, … Reed DR. Genetic analysis of chemosensory traits in human twins. Chemical Senses. 2012;37:869–881. doi: 10.1093/chemse/bjs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda M, Kutalik Z, Souza Destito MC, Souza MM, Cirillo CA, Zamboni A, … Genick UK. GWAS of human bitter taste perception identifies new loci and reveals additional complexity of bitter taste genetics. Human Molecular Genetics. 2014;23:259–267. doi: 10.1093/hmg/ddt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RJ, Xiong G, Kofonow JM, Chen B, Lysenko A, Jiang P, … Cohen NA. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. The Journal of Clinical Investigation. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchock SV, Mennella JA, Spielman AI, Reed DR. Human bitter perception correlates with bitter receptor messenger RNA expression in taste cells. American Journal of Clinical Nutrition. 2013;98:1136–1143. doi: 10.3945/ajcn.113.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- Mela DJ, Mattes RD, Tanimura S, Garcia-Medina MR. Relationships between ingestion and gustatory perception of caffeine. Pharmacology Biochemistry and Behavior. 1992;43:513–521. doi: 10.1016/0091-3057(92)90186-j. [DOI] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Duke FF, Reed DR. Psychophysical dissection of genotype effects on human bitter perception. Chemical Senses. 2010;36:161–167. doi: 10.1093/chemse/bjq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Reed DR. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115:e216–e222. doi: 10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella JA, Spector AC, Reed DR, Coldwell SE. The bad taste of medicines: Overview of basic research on bitter taste. Clinical Therapeutics. 2013;35:1225–1246. doi: 10.1016/j.clinthera.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, … Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chemical Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Miller IJ, Jr, Reedy FE., Jr Variations in human taste bud density and taste intensity perception. Physiology & Behavior. 1990;47:1213–1219. doi: 10.1016/0031-9384(90)90374-d. [DOI] [PubMed] [Google Scholar]
- Newcomb RD, Xia MB, Reed DR. Heritable differences in chemosensory ability among humans. Flavour. 2012;1:9. [Google Scholar]
- Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proceedings of the National Academy of Sciences USA. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Butnaru M, von Buchholtz L, Ryba NJ, Zuker CS. High salt recruits aversive taste pathways. Nature. 2013;494:472–475. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DR, Rhodes DG, Kramer M, Baer DJ, Rumpler WV. Validation of a food frequency questionnaire by direct measurement of habitual ad libitum food intake. American Journal of Epidemiology. 2005;162:806–814. doi: 10.1093/aje/kwi279. [DOI] [PubMed] [Google Scholar]
- Prodi DA, Drayna D, Forabosco P, Palmas MA, Maestrale GB, Piras D, … Angius A. Bitter taste study in a Sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chemical Senses. 2004;29:697–702. doi: 10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Current Biology. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, … Wright MJ. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Human Molecular Genetics. 2010;19:4278–4285. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnitzky N, Behrens M, Engel A, Kohl S, Thalmann S, Hubner S, … Meyerhof W. Receptor polymorphism and genomic structure interact to shape bitter taste perception. PLoS Genetics. 2015;11:e1005530. doi: 10.1371/journal.pgen.1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnitzky N, Bufe B, Thalmann S, Kuhn C, Gunn HC, Xing C, … Wooding SP. Genomic, genetic, and functional dissection of bitter taste responses to artificial sweeteners. Human Molecular Genetics. 2011;20:3437–3449. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- Roura E, Aldayyani A, Thavaraj P, Prakash S, Greenway D, Thomas WG, … Foster SR. Variability in human bitter taste sensitivity to chemically diverse compounds can be accounted for by differential TAS2R activation. Chemical Senses. 2015;40:427–435. doi: 10.1093/chemse/bjv024. [DOI] [PubMed] [Google Scholar]
- Spielman AI, Pepino MY, Feldman R, Brand JG. Technique to collect fungiform (taste) papillae from human tongue. Journal of Visualized Experiment. 2010;18:2201. doi: 10.3791/2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suitor CJ, Gardner J, Willett WC. A comparison of food frequency and diet recall methods in studies of nutrient intake of low-income pregnant women. Journal of the American Dietetic Association. 1989;89:1786–1794. [PubMed] [Google Scholar]
- Timpson NJ, Heron J, Day IN, Ring SM, Bartoshuk LM, Horwood J, … Davey-Smith G. Refining associations between TAS2R38 diplotypes and the 6-n-propylthiouracil (PROP) taste test: Findings from the Avon Longitudinal Study of Parents and Children. BMC Genetics. 2007;8:51. doi: 10.1186/1471-2156-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff MG, Ellis HT, Aleman TR, Downing A, Marambaud P, Foskett JK, … McCaughey SA. Salty taste deficits in CALHM1 knockout mice. Chemical Senses. 2014;39:515–528. doi: 10.1093/chemse/bju020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, … Lam WL. A comprehensive analysis of common copy-number variations in the human genome. The American Journal of Human Genetics. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding SP, Atanasova S, Gunn HC, Staneva R, Dimova I, Toncheva D. Association of a bitter taste receptor mutation with Balkan Endemic Nephropathy (BEN) BMC Medical Genetics. 2012;13:961. doi: 10.1186/1471-2350-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.