The double stranded structure of DNA suggested a mechanism for replication. Overlooked was that it also served to maintain genome stability by providing a template for the repair of damage and mistakes in replication...
Keywords: DNA excision repair, double stranded, mismatch repair, base excision repair, double-strand break repair
Abstract
The persistence of hereditary traits over many generations testifies to the stability of the genetic material. Although the Watson–Crick structure for DNA provided a simple and elegant mechanism for replication, some elementary calculations implied that mistakes due to tautomeric shifts would introduce too many errors to permit this stability. It seemed evident that some additional mechanism(s) to correct such errors must be required. This essay traces the early development of our understanding of such mechanisms. Their key feature is the cutting out of a section of the strand of DNA in which the errors or damage resided, and its replacement by a localized synthesis using the undamaged strand as a template. To the surprise of some of the founders of molecular biology, this understanding derives in large part from studies in radiation biology, a field then considered by many to be irrelevant to studies of gene structure and function. Furthermore, genetic studies suggesting mechanisms of mismatch correction were ignored for almost a decade by biochemists unacquainted or uneasy with the power of such analysis. The collective body of results shows that the double-stranded structure of DNA is critical not only for replication but also as a scaffold for the correction of errors and the removal of damage to DNA. As additional discoveries were made, it became clear that the mechanisms for the repair of damage were involved not only in maintaining the stability of the genetic material but also in a variety of biological phenomena for increasing diversity, from genetic recombination to the immune response.
THE Austrian theoretical physicist, Erwin Schrödinger, one of the inventors of wave mechanics, was fascinated by the Hapsburg lip, a distinctive facial feature of the Hapsburg imperial family. This was not only because he was Austrian but, as a physicist trying to understand biology, he was fascinated by the stability of this trait over the centuries, something that seemed to defy the laws of thermodynamics (Schrödinger 1945). Geneticists and biochemists in the 1940s were comparably impressed by the apparent removal of DNA from the hurly-burly of cellular metabolism, a property that one might associate with such hereditary stability (Mazia 1952).
A major step forward in understanding the properties of the genetic material was the formulation of the double-stranded structure of DNA by James Watson and Francis Crick in 1953, which suggested a mechanism for its replication and accordingly its perpetuation. In one of the more famous understatements in the scientific literature they wrote: “It has not escaped our attention that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material” (Watson and Crick 1953a,b). What apparently did escape their attention, and that of the early molecular biologists, was that this double-stranded structure also served as a safety device, permitting the repair of damage to one or the other of the strands. Even more surprising, in hindsight at least, was that this recognition first came from what was then the unfashionable field of radiation biology.
Today the subject of DNA repair is a fully accepted part of the body of contemporary molecular knowledge. Current textbooks of molecular biology, genetics, and biochemistry list DNA repair mechanisms comfortably among the multitude of metabolic pathways. Table 1 summarizes the ones discussed in this article. Manipulation of these pathways is central to the application of CRISPR, perhaps the most productive of recent biological technologies and the latest major addition to the field of DNA repair. The Nobel Prize in chemistry for 2015 was awarded to Tomas Lindahl, Paul Modrich, and Aziz Sancar for their detailed mechanistic studies on repair, which is confirmation of the current respectability of studies on DNA repair.
Table 1. Some DNA repair mechanisms.
| Type | Substrate | Double-stranded DNA required | Mechanism | Key reference(s) and/or reviews |
|---|---|---|---|---|
| Nucleotide excision repair | UV-induced pyrimidine dimers, bulky adducts | Yes | Distortions in DNA helix are recognized. Nucleases cut the nucleotide chain above and below the damage. DNA helicase removes a 12-nt segment. The gap is filled in by DNA polymerase and ligase. | Boyce and Howard-Flanders (1964); Pettijohn and Hanawalt (1964); Setlow and Carrier (1964); Hanawalt and Haynes (1967) |
| Transcription-coupled NER | As above | Yes | A variant of NER. Acts at site of stalled RNA polymerase | Mellon et al. (1987) |
| Base excision repair | Unusual or mispairing damaged bases (oxidized, methylated, deaminated, uracil, single-strand breaks) | Yes | DNA glycosylases remove bases forming abasic sites which are cleaved by an endonuclease. The single-strand break is processed and a patch of from 1 to 2–10 nt is inserted. | Lindahl (1974) |
| Mismatch repair | Mismatched bases produced by replication or recombination | Yes | Mismatches are detected in the newly synthesized strand by methylation of the parent strand in enteric bacteria, single-strand nicks in other species. The mismatched strand is then cleaved and a segment of variable length including the mismatch is removed. The single-strand gap is repaired by the replicative polymerase and sealed by ligase. | Wagner and Meselson (1976); Lahue and Modrich (1988) |
| Double-strand break repair, homologous recombination | DNA with both strands broken by external agents (e.g., radiation) or during genetic recombination | Yes (a homologous chromosome or chromatid) | The double-strand break is enlarged by nucleases to leave overhanging single strands. An undamaged strand from the double-stranded homolog pairs with the resected region and serves as a template for DNA synthesis. The crossed over strands form a Holliday junction which can be resolved in different ways. | Szostak et al. (1983) |
| Nonhomologous end joining | DNA with both strands broken by external agents or during the immune response | ? (short microhomologies) | The broken ends are brought together with the deletion or addition of a few bases and then patched together by specific proteins. | Moore and Haber (1996) |
| Photoreactivation | DNA with pyrimidine dimers produced by UV radiation | No? | The bonds connecting adjacent pyrimidines are enzymatically broken using energy from visible light. The repaired strand is not broken. | Kelner (1949); Wulff and Rupert (1962) |
| Methyl removal from O6 methylguanine | O6 methylguanine in DNA | No | The methyl from O6 methylguanine is transferred to O6 methylguanine DNA transferase. The protein is inactivated as a result. The repaired strand is not broken. | Karran et al. (1979) |
A listing of (human) proteins involved in DNA repair processes can be found in Wood et al. (2005). NER, nucleotide excision repair.
Yet it is clear that the early workers in this field were justified in feeling that their work was not given the recognition it deserved as a key factor in the DNA-centered view of life that became the science of molecular biology. John Cairns, a key figure in that development, writing as late as in 2008, was able to trace the foundations of molecular biology and list its exciting discoveries without mentioning the fact that DNA could be repaired (Cairns 2008). A review on the history of “target theory” (a pioneering, somewhat earlier, attempt to understand the biological effects of radiation) (Box 1) reports: “Around 30 years ago, a very prominent molecular biologist confidently proclaimed that nothing of fundamental importance has ever been learned by irradiating cells!” (Bedford and Dewey 2002; J. S. Bedford, personal communication). What was the basis for this attitude and what produced the change?
BOX 1.
The introduction of ionizing radiation as a tool in the 1920s and 1930s led to major advances in our understanding of the gene. The discovery by Muller (1927) and almost simultaneously by Stadler (1928a,b) that ionizing radiation could produce mutations in what had hitherto been an impenetrable gene opened up the possibility of actually investigating the properties of this biological entity by physical means. Further investigations by Timofeeff-Ressovsky et al. (1935) led to the hypothesis that the “gene” was a molecule and to a calculation of its possible size that was/is reasonable. This “three-man article” found its way to Schrödinger (1945) who made Delbrück’s model a key feature of his book What is Life?, a work which enticed many of its founders into what became molecular biology.
The advantage and disadvantage of radiation is that it lent itself to quantitative studies and to a mathematical analysis of the results obtained. The result was target theory: the idea that the gene, or virus, was a target at which quantum bullets could be shot. There was then a relationship between the size of the target and the number (dose) of bullets that needed to be shot at random to hit the target. The hypothesis was reasonable as a first approximation and was developed to a high degree of sophistication (Lea 1946). The hypothesis had many failings but, to my mind, a major one was the concentration of research on the absolute linear dependence of the mutation rate on dose. There were political and social reasons for this concentration in a world attempting to come to terms with the development of atomic energy. One scientific result was a concentration of radiation research on the interpretation of killing curves with different types of radiation being applied at differing dose rates and with different end points. At no point was this research able to identify the target molecule. Notwithstanding really sophisticated analysis, this research did not provide as much insight as subsequent biochemical analysis.
Replication as the Central Problem in Biology
In the 1930s, Max Delbrück, a brilliant young German physicist, became interested in radiation biology as a tool for discerning the nature of the gene (Strauss 2017). He was looking for some way to validate Niels Bohr’s speculation that understanding biology required the recognition of unique processes that could not be explained by the application of (known) physical and chemical principles (Bohr 1933). Replication of the genetic material appeared to be the most likely place in which such new principles might be found. Delbrück’s work on bacterial viruses started with the expectation that these entities might be the simplest objects to study “pure” replication without the distractions of metabolism. Unfortunately, the viruses turned out to be not nearly as simple as Delbrück had imagined. They possessed multiple genes, a complex recombination mechanism, and even a sequential developmental pattern. Accordingly, the problem of replication remained.
At almost the same time as these investigations on bacteriophage were beginning, Oswald Avery and his co-workers were demonstrating that (at least some) genetic information was carried by DNA (Avery et al. 1944). By 1952 it was generally recognized that it was DNA rather than protein that carried the genetic message (Mazia 1952). The culmination of these efforts was the elucidation of the DNA structure by Watson and Crick (1953a,b).
Crick, at least in hindsight, recognized that their proposed mechanism had a potentially fatal flaw (Crick 1974). DNA could not be the carrier of stable genetic information since the calculated rate of errors in its replication based on the rate of tautomeric shifts in the nucleotide bases would make such hereditary stability impossible. Yet, in spite of that theoretical objection, DNA is the genetic material. Therefore, there must be mechanisms for correcting errors introduced in the normal replication process.
For most of the founders of molecular biology, the exact nature of such mechanisms were just secondary details that could be worked out later compared with the really important questions of how DNA replicated and functioned. Their rationale was outlined by Crick much later (Crick 1974, 1988):
Surely then, DNA cannot be the genetic material since its replication would produce too many errors.... Fortunately, we never took this argument seriously…. DNA is, in fact, so precious and so fragile that we now know that the cell has evolved a whole variety of repair mechanisms to protect its DNA from assaults by radiation, chemicals and other hazards… (Crick 1988, p. 111).
This clearly reflects the wisdom of hindsight. Replication using complementary base pairing is possible without a permanent double-stranded DNA structure: consider the single-stranded viruses. However, the stability of the genome requires repair and at least four separate pathways—nucleotide excision repair and its variation, transcription-coupled nucleotide excision repair, base excision repair, and mismatch repair—have evolved to accomplish it (Table 1).
The “Stability of DNA”
By 1952, as noted above, it was pretty well established that DNA was the genetic material. A review by Mazia (1952) summarizes the reasons for this belief. Mazia emphasized the finding that DNA, but not protein, met the quantitative expectations for the hereditary substance. The amount of DNA was the same in all diploid cells and was halved in haploid cells. The DNA content of the nucleus (exactly) doubled during the mitotic cycle. An interesting additional argument was based on the apparent stability of DNA. It had recently been shown that body constituents were in a constant state of turnover (Schoenheimer 1942), but there was an important exception. DNA seemed to be different and unusually stable. Its constituent atoms did not seem to be replaced to the same extent as other molecules, particularly RNA (then called PNA for pentose nucleic acid) (e.g., Furst et al. 1950). This was in accord with the (supposed) requirements of the genetic material, the guardian of the cell’s history, protected from the vicissitudes of metabolism, and able to remain stable for hundreds of years as Schrödinger had pointed out in his 1945 essay.
Mazia wrote his review just before publication of either the Hershey–Chase experiment (Hershey and Chase 1952) or the Watson and Crick model (Watson and Crick 1953b). He was aware of the Avery experiments (Avery et al. 1944) but, like most cell physiologists, was not sure what to make of experiments with bacteria. Mazia was reasonably sure that transformation was not directed mutation and thought it unlikely to be due to protein contamination, but he was not sure where the specificity of DNA came from. He quotes Chargaff’s work showing the different base composition of DNAs from various bacteria (Chargaff 1950), but he also quotes work from Mirsky’s laboratory (Daly et al. 1950) showing that the DNAs from a variety of vertebrates and Pneumococcus had essentially the same base composition. Nonetheless, Mazia came down on the side of DNA as the genetic material and identified what he saw as the two remaining problems: how did DNA reproduce and how did it function? The Watson–Crick structure pointed to the solution of the first question. The second was Crick’s major question (how genes functioned) and occupied him and many prominent molecular biologists for the decades of the 1950s and 1960s. The discovery of messenger RNA, elucidation of the role of repressors and promoters, and an increased understanding of how DNA coded information early in the 1960s made it possible to think that Crick’s second question had been solved in principle.
In parallel with these developments, but carried out by a separate set of investigators, were developments in the study of mutation. It was demonstrated by bona fide members of the “phage group” that a variety of nucleic acid base analogs could induce mutations in phage, and Ernst Freese and Seymour Benzer had developed a molecular explanation for their action (Benzer and Freese 1958). The earlier pioneering work of Charlotte Auerbach (Auerbach and Robson 1947; Auerbach et al. 1947) had spawned a series of experiments showing the production of mutations by chemical agents, particularly the alkylating agents. A group of investigators at The University of Texas had also demonstrated that UV irradiation of the medium made it mutagenic for bacteria, suggesting that at least some radiation effects might be indirect (Stone et al. 1947). Furthermore, there was increasing evidence that protein synthesis was required to fix or “cement” mutations (Witkin 1956). Therefore, by the mid-1950s, there was evidence indicating that biochemistry intervened between an insult to DNA and the production of a mutation.
The Contribution of Radiation Biology
The development of atomic weapons remains both a (perhaps “the”) major problem for our times and also represents the practical achievement of a half century of research in physics. This development could not help but affect biological research, and it did so in at least two ways. It certainly persuaded many physical scientists to look for a field they could pursue without qualms of conscience and it persuaded the United States government, anxious to promote the peaceful uses of atomic energy, to establish a number of National Laboratories and to spend relatively large sums, for those times, on biological research to ascertain the safety of such applications. Errol Friedberg (1997) and some other key investigators of DNA repair are convinced that “This situation did little to endear the community of radiobiologists to the ‘aristocrats’ of molecular biology, who not only labored under more restrictive financial conditions, but additionally considered much of the research done in these Laboratories as frankly pedestrian.” (The data don’t actually support the reality of the relative lavishness of support for radiation research but the feeling was certainly there.)
By 1960 there was an awareness that cells could recover from the effects of radiation even though the nature of the recovery process(es) was unknown. It was also recognized that any explanation of the origin and evolution of life would have to take into account damage to the genetic material from radiation in the prebiotic and early postbiotic eras. The astrophysicist Carl Sagan made some calculations for the Radiation Research Society and came to the conclusion that the major problem would be UV radiation (Sagan 1961). The flux of UV light would have resulted in a mutation rate too high to permit stable transmission of hereditary information. Sagan’s 1961 calculations suggested that the original life forms would have to have been benthic, i.e., living on the ocean bottom, but even then there would be a problem resulting from the production of peroxides. His suggestion as to how primitive organisms solved this problem reflected current knowledge and involved the development of catalases that would detoxify the peroxides. These speculations ignored the possibility that damaged DNA itself might be repaired. By the start of the 1960s it was recognized that radiation, particularly UV radiation, would have posed a significant problem for early (as well as later) organisms.
By this time there was sufficient information indicating genetic recovery processes to permit a Symposium to be held in Leiden in August 1962 with the title Repair from Genetic Radiation Damage (Sobels 1963). By 1962 it was recognized that there was a repair process(es) acting on DNA and that this process acted in the dark and was distinct from a previously discovered light-dependent form of DNA repair: photoreactivation. The phenomenon of photoreactivation had been described in the late 1940s (Kelner 1949) and had even been subsequently accomplished in vitro (Wulff and Rupert 1962). It had been discovered that neighboring thymines in DNA were dimerized by UV radiation (Beukers and Berends 1960; Wulff and Fraenkel 1961) and that these dimers disappeared during photoreactivation (Wulff and Rupert 1962) (for review of this phenomenon see Friedberg 1997). In some ways, the discovery may have diverted attention from the role of the double-stranded DNA structure in repair, since photoreactivation involves the direct reversal of a lesion without breaking the DNA chain. The mechanism of the dark repair was unknown. No one at the 1962 Symposium recognized (or wrote about) the importance of having the DNA be double stranded to permit repair of any kind.
Excision Repair
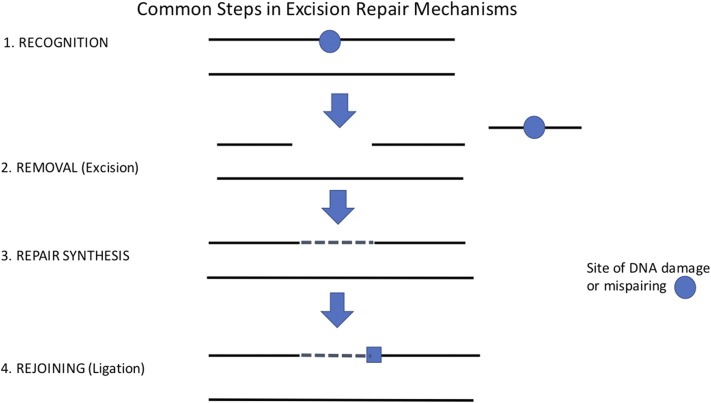
The discovery of thymine dimerization by radiation was the key to an understanding of how DNA could be repaired. Within 2–3 years from the report of the UV-induced dimerization of thymine, a general scheme of excision repair had been established as the result of work by Richard Setlow, Paul Howard-Flanders, Philip Hanawalt, and their co-workers. The demonstration depended on the isolation of radiation-sensitive mutants in Escherichia coli, first by Ruth Hill (1958) (who tragically died at an early age in 1973) and on some astute biochemical intuition by Richard Setlow about the properties and analysis of the excised thymine dimer-containing fragments (Friedberg 1997). The precise timing of the discoveries by the three laboratories is not clear since the key articles were published within weeks of one another (Boyce and Howard-Flanders 1964; Pettijohn and Hanawalt 1964; Setlow and Carrier 1964). An earlier result of Hanawalt (Pettijohn and Hanawalt 1963) included evidence for what is now known as repair synthesis but without that interpretation. I believe that all subsequent studies on DNA repair mechanisms can be traced to these three articles. The elucidation of the general scheme of excision repair (Figure 1) provided a paradigm for subsequent work.
Figure 1.
Common steps in excision repair mechanisms.
At least one of the founders of molecular biology did immediately appreciate the importance of the discovery. Philip Hanawalt (personal communication) recalls that Max Delbrück was so excited about the discovery of thymine dimers that he decided to offer a course on photobiology and during one lecture he “became so excited that he lapsed into German and didn’t notice for a few minutes, before laughing and switching back to English.” Delbrück was the contributing editor of the Boyce–Howard-Flanders excision repair article. He was well known for his frequent, devastating comments at seminars (Strauss 2017), but had a different attitude about the discovery of excision repair. In retrospect, this may not have been so strange. Delbrück had a lifelong interest in photobiology stemming from Niels Bohr’s lecture on Light and Life (Bohr 1933), a lecture that influenced all his future work. I have written about Delbrück before (Strauss 2017) but it is now clear to me that I had not sufficiently appreciated his interest in photobiology and repair. He moderated the final session of the first major meeting on DNA repair (Haynes et al. 1965), but I did not then understand why he bothered to attend.
By 1967 the accumulating evidence for a generalized DNA repair mechanism was great enough to merit a Scientific American article describing the system and arguing for a generalized DNA repair mechanism (Hanawalt and Haynes 1967). A great boon to research in the field was the discovery of a disease linked to cancer and resulting from a deficiency in DNA repair. This was the demonstration by James Cleaver that the sun-sensitive, cancer-prone genetic disease xeroderma pigmentosum was associated with a deficiency of DNA repair (Cleaver 1968). Besides the obvious fundamental scientific importance of the discovery, it also established the health relevance of DNA repair, thereby providing an important rationale for securing financial support from the National Institutes of Health.
The fidelity of normal DNA replication is maintained by several processes. The overall error rate in vivo for undamaged DNA is ∼1 in 109 (error rates vary in different portions of the genome). Physical base pairing and stacking alone result in an error rate of ∼1 in 102. Polymerase structure, the fitting of incoming nucleotides and template into the enzyme, contribute a factor of specificity of an additional 103 (McCulloch and Kunkel 2008). Replicative polymerases have associated with them a “proofreading” exonuclease, which examines the incoming nucleotide for fit and which contributes an additional factor of ∼102 to the specificity (Fersht et al. 1982). The additional factor of ∼102 for normal replication is contributed by the mismatch (excision) repair system. In addition, there are three excision repair mechanisms to deal with DNA damage and one distinct mechanism (homologous recombination) for the error-free repair of double-strand breaks in DNA (Table 1). The pathways of the four excision repair mechanisms (including mismatch repair) are remarkably similar involving a recognition step, removal of the incorrect base, which involves a break in the phosphodiester chain, and a resynthesis step using the normal DNA strand as a template (Figure 1). There is a second process for the repair of double-strand breaks (nonhomologous end joining) that is somewhat different but with a variant that does involve the two strands. To my knowledge, there has been no report of homologies in the proteins involved, which suggests that all have been independently evolved. However, it has been proposed that one or more of the mismatch repair proteins have been coopted to play a regulatory role in some of the excision repair mechanisms (Mellon and Champe 1996; Polosina and Cupples 2010). The common theme of all the excision mechanisms is that DNA must be double stranded to provide a template for the repair. Separate from these, there are proteins involved in the direct reversal of damage (e.g., photoreactivation, O6 methylguanine DNA methyl transferase).
Once the basic principles of nucleotide excision repair had been established, it became somewhat easier to think of variations on the process. For example, I had been studying the effect of the monofunctional alkylating agent, methyl methanesulfonate (MMS), on Bacillus subtilis and had observed a recovery process which, as a result of some convoluted arithmetical calculations, I attributed to residual DNA synthesis (Strauss 1963). Although then a member of the Department of Microbiology at The University of Chicago, my laboratory was in the Research Institutes that also housed the Committee on Biophysics. Bob Haynes, who was a collaborator of Phil Hanawalt, was a member of that unit and we met more or less regularly at the mail boxes (this was in the period when “snail mail”—the regular postal delivery of paper—was still an important means of communication). Haynes was one of the organizers of the first meeting devoted to DNA repair held in Chicago in 1965 (Haynes et al. 1965). One day I learned from Bob that there was a process called DNA repair. As a result, we then tested the different stocks of B. subtilis in our freezer and discovered that we had been using a UV-sensitive strain: origin unknown. We used this strain to show that there were variations in excision repair (Reiter and Strauss 1965) since, along with other evidence, this strain did not repair UV-induced or nitrogen mustard-induced damage but did still repair MMS-induced damage. We concluded: “The repair of damage induced by ultraviolet irradiation differs by at least one step from the repair of damage induced by methyl methanesulphonate.” (I sent a copy of our manuscript to Richard Setlow asking for comment and he tried to dissuade me from this conclusion since it contradicted the hypothesis that excision repair was a general error-correcting mechanism.) In retrospect, what we were seeing, but did not understand, is now recognized as base excision repair; a system using different enzymes and a smaller patch.
Base Excision and Mismatch Repair
Two other discoveries were needed to make the study of DNA repair processes part of the catechism of molecular biology. First, it needed to be shown that DNA repair was not a phenomenon limited to radiation- or alkylating agent-induced damage, but that there was a significant natural rate of damage or mismatch that required repair over and above the exonucleolytic proofreading activities of the replicative DNA polymerases. Second, it needed to be demonstrated that DNA recombination was a process that involved using some of the same tactics used for repair. To resolve Crick’s conundrum, the result of all the repair processes should be found to be a reduction of the overall replication error rate to a level consistent with the stability of the genome.
That some such system(s) was required was made empirically clear by a set of studies by Tomas Lindahl in the early 1970s. One of these studies was a measurement of the spontaneous loss of purines by DNA (Lindahl and Nyberg 1972). Lindahl showed that, without any intervention whatsoever, a typical human cell might be expected to lose ∼600 purines/hr (Lindahl and Nyberg 1972). This calculation, coupled with the discovery of an enzyme to remove spontaneously produced uracil from DNA (Lindahl 1974), was evidence that the genetic material needed continuing surveillance and repair to maintain its integrity.
In hindsight, one can argue that the basic principles of mismatch repair could, and should, have been deduced by the early 1960s from the new data about the mechanisms of meiosis coming from the studies on Neurospora and yeast. In these studies, all the products of an individual meiosis could be recovered; hence, in these organisms it was possible to follow the fate of each DNA strand through the meiotic process rather than depending on statistical analysis of the results of mass crosses. Explanations of the events in meioisis involving mismatch repair had been provided based on this new data by Harold Whitehouse (1963) and his student Robin Holliday (1964). These investigators argued that in the process of meiosis there was produced a region of DNA in which the two strands were not (necessarily) complementary but were derived from the two different parents, i.e., the molecule itself was heterozygous. Completion of meiosis almost always involved conversion of the mismatch to a homozygous state by a process that Holliday identified as requiring enzymatic intervention and which consequently deviated from the expected Mendelian ratios. Holliday’s article was rejected by both Nature and GENETICS and was eventually published in Genetical Research (Holloman 2014)!
The (bio-) chemists seemed unable to respond. Genetics, as taught then (and unfortunately often still taught), started and ended with Mendel and his peas and the conventional ratios derived from so-called monohybrid and dihybrid crosses. The newer studies used ascomycete fungi (e.g., brewer’s yeast, Neurospora) in which “all the products of a single meiotic event” could be analyzed. I believe that this phrase itself was mysterious to both biochemists and many (but not all) of the new molecular biologists. That the difference between a 4:4 ratio and a 5:3 ratio derived from a single meiotic event indicated the operation of a new biochemical pathway was not something easily accepted by those unfamiliar with these genetic systems.
As an example of the disconnect between genetics and biochemistry, consider the following: At a retreat, probably in the 1980s, for faculty and students in the Department of Molecular Genetics and Cell Biology at the University of Chicago, a student in Rochelle Esposito’s laboratory was explaining his research. He displayed Northern and Western blots without comment, knowing his audience would understand these, but stopped to explain the details of meiosis on the (probably correct) supposition that the details of this arcane biological process were probably unknown to these graduate students in biology, who would be unable to understand his research without such instruction.
Some prominent molecular biologists seemed quite willing to express their thoughts about genetics (or geneticists). Here, in 1968, is James Watson’s statement:
That was not to say that the geneticists themselves provided any intellectual help. You would have thought that with all their talk about genes they should worry about what they were. Yet almost none of them seemed to take seriously the evidence that genes were made of DNA. This fact was unnecessarily chemical. All that most of them wanted out of life was to set their students onto uninterpretable details of chromosome behavior or to give elegantly phrased, fuzzy-minded speculations over the wireless on topics like the role of genetics in this transitional age of changing values (Watson 1968, p. 74, my italics) (Watson 1968).
Forty years later, consider this comparable view from a sketch of the history of molecular biology:
Geneticists seem to have been less pessimistic, perhaps because theirs was a subject that rejoiced in a multitude of essentially abstract words (dominant, recessive, epistatic and so on) — the kind of words that are designed to avoid the need for further thought (Cairns 2008).
It is only fair to point out that the miscomprehension between these two fields is a two-way street. I can remember, as a graduate student, one cytologist being rather pleased that his subject could be handled in purely biological terms without reference to physics or chemistry. Further along, in the late 1960s, my graduate students insisted that we had to include an experiment with UV light in any article dealing with the repair of alkylation damage or the investigators studying DNA repair wouldn’t pay any attention!
It took almost 15 years to add biochemistry to Holliday’s 1964 suggestion, possibly because, as pointed out above, biochemists did not appreciate the problem or, more likely, because the tools were not available. An essential biochemical element was the discovery of DNA ligases: enzymes that joined broken DNA molecules. This finding was made in 1967 by at least four different laboratories (Gefter et al. 1967; Olivera and Lehman 1967; Weiss and Richardson 1967; Zimmerman et al. 1967).
There was now good reason to suppose that recombination might actually be amenable to biochemical study. It was Matthew Meselson, using phage λ, who demonstrated that genetic recombination was accompanied by actual physical exchange of sections of DNA (Meselson and Weigle 1961; Meselson 1964). Recombination therefore necessarily involved breaks in DNA and the models of recombination that were developed are similar to, and can be traced to, an understanding of how cells avoid the lethality associated with the production of double-strand breaks in DNA, a major consequence of ionizing radiation. I suggest that they derive from a recovery mechanism not involving excision called postreplication repair or replication repair first discovered by Rupp and Howard-Flanders (1968) and later shown to involve the product of the gene recA (see Smith and Wang 1989). When the DNA synthetic apparatus encounters a replication blocking lesion on one strand, it continues synthesis on the undamaged strand. Later, after synthesis of one double-stranded region, synthesis proceeds past the critical site on the damaged strand using the newly synthesized complementary strand as a template (see Higgins et al. 1976 for one way this might be done).
Haploid organisms such as bacteria require replication to provide such a template. Diploid organisms come equipped with a template for both strands as part of the homologous chromosome. The realization that an undamaged sister chromatid (or chromosome) could provide a template for the repair of both strands of DNA led to the general incorporation of repair concepts in the models of recombination that are the basis for current research (Szostak et al. 1983).
The genetic data still required some sort of mismatch repair for a satisfactory interpretation of recombination (see above) but such data had an esoteric and nonconvincing air for biochemists who needed to know, among other things, how organisms recognized which strand needs correction. The discovery of methyl-directed mismatch repair in E. coli solved that problem (Meselson and Radding 1975; Wagner and Meselson 1976) although we now know that these bacteria employ a sophisticated mechanism to discriminate between the old and newly synthesized DNA strand. The elegant work of the Modrich laboratory in particular then provided the specific mechanistic details (Modrich 1986).
Recognition of the role of deficient mismatch repair in certain colon cancers (Parsons et al. 1993) helped (in my opinion) to make the study of repair popular enough for Science magazine to decide that DNA repair enzymes were the 1994 “Molecule of the Year.” In 2000, a Cold Spring Harbor Symposium was devoted to Biological Responses to DNA Damage with the explanation on its website:
Discussion of the nature of mutations and genetic damage was pervasive at Cold Spring Harbor in the 1940s and 1950s due to the presence of Demerec, Max Delbrück, Salvador Luria, and Barbara McClintock and can be traced to the 1941 Symposium on “Genes and Chromosomes: Structure and Organization,” which occurred long before the double helix was revealed. We have come a long way in our understanding since then, and it was about time to correct the long absence of this topic from these Symposia (Witkowski 2000).
Conclusion
This is a personal view of the early days of DNA repair studies. I have thought it useful to trace the development of our ideas about (mainly) excision repair to emphasize the importance of the double-stranded structure of DNA in the maintenance of genetic integrity. I have said nothing about the mutagenic SOS pathways studied so imaginatively by Evelyn Witkin and Miroslav Radman.
Without the existence of an active repair mechanism early in the history of life on earth and given the radiation flux in the prebiotic and early biotic environments, it would have been impossible for a molecule like DNA to have survived, let alone to have served as a stable repository of genetic information. Both single- and double-strand breaks in a polynucleotide chain would be necessarily lethal without repair, but the pathway(s) of restoration without introducing error is based on finding an undamaged homologous strand. It might therefore be argued that the double-stranded DNA structure was so successful in the evolution of living systems precisely because it provided a way to conserve genetic information as well as furnishing a mechanism for its copying.
It is hardly a surprise that students of a biological process should argue for the fundamental importance of the subject of their investigations. And it is therefore not surprising that many of us working on DNA repair should have felt that, although our studies seemed to us of vital interest, they were largely ignored by the molecular biologists we were trying to impress (see Friedberg 1997). Two questions can be posed: first, can anything of general interest can be learned from this history? Second, does the history itself actually matter?
This second question is easier to answer. The general “vote” on what is important in science has material consequences. Where should we put the money, what young person in what field should we hire? Once molecular biology was established as a scientific discipline in its own right, the opinions of the pioneers were hard to ignore. It is clear, as illustrated above, that many of them did think the mechanisms for correcting errors were mere details. Luckily, all of this happened in a period when support for science was (in retrospect) relatively lavish and the different fields were not in serious financial conflict, so the field of DNA repair could develop in part because of the link to cancer research and its funding provided by Cleaver’s discovery (Cleaver 1968).
The history of the studies on mismatch repair illustrates another, possibly related, difficulty. Reading the studies on gene conversion and the articles of Whitehouse and Holliday with all the benefit of hindsight, it would seem that their connection to the newly described excision repair mechanisms would have been obvious. Yet it took at least a decade for mismatch repair as a separate process to be described. This must partly reflect the lag in discovering the enzymes that synthesized and glued together DNA chains. But it seems clear that both biochemists and (many of) the new molecular biologists were unable to take the genetic evidence seriously. I have copied some appropriate comments from James Watson, but the biochemists were equally parochial in their reluctance to accept a phenomenon as established without demonstrated mechanistic (enzymatic) detail (Kornberg 2000). There was just no mutual understanding. It would be pleasant to believe that things are different today and that the emphasis on multi-disciplinary teams of investigators ensures that all avenues of potential interest will be explored. Perhaps.
Practical application of discovery follows unexpected paths. One of the most useful new biological technologies is the use of CRISPR-Cas9 and its modifications. The technology depends on the ability to make double-strand breaks at specific locations. What follows depends on the investigators making clever use of the various mechanisms for the repair of double-strand breaks along with mismatch repair. A Danish proverb attributed to Niels Bohr (and also to Yogi Berra) may be appropriate: “prediction is difficult, especially about the future.” Modesty about the relative importance of different approaches might be a desirable quality to cultivate at all times.
Acknowledgments
Errol Friedberg’s record of his many interviews with central participants (Friedberg 1997) was a great help in preparation of this essay. I thank Phil Hanawalt for some insights into the exciting early days of nucleotide excision repair research.
Footnotes
Communicating editor: A. Wilkins
Literature Cited
- Auerbach C., Robson J. M., 1947. The production of mutations by chemical substances. Proc. R. Soc. Edinb. Biol. 62: 271–283. [PubMed] [Google Scholar]
- Auerbach C., Robson J. M., Carr J. G., 1947. The chemical production of mutations. Science 105: 243–247. 10.1126/science.105.2723.243 [DOI] [PubMed] [Google Scholar]
- Avery O. T., Macleod C. M., McCarty M., 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79: 137–158. 10.1084/jem.79.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford J. S., Dewey W. C., 2002. Radiation research society. 1952–2002. Historical and current highlights in radiation biology: has anything important been learned by irradiating cells? Radiat. Res. 158: 251–291. 10.1667/0033-7587(2002)158[0251:HACHIR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Benzer S., Freese E., 1958. Induction of specific mutations with 5-Bromouracil. Proc. Natl. Acad. Sci. USA 44: 112–119. 10.1073/pnas.44.2.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukers R., Berends W., 1960. Isolation and identification of the irradiation product of thymine. Biochim. Biophys. Acta 41: 550–551. 10.1016/0006-3002(60)90063-9 [DOI] [PubMed] [Google Scholar]
- Bohr N., 1933. Light and life*. Nature 131: 457–459. 10.1038/131421a0 [DOI] [Google Scholar]
- Boyce R. P., Howard-Flanders P., 1964. Release of ultraviolet light-induced thymine dimers from DNA in E. Coli K-12. Proc. Natl. Acad. Sci. USA 51: 293–300. 10.1073/pnas.51.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., 2008. The foundations of molecular biology: a 50th anniversary. Curr. Biol. 18: R234–R236. 10.1016/j.cub.2008.02.016 [DOI] [PubMed] [Google Scholar]
- Chargaff E., 1950. Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia 6: 201–209. 10.1007/BF02173653 [DOI] [PubMed] [Google Scholar]
- Cleaver J. E., 1968. Defective repair replication of DNA in xeroderma pigmentosum. Nature 218: 652–656. 10.1038/218652a0 [DOI] [PubMed] [Google Scholar]
- Crick F., 1974. The double helix: a personal view. Nature 248: 766–769. 10.1038/248766a0 [DOI] [PubMed] [Google Scholar]
- Crick F. H., 1988. What Mad Pursuit. Basic Books, New York. [Google Scholar]
- Daly M. M., Allfrey V. G., Mirsky A. E., 1950. Purine and pyrimidine contents of some desoxypentose nucleic acids. J. Gen. Physiol. 33: 497–510. 10.1085/jgp.33.5.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C., 1982. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J. Mol. Biol. 156: 37–51. 10.1016/0022-2836(82)90457-0 [DOI] [PubMed] [Google Scholar]
- Friedberg E., 1997. Correcting the Blueprint of Life: An Historical Account of the Discovery of DNA Repair Mechanisms. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Furst S. S., Roll P. M., Brown G., 1950. On the renewal of the purines of the desoxypentose and pentose nucleic acids. J. Biol. Chem. 183: 251–266. [Google Scholar]
- Gefter M. L., Becker A., Hurwitz J., 1967. The enzymatic repair of DNA. I. Formation of circular lambda-DNA. Proc. Natl. Acad. Sci. USA 58: 240–247. 10.1073/pnas.58.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P. C., Haynes R. H., 1967. The repair of DNA. Sci. Am. 216: 36–43. 10.1038/scientificamerican0267-36 [DOI] [PubMed] [Google Scholar]
- Haynes R. H., Wolff S., Till J., 1965. Structural Defects in DNA and Their Repair in Microorganisms, edited by Haynes R. H., Wolff S., Till J. Radiation Research, Chicago. [Google Scholar]
- Hershey A. D., Chase M., 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36: 39–56. 10.1085/jgp.36.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Kato K., Strauss B., 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101: 417–425. 10.1016/0022-2836(76)90156-X [DOI] [PubMed] [Google Scholar]
- Hill R. F., 1958. A radiation-sensitive mutant of Escherichia coli. Biochim. Biophys. Acta 30: 636–637. 10.1016/0006-3002(58)90112-4 [DOI] [PubMed] [Google Scholar]
- Holliday R., 1964. A mechanism for gene conversion in fungi. Genet. Res. 5: 282–304. 10.1017/S0016672300001233 [DOI] [PubMed] [Google Scholar]
- Holloman B., 2014. Robin Holliday: 1932–2014. Cell 157: 1001–1003. 10.1016/j.cell.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B., 1979. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature 280: 76–77. 10.1038/280076a0 [DOI] [PubMed] [Google Scholar]
- Kelner A., 1949. Effect of visible light on the recovery of Streptomyces griseus conidia from ultra-violet irradiation injury. Proc. Natl. Acad. Sci. USA 35: 73–79. 10.1073/pnas.35.2.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., 2000. Ten commandments: lessons from the enzymology of DNA replication. J. Bacteriol. 182: 3613–3618. 10.1128/JB.182.13.3613-3618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Modrich P., 1988. Methyl-directed DNA mismatch repair in Escherichia coli. Mutat. Res. 198: 37–43. 10.1016/0027-5107(88)90037-1 [DOI] [PubMed] [Google Scholar]
- Lea D. E., 1946. Actions of Radiations on Living Cells. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Lindahl T., 1974. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 71: 3649–3653. 10.1073/pnas.71.9.3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B., 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11: 3610–3618. 10.1021/bi00769a018 [DOI] [PubMed] [Google Scholar]
- Mazia D., 1952. Physiology of the cell nucleus, pp. 77–122 in Modern Trends in Physiology and Biochemistry, edited by Guzman-Barron E. Academic Press, New York. [Google Scholar]
- McCulloch S. D., Kunkel T. A., 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18: 148–161. 10.1038/cr.2008.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Champe G. N., 1996. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 93: 1292–1297. 10.1073/pnas.93.3.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C., 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51: 241–249. 10.1016/0092-8674(87)90151-6 [DOI] [PubMed] [Google Scholar]
- Meselson M., 1964. On the mechanism of genetic recombination between DNA molecules. J. Mol. Biol. 9: 734–745. 10.1016/S0022-2836(64)80178-9 [DOI] [PubMed] [Google Scholar]
- Meselson M., Weigle J. J., 1961. Chromosome breakage accompanying genetic recombination in bacteriophage. Proc. Natl. Acad. Sci. USA 47: 857–868. 10.1073/pnas.47.6.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M., 1975. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 72: 358–361. 10.1073/pnas.72.1.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., 1986. Mismatch correction. Basic Life Sci. 38: 303–310. [DOI] [PubMed] [Google Scholar]
- Moore J. K., Haber J. E., 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. 10.1128/MCB.16.5.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1927. Artificial transmutation of the gene. Science 66: 84–87. 10.1126/science.66.1699.84 [DOI] [PubMed] [Google Scholar]
- Olivera B. M., Lehman I. R., 1967. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 57: 1426–1433. 10.1073/pnas.57.5.1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., et al. , 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75: 1227–1236. 10.1016/0092-8674(93)90331-J [DOI] [PubMed] [Google Scholar]
- Pettijohn D., Hanawalt P., 1964. Evidence for repair-replication of ultraviolet damaged DNA in bacteria. J. Mol. Biol. 9: 395–410. 10.1016/S0022-2836(64)80216-3 [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E., Hanawalt P. C., 1963. Deoxyribonucleic acid replication in bacteria following ultraviolet irradiation. Biochim. Biophys. Acta 72: 127–129. 10.1016/0926-6550(63)90324-4 [DOI] [PubMed] [Google Scholar]
- Polosina Y. Y., Cupples C. G., 2010. MutL: conducting the cell’s response to mismatched and misaligned DNA. BioEssays 32: 51–59. 10.1002/bies.200900089 [DOI] [PubMed] [Google Scholar]
- Reiter H., Strauss B., 1965. Repair of damage induced by a monofunctional alkylating agent in a transformable, ultraviolet-sensitive strain of Bacillus subtilis. J. Mol. Biol. 14: 179–194. 10.1016/S0022-2836(65)80239-X [DOI] [PubMed] [Google Scholar]
- Rupp W. D., Howard-Flanders P., 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31: 291–304. 10.1016/0022-2836(68)90445-2 [DOI] [PubMed] [Google Scholar]
- Sagan C., 1961. On the origin and planetary distribution of life. Radiat. Res. 15: 174–192. 10.2307/3571249 [DOI] [PubMed] [Google Scholar]
- Schoenheimer R., 1942. The Dynamic State of Body Constituents. Harvard University Press, Cambridge, MA. [Google Scholar]
- Schrödinger E., 1945. What Is Life? The Physical Aspect of the Living Cell The Macmillan Company, New York. [Google Scholar]
- Setlow R. B., Carrier W. L., 1964. The disappearance of thymine dimers from DNA: an error-correcting mechanism. Proc. Natl. Acad. Sci. USA 51: 226–231. 10.1073/pnas.51.2.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. C., Wang T. C., 1989. recA-dependent DNA repair processes. BioEssays 10: 12–16. 10.1002/bies.950100104 [DOI] [PubMed] [Google Scholar]
- Sobels F. H. (Editor), 1963. Repair from Genetic Radiation Damage. The MacMillan Company, New York. [Google Scholar]
- Stadler L. J., 1928a Genetic effects of X-rays in maize. Proc. Natl. Acad. Sci. USA 14: 69–75. 10.1073/pnas.14.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler L. J., 1928b Mutations in barley induced by X rays and radium. Science 68: 186–187. 10.1126/science.68.1756.186 [DOI] [PubMed] [Google Scholar]
- Stone W. S., Wyss O., Haas F., 1947. The production of mutations in Staphylococcus aureus by irradiation of the substrate. Proc. Natl. Acad. Sci. USA 33: 59–66. 10.1073/pnas.33.3.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B. S., 1963. Recovery of deoxyribonucleic acid from the effects of alkylation. J. Gen. Microbiol. 30: 89–103. 10.1099/00221287-30-1-89 [DOI] [PubMed] [Google Scholar]
- Strauss B. S., 2017. A physicist’s quest in biology: Max Delbrück and “complementarity.” Genetics 206: 641–650. 10.1534/genetics.117.201517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. 10.1016/0092-8674(83)90331-8 [DOI] [PubMed] [Google Scholar]
- Timofeeff-Ressovsky N. W., Zimmer K. G., Delbrück M., 1935. Über die Natur der Genmutation und der Genstruktur. Nachrichten von der Gesellschaft der Wissenschaften zu Göttingen, Math.-. Phys. Kl Fachgruppe 6: 190–245. [Google Scholar]
- Wagner R., Jr., Meselson M., 1976. Repair tracts in mismatched DNA heteroduplexes. Proc. Natl. Acad. Sci. USA 73: 4135–4139. 10.1073/pnas.73.11.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., 1968. The Double Helix. A Personal Account of the Discovery of the Structure of DNA. Atheneum, New York. [Google Scholar]
- Watson J. D., Crick F. H., 1953a Genetical implications of the structure of deoxyribonucleic acid. Nature 171: 964–967. 10.1038/171964b0 [DOI] [PubMed] [Google Scholar]
- Watson J. D., Crick F. H., 1953b Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171: 737–738. 10.1038/171737a0 [DOI] [PubMed] [Google Scholar]
- Weiss B., Richardson C. C., 1967. Enzymatic breakage and joining of deoxyribonucleic acid, I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. Proc. Natl. Acad. Sci. USA 57: 1021–1028. 10.1073/pnas.57.4.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse H. L., 1963. A theory of crossing-over by means of hybrid deoxyribonucleic acid. Nature 199: 1034–1040. 10.1038/1991034a0 [DOI] [PubMed] [Google Scholar]
- Witkin E. M., 1956. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harb. Symp. Quant. Biol. 21: 123–140. 10.1101/SQB.1956.021.01.011 [DOI] [PubMed] [Google Scholar]
- Witkowski, J. A., 2000 Introduction to Biological Responses to DNA Damage, Vol. LXV Cold Spring Harbor Symposia on Quantitative Biology. Available at: http://symposium.cshlp.org/site/misc/topic65.xhtml. [DOI] [PubMed]
- Wood R. D., Mitchell M., Lindahl T., 2005. Human DNA repair genes, 2005. Mutat. Res. 577: 275–283. 10.1016/j.mrfmmm.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Wulff D. L., Fraenkel G., 1961. On the nature of thymine photo-product. Biochim. Biophys. Acta 51: 332–339. 10.1016/0006-3002(61)90174-3 [DOI] [PubMed] [Google Scholar]
- Wulff D. L., Rupert C. S., 1962. Disappearance of thymine photodimer in ultraviolet irradiated DNA upon treatment with a photoreactivating enzyme from baker’s yeast. Biochem. Biophys. Res. Commun. 7: 237–240. 10.1016/0006-291X(62)90181-X [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Little J. W., Oshinsky C. K., Gellert M., 1967. Enzymatic joining of DNA strands: a novel reaction of diphosphopyridine nucleotide. Proc. Natl. Acad. Sci. USA 57: 1841–1848. 10.1073/pnas.57.6.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]