Abstract
The histone demethylase LSD1 was originally discovered by removing methyl groups from di- and monomethylated histone H3 lysine 4 (H3K4me2/1). Several studies suggest that LSD1 plays roles in meiosis as well as in the epigenetic regulation of fertility given that, in its absence, there is evidence of a progressive accumulation of H3K4me2 and increased sterility through generations. In addition to the progressive sterility phenotype observed in the mutants, growing evidence for the importance of histone methylation in the regulation of DNA damage repair has attracted more attention to the field in recent years. However, we are still far from understanding the mechanisms by which histone methylation is involved in DNA damage repair, and only a few studies have focused on the roles of histone demethylases in germline maintenance. Here, we show that the histone demethylase LSD1/CeSPR-5 interacts with the Fanconi anemia (FA) protein FANCM/CeFNCM-1 using biochemical, cytological, and genetic analyses. LSD1/CeSPR-5 is required for replication stress-induced S phase-checkpoint activation, and its absence suppresses the embryonic lethality and larval arrest observed in fncm-1 mutants. FANCM/CeFNCM-1 relocalizes upon hydroxyurea exposure and colocalizes with FANCD2/CeFCD-2 and LSD1/CeSPR-5, suggesting coordination between this histone demethylase and FA components to resolve replication stress. Surprisingly, the FA pathway is required for H3K4me2 maintenance, regardless of the presence of replication stress. Our study reveals a connection between FA and epigenetic maintenance and therefore provides new mechanistic insight into the regulation of histone methylation in DNA repair.
Keywords: LSD1/SPR-5, FANCM/FNCM-1, FANCD2/FCD-2, histone demethylation and DNA repair, germline
MOST eukaryotes package their DNA around histones and form nucleosomes to compact the genome. A nucleosome is the basic subunit of chromatin and comprises ∼147 bp of DNA wrapped around a protein octamer which consists of two molecules each of four highly conserved core histones: H2A, H2B, H3, and H4. Core histones can be replaced by various histone variants, each of which is associated with dedicated functions such as packaging the genome, gene regulation, DNA repair, and meiotic recombination (Talbert and Henikoff 2010). Both the N- and C-terminal tails of core histones are subjected to various types of post-translational modifications including acetylation, methylation, SUMOylation, phosphorylation, ubiquitination, ADP-ribosylation, and biotinylation.
Histone demethylases have been linked to a wide range of human carcinomas (Pedersen and Helin 2010). Dynamic histone methylation patterns influence DNA double-strand break (DSB) formation and DNA repair, meiotic crossover events, and transcription levels (Zhang and Reinberg 2001; Clément and de Massy 2017). However, the mechanisms by which histone-modifying enzymes coordinate their efforts to signal for the desired outcome are not well understood, and even less is known about the role of histone demethylases in promoting germline maintenance.
The mammalian histone demethylase LSD1 was originally discovered as a di- and monomethylated histone H3 lysine 4 (H3K4me2/1)-specific demethylase (Shi et al. 2004). Studies in flies and fission yeast revealed increased sterility in the absence of LSD1; however, the underlying mechanism of function by which LSD1 promotes fertility remained elusive (Di Stefano et al. 2007; Lan et al. 2007; Rudolph et al. 2007). Caenorhabditis elegans studies suggested that it plays a role in meiosis, and LSD1/CeSPR-5 mutant analysis revealed a progressive sterility accompanied by a progressive accumulation of H3K4me2 on a subset of genes, including spermatogenesis genes (Katz et al. 2009). In addition to transgenerational sterility, our previous studies discovered that this histone demethylase is important for DSB repair (DSBR) as well as p53-dependent germ cell apoptosis in the C. elegans germline (Nottke et al. 2011), linking H3K4me2 modulation via SPR-5 to proper repair of meiotic DSBs for the first time. Other studies supporting the importance of histone methylation in the regulation of DNA damage repair have attracted more attention to the field in recent years (Huang et al. 2007; Katz et al. 2009; Black et al. 2010; Mosammaparast et al. 2013; Peng et al. 2015). However, the mechanisms by which histone demethylation is involved in DNA damage repair remain unclear and only a few studies have been focused on its roles in germline maintenance.
A growing body of work supports a role for components from the Fanconi anemia (FA) pathway in response to DNA replication-fork arrest and interstrand cross-link (ICL) repair (Adamo et al. 2010; Schlacher et al. 2012; Raghunandan et al. 2015; Lachaud et al. 2016). FANCM guides the FA core complex to DNA lesions and displays a strong preference for binding branched DNA structures, such as replication forks, in vitro (Gari et al. 2008). The FA core complex monoubiquitinates a heterodimer of FANCD2/FANCI at ICL-induced stalled replication forks, which in turn recruits and activates downstream FA proteins and participates with BRCA1 and RAD51 in repair during S phase (Taniguchi et al. 2002; Xue et al. 2008). C. elegans FA proteins—FNCM-1, FCD-2, and FNCI-1—are required for ICL repair (Lee et al. 2010), however their function remains to be investigated for DSBR.
Here, we show that the histone demethylase LSD1/CeSPR-5 interacts with the FA FANCM/CeFNCM-1 protein using biochemical, cytological, and genetic analyses. LSD1/CeSPR-5 is required for hydroxyurea (HU)-induced S phase DNA damage checkpoint activation, and its absence suppresses the embryonic lethality and larval arrest displayed in fncm-1 mutants. We show that FANCM/CeFNCM-1 relocalizes upon HU exposure and colocalizes with FANCD2/CeFCD-2 and LSD1/CeSPR-5. We also show that the potential helicase/translocase domain of FANCM/CeFNCM-1 is necessary for recruiting FANCD2/CeFCD-2 to the site of replication arrest. Surprisingly, the FA pathway is required for H3K4me2 maintenance regardless of the presence of replication stress. Our study reveals a link between FA and epigenetic maintenance, therefore providing new insights into the functions of the FA pathway and the regulation of histone methylation in DNA repair.
Materials and Methods
Strains and alleles
C. elegans strains were cultured at 20° under standard conditions as described in Brenner (1974). The N2 Bristol strain was used as the wild-type background. The following mutations and chromosome rearrangements were used: linkage group I (LGI): fncm-1(tm3148), spr-5(by101), and hT2[bli-4(e937) let-?(q782) qIs48] (I; III); LGIV: spo-11(ok79), nT1 [unc-?(n754) let-?(m435)] (IV; V), fcd-2 (tm1298), and opIs406 [fan-1p::fan-1::GFP::let-858 3′UTR + unc-119(+)] (Kratz et al. 2010).
Transgenic animals
The following set of transgenic worms was generated with CRISPR-Cas9 technology as described in Kim and Colaiácovo (2014, 2016) and Norris et al. (2015). In brief, the conserved potential helicase motifs were mutated in FNCM-1 animals (fncm-1(rj43[S154Q]) and fncm-1(rj44[M247N E248Q K250D])) as described in Kim and Colaiácovo (2014, 2015b, 2016). The FNCM-1-tagged animal (rj45[fncm-1::GFP::3xFLAG]) was created with a few modifications of the CRISPR-Cas toolkit as described in Norris et al. (2015). The SPR-5-tagged animal (rj18[spr-5::GFP::HA + loxP unc-119(+) loxP] I; unc-119(ed3) III) was generated as described in Dickinson et al. (2013). All transgenic lines were outcrossed with wild type between four and six times.
Analysis of FNCM-1 protein conservation and motifs
FNCM-1 homology searches and alignments were performed using T-COFFEE (http://tcoffee.crg.cat/) (Di Tommaso et al. 2011). Pfam and Prosite (release 20.70) were used for zinc-finger motif predictions (Sonnhammer et al. 1997).
Plasmids
Single-guide RNAs (sgRNAs) targeting fncm-1 were created as described in Norris et al. (2015) and Kim and Colaiácovo (2016). In brief, the top and bottom strands of the sgRNA-targeting oligonucleotides (5 μl of 200 μM each) were mixed and annealed to generate double-stranded DNA which then replaced the BamHI and NotI fragment in an empty sgRNA expression vector (pHKMC1, #67720; Addgene) using Gibson assembly (Norris et al. 2015; Kim and Colaiácovo 2016).
To build the fncm-1::GFP::FLAG donor plasmid, genomic DNA containing up- and downstream homology arms of ∼1 kb were PCR amplified and cloned into the multi-cloning site of the pUC18 plasmid along with GFP and FLAG tags synthesized by Integrated DNA Technologies (IDT). To build the spr-5::GFP::HA donor vector, spr-5 genomic DNA containing up- and downstream ∼1-kb homology arms together with GFP::HA + loxP unc-119(+) loxP were cloned into the ZeroBlunt Topo vector as described in Dickinson et al. (2013).
DNA micro-injection
Plasmid DNA was micro-injected into the germline as described in Friedland et al. (2013), Tzur et al. (2013), and Kim and Colaiácovo (2016). Injection solutions were prepared to contain 5 ng/µl of pCFJ90 (Pmyo-2::mCherry; Addgene), which was used as the co-injection marker; 50–100 ng/µl of the sgRNA vector; 50 ng/µl of the Peft-3Cas9-SV40 NLStbb-2 3′UTR; and 50–100 ng/µl of the donor vector.
Monitoring S-phase progression in the germline
Nuclei in the C. elegans germline are positioned in a temporal-spatial manner and both mitotic as well as meiotic S-phase progression can be monitored at the distal tip (Jaramillo-Lambert et al. 2007). To monitor S-phase progression in the germline, ∼200 pmol/µl Cyanine 3-dUTP (ENZO Cy3-dUTP) was injected into the distal tip of the gonad of 20- to 24-hr post-L4 worms. Worms were dissected and immunostained 2.5 hr after injection.
DNA damage sensitivity experiments
Young adult homozygous fncm-1 animals were picked from the progeny of fncm-1/hT2 parent animals. To assess for ionizing irradiation (IR) sensitivity, animals were treated with 0 and 50 Gy of γ-IR from a 137Cs source at a dose rate of 1.8 Gy/min. HU sensitivity was assessed by placing animals on seeded NGM plates containing 0, 3.5, and 5.5 mM HU for 12–16 hr. For ICL sensitivity, animals were treated with 0 and 25 µg/ml of Trioxsalen (trimethylpsoralen; Sigma Chemical, St. Louis, MO) in M9 buffer with slow agitation in the dark for 30 min. Worms were exposed to 200 J/m2 of UVA. For all embryonic hatching assays, >36 animals were plated, 6 per plate, and hatching was monitored 60–72 hr after treatment as a readout of mitotic effects given how long it takes to proceed from the premeiotic region to egg laying (Jaramillo-Lambert et al. 2010; Kim and Colaiácovo 2015a,b).
For larval arrest assays, L1 worms were plated on NGM plates with either 0 or 5.5 mM HU and incubated for 12–16 hr. The number of hatched worms and live adults were counted. Each damage condition was replicated at least twice in independent experiments as described in Kim and Colaiácovo (2015a).
Immunofluorescence and Western blot analysis
Whole mount preparations of dissected gonads, fixation, and immunostaining procedures were carried out as described in Colaiácovo et al. (2003). Primary antibodies were used at the following dilutions: rabbit anti-SPR-5 (sc-98749, 1:500; Santa Cruz), rabbit anti-SPR-5 (1:1000 for Western blot; Nottke et al. 2011), rabbit anti-RAD-51 (1:20,000; SDI), rat anti-FCD-2 (1:300; Lee et al. 2010), rat anti-RPA-1 (1:200; Lee et al. 2010), rabbit anti-pCHK-1 (sc17922, 1:50; Santa Cruz), chicken anti-GFP (ab13970, 1:400; Abcam), and mouse anti-H3K4me2 (CMA303, 1:200; Millipore, Bedford, MA). Secondary antibodies used were: Cy3 anti-rabbit, FITC anti-rabbit, Cy3 anti-rat, Alexa 488 anti-chicken, and FITC anti-mouse (all from Jackson Immunochemicals), each at 1:250. Immunofluorescence images were collected at 0.2-μm intervals with an IX-70 Microscope (Olympus) and a CoolSNAP HQ CCD Camera (Roper Scientific) controlled by the DeltaVision system (Applied Precision). Images were subjected to deconvolution by using the SoftWoRx 3.3.6 software (Applied Precision).
For Western blot analysis, age-matched 24-hr post-L4 young adult worms were washed off of plates with M9 buffer. SDS buffer (6×) was added to the worm pellets which were then flash frozen in liquid nitrogen and boiled before equal amounts of samples were loaded on gels for SDS-PAGE separation.
Colocalization analysis
The colocalization tool in SoftWoRx from Applied Precision was employed for colocalization analysis (Adler and Parmryd 2010).
Mass spectrometry analysis
Pellets of age-matched 24-hr post-L4 young adult worms (wild type or spr-5::GFP::HA) were flash frozen in lysis buffer (50 mM HEPES, pH 7.4, 1 mM EGTA, 3 mM MgCl2, 300 mM KCl, 10% glycerol, 1% NP-40) with protease inhibitors (11836153001; Roche) using liquid nitrogen. They were then ground to a fine powder with a mortar and pestle. Lysis buffer was added to the thawed worms and samples were sonicated for 30 cycles of 20 sec each. The soluble fraction of the lysate was applied to a 0.45-µm filter and applied to either anti-HA beads (E6779; Sigma Chemical) or GFP-Trap (gta-20; Chromotek) that were incubated at 4° overnight. After three washes with lysis buffer lacking NP-40, the bound proteins were eluted with either 1 mg/ml HA peptide (I2149; Sigma Chemical) or 0.1 M glycine and precipitated using the Proteo Extract Protein Precipitation Kit (539180; Calbiochem, San Diego, CA). The dry pellet was submitted to the Taplin Mass Spectrometry Facility (Harvard Medical School) for analysis. The wild-type sample was used as a negative control to remove false positive hits.
Co-immunoprecipitation
Co-immunoprecipitations (co-IPs) were performed with worm lysates from FNCM-1-tagged animals (rj45[fncm-1::GFP::3xFLAG]). Lysis buffer was added to the worm lysates and they were sonicated for 30 cycles of 20 sec each. The soluble fraction of the lysates was applied to anti-flag M2 magnetic beads (Sigma Chemical) that were incubated at 4° overnight. Interacting proteins were eluted with glycine buffer (pH 2). Eluates were used for Western blot analysis to confirm the interaction of SPR-5 and FNCM-1 proteins.
Statistical analysis
Statistical comparisons between mutants and control worms were carried out using the two-tailed Mann–Whitney U-test with a 95% confidence interval. A significance value of P < 0.05 was used.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results
Mass spectrometry and co-IP analyses reveal that SPR-5 interacts with FNCM-1
The histone demethylase SPR-5 in C. elegans as well as its orthologs in humans have been reported to function in DSBR (Huang et al. 2007; Katz et al. 2009; Black et al. 2010; Nottke et al. 2011; Mosammaparast et al. 2013; Peng et al. 2015). To better understand the roles played by SPR-5 in DNA damage repair throughout the germline, we applied a proteomic approach to search for its interacting partners. Specifically, we performed pull-downs with a CRISPR-Cas9-engineered transgenic line expressing the endogenous SPR-5 tagged with GFP and HA (spr-5::GFP::HA), which did not display either the embryonic lethality or DSB sensitivity observed in spr-5 null mutants (Supplemental Material, Figure S1), followed by liquid chromatography–mass spectrometry (LC-MS) analysis. The FA FANCM homolog in C. elegans, FNCM-1, was identified in two independent samples using this strategy, each processed with α-HA and α-GFP antibodies (Table 1). The SPR-5 and FNCM-1 interaction was not detected in control worms with untagged SPR-5 (Table 1), suggesting that SPR-5’s interaction with FANCM/FNCM-1 is specific. Proteins previously shown to interact with SPR-5, such as SPR-1 (the ortholog of human corepressor CoREST) and RCOR-1 [an ortholog of human REST corepressors 2 and 3 (RCOR2 and RCOR3)] (Jarriault and Greenwald 2002; Lee et al. 2008), were also identified, indicating that the pull-down followed by LC-MS worked efficiently.
Table 1. SPR-5-interacting proteins identified by LC-MS analysis.
| Protein name | SPR-5::GFP::HA | Control |
|---|---|---|
| RCOR-1 | 45 | Not detected |
| SPR-1 | 18 | Not detected |
| SPR-5 | 268 | Not detected |
| FNCM-1 | 3 | Not detected |
Immunoprecipitations from spr-5::GFP::HA and wild-type (N2) whole-worm extracts with antibodies against either HA or GFP were analyzed by LC-MS. The FA FANCM homolog in C. elegans, FNCM-1, was identified with both anti-HA and anti-GFP antibodies. The wild-type (N2) extract was used as a negative control to remove false positive hits. SPR-5-interacting proteins that were identified in both anti-HA and anti-GFP pull-downs are listed. Numbers indicate the total mass spectra collected from two samples.
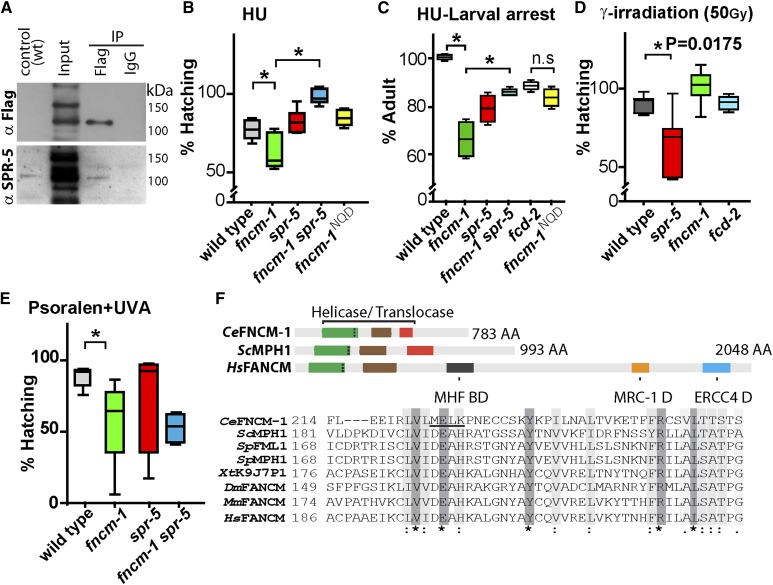
To further validate the interaction between SPR-5 and FNCM-1, we used a functional CRISPR-Cas9-engineered transgenic line expressing endogenous FNCM-1 tagged with GFP and FLAG (fncm-1::GFP::FLAG; Figure S2B) in co-IP experiments. We detected SPR-5 in pull-downs done from fncm-1::GFP::FLAG worm lysates with an α-FLAG antibody, further supporting an SPR-5 and FNCM-1 interaction in vivo (Figure 1A).
Figure 1.
FANCM/CeFNCM-1 interacts with histone demethylase LSD1/CeSPR-5 and fncm-1 mutants display HU-induced replication-stress sensitivity that is suppressed in spr-5 mutants. (A) Western blots showing co-IP of FNCM-1 and SPR-5 from fncm-1::GFP::FLAG transgenic whole worm lysates with anti-FLAG and anti-SPR-5 antibodies, respectively. Input represents a concentrated whole-worm lysate sample prepared for co-IP. A wild-type (wt; N2) worm lysate is shown as a control for anti-SPR-5 and anti-FLAG antibodies. IgG is used as a control for the IP. (B and C) Relative percentage of hatching and larval arrest for the indicated genotypes after treatment with 3.5 and 5.5 mM HU, respectively. Relative values are calculated against the absence of treatment. (D) fncm-1 and fcd-2 are not hypersensitive to γ-IR. fncm-1 and fcd-2 mutants did not exhibit a decrease in embryonic viability (shown as % hatching) compared to wild type following exogenous DSB formation by γ-IR exposure (P = 0.0530, 100% hatching for fncm-1 and P = 0.8357, 90% hatching for fcd-2). (E) Relative percentage of hatching for the indicated genotypes after treatment with 0 and 25 µg/ml trimethylpsoralen-UVA. P-values were calculated by the two-tailed Mann–Whitney test, 95% C.I. More than 36 animals were plated for assays (B–E). (F) Representation of the helicase/translocase amino acid sequence conservation of C. elegans FNCM-1 and its homologs in Homo sapiens, Mus musculus, Drosophila melanogaster, Xenopus tropicalis, Schizosaccharomyces pombe, and Saccharomyces cerevisiae. Alignment was performed using T-COFFEE and Pfam (http://xfam.org/). Dark gray boxes (*) indicate amino acid identity and light gray boxes (:) indicate similarity. Three vertical dots inside the green boxes indicate the position of the represented amino acid sequence. The location of the MEK to NQD mutation in the C. elegans sequence is underlined (MELK).
SPR-5 and FNCM-1 cooperate upon DNA replication-fork arrest
Since our analysis supports the interaction of SPR-5 with FANCM/FNCM-1 and we previously demonstrated that SPR-5 is required for DSBR (Nottke et al. 2011), we set out to gain insight into the link between SPR-5 and the FA pathway during DNA repair. To this end, we examined the sensitivity of fncm-1 and spr-5 null mutants to different types of DNA damage (Lee et al. 2010; Nottke et al. 2011). Given that spr-5 mutants exhibit progressive deterioration of germline functions, which is associated with increased H3K4me2 (Nottke et al. 2011), we only used early generation (F1–F5) spr-5 mutants throughout the experiments in this study. First, we found that fncm-1 mutants displayed sensitivity to HU treatment, which results in replication arrest (Figure 1, B and C). Specifically, only 61% of embryos hatched in fncm-1 mutants compared to 75% for wild type (P = 0.0367 by the two-tailed Mann–Whitney U-test, 95% C.I.) following an exposure to 3.5 mM HU. Moreover, the HU sensitivity observed in fncm-1 mutants was suppressed in fncm-1spr-5 double mutants (P = 0.0006), while spr-5 single mutants did not exhibit any sensitivity compared to wild type (P = 0.1120). Similarly, the increased larval arrest observed in fncm-1 mutants following HU treatment was also suppressed in fncm-1spr-5 double mutants (Figure 1C). Taken together, these observations suggest that FNCM-1 and SPR-5 play a role in DNA repair following collapse of stalled replication forks.
Next we examined the DNA damage sensitivity of spr-5 and FA pathway mutants to exogenous DSBs generated by γ-IR. A significant reduction in the levels of hatched embryos was observed in spr-5 null mutants compared to wild-type animals (61 and 89%, respectively, at a dose of 50 Gy; P = 0.0175 by the two-tailed Mann–Whitney U-test, 95% C.I.; Figure 1D). However, both fncm-1 and fcd-2 null mutants, which lack the FANCD2 homolog in worms, were not sensitive to exogenous DSBs (100 and 90% hatching, respectively) suggesting that the FA pathway is not involved in DSB repair.
Analysis of the sensitivity to DNA ICLs revealed that spr-5 mutants were not sensitive to ICLs induced by psoralen-UVA in germline nuclei (Figure 1E). Specifically, 75% of embryos laid by spr-5 mutants hatched, compared to 94% in wild type (P = 0.2307 by the two-tailed Mann–Whitney U-test, 95% C.I.). However, fncm-1 mutants exhibited significant sensitivity with only 55% hatching (P = 0.0087), which is expected given that FNCM-1 is required for ICL repair (Collis et al. 2006). fncm-1spr-5 double mutants did not alter the sensitivity observed in fncm-1 single mutants (57%, P = 0.9176), indicating that SPR-5 does not play a role in ICL repair in germline nuclei (Figure 1E). Altogether, these observations suggest that the FA pathway may not be involved in DSBR in conjunction with SPR-5 and that SPR-5 does not participate in ICL repair along with the FA pathway, but that instead their interaction is necessary upon DNA replication-fork arrest.
A potential helicase/translocase domain in FNCM-1 is important for somatic repair
The FANCM C. elegans homolog, FNCM-1, contains well-conserved helicase/translocase domains which are also present from budding yeast to humans (Figure 1F). We generated a helicase/translocase dead mutant by CRISPR-Cas9 engineering based on helicase/translocase dead mutants produced in the MPH1 gene in yeast (Scheller et al. 2000). The mutant contains the following amino acid changes: M to N, E to Q, and K to D at positions 247, 248, and 250. Interestingly, the fncm-1NQD mutant exhibited larval arrest levels similar to that observed in fcd-2 null mutants (P = 0.0247; 84% adults for NQD and 100% for wild type, values are normalized against untreated controls; Figure 1, C and F), suggesting that the potential helicase/translocase domain (M247E248K250) is important for somatic repair. However, this helicase/translocase domain is not necessary for DNA repair upon replication-fork arrest in the germline (P = 0.0017 and P = 0.7206, compared to fncm-1 and wild type, respectively; Figure 1B).
FNCM-1 promotes replication-fork progression and SPR-5 is required for the formation of single-stranded DNA regions induced by FNCM-1 deficiency
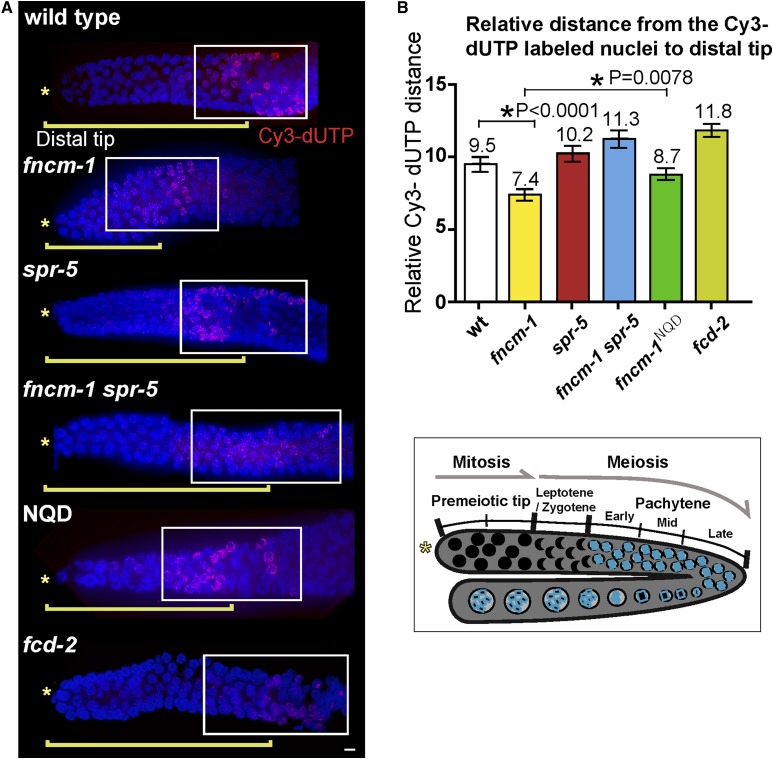
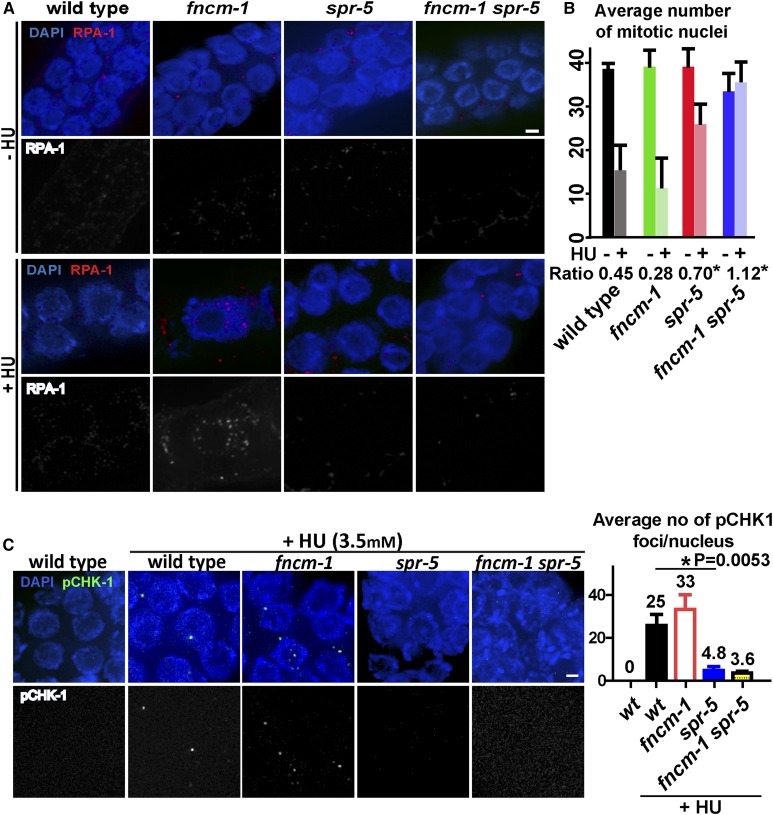
Since FANCM has been implicated in promoting S-phase progression (Whitby 2010), we hypothesized that FNCM-1 might have a similar role. To address FNCM-1’s potential role in S-phase progression, we monitored the incorporation of a fluorescent nucleotide during S phase by injecting Cyanine-3-dUTP into the C. elegans gonad. Although we did not observe overt differences in the overall length of the gonads in the mutants compared to wild type, we accounted for this possibility by assessing the relative distance of Cy3-labeled nuclei. We divided the distance of Cy3-labeled nuclei from the distal tip by the length of the specific gonad from distal tip to late pachytene. The relative distance between the Cy3-labeled nuclei and the distal tip was reduced significantly in fncm-1 mutant germlines compared to wild type, suggesting a slowdown in the rate of S-phase progression in the fncm-1 mutants (relative distance of 7.4 for fncm-1 and 9.5 for wild type, P < 0.0001; Figure 2). Consistent with the HU-sensitivity assay, the slowdown in S-phase progression observed in fncm-1 single mutants was suppressed in fncm-1spr-5 double mutants (P < 0.0001). Furthermore, fncm-1NQD mutants also displayed a slowdown in S-phase progression, albeit not as severe as that observed for fncm-1 null mutants; suggesting that the fncm-1NQD mutant is likely a hypomorphic allele (7.4 for fncm-1 and 8.7 for fncm-1NQD, P = 0.0078; relative distance of 9.5 for wild type and 8.7 for fncm-1NQD, P = 0.3165; Figure 2). To further validate the Cy3-labeling results, we examined the formation of single-stranded DNA regions as a result of replication blockage by assessing the presence of RPA-1 signal, which localizes to single-stranded DNA. RPA-1 signal was detected following treatment with 3.5 mM HU in fncm-1 mutants but not in either wild type or spr-5 null mutants (Figure 3A). Moreover, the RPA-1 signal observed in fncm-1 mutants was suppressed in the fncm-1spr-5 double mutants. Taken together, these observations suggest that FNCM-1 is required for replication-fork progression upon DNA damage and that SPR-5 may be involved in the formation of the single-stranded DNA regions induced upon absence of FNCM-1 function.
Figure 2.
FNCM-1 is required for S-phase progression, and impaired S-phase progression in fncm-1 mutants is suppressed by lack of SPR-5. (A) Cyanine-3-dUTP was injected into C. elegans gonads to monitor S-phase progression. The distance between the Cy3-labeled nuclei and the distal tip (*) was measured for at least four gonads for each of the indicated genotypes. Bar, 2 µm. (B) Top: Quantitation of the relative distance between Cy3-labeled nuclei and the distal tip in the germlines of the indicated genotypes. To account for potential variations in gonad size, the distance of Cy-3-labeled nuclei from the distal tip is divided by the length of the specific gonad from distal tip to late pachytene. Relative distance of Cy3-labeled nuclei = the distance of Cy3-labeled nuclei from the distal tip/the length of the specific gonad from distal tip to late pachytene × 100. At least four gonads were scored for each. n = 4–6 gonads. P-values calculated by the two-tailed Mann–Whitney test, 95% C.I. Bottom: Diagram of the C. elegans germline indicating the mitotic (premeiotic tip) and meiotic stages represented in (A). wt, wild type; *, distal tip.
Figure 3.
FNCM-1 promotes replication-fork progression and SPR-5 is required for the S-phase checkpoint sensing the single-stranded DNA region formed upon lack of FNCM-1. (A) Immunolocalization of single-stranded DNA binding protein RPA-1 upon 3.5 mM HU treatment in the PMT for the indicated genotypes. Bar, 2 µm. (B) Quantitation of the average number of mitotic nuclei within 40 µm in the PMT region of the germlines from the indicated genotypes. Ratio represents the number of nuclei observed following HU treatment (+HU) divided by the number observed without treatment (−HU). * indicates statistical significance compared to wild-type control. P = 0.0422 for spr-5, P = 0.0095 for fncm-1 spr-5. P-values calculated by the two-tailed Mann–Whitney U-test, 95% C.I. (C) S-phase DNA damage checkpoint activation is impaired in spr-5 single and fncm-1 spr-5 double mutants. Left: Immunostaining for pCHK-1 on germline nuclei at the PMT following 3.5 mM HU treatment. Bar, 2 µm. Right: Quantitation of pCHK-1 foci. n = 4–6 gonads. P-values calculated by the two-tailed Mann–Whitney U-test, 95% C.I.
S-phase DNA damage checkpoint activation is dependent on SPR-5
The DNA replication-dependent S-phase checkpoint is activated upon stress—such as HU treatment, DNA damage, and the presence of abnormal DNA structures—and results in S-phase arrest, which is characterized by a premeiotic tip (PMT) exhibiting enlarged nuclear diameters as well as a reduced number of nuclei in the C. elegans germline (Bartek et al. 2004; Garcia-Muse and Boulton 2005; Kim and Colaiácovo 2014). Since the spr-5 null mutation suppressed the single-stranded DNA formed in fncm-1, we examined whether SPR-5 is required for the activation of the S-phase DNA damage checkpoint.
The ratio of mitotic nuclei (+HU:−HU) was not significantly changed in fncm-1 mutants compared to wild type, suggesting that the S-phase checkpoint is intact (0.45 and 0.28, respectively; Figure 3B). However, a significant increase in the number of nuclei was observed in both spr-5 single (0.70) and fncm-1spr-5 (1.126) double mutants compared to wild type (P = 0.0422 and P = 0.0095, respectively), indicating that SPR-5 is required for the S-phase DNA damage checkpoint and that lack of SPR-5, which causes accumulation of active chromatin (Katz et al. 2009; Nottke et al. 2011), circumvents proper activation of the S-phase checkpoint.
Single-stranded DNA formed at a stalled replication fork is recognized by RPA and this triggers ATR kinase activation, which results in S phase-checkpoint activation by phosphorylating its downstream target checkpoint kinase 1 (Chk-1) (Cimprich and Cortez 2008). Consistent with our observations of an impaired S-phase checkpoint, such as the increased number of mitotic germline nuclei as well as suppressed detection of single-stranded DNA, we detected a decrease in the levels of phosphorylated CHK-1 (pCHK-1) in these nuclei in spr-5 mutants compared to wild type upon 3.5 mM HU treatment (P = 0.0053; Figure 3C). Altogether, these data indicate that SPR-5 is required for S-phase DNA damage checkpoint activation.
The localization of SPR-5 and the FA pathway components FCD-2, FAN-1, and FNCM-1 is altered in response to replication stress
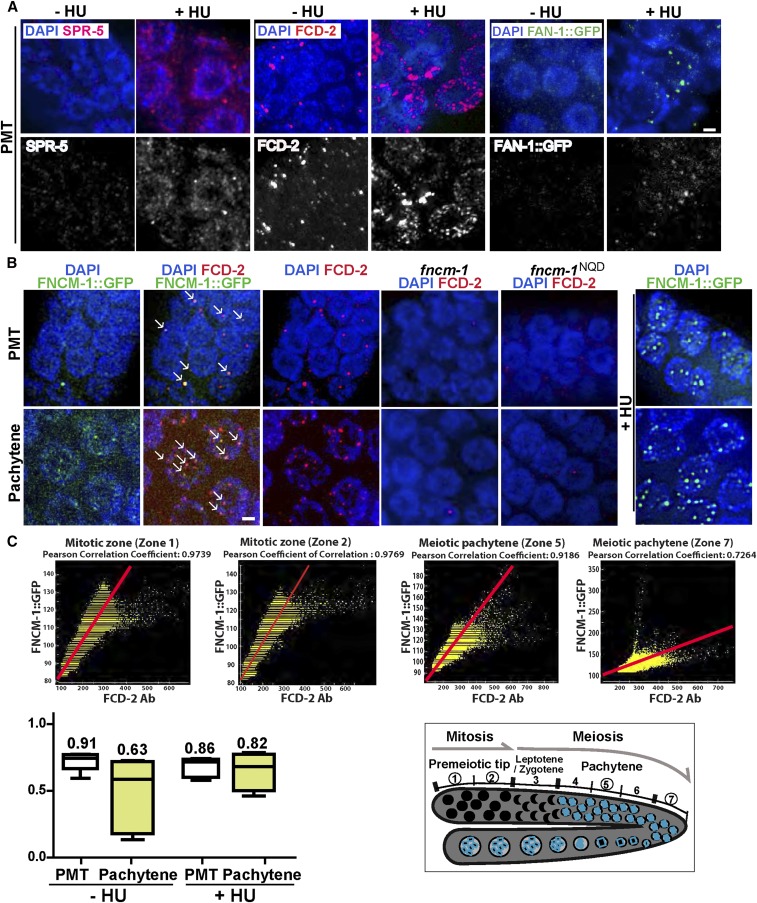
Since our analysis links the functioning of SPR-5 with the FA pathway via FNCM-1 at stalled replication forks, we examined the localization of SPR-5 and factors acting in the FA pathway by immunostaining. Consistent with our previous observation, SPR-5 shows a nuclear-associated pattern (Nottke et al. 2011). Interestingly, upon HU treatment, we observed an increase in both peri-chromosomal SPR-5 signal as well as bright foci on chromatin compared to untreated (−HU) control wild type (Figure 4A), suggesting a role for the histone demethylase at replication-fork arrest during S phase. We also observed a brighter and elevated number of FANCD2/FCD-2 as well as FAN1/FAN-1 chromatin-associated foci following HU treatment, which supports the function of the C. elegans FA pathway at stalled DNA replication forks, analogous to recent reports in other species (Figure 4A) (Lachaud et al. 2016; Michl et al. 2016). FNCM-1::GFP::FLAG signal was observed as a combination of foci associated with the DAPI-stained chromosomes as well as a diffuse haze throughout the germline, which was not detected in the control wild type (Figure 4B and Figure S2). FNCM-1::GFP::FLAG partly colocalized with FCD-2 in the absence of any stress (−HU). However, its localization was altered upon replication-fork arrest (+HU), as shown by the reduction of the diffuse germline signal and increase in bright chromatin-associated foci; suggesting that FNCM-1 responds to replication stress similar to FCD-2 and FAN-1, consistent with reports in other species (Figure 4B) (Xue et al. 2008). Furthermore, we observed a higher level of colocalization between FNCM-1 and FCD-2 in the mitotically dividing nuclei at the PMT and a reduction in the level of colocalization at the pachytene stage, which supports the role of FNCM-1 and FCD-2 in replication-fork arrest at the mitotic stage (Figure 4C).
Figure 4.
Proteins in the FA pathway and the histone demethylase SPR-5 display a dynamic localization upon HU treatment and colocalize. (A) Immunolocalization of SPR-5, FCD-2, and FAN-1::GFP (FAN-1::GFP was detected with an anti-GFP antibody) upon 3.5 mM HU treatment in the mitotically dividing germline nuclei (PMT). The localization pattern of SPR-5 and of the FA pathway components FCD-2 and FAN-1 is altered in response to replication stress (+HU). (B) Immunolocalization of FNCM-1::GFP and FCD-2 in the PMT. FNCM-1 and FCD-2 colocalize on chromatin-associated foci (indicated by →) in the absence of any stress (−HU; left panels). Panel on the right shows that FNCM-1 relocalizes in response to replication stress (+HU), changing from a more diffuse to a more focal localization. The dispersed FNCM-1::GFP signal was not detected in control wild type (Figure S2). Bars, 2 µm. (C) Top: Graphs showing Pearson colocalization correlation coefficient values indicate higher colocalization levels between FCD-2 and FNCM-1::GFP starting at the PMT and slowly decreasing throughout meiosis (zones 1 and 2 = mitotic zone; zone 5 = midpachytene; zone 7 = late pachytene). Bottom left: Mean numbers of Pearson colocalization correlation coefficient values between FCD-2 and FNCM-1::GFP for both mitotic (PMT) and meiotic (pachytene) zones with or without HU exposure. n > 5 gonads. A value of 1 indicates that the patterns are perfectly similar, every pixel that contains Cy3 (FCD-2, red) also contains GFP (FNCM-1::GFP, green); while a value of −1 would mean that the patterns are perfectly opposite, every pixel that contains Cy3 does not contain GFP and vice versa. Bottom right: Diagram of the C. elegans germline indicating the mitotic (zones 1 and 2) and meiotic stages (zones 5 and 7) represented in the top panel.
While FNCM-1 is known to be required for FCD-2 localization (Figure 4B) (Collis et al. 2006), analysis of our helicase-dead fncm-1NQD mutant revealed a lack of FCD-2 localization, suggesting that the helicase/translocase domain is required for recruiting FCD-2 (Figure 4B). This is further supported by the observation that both fcd-2 null and fncm-1NQD mutants displayed similar levels of larval arrest (Figure 1C), potentially due to the lack of FCD-2 localization in fncm-1NQD mutants mimicking fcd-2 mutants. Taken together, these data support the idea that FNCM-1 responds to replication-fork arrest and recruits the downstream players FCD-2 and FAN-1, consistent with previous reports from other species. Also, we show for the first time that the helicase/translocase domain of FNCM-1 is necessary for recruiting FCD-2.
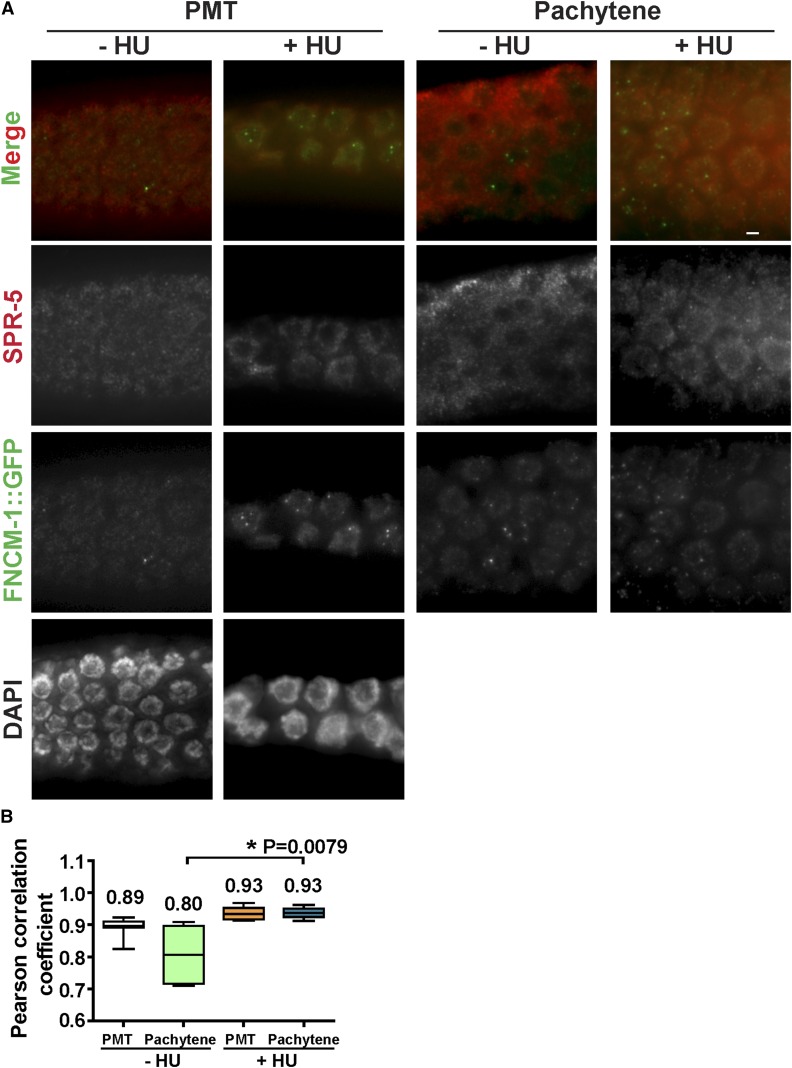
SPR-5 colocalizes with FNCM-1
Both SPR-5 and FNCM-1 localize from the PMT (mitotic zone) to pachytene (Figure 5A). We investigated whether SPR-5 and FNCM-1 colocalize on germline nuclei. However, since SPR-5 exhibits a dispersed localization, not limited to distinct foci, it is not possible to assess the colocalization of SPR-5 and FNCM-1 by scoring levels of superimposed foci. To circumvent this issue, we applied a Pearson correlation coefficient method (Adler and Parmryd 2010). Consistent with their interaction by co-IP and LC-MS analysis, we found a high level of colocalization for FNCM-1 and SPR-5 (Figure 5B). The average Pearson correlation coefficient was 0.89 at the PMT and 0.80 at the pachytene stage in the germline. Interestingly, upon replication arrest following HU treatment, we found a high level of colocalization between SPR-5 and FNCM-1 persisting from the PMT to the pachytene stage, unlike in the control where this was progressively reduced. These observations support the idea that cooperation between the H3K4me2 histone demethylase and the FA pathway is reinforced to deal with replication-fork blockage.
Figure 5.
SPR-5 and FNCM-1 colocalization is extended upon replication-fork stalling. (A) Immunostaining of SPR-5 and FNCM-1::GFP (endogenous signal) in nuclei at either the PMT or at pachytene in the presence or the absence of 3.5 mM HU treatment. Bar, 2 µm. (B) Quantitation of Pearson colocalization correlation coefficient observed in (A) indicates that colocalization between SPR-5 and FNCM-1::GFP extends into the pachytene stage upon replication-fork stalling. A value of 1 indicates a perfect positive linear relationship between variables. P = 0.0079 for − and +HU treatment in the pachytene stage. n = 4–6 gonads. P-values calculated by the two-tailed Mann–Whitney U-test, 95% C.I.
Two-way interaction of FANCM/FNCM-1 and LSD1/SPR-5: FNCM-1 and FCD-2 are necessary for maintaining proper H3K4 dimethylation levels
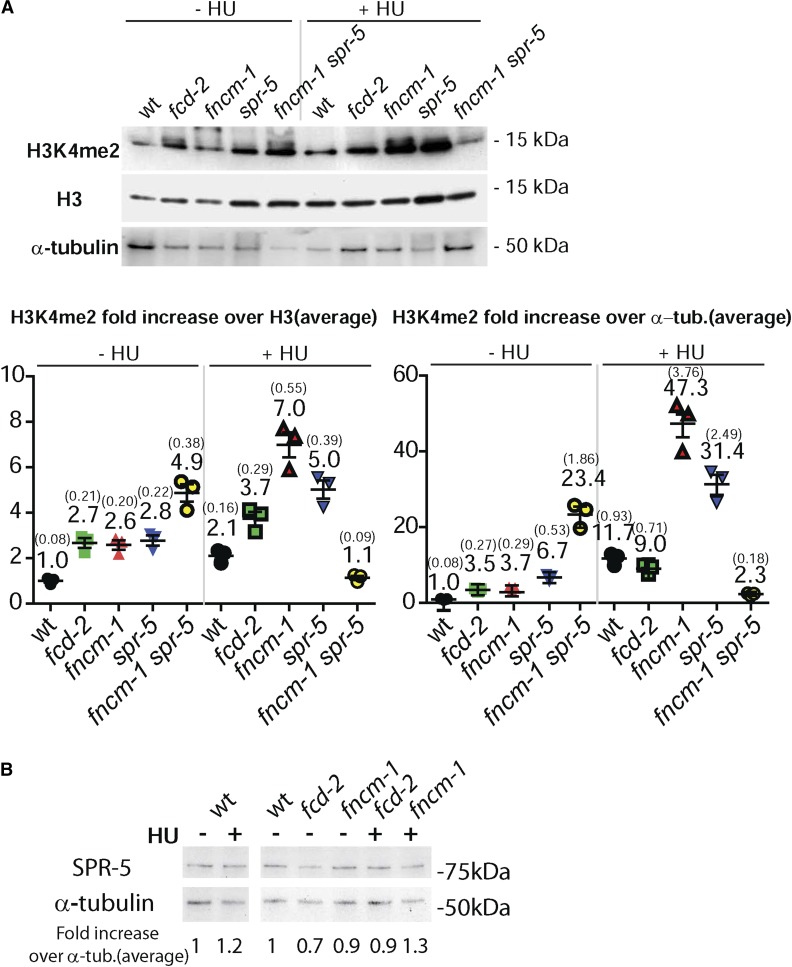
Since lack of LSD1/SPR-5 suppresses the HU sensitivity observed in fncm-1 mutants, we next examined whether H3K4me2, which is regulated by the SPR-5 histone demethylase (Katz et al. 2009; Nottke et al. 2011), was altered by the lack of FNCM-1. Surprisingly, we observed an increase in the levels of H3K4me2 in mutants lacking fncm-1, suggesting a bidirectional functional interaction between SPR-5 and FNCM-1 (Figure 6A, numbers represent mean data from three independent experiments). Furthermore, H3K4me2 levels are also increased in fcd-2 mutants, indicating that not only FANCM/FNCM-1 but also the FA pathway is necessary for maintaining histone demethylation together with LSD1/SPR-5. However, we cannot rule out the possibility that the increase in H3K4me2 levels could be an indirect consequence resulting, for example, from alterations to cell-cycle progression in the mutants.
Figure 6.
FNCM-1 and FCD-2 are necessary for maintaining H3K4 dimethylation levels. (A) Top: Western blot analysis comparing H3K4me2 levels with histone H3 and α-tubulin antibodies for the indicated genotypes either in the absence or presence of HU (3.5 mM). Bottom: Quantitation of H3K4me2 levels normalized against either histone H3 or α-tubulin. Signal intensity was measured with GelQuant.NET. Numbers represent average for data from three independent experiments. SEM values are presented in parentheses. (B) Western blot analysis comparing the levels of SPR-5 normalized against α-tubulin for the indicated genotypes upon absence or presence of 3.5 mM HU treatment. α-tub, α-tubulin; wt, wild type.
A previous study reported that human FANCD2 and FANCI are required for histone H3 exchange when cells are saturated with mitomycin C-induced DNA ICLs (Sato et al. 2012). Since defective H3 mobility possibly interferes with the accurate interpretation of H3K4 dimethylation levels, we normalized the H3K4me2 value to α-tubulin in addition to H3. Although we observed changes in the normalized level of H3K4me2, the overall conclusion from this analysis was not altered.
Since we observed an inverse correlation between the FA components and the levels of H3K4me2, we also examined the level of SPR-5 protein expression in fncm-1and fcd-2 mutants in the absence or presence of HU exposure. However, the normalized expression level of SPR-5 against α-tubulin was not altered in wild type with or without HU exposure (Figure 6B). Also, FA mutants did not affect the level of SPR-5 expression, regardless of HU exposure. These observations show that the level of H3K4me2 is not regulated by the level of expression of SPR-5 protein when replication forks stall. Taken together, our data support a two-way functional interaction between SPR-5 and the FA pathway in the germline: (1) in the activation of the S-phase DNA damage checkpoint in response to stalled replication forks, and (2) in the regulation of H3K4me2.
Discussion
Several studies have investigated the connections between epigenetic marks and DNA repair; however, the mechanisms by which epigenetic marks work in DNA repair remained unclear. Here, we show that the histone demethylase LSD1/CeSPR-5 interacts with the FA FANCM/CeFNCM-1 protein by using biochemical, cytological, and genetic analyses. LSD-1/CeSPR-5 is required for activation of the S-phase DNA damage checkpoint. Surprisingly, the FA pathway is required for H3K4me2 maintenance. Although a previous mouse study reported that FANCD2 modulates H3K4me2 at the sex chromosome, their analysis was confined to immunostaining (Alavattam et al. 2016). With biochemical, cytological, and genetic analyses, our study reveals that the FA pathway is necessary for epigenetic maintenance and sheds light on understanding the epigenetic mechanisms underlying FA.
The FA pathway responds to HU-induced replication-fork arrest
The FA pathway has been mainly studied in mitotically dividing cells but not in germline nuclei. In this study, we identified a dynamic localization pattern for FNCM-1, FCD-2, FAN-1, and LSD-1/CeSPR-5 upon replication-fork arrest induced by HU exposure (Figure 4, A and B). In addition, colocalization, supported by an increased colocalization correlation coefficient, and co-IP results suggest that SPR-5 and FNCM-1 work together in response to replication-fork arrest (Figure 1 and Figure 5). Interestingly, spr-5 mutants displayed DSB sensitivity but not the HU-induced replication-fork sensitivity observed in fncm-1 mutants (P = 0.0175 and P = 0.1720, respectively; Figure 1, B and D). However, a mild but significant reduction in larval arrest was observed (P = 0.0285, 100% for wild type and 79% for spr-5; Figure 1C), suggesting a role for SPR-5 in mitotic cell division upon DNA replication stress. The interaction between these two proteins, as well as their altered localization upon HU stress, suggest that SPR-5 and FNCM-1 work together upon replication-fork arrest.
SPR-5 is necessary for activation of the DNA damage checkpoint
The S-phase checkpoint failure observed in spr-5 mutants can be due to an impaired checkpoint signaling pathway per se. Suppression of the formation of a single-stranded DNA region in the fncm-1spr-5 double mutants (Figure 3A) suggests that SPR-5 may function in replication-fork stalling/pause and that being deficient for SPR-5 prevents fork stalling, which then circumvents S-phase checkpoint activation. The defective checkpoint was observed at a lower (3.5 mM) but not at a higher (5.5 mM) dose of HU, suggesting that an alternative/redundant mechanism for S-phase checkpoint activation is triggered under severe replication stress conditions (Figure S3). It is worth noting that a similar role in checkpoint function was proposed in fission yeast for the Lsd1/2 histone demethylases, which are indispensable for replication-fork pause within the ribosomal DNA region (Holmes et al. 2012).
FA components are required for proper H3K4me2 levels regardless of replication-fork arrest
Surprisingly, FNCM-1 and FCD-2 were necessary to maintain proper H3K4me2 levels regardless of replication-fork arrest (Figure 6). Since HU-induced replication arrest accumulates active chromatin marks during S phase, the slowing down of S phase observed in fncm-1 mutants may result in H3K4me2 accumulation (Figure 2A, Figure 3A, and Figure 6A) (Alper et al. 2012). However, this does not explain how FCD-2, which did not alter S-phase progression, is required for H3K4me2 with or without replication stress (Figure 2B and Figure 6A). This suggests that, in addition to promoting the S phase-induced euchromatic state, the FA pathway may have an alternative role in maintaining histone methylation.
Although the FA pathway is connected to the regulation of histone demethylation regardless of the presence of stalled replication, a direct role for the FA pathway in histone demethylation became more evident when fncm-1spr-5 double mutants suppressed H3K4me2 upon HU arrest, unlike either single mutant (Figure 6A). One possible reason for this is that a defective checkpoint in spr-5 somehow gains synergy in fncm-1 mutants. Alternatively, a severe accumulation of dimethylation displayed in the double mutants may trigger/activate other histone demethylases. In fact, the LSD2 ortholog in C. elegans, amx-1, has been reported to be upregulated over fivefold in spr-5 mutants, thus supporting this idea (Katz et al. 2009; Nottke et al. 2011).
The potential helicase domain (MEK) of FNCM-1 is necessary for recruiting FCD-2
Although C. elegans FNCM-1 displayed relatively less conservation of its DExD/H domain compared to other species, its flanking sequences are still well conserved (Figure 1F). Previous studies reported that the helicase domain of budding yeast, Mph1 (an ortholog of human FANCM), was required for mitotic crossover formation (Prakash et al. 2009). Interestingly, mutations in the potential helicase domain (MEK to NQD) of FNCM-1 resulted in loss of FCD-2 localization and a slowdown of S-phase progression (Figure 2). Moreover, it also led to larval arrest upon replication-fork arrest comparable to that observed in fcd-2 mutants, albeit not as severe as observed in fncm-1 mutants (Figure 1C), which supports our observation that this domain in FNCM-1 is necessary to recruit the downstream FA pathway component FCD-2 (Figure 4B).
FA is a rare genetic disorder but it is still the most frequent inherited instability syndrome. It is characterized by bone marrow failure; hypersensitivity to cross-linking agents; and a high risk for acute myeloid leukemia, ataxia aelangiectasia, xeroderma pigmentosum, and Bloom, Werner, Nijmegen, Li–Fraumeni, and Seckel syndromes (Schroeder 1982). Recent studies emphasize the role of FA components in DNA replication arrest in addition to ICL repair (Blackford et al. 2012; Lachaud et al. 2016). Our finding that the FA pathway has a role in maintaining histone H3K4 dimethylation regardless of replication stress supplies an important connection between DNA damage repair and epigenetic regulation (Figure 7). Furthermore, fncm-1 mutants that are defective in recruiting FCD-2 will assist in defining the precise contribution of the FA genes in this regulation.
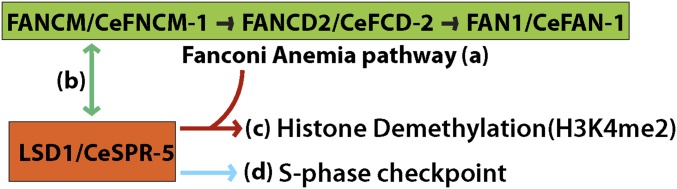
Figure 7.
The FA pathway and SPR-5 cooperate in DNA repair and regulation of histone demethylation. (a) C. elegans FNCM-1 is required for recruiting FCD-2 and its downstream nuclease FAN-1 in the germline. The potential helicase/translocase domain in FNCM-1 is necessary for this process. (b) SPR-5 and FNCM-1 interact with each other and their colocalization in the germline is extended under conditions leading to stalled replication forks. (c) FNCM-1 and FCD-2 are necessary for maintaining proper H3K4 dimethylation levels. (d) SPR-5-dependent S-phase checkpoint activation is required in response to the single-stranded DNA region formed in the absence of FNCM-1 in germline nuclei.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.118.300823/-/DC1.
Acknowledgments
We thank Doris Lui for comments on the manuscript and members of the Colaiácovo laboratory for discussions. We thank H.S. Koo for the RPA-1 and FCD-2 antibodies. This work was supported by a Ruth L. Kirschstein National Research Service award to S.E.B.-S. (F32 GM-100515) and a National Institutes of Health grant R01 GM-105853 to M.P.C.
Footnotes
Communicating editor: J. Engebrecht
Literature Cited
- Adamo A., Collis S. J., Adelman C. A., Silva N., Horejsi Z., et al. , 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell 39: 25–35. 10.1016/j.molcel.2010.06.026 [DOI] [PubMed] [Google Scholar]
- Adler J., Parmryd I., 2010. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A 77: 733–742. 10.1002/cyto.a.20896 [DOI] [PubMed] [Google Scholar]
- Alavattam K. G., Kato Y., Sin H. S., Maezawa S., Kowalski I. J., et al. , 2016. Elucidation of the Fanconi Anemia protein network in meiosis and its function in the regulation of histone modifications. Cell Rep. 17: 1141–1157. 10.1016/j.celrep.2016.09.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper B. J., Lowe B. R., Partridge J. F., 2012. Centromeric heterochromatin assembly in fission yeast–balancing transcription, RNA interference and chromatin modification. Chromosome Res. 20: 521–534. 10.1007/s10577-012-9288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Lukas C., Lukas J., 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5: 792–804. 10.1038/nrm1493 [DOI] [PubMed] [Google Scholar]
- Black J. C., Allen A., Van Rechem C., Forbes E., Longworth M., et al. , 2010. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell 40: 736–748. 10.1016/j.molcel.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Blackford A. N., Schwab R. A., Nieminuszczy J., Deans A. J., West S. C., et al. , 2012. The DNA translocase activity of FANCM protects stalled replication forks. Hum. Mol. Genet. 21: 2005–2016. 10.1093/hmg/dds013 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D., 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627. 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément J., de Massy B., 2017. Birth and death of a protein. eLife 6: e29502 10.7554/eLife.29502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. 10.1016/S1534-5807(03)00232-6 [DOI] [PubMed] [Google Scholar]
- Collis S. J., Barber L. J., Ward J. D., Martin J. S., Boulton S. J., 2006. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair (Amst.) 5: 1398–1406. 10.1016/j.dnarep.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L., Ji J. Y., Moon N. S., Herr A., Dyson N., 2007. Mutation of Drosophila Lsd1 disrupts H3–K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 17: 808–812. 10.1016/j.cub.2007.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., et al. , 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39: W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., Boulton S. J., 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24: 4345–4355. 10.1038/sj.emboj.7600896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Decaillet C., Stasiak A. Z., Stasiak A., Constantinou A., 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29: 141–148. 10.1016/j.molcel.2007.11.032 [DOI] [PubMed] [Google Scholar]
- Holmes A., Roseaulin L., Schurra C., Waxin H., Lambert S., et al. , 2012. Lsd1 and lsd2 control programmed replication fork pauses and imprinting in fission yeast. Cell Rep. 2: 1513–1520. 10.1016/j.celrep.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., et al. , 2007. p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108. 10.1038/nature06092 [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221. 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., Engebrecht J., 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20: 2078–2089. 10.1016/j.cub.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Greenwald I., 2002. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 16: 2713–2728. 10.1101/gad.1022402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V., Kelly W. G., 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiácovo M. P., 2014. ZTF-8 interacts with the 9–1-1 complex and is required for DNA damage response and double-strand break repair in the C. elegans germline. PLoS Genet. 10: e1004723 10.1371/journal.pgen.1004723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiácovo M. P., 2015a DNA damage sensitivity assays in Caenorhabditis elegans. Bio Protoc. 5: e1487 10.21769/BioProtoc.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiácovo M. P., 2015b New insights into the post-translational regulation of DNA damage response and double-strand break repair in Caenorhabditis elegans. Genetics 200: 495–504. 10.1534/genetics.115.175661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiácovo M. P., 2016. CRISPR-Cas9-guided genome engineering in C. elegans. Curr. Protoc. Mol. Biol. 115: 31.7.1–31.7.18. 10.1002/cpmb.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K., Schopf B., Kaden S., Sendoel A., Eberhard R., et al. , 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88. 10.1016/j.cell.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Lachaud C., Moreno A., Marchesi F., Toth R., Blow J. J., et al. , 2016. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 351: 846–849. 10.1126/science.aad5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Zaratiegui M., Villen J., Vaughn M. W., Verdel A., et al. , 2007. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol. Cell 26: 89–101. 10.1016/j.molcel.2007.02.023 [DOI] [PubMed] [Google Scholar]
- Lee I., Lehner B., Crombie C., Wong W., Fraser A. G., et al. , 2008. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat. Genet. 40: 181–188. 10.1038/ng.2007.70 [DOI] [PubMed] [Google Scholar]
- Lee K. Y., Chung K. Y., Koo H. S., 2010. The involvement of FANCM, FANCI, and checkpoint proteins in the interstrand DNA crosslink repair pathway is conserved in C. elegans. DNA Repair (Amst.) 9: 374–382. 10.1016/j.dnarep.2009.12.018 [DOI] [PubMed] [Google Scholar]
- Michl J., Zimmer J., Buffa F. M., McDermott U., Tarsounas M., 2016. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Struct. Mol. Biol. 23: 755–757. 10.1038/nsmb.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Kim H., Laurent B., Zhao Y., Lim H. J., et al. , 2013. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J. Cell Biol. 203: 457–470. 10.1083/jcb.201302092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Kim H. M., Colaiacovo M. P., Calarco J. A., 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201: 449–458. 10.1534/genetics.115.180679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke A. C., Beese-Sims S. E., Pantalena L. F., Reinke V., Shi Y., et al. , 2011. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc. Natl. Acad. Sci. USA 108: 12805–12810. 10.1073/pnas.1102298108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. T., Helin K., 2010. Histone demethylases in development and disease. Trends Cell Biol. 20: 662–671. 10.1016/j.tcb.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Peng B., Wang J., Hu Y., Zhao H., Hou W., et al. , 2015. Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res. 43: 5936–5947. 10.1093/nar/gkv528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Satory D., Dray E., Papusha A., Scheller J., et al. , 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23: 67–79. 10.1101/gad.1737809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunandan M., Chaudhury I., Kelich S. L., Hanenberg H., Sobeck A., 2015. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle 14: 342–353. 10.4161/15384101.2014.987614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T., Yonezawa M., Lein S., Heidrich K., Kubicek S., et al. , 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26: 103–115. 10.1016/j.molcel.2007.02.025 [DOI] [PubMed] [Google Scholar]
- Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., et al. , 2012. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 31: 3524–3536. 10.1038/emboj.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Schurer A., Rudolph C., Hettwer S., Kramer W., 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., Jasin M., 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22: 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. M., 1982. Genetically determined chromosome instability syndromes. Cytogenet. Cell Genet. 33: 119–132. 10.1159/000131736 [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., et al. , 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953. 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., Eddy S. R., Durbin R., 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2010. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11: 264–275. 10.1038/nrm2861 [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., et al. , 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100: 2414–2420. 10.1182/blood-2002-01-0278 [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. 10.1534/genetics.113.156075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2010. The FANCM family of DNA helicases/translocases. DNA Repair (Amst.) 9: 224–236. 10.1016/j.dnarep.2009.12.012 [DOI] [PubMed] [Google Scholar]
- Xue Y., Li Y., Guo R., Ling C., Wang W., 2008. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 17: 1641–1652. 10.1093/hmg/ddn054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Reinberg D., 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15: 2343–2360. 10.1101/gad.927301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.