Increased expression of mismatch repair proteins often correlates with tumor aggressiveness. Chakraborty, Dinh, and Alani report that co-overexpression of Msh2 and Msh6 in yeast results in genome instability phenotypes that are dependent on interaction with...
Keywords: DNA mismatch repair, Msh2-Msh6 and Msh6 overexpression, heteroduplex rejection, Sgs1, PCNA
Abstract
Mismatch repair (MMR) proteins act in spellchecker roles to excise misincorporation errors that occur during DNA replication. Curiously, large-scale analyses of a variety of cancers showed that increased expression of MMR proteins often correlated with tumor aggressiveness, metastasis, and early recurrence. To better understand these observations, we used The Cancer Genome Atlas and Gene Expression across Normal and Tumor tissue databases to analyze MMR protein expression in cancers. We found that the MMR genes MSH2 and MSH6 are overexpressed more frequently than MSH3, and that MSH2 and MSH6 are often cooverexpressed as a result of copy number amplifications of these genes. These observations encouraged us to test the effects of upregulating MMR protein levels in baker’s yeast, where we can sensitively monitor genome instability phenotypes associated with cancer initiation and progression. Msh6 overexpression (two- to fourfold) almost completely disrupted mechanisms that prevent recombination between divergent DNA sequences by interacting with the DNA polymerase processivity clamp PCNA and by sequestering the Sgs1 helicase. Importantly, cooverexpression of Msh2 and Msh6 (∼eightfold) conferred, in a PCNA interaction-dependent manner, several genome instability phenotypes including increased mutation rate, increased sensitivity to the DNA replication inhibitor HU and the DNA-damaging agents MMS and 4-nitroquinoline N-oxide, and elevated loss-of-heterozygosity. Msh2 and Msh6 cooverexpression also altered the cell cycle distribution of exponentially growing cells, resulting in an increased fraction of unbudded cells, consistent with a larger percentage of cells in G1. These novel observations suggested that overexpression of MSH factors affected the integrity of the DNA replication fork, causing genome instability phenotypes that could be important for promoting cancer progression.
MISMATCH repair (MMR), a highly conserved mechanism, plays a critical role in faithfully replicating genetic material by excising DNA mismatches resulting from DNA polymerase misincorporation errors. In Saccharomyces cerevisiae, mismatches in newly replicated DNA are recognized by the MSH complexes Msh2-Msh6 (MutSα) and Msh2-Msh3 (MutSβ). Msh2-Msh6 primarily recognizes base–base and small (1–2-nt) insertion–deletion mismatches, and Msh2-Msh3 primarily recognizes small and large (up to ∼17-nt) insertion–deletion loop mismatches [reviewed in Chakraborty and Alani (2016)]. Msh6 interacts with the polymerase processivity clamp PCNA (Flores-Rozas et al. 2000), resulting in a fraction of Msh2-Msh6 colocalizing with the replication fork, presumably facilitating the efficient detection of DNA mismatches generated during replication (Hombauer et al. 2011). Upon mismatch recognition by MSH heterodimers, MLH heterodimers (primarily Mlh1-Pms1) are recruited to the MSH–mismatch complex, which in turn recruits downstream MMR proteins such as Replication factor C (RFC), PCNA, Exo1, single-strand-binding protein RPA, DNA polymerase δ and ε, and DNA ligase to promote MMR through excision, resynthesis, and ligation steps [reviewed in Kunkel and Erie (2005, 2015), Chakraborty and Alani (2016), and Bowen and Kolodner (2017)].
Subsets of MMR proteins also play vital roles in preventing homologous recombination (HR) between divergent DNA sequences through a process known as heteroduplex rejection. During HR, cells repair double-strand breaks (DSBs) in DNA using a homologous template. Recombination involving repair though nonallelic, divergent sequences (also known as homeologous recombination) can cause mutations, loss-of-heterozygosity (LOH), and chromosomal rearrangements that are often associated with diseases such as cancer (George and Alani 2012; Liu et al. 2012; Zhang et al. 2013). Heteroduplex rejection is a mechanism by which cells prevent deleterious recombination events involving divergent DNA sequences. In this process, MSH complexes recognize mismatches during strand invasion and recruit the Sgs1-Top3-Rmi1 helicase–topoisomerase complex to unwind the heteroduplex DNA, resulting in a new search for a homologous donor sequence (Datta et al. 1996; Chen and Jinks-Robertson 1999; Nicholson et al. 2000; Myung et al. 2001; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Goldfarb and Alani 2005; Chakraborty et al. 2016).
MMR proteins have also been shown to play genome stability-promoting roles in other DNA repair pathways such as base excision repair, nucleotide excision repair, interstrand cross-link repair, single-strand annealing, and alternative nonhomologous end joining [reviewed in Liu et al. (2017)]. Interestingly, MMR proteins can also promote genome instability. For example, MutSβ promotes trinucleotide repeat expansions that are implicated in several neurodegenerative diseases [reviewed in Iyer et al. (2015), Zhao and Usdin (2015), and Zhao et al. (2016)]. Moreover, in higher eukaryotes, MMR plays a mutagenic role in somatic hypermutation and class switch recombination, leading to immunoglobulin diversity [reviewed in Chahwan et al. (2011) and Peña-Diaz and Jiricny (2012)].
Given the many roles of MMR proteins in DNA repair and recombination, it is not surprising that mutations or downregulation of several MMR proteins cause microsatellite instability (MSI) and defects in MMR and are linked to cancer [Kansikas et al. 2014; reviewed in Li (2008)]. On the other hand, overexpression of certain MMR proteins has been shown to have both beneficial and deleterious effects on genome stability. For example, (1) increased expression of Msh3 was shown to sequester Msh2 and cause a defect in Msh2-Msh6-dependent repair of base–base mismatches (Drummond et al. 1997; Marra et al. 1998); (2) overexpression of Msh6 in yeast improved heteroduplex rejection during a single-strand annealing repair mechanism, but severely compromised rejection in an inverted repeat recombination system that measures levels of spontaneous recombination thought to be initiated during S-phase (Chakraborty et al. 2016); (3) high expression levels of Mlh1 in yeast conferred a strong mutator phenotype that was partly suppressed by the overexpression of Pms1 (Shcherbakova et al. 2001); and (4) overexpression of PMS2, the mammalian homolog of yeast Pms1, in a mouse fibroblast cell line caused hypermutability and increased tolerance to DNA damage (Gibson et al. 2006).
Levels of MMR proteins such as MSH2, MSH6, MLH1, PMS2, and EXO1 are often increased in a variety of cancers, and are correlated with high levels of genomic instability that include genomic deletions and MSI, increased tumor aggressiveness, increased cancer recurrence, and poor survival (Velasco et al. 2002; Kauffmann et al. 2007; Li et al. 2008; Wagner et al. 2016; Wilczak et al. 2017). However, a cause–effect relationship between these deleterious characteristics and increased levels of MMR proteins has not been established. Increased expression of MMR proteins in cancers is currently thought to occur in response to increased proliferation of cells, as a mechanism to cope with the higher mutation and DNA damage loads (Wilson et al. 1995; Leach et al. 1996; Marra et al. 1996; Chang et al. 2000; Hamid et al. 2002). Presumably, altered expression of MMR proteins in cancers can also occur as a result of genetic or epigenetic changes that affect the levels of these proteins by altering their DNA copy number, transcription, translation, or stability. For example, histone deacetylase 6 (HDAC6) interacts with Msh2 and regulates its cellular levels by sequential deacetylation and ubiquitination, which ultimately leads to its degradation (Zhang et al. 2014). In contrast, ubiquitin-specific peptidase 10 (USP10) counteracts the effect of HDAC6 by stabilizing Msh2 (Zhang et al. 2016). Protein kinase C (PKC) has also been shown to regulate levels of human MMR proteins such as MSH2, MSH6, and PMS2, and increased PKC activation leads to higher expression of MMR proteins (Humbert et al. 2002).
Based on the above observations, we hypothesized that overexpression of subsets of MMR proteins could directly impact genome stability and contribute to cancer progression. To test this, we first looked at publicly available data sets from a variety of cancers in the TCGA (The Cancer Genome Atlas) and GENT (Gene Expression across Normal and Tumor tissue; Shin et al. 2011) databases, and found that MSH2 and MSH6 are frequently overexpressed in cancer tissues compared to normal tissues. However, MSH3 was less frequently overexpressed in cancers. In addition, using data available in the cBioPortal database (Cerami et al. 2012; Gao et al. 2013), we found that MSH2 and MSH6 genes often exhibit copy number amplifications either individually or simultaneously. To experimentally test our hypothesis, we overexpressed MMR proteins in a genetically tractable yeast model, where highly sensitive assays exist to measure recombination rates and fidelity, mutation rates, sensitivity to DNA-damaging agents, and LOH. Our results indicated that overexpression of MSH proteins conferred several genomic instability phenotypes. In particular, increased levels of Msh6 caused a severe decrease in heteroduplex rejection of events that occur during the replication phase of the cell cycle by sequestering Sgs1 to the replication fork. In contrast, increased levels of Msh2-Msh6 improved heteroduplex rejection, but caused increased mutation and HR rates, increased LOH, and increased sensitivity to compounds that disrupt replication fork progression. These data, in conjunction with the bioinformatic analyses, strongly suggest that increased levels of MSH proteins in higher eukaryotes are likely to have deleterious effects on genome stability and may play a causal role in cancer progression.
Materials and Methods
Yeast strains
Yeast strains used in this study are shown in Supplemental Material, Table S2, and were constructed and grown using standard techniques (Rose et al. 1990; Gietz and Schiestl 1991).
Construction of 2μ plasmids
S288c-derived genes listed in Table S2 were inserted into 2μ pRS vectors (Christianson et al. 1992). Briefly, these plasmids were constructed using existing Alani laboratory plasmids (cloning details are provided upon request). In all cases, at least 300 bp of DNA sequence upstream of the start codon was included.
Homeologous recombination using an inverted repeat reporter assay
Strains used to measure homeologous recombination are listed in Table S2. Strains containing 2μ plasmids were struck onto minimal dropout media plates. A total of 9–21 single colonies per strain were then inoculated into 5 ml of minimal dropout medium containing 4% galactose and 2% glycerol and grown for 2 days at 30°. Appropriate dilutions of cells were plated onto minimal media (2% galactose and 2% glycerol) plates lacking histidine and the amino acid required to maintain the 2μ plasmid (selective), and onto minimal media (2% glucose) plates lacking the amino acid required to maintain the 2μ plasmid (permissive). Plates were incubated for 4 days at 30° and then scored for frequency of His+ colonies. The rate of homeologous recombination was calculated as described (Nicholson et al. 2000). Pairwise Mann–Whitney U-tests were performed between mutants and the corresponding wild-type of each strain. Differences were considered significant when P < 0.05.
Phenotypic analysis of strains overexpressing MMR proteins in MMS, HU, and 4-nitroquinoline N-oxide sensitivity assays
Yeast strain SJR769 transformed with either pRS426 (wild-type) or 2μ plasmids containing the indicated gene were analyzed. Single colonies were inoculated into 5 ml minimal dropout media lacking the amino acid required to maintain the plasmid and incubated for 24 hr at 30° to bring the cultures to saturation. Saturated cultures were diluted in sterile distilled water to OD600 of 1. They were subsequently serially diluted four times in 1:10 increments, and 4 μl of each were spotted onto minimal dropout plates containing 0.02% MMS, 200 mM HU, 0.25 µg/ml 4-nitroquinoline N-oxide (4-NQO), or no drug. Plates were incubated at 30° for 2–4 days.
Cell cycle distribution analysis of strains overexpressing MMR proteins
Midlog-phase cultures of yeast strain SJR769 transformed with either pRS426 (wild-type) or 2μ plasmids containing the indicated gene were examined by light microscopy. Cells were categorized and counted as unbudded, containing a small bud, or containing a large bud.
Measuring LOH
Yeast cells containing 2μ plasmids were struck to single colonies on minimal dropout plates and incubated at 30° for 4 days. Single whole colonies were picked, resuspended in 1 ml of sterile distilled water, and serially diluted, and appropriate dilutions were plated on minimal dropout medium lacking the amino acid required to maintain the plasmid, containing 1 g/liter of 5-FOA (selective) and minimal dropout medium lacking the amino acid required to maintain the plasmid (permissive). Plates were incubated at 30° and colonies were counted after 2 days. Recombination rates and 95% C.I.s were calculated using the Lea and Coulson method of the median within the FALCOR web application (http://www.keshavsingh.org/protocols/FALCOR.html; Lea and Coulson 1949; Hall et al. 2009). Pairwise Mann–Whitney U-tests were performed between each mutant and wild-type strain. Differences were considered significant when P < 0.05.
Measuring mutation rates using the lys2-A14 reversion assay
The lys2-A14 reversion assay was performed as described (Heck et al. 2006). Pairwise Mann–Whitney U-tests were performed between each mutant and wild-type strain. Differences were considered significant when P < 0.05.
Western blot analysis
Cell pellets from saturated 5 ml cell cultures of BJ5464 containing the indicated plasmids were resuspended in 0.5 ml lysis buffer (150 mM NaCl, 25 mM Tris pH 8.0, 1 mM EDTA, 10 mM β-mercaptoethanol, and 1 mM PMSF) and lysed by vortexing with glass beads. Unless otherwise indicated, 20 µg of each protein lysate, measured using the Bradford (1976) assay, were run on each lane of an 8% SDS/PAGE gel. Contents of the gel were transferred onto a Bio-Rad nitrocellulose membrane using a Mini Trans-Blot cell (Bio-Rad, Hercules, CA). The membrane was then blocked overnight at 4° and probed with 1:4000 diluted rabbit anti-Msh6 (Studamire et al. 1998; Kumar et al. 2011) for 1 hr and 1:15,000 diluted horseradish peroxidase-conjugated goat anti-rabbit antibody for 1 hr. HRP signal was detected using the Bio-Rad Clarity Western ECL substrate kit and exposed to CL-XPosure film (Thermo Scientific).
TCGA RNA sequencing and reverse phase protein arrays
Normalized (quartile normalization) data for gene expression analysis across 21 tumor types were obtained from the TCGA Data Portal (https://cancergenome.nih.gov/). Differential gene expression was compared using Mann–Whitney U-tests and corrected for multiple testing using the Bonferroni correction. Differences were considered significant when corrected P < 0.05. When using reverse phase protein arrays (RPPAs) (http://tcpaportal.org/tcpa/), correlations and P-values were calculated using Spearman’s correlation and Spearman’s rank test, respectively.
Repetition of experiments
All wet laboratory experiments presented (inverted repeat recombination, lys2-A14 reversion, DNA damage sensitivity, LOH, cell cycle distribution, and western blots) were repeated on at least two separate days.
Data availability
Strains and plasmids are available upon request. Supporting information contains detailed descriptions of all supplemental files. The following figures and tables can be found in the Genetics Society of America Figshare portal. Figure S1, a comparison of gene expression patterns of MSH2, MSH3, and MSH6 across diverse human cancer and normal tissues using the GENT database. Figure S2, correlation of MSH2 and MSH6 protein expression in TCGA tumor types with significantly upregulated MSH2 and MSH6 mRNA expression. Figure S3, western blot analysis of Msh2 and Msh6 levels; independent measurements. Figure S4, western blot analysis of Msh2 and Msh6 levels in strains containing ARS-CEN plasmids. Table S1, literature review of MMR proteins overexpressed in cancers. Table S2, strains and plasmids used in this study. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5991208.
Results
MSH2 and MSH6 are often overexpressed in a variety of cancers
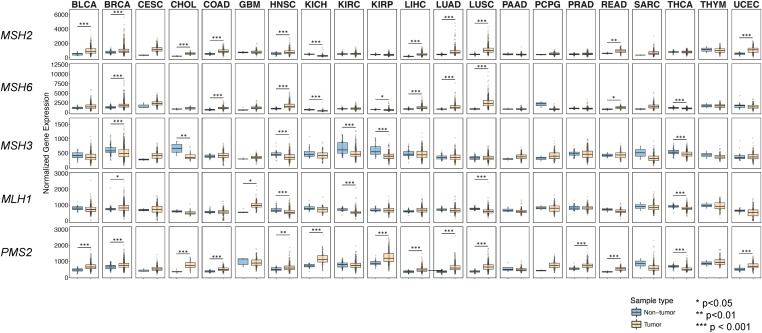
MMR proteins are often overexpressed in cancers, and in some cases this overexpression has been correlated with genome instability (summarized in Table S1; Velasco et al. 2002; Kauffmann et al. 2007; Li et al. 2008; Wagner et al. 2016; Wilczak et al. 2017). These observations encouraged us to compare expression levels of MMR genes, using data available in cancer databases such as TCGA and GENT (Shin et al. 2011). Using data from TCGA, RNA sequencing expression levels for MSH2, MSH6, MSH3, MLH1, and PMS2 were compared between tumor samples and nontumor samples in 21 tumor types (Figure 1). We found that MSH2 and MSH6 were significantly overexpressed in 10 of 21 and 7 of 21 tumor types, respectively, when compared to their nontumor tissue counterparts (P < 0.05, Mann–Whitney U-test), and all tumor types with increased MSH6 expression showed increased MSH2 expression. MSH3, on the other hand, was not significantly overexpressed in any of the 21 tumor types analyzed, compared to nontumor samples. Thus, MSH2 and MSH6 are more frequently overexpressed than MSH3 (P < 0.01, Fisher’s exact test). MLH1 was found to be overexpressed in 2 out of the 21 tumor types and PMS2 was overexpressed in 13 out of the 21 tumor types compared to the corresponding normal tissues (P < 0.05, Mann–Whitney U-test).
Figure 1.
Comparison of gene expression patterns of MSH2, MSH3, and MSH6 between different human cancer and normal tissues using The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). mRNA expression levels of MSH2, MSH6, MSH3, MLH1, and PMS2 were compared in cancer vs. normal tissues using RNA sequencing (RNA-seq) data from the TCGA across 21 different tumor types. The data are shown as box plots, circles represent individual samples, and the y-axis corresponds to normalized RNA-seq expression values. TCGA tumor codes are: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; THCA, thyroid carcinoma; THYM, thymoma; and UCEC, uterine corpus endometrial carcinoma.
We then compared gene expression of MSH2, MSH3, and MSH6 across > 24,000 samples from different tissue types using the GENT database (Shin et al. 2011). Several cancer tissues showed overexpression of MSH2 and MSH6 genes when compared to their normal tissue counterparts (Figure S1). In total, 13/25, 11/25, and 7/25 of cancers showed increased expression of the MSH2, MSH6, and MSH3 genes, respectively. Together, these data show that MSH2 and MSH6 are often cooverexpressed in a variety of cancers. However, MSH3 is less frequently overexpressed.
Lastly, we compared protein expression of MSH2 and MSH6 for the seven tumor types with significantly upregulated MSH2 and MSH6 mRNA expression in TCGA (Figure 1) using RPPAs (http://tcpaportal.org/tcpa/index.html). We found that MSH2 and MSH6 were expressed in a correlated manner for six of the tumor types (Figure S2). Unfortunately, this analysis only contains information from tumor samples, and thus one cannot perform tumor–normal comparisons as was done for the mRNA expression data set.
Copy number amplifications of MSH2 and MSH6 are observed in a variety of cancers
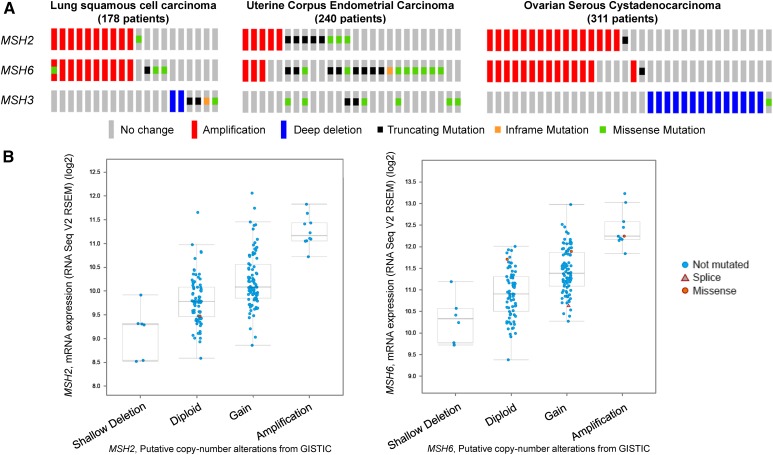
To test if genomic alterations can lead to increased expression of certain MMR genes in cancers, we analyzed genomic alterations seen in MSH2, MSH6, and MSH3 genes in a variety of cancers using data available in the cBioPortal database (Cerami et al. 2012; Gao et al. 2013). In Figure 2A, three representative images of genomic alterations in different cancer data sets are shown. These images show that the MSH2 and MSH6 genes were coamplified or individually amplified in a number of patients. However, MSH3 was not amplified in any of the data sets shown and, in general, seemed to be amplified less frequently when compared to MSH2 or MSH6. MSH2 and MSH6, on the other hand, were mostly coamplified together, possibly because the two genes are located near each other on chromosome 2. Next, we tested if DNA copy number amplifications of the MSH2 and MSH6 genes resulted in higher mRNA levels. As shown in Figure 2B, increases in copy numbers of MSH2 and MSH6 resulted in higher mRNA levels. Together, these data indicate that MSH2 and MSH6 are often coamplified, resulting in cooverexpression in a variety of cancers.
Figure 2.
Genomic alterations in MSH2, MSH3, and MSH6. (A) Mutations and copy number alterations in MSH2, MSH3, and MSH6 are shown for patient samples from three different types of cancers, obtained from cBioPortal (http://www.cbioportal.org/index.do). Only tumor samples from patients that show alterations in MSH2, MSH6, or MSH3 are shown, and each patient is represented by a single column that includes the analysis for all three genes. Red bars represent amplifications and blue bars represent homozygous deletions. Shallow deletions refer to heterozygous deletions and deep deletions refer to homozygous deletions. A truncating mutation (black square) is one that results in a truncated protein. An in-frame mutation (orange square) is an insertion or a deletion that does not cause a shift in the triplet reading frame but can result in an abnormal protein product. A missense mutation (green square) is a point mutation that results in a single amino acid change. Note that the truncating, missense, and in-frame mutations are shown as squares so that multiple mutations can be shown if they exist in a single gene. For example, the MSH6 gene amplified in lung squamous cell carcinoma patient 1 (from left to right) has both an amplification and a missense mutation in the MSH6 gene, and the MSH2 gene in lung squamous cell carcinoma patient 11 is not amplified but contains a missense mutation. (B) mRNA expression levels of MSH2 (left panel) and MSH6 (right panel) are shown as a function of putative copy number alterations for the lung squamous cell carcinoma data set of 178 patients shown in (A). The mRNA expression data are shown as boxplots and are given in RNA Seq V2 RSEM (RNA-seq by expectation maximization) in log2 scale.
Msh6 overexpression in wild-type strains disrupts heteroduplex rejection by sequestering Sgs1
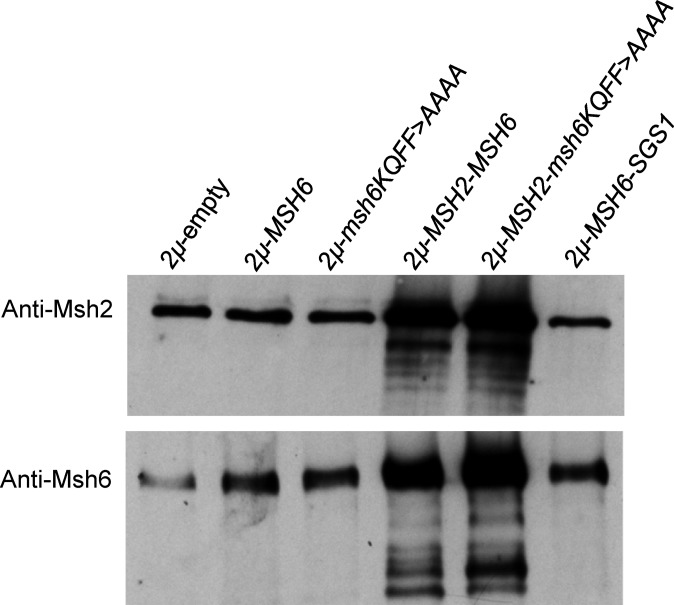
The above observations encouraged us to determine if overexpression of MMR proteins in wild-type strains affects genome stability in yeast. Overexpression of all proteins presented in this study was achieved by cloning the native gene and its promoter into a 2μ vector that is present at roughly 20 copies per cell (Christianson et al. 1992; Chakraborty et al. 2016; Materials and Methods). We performed western blot analysis to assess overexpression of the Msh2 and Msh6 protein. As shown in Figure 3 and Figure S3, Msh6 is overexpressed by no greater than fourfold in strains containing 2µ-MSH6. We also observed that 2μ overexpression of msh6-KQFF > AAAA, a mutant form of the Msh6 protein that is compromised for Msh2-Msh6 interactions with PCNA (Clark et al. 2000), occurred at a level comparable to MSH6, indicating that msh6-KQFF > AAAA is stably expressed. Interestingly, strains harboring the 2µ-MSH2-MSH6 or 2µ-MSH2-msh6-KQFF > AAAA plasmids showed similar levels of overexpression of Msh6. Expression of each MSH subunit in these strains was ∼eightfold higher than in strains containing empty vectors (Figure S3). This result suggests that formation of the Msh2-Msh6 heterodimer improves the stability of each subunit [see Chang et al. (2000)].
Figure 3.
Western blots to measure levels of overexpression of mismatch repair proteins. Western blot analysis (Materials and Methods) using Msh6 and Msh2 antibodies was performed on cell extracts derived from the BJ5464 (wild-type) strain containing either pRS426 or 2μ vectors with the indicated genes. Lane 1, wild-type containing a 2μ empty vector. Lane 2, wild-type containing 2μ-MSH6. Lane 3, wild-type containing 2μ-msh6-KQFF > AAAA. Lane 4, wild-type containing 2μ-MSH2-MSH6. Lane 5, wild-type containing 2μ-MSH2-msh6-KQFF > AAAA. Lane 6, wild-type containing 2μ-MSH6-SGS1. msh2Δ and msh6Δ in the EAY1597 background are shown in Figure S3. In each lane, 15 μg of each protein extract was loaded.
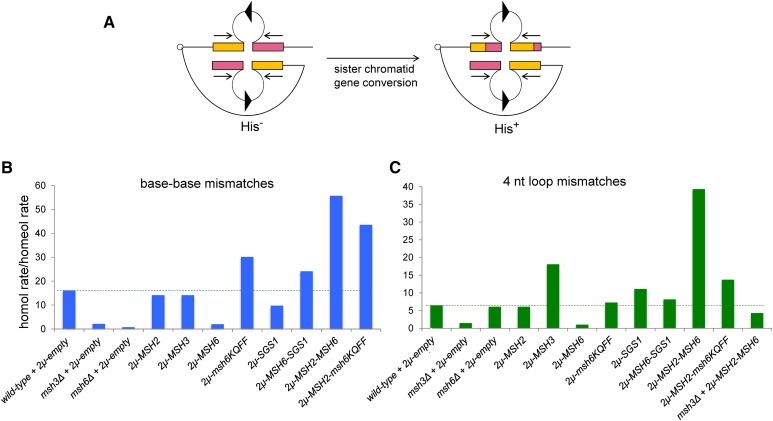
To determine if overexpression of MMR proteins causes genome instability, we utilized several sensitive assays in the S. cerevisiae model organism that measure genetic recombination, mutation rate, LOH, and sensitivity to DNA-damaging agents (Miller 1970; Slater 1973; Prakash and Prakash 1977; Tran et al. 1997; Chen and Jinks-Robertson 1998; Conover et al. 2015). Initially, we used an inverted repeat recombination assay to assess recombination frequency and fidelity in baker’s yeast (Chen and Jinks-Robertson 1998; Nicholson et al. 2000; Figure 4A). In this assay, recombination events occur at different rates between identical and divergent (either from single-nt base–base or 4-nt insertion–deletion substitutions) DNA sequences. Such recombination events, which reorient the HIS3 and intron sequences to yield a functional HIS3 gene, are thought to be initiated by DNA lesions that arise during or shortly after the replication of the recombination substrates. Repair of these lesions using donor sequences occurs at a much lower rate between divergent sequences than between homologous sequences (Chen and Jinks-Robertson 1998; Nicholson et al. 2000).
Figure 4.
Overexpression of MSH proteins in wild-type strains alters rejection efficiency in an inverted repeat recombination assay. (A) Intron-based intramolecular recombination assay involving inverted repeat sequences (blue and green boxes) that generate a functional HIS3 reporter following homologous recombination [adapted from Nicholson et al. (2000)]. In this assay, His+ recombinants are thought to result from gene conversion events between inverted repeat sequences present on sister chromatids. Identical or divergent substrates predicted to form base–base (cβ2/cβ2-ns) and 4-nt loop (cβ2/cβ2-4L) mismatches in heteroduplex DNA were analyzed in this study. Homologous and homeologous recombination rates for both base–base (B) and 4-nt loop (C) mismatches were calculated for strains containing the indicated plasmids, as described in the Materials and Methods. The ratio of homologous to homeologous recombination rates is shown for both types of mismatch as a measure of heteroduplex rejection efficiency. Data for overexpression of MSH2, MSH3, MSH6, and msh6-KQFF > AAAA and the matched wild-type were obtained from Chakraborty et al. (2016). The dashed lines indicate the ratio of homologous to homeologous recombination rates for wild-type strains containing the pRS426 vector. See Table 1 for the presentation of quantitative measurements and statistical analysis.
Previous work in our laboratory showed that overexpression of Msh6 severely compromised heteroduplex rejection of substrates containing base–base or 4-nt insertion–deletion sequence divergence (Chakraborty et al. 2016; Figure 4, B and C and Table 1). However, overexpression of msh6-KQFF > AAAA improved heteroduplex rejection in the base–base mismatch substrate and restored it to wild-type levels in the 4-nt loop substrate (Chakraborty et al. 2016; Figure 4, B and C and Table 1). These observations suggested that overexpression of the Msh6 subunit disrupted heteroduplex rejection through steps involving interactions with PCNA. An alternative possibility is that some of the phenotypes observed resulted from Msh6 overexpression sequestering Msh2 and thus depleting Msh2-Msh3 levels. However, the finding that overexpression of msh6-KQFF > AAAA restored heteroduplex rejection for both base–base and 4-nt loop substrates provides support for Msh6 subunit overexpression disrupting heteroduplex rejection through interactions with PCNA.
Table 1. Recombination rates in strains overexpressing MMR proteins as measured in the inverted repeat reporter assay.
| Strain | Relevant genotype | Rate of His+ recombination (×10−6) | Homologous rate/Homeologous ratea |
|---|---|---|---|
| Cβ2-Cβ2 | pRS426b | 0.64 (0.47–0.90)c | |
| msh6Δ + pRS426 | 0.62 (0.54–1.7) | ||
| msh3Δ + pRS426 | 2.5 (2.1–3.3)d | ||
| 2µ-MSH6b | 1.5 (0.68–2.44)e | ||
| 2µ-msh6-KQFF > AAAAb | 2.1 (1.38–2.96)d | ||
| 2µ-MSH3b | 1.1 (0.66–1.65)d | ||
| 2µ-MSH2b | 0.54 (0.26–0.72) | ||
| 2µ-MLH1-PMS1 | 1.57 (0.83–3.11)d | ||
| 2µ-EXO1 | 0.63 (0.42–1.09) | ||
| 2µ-MSH2-MSH6 | 6.0 (4.09–9.40)d | ||
| 2µ-MSH2-msh6-KQFF > AAAA | 1.2 (0.90–1.8)d | ||
| 2µ-POL30 | 3.6 (1.0–5.5)d | ||
| msh3Δ + 2µ-MSH2-MSH6 | 12.7 (10.4–21.8)d | ||
| 2µ-SGS1 | 1.3 (0.61–4.41)e | ||
| 2µ-MSH6-SGS1 | 2.6 (2.1–3.9)d | ||
| Cβ2/Cβ2-ns | pRS426b | 0.04 (0.02–0.08) | 16 |
| msh6Δ + pRS426 | 1.0 (0.82–4.5)d | 0.61 | |
| msh3Δ + pRS426 | 1.3 (0.90–2.4)d | 2.0 | |
| 2µ-MSH6b | 0.80 (0.60–1.31)d | 1.9 | |
| 2µ-msh6-KQFF > AAAAb | 0.07 (0.03–0.12) | 30 | |
| 2µ-MSH3b | 0.08 (0.03–0.16) | 14 | |
| 2µ-MSH2b | 0.04 (0.01–0.06) | 14 | |
| 2µ-MLH1-PMS1 | 0.08 (0.03–0.35) | 20 | |
| 2µ-EXO1 | 0.04 (0.02–0.09) | 16 | |
| 2µ-MSH2-MSH6 | 0.11 (0.05–0.23)d | 56 | |
| 2µ-MSH2-msh6-KQFF > AAAA | 0.03 (0.02–0.05) | 43 | |
| 2µ-POL30 | 0.26 (0.13–0.54)d | 14 | |
| 2µ-SGS1 | 0.14 (0.10–0.41)d | 9.6 | |
| 2µ-MSH6-SGS1 | 0.11 (0.02–0.57) | 24 | |
| Cβ2/Cβ2-4L | pRS426b | 0.10 (0.05–0.56) | 6.4 |
| msh6Δ + pRS426 | 0.10 (0.04–0.16) | 6.0 | |
| msh3Δ + pRS426 | 1.8 (1.3–2.6)d | 1.4 | |
| 2µ-MSH6b | 1.57 (1.12–1.81)d | 0.96 | |
| 2µ-msh6-KQFF > AAAAb | 0.29 (0.11–0.36) | 7.2 | |
| 2µ-MSH3b | 0.06 (0.05–0.09) | 18 | |
| 2µ-MSH2b | 0.09 (0.04–0.13) | 6.0 | |
| 2µ-MLH1-PMS1 | 0.15 (0.10–0.29) | 11 | |
| 2µ-MSH2-MSH6 | 0.15 (0.09–0.44) | 39 | |
| msh3Δ + 2µ-MSH2-MSH6 | 3.0 (2.3–5.0)d | 4.2 | |
| 2µ-MSH2-msh6-KQFF > AAAA | 0.09 (0.07–0.17) | 14 | |
| 2µ-POL30 | 0.35 (0.26–0.51) | 10 | |
| 2µ-SGS1 | 0.12 (0.06–0.21) | 11 | |
| 2µ-MSH6-SGS1 | 0.32 (0.12–0.81) | 8.1 |
Homeologous recombination rates and 95% C.I.s were calculated as described in the Materials and Methods. The genotypes of the strains are shown in Table S2. Cβ2/Cβ2, homologous substrate; Cβ2/Cβ2-ns, base–base mismatch substrate; Cβ2/Cβ2-4L, 4-nt loop mismatch substrate.
Homologous rate (Cβ2-Cβ2)/homeologous rate for strains with the same overexpression plasmid.
Published data from Chakraborty et al. (2016).
Numbers in parentheses indicate 95% C.I.s.
Significantly different from wild-type of the same strain (P < 0.01, Mann–Whitney U-test).
Significantly different from wild-type of the same strain (P < 0.05, Mann–Whitney U-test).
Based on the above observations, we hypothesized that Msh6 overexpression sequestered proteins critical for heteroduplex rejection. We focused on Sgs1, a critical component in heteroduplex rejection, because it was shown to directly interact with Msh2-Msh6 in co-immunoprecipitation experiments (Chakraborty et al. 2016). As shown in Figure 4, B and C and Table 1, cooverexpression of Msh6 and Sgs1 suppressed the disruption of heteroduplex rejection caused by Msh6 overexpression and, compared to wild-type strains containing an empty vector, increased the efficiency of heteroduplex rejection by two- and 1.4-fold in the base–base and 4-nt loop mismatch substrates, respectively. However, Sgs1 overexpression alone did not affect the heteroduplex rejection efficiency in the base–base mismatch substrates but increased it by 1.8-fold in the 4-nt loop mismatch substrates. This result is not surprising because in strains containing the 4-nt loop mismatch substrates, rejection relies primarily on Msh2-Msh3 and is improved similarly by the overexpression of Sgs1 or cooverexpression of Msh6 and Sgs1. These observations are consistent with excess Msh6 inhibiting heteroduplex rejection by associating with PCNA and sequestering Sgs1 from participating in antirecombination activities that require a functional Msh2-Msh6 complex. An alternative explanation for the improvement of heteroduplex rejection observed in wild-type strains cooverexpressing Msh6 and Sgs1 is that Msh6 levels were reduced compared to strains overexpressing only Msh6. However, this does not appear to be the case; Msh6 levels were similar in wild-type strains containing 2µ-MSH6-SGS1 or 2µ-MSH6 (Figure 3 and Figure S3). These results indicate that overexpression of MSH6 alone could compromise heteroduplex rejection and could thus promote genomic rearrangements often associated with cancers.
The above observations suggest that Msh2 and Msh6 cooverexpression should suppress the Msh6 overexpression effect because a functional Msh2-Msh6 complex would be available to promote Sgs1 function. In fact, compared to a wild-type strain containing an empty vector, cooverexpression of Msh2 and Msh6 improved heteroduplex rejection for both the base–base and 4-nt loop substrates by 3.5- and 6.1-fold, respectively (Figure 4, B and C and Table 1). Curiously, cooverexpression of Msh2 and Msh6 in wild-type increased the rate of HR nearly 10-fold, to 6.0 × 10−6/cell/division from 0.64 × 10−6/cell/division (Table 1). However, when Msh2 and msh6-KQFF > AAAA were cooverexpressed, this rate dropped to 1.2 × 10−6/cell/division, suggesting that the increase in recombination rate in strains with high Msh2-Msh6 expression was due to excess amounts of Msh2-Msh6 associating with PCNA. Moreover, cooverexpression of Msh2 and msh6-KQFF > AAAA improved rejection efficiency in both base–base and 4-nt loop mismatch strains, consistent with previous work showing that the Msh6-PCNA interaction is not essential for rejection, and that the msh6-KQFF > AAAA protein is stably expressed (Stone et al. 2008; Chakraborty et al. 2016). Overexpression of other MMR/replication factors such as Mlh1-Pms1, Exo1, or PCNA did not alter heteroduplex rejection efficiency (Table 1).
The above results led us to hypothesize that excessive association of Msh2-Msh6 with PCNA caused replication fork instability, giving rise to a higher frequency of replication fork stalling that results in DNA lesions that are repaired by HR. In this model, cooverexpression of Msh2 and msh6-KQFF > AAAA does not confer a similar deleterious effect because this complex is defective in its interaction with PCNA. Interestingly, increased levels of Msh2-Msh6 did not appear to sequester Sgs1 from heteroduplex DNA because heteroduplex rejection was improved. We believe that this is due to the presence of higher levels of functional MSH complexes that disrupt the replication fork due to Msh2-Msh6-PCNA interactions, yet are also available to productively interact with Sgs1 for heteroduplex rejection (see Discussion). In this model, cooverexpression of Msh2 and msh6-KQFF > AAAA results in improved heteroduplex rejection because interactions between PCNA and Msh6 are not required for rejection (Stone et al. 2008; Chakraborty et al. 2016).
Msh2-Msh6 can reject divergent substrates with 4-nt loop mismatches when present at high concentrations in the cell
The fact that cooverexpression of Msh2-Msh6 improved rejection in strains containing the 4-nt loop mismatch substrates was surprising because Msh2-Msh6 primarily recognizes base–base mismatches and small (1-nt) insertion–deletion loop mismatches during heteroduplex rejection (Nicholson et al. 2000). Msh2-Msh3, on the other hand, recognizes small and large insertion–deletion loops and has been shown to be required for the rejection of substrates with 4-nt loops (Lee et al. 2007; Table 1). To test if the increase in rejection conferred by cooverexpressing Msh2 and Msh6 was dependent on the Msh3 protein, we cooverexpressed Msh2 and Msh6 in a msh3Δ background (Table 1). The heteroduplex rejection ratio involving 4-nt loop mismatch substrates was sixfold higher in a wild-type strain cooverexpressing Msh2 and Msh6 compared to the same strain containing an empty vector. When Msh2 and Msh6 were cooverexpressed in a msh3Δ background, the reject-on ratio was threefold higher than in a msh3Δ strain containing an empty vector. The msh3Δ strain showed a 4.6-fold decrease in the heteroduplex rejection ratio compared to wild-type strains containing an empty vector (Table 1). These findings indicate that the deficiency in rejecting 4-nt loop mismatches observed in msh3Δ strains is partially suppressed by the overexpression of Msh2-Msh6, and suggests that Msh2-Msh6 overexpression confers some of its heteroduplex rejection activity in the absence of Msh3.
Cooverexpression of Msh2-Msh6 causes sensitivity to MMS, HU, and 4-NQO
The increased level of HR and improved heteroduplex rejection seen in strains overexpressing Msh2 and Msh6 suggested that multiple genome stability pathways were affected. We further tested this idea by determining if cooverexpression of Msh2 and Msh6 conferred sensitivity to the DNA-damaging drugs MMS, HU, and 4-NQO. MMS is a DNA alkylating agent and a carcinogen that predominantly modifies both guanine (to 7-methylguanine) and adenine (to 3-methyladenine) residues, causing both base mispairing and replication blocks (Beranek 1990). HU inhibits the ribonucleotide reductase enzyme, and thus depletes deoxynucleoside triphosphate (dNTP) pools and disrupts DNA replication. At high concentrations (200 mM), HU reduces cellular dNTPs that normally accumulate as cells enter S phase and impedes S-phase progression (Chabes et al. 2003; Koç et al. 2004). 4-NQO is a UV mimetic agent that leads to DNA damage by generating stable quinoline monoadducts such as 3-(deoxyadenosin-N6-yl)-4AQO and N4-(guanosin-7-yl-4AQO), that stall DNA polymerase progression (Miller 1970; Kohda et al. 1991). MMS, HU, and 4-NQO activate the intra-S checkpoint (Iyer and Rhind 2017).
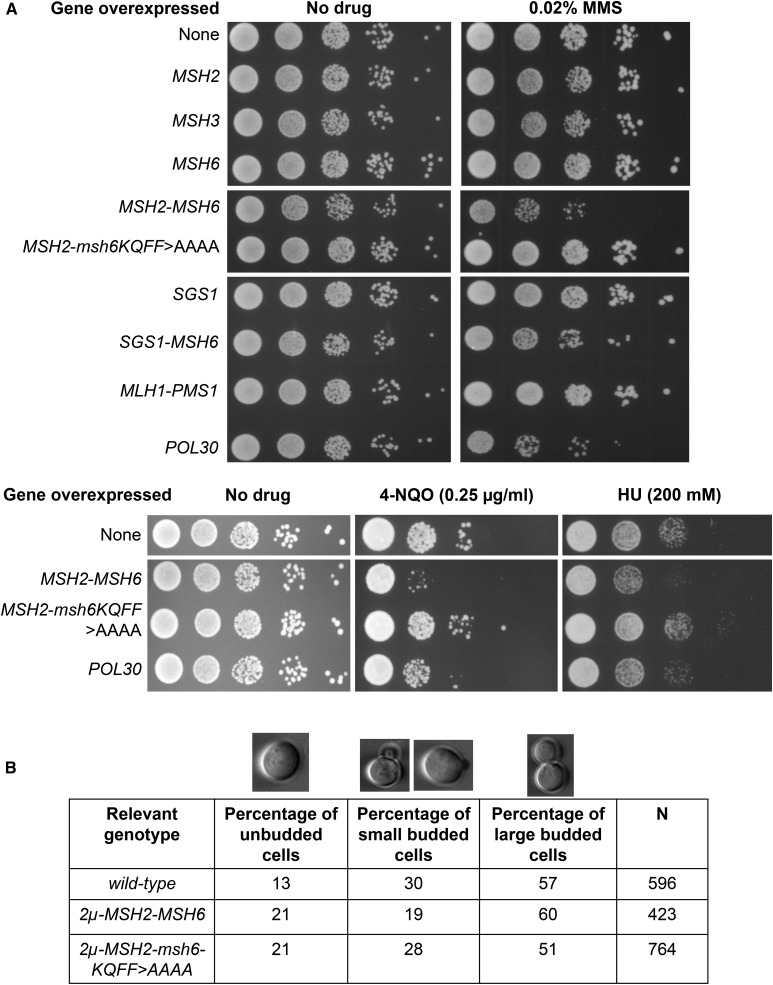
As shown in Figure 5A, overexpression of Msh2, Msh3, or Msh6 did not confer a difference in sensitivity to MMS (0.02%) compared to the wild-type control. However, cooverexpression of Msh2 and Msh6 conferred a moderate increase in sensitivity (Figure 5A), but cooverexpression of Msh2 and msh6-KQFF > AAAA did not cause such an effect. The former observation was consistent with high levels of Msh2-Msh6 altering interactions of PCNA with other factors. Consistent with this idea, overexpression of PCNA conferred moderate sensitivity to MMS (Figure 5A). We find this observation interesting because PCNA overexpression has been observed in many cancers (Table S1). Also, recent work by Johnson et al. (2016) showed that overexpression of PCNA (by fusing it to a galactose inducible promoter) caused MMS sensitivity due to accumulation of PCNA on DNA. Overexpression of Sgs1 or cooverexpression of Sgs1 and Msh6 had no significant effect on MMS sensitivity (Figure 5A). We also tested if cooverexpression of Mlh1 and Pms1 affected MMS sensitivity. We were interested in testing this because the human homologs of Mlh1 and Pms1 are often overexpressed in a variety of cancers (Figure 1 and Table S1). However, unlike the MSH complex, cooverexpression of Mlh1 and Pms1 did not affect MMS sensitivity (Figure 5A).
Figure 5.
Effect of overexpression of mismatch repair proteins on sensitivity to DNA-damaging agents and cell cycle progression. (A) Serial dilutions (10-fold) of the strain SJR769 containing 2μ plasmids with the indicated genes were spotted on minimal dropout media lacking the amino acid required to maintain the 2μ plasmid, with or without the indicated drug (MMS, HU, and 4-NQO). Final concentrations of the drugs are indicated in parentheses. The wild-type strain contains the pRS426 plasmid. (B) Distributions of cells in the different cell cycle stages were measured by counting the percentages of cells in midlog-phase cultures that were unbudded, had a small bud, or had a large bud. The percentages and total numbers of cells counted (N) are shown.
As shown in Figure 5A, cooverexpression of Msh2 and Msh6 also conferred sensitivity to HU and 4-NQO, providing additional evidence indicating that high levels of Msh2-Msh6 make cells susceptible to replication stress and are likely to cause genomic instability during replication that can lead to harmful genomic rearrangements. The sensitivity to both HU and 4-NQO, like MMS, was relieved when Msh2 and msh6-KQFF > AAAA were cooverexpressed. However, Overexpression of PCNA did not seem to cause a significant sensitivity to 4-NQO or HU.
We also tested if overexpressing Msh2-Msh6 alters the distribution of cells in the cell cycle. As shown in Figure 5B, we found that the percentage of unbudded cells (G1) was significantly higher (P = 0.002, Fisher’s exact test) when Msh2-Msh6 was cooverexpressed (21%, N = 423) compared to wild-type strains containing an empty vector (13%, N = 596), and the percentage of small-budded cells was significantly lower (P = 0.0001, Fisher’s exact test) when Msh2-Msh6 was overexpressed (19%, N = 423) compared to wild-type strains (30%, N = 596). The percentage of large-budded cells remained relatively unchanged between the two strains. Interestingly, HU has been known to synchronize mammalian cells at G1/S phase (Rosner et al. 2013). We saw a similar effect in cells overexpressing Msh2-Msh6, which seem to have difficulty in transitioning from G1 (unbudded) to S (small-budded) phase. Consistent with this effect involving a replication fork defect, overexpressing Msh2-msh6-KQFF > AAAA restored the percentage of small-budded cells to wild-type levels (28%, N = 764); however, there were also changes in the proportion of cells in other stages.
Cooverexpressing Msh2 and Msh6 causes an increase in the rate of LOH
As shown above, overexpression of the Msh2-Msh6 complex conferred higher recombination rates and increased sensitivity to MMS, HU, and 4-NQO. These observations suggested that overexpression of Msh2-Msh6 might increase the rate of other types of genome instability, such as LOH. We utilized an assay involving isogenic strains, which contain a single hemizygous copy of the CORE2 counterselectable cassette with two diverged orthologous copies of the URA3 gene and a marker for geneticin resistance, at chromosome 7. Spontaneous mitotic interhomolog allelic recombination events result in LOH of the CORE2 cassette (Conover et al. 2015; Figure 6A). Since we select for homozygosity of the homolog lacking the CORE2 insertion, the recombination rates measured represent half of the LOH events that occur in the region. Overexpression of Msh2 or Msh6, or cooverexpression of Mlh1 and Pms1, did not change the rate of LOH in comparison with wild-type strains. However, cooverexpression of Msh2 and Msh6 increased the rate of LOH by > 2.8-fold (P = 0.001, Mann–Whitney U-test; Figure 6B). This observation is consistent with the genome instability phenotypes associated with elevated levels of Msh2-Msh6. LOH is often a common genetic event in cancer development and, thus, our data suggest that cooverexpression of Msh2-Msh6 can promote genomic instability. It is important to note that Conover et al. (2015) provided evidence that the LOH events in their assay occurred almost exclusively from interhomolog recombination rather than chromosome loss. However, the genome instability phenotypes conferred by Msh2 and Msh6 overexpression do not exclude the possibility that all or part of the increase in LOH rate seen in Msh2 and Msh6 overexpression strains is due to chromosome loss.
Figure 6.
Effect of overexpression of mismatch repair proteins on loss-of-heterozygosity (LOH). (A) Assay to measure LOH in isogenic diploids [adapted from Conover et al. (2015)]. The strain is hemizygous for the counterselectable CORE2 cassette present on one homolog of chromosome 7, as shown. The CORE2 cassette can be lost as a result of recombination between the two homologs of chromosome 7, resulting in LOH in the region. The resulting recombinants are resistant to 5-FOA and sensitive to geneticin. (B) LOH rates are shown for the JAY1201 strain containing 2μ plasmids that encode the indicated genes. The wild-type strain contains the pRS424 plasmid. For 14–22 samples, 95% C.I.s are shown in parentheses. Pairwise Mann–Whitney U-tests were performed between the mutant and wild-type strains. Differences were considered significant when P < 0.05. **Significantly different from wild-type (P < 0.01, Mann–Whitney U-test).
Cooverexpression of Msh2 and Msh6 causes a mutator phenotype
The above observations suggested that overexpression of Msh2-Msh6 would likely confer a mutator phenotype. We tested this using a lys2-A14 reversion assay (Heck et al. 2006). As shown in Table 2, cooverexpression of Msh2 and Msh6 increased the rate of reversion to Lys+ by 44-fold, approaching the level seen in msh6Δ strains (59-fold higher than wild-type). Cooverexpression of Msh2 and msh6-KQFF > AAAA did not confer a mutator phenotype (Table 2), indicating that overexpression of Msh2-Msh6 causes a mutator phenotype as a consequence of its interaction with PCNA. Overexpression of Msh2 had no effect on the reversion rate (Table 2), whereas overexpression of Msh6 caused an 11-fold increase (Chakraborty et al. 2016; Table 2). Overexpression of other MMR proteins, such as Msh3 or the Mlh1-Pms1 complex, did not affect the lys2-A14 reversion rate. Also, overexpression of PCNA did not have any effect despite the fact that such overexpression conferred sensitivity to MMS and an increase in HR (Figure 5 and Tables 1 and 2).
Table 2. Mutation rates using lys2-A14 reversion assay.
| Strain | Reversion rate (×10−7) (95% C.I.) | Normalized |
|---|---|---|
| 2μ plasmids | ||
| wild-type + pRS424a | 9.9 (5.8–67) | 1 |
| wild-type + MSH6-2µa | 110 (31–647)b | 11.1 |
| wild-type + 2μ-msh6-KQFF > AAAAa | 92.5 (67.6–149)b | 9.3 |
| msh6Δa | 585 (207–2030)b | 59.1 |
| msh3Δa | 37.7 (22.3–49.4)c | 3.8 |
| wild-type + 2μ-MSH2 | 9.2 (7.2–13.8) | 0.93 |
| wild-type + 2μ-MSH3 | 8.3 (4.8–25.0) | 0.84 |
| wild-type + 2μ-MSH2-MSH6 | 433 (301–537)b | 44 |
| wild-type + 2μ-MLH1-PMS1 | 13.3 (10.0–25.4) | 1.3 |
| wild-type + 2μ-POL30 | 17.4 (10.4–21.3) | 1.8 |
| wild-type + pRS426 | 7.3 (4.9–18.4) | 1 |
| wild-type + 2μ-MSH2-msh6-KQFF > AAAA | 12.6 (8.2–21.2) | 1.7 |
| ARS-CEN plasmids | ||
| wild-type + pRS316, pRS414 | 9.0 (6.9–17.5) | 1 |
| wild-type + MSH2, pRS414 | 13.3 (8.2–16.0) | 1.5 |
| wild-type + MSH6, pRS316 | 15.7 (10.9–18.3) | 1.7 |
| wild-type + MSH2, MSH6 | 16.0 (11.5–21.8)c | 1.8 |
FY23-derived strains were analyzed for lys2-A14 reversion as described in the Materials and Methods and Table S2. Rates are presented per cell per division. Numbers in parentheses indicate 95% C.I.s. Rates and 95% C.I.s were calculated from 10 to 22 independent cultures.
Published data from Chakraborty et al. (2016).
Significantly different from wild-type using Mann–Whitney U-test (P < 0.01).
Significantly different from wild-type using Mann–Whitney U-test (P < 0.05).
To determine if more modest overexpression of Msh2-Msh6 caused genome instability, we tested if introducing an additional copy of the MSH2 and/or MSH6 genes expressed on an ARS-CEN plasmid would impact mutation rate in a wild-type strain background. The wild-type strain expressing an additional copy of both MSH2 and MSH6 showed a modest increase in mutation rate compared to the wild-type strain containing empty vectors [1.8-fold (P < 0.05); Table 2] and such strains showed a subtle increase in Msh2 and Msh6 protein levels as detected by western blotting (Figure S4). Together, these data provide further support for our hypothesis that elevated levels of Msh2-Msh6 promote genome instability.
Discussion
Overexpression of MutSα likely interferes with the replication fork function
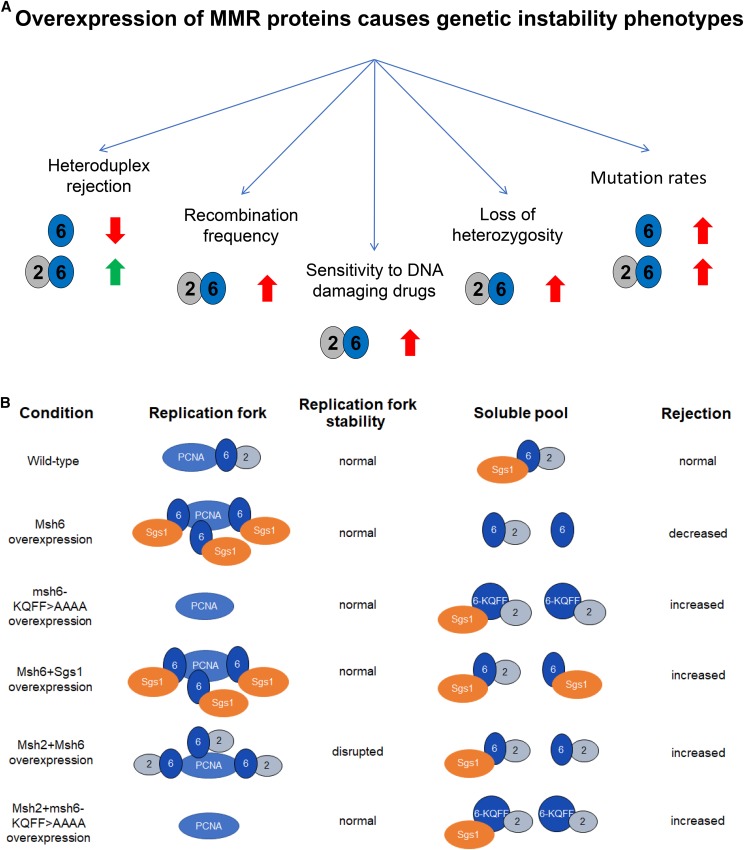
We showed that overexpression of the Msh6 protein disrupted mechanisms that prevent recombination between divergent DNA sequences. This disruption appeared to be due to the sequestration of Sgs1 through Sgs1-Msh6-PCNA interactions. In contrast, overexpression of both Msh2 and Msh6 improved heteroduplex rejection; however, this overexpression increased genome instability in steps dependent on Msh2-Msh6-PCNA interactions. The genome instability defects included increased rates of HR and LOH, increased sensitivity to DNA-damaging drugs such as MMS, HU, and 4-NQO, an increased proportion of unbudded cells in an unsynchronized midlog-phase population, and increased mutation rates. The effects of overexpressing Msh6 or cooverexpressing Msh2-Msh6 on various genome stability pathways are summarized in Figure 7A. It is important to note that Msh3 overexpression did not confer similar genome instability phenotypes, as shown in mutator and MMS sensitivity assays.
Figure 7.
Model of Msh6 and MutSα overexpression-mediated genome instability. (A) Summary of outcomes resulting from the overexpression of mismatch repair (MMR) proteins. Upward or downward block arrows indicate an increase or decrease, respectively, in the indicated phenotype. The green and red colors of the block arrows indicate if the change in phenotype is likely to be beneficial (green) or deleterious (red) for the cell. (B) A model to explain how overexpression of MSH proteins changes the distribution of MMR and other interacting proteins in the cell to impact rejection and replication fork stability. In wild-type cells, a fraction of the Msh2-Msh6 complex interacts with the replication fork via PCNA and the remaining Msh2-Msh6 exists as a soluble fraction, which is responsible for heteroduplex rejection (Hombauer et al. 2011). Upon Msh6 overexpression, an excess of Msh6 protein associates with PCNA at the replication fork and sequesters Sgs1, making it unavailable for rejection. Overexpressing msh6-KQFF > AAAA does not deplete the soluble pool of Sgs1 since msh6-KQFF > AAAA does not interact with PCNA, thereby improving rejection due to an increased availability of MSH proteins and the Msh6-PCNA interaction being dispensable for rejection. Cooverexpressing Msh6 and Sgs1 restores Sgs1 levels in the soluble pool of proteins and thus shows an increased rejection efficiency due to an increased availability of rejection proteins. Cooverexpression of Msh2 and Msh6 does not sequester Sgs1 in the same way that overexpressing Msh6 alone does, and hence results in an increase of the Msh2-Msh6 complex in both the replication fork pool and soluble pool. Increased levels of Msh2-Msh6 in the soluble pool allows for increased rejection. However, increased Msh2-Msh6 at the replication fork destabilizes proper fork function and results in hyperrecombination, loss-of-heterozygosity, increased sensitivity to MMS, HU, and 4-NQO, and a mutator phenotype, possibly by blocking other critical proteins from efficiently interacting with PCNA at the replication fork. Cooverexpressing Msh2 and msh6-KQFF > AAAA instead relieves the fork destabilization phenotype and is also proficient in heteroduplex rejection.
How does Msh2-Msh6 overexpression cause genome instability phenotypes? The finding that Msh6-PCNA interactions were critical for the vast majority of genome instability phenotypes leads us to three possible mechanisms. First, elevated levels of Msh2-Msh6 saturate PCNA-binding sites and prevent PCNA from interacting with other critical repair pathways. In support of this idea, PCNA and PCNA modifications (e.g., ubiquitination) have been shown to be directly involved in a variety of DNA repair mechanisms that are involved in repairing lesions resulting from MMS- and 4-NQO-induced DNA damage and HU-induced fork stalling; these include template switching, translesion bypass, HR, base excision repair, and nucleotide excision repair (Ikenaga et al. 1975; Ishii and Kondo 1975; Xiao et al. 1996; Fasullo and Sun 2017). Similar to Msh2-Msh6, many translesion polymerases interact with PCNA via the PCNA Interaction Protein motif (PIP) box motif that was mutated in the msh6-KQFF > AAAA mutation. Additionally, PCNA interacts with a host of factors involved in base excision repair and nucleotide excision repair (Moldovan et al. 2007; Dou et al. 2008; Naryzhny 2008; Burkovics et al. 2009; Strzalka and Ziemienowicz 2011). Thus, it is easy to imagine that overexpression of Msh2-Msh6 interferes with the ubiquitination of PCNA, polymerase switching, or interactions with base excision and nucleotide excision repair factors, resulting in the genome instability phenotypes observed. A second possibility is that interactions between Msh2-Msh6 and PCNA directly inhibit the replication machinery (e.g., inhibit interactions with clamp loader-specific DNA polymerases), leading to replication stress and/or error prone repair. A third explanation is that overloading PCNA with Msh2-Msh6 interferes with the activation of the intra-S-phase checkpoint, thus resulting in sensitivity to DNA-damaging drugs. It is likely that cooverexpression of Msh2 and Msh6 interferes with replication fork function in more than one way, given that PCNA has many interactors and plays a role in many pathways.
Overexpression of Msh2-Msh6 resulted in a 44-fold increase in the rate of lys2-A14 reversion, approaching levels seen in a msh6Δ mutant (59-fold). Furthermore, this increase was almost entirely dependent on Msh2-Msh6-PCNA interactions. The mutator phenotype observed in strains overexpressing Msh2-Msh6 could result from disruption in the coordination of Msh2-Msh6 with the replication fork or the sequestration of critical downstream MMR proteins. Another possibility is that Msh2-Msh6 overexpression impairs replication functions, which is consistent with the pleiotropic genome instability phenotypes that were observed.
It is important to note that overexpression of Msh6 improved heteroduplex rejection in a single-strand annealing recombination assay that does not appear to be linked to DNA replication (Chakraborty et al. 2016). In this situation, Msh6 overexpression sequestered Msh2 away from Msh3, thus preventing Msh2-Msh3 from initiating commitment steps to the single-strand annealing pathway. These observations provide evidence that MSH protein expression can affect recombination events in mechanisms independent of the replication fork.
Sgs1 interacts with Top3 and Rmi1 in a variety of DNA transactions including 5′ to 3′ strand resection during HR, dissolution of double Holliday junctions, and heteroduplex rejection, and Sgs1 and Top3-Rmi1 have interdependent and independent functions in meiosis [reviewed in Chakraborty et al. (2016)]. Why does overexpression of Sgs1 without overexpression of Top3-Rmi1 suppress the defect in heteroduplex rejection resulting from Msh6 overexpression? One possibility is that Msh6 and Sgs1 directly interact and that overexpression of Msh6 sequesters Sgs1 in a nonfunctional complex that prevents it from interacting with Top3-Rmi1. Alternatively, Sgs1 could be the limiting component of the Sgs1-Top3-Rmi1 complex and thus overexpression of Sgs1 is sufficient to confer a phenotype that requires the functional Sgs1-Top3-Rmi1 complex. Unfortunately, we did not test these possibilities because of challenges in obtaining yeast transformants that cooverexpress Sgs1, Top3, and Rmi1.
Another unresolved question is why Sgs1 would be sequestered by overexpressing Msh6, but not by cooverexpressing Msh2 and Msh6? One possibility is that the strength/dynamics of the interaction between PCNA, Msh2-Msh6, and other DNA repair and replication factors are different when functional complexes vs. single subunits (e.g., Msh6) are overexpressed. For example, Sgs1 may interact more dynamically with a Msh2-Msh6-PCNA complex compared to a Msh6-PCNA complex. In the former case, such a dynamic interaction would allow Sgs1 to displace PCNA and participate in heteroduplex rejection. Alternatively, Sgs1 is occluded from interaction with a PCNA-Msh2-Msh6 complex but such a block does not exist with a PCNA-Msh6 complex.
Changing the relative cellular distribution of MSH proteins between chromatin-bound and soluble pools can affect different genome stability pathways
In yeast, Msh2-Msh6 appears to colocalize with replication centers irrespective of the presence of mispaired bases and is thought to move with the replication fork via its interaction with PCNA (Kleczkowska et al. 2001; Hombauer et al. 2011; Haye and Gammie 2015). Curiously, the replication fork-localized pool of Msh2-Msh6 proteins accounts for only 10–15% of Msh2-Msh6-dependent MMR events (Hombauer et al. 2011). This observation indicates that two pools of Msh2-Msh6 exist in the cell during the replication phase of the cell cycle: one that interacts with PCNA and moves with the replication fork, and a second unassociated/soluble pool. Our data suggest that overexpression of MSH proteins is likely to change the distribution in the two pools. In the case of Msh6 overexpression, our data are consistent with a higher level of Msh6 accumulating at replication forks, where it sequesters Sgs1 from acting in heteroduplex rejection (Figure 7B). In the case of Msh2-Msh6, overexpression of this complex causes genome instability defects that are consistent with its overloading at replication forks (see above, Figure 7B). This observation could provide a possible explanation for why yeast cells, despite likely benefiting from Msh2-Msh6 linkage to the replication fork with respect to increased efficiency of mismatch detection, have evolved such that only a small fraction of MSH protein associates with the replication fork.
Implications for understanding cancer progression
In higher eukaryotes, increases in the rate of recombination, LOH, and mutation rate can lead to genetic alterations such as genomic rearrangements that can play primary roles in carcinogenesis [reviewed in Bishop and Schiestl (2002) and Fox et al. (2013)]. Multiple studies have linked overexpression of MMR proteins to deleterious cancer outcomes (listed in Table S1):
In a study of 11,152 prostate cancer specimens, Wilczak et al. (2017) observed that MSH6, MLH1, and PMS2 expression were high in cancers with advanced pathological tumor stage, high Gleason grade, nodal metastasis, and early biochemical recurrence. They also found that high levels of MMR gene expression paralleled features of genetic instability, such as the number of genomic deletions per cancer.
Wagner et al. (2016) showed, in an analysis of 115 oral squamous cell carcinomas, that MSH6 and MSH2-MSH6 overexpression were associated with poor survival rates.
In a study involving 33 tumors from prostate cancer patients, Norris et al. (2007) showed that tumors with elevated levels of PMS2 caused MSI, and that this MSI phenotype was corrected by increasing expression of MLH1.
Velasco et al. (2002) showed that MSI was detected in 26% of prostate carcinoma specimens with high levels of MSH2 in a study with 101 prostate cancer specimens.
PCNA overexpression has been observed in many cancers (Table S1), and we found that overexpression of PCNA conferred moderate sensitivity to MMS (Figure 5) and increased the rate of HR by fivefold compared to wild-type strains that that did not overexpress PCNA (Table 1).
The above examples indicate that overexpression of certain MMR proteins is associated with deleterious outcomes and phenotypes in a variety of cancers. These studies, in conjunction with our data, suggest that high levels of certain MMR proteins can confer genomic instability.
Although several studies have observed correlations between increased MMR protein levels and genomic instabilities, a cause–effect relationship has not been established (Velasco et al. 2002; Norris et al. 2007; Wilczak et al. 2017). A current model in the field is that certain MMR proteins are upregulated in cancers in response to the higher proliferation rates in cells that have an increased need to correct mismatches, which would presumably arise at higher rates (Wilson et al. 1995; Leach et al. 1996; Marra et al. 1996; Chang et al. 2000; Hamid et al. 2002). In fact, a recent study analyzing the levels of different MSH proteins in mice showed that different tissues show wide variability in the expression of these proteins (Tomé et al. 2013). The authors found that most tissues had higher levels of MSH3 than MSH6, except proliferative tissues where MSH6 protein levels were higher than MSH3. Measurements of steady-state levels of MMR proteins in various MMR-proficient human tumor cell lines showed that hMSH6 protein expression was 4–12 times higher than hMSH3 (Drummond et al. 1997; Chang et al. 2000). Additionally, viral transformation of primary fibroblast cells resulted in increased cell proliferation, chromosome instability, and expression levels of MMR proteins. These data indicate that increased rates of proliferation likely upregulate MMR proteins. However, in normal cells, genome stability is likely maintained by balancing the levels of other proteins in the cell to offset possible deleterious effects of increased MMR proteins.
While cell proliferation may be partially responsible for increased MMR expression in cancer cells, we believe that it is not the only reason for why MMR overexpression is observed in cancers. We hypothesize that overexpression of MMR components, specifically MSH6 and/or MSH2-MSH6, can result from increased proliferation rates in cancer cells as well as from independent mutations, copy number variations, and epigenetic changes that arise prior to or after cellular transformation. We further hypothesize that such overexpression causes genomic instabilities and can potentially act as “drivers” that contribute to the development/progress of cancers. We present the following arguments in support of this idea:
Chang et al. (2000) observed that transformation of lung fibroblasts with simian virus 40 led to an increased proliferation rate and resulted in increased levels of MSH2, MSH3, and MSH6 proteins. This suggested that increased proliferation causes upregulation of all three MSH components. Curiously, as shown in Figure 1 and Figure S1, when we compared gene expression of MSH2, MSH3, and MSH6 across a large number of samples from TCGA and the GENT web database (Shin et al. 2011), several cancer tissues showed overexpression of MSH2 and MSH6 genes but not the MSH3 gene when compared to their normal tissue counterparts. MSH2 and MSH6 showed a similar pattern of overexpression across different cancer tissues, unlike MSH3, indicating that the overexpression of MSH2 and/or MSH6 may have arisen independently in many of these cancers, and not just as a result of increased proliferation, since the latter would likely have resulted in the overexpression of MSH3 as well.
Analysis of genomic alterations in MSH2, MSH6, and MSH3 using data available in cBioPortal shows that these genes often undergo copy number amplifications that result in higher levels of expression of these genes (Figure 2).
Rass et al. (2001) found, in a study of melanocytic tumors, that overexpression of MSH2 was not correlated with the proliferation marker Ki67, indicating that the high MSH2 levels may not have arisen as a result of increased proliferation.
Our data show that overexpressing Msh6 or Msh2-Msh6 in baker’s yeast cause several genome instability phenotypes that appear similar to those observed in cancers.
Thus, it is likely that genomic and epigenomic alterations in cancers further increase MMR protein expression levels beyond the increase due to proliferation. For example, Figure 2B shows that DNA copy number amplifications resulted in increased mRNA levels beyond the normal diploid levels found in cancers.
The genome instability phenotypes in yeast overexpressing Msh6 can provide an explanation for the observation that overexpression of MSH6 correlated to an increase in genomic deletions in prostate cancer specimens (Wilczak et al. 2017). Unfortunately, the authors did not test if MSH2 was also overexpressed in those cancer specimens, and whether the cooverexpression of MSH2 and MSH6 also correlated to increases in the number of genomic deletions. Our data showing that overexpression of Msh2-Msh6 in yeast causes a mutator phenotype provide a possible explanation for why Velasco et al. (2002) observed a significant fraction of cancers overexpressing MSH2 displaying an MSI phenotype. Again, the authors did not test if MSH6 was overexpressed as well. However, a number of studies suggest that MSH2 and MSH6 mRNA and protein expression in cancers are correlated to each other (Vageli et al. 2013; Wagner et al. 2016), as we found in certain tumors overexpressing both MSH2 and MSH6 (Figure S2). Additionally, in their study of 115 oral squamous cell carcinoma samples, Wagner et al. (2016) showed that many of these cancers exhibited overexpression of MSH2 or MSH6 alone, and a significant fraction showed cooverexpression of MSH2 and MSH6. They also showed that MSH6 or MutSα overexpression was associated with poor survival rates. Our data in yeast, showing distinct deleterious genomic instabilities caused by overexpression of Msh6 or the MutSα complex, provides an explanation for why such cancers show increased expression of Msh6 alone or the Msh2-Msh6 complex, and how they could be associated with specific deleterious outcomes.
Recent studies have implicated MMR deficiency as being critical for the effective immunotherapeutic treatment of cancers by immune checkpoint blockade using anti-PD-1 antibodies (Le et al. 2015, 2017). The effectiveness of this approach relies on the fact that MMR-deficient cancers produce a large number of mutant neoantigens that can be recognized by the immune system, which make them sensitive to immune checkpoint blockade. It is interesting to speculate that cancers overexpressing MSH2-MSH6 may also be sensitive to immune checkpoint blockade, as they would likely have a high load of neoantigens due to the high rate of mutations and genetic instability associated with increased MSH2-MSH6 expression. Taken together, our data suggest that overexpression of MSH6 or MSH2-MSH6 destabilize the genome and may contribute to cancer formation or progression in higher eukaryotes, and that such overexpression may also provide novel therapeutic approaches.
Acknowledgments
We thank Praveen Sethupathy, Lucas Argueso, Robert Weiss, Mike Fasullo, Sue Jinks-Robertson, Linda Nicholson, and the Alani laboratory for fruitful discussions, analyses, and reagents. U.C. and E.A. were supported by National Institutes of Health (NIH) grant GM53085. T.A.D. is supported by a grant from the Fibrolamellar Cancer Foundation awarded to Praveen Sethupathy (Department of Biomedical Sciences, Cornell University). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the NIH.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5991208.
Communicating editor: D. Bishop
Literature Cited
- Beranek D. T., 1990. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 231: 11–30. 10.1016/0027-5107(90)90173-2 [DOI] [PubMed] [Google Scholar]
- Bishop A. J. R., Schiestl R. H., 2002. Homologous recombination and its role in carcinogenesis. J. Biomed. Biotechnol. 2: 75–85. 10.1155/S1110724302204052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen N., Kolodner R. D., 2017. Reconstitution of Saccharomyces cerevisiae DNA polymerase ε-dependent mismatch repair with purified proteins. Proc. Natl. Acad. Sci. USA 114: 3607–3612. 10.1073/pnas.1701753114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Burkovics P., Hajdú I., Szukacsov V., Unk I., Haracska L., 2009. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′–5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 37: 4247–4255. 10.1093/nar/gkp357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., et al. , 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., et al. , 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. 10.1016/S0092-8674(03)00075-8 [DOI] [PubMed] [Google Scholar]
- Chahwan R., Edelmann W., Scharff M. D., Roa S., 2011. Mismatch-mediated error prone repair at the immunoglobulin genes. Biomed. Pharmacother. 65: 529–536. 10.1016/j.biopha.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty U., Alani E., 2016. Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Res. 16: fow071 10.1093/femsyr/fow071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty U., George C. M., Lyndaker A. M., Alani E., 2016. A delicate balance between repair and replication factors regulates recombination between divergent DNA sequences in Saccharomyces cerevisiae. Genetics 202: 525–540. 10.1534/genetics.115.184093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. K., Ricciardiello L., Goel A., Chang C. L., Boland C. R., 2000. Steady-state regulation of the human DNA mismatch repair system. J. Biol. Chem. 275: 18424–18431. 10.1074/jbc.M001140200 [DOI] [PubMed] [Google Scholar]
- Chen W., Jinks-Robertson S., 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18: 6525–6537. 10.1128/MCB.18.11.6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jinks-Robertson S., 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A., 2000. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J. Biol. Chem. 275: 36498–36501. 10.1074/jbc.C000513200 [DOI] [PubMed] [Google Scholar]
- Conover H. N., Lujan S. A., Chapman M. J., Cornelio D. A., Sharif R., et al. , 2015. Stimulation of chromosomal rearrangements by ribonucleotides. Genetics 201: 951–961. 10.1534/genetics.115.181149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Adjiri A., New L., Crouse G. F., Jinks Robertson S., 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 1085–1093. 10.1128/MCB.16.3.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., et al. , 2008. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen: the potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 283: 3130–3140. 10.1074/jbc.M709186200 [DOI] [PubMed] [Google Scholar]
- Drummond J. T., Genschel J., Wolf E., Modrich P., 1997. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc. Natl. Acad. Sci. USA 94: 10144–10149. 10.1073/pnas.94.19.10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M. T., Sun M., 2017. Both RAD5-dependent and independent pathways are involved in DNA damage-associated sister chromatid exchange in budding yeast. AIMS Genet. 4: 84–102. 10.3934/genet.2017.2.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E. J., Prindle M. J., Loeb L. A., 2013. Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev. 32: 353–361. 10.1007/s10555-013-9426-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., et al. , 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C. M., Alani E., 2012. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit. Rev. Biochem. Mol. Biol. 47: 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. L., Narayanan L., Hegan D. C., Buermeyer A. B., Liskay R. M., et al. , 2006. Overexpression of the DNA mismatch repair factor, PMS2, confers hypermutability and DNA damage tolerance. Cancer Lett. 244: 195–202. 10.1016/j.canlet.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7: 253–263. 10.1002/yea.320070307 [DOI] [PubMed] [Google Scholar]
- Goldfarb T., Alani E., 2005. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics 169: 563–574. 10.1534/genetics.104.035204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. M., Ma C. X., Liang P., Singh K. K., 2009. Fluctuation analysis calculator: a web tool for the determination of mutation rate using Luria–Delbrück fluctuation analysis. Bioinformatics 25: 1564–1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid A. A., Mandai M., Konishi I., Nanbu K., Tsuruta Y., et al. , 2002. Cyclical change of hMSH2 protein expression in normal endometrium during the menstrual cycle and its overexpression in endometrial hyperplasia and sporadic endometrial carcinoma. Cancer 94: 997–1005. 10.1002/cncr.10341 [DOI] [PubMed] [Google Scholar]
- Haye J. E., Gammie A. E., 2015. The eukaryotic mismatch recognition complexes track with the replisome during DNA synthesis. PLoS Genet. 11: e1005719 10.1371/journal.pgen.1005719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. A., Argueso J. L., Gemici Z., Reeves R. G., Bernard A., et al. , 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261. 10.1073/pnas.0510998103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., 2011. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair Intermediates. Cell 147: 1040–1053. 10.1016/j.cell.2011.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert O., Hermine T., Hernandez H., Bouget T., Selves J., et al. , 2002. Implication of protein kinase C in the regulation of DNA mismatch repair protein expression and function. J. Biol. Chem. 277: 18061–18068. 10.1074/jbc.M103451200 [DOI] [PubMed] [Google Scholar]
- Ikenaga M., Ishii Y., Tada M., Kakunaga T., Takebe H., 1975. Excision-repair of 4-nitroquinolin-1-oxide damage responsible for killing, mutation, and cancer. Basic Life Sci. 5B: 763–771. [DOI] [PubMed] [Google Scholar]
- Ishii Y., Kondo S., 1975. Comparative analysis of deletion and base-change mutabilities of Escherichia coli B strains differing in DNA repair capacity (wild-type, uvrA-, polA-, recA-) by various mutagens. Mutat. Res. 27: 27–44. 10.1016/0027-5107(75)90271-7 [DOI] [PubMed] [Google Scholar]
- Iyer D., Rhind N., 2017. The intra-S checkpoint responses to DNA damage. Genes 8: 74 10.1146/annurev-biochem-060614-034010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R. R., Pluciennik A., Napierala M., Wells R. D., 2015. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 84: 199–226. 10.1146/annurev-biochem-060614-034010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Gali V. K., Takahashi T. S., Kubota T., 2016. PCNA retention on DNA into G2/M phase causes genome instability in cells lacking Elg1. Cell Rep. 16: 684–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansikas M., Kasela M., Kantelinen J., Nyström M., 2014. Assessing how reduced expression levels of the mismatch repair genes MLH1, MSH2, and MSH6 affect repair efficiency. Hum. Mutat. 35: 1123–1127. 10.1002/humu.22605 [DOI] [PubMed] [Google Scholar]
- Kauffmann A., Rosselli F., Lazar V., Winnepenninckx V., Mansuet-Lupo A., et al. , 2007. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 27: 565–573. 10.1038/sj.onc.1210700 [DOI] [PubMed] [Google Scholar]
- Kleczkowska H. E., Marra G., Lettieri T., Jiricny J., 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15: 724–736. 10.1101/gad.191201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koç A., Wheeler L. J., Mathews C. K., Merrill G. F., 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279: 223–230. 10.1074/jbc.M303952200 [DOI] [PubMed] [Google Scholar]
- Kohda K., Kawazoe Y., Minoura Y., Tada M., 1991. Separation and identification of N4- (guanosin-7-yl)-4-aminoquinoline 1-oxide, a novel nucleic acid adduct of carcinogen 4-nitroquinoline 1-oxide. Carcinogenesis 12: 1523–1525. 10.1093/carcin/12.8.1523 [DOI] [PubMed] [Google Scholar]
- Kumar C., Piacente S. C., Sibert J., Bukata A. R., O’Connor J., et al. , 2011. Multiple factors insulate Msh2–Msh6 mismatch repair activity from defects in Msh2 domain I. J. Mol. Biol. 411: 765–780. 10.1016/j.jmb.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710. 10.1146/annurev.biochem.74.082803.133243 [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2015. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49: 291–313. 10.1146/annurev-genet-112414-054722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Uram J. N., Wang H., Bartlett B. R., Kemberling H., et al. , 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372: 2509–2520. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D. T., Durham J. M., Smith K. N., Wang H., Bartlett B. R., et al. , 2017. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285. 10.1007/BF02986080 [DOI] [PubMed] [Google Scholar]
- Leach F. S., Polyak K., Burrell M., Johnson K. A., Hill D., et al. , 1996. Expression of the human mismatch repair gene hMSH2 in normal and neoplastic tissues. Cancer Res. 56: 235–240. [PubMed] [Google Scholar]
- Lee S. D., Surtees J. A., Alani E., 2007. Saccharomyces cerevisiae MSH2–MSH3 and MSH2–MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J. Mol. Biol. 366: 53–66. 10.1016/j.jmb.2006.10.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. M., 2008. Mechanisms and functions of DNA mismatch repair. Cell Res. 18: 85–98. 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- Li M., Liu L., Wang Z., Wang L., Liu Z., et al. , 2008. Overexpression of hMSH2 and hMLH1 protein in certain gastric cancers and their surrounding mucosae. Oncol. Rep. 19: 401–406. [PubMed] [Google Scholar]
- Liu D., Keijzers G., Rasmussen L. J., 2017. DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. 773: 174–187. 10.1016/j.mrrev.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Liu P., Carvalho C. M. B., Hastings P. J., Lupski J. R., 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22: 211–220. 10.1016/j.gde.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra G., Chang C. L., Laghi L. A., Chauhan D. P., Young D., et al. , 1996. Expression of human MutS homolog 2 (hMSH2) protein in resting and proliferating cells. Oncogene 13: 2189–2196. [PubMed] [Google Scholar]
- Marra G., Iaccarino I., Lettieri T., Roscilli G., Delmastro P., et al. , 1998. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl. Acad. Sci. USA 95: 8568–8573. 10.1073/pnas.95.15.8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. A., 1970. Carcinogenesis by chemicals: an overview—G. H. A. Clowes memorial lecture. Cancer Res. 30: 559–576. [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S., 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679. 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R. D., 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. 10.1038/83673 [DOI] [PubMed] [Google Scholar]
- Naryzhny S. N., 2008. Proliferating cell nuclear antigen: a proteomics view. Cell. Mol. Life Sci. 65: 3789–3808. 10.1007/s00018-008-8305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A., Hendrix M., Jinks-Robertson S., Crouse G. F., 2000. Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics 154: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. M., Woodruff R. D., D’Agostino R. B., Clodfelter J. E., Scarpinato K. D., 2007. Elevated levels of the mismatch repair protein PMS2 are associated with prostate cancer. Prostate 67: 214–225. 10.1002/pros.20522 [DOI] [PubMed] [Google Scholar]
- Peña-Diaz J., Jiricny J., 2012. Mammalian mismatch repair: error-free or error-prone? Trends Biochem. Sci. 37: 206–214. 10.1016/j.tibs.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Prakash L., Prakash S., 1977. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86: 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass K., Gutwein P., Welter C., Meineke V., Tilgen W., et al. , 2001. DNA mismatch repair enzyme hMSH2 in malignant melanoma: increased immunoreactivity as compared to acquired melanocytic nevi and strong mRNA expression in melanoma cell lines. Histochem. J. 33: 459–467. 10.1023/A:1014472314354 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rosner M., Schipany K., Hengstschläger M., 2013. Merging high-quality biochemical fractionation with a refined flow cytometry approach to monitor nucleocytoplasmic protein expression throughout the unperturbed mammalian cell cycle. Nat. Protoc. 8: 602–626. 10.1038/nprot.2013.011 [DOI] [PubMed] [Google Scholar]
- Shcherbakova P. V., Hall M. C., Lewis M. S., Bennett S. E., Martin K. J., et al. , 2001. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol. Cell. Biol. 21: 940–951. 10.1128/MCB.21.3.940-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin G., Kang T. W., Yang S., Baek S. J., Jeong Y. S., et al. , 2011. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 10: 149–157. 10.4137/CIN.S7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M. L., 1973. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J. Bacteriol. 113: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell R. M., Jinks-Robertson S., 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168: 1855–1865. 10.1534/genetics.104.032771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. E., Ozbirn R. G., Petes T. D., Jinks-Robertson S., 2008. Role of proliferating cell nuclear antigen interactions in the mismatch repair-dependent processing of mitotic and meiotic recombination intermediates in yeast. Genetics 178: 1221–1236. 10.1534/genetics.107.085415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzalka W., Ziemienowicz A., 2011. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 107: 1127–1140. 10.1093/aob/mcq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B., Quach T., Alani E., 1998. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 18: 7590–7601. 10.1128/MCB.18.12.7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J. E., 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. 10.1073/pnas.0305749101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé S., Simard J. P., Slean M. M., Holt I., Morris G. E., et al. , 2013. Tissue-specific mismatch repair protein expression: MSH3 is higher than MSH6 in multiple mouse tissues. DNA Repair (Amst.) 12: 46–52. 10.1016/j.dnarep.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Tran H. T., Keen J. D., Kricker M., Resnick M. A., Gordenin D. A., 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865. 10.1128/MCB.17.5.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vageli D. P., Giannopoulos S., Doukas S. G., Kalaitzis C., Giannakopoulos S., et al. , 2013. Mismatch repair hMSH2, hMLH1, hMSH6 and hPMS2 mRNA expression profiles in precancerous and cancerous urothelium. Oncol. Lett. 5: 283–294. 10.3892/ol.2012.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco A., Albert P. S., Rosenberg H., Martinez C., Leach F. S., 2002. Clinicopathologic implications of hMSH2 gene expression and microsatellite instability in prostate cancer. Cancer Biol. Ther. 1: 362–367. 10.4161/cbt.1.4.7 [DOI] [PubMed] [Google Scholar]
- Wagner V. P., Webber L. P., Salvadori G., Meurer L., Fonseca F. P., et al. , 2016. Overexpression of MutSα complex proteins predicts poor prognosis in oral squamous cell carcinoma. Medicine (Baltimore) 95: e3725 10.1097/MD.0000000000003725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczak W., Rashed S., Hube-Magg C., Kluth M., Simon R., et al. , 2017. Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. Carcinogenesis 38: 19–27. 10.1093/carcin/bgw116 [DOI] [PubMed] [Google Scholar]
- Wilson T. M., Ewel A., Duguid J. R., Eble J. N., Lescoe M. K., et al. , 1995. Differential cellular expression of the human MSH2 repair enzyme in small and large intestine. Cancer Res. 55: 5146–5150. [PubMed] [Google Scholar]
- Xiao W., Chow B. L., Rathgeber L., 1996. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 30: 461–468. 10.1007/s002940050157 [DOI] [PubMed] [Google Scholar]
- Zhang C. Z., Leibowitz M. L., Pellman D., 2013. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 27: 2513–2530. 10.1101/gad.229559.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Xiang S., Joo H. Y., Wang L., Williams K. A., et al. , 2014. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol. Cell 55: 31–46. 10.1016/j.molcel.2014.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Hu C., Tong D., Xiang S., Williams K., et al. , 2016. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage. J. Biol. Chem. 291: 10783–10791. 10.1074/jbc.M115.700047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. N., Usdin K., 2015. The repeat expansion diseases: the dark side of DNA repair. DNA Repair (Amst.) 32: 96–105. 10.1016/j.dnarep.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. N., Lokanga R., Allette K., Gazy I., Wu D., et al. , 2016. A MutSβ-dependent contribution of MutSα to repeat expansions in fragile X premutation mice? PLoS Genet. 12: e1006190 10.1371/journal.pgen.1006190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supporting information contains detailed descriptions of all supplemental files. The following figures and tables can be found in the Genetics Society of America Figshare portal. Figure S1, a comparison of gene expression patterns of MSH2, MSH3, and MSH6 across diverse human cancer and normal tissues using the GENT database. Figure S2, correlation of MSH2 and MSH6 protein expression in TCGA tumor types with significantly upregulated MSH2 and MSH6 mRNA expression. Figure S3, western blot analysis of Msh2 and Msh6 levels; independent measurements. Figure S4, western blot analysis of Msh2 and Msh6 levels in strains containing ARS-CEN plasmids. Table S1, literature review of MMR proteins overexpressed in cancers. Table S2, strains and plasmids used in this study. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5991208.