Summary
The antagonistic pleiotropy theory hypothesizes that evolutionary adaptations maximizing the fitness in early age increase disease burden after reproduction. This theory remains largely untested at the molecular level. Here, we analyzed enhancer evolution in primates to investigate the relationships between aging-related diseases and enhancers acquired after the human–chimpanzee divergence. We report a 5-fold increased evolutionary rate of enhancers that are activated in neural tissues, leading to fixation of ~100 human-specific enhancers potentially under adaptation. These enhancers show prognostic expression levels and correlations with driver genes in cancer, and their nearby genes are enriched in known loci associated with aging-related diseases. Using CRISPR/Cas9, we further functionally validated an enhancer on chr8p23.1 as activator counteracting REST, a master regulator known to be a transcriptional suppressor of Alzheimer’s disease. Our results suggest an evolutionary origin of aging-related diseases: the side effects of human-specific, neural-tissue expressed enhancers. Thus, adaptive molecular changes in human macroevolution may introduce vulnerabilities to disease development in modern populations.
Graphical Abstract
Introduction
Diseases of aging, such as cancer and Alzheimer’s, represent a major health burden and are collectively responsible for nearly 90% of deaths nationwide (Grey, 2007). With longer expected lifespans, this burden is increasing. Over recent decades, tremendous efforts have characterized the molecular basis of diseases related to aging in order to develop effective therapies. Advances in high-throughput technologies have enabled systematic approaches to identify hundreds of related vulnerability loci in the human genome (Jeck et al., 2012; Ramos et al., 2014). However, our understanding of the evolutionary origin of aging-related diseases remain very limited (Daniel Fabian, 2011).
Along with the cell division limit theory (Weismann, 1889) and mutation accumulation theory (Medawar, 1952), antagonistic pleiotropy is one of the major theories proposed to address the origin of aging-related diseases (Williams, 1957), in which trade-off is the key. As first suggested by Charles Darwin, compromise is inherent in every adaptation as a trade-off (Garland, 2014). George Williams further formulated the hypothesis and argued that if a change caused both increased reproduction in early life and aging in later life, then that change would be adaptive in evolution (Williams, 1957). If true, this theory should particularly apply to the fast adaptations humans have recently experienced, which should be accompanied by a series of trade-offs because of limited evolutionary time for fine tuning. These trade-offs then would have more profound impacts on the aging-related phenotypes of modern human populations than in any time of macroevolution due to our substantially increased lifespan over the last two centuries (Finch, 2010) and limited purifying selection, which has been weakened by the use of advanced medications (Daniel Fabian, 2011). Following this logic, we hypothesize that the adaptive molecular changes recently acquired in humans may contribute to the development of aging-related diseases. Since the classic paper by King and Wilson in 1975 (King and Wilson, 1975), increasing evidence has indicated that our evolution since the human-chimpanzee divergence has been driven more frequently by changes in gene regulatory elements than in protein sequences (Carroll, 2005; Haygood et al., 2007; Liang et al., 2008; Shi et al., 2006). Therefore, we focused on enhancers, a key class of noncoding regulatory elements (Andersson et al., 2014; Encode Project Consortium, 2012), to test our hypothesis.
Results
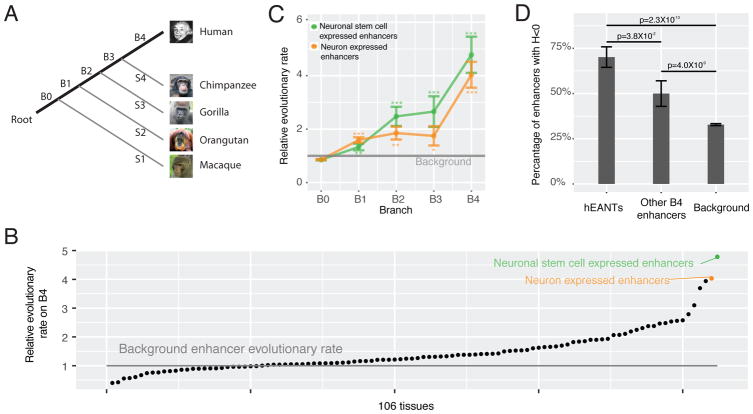
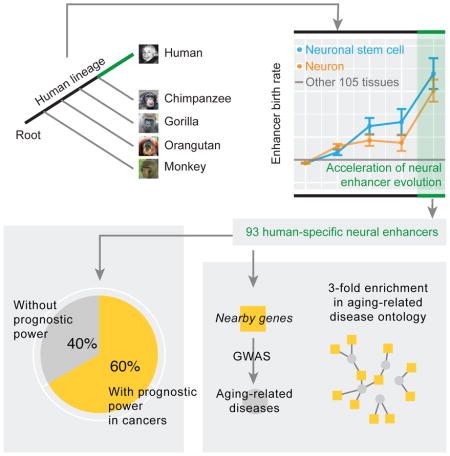
Accelerated evolution generates human-specific enhancers activated in nervous tissues
Our analysis took advantage of high-resolution, systematic annotation of human tissue-specific enhancers recently available from the FANTOM project (Andersson et al., 2014), which annotated 32748 enhancers showing tissue-biased expression patterns. We aligned each enhancer to the other four primate genomes (chimpanzee, gorilla, orangutan and macaque, as shown in Figure 1A) and inferred its origin based on the presence/absence of its orthologs in these primate species (Table S1, STAR Methods). Then, for a given tissue/cell type (n = 106) and a given branch segment (B0, …, B4) along the phylogenetic tree, we first calculated the proportion of the enhancers expressed in that tissue and evolved in that branch among the ones evolved in that branch, then calculated the proportion of the enhancers expressed in that issue among all the enhancers surveyed, and defined their ratio as the relative evolutionary rate of enhancers (STAR Methods). In this way, we used the birth rate of enhancers in all tissues as the internal control to normalize the effect of branch length, allowing a fair comparison across different tissues. Interestingly, we found that the neuronal stem cell expressed enhancers and the neuron expressed enhancers were the two fastest evolving categories on B4, the branch representing human-specific evolution (Figure 1B, Table S2). Because of their similar physiological origins, we combined these two groups of enhancers in the subsequent analyses. The evolution of these enhancers accelerated gradually and reached a >4-fold increased rate after the human-chimpanzee divergence (B4), suggesting that >75% of the enhancers that evolved on B4 are due to their elevated evolutionary rate. Importantly, this observed pattern remains the same when considering confounding factors (e.g., nucleotide composition, SNP density, and enhancer length) or technical issues (e.g., genome incompleteness and alignment errors) that could potentially compromise the accuracy of our phylogenetic analysis (Chen et al., 2015; Moyers and Zhang, 2016) (Table S3, Figure S1). In total, our analysis revealed 93 non-redundant human–specific enhancers activated in nervous tissues (47 and 66 in neuronal-stem-cells and neurons, respectively), which hereafter we refer to as hEANTs (Figure 1C, Table S1).
Figure 1. Accelerated gain of hEANTs during human-specific evolution.
(A) The primate phylogeny. B0… B4 donate the branch segments of different evolutionary ages. (B) Relative B4 evolutionary rate of all tissues. All enhancers (n = 32748) were categorized to be expressed in one or more of the tissue types annotated by the FANTOM project. Only the tissue types with >100 enhancers were considered (n = 106). The background rate was calculated based on enhancers evolved other than B4. (C) Accelerated gain of hEANTs in the primate evolution. P values were calculated using a chi-squared test (* for p < 0.05, ** for p < 0.01, and *** for p < 0.0001). Error bars represent mean ± SE. (D) Proportion of enhancers with a negative Fay & Wu’s H value (an indication of an excess of high-frequency derived alleles). A chi-squared test was used to compare the difference between any two groups. Error bars represent mean ± SE. See also Figures S1, S2 and Tables S1, S2, S3.
The evolutionary acceleration of hEANTs may be due to relaxed purifying selection or positive selection. We next examined their functional relevance and selective forces. Gene set enrichment analysis indicates that the hEANTs’ closest genes are functionally enriched in axon guidance (p =1.8×10−5, FDR = 0.01), a critical function for human brain development (Suarez et al., 2014). Since there are no hEANT orthologs in other primate genomes, the common methods for assessing the selective force in macroevolution such as dN/dS test and McDonald-Kreitman test could not be applied to these human-specific enhancers. However, given that the acceleration trend was observed across the whole human evolution in the primate lineage (Figure 1C), there is a possibility to detect some signals within human populations if some hEANTs were fixed by adaptation recently. For example, the BRCA1 gene under positive selection after the human-gorilla divergence still shows Hardy-Weinberg disequilibrium in the European population (Huttley et al., 2000). We therefore performed a population genetic analysis based on the 1000 Genomes Project (1000 Genomes Project et al., 2012) and found that 71% of these hEANTs have negative Fay & Wu’s H indexes, a well-established signature of recent positive selection (Fay and Wu, 2000). This proportion is significantly higher than that for other human-specific enhancers or the background enhancers (71% vs. 50%, p = 3.8×10−2; 71% vs. 32%, p = 2.3×10−10; Figure 1D), suggesting that positive selection may have played a key role in recruiting the hEANTs into the human genome. Interestingly, this adaptation appeared to be specific to enhancers, as protein-coding genes specifically expressed in the nervous tissues are substantially more conserved than those expressed in other tissues (Figure S2).
Besides the introduction of new enhancers into the human genome, another way to achieve evolutionary innovations is to modify existing enhancers by substitutions. We performed a comparative analysis between human and chimpanzee enhancers using the macaque as the out-group. However, given a very low enhancer substitution rate (dEnhancer = ~0.003) and an average enhancer length of ~300 bp, we only detected ~1.2 human-specific substitutions per enhancer and thus could not identify specific enhancers that may have undergone human-specific acceleration.
Evolutionary mechanisms of hEANTs
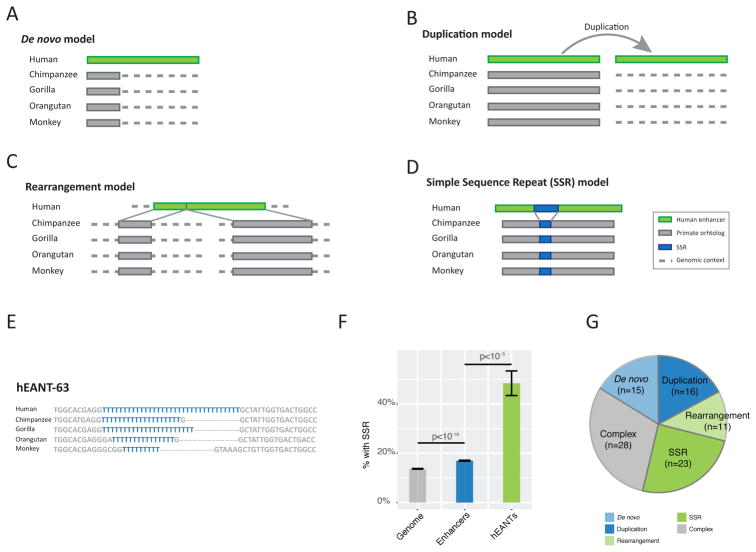
To understand how hEANTs originated during the macroevolution, we searched the homologous/orthologous sequences (orthologous sequences broken or missing according to the UCSC whole-genome alignments) of the 93 hEANTs and their flanking regions (500 bp) in other primate genomes using BLAST. We identified four major models of hEANT origination (Figure 2). (i) De novo model: for 15 hEANTs, at least 20% of the enhancer sequences and the corresponding flanking 500 bp regions have no homologous sequence in any of the primate genomes (BLAST e < 10−10), indicating their de novo origin or gene transfer from species far apart (Figure 2A). (ii) Duplication model: consistent with the notion that duplication is a major mechanism through which new genetic elements originate, 16 hEANTs are not the best hits when we BLAST the human genome with their best hits in other primate genomes as inputs, suggesting them to be recent duplications (Figure 2B). This model well explains why they have highly homologous sequences in other primates but no orthologs detected in the whole-genome alignment. (iii) Rearrange model: for 11 hEANTs, their orthologs were separated into at least two fragments locating separately in the primate genomes (e.g., on different chromosomes), indicating a human-specific rearrangement jointing them into one enhancer (Figure 2C). (iv) Simple sequence repeat (SSR) model: we also observed SSR extensions in 23 hEANTs which are often detected as breaks by the whole-genome alignment (Figure 2D); hEANT-63 serves as a good example of SSR extension in the primate lineage (SSR highlighted in blue Figure 2E).
Figure 2. Mechanistic models of hEANT origination.
(A) De novo model. (B) Duplication model. (C) Rearrangement model in which an enhancer is created by joining distant genome sequences. (D) Simple sequence repeat (SSR) model in which a hEANT evolves through SSR extension. (E) Alignments of hEANT-63 in the primate lineage. (F) Enrichment of SSR in the hEANTs. The genome background was calculated by randomly generating pseudo-enhancers with the same length distribution (STAR Methods). P values were calculated with permutation tests for enhancer/background comparison and with chi-squared test for hEANTs/enhancer comparison. Error bars represent mean ± SE. (G) A summary of hEANTs evolved through different models.
Interestingly, previous studies have indicated that SSRs usually locate in regulatory elements (Usdin, 2008), are unstable and confer hyper transcriptional evolvability in macroevolution (Vinces et al., 2009) and diversity within the human population (Gymrek et al., 2016). Consistently, we found that the hEANTs strongly enrich SSR, compared with the genome-wide background or all enhancers (Figure 2F), well explaining our findings of accelerated enhancer evolution. Besides the four simple evolutionary models, 28 hEANTs were classified as complex events since they could not be explained by single events, or the homologous sequences were deleted in some primates (Figure 2G).
Association of hEANTs with aging-related diseases
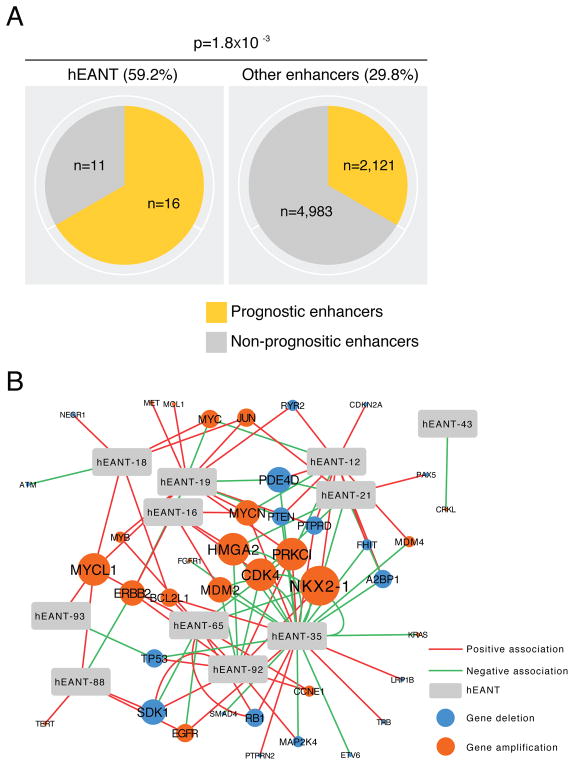
According to the antagonistic pleiotropy theory, we predicted that the hEANTs’ trade-offs can cause some deleterious effects on other phenotypic traits and contribute to aging-related diseases. To test this prediction, we first focused on cancer, a major group of aging-related diseases with the most comprehensive genomic data available through The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research et al., 2013). We used the expression levels of an enhancer calculated from TCGA RNA-seq data as an approximation of its activity in patient samples (Andersson et al., 2014) since ChIP-seq data are not available for the large patient cohorts. Across 23 TCGA cancer types (Table S4), we focused on 7131 enhancers with detectable expression and identified the prognostic enhancers, those whose expression was significantly associated with patient survival times (log-rank test or univariate Cox model, FDR < 0.05; Table S1, STAR Methods). We observed a 2.0-fold enrichment of prognostic enhancers in the hEANTs relative to all enhancers (59.2 vs. 29.8%, Fisher’s exact test p = 1.8×10−3 and Binomial test = 1.5×10−3; Figure 3A). Indeed, the number of cancer types prognostically associated with a hEANT is much higher than that for an enhancer otherwise (0.70 vs. 0.36, p = 0.013). We also examined the associations between the hEANTs and somatic copy number alterations (SCNAs) of known cancer genes in the TCGA cohorts and identified many associations of hEANTs with the amplifications or deletions of cancer driver genes such as TP53, PTEN and EGFR in multiple cancer types (Figure 3B). Collectively, these results strongly support active roles of hEANTs in cancer development.
Figure 3. The association of hEANTs with cancer.
(A) Enrichment of hEANTs in enhancers whose expression levels were significantly correlated with cancer patients’ survival times. The analysis only included 7131 out of the 32748 enhancers on genomic regions without overlaps with any previously annotated transcriptional events (e.g., genes, lincRNAs, and microRNAs). Prognostically relevant enhancers were identified in at least one of the 23 TCGA cancer types surveyed (q < 0.05, Table S4). P value was calculated using a Fisher’s exact test. (B) A network of associations between hEANTs and SCNAs of caner driver genes. See also Table S4.
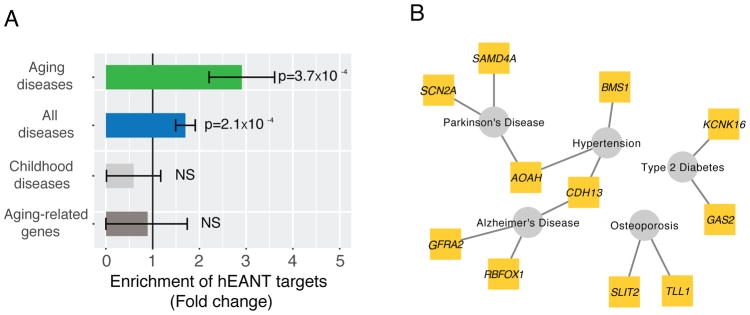
To more generally assess the impact of hEANTs on other aging-related diseases, we selected five well-known diseases from the PheGenI project (Ramos et al., 2014), including Alzheimer’s disease, Parkinson’s disease, hypertension, type-II diabetes and osteoporosis, and obtained their vulnerability loci identified from related genome-wide association studies. Interestingly, 11 hEANTs’ closest genes are associated with the loci identified for these diseases, leading to a 3-fold enrichment (p < 3.7×10−4, Figure 4, Table S5, Table S6). In comparison, there is even a depletion (although not significant) of the hEANTs’ closest genes for the susceptibility loci of five typical childhood diseases, including asthma, autism, juvenile arthritis, sickle cell anemia and type-I diabetes (Figure 4A). As previously reported, aging-related disease genes and aging-related genes have limited overlaps in most model organisms, indicating their functional differences (Fernandes et al., 2016). Consistently, the hEANT’s closet genes show no enrichment for aging-related genes (Figure 4A), suggesting that their associations with aging-related diseases were not due to the involvement of the general aging process (de Magalhaes et al., 2009).
Figure 4. The association of hEANTs with other aging-related diseases.
(A) Enrichment of hEANTs’ closest coding genes in genes associated with aging-related diseases and aging-related genes. Aging-related diseases included Alzheimer’s disease, Parkinson’s disease, hypertension, type-II diabetes and osteoporosis; childhood diseases included including asthma, autism, juvenile arthritis, sickle cell anemia and type-I diabetes. For disease association analysis, all genes included in the PheGenI database were used as the background gene set. The list of aging-related genes was obtained from Human Ageing Genomic Resources. P value was calculated using a chi-squared test. Error bars represent mean ± SE. (B) A network showing the connections between hEANTs’ closest genes and aging-related diseases. See also Table S5 and Table S6.
hEANT-8 promotes Alzheimer’s disease by counteracting REST
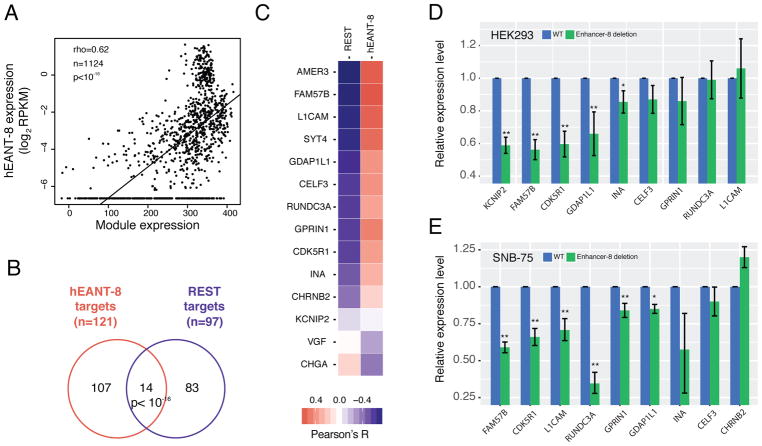
To elucidate the molecular mechanism by which a hEANT plays a critical role in aging-related diseases, we constructed a network of genes co-expressed with each hEANT using the expression data of TCGA brain tumors (i.e., glioblastomas and low grade glioma), as the brain is the organ in which hEANTs are supposed to function. Interestingly, we identified a well-organized co-expression module containing 121 potential target genes for a hEANT on the 8q23.1 region of chromosome 8 (hereafter referred to as hEANT-8) in both cancer types (Pearson’s R > 0.3 and FDR < 0.01, STAR Methods, Table S7). We further validated the association between the module expression and hEANT-8 using the GTEx dataset of normal brain tissues (GTEx Consortium, 2015) (Figure 5A). Further analysis of TCGA copy number data revealed that copy number variations of hEANT-8 can lead to module activity changes, suggesting its role as an upstream regulator of the module (Figure S3). This module is associated with a panel of neural or mental diseases (Schriml et al., 2012) (Figure S4). Intriguingly, through the search for motifs enriched on the module genes, we found enrichment of the binding motif of REST (Figure 5B, >24 fold, p < 10−16), a well-studied transcription suppressor in neurons (Schoenherr et al., 1996), indicating the module genes’ functional similarity. To further validate its effects on enhancing the module activity, we generated HEK293T cell lines with homozygous hEANT-8 deletion using the CRISPR/Cas9 genetic perturbation system (STAR Methods) and measured the expression levels of 11 module genes positively correlated with the enhancer (Figure 5C; top 11 genes, Table S8). Out of the 9 genes with a detectable mRNA expression level, 7 genes showed a reduced expression after hEANT-8 deletion, and the reductions in 5 of them were statistically significant, ranging from 0.60~0.85 (Figure 5D). We also observed similar effects in SNB-75, a human brain tumor cell line: 6 out of the 9 hEANT-8 positively correlated genes with a detectable mRNA expression level showed a significant reduction after hEANT-8 deletion (Figure 5E). These results confirmed the causal effects of hEANT-8 on the module activity.
Figure 5. Regulation of hEANT-8 on a downstream module enriched with REST target genes.
(A) The correlation between hEANT-8 expression and the module expression using the GTEx samples of the central nervous system (n = 1124). The module’s overall expression activity was calculated as the sum of log2RPKM across all 121 module genes in a sample. The correlation was measured as Spearman’s correlation coefficient (rho). (B) Enrichment of the hEANT-8 target genes (co-expressed module genes) in the REST target genes (genes with a REST binding motif). P value was calculated using a chi-squared test. (C) Co-expression between hEANT-8 and the 14 genes identified in (B). The colors indicate the correlations with REST or hEANT-8 expression level. (D) and (E) Expression changes of the 9 detectable genes whose expression is positively correlated with hEANT-8 in (C) after its deletion in HEK293T (D) and SNB-75 cells (E). Relative expression levels were calculated as the fold change of the WTs. Error bars show mean ± SE. P values were calculated by t-test with Storey’s correction of multiple comparison (* for p < 0.05, ** for p < 0.01). See also Figure S3, Figure S4, Table S7, Table S8.
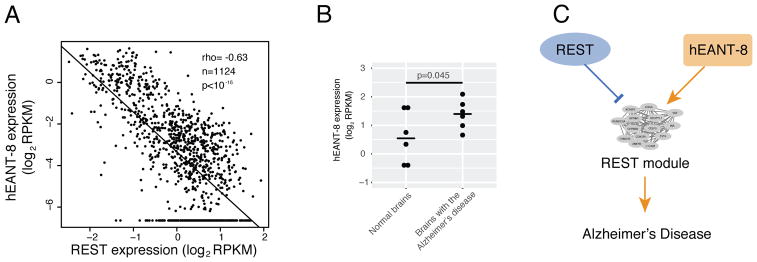
Importantly, REST is a key checkpoint for avoiding Alzheimer’s disease when aging. The loss of the function of this gene in aged brains would abnormally elevate the expression level of its downstream targets, eventually leading to Alzheimer’s disease (Lu et al., 2014). We found that hEANT-8 showed expression patterns that were the reverse of those of REST and their shared targets, suggesting their opposite cellular functions (Figure 6A). Thus, hEANT-8 was predicted to be activated in diseased brains, and indeed this pattern was observed in an independent dataset (Scheckel et al., 2016) (one-tail t-test, p < 0.045, Figure 6B). With its reverse effects of REST, hEANT-8 may contribute to the susceptibility to Alzheimer’s disease by abnormally activating the module, which should have been suppressed by REST (Figure 6C). These results provide a good illustration of how a hEANT is involved in the pathobiology of an aging-related disease.
Figure 6. The role of hEANT-8 in promoting Alzheimer’s disease by counteracting REST.
(A) Correlation between hEANT-8 and REST using the same data as Figure 5A. (B) A higher expression of hEANT-8 in diseased tissues (n = 6) than in normal brain tissues (n = 6). P value was based on a one-sided t-test. (C) Schematic representation of the hEANT-8/REST counteractions in the development of Alzheimer’s disease.
Discussion
George Williams proposed the antagonistic pleiotropic theory that human aging-related diseases originate because natural selection would recruit molecular changes benefiting in the reproductive age even with late life costs. This theory has not been rigorously tested and its molecular mechanism remains largely unclear, even though some interesting cases of balancing selection have been reported. For example, the APOE gene has an allele generating the Apoe4 isoform that benefits young humans with heighten immune response to viruses while contributing to risks of aging-related diseases such as the Alzheimer’s and higher blood cholesterol (Finch, 2010). A recent systematic analysis on aging-related genes and aging-related diseases genes found that for most of the model organisms studied (Fernandes et al., 2016), aging-related genes and aging-related diseases genes have few overlaps. The genetic links between aging and aging-related diseases are due to a small fraction of aging-related genes that tend to have a high network connectivity. This findings fit the antagonistic pleiotropic theory well and could be explained as that the general aging process is conducted by the related genes’ major functions, while the aging-related diseases are usually associated with the genes’ negatively pleiotropic functions. Moreover, the analysis found that while the aging-related genes tend to be more conserved than the overall genome (median dN/dS = 0.091), the aging-diseases genes show a higher dN/dS ratio (median = 0.137). The higher dN/dS ratio could be explained by either positive selection or relaxation, both of which don’t reject the concept of antagonistic pleiotropic.
In the present study, we followed the famous hypothesis of King and Wilson that human adaptations are largely driven by the molecular changes in regulatory elements and tested the antagonistic pleiotropic theory in terms of enhancer evolution. Our study identifies a novel evolutionary mechanism of how aging-related diseases may originate as the trade-off effects of adaptive enhancers recently acquired in human evolution, strongly supporting the theory. Our study links the molecular changes that once conferred some advantages in our long-term evolution with the vulnerability loci that contribute to the development of aging-related diseases in the modern human population, contributing to a deeper understanding of human evolution. Our results have several implications. First, a common practice in biomedical research is to focus on genetic elements with high conservation. But our observation emphasizes the pathological significance of elements newly evolved in our genome when studying aging-related diseases, especially those with a signature of positive selection (e.g., a non-conserved lncRNA). This notion helps prioritize candidate targets for functional and clinical investigations. Second, our study suggests that the genetic network underlying aging-related diseases is perturbed by many molecular changes without being subject to purifying selection after reproductive age, thereby creating complicated epistasis and endless complexity (Chen et al., 2017). This may help to explain the contrasting genetic patterns between many aging-related diseases and Mendelian diseases (Antonarakis and Beckmann, 2006). Finally, because the trade-offs introduced by molecular changes in a gene are not necessarily related to the primary function of the gene, our results suggest that the pleiotropic effects (sub-functions) of a gene, rather than its major function, should be considered when studying aging-related diseases.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Han Liang (hliang1@mdanderson.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK (human embryonic kidney) 293T cell line and SNB-75 cell line were obtained from the Characterized Cell Line Core Facility (CCLC) at MD Anderson Cancer Center. Both cell lines were fingerprinted by short tandem repeat (STR) DNA profiling at CCLC. The sex of these two cell lines were female. HEK293T cells and the cell lines established based on HEK293T were cultured in Dulbecco’s modification of Eagle’s medium (DMEM) with 10% fetal bovine serum (Invitrogen) at 37°C and 5% CO2. SNB-75 cells and the cell lines established from SNB-75 were maintained in RPMI-1640 with 10% fetal bovine serum at 37°C and 5% CO2.
METHOD DETAILS
Evolutionary rate analysis of enhancers
The FANTOM project annotated the expression patterns of enhancers in the human genome (Andersson et al., 2014). We obtained 32748 enhancers showing a tissue-specific expression pattern from http://enhancer.binf.ku.dk/presets/facet_expressed_enhancers.tgz. Using the UCSC whole genome alignment chain files (which considered the synteny and orientation) and the liftOver tool, we aligned these enhancers to four primate genomes, including the chimpanzee (Pan troglodytes, PanTro4), gorilla (Gorilla gorilla, GorGor3), orangutan (Pongo pygmaeus abelii, PonAbe2) and monkey (Macaca mulatta, RheMac3) (Rosenbloom et al., 2015). The primate phylogeny is ((((Human, Chimpanzee), Gorilla), Orangutan), Monkey) (Locke et al., 2011). Enhancers unmapped by the liftOver were considered as missing in a genome of interest. We then defined the origin of an enhancer on the deepest branch ahead of the last common ancestry that descended to all species with its ortholog. For example, an enhancer only aligned to the gorilla genome was considered to originate on B2 (Figure 1A). Table S1 provides the evolutionary ages (branch) of all 32748 enhancers. For the given tissue TY and branch BX, the relative evolutionary rate was calculated as the ratio of the proportion of TY enhancers on BX to the proportion of enhancers expressed in TY among all enhancers. We only considered the tissue or cell types with >100 enhancers (n = 106). We combined the neuronal stem cell expressed enhancers (FANTOM ID CL:0000047) and neuron expressed enhancers (CL:0000540) and termed them as enhancers activated in nervous tissue (EANT). EANTs evolved on B4 were referred to as human-specific ENATs, or hEANTs. Table S2 provides the details about the calculation of relative evolutionary rates in Figure 1B and C.
We used the analysis of covariance (ANCOVA) to study the dependency of evolutionary age on a series of factors, including EANT tissue type, GC content, enhancer length, and expression level. The evolutionary age was the response variable, where 1 was for enhancers on B4 or 0 for otherwise. The independent variable included tissue type (1 for EANTs or 0 for otherwise), GC content, SNP density and enhancer length. SNP density was calculated based on the 1000 Genomes Project (1000 Genomes Project et al., 2012). The other factors were transformed into Z-scores first. The ANCOVA was performed using the R package anova. We also considered the effects of genome completeness and false alignments. The genome completeness data were obtained from the BUSCO database (Simao et al., 2015). We artificially introduced a 10% false positive/negative alignment rate by randomly adding/removing 10% of the human enhancers missed/present in each of the four genomes. The resulting data were used to recalculate a relative evolutionary rate on B4. This procedure was repeated 100 times to estimate the impact of a 10% false alignment rate on our results.
We used SNP allele frequencies in the 1000 Genomes Database to detect positive selection on the enhancers. Specifically, we obtained from the reference genome, for each enhancer, a 2-kb sequence in which the enhancer is located in the middle. The allele frequencies of all SNPs on this sequence were calculated using the 1000 Genomes Project VCF files (http://www.1000genomes.org/data#download) and then subjected to Fay & Wu’s H-index calculation (Fay and Wu, 2000). For the H-index, the simulation was performed using the default settings (http://www.genetics.wustl.edu/jflab/htest.html).
For each protein-coding gene, we measured its conservation level as the (1-dN/dS) after the human orangutan divergence (the rhesus and orangutan orthologs were used to infer the ancestry status), where dN/dS represents the ratio of the number of nonsynonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS). Only genes with alignment >100 amino acids and dS >0.01 (n = 16001) were considered to ensure the accuracy of the conservation calculation. For each of the 54 GTEx tissues, we measured a given gene’s expression bias as the ratio of its expression (log2RPKM) in the tissue to its median level across all tissues. We sorted all the protein-coding genes by their conservation and evenly divided them into 100 groups. Then we calculated the means of conservation and expression bias for genes in these groups and examined the correlation (Pearson’s R in Figure S2) between the genes’ conservation levels and their expression bias in a tissue.
Evolutionary mechanism analysis of hEANTs
Since the LiftOver tool could not provide detailed information on how the hEANTs originated, we searched the hEANTs and their flanking 500bp sequences in the four primate genomes using BLAST in order to examine their evolutionary mechanisms. We used the e-value of 10−25 as the cutoff of positive BLAST results and manually curated all BLAST results in the following five steps. (1) We classified the hEANTs (n = 15) without BLAST results in the primate genomes as De novo model (Figure 2A). (2) We determined hEANTs (n = 11) generated by rearrangement as indicated by their flanking sequences being BLAST to separated genomic regions in the primate genomes (Figure 2B). (3) For the other hEANTs, we picked their best hits in the four primate genomes and blasted them back in the human genome. If none of the best hits in the human genome is the hEANT itself, we classified it as duplication model (n = 16; Figure 2C). (4) We subjected the remaining hEANTs to Tandem Repeats Finder (TRF; default settings; https://tandem.bu.edu/trf/trf.html) for SSR identification. For 23 hEANTs with SSRs in all the genomes involved, we manually aligned them and found that all these hEANTs have their SRRs extended in the human genome, explaining why they could not be mapped to the primate genomes by LiftOver. Thus, they were classified as SSR model (Figure 2D, E). (5) The remaining 28 hEANTs could not be explained by any simple model and usually involved more than one of the four models, thereby being classified as complex model (Figure 2G). We calculated the SSR rate of hEANTs, total enhancers, and the genome background using TRF with default settings. The genome background was calculated by randomly generating pseudo-enhancers (n = 32748) with the same length distribution for 100 times.
Analysis of cancer genomic data
For enhancer expression analysis, we first re-annotated the 32748 FANTOM enhancers according to the UCSC gene annotation file (refgene.txt) and removed the enhancers that overlapped with known genes or intron regions, resulting in a subset of 7,131 enhancers for which the RNA-seq reads can be confidently assigned (Table S1). We obtained TCGA RNA-seq BAM files from UCSC CGHub and our dataset contained >8,000 tumor samples of 23 cancer types. We obtained patient survival data from the Broad Firehose website, from which 8,380 samples have available overall patient survival data and 6,837 samples have progression-free survival data for the analysis (Cancer Genome Atlas Research et al., 2013) (Table S4). We calculated the RPKM values of the enhancers in TCGA samples as previously described (Li et al., 2015). For each cancer type, we further filtered enhancers without detectable expression in >10% of the samples. For each cancer type, we identified prognostic enhancers (overall survival or progression-free survival) using the Cox-regression model and the log-rank test. In the analysis, q < 0.05 (John Storey’s correction) in any cancer type were considered statistically significant.
For the SCNA-enhancer association analysis, we obtained the copy number information of 586 cancer driver genes (Cancer Gene Census) in 8948 tumors of 23 cancer types. The association between a hEANT expression and a CGC gene was examined using Spearman’s rank correlation within each cancer type. Significant correlations were identified (q < 0.01; John Storey’s correction) in a given cancer type.
Gene set enrichment and disease-associated gene analysis
We identified the nearest coding gene for each of the 93 hEANTs and pooled them as the input for enrichment set. Gene set enrichment analysis was based on the online tools on GSEA website (http://software.broadinstitute.org/gsea/index.jsp), using the default setting (Subramanian et al., 2005). Gene set collections included in this analysis were CP:BIOCARTA, CP:KEGG and CP:REACTOME. For hEANTs-disease analysis on Figure 4, we first obtained a gene-disease association network in the PheGenI database (Ramos et al., 2014), from which we manually selected (i) five typical aging-related diseases, including Alzheimer’s disease, Parkinson’s disease, hypertension, type-II diabetes, and osteoporosis; and (ii) five childhood diseases as a control, including asthma, autism, juvenile arthritis, sickle cell anemia and type-I diabetes. The aging-related gene list was obtained from Human Ageing Genomic Resources (Tacutu et al., 2017). We used a chi-squared test to compare hEANT neighbor genes with known genes associated with these two groups of diseases to determine the significance of the enrichment. Table S5 and Table S6 provide the data related to this section.
Case study of hEANT-8 and Alzheimer’s disease
We constructed hEANT co-expression networks using TCGA glioblastoma (GBM) and low grade glioma (LGG) datasets (Table S4). For each hEANT, we identified genes co-expressed (absolute Pearson R > 0.3 and FDR < 0.01) in both datasets to form a hEANT-centered co-expression module. We found 121 co-expressed genes for the hEANT-8 on chr8:9764051–9764441, while no other hEANT had a module size of >5 (Table S7). Disease ontology analysis on the hEANT-8 module was based on the R package DOSE using the default setting (Figure S4) (Schriml et al., 2012; Yu et al., 2015). We further obtained 1124 samples of different brain regions from the GTEx dataset to validate the co-expression module (GTEx Consortium, 2015). The overall activity of the module (the x-axis in Figure 5A) in a given sample was calculated as the sum of all genes’ log2RPKM values. For the copy number analysis in Figure S3, we classified the patients by the copy number of hEANT-8, where patients with <0.9, 0.9~1.1 and >1.1 copies of hEANT-8 were grouped respectively as copy number loss, copy number neutral and copy number gain. The overall activity of the module was calculated as previously described. For the motif analysis, the REST target genes were obtained from the GSEA database under the gene set ID V$NRSF_01 which was curated from previous studies (Schoenherr and Anderson, 1995; Schoenherr et al., 1996). To compare the hEANT-8 expression between normal and diseased brains, we obtained the dataset from the GEO (accession number, GSE53697) (Scheckel et al., 2016). The original dataset contains brain samples representing different pathological stages of Alzheimer’s disease (AD) from a normal condition to serious AD. We filtered two samples from patients of extremely old age (≥100 years). For the remaining samples, six with Braak stage = 0 were used as the normal control group, while six with Braak stage ≥5 were used as the disease group (Braak and Braak, 1991). We used Student’s t-test to assess if the expression of hEANT-8 is higher in the disease group.
CRISPR/Cas9 perturbation of hEANT-8
We designed 2/3 gRNA sequences using Cas-Designer (http://www.rgenome.net/cas-designer/) within 150 bp sequences up/down stream of hEANT-8, generating 6 gRNA combinations. We chose up/downstream gRNA targeting the genomic sequence CCTCTTCTTCCACCTCCCCG/CGCAGAAGTGCGCTCCACGA to generate hEANT-8 deletion clones. The up/down stream gRNAs were cloned into cas9 plasmid (Addgene 48138/64324) with GFP/mCherry as reporters. In day 0, each combination of gRNAs was transfected, using lipofectamin 3000, into HEK293T cells grown in Dulbecco’s modified Eagle’s medium (DMEM) or SNB-75 cells (human brain origin) grown in RPMI-1640, both with 10% fetal bovine serum (Invitrogen) at 37 °C and 5% CO2. In the day 3, the cells were collected for gRNA efficiency examination using genomic PCR. We transfected the gRNA plasmid mixture, and then sorted the GFP/mCherry positive single cells into 96-well plates two days after transfection using MoFlo Astrios Cell Sorters (Beckman Coulter, for HEK293T cells), or three days after transfection using BD FACSAria II Cell Sorter (BD Biosciences, for SNB-75 cells) for colony expansion. The hEANT-8 deletion clones were examined using genomic PCR after ~1 month (HEK293T cells) or ~2 months (SNB-75 cells). Their RNAs were extracted using RNeasy Plus Mini Kit (Qiagen), and 1 μg (for HEK293T cells) or 2 μg (for SNB-75 cells) total RNAs were reversely transcribed into cDNA with High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT-PCR was performed with SYBR Select master mix (Thermo Fisher Scientific) and in Mastercycler RealPlex (Eppendorf). Table S8 lists the qRT-PCR primer sequences of hEANT-8 target genes. For HEK293T cells, we obtained 4 single-cell clones with homologous hEANT-8 deletion, and for each single-cell clone, 3~4 experimental replicates were carried out; and for each gene in each replicate, qRT-PCR measurements were repeated for 3 times. For SNB-75 cells, we obtained one single-cell clone with heterozygous hEANT-8 deletion, and for each gene, qRT-PCR measurements were repeated for 4 times. For both HEK293T and SNB-75 cell, 2 out of the 11 hEANT-8 positively correlated genes had a baseline expression level lower than our qRT-PCR detection limit (Ct > 40) and were excluded from the analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
The definitions of significance for the various statistical tests are described and referenced in the respective Method Details sections.
DATA AND SOFTWARE AVAILABILITY
Data and Software availability are described in Key Resources Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Reference genomes (hg19, PanTro4, GorGor3, PonAbe2, and Rh3Mac3) | UCSC Genome Browser | http://hgdownload.soe.ucsc.edu/downloads.html |
| TCGA RNA-seq BAM files | Genomic Data Commons | https://portal.gdc.cancer.gov/legacy-archive/search/f |
| TCGA somatic copy number alteration thresholded data | Firehose Broad GDAC | https://gdac.broadinstitute.org/ |
| TCGA gene expression data | Firehose Broad GDAC | https://gdac.broadinstitute.org/ |
| TCGA patient clinic data | cBioPortal | http://www.cbioportal.org/ |
| FANTOM enhancer annotation | (Anderson et al., 2014) | http://fantom.gsc.riken.jp/5/datafiles/latest/extra/Enhancers/human_permissive_enhancers_phase_1_and_2.bed |
| Genome completeness data | (Simao et al., 2015) | http://busco.ezlab.org/ |
| 1000 genome project SNP data | 1000 Genomes Project Consortium | http://www.1000genomes.org/data#download |
| Phenotype–Genotype Integrator (PheGenI) database | (Erin M Ramos et al., 2014) | https://www.ncbi.nlm.nih.gov/gap/phegeni |
| The Alzheimer’s Disease RNAseq dataset | (Scheckel C. et al., 2016) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53697 |
| GTEx RNAseq dataset | dbGAP | https://www.gtexportal.org/home/ |
| Software and Algorithms | ||
| LiftOver | UCSC Genome Browser | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| Samtools | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| Tandem Repeats Finder | (G Benson 1999) | https://tandem.bu.edu/trf/trf.html |
| Disease ontology analysis | (Guangchuang Yu et al., 2015) | https://www.bioconductor.org/packages/3.7/bioc/vignettes/DOSE/inst/doc/DOSE.html |
| Fay & Wu H-test | (Fay and Wu, 2000) | http://www.genetics.wustl.edu/jflab/htest.html |
| Gene Set Enrichment Analysis (GSEA) | (Mootha et al., 2003; Subramanian et al., 2005) | http://software.broadinstitute.org/gsea/index.jsp |
| Experimental Models: Cell Lines | ||
| HEK293T | MD Anderson Characterized Cell Line Core Facility | HEK293T |
| SNB-75 | MD Anderson Characterized Cell Line Core Facility | SNB75 |
| Plasmids | ||
| pSpCas9(BB)-2A-GFP (PX458) | This study | Addgene: Plasmid #48138 |
| pU6-(BbsI)CBh-Cas9-T2A-mCherry | This study | Addgene: Plasmid #64324 |
| Oligonucleotides | ||
| sgRNA-L2 target: CCTCTTCTTCCACCTCCCCG | This study | NA |
| sgRNA-L3 target: ACTCCCGGTCTCCACGGCTT | This study | NA |
| sgRNA-R1 target: CGCAGAAGTGCGCTCCACGA | This study | NA |
| sgRNA-R2 target: TAGGTCTGATGTCCGCGGGA | This study | NA |
| sgRNA-R3 target: GAAAGACACAAACACCGCCA | This study | NA |
| 5′ PCR primer for detecting hEANT-8 deletion: GCTGAGGTCGAGCTCCTTTT | This study | NA |
| 3′ PCR primer for detecting hEANT-8 deletion: AAATGGCCTCGAAGCGAGAA | This study | NA |
| PCR primer sequences for hEANR-8 coexpression module genes | This study | Table S8 |
Supplementary Material
Table S1. Information about enhancers surveyed in this study, Related to Figure 1
Table S6. Information about hEANTs and genes related to diseases based on PheGenI, Related to Figure 4
Table S7. List of hEANT-8 co-expression module genes, Related to Figure 5
Highlights.
Identify a group of human-specific enhancers that are activated in neural tissues
Extensive associations of these enhancers with cancer and other aging diseases
One such enhancer potentially promotes Alzheimer’s disease by counteracting REST
Provide systematic evidence of antagonistic pleiotropy in human evolution
Acknowledgments
This study was supported by the National Institutes of Health (CA175486, CA209851 and CCSG grant CA016672), a grant from the Cancer Prevention and Research Institute of Texas (RP140462), a University of Texas System STARS award, the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine to H.L.; a training fellowship from the Gulf Coast Consortia, on the NLM Training Program in Biomedical Informatics and Data Science T15 LM007093 to H.C., CAS-Sponsored Scholarship Program for Visiting Scholars (2015-40) to C.L. We thank the MD Anderson high-performance computing core facility for computing, Dr. Gordon Mills and Characterized Cell Line Core Facility for providing the cell line SNB-75, Flow Cytometry and Cellular Imaging Facility for cell sorting, and LeeAnn Chastain for editorial assistance.
Footnotes
Author Contributions
H.L. supervised the whole project. H.C. and H.L. conceived of and designed the research. H.C. and H.L. contributed to the data analysis. C.L. and Z.Z. performed the experiments. H.C., C.L. and H.L. wrote the manuscript, with input from all other authors.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Beckmann JS. Mendelian disorders deserve more attention. Nature reviews Genetics. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta neuropathologica. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS biology. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin F, Xing K, He X. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nature communications. 2015;6:6367. doi: 10.1038/ncomms7367. [DOI] [PubMed] [Google Scholar]
- Chen H, Wu CI, He X. The genotype-phenotype relationships in the light of natural selection. Molecular Biology and Evolution. 2017 doi: 10.1093/molbev/msx288. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel Fabian TF. The Evolution of Aging. Nature Education Knowledge. 2011;3(10) [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Wan C, Tacutu R, Barardo D, Rajput A, Wang J, Thoppil H, Thornton D, Yang C, Freitas A, et al. Systematic analysis of the gerontome reveals links between aging and age-related diseases. Human molecular genetics. 2016;25:4804–4818. doi: 10.1093/hmg/ddw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T. Trade-offs. Current Biology. 2014;24:R60–R61. doi: 10.1016/j.cub.2013.11.036. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey ADNJd. Life Span Extension Research and Public Debate: Societal Considerations. Studies in Ethics, Law, and Technology. 2007;1 [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nature genetics. 2016;48:22–29. doi: 10.1038/ng.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nature genetics. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Huttley GA, Easteal S, Southey MC, Tesoriero A, Giles GG, McCredie MR, Hopper JL, Venter DJ. Adaptive evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Australian Breast Cancer Family Study. Nature genetics. 2000;25:410–413. doi: 10.1038/78092. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging cell. 2012;11:727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer research. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Lin YS, Li WH. Fast evolution of core promoters in primate genomes. Molecular biology and evolution. 2008;25:1239–1244. doi: 10.1093/molbev/msn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. Comparative and demographic analysis of orangutan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar PB. An Unsolved Problem of Biology 1952 [Google Scholar]
- Moyers BA, Zhang J. Evaluating Phylostratigraphic Evidence for Widespread De Novo Gene Birth in Genome Evolution. Molecular biology and evolution. 2016;33:1245–1256. doi: 10.1093/molbev/msw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, Feolo M, Hindorff LA. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. European journal of human genetics: EJHG. 2014;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. The UCSC Genome Browser database: 2015 update. Nucleic acids research. 2015;43:D670–681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckel C, Drapeau E, Frias MA, Park CY, Fak J, Zucker-Scharff I, Kou Y, Haroutunian V, Ma’ayan A, Buxbaum JD, et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife. 2016:5. doi: 10.7554/eLife.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease Ontology: a backbone for disease semantic integration. Nucleic acids research. 2012;40:D940–946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Bakewell MA, Zhang J. Did brain-specific genes evolve faster in humans than in chimpanzees? Trends in genetics: TIG. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Suarez R, Gobius I, Richards LJ. Evolution and development of interhemispheric connections in the vertebrate forebrain. Frontiers in human neuroscience. 2014;8:497. doi: 10.3389/fnhum.2014.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, Craig T, Diana E, Lehmann G, Toren D, Wang J, et al. Human Ageing Genomic Resources: new and updated databases. Nucleic acids research. 2017 doi: 10.1093/nar/gkx1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome research. 2008;18:1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. Essays Upon Heredity and Kindred Biological Problems 1889 [Google Scholar]
- Williams GC. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution. 1957;11:398–411. [Google Scholar]
- Yu G, Wang LG, Yan GR, He QY. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information about enhancers surveyed in this study, Related to Figure 1
Table S6. Information about hEANTs and genes related to diseases based on PheGenI, Related to Figure 4
Table S7. List of hEANT-8 co-expression module genes, Related to Figure 5