Abstract
Due to issues of practicality and confidentiality of genomic data sharing on a large scale, typically only meta- or mega-analyzed genome-wide association study (GWAS) summary data, not individual-level data, are publicly available. Reanalyses of such GWAS summary data for a wide range of applications have become more and more common and useful, which often require the use of an external reference panel with individual-level genotypic data to infer linkage disequilibrium (LD) among genetic variants. However, with a small sample size in only hundreds, as for the most popular 1000 Genomes Project European sample, estimation errors for LD are not negligible, leading to often dramatically increased numbers of false positives in subsequent analyses of GWAS summary data. To alleviate the problem in the context of association testing for a group of SNPs, we propose an alternative estimator of the covariance matrix with an idea similar to multiple imputation. We use numerical examples based on both simulated and real data to demonstrate the severe problem with the use of the 1000 Genomes Project reference panels, and the improved performance of our new approach.
Keywords: 1000 Genomes Project, COJO analysis, gene-based testing, multiple imputation, multiple SNPs, type I error, Wald test
DUE to logistic reasons and privacy concerns, individual-level genotypic and phenotypic data from large genome-wide association studies (GWAS) are often not publicly available; in contrast, GWAS summary statistics, in the form of the z-scores and/or P-values of single SNPs based on their univariate/marginal associations with a GWAS trait, are publicly available. Interestingly, as first demonstrated by Yang et al. (2012), one can combine GWAS summary statistics with a reference panel with individual-level genotypic data (that mimic the original population for the GWAS summary data) to conduct conditional and joint (COJO) analyses; that is, one can estimate and test the joint effects of multiple SNPs in a genomic region such as a gene or a pathway, which may be more powerful and/or interpretable than the standard/marginal single SNP-based analysis. In addition, based on a joint regression model for multiple SNPs, one can conduct COJO analysis: conditional on (i.e., after accounting for the effects of) other SNPs, one can estimate and test for possible association of one SNP (or multiple SNPs), which is useful in sorting out multiple causal SNPs as in fine mapping (Hormozdiari et al. 2014; Kichaev et al. 2014; Chen et al. 2015). The usefulness of such analyses and other ones on some publicly available GWAS summary data sets has been nicely reviewed in Pasaniuc and Price (2017).
A critical issue in these approaches with GWAS summary statistics is to estimate linkage disequilibrium (LD) among the SNPs in a genomic region using a reference panel, which is necessary for estimating the correlation or covariance matrices of various parameter estimates and their associated testing statistics in any subsequent conditional or joint analysis. As pointed out by Pasaniuc and Price (2017), “Conditional association and imputation using summary statistics crucially rely on accurate LD information from a population reference panel. Even in the best case, when the reference population closely matches the GWAS population, the relatively small size of reference panels for which LD information is publicly available (typically hundreds or at most thousands of individuals) makes accurate estimation of a large number of LD parameters a challenge.” Although regularization-based methods for estimating LD or covariance matrices have been discussed in other related contexts, as to be shown, it is unclear how to choose tuning parameters associated with any regularization-based method in the current context of statistical inference, not of estimation or prediction (Pasaniuc et al. 2014; Kichaev and Pasaniuc 2015; Shi et al. 2016). However, in spite of the obvious importance of the issue, it has been largely ignored in practice. Although a small reference panel with a sample size in the hundreds, such as any of the 1000 Genomes Project (1000G) racial/ethnic group-specific panels, is often used, it is striking that there has been barely any assessment on its effects on subsequent statistical inference. As will be shown here, both expectedly and surprisingly, it often leads to dramatically inflated type I error rates and thus large numbers of false positives. We also point out that Yang et al. (2012) used a reference sample size of over 6000, > 10 times larger than that of the popular choice with the 1000G data in practice. Even though the total sample size of the 1000G data (1000 Genomes Project Consortium et al. 2015) is > 3000, due to the presence of multiple populations or ethnic groups, its sample size for a single population is no more than a few hundreds, which is often used in practice.
We emphasize that the underlying issue discussed here is quite general and wide-ranging: although our focus is on COJO analyses of multiple SNPs such as in gene-based testing and fine mapping, any approach using GWAS summary data and a reference panel may suffer from the same problem, no matter it is polygenic risk prediction (Vilhjalmsson et al. 2015), or inferring genetic correlations among complex traits (Bulik-Sullivan et al. 2015), or Mendelian randomization for causal inference (Burgess et al. 2013). Very recently Benner et al. (2017) demonstrated the severe problem in the context of fine-mapping, while we consider both conditional and global testing with a group of SNPs. More importantly, we propose a new method to alleviate the problem. We note that, even if a reference panel comes from the same population of the GWAS data, using the reference data with a small sample size may still lead to increased numbers of false positives. Of course, if the reference sample is from a different population, the situation becomes worse; here we mainly focus on the former case. The main issue is the ignorance of the small sample size of the reference panel, and thus its associated estimation errors or uncertainties. Accordingly, we propose using an idea similar to multiple imputations (MIs) (Rubin 1996) to alleviate the problem. We provide numerical examples based on both simulated and real data to show the impact of small reference panels, even when they are drawn from the same GWAS population, and the effectiveness of our proposed MI-type approach.
Methods
To be concrete and general, we focus on the joint analysis of a group of SNPs in a genomic region and on the COJO analysis of one of the SNPs (after accounting for the effects of other SNPs) with a single quantitative trait. For a quantitative trait with a normal distribution, although an F-test is exact, due to the large sample size of a typical GWAS, we restrict our attention to the asymptotically equivalent χ2 test.
Suppose that we are interested in SNPs and one trait, denoted by and which are vectors for subjects. Given the summary statistics and in marginal analysis
| (1) |
as well as an estimate of the correlation matrix of the SNPs from a reference panel, we can obtain the regression coefficient estimates and their covariance matrix, denoted as and , respectively (Yang et al. 2012), in the joint model
| (2) |
The Wald test statistic is
| (3) |
Under the global/overall null hypothesis H0: asymptotically (approximately) follows a χ2 distribution with d.f. When the covariance matrix is not invertible, we use the Moore–Penrose generalized inverse and modify the d.f. as the rank of the covariance matrix.
From our experience, using the estimated correlations of the SNPs based on a small reference panel may lead to inflated type I errors, since may not be accurate enough. From a different angle, we can regard that using the reference sample only once to estimate LD among the SNPs ignores the nonnegligible uncertainty in the resulting estimate due to the small sample size. To account for the estimation uncertainty, we borrow the idea of MIs (Rubin 1996) and propose a MI-type method. Specifically, in addition to using the reference panel to build one model and obtain and we can use the data more times to get estimates of the coefficients, and then inflate the covariance estimates for a more conservative inference (to battle the issue of inflated type I errors). We denote the parameter estimate and its covariance matrix based on the complete reference panel as and In each imputation sample subjects from the subjects in the reference panel with replacement. Take these subjects as the reference sample to build a joint model. Estimate the coefficients as and its covariance matrix as Calculate and using the following formulas, then carry out the Wald test (3):
| (4) |
From our experience, as in MI, usually setting ≤ 50 should be enough. We can regard the existing approach of using the complete reference panel only once as a special case of our proposed MI-type approach with .
COJO analysis on an individual SNP can be conducted based on and which may be based on individual-level data, the complete reference sample, or MI-type estimation. For example, if we would like to test H0j: against H1j: then the Wald test statistic is
| (5) |
where is the th diagonal element of Under the null hypothesis H0j, the test statistic follows a χ2 distribution with 1 d.f.
There are four ways to conduct a COJO analysis on a SNP (conditioning on other SNPs) or a group of SNPs. The first, denoted “Ind,” is based on using individual-level data. The second uses summary statistics but the LD matrix X’X is estimated from the original data (i.e., assuming the availability of LD from the original data); it was confirmed to give exactly the same results as that of the first method Ind, and hence is omitted in the sequel. The third one is the naive method of using summary statistics with a reference panel to estimate LD or the matrix X’X only once, i.e., with The fourth method, denoted “Sum-MI,” is our proposed new MI-type method with
Alternatively, a general class of approaches to better estimate LD or covariance matrices is to apply regularizations: one is to truncate the eigenvalues of a matrix based on its singular value decomposition (SVD) (Shi et al. 2016), while the other is to impose a penalty like the ridge penalty (Pasaniuc et al. 2014; Kichaev and Pasaniuc 2015), which has been studied in other contexts. The general ideas can be applied here. Specifically, we can decompose the covariance matrix of the SNPs, estimated from the complete reference panel (with T = 1), as
| (6) |
where are the th largest eigenvalue and the th eigenvector of Applying the ridge penalty is equivalent to replacing with while the truncation is to replace with and are the corresponding tuning parameters. Instead of one can also apply each of the two regularization methods to the estimated X’X, the LD matrix. Then we can carry out the Wald test as usual. As expected and to be shown, the performance of the regularization methods critically depends on the choice of the tuning parameters; however, differing from estimation and prediction, it is quite difficult and largely unknown how to choose tuning parameters for a regularization method in the current context of hypothesis testing.
Data availability
The 2013 lipid data (Willer et al. 2013) is publicly available at http://csg.sph.umich.edu/abecasis/public/lipids2013/. The Lung Health Study (LHS) data can be downloaded from the database of Genotypes and Phenotypes (dbGaP) database (accession: phs000335.v3.p2) by request. Information on the WTCCC (Wellcome Trust Case Control Consortium) data and how to apply for access can be found at https://www.wtccc.org.uk/info/access_to_data_samples.html. The method is implemented and freely available in R package jointsum at https://github.com/yangq001/conditional. The package will also be available on CRAN soon.
Results
Simulations
To investigate the reference panels’ impact on the testing performance, we first did some simulation studies. To be as realistic as possible, we used the individual-level genotypic data of 2938 subjects in the control group from the WTCCC data (Wellcome Trust Case Control Consortium 2007). We randomly chose some SNPs in genomic regions on chromosome 19 so that none of the pair-wise (absolute) correlations were > 0.9. For power study, we needed to specify effect sizes, so we used the lipid data (Willer et al. 2013) to build a joint model for triglycerides (TGs) vs. SNPs. Denote its coefficients by Since the lipid data only contains summary statistics, we used the correlations of the SNPs estimated from the WTCCC data. We scaled the significant effects (P-value < 5e−8) while forcing insignificant effects as zero to obtain a true regression model. Then, we generated a quantitative trait for the 2938 subjects using the model (2) with no intercept and otherwise, and the error term was an independent normal random variable with mean 0 and variance obtained from the joint model. For each replication, we randomly chose and subjects from subjects as the reference panel and the GWAS sample, respectively.
For the approaches based on summary statistics, in addition to a subsample of the WTCCC data, we also chose the 1000G data as our reference panel. We used the 379 CEU (Utah Residents with Northern and Western Ancestry) samples from the 1000G phase I version 3 Shapeit2 Reference data from the KGG software website (Li et al. 2012), denoted as 1000G A. We calculated the rejection rates based on 10,000 replications for the null case.
In a representative region with eight SNPs, as shown in Table 1, in both COJO analyses, using a reference sample of size 500 or 900 drawn from the same population led to inflated type I error rates, while our proposed approach largely corrected the problem with a small T = 10. It is noteworthy that using the 1000G reference panel also gave an inflated type I error rate for the COJO analysis in the naive approach, but yielded very conservative global testing. One possible explanation for the latter is the possible difference inherent between the 1000G data and the WTCCC data: the correlation structures of the eight SNPs in the reference data were different from that of the WTCCC data, leading to a huge difference in the test statistics. Since the Wald test statistic involves the inverse of the correlation matrix, we examined the eigenvalues of the inverse correlation matrices estimated from the individual-level WTCCC data and that from the 1000G reference data: their largest eigenvalues were 17.1 and 14.6, respectively, explaining why using the 1000G reference data led to a lower rejection rate than that of the nominal level. For this situation, it is unknown how to avoid conservative inference; our method cannot avoid it either.
Table 1. Type I error rates for simulations in a region with eight SNPs, none of which was associated with the trait.
| 900 | 0.0531 | 0.0504 | 0.0502 | 0.0621 | 0.0518 | 0.0507 | 0.0512 | 0.0509 | 0.0510 |
| 500 | 0.0559 | 0.0504 | 0.0506 | 0.0694 | 0.0520 | 0.0500 | 0.0507 | 0.0499 | 0.0507 |
| 379 (1000G A) | 0.0548 | 0.0516 | 0.0520 | 0.0092 | 0.0062 | 0.0055 | 0.0057 | 0.0057 | 0.0057 |
| Ind | 0.0515 | 0.0515 | |||||||
The nominal significance level was at 0.05; in each set-up, there were 10,000 replications; the genome-wide association study sample size was Ind, individual.
We also considered the two regularization methods. Table 2 shows the results for regularizing X’X. As expected, the performance critically depends on the choice of the tuning parameter, which is unknown. The same conclusion can be drawn on regularizing the covariance matrix For example, for the SVD-truncation method keeping S = 3, 5, and 7 top eigenvalues: the empirical type I error rates were (1) 0.002, 0.012, and 0.039, respectively, with the reference panel of 900 subjects and (2) 2e−4, 0.001, and 0.005, respectively, with the 1000G A reference panel.
Table 2. Type I error rates of the regularization methods for simulations in a region with eight SNPs, none of which was associated with the trait.
| Ridge penalty | SVD truncation (number of eigenvalues) | ||||||
|---|---|---|---|---|---|---|---|
| 900 | 0.0621 | 0.0248 | 0.0164 | 0.0108 | 0.920 | 0.0140 | 0.0362 |
| 500 | 0.0694 | 0.0270 | 0.0175 | 0.0115 | 0.833 | 0.0162 | 0.0400 |
| 379 (1000G A) | 0.0092 | 0.0048 | 0.0048 | 0.0042 | 0.127 | 0.0022 | 0.0046 |
| Ind (individual) | 0.0515 | ||||||
The null hypothesis tested was with the nominal significance level at 0.05; in each set-up there were 10,000 replications; the genome-wide association study sample size was SVD, singular value decomposition; Ind, individual.
For empirical power, as shown in Table 3, in all situations corresponding to the anticonservative inference of the naive approach, our proposed method barely lost power as compared to the individual-level data-based method. On the other hand, for the global testing, due to the its conservativeness with the use of the 1000G A reference panel (for its possible difference from the WTCCC data), there was some power loss from the naive and our new methods based on summary statistics as compared to the individual-level data-based method; nevertheless, at least compared to the naive method, our method lost only minimal power.
Table 3. Empirical power for simulations in a region with eight SNPs.
| 0.01 | 900 | 0.738 | 0.739 | 0.739 | 0.408 | 0.395 | 0.393 | 0.398 | 0.399 |
| 500 | 0.738 | 0.737 | 0.737 | 0.415 | 0.388 | 0.388 | 0.388 | 0.389 | |
| 379 (1000G A) | 0.736 | 0.734 | 0.738 | 0.279 | 0.259 | 0.257 | 0.249 | 0.250 | |
| Ind | 0.731 | 0.405 | |||||||
| 0.015 | 900 | 0.970 | 0.970 | 0.970 | 0.808 | 0.798 | 0.800 | 0.798 | 0.798 |
| 500 | 0.971 | 0.970 | 0.971 | 0.810 | 0.793 | 0.794 | 0.793 | 0.792 | |
| 379 (1000G A) | 0.970 | 0.970 | 0.970 | 0.722 | 0.711 | 0.710 | 0.702 | 0.702 | |
| Ind | 0.970 | 0.806 | |||||||
The nominal significance level was at 0.05; in each set-up there were 3000 replications; the genome-wide association study sample size was n = 1000. 1000G, 1000 Genomes Project; Ind, individual.
We did another simulation with 100 randomly selected regions, each including 5–37 SNPs. Most of the regions were larger than the region in Table 1. As shown in Table 4, again the naïve method could not control the type I error rate while the new method performed much better, although the new method became conservative as went up. A possible explanation is that the sample size needed to estimate the LD accurately for a larger number of SNPs should be larger. With relatively small reference samples, the estimation of the regression coefficients is unstable, leading to large and thus less significant test statistics. Nevertheless, the performance improved as the reference sample size increased from 500 to 900 with little loss of power.
Table 4. Empirical type I error rate (with =0) and power (with for simulations in 100 regions with 5–37 SNPs (17.8 on average), none or only the first one of which was associated with the trait.
| 0 | 900 | 0.089 | 0.024 | 0.022 | 0.162 | 0.020 | 0.016 | 0.014 | 0.013 |
| 500 | 0.107 | 0.022 | 0.020 | 0.197 | 0.022 | 0.014 | 0.012 | 0.011 | |
| 379 (1000G A) | 0.171 | 0.048 | 0.045 | 0.352 | 0.085 | 0.072 | 0.064 | 0.063 | |
| Ind | 0.053 | 0.055 | |||||||
| 0.001 | 900 | 0.580 | 0.579 | 0.577 | 0.827 | 0.742 | 0.736 | 0.732 | 0.733 |
| 500 | 0.605 | 0.577 | 0.576 | 0.847 | 0.740 | 0.732 | 0.727 | 0.724 | |
| 379 (1000G A) | 0.596 | 0.597 | 0.589 | 0.822 | 0.759 | 0.745 | 0.738 | 0.735 | |
| Ind | 0.495 | 0.794 | |||||||
The nominal significance level was at 0.05; in each set-up there were 3000 replications (30 per region); the genome-wide association study sample size was 1000G, 1000 Genomes Project; Ind, individual.
LHS data
Next, we applied the methods to the LHS data with 4387 subjects and 5112 SNPs on chromosome 19, downloaded from the dbGaP database (accession: phs000335.v3.p2). Our trait of interest was forced expiratory volume (FEV) at the baseline, FEVAC112. First, to adjust for nongenetic covariates, we built a linear model: FEVAS112 ∼ AGE + SEX + PACKYEAR. Then we treated the residuals as the quantitative trait Y for the SNPs. We obtained the summary statistics of the marginal effects for each individual SNP on Y after centering the data at 0.
After choosing 4132 subjects with complete outcomes and 5111 SNPs that were present in both the LHS and 1000G data, we tested each single SNP and found none of them to be marginally significant. Then, we used a sliding window approach to test the association between the trait and the SNPs inside each sliding window in a joint linear model (with the trait vs. multiple SNPs). In each window, we selected SNPs so that none of their pair-wise correlation absolute values was > 0.95. We used two window sizes of 20 and 50 with two moving step sizes/gaps of 1 and 20, respectively.
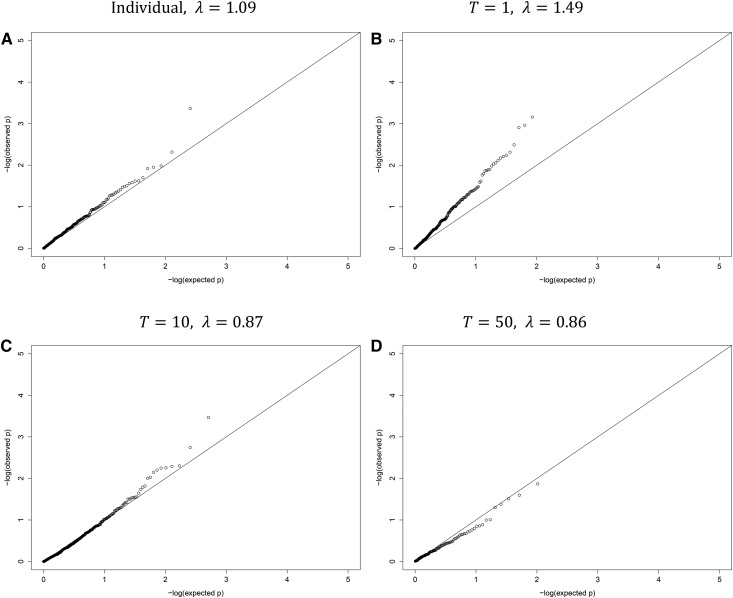
For the global/overall testing, as shown in Table 5, the Wald test based on the individual-level data detected no significant association regardless of the window size and moving step size; in contrast, the naive method based on the summary statistics (T = 1) reported many significant associations, which (or at least most of which) are most likely to be false positives. Our new method with T = 30 or larger eliminated all the false positives. The QQ (quantile-quantile) plot in Figure 1 also demonstrates the problem of the naive method with an inflation factor λ = 1.49, much larger than 1, while the new method might be a bit conservative with an inflation factor < 1 (Devlin and Roeder 1999).
Table 5. Numbers of the significant sliding windows for global testing with the LHS data.
| U | ξ | Gap | f | Ind | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5111 | 20 | 1 | 5092 | 0 | 20 | 4 | 0 | 0 | 0 | 0 |
| 20 | 255 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | ||
| 50 | 1 | 5062 | 0 | 103 | 51 | 17 | 4 | 0 | 0 | |
| 50 | 102 | 0 | 7 | 3 | 2 | 1 | 0 | 0 |
The nominal significance was at 0.05 with the Bonferroni adjustment, with a cutoff 0.05/# windows. The reference sample size was based on the 1000G A reference panel. , and Gap were the total number of SNPs, window size, the number of windows, and the moving-step/gap size, respectively. 1000G, 1000 Genomes Project; Ind, individual.
Figure 1.
QQ (quantile-quantile) plots for the Lung Health Study data. ξ and the gap size are both 20.
Similarly, Table 6 shows that in the COJO analysis on the first SNP inside each window, the individual-level data-based method identified no significant association. Again, the naive method with summary statistics detected three significant ones, most likely false positives; two or all three could be eliminated by the new method.
Table 6. Numbers of the windows with the first SNP being significant in COJO analysis with the LHS data.
| U | ξ | Gap | f | Ind | T = 1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| 5111 | 20 | 1 | 5092 | 0 | 1 | 0 | 0 | 0 | 0 |
| 20 | 255 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 50 | 1 | 5062 | 0 | 2 | 0 | 1 | 1 | 1 | |
| 50 | 102 | 0 | 0 | 0 | 0 | 0 | 0 |
The statistical significance cutoff was 0.05/# windows. , and Gap were the total number of SNPs, window size, the number of windows, and the moving-step/gap size, respectively. Ind, individual.
Lipid data
We applied the methods to the 2010 and 2013 lipid data (Teslovich et al. 2010; Willer et al. 2013), testing the association between TGs and SNPs on chromosome 19 that are present in both data sets. To save space, we only present the results for the 2013 lipid data in the following. We chose 7366 SNPs that were present in both the lipid data and the 1000G phase 3 data with 503 subjects from the European population as the reference panel (denoted as the 1000G B reference panel in the following), with minor allele frequencies larger than 0.01. First, we looked at the marginal P-values of each SNP, and found 86 of the 7366 SNPs with P-values < 0.05/7366, and 911 with P-values < 0.05. The estimated inflation factor was 1.0.
In addition to the 1000G B reference panel, we also used various subsets of the LHS data as a reference panel. We randomly sampled subjects from the 4136 subjects in the LHS data as the reference data before applying the sliding window approach to the 4364 overlapping SNPs in the 2013 lipid data. As shown in Table 7 for global testing, as expected, the naive method gave much larger numbers of significant associations than that of the proposed new method, in which T = 30 or larger seemed to give stable results. The same conclusion can be drawn for the COJO analysis as shown in Table 8. In summary, we expect that the naive method gave too many false positives.
Table 7. Numbers of the significant sliding windows for global testing with the 2013 lipid data, using subsamples of the LHS data as reference with or using the 1000G B data with .
| Chromosome | ξ | () | ||||||
|---|---|---|---|---|---|---|---|---|
| 19 | 10 | 503 (1000G B) | 7366 (735) | 57 | 22 (22) | 20 (20) | 22 (22) | 22 (22) |
| 1000 | 4364 (435) | 38 | 19 (19) | 15 (15) | 14 (14) | 15 (15) | ||
| 2000 | 37 | 19 (19) | 17 (17) | 15 (15) | 15 (15) | |||
| 4000 | 40 | 17 (17) | 16 (16) | 15 (15) | 15 (15) | |||
| 20 | 503 (1000G B) | 7366 (367) | 59 | 27 (27) | 20 (20) | 17 (17) | 17 (17) | |
| 1000 | 4364 (217) | 35 | 23 (23) | 14 (14) | 10 (10) | 9 (9) | ||
| 2000 | 35 | 17 (17) | 12 (12) | 10 (10) | 10 (10) | |||
| 4000 | 33 | 18 (18) | 10 (10) | 11 (11) | 11 (11) |
The statistical significance cut-off was 0.05/# windows ξ, and were the total number of SNPs, window size, and the number of windows, respectively; the moving-step size or gap size was equal to the window size. The numbers of overlapping SNPs between T = 1 and others are shown in parentheses. 1000G, 1000 Genomes Project.
Table 8. Numbers of the windows with the first SNP being significant in COJO analysis with the 2013 lipid data, using subsamples of the LHS data as reference with .
| Chromosome | U | () | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 19 | 4364 | 10 (435) | 1000 | 6 | 2 (2) | 1(1) | 1 (1) | 2 (2) | 2 (2) |
| 2000 | 8 | 4 (4) | 2 (2) | 2 (2) | 3 (3) | 3 (3) | |||
| 4000 | 8 | 2 (2) | 2 (2) | 2 (2) | 2 (2) | 2 (2) | |||
| 20 (217) | 1000 | 6 | 3 (3) | 3 (3) | 1 (1) | 2 (2) | 2 (2) | ||
| 2000 | 6 | 4 (4) | 3 (3) | 3 (3) | 3 (3) | 3 (3) | |||
| 4000 | 6 | 2 (2) | 2 (2) | 3 (3) | 2 (2) | 2 (2) |
The statistical significance cut-off was 0.05/# windows; , and were the total number of SNPs, window size, and the number of windows, respectively; the moving-step size or gap size was equal to the window size. The numbers of overlapping SNPs between T = 1 and others are shown in parentheses.
Discussion
Using simulated and real data, we have convincingly shown the severe problem of inflated type I error rates in integrating GWAS summary data with small reference panels for COJO analyses, which have been widely applied in the last few years, ranging from gene-based testing with one or more traits (Kwak and Pan 2016, 2017; Deng and Pan 2017) to fine mapping. In particular, as a gene-based testing approach to integrating eQTL (expression quantitative trait loci) data with GWAS summary data, the recently proposed transcriptome-wide association studies are expected to share the same problem with small reference panels (Gamazon et al. 2015; Gusev et al. 2016; Xu et al. 2017). We emphasize that, although we have focused on conditional and global testing on a group of SNPs, the same issue of using small reference panels persists in many new and existing applications: to name a few, fine mapping (Benner et al. 2017), polygenic risk prediction (Vilhjalmsson et al. 2015), inferring genetic correlations among complex traits (Bulik-Sullivan et al. 2015), and Mendelian randomization for causal inference (Burgess et al. 2013). Although standard reference panel samples, as for the 1000G data, are continuing to grow with increasing sample sizes, the current and almost exclusive use of the popular 1000G reference panels is expected to suffer from the small sample issue as demonstrated here. Furthermore, even with a larger reference panel, if a GWAS sample size is larger (Benner et al. 2017) or if we expand the SNPs to be tested to cover less frequent or rare ones and/or those in high LD, as in fine mapping with sequencing data, the problem may still arise. Our proposed method, or its idea, could be applied (possibly after suitable modifications) to at least check whether the problem is severe in a given situation. Finally, we note that it is unclear how to deal with the problem if there are genotypic discrepancies between the reference panel and the GWAS data, which may happen in practice, especially with meta-analyzed GWAS summary statistics with multiple racial/ethnic subpopulations, for which any reference sample from a single population may not suffice (for the mixed GWAS population). In this case, perhaps the most straightforward solution is to conserve and share the LD structure from the original GWAS data. This problem is similar to meta-analysis of rare variants with sequencing data (Lee et al. 2013). We hope that this study, along with Benner et al. (2017), will raise the awareness of and attention to this important and urgent problem in light of the increasing use of GWAS summary data and (small) reference panels.
Acknowledgments
The authors thank the reviewers for many helpful comments and Chong Wu for help with the data. We downloaded the LHS data from dbGaP. This research was supported by National institutes of Health grants R21 AG-057038, R01 HL-116720, R01 GM-113250, and R01 HL-105397; by National Science Foundation grant DMS1711226, and by the Minnesota Supercomputing Institute.
Footnotes
Communicating editor: E. Eskin
Literature Cited
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C., Havulinna A. S., Järvelin M. R., Salomaa V., Ripatt S., et al. , 2017. 2017 prospects of fine-mapping trait-associated genomic regions by using summary statistics from genome-wide association studies. Am. J. Hum. Genet. 101: 539–551. 10.1016/j.ajhg.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B., Finucane H. K., Anttila V., Gusev A., Day F. R., et al. , 2015. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47: 1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Butterworth A., Thompson S. G., 2013. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37: 658–665. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Larrabee B. R., Ovsyannikova I. G., Kennedy R. B., Haralambieva I. H., et al. , 2015. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200: 719–736. 10.1534/genetics.115.176107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Pan W., 2017. Conditional analysis of multiple quantitative traits based on marginal GWAS summary statistics. Genet. Epidemiol. 41: 427–436. 10.1002/gepi.22046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K., 1999. Genomic control for association studies. Biometrics 55: 997–1004. 10.1111/j.0006-341X.1999.00997.x [DOI] [PubMed] [Google Scholar]
- Gamazon E. R., Wheeler H. E., Shah K. P., Mozaffari S. V., Aquino-Michaels K., et al. , 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47: 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Ko A., Shi H., Bhatia G., Chung W., et al. , 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48: 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., Kostem E., Kang E. Y., Pasaniuc B., Eskin E., 2014. Identifying causal variants at loci with multiple signals of association. Genetics 198: 497–508. 10.1534/genetics.114.167908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Pasaniuc B., 2015. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97: 260–271 (erratum: Am. J. Hum. Genet. 97: 353) 10.1016/j.ajhg.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Yang W. Y., Lindstrom S., Hormozdiari F., Eskin E., et al. , 2014. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10: e1004722 10.1371/journal.pgen.1004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak I., Pan W., 2016. Adaptive gene- and pathway-trait association testing with GWAS summary statistics. Bioinformatics 32: 1178–1184. 10.1093/bioinformatics/btv719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak I., Pan W., 2017. Gene- and pathway-based association tests for multiple traits with GWAS summary statistics. Bioinformatics 33: 64–71. 10.1093/bioinformatics/btw577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Teslovich T. M., Boehnke M., Lin X., 2013. General framework for meta-analysis of rare variants in sequencing association studies. Am. J. Hum. Genet. 93: 42–53. 10.1016/j.ajhg.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-X., Kwan J. S. H., Sham P. C., 2012. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am. J. Hum. Genet. 91: 478–488. 10.1016/j.ajhg.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Price A. L., 2017. Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet. 18: 117–127. 10.1038/nrg.2016.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Zaitlen N., Shi H., Bhatia G., Gusev A., et al. , 2014. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics 30: 2906–2914. 10.1093/bioinformatics/btu416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D., 1996. Multiple imputation after 18 years. J. Am. Stat. Assoc. 91: 473–489. 10.1080/01621459.1996.10476908 [DOI] [Google Scholar]
- Shi H., Kichaev G., Pasaniuc B., 2016. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 99: 139–153. 10.1016/j.ajhg.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., et al. , 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjalmsson B. J., Yang J., Finucane H. K., Gusev A., Lindstrom S., et al. , 2015. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97: 576–592. 10.1016/j.ajhg.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., et al. , 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wu C., Wei P., Pan W., 2017. A powerful framework for integrating eQTL and GWAS summary data. Genetics 207: 893–902. 10.1534/genetics.117.300270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ferreira T., Morris A. P., Medland S. E., GIANT Consortium et al. , 2012. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012 18: 44(4):369-75, S1-3 10.1038/ng.2213 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 2013 lipid data (Willer et al. 2013) is publicly available at http://csg.sph.umich.edu/abecasis/public/lipids2013/. The Lung Health Study (LHS) data can be downloaded from the database of Genotypes and Phenotypes (dbGaP) database (accession: phs000335.v3.p2) by request. Information on the WTCCC (Wellcome Trust Case Control Consortium) data and how to apply for access can be found at https://www.wtccc.org.uk/info/access_to_data_samples.html. The method is implemented and freely available in R package jointsum at https://github.com/yangq001/conditional. The package will also be available on CRAN soon.