Filamentous fungi depend on asexual cell-to-cell communication and cell fusion to establish and maintain an interconnected mycelial colony. In Neurospora crassa, genetically identical asexual spores undergo chemotropic interactions resulting in cell fusion and colony establishment...
Keywords: Neurospora crassa, MAP kinase, regulatory networks, DAP-seq, cell-to-cell communication, cell fusion
Abstract
Maintenance of cell integrity and cell-to-cell communication are fundamental biological processes. Filamentous fungi, such as Neurospora crassa, depend on communication to locate compatible cells, coordinate cell fusion, and establish a robust hyphal network. Two MAP kinase (MAPK) pathways are essential for communication and cell fusion in N. crassa: the cell wall integrity/MAK-1 pathway and the MAK-2 (signal response) pathway. Previous studies have demonstrated several points of cross-talk between the MAK-1 and MAK-2 pathways, which is likely necessary for coordinating chemotropic growth toward an extracellular signal, and then mediating cell fusion. Canonical MAPK pathways begin with signal reception and end with a transcriptional response. Two transcription factors, ADV-1 and PP-1, are essential for communication and cell fusion. PP-1 is the conserved target of MAK-2, but it is unclear what targets ADV-1. We did RNA sequencing on Δadv-1, Δpp-1, and wild-type cells and found that ADV-1 and PP-1 have a shared regulon including many genes required for communication, cell fusion, growth, development, and stress response. We identified ADV-1 and PP-1 binding sites across the genome by adapting the in vitro method of DNA-affinity purification sequencing for N. crassa. To elucidate the regulatory network, we misexpressed each transcription factor in each upstream MAPK deletion mutant. Misexpression of adv-1 was sufficient to fully suppress the phenotype of the Δpp-1 mutant and partially suppress the phenotype of the Δmak-1 mutant. Collectively, our data demonstrate that the MAK-1/ADV-1 and MAK-2/PP-1 pathways form a tight regulatory network that maintains cell integrity and mediates communication and cell fusion.
CELL-TO-CELL communication is a fundamental biological process across the tree of life. There is abundant work detailing mechanisms that mediate communication processes in diverse organisms, such as quorum sensing in bacteria, neurotransmission in mammals, or pheromone sensing in Saccharomyces cerevisiae (Perbal 2003; Merlini et al. 2013). All of these systems share a general mechanism of signal release and reception that subsequently initiates specific molecular responses that often lead to changes in transcription.
Filamentous fungi, such as Neurospora crassa, depend on cell-to-cell communication to locate compatible cells and coordinate the process of cell fusion. N. crassa is a well-developed model for investigating mechanisms of cell-to-cell communication, chemotropic growth, and cell fusion at several points throughout the life cycle. During germination of genetically identical asexual spores, individual cells (germlings) collaborate to establish a new colony by engaging in cell-to-cell communication and chemotropic growth that ultimately results in cell fusion (Glass 2004; Fleissner et al. 2009; Richard et al. 2012; Leeder et al. 2013; Bastiaans et al. 2015). Germling fusion is an important aspect of colony establishment, and as a mature colony develops, hyphae within a colony also undergo chemotropic growth and fusion to further reinforce a robust hyphal network (Hickey et al. 2002). Hyphal fusion also occurs between different colonies, which is important for mediating postfusion self/nonself recognition mechanisms and heterokaryosis (Garnjobst and Wilson 1952; Saupe 2000; Glass and Kaneko 2003; Simonin et al. 2012). Lastly, cell-to-cell communication and fusion are necessary for fertilization during the process of sexual reproduction. In filamentous ascomycete species such as N. crassa, fertile hyphae (trichogynes) from protoperithecia chemotropically grow toward the male gamete with the goal of cell fusion and the initiation of perithecium development, karyogamy, meiosis, and finally, ascospore development (Kim and Borkovich 2004; Engh et al. 2010).
More than 60 genes have been identified as required for the process of germling communication and cell fusion (Fu et al. 2011; Read et al. 2012; Palma-Guerrero et al. 2013; Dettmann et al. 2014; Fleißner and Herzog 2016). Strains carrying mutations in many of the genes involved in mediating germling or hyphal fusion show a pleiotropic vegetative and female sexual development phenotype. For example, in both N. crassa and Sordaria macrospora, most fusion mutants fail to develop female reproductive structures (protoperithecia) (Engh et al. 2010; Fu et al. 2011). Some of these genes are also essential for sexual reproduction, but many are not, indicating that asexual (germling and hyphal) cell fusion is mechanistically distinct from sexual (gamete) fusion. Among the >60 known genes, there are many encoding hypothetical proteins of unknown function, but also genes/proteins in characterized pathways, including two ERK-like mitogen activated protein kinase (MAPK) pathways and pathways involved in the production of reactive oxygen species, actin organization, and calcium signaling.
Two of the three predicted MAPK pathways in N. crassa are required for germling communication and cell fusion (Colot et al. 2006; Park et al. 2008). In general, MAPK pathways are intermediaries that transduce information from one part of the cell (i.e., a cell-surface sensor; input) to factors in the nucleus that alter gene transcription (output). The two MAPK pathways essential for communication and cell fusion are the MAK-2 pathway, which is homologous to the pheromone response pathway in S. cerevisiae, and the MAK-1 pathway, which is homologous to the cell wall integrity pathway in S. cerevisiae. In N. crassa, the core conserved components of the MAK-1 pathway are MIK-1 (MAPKKK), MEK-1 (MAPKK), MAK-1 (MAPK), and SO, a scaffold important for some functions of the MAK-1 pathway (Teichert et al. 2014; Weichert et al. 2016). It is currently unknown which transcription factors are targets of the MAK-1 pathway, although transcription of several genes is dependent on MAK-1, and the rhythmic pattern of expression for a few MAK-1-dependent genes mirrors the rhythmic expression pattern of ADV-1-dependent genes (Bennett et al. 2013; Dekhang et al. 2017). The adv-1 gene encodes a Zn(II)2Cys6 transcription factor, and both mak-1 and adv-1 are targets of the circadian clock in N. crassa (Bennett et al. 2013; Dekhang et al. 2017). The core components of the conserved N. crassa MAK-2 pathway are NRC-1 (MAPKKK), MEK-2 (MAPKK), MAK-2 (MAPK), and HAM-5, a scaffold protein important during cell fusion (Dettmann et al. 2014; Jonkers et al. 2014). PP-1 is the predicted downstream transcription factor target of MAK-2; a microarray expression study demonstrated that there is overlap between MAK-2-dependent and PP-1-dependent gene expression (Leeder et al. 2013). Several other studies have documented cross talk between the MAK-1 and MAK-2 pathways in N. crassa and in other filamentous ascomycete fungi (Maerz et al. 2008; Dettmann et al. 2012, 2013; Maddi et al. 2012; Leeder et al. 2013; Fu et al. 2014).
Transcription is a common point of regulation for biological processes, and two conserved transcription factors in N. crassa (ADV-1 and PP-1) are essential for germling communication and fusion. Both the Δadv-1 and Δpp-1 deletion mutants have a pleiotropic phenotype and were initially identified as female-sterile mutants (Li 2005; Colot et al. 2006; Fu et al. 2011). PP-1 is a C2H2 zinc finger transcription factor that is homologous to the S. cerevisiae pheromone response pathway transcription factor, STE12 (Leeder et al. 2013). The core component of this pathway in S. cerevisiae is the Fus3p MAPK cascade, that once activated, leads to de-repression of Ste12p (Merlini et al. 2013). Ste12-like proteins, including PP-1, have two C2H2-Zn2+ motifs and a homeobox-like STE domain involved in binding DNA (Errede and Ammerer 1989). In N. crassa the STE domain is essential for PP-1 function, but the C2H2-Zn2+ motifs are dispensable (Leeder et al. 2013). Ste12-like transcription factors in fungi are regulated by direct phosphorylation and phosphorylation of associated regulatory proteins (Blackwell et al. 2007). Several phosphorylation sites have been identified on PP-1 in N. crassa, but the biological significance of these sites remains unknown (Leeder et al. 2013; Jonkers et al. 2014; Xiong et al. 2014).
ADV-1, like PP-1, regulates growth, and asexual and sexual development. ADV-1 is not as broadly conserved as PP-1, and clear adv-1 homologs are restricted to the filamentous Ascomycete species (Pezizomycotina). In the self-fertile (homothallic) species S. macrospora, the adv-1-ortholog, pro1, is required for protoperithecial development, while heterothallic N. crassa does not require adv-1 for protoperithecia development (Masloff et al. 1999). In N. crassa, adv-1 is essential for postmating perithecial development, asexual cell fusion, and wild-type-like growth rate. Both ADV-1 and Pro1 have a GAL4-like DNA binding domain and a transcription-activation domain. In contrast to the S. cerevisiae protein GAL4p, Pro1 lacks a coiled-coil dimerization domain, indicating that Pro1 (and ADV-1) likely function independently (Masloff et al. 2002). Unlike PP-1, upstream factors that influence ADV-1–regulated transcription are largely unknown, except for a chromatin immunoprecipitation sequencing (ChIP-seq) experiment that identified adv-1 as a target of the circadian clock master regulator, the White Collar Complex (Smith et al. 2010). ADV-1 is essential for developmental oscillations (i.e., conidiation) and the quantity of adv-1 mRNA and ADV-1 protein oscillates with the clock (Smith et al. 2010). Furthermore, the expression of ADV-1 target genes in a mature colony matches the rhythm of other clock-controlled genes. These data led to the hypothesis that hyphal fusion is a clock-regulated developmental process (Dekhang et al. 2017).
The Δadv-1 and Δpp-1 mutants share many phenotypes across filamentous fungi, yet the relationship between these two transcription factors remains unclear. Independent expression profiling studies investigating ADV-1- or PP-1-dependent transcription indicate that at least some genes require ADV-1 and PP-1 for wild-type levels of expression (Li 2005; Nowrousian et al. 2007; Leeder et al. 2013; Dekhang et al. 2017). Here, we compared expression profiles using RNA sequencing (RNA-seq) on Δadv-1 and Δpp-1 germlings relative to expression patterns in wild-type germlings. To identify genes that were regulated and bound by ADV-1 or PP-1, we developed an in vitro method for identifying transcription factor binding sites in N. crassa and other fungi called DNA-affinity purification sequencing (DAP-seq). DAP-seq is similar to ChIP-seq, except that in vitro synthesized transcription factors are incubated with native genomic DNA. DNA fragments bound by the transcription factor are then identified by high-throughput sequencing methods. DAP-seq has been used in a global analysis of transcription factor binding sites in Arabidopsis thaliana (O’Malley et al. 2016). Our data showed that PP-1 directly regulates adv-1; ADV-1 is the primary regulator of many genes that are important for asexual growth and cell fusion and development. To investigate the linkage between mak-1, mak-2, adv-1, and pp-1, we used misexpression and phenotypic analyses. Our data showed that mak-1 primarily functions upstream of adv-1, and mak-2 primarily functions upstream of pp-1. However, the MAK-1/ADV-1 pathway and the MAK-2/PP-1 pathway engage in cross-talk and both pathways form a regulatory network that mediates growth, communication, fusion, and the response to cell wall stress.
Materials and Methods
RNA isolation
We used strains FGSC 2489 (wild type), FGSC 11042 (Δadv-1), and Δpp-1 (Leeder et al. 2013). Each strain was initially grown on Vogel’s Minimal Medium (VMM) (Vogel 1956) agar slant tubes for 5 days. Conidia were harvested, filtered through cheesecloth, and then inoculated into 100 ml of liquid VMM in a 250 ml flask, to a final concentration of 106 conidia/ml. Flasks were incubated at 30° in constant light for 2.5 hr shaking at 220 rpm to induce germination, followed by 2.5 hr stationary incubation to allow communication to occur. Δpp-1 conidia have slightly delayed germination, thus these conidia were shaken for 3 hr, followed by 2.5 hr stationary incubation. Germlings were harvested by vacuum filtration over nitrocellulose paper and transferred to a 2 ml screw cap tube that was immediately frozen with liquid nitrogen. Experimental design for each strain: conidia from one VMM slant tube was used to inoculate eight flasks of liquid VMM, then two flasks were pooled during germling harvest, resulting in a total of four samples per strain. RNA was extracted using a previously described TRIzol-based method (Leeder et al. 2013). RNA quality and concentration were quantified via Bioanalyzer at the qb3 facility at University of California, Berkeley. Three samples per strain with the highest quality and concentration of RNA were submitted for library preparation and sequencing on an Illumina HiSeq3000 at the University of California, Davis, DNA Technologies Core.
RNA-seq data analysis and visualization
Fast-X Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used to filter out low quality raw reads (∼11–12% of all reads) and Tophat (Langmead et al. 2009) mapped high quality reads to the N. crassa transcriptome version 12 (ftp://ftp.broadinstitute.org/pub/annotation/fungi/neurospora_crassa/assembly/NC12). Differential expression was calculated using three independent methods: Cuffdiff (Roberts et al. 2011), DESeq2 (Love et al. 2014), and EdgeR (Robinson et al. 2010; McCarthy et al. 2012). We defined the threshold for significant differential expression to be −2 < log2 fold change < 2 and p.adj < 0.01. RNA-seq raw data (fastq) is available at the NCBI Sequence Read Archive under accession number SRP133239.
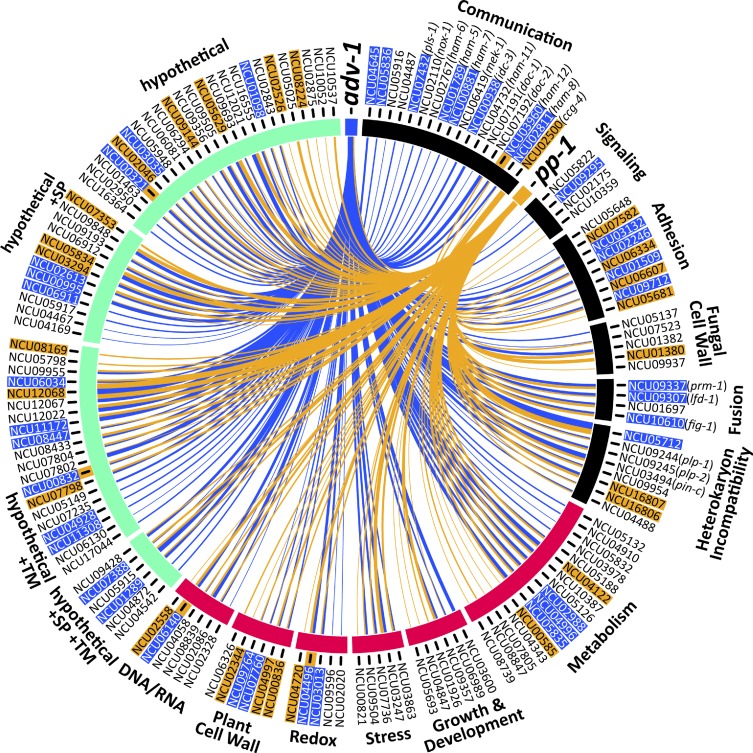
We used the Circos data visualization tool (Krzywinski et al. 2009) to generate Figure 2. Genes are grouped according to known function or predicted function based on homology (BLASTp, FungiDB, or the Broad Institute’s Fungal Orthogroups Repository v1.1). Highlighted gene IDs indicate genes that have an ADV-1 binding site or a PP-1 binding site 2 kb upstream from the start codon. PP-1 binding sites were identified solely via DAP-seq, whereas ADV-1 binding sites are the consensus between DAP-seq data and a previously published ChIP-seq data set (Dekhang et al. 2017).
Figure 2.
Genes that are positively regulated by ADV-1 and PP-1 in germlings. Circos plot depicting the 155 genes that are significantly downregulated in either Δadv-1 or Δpp-1 germlings as compared to parental wild-type germlings (log2 fold change < −2 and P < 0.01, consensus among Cuffdiff, DESeq2, and EdgeR). Genes are organized based on function that is either known from previous work, or inferred via homology and protein prediction. Gene function is clustered into three major groups: communication and fusion (black), basic cellular processes (magenta), and hypothetical proteins (green). Blue lines indicate genes that are regulated by ADV-1, orange lines indicate genes that are regulated by PP-1, and line thickness is proportional to the fold-change difference in expression between the transcription factor mutant and wild-type germlings. Gene IDs are highlighted to indicate the presence of at least one ADV-1 (blue) or PP-1 (orange) binding site in the promoter region within 2 kb upstream of ATG. Five genes were bound by both ADV-1 and PP-1; these gene IDs are highlighted in blue and have an additional orange highlight immediately adjacent to the gene ID. PP-1 binding sites were determined by consensus between DAP-seq and RNA-seq, and ADV-1 binding sites are the consensus between DAP-seq, RNA-seq, and ChIP-seq data sets. ChIP-seq data are available from Table S3 in Dekhang et al. (2017), in which ChIP-seq was performed at four different time points (24 hr dark, 24 hr dark + 15 min light, 25 hr dark + 30 min light, and 24 hr dark + 60 min light). Genes were included here if they were bound by ADV-1 during at least one time point. This list of 155 genes is significantly enriched for ADV-1 binding sites (P = 0.0004, Fisher’s exact test), but not PP-1 binding sites (P = 0.02, Fisher’s exact test).
Genomic DNA library preparation
DAP-seq was originally developed for A. thaliana (O’Malley et al. 2016). Here we adapted the protocol in O’Malley et al. for use with N. crassa. FGSC 2489 was grown in liquid VMM for 24 hr at 25° and shaking at 220 rpm. Mycelia were harvested using vacuum filtration over Whatman #1 filter papers, and transferred into 2 ml tubes for flash freezing in liquid nitrogen. Cells were ruptured by bead beating for 1 min with 1 mm silica beads and DNA lysis buffer (0.05 M NaOH, 1 mM EDTA, 1% Triton X-100) was added to each sample tube. DNA was purified using DNeasy Blood & Tissue kit (Qiagen, Valencia, CA), and sheared to 300 bp peak using Covaris LE220 sonicator. AMPure XP beads were used to remove DNA above and below target molecular weight. Initially, sheared DNA was mixed in with AMPure XP beads (in PEG-8000) at a ratio of 100:60. At this ratio, beads bind DNA with molecular weight above 700 bp. Supernatant from this primary binding was taken and added to new beads where the final ratio of DNA solution to PEG-8000 was 100:90. At this ratio, DNA below ∼300 bp do not bind to AMPure XP beads, and remaining DNA was eluted for library preparation. KAPA library kit for Illumina sequencing was used to prepare final libraries and stored at −20° for later use.
Transcription factor cloning, transcription, translation, and DAP
Transcription factor open reading frames (ORFs) were amplified from complementary DNA (cDNA) using RNA to cDNA ecodry premix (Clonetech). Amplified transcription factors sequences were inserted into pIX in vitro expression vector modified to contain an N-terminal HALO-Tag (O’Malley et al. 2016). Vector backbone was amplified and assembled with transcription factor ORFs using Gibson assembly, and then transformed into competent Escherichia coli cells for storage and production.
In vitro transcription and translation of transcription factors was achieved by using Promega TnT T7 Rabbit Reticulocyte Quick Coupled Transcription/Translation System. We combined 1 μg of plasmid DNA, 60 μl of TnT Master Mix, and 1.5 μl of 1 mM methionine and incubated the mixture overnight at room temperature. Expression was verified using Western blot analysis with Promega HaloTag monoclonal antibody. Completed TnT reactions were incubated with 20 ng of genomic DNA library, 1 μg salmon sperm for blocking, and 20 μl Promega Magne HaloTag Beads on a rotator for 1 hr at room temperature. Bead-bound proteins and protein-bound DNA were washed three times with 2.5% Tween20 in PBS. HaloTag beads were resuspended in 30 μl double-distilled water and heated to 98° for 10 min to denature protein and release DNA fragments into solution. Supernatant was transferred to a new tube for PCR amplification. DNA was amplified for final libraries using KAPA Hifi polymerase for 12−16 cycles of PCR.
DAP-seq data analysis
Filtered reads were mapped against N. crassa OR74A genome (v12) using bowtie2 v2.3.2 (Kim et al. 2013). SamTools (Li et al. 2009) was used to convert .sam to .bam files and to create .bai index files for viewing reads on Integrated Genomics Viewer. MACS2 (Zhang et al. 2008) with P-value cutoff at 0.001 was used for calling peaks. A negative control data set consisting of DAP pull-down with Promega TnT master mix with no plasmid added, salmon sperm, and genomic DNA was also input into MACS2 as the control condition. We repeated the ADV-1 DAP-seq once and pooled the results of both DAP-seq runs. DAP-seq data are available at the NCBI Sequence Read Archive under accession number SRP133627.
Motif construction
To construct biologically meaningful transcription factor DNA binding motifs, we used DAP-seq peaks associated with differentially expressed genes according to corresponding RNA-seq data. Sequences of these true-positive peaks were collected using a custom python script that reads in genomic position of each peak from the MACS2 output file, and ascertains the sequence from N. crassa OR74A Broad Institute v12 genome FASTA file. The script output is a FASTA file with sequences from all true-positive peaks. Output FASTA files were input into MEME or DREME v4.12.0 with flags maxw = 20, minsites = 5, nmotifs = 8, denoting max width of motif, minimum number of sites for each motif and number of motifs to generate, respectively. For ADV-1 specifically, we used set of 41 DAP binding peak sequences. These peaks were within the promoter regions of from genes that fit two parameters: within 1 kbp upstream of the ATG start site according to DAP-seq, as well as four-fold downregulated in the ∆adv-1 mutant as compared to wild type. Nucleotide sequences for these 41 peaks were fed into MEME v4.12.0 to build the ADV-1 binding motif. For PP-1, we loosened the parameters slightly to two-fold downregulated in Δpp-1 as compared to wild type, due to the small number of genes both directly bound according to DAP-seq and differentially expressed at a four-fold level. Nucleotide sequences for 22 peaks were fed into MEME v4.12.0 to build the PP-1 binding motif.
Quantitative RT-PCR
Germlings were prepared and RNA extracted as described for RNA-seq samples. Quantitative RT-PCR (qRT-PCR) reactions were prepared following the manufacturer protocols for the Bioline SensiFast SYBR no-ROX One-Step kit and Bio-Rad CFX Connect Real-Time PCR Detection System. Each sample was replicated at least four times within a 96-well plate and total reaction volume was 20 μl. Expression data were normalized to both actin and wild type following the 2−ΔΔCt method (Livak and Schmittgen 2001).
Strain construction
Misexpression strains were made by transforming the his-3-targeted vectors described below into his-3− deletion strains as previously described (Colot et al. 2006). Positive transformants were selected for histidine prototrophy and hygromycin resistance. To avoid any off-target affects that may have resulted from the transformation process, we backcrossed each transformant to his-3 (FGSC 9716 or FGSC 6103). Transformants with a mutant background that is defective at any stage of the sexual cycle and thus unable to be backcrossed were purified via microconidial isolation (Pandit and Maheshwari 1994). All genotypes were confirmed via PCR.
The Ptef1-adv-1-v5 his-3 vector was constructed by amplifying adv-1 from genomic DNA with primers that omitted the native STOP codon and added XbaI and PacI cut sites on either end of the adv-1 ORF. PCR products were gel purified and blunt ligated into pCR-Blunt. The adv-1 sequence was ligated into an in-house vector containing V5 using XbaI and PacI sites. adv-1-v5 was amplified using a reverse primer that added a TGA stop codon at the end of the V5 sequence, in addition to EcoRI and ApaI cut sites. This PCR product was gel purified and blunt ligated into pCR-Blunt, digested and ligated into an in-house vector based on pMF272 (Freitag et al. 2004) containing the tef-1 promoter (Ptef1) using XbaI and ApaI cut sites. The Ptef1-pp-1-v5 vector was constructed by amplifying pp-1 from the Pccg1-pp-1-gfp his-3 vector (Leeder et al. 2013) with primers that added BamHI and PacI sites. This PCR product was ligated into pCR-Blunt, digested and ligated into the Ptef1-adv-1-v5 his3 vector using BamHI and PacI cut sites, which resulted in replacement of the adv-1 sequence with pp-1 coding sequences. The sequence of Ptef1-adv-1-v5 and Ptef1-pp-1-v5 in the his-3 vectors was confirmed via Sanger sequencing prior to transforming into his3− deletion strains.
Cosegregation analyses were performed on mutants acquired from the FGSC deletion collection to confirm that the observed phenotype cosegregated with the hygromycin-resistance marker. The ΔNCU05836; mat A, ΔNCU04487; mat A, and ΔNCU05916; mat A mutants were crossed to the wild-type mat a strain, FGSC 4200. The ΔNCU04645; mat-a mutant was crossed to the wild-type mat A strain, FGSC 2489. Twenty ascospores from each cross were isolated and tested for hygromycin resistance and germling fusion phenotype. Approximately one half of the progeny from each cross were hygromycin resistant, and hygromycin resistance clearly segregated with the germling fusion phenotype of each parental strain.
Growth, fusion, mating, and cell wall stress assays
Standard N. crassa growth conditions, media, and protocols are available at http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm. For assays to quantify aerial hyphae height, growth rate, and germling communication, conidia were grown up and harvested as described for RNA-seq. Conidial suspensions were diluted to specified concentrations and immediately used for phenotyping assays. Aerial hyphae were grown up from 106 conidia in 1 ml of liquid VMM and incubated at 30° in the dark for 72 hr (n = 6). Growth rate was measured by inoculating 100 μl of 106 conidia/ml onto one end of a glass race tube containing 25 ml of VMM 1.5% agar. Linear growth in race tubes was measured every 24 hr for at least 4 days (n = 3). Germling communication assays consisted of spreading 300 μl of 107 conidia/ml on a 9 cm VMM 1.5% agar plate. Plates were incubated at 30° in the dark for 3.5 hr. To visualize germlings, agar squares (∼1 cm2) were excised and observed with a Zeiss Axioskop 2 equipped with a Q Imaging Retiga-2000R camera (Surrey), using a 40×/1.30 Plan-Neofluar oil immersion objective and the iVision Mac4.5 software (Heller et al. 2016). Germling communication frequency was determined with the ImageJ Cell Counter tool (http://imagej.nih.gov/ij/). For mating assays, female strains were inoculated onto Synthetic Cross (SC) agar 5.4 cm plates (Westergaard and Mitchell 1947). These plates were incubated at 30° in the dark for 2 days, and then moved to 25° and light for an additional 5 days to allow for full production of protoperithecia. Male strains were grown up on VMM as described above. Mating was initiated by inoculating female plates with 150 μl of undiluted conidial suspension of male strains of the opposite mating type. Cell wall stress assays were conducted on large petri plates containing 45 ml of VMM with 1.5% agar and 2% FGS. The following cell wall stress drugs were added and mixed to VMM+FGS immediately prior to pouring each plate (one drug per plate): 1.3 μg/ml Caspofungin, 1.5 mg/ml Calcofluor White, and 1 mg/ml Congo Red. Conidia for cell wall stress tests were grown up and harvested as described above, then diluted to 106 spores/ml. A 1:5 dilution series was prepared starting with 106 spore/ml as the most concentrated dilution. Conidial solutions were then spotted onto freshly poured plates at 5 μl per spot.
Data availability
Strains and plasmids are available upon request. Some strains are also publicly available through the FGSC. DAP-seq raw data are available at the NCBI Sequence Read Archive with accession number (SRP133627). RNA-seq raw data (.fastq) is available at the NCBI Sequence Read Archive, accession number SRP133239. Differential expression analysis outputs are included with this publication as Supplemental Material, File S1. File S2 includes detailed information about the 155 differentially expressed genes that we focused on. Results from DAP-seq data analysis are in File S3. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6146681.
Results
ADV-1 and PP-1 have a shared regulon in germlings
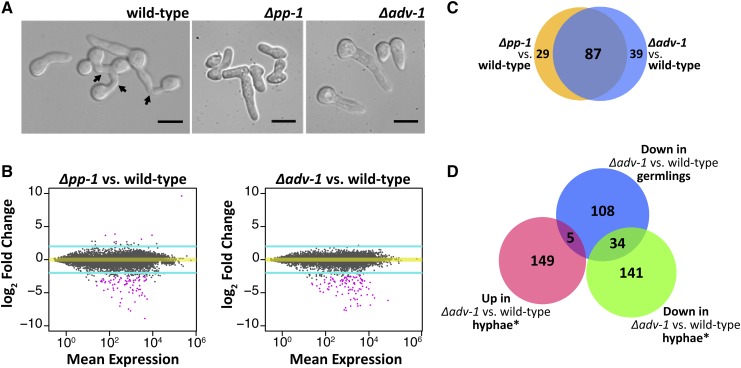
To investigate the how ADV-1 (NCU07392) and PP-1 (NCU00340) regulate germling communication and fusion, we compared expression profiles of Δadv-1, Δpp-1, and wild-type germlings. We extracted RNA from germlings 2.5 hr after germination, which is when the majority of wild-type germlings were actively engaging in chemotropic growth and cell fusion (Figure 1A). Our RNA-seq data confirm previous studies that implicated PP-1 and ADV-1 as transcriptional activators (Masloff et al. 2002; Leeder et al. 2013). Figure 1B illustrates that the vast majority of differentially expressed genes were downregulated in Δadv-1 and Δpp-1 mutants compared to wild-type germlings (Figure 1B). We defined significant differential expression in each mutant compared to wild type as −2 > log2 fold change > 2, adjusted P-value <0.01, and consensus among three different programs that calculate differential expression; DESeq2, EdgeR, and Cuffdiff (File S1 and File S2).
Figure 1.
ADV-1 and PP-1 have a shared regulon in germlings. (A) Microscopic images showing germling morphology at the time point when we extracted RNA for RNA-seq. Arrows indicate fusion events. (B) MA plots summarizing genome-wide RNA-seq data for each transcription factor mutant compared to wild-type. The y-axis represents the log fold change difference in expression (M) between the mutant and wild-type for each gene. The x-axis represents the level of expression for each gene averaged between the mutant and wild-type (A). Turquoise lines denote threshold of 2 < log2 fold change < −2, and pink points indicate genes with significant differential expression (P < 0.01, DESeq2). (C) Number of genes significantly down regulated in each mutant as compared to the wild-type parental strain (consensus between Cuffdiff, EdgeR, and DESeq2, log2 fold change < −2 and adjusted P-value <0.01). The blue circle is Δadv-1 compared to wild-type germlings, and the orange circle is Δpp-1 compared to wild-type germlings. (D) Number of significantly differentially expressed genes in Δadv-1 as compared to the parental wild-type strain in hyphae compared to germlings (−2 > log2 fold change > 2 and P < 0.01, Cuffdiff). *Hyphal data from Table S5 in Dekhang et al. (2017), where RNA-seq data were collected from three different time points (24 hr dark, 24 hr dark + 15 min light, and 24 hr dark + 60 min light). Genes were included if they were differentially expressed during at least one time point. Scale bars=10 μm.
We did not identify any significantly upregulated genes in the Δadv-1 mutant as compared to wild-type germlings; 17 genes were upregulated in Δpp-1 cells as compared to wild-type germlings (File S1). The a-pheromone precursor gene, mfa-1, was the most highly expressed gene in Δpp-1 (mat a) germlings (File S1); it is a clear outlier in the top right corner of Figure 1B. These data complement a previous study that used qRT-PCR to show that both ccg-4 (A-pheromone precursor) and mfa-1 are substantially overexpressed in Δpp-1 cells as compared to wild type (Leeder et al. 2013), indicating that PP-1 specifically represses expression of the mating pheromones in germlings.
Analyses of RNA-seq data identified 155 significantly downregulated genes in either Δadv-1 or Δpp-1 as compared to wild-type germlings (Figure 2 and File S2). There was substantial overlap between the downregulated genes in both mutants (Figure 1C). Of the downregulated genes in Δpp-1 germlings, 75% (87 out of 116) were also downregulated in Δadv-1 cells, while 69% (87 out of 126) of the genes downregulated in Δadv-1 germlings were also downregulated in Δpp-1 cells. Before imposing a threshold of significant differential expression, we first calculated the distance between samples on our entire RNA-seq data set. These data further demonstrated that gene expression was more similar between Δadv-1 and Δpp-1 germlings than either mutant compared to wild-type germlings (Figure S1). We also did not observe any pattern in the genomic location of genes that are regulated by ADV-1 and/or PP-1 (Figure S2).
In an independent study, expression patterns in the hyphal stage of the Δadv-1 mutant were compared to those in wild-type hyphae at three different circadian time points; 24 hr of growth in the dark, and 24 hr of darkness plus an additional 15 or 60 min of light (Dekhang et al. 2017). We compared this Δadv-1 hyphal data set with our Δadv-1 germling data set (−2 < log2 fold change < 2, adjusted P-value <0.01, Cuffdiff); all genes differentially expressed during at least one time point were included. We found a modest overlap (26.5%, 39 out of 147) between our Δadv-1 germling data set and the Δadv-1 hyphal data set (Figure 1D). Of the 39 genes regulated by ADV-1 in both hyphae and germlings (Table S1), 22 are either predicted or known to be involved in the processes of communication, cell fusion, development, or metabolism including ham-6, ham-8, ham-11, doc-2, lfd-1, prm-1, mat A-1, and esd-C (Glass et al. 1988; Fleissner et al. 2008; Han et al. 2008; Fu et al. 2011; Leeder et al. 2013; Palma-Guerrero et al. 2014; Heller et al. 2016). The remaining 19 genes encode hypothetical proteins. Although the overlap between Δadv-1 RNA-seq data sets can be explained by the fact that chemotropic growth and fusion occur during both germling and hyphal stages, there are additional developmental and morphological differences between germlings and hyphae that could explain reduced overlap between the germling and hyphal Δadv-1 data sets. Consistent with this hypothesis was the observation that 58% the genes of that were downregulated in our Δpp-1 germling data set were also downregulated in a previous single-sample RNA-seq experiment on Δpp-1 vs. wild-type germlings (Leeder et al. 2013).
Identification of ADV-1 and PP-1 binding sites by DAP-seq
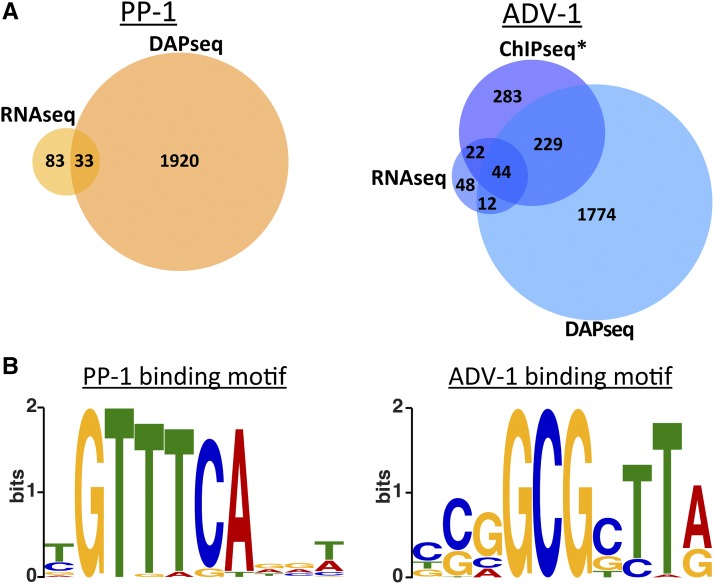
DAP-seq was recently developed as a high-throughput in vitro method for identifying transcription factor binding sites using genomic DNA (O’Malley et al. 2016). We adapted this method for N. crassa. Briefly, we amplified the adv-1 and pp-1 ORFs from cDNA and added a N-terminal HALO-tag to each gene (see Materials and Methods). The HALO-ADV-1 and HALO-PP-1 in vitro synthesized proteins were used for immunoprecipitation experiments using sheared N. crassa genomic DNA that contained adaptors for sequencing. From DAP-seq, we identified PP-1 binding sites that were significantly enriched 2 kb upstream of the predicted ATG translation start site for 1953 genes, and ADV-1 binding sites that were significantly enriched within 2 kb upstream of the ATG site for 2059 genes (P < 0.0001, MACS2) (Figure 3A and File S3).
Figure 3.
DAP-seq identifies promoters bound by ADV-1 or PP-1. (A) Number of genes that are downregulated in Δpp-1 germlings (RNA-seq) or bound by PP-1 (DAP-seq) (left panel). The number of genes that are down regulated in Δadv-1 germlings (RNA-seq) or bound by ADV-1 (DAP-seq and ChIP-seq) (right panel). Downregulated genes were identified by consensus between Cuffdiff, EdgeR, and DESeq2, log2 fold change < −2 and adjusted P-value <0.01 (compare with Figure 1C). Genes bound by each transcription factor were counted if the transcription factor was bound within 2 kb upstream of the ATG (P < 0.001). *ADV-1 ChIP-seq data are available from Table S3 in Dekhang et al. (2017), in which ChIP-seq was performed at four different circadian time points (24 hr dark, 24 hr dark + 15 min light, 24 hr dark + 30 min light, and 24 hr dark + 60 min light). Genes were included here if they were bound by ADV-1 during at least one time point. (B) Consensus DNA binding motif for PP-1 or ADV-1 based on DAP-seq data.
All in vitro assays, such as DAP-seq, contain false positives and false negatives because biologically relevant factors are excluded from the assay. For example, a false negative could arise if a transcription factor requires a cofactor to bind a particular sequence of DNA. A false positive could occur if a factor normally blocks a transcription factor from binding a particular DNA sequence. It is also impossible to know from DAP-seq how environmental or biological conditions affect transcription factor binding. In contrast, in vivo methods such as ChIP-seq provide a snapshot of where a transcription factor binds under the precise environmental and biological conditions of the assay. Comparing data from both DAP-seq and ChIP-seq experiments eliminates some of the error associated with each assay. Additional comparison with expression data sets such as RNA-seq can elucidate true-positive peaks associated with genes that are directly bound and regulated by a transcription factor.
Of the 2059 genes bound by ADV-1 identified with DAP-seq, 273 genes were in consensus with a previous ADV-1 ChIP-seq data set (Dekhang et al. 2017). Furthermore, 44 of these 273 genes were differentially expressed in Δadv-1 germlings vs. wild-type germlings (Figure 2A). The ADV-1 germling regulon (126 genes) (Figure 2) was enriched for ADV-1 binding sites (consensus between ChIP-seq and DAP-seq data sets) as compared to ADV-1 binding sites across the entire genome; 35% of the ADV-1 regulon vs. 21% of the genome (P = 0.0004, Fisher’s exact test). Of the 1953 genes bound by PP-1, only 33 genes are differentially expressed in Δpp-1 vs. wild-type germlings. These 33 genes bound by PP-1 represented 28% of the PP-1 germling regulon (Figure 2), which is what we would expect by chance as compared to the 20% of the entire genome bound by PP-1 (P = 0.02, Fisher’s exact test).
To construct biologically meaningful transcription factor DNA binding motifs, we analyzed only the strongest DAP-seq peaks that were within a 2 kb region upstream of differentially expressed genes. These true-positive peaks were enriched for consensus binding motifs (Figure 3B). The ADV-1 binding motif mirrored previously reported ADV-1 binding motifs (Weirauch et al. 2014; Dekhang et al. 2017). In contrast, our PP-1 motif differed from the PP-1 motif identified using peptide binding arrays (Weirauch et al. 2014).
PP-1 and ADV-1 regulate genes required for communication, fusion, growth, and development
The Δadv-1 and Δpp-1 mutants are impaired in several morphological processes including conidial germination, germling communication, growth rate, aerial hyphae extension, female sexual development, and ascospore germination (Fu et al. 2011; Leeder et al. 2013; Dekhang et al. 2017) (Figure 4, Figure S3, and Table S2). The pleiotropic nature of Δpp-1 and Δadv-1 mutants was reflected in the broad functional diversity of the 155 genes that were positively regulated by ADV-1 and/or PP-1 (Figure 2 and File S2). These 155 genes fell into three major groups: basic cellular processes (44 genes), communication/fusion/nonself recognition (49 genes), and hypothetical proteins (62 genes). ADV-1 and PP-1 regulate at least some genes in each group, and no group was uniquely regulated by one transcription factor (Figure 2). The significant reduction in growth rate and aerial hyphae in Δadv-1 and Δpp-1 cells (Figure 4, B and C) can be at least partially explained by the 44 genes whose function was implicated in basic cellular processes such as metabolism, nutrition, growth, development, and general stress response (Figure 2 and File S2). Of particular interest to this study are the 49 genes involved in the process of communication, adhesion, fusion, and nonself recognition (Figure 2 and File S2). Twenty two of these 49 genes have been reported previously as being required for normal germling communication, cell fusion, or nonself recognition via heterokaryon incompatibility (Kaneko et al. 2006; Kim and Borkovich 2006; Fleissner et al. 2008; Fu et al. 2011; Leeder et al. 2013; Palma-Guerrero et al. 2014; Hernández-Galván et al. 2015; Heller et al. 2016, 2018; Lalucque et al. 2016). The remaining 27 genes either have predicted protein domains or homology with proteins implicated in the processes of signaling, adhesion, fusion, cell wall remodeling, or heterokaryon incompatibility.
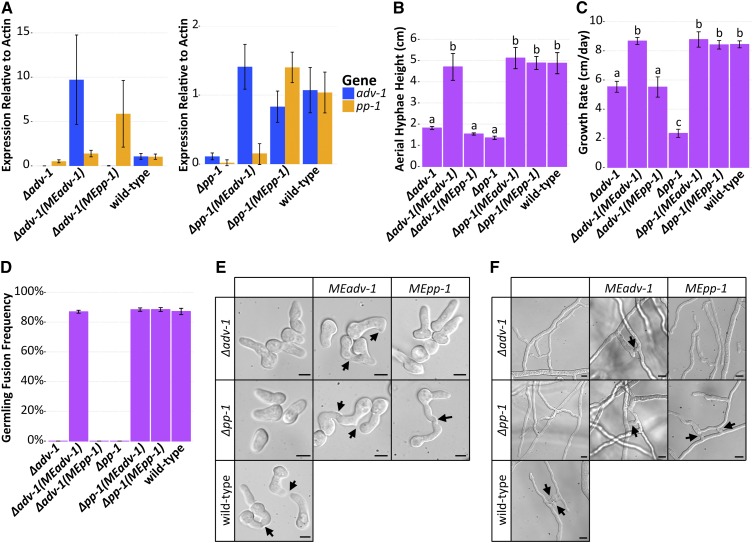
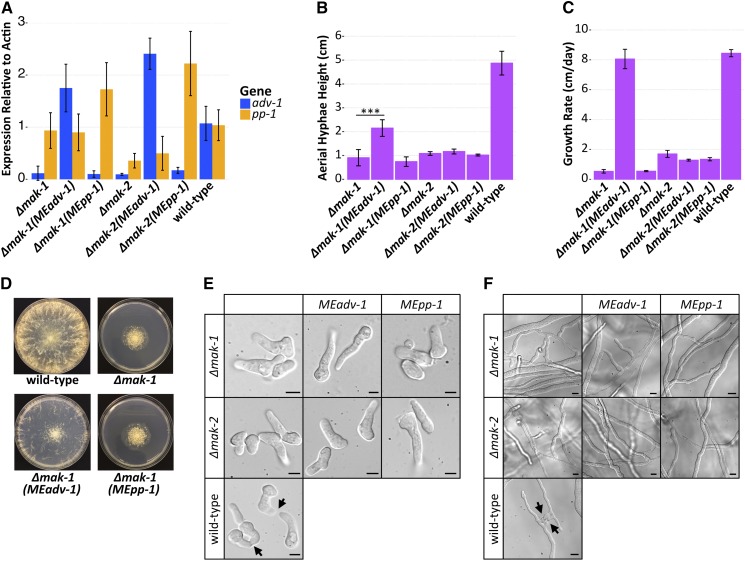
Figure 4.
Misexpression of adv-1 suppresses the phenotype of the Δpp-1 mutant. (A) qRT-PCR data showing mRNA expression levels of adv-1 and pp-1 in Δadv-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)) and Δadv-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)) germlings (left panel) and in Δpp-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)) and Δpp-1 (Ptef1-pp1-v5;his-3 (MEpp-1)) germlings (right panel) compared to the wild-type parental strain. (B) Mean height of aerial hyphae of strains in (A) 3 days after inoculation (ANOVA + Tukey HSD, P < 0.0001, n = 6). (C) Mean growth rate per 24 hr of strains in (A) measured over 4 days (ANOVA + Tukey HSD, P < 0.01, n = 3). (D) Mean frequency of communication and fusion between pairs of germlings for each strain in A (n = 3, 400–700 germling pairs counted per sample). For all bar plots, error bars indicate SD. Photos of the (E) germlings (Bar, 5 μm) and (F) hyphae (Bar, 10 μm) are shown for each strain in A. Arrows indicate chemotropic interactions and successful cell fusion. Lowercase letters in panels B and C denote the result of the ANOVA + Tukey HSD statistical tests.
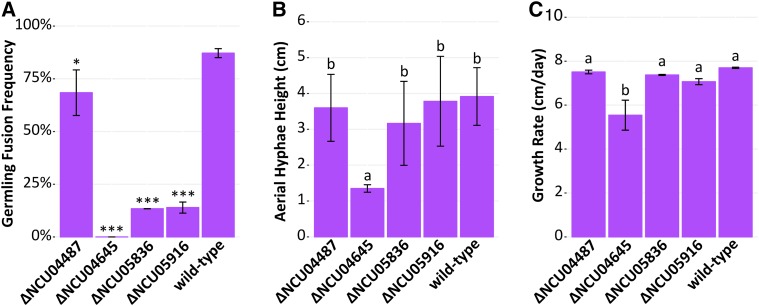
We assessed the germling fusion phenotype for each differentially expressed gene that had an available deletion mutant and that had not previously been described (110 deletion mutants available, 23 mutants unavailable or heterokaryotic, and 22 mutants previously described). Qualitatively, 106 of these mutants had a wild-type-like germling fusion phenotype. However, four mutants (ΔNCU04645, ΔNCU05836, ΔNCU05916, and ΔNCU04487) had obviously reduced levels of germling fusion. We therefore confirmed that the germling fusion phenotype was due to the deleted gene by cosegregation analysis of 20 independent progeny from each mutant backcrossed to wild type (data not shown). After confirming the genotype of these four mutants, we quantified their frequency of germling fusion (Figure 5A). All four mutants have a significantly reduced frequency of germling fusion compared to the wild-type parental strain (P < 0.01, ANOVA with Tukey’s Honest Significant Difference (HSD), n = 3, ∼200–400 germling pairs per sample). ΔNCU04645 was the only mutant that completely lacked any germling communication or fusion. Similarly, ΔNCU04645 was also the only mutant with significantly reduced height of aerial hyphae (Figure 5B, ANOVA with Tukey HSD, P < 0.0001, n = 6) (Figure 5B and Figure S4) and growth rate compared to the wild-type parental strain (Figure 5C, ANOVA with Tukey HSD, P < 0.01, n = 2) (Figure 5C). NCU05836 is predicted to be an ER mannosidase and NCU05916 is a predicted α-1,3-mannosyltransferase with homology to the Cu-responsive virulence protein CMT1 in Cryptococcus neoformans (Ding et al. 2013). Both NCU04487 and NCU04645 are uncharacterized hypothetical proteins. The NCU04487 protein sequence has one predicted C-terminal transmembrane domain and no characterized orthologs. The NCU04645 protein sequence contains a predicted C-terminal AIM24 domain (E-value = 1.9E−51, pfam). The AIM24 domain is named after the S. cerevisiae protein Aim24p, which is a nonessential mitochondrial inner-membrane protein. In S. cerevisiae, Δaim24 mutant shows decreased growth, and Aim24p coordinates the assembly of the MICOS (mitochondrial contact site and cristae organizing system) protein complex (Harner et al. 2014).
Figure 5.
Four ADV-1-regulated genes are required for normal germling fusion and growth. A screen of 110 deletion mutants of the 155 genes regulated by ADV-1 and/or PP-1 revealed that strains carrying mutations in four genes have cell fusion defects. The remaining 151 mutants either have a previously described germling fusion defect, wild-type-like germling fusion phenotype, or homokaryotic deletion mutants are not available in the deletion collection. (A) ΔNCU04487, ΔNCU04645, ΔNCU05836, and ΔNCU05916 mutants showed reduced germling fusion. Mean germling fusion frequency of each mutant and wild type (n = 3, ∼200–400 germling pairs counted per sample). Stars indicate a significant difference compared to the parental wild-type strain (* P = 0.007, *** P < 1E−7, ANOVA + Tukey HSD). (B) Mean height of aerial hyphae of ΔNCU04487, ΔNCU04645, ΔNCU05836, and ΔNCU05916 mutants 3 days after inoculation (ANOVA + Tukey HSD, P < 0.0001, n = 6). (C) Mean growth rate of ΔNCU04487, ΔNCU04645, ΔNCU05836, and ΔNCU05916 mutants per day measured over 4 days (ANOVA + Tukey HSD, P < 0.01, n = 2). For all bar plots, error bars indicate SD. Lowercase letters in panels B and C denote the result of the ANOVA + Tukey HSD statistical tests.
Misexpression of adv-1 suppresses the pleiotropic phenotype of the Δpp-1 mutant
There are four lines of evidence indicating that PP-1 activates transcription of adv-1, with ADV-1 as the primary regulator of the PP-1/ADV-1 regulon. First, adv-1 is downregulated in the Δpp-1 mutant, but the inverse is not true. Second, our list of 155 ADV-1/PP-1 regulated genes is significantly enriched for ADV-1 binding sites (P = 0.0004, Fisher’s exact test), whereas the same is not true for PP-1 binding sites (P = 0.02 Fisher’s exact test). Third, coregulated genes are more strongly downregulated in the Δadv-1 mutant than the Δpp-1 mutant (Figure 2 and File S2). Fourth, there are no potential ADV-1 binding motifs upstream of pp-1, but there are several potential PP-1 binding motifs upstream of adv-1, despite the fact that DAP-seq did not identify PP-1 bound to the adv-1 promoter. These observations led to the hypothesis that PP-1 regulates adv-1 and ADV-1 directly regulates downstream gene expression.
To test this hypothesis, we made epitope-tagged misexpression constructs (Ptef1-adv-1-v5 and Ptef1-pp-1-v5) and transformed them into Δpp-1 and Δadv-1 cells. Successful misexpression in each mutant was verified by qRT-PCR (Figure 4A and Figure S3A). As predicted, expression of adv-1 was restored in Δpp-1 (Ptef1-pp-1-v5) germlings as compared to the Δpp-1 mutant itself (P = 1.8e−7, Welch’s t-test, n = 8). In contrast, expression levels of pp-1 were equivalent between Δadv-1 (Ptef1-adv-1-v5) and Δadv-1 germlings (P = 0.0045, Welch’s t-test, n = 4) (Figure 4A). Both the Δadv-1 mutant and the Δpp-1 mutant have a pleiotropic phenotype when compared with wild-type cells (Figure 4, Figure S3, and Table S2). The introduction of Ptef1-adv-1-v5 fully complemented the pleiotropic phenotype of both Δadv-1 and Δpp-1 mutants, while the introduction of Ptef1-pp-1-v5 only complemented the phenotype of the Δpp-1 mutant, but not the Δadv-1 mutant (Figure 4 and Figure S3). Collectively, these misexpression experiments support the RNA-seq data and the hypothesis that PP-1 controls expression of adv-1; ADV-1 is the primary transcriptional regulator of genes involved in cell communication, fusion, protoperithecial development, and growth.
PP-1 binds the promoter of adv-1
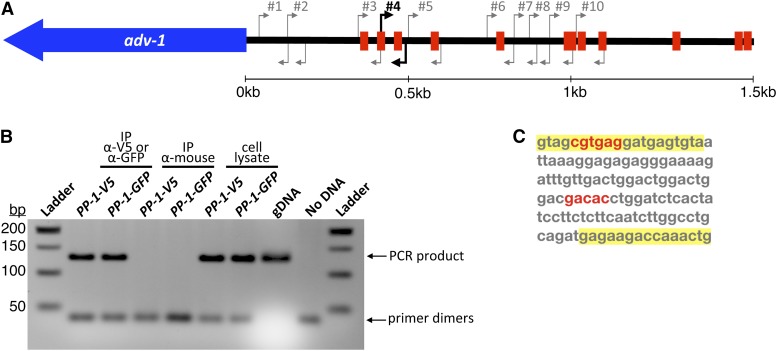
Given that PP-1 was required for expression of adv-1, and misexpression of adv-1 was sufficient to suppress the phenotype of Δpp-1 cells, we were surprised that our DAP-seq data failed to identify PP-1 binding to the promoter of adv-1. We used the consensus DNA binding motif for each transcription factor (Figure 3B) to search for potential binding sites 2 kb upstream of both adv-1 and pp-1. Potential ADV-1 binding sites were not identified in the promoter region of either adv-1 or pp-1; however, several potential PP-1 binding sites were identified within 2 kb of the adv-1 ORF (Figure 6A). Using antibodies to V5 or GFP, PP-1-bound chromatin was immunoprecipitated from Δpp-1 (Ptef1-pp-1-v5) and Δpp-1 (Pccg1-pp-1-gfp) germlings. Ten different primer sets in the adv-1 promoter region were used to interrogate this pool of PP-1-bound DNA. One primer set successfully amplified a region of DNA ∼500 bp upstream of the adv-1 ORF (Figure 6B and Figure S5). This immunoprecipitated and amplified region is 127 bp long and contains two potential PP-1 binding sites (Figure 6C). These data confirm our hypothesis that PP-1 binds the promoter and regulates expression of adv-1.
Figure 6.
ChIP-PCR identifies a PP-1 binding site ∼500 bp upstream of the adv-1 ORF. (A) Diagram of predicted PP-1 binding sites within 1.5 kb upstream of adv-1 ORF, based on the motif depicted in Figure 2B. Arrows indicate 10 different PCR primer sets used to interrogate immunoprecipitated chromatin. (B) Agarose DNA gel showing results of ChIP-PCR with primer set #4. The remaining PCR results are included in Figure S5. Immunoprecipitation with Δpp-1(Ptef1-pp-1-v5; his-3) and Δpp-1(Pccg1-pp-1-gfp; his-3) strains was performed using α-V5 or α-GFP antibodies. α-mouse antibodies were used as a negative control. Whole-cell lysate and independent N. crassa genomic DNA were included as positive PCR controls, with a PCR reaction lacking DNA as an additional negative control. (C) The sequence of the PCR product in B. Primers highlighted in yellow correspond to primer set #4 in A, and predicted PP-1 binding sites are in red.
Misexpression of adv-1 rescues growth defects of Δmak-1, but not Δmak-2 mutants
We reasoned that because misexpression of adv-1 complemented the phenotype of Δpp-1, then (misexpressed) MEadv-1 or MEpp-1 might also complement the phenotype of MAPK mutants predicted to function upstream of these transcription factors. Previous data indicates that the MAK-1 and MAK-2 MAPK pathways engage in cross-talk and likely function upstream of both PP-1 and ADV-1 (Maerz et al. 2008; Dettmann et al. 2012; Maddi et al. 2012; Leeder et al. 2013; Fu et al. 2014). In an effort to elucidate this MAP -transcription factor network, we misexpressed adv-1 or pp-1 in the terminal MAPK mutant (Δmak-1 and Δmak-2) for each MAPK pathway; misexpression was confirmed by qRT-PCR (Figure 7A).
Figure 7.
Misexpression of adv-1 suppresses the growth phenotype of the Δmak-1 mutant. (A) qRT-PCR data showing mRNA expression levels of adv-1 and pp-1 in Δmak-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)), Δmak-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)), Δmak-2 (Ptef1-adv-1-v5; his-3 (MEadv-1)), and Δmak-2 (Pccg1-pp-1-gfp; his-3 (MEpp-1)) strains compared to Δmak-1, Δmak-2, and wild-type cells. (B) Mean height of aerial hyphae of strains in A 3 days after inoculation (ANOVA + Tukey HSD, *** P = 3.6E−8, n = 6). (C) Mean growth rate per day of strains in A measured over 4 days (ANOVA + Tukey HSD, P < 0.01, n = 3). For all bar plots, error bars indicate SD. (D) Colony morphology of the Δmak-1 mutant relative to the wild-type strain and the Δmak-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)) and Δmak-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)) strains. Photos showing a lack of (E) germling fusion (Bar, 5 μm) or (F) hyphal fusion (Bar, 10 μm) for each of the strains shown in A. Arrows indicate chemotropic interactions in the wild-type strain. Plates shown in D are 8 cm in diameter.
Expression of adv-1 was effectively zero in Δmak-1 and Δmak-2 germlings as compared to wild-type germlings (P = 3.9e−19 and P = 1.4e−23, respectively, Welch’s t-test, n = 8), while expression of pp-1 was reduced only in Δmak-2 cells (P = 1.15e−9, Welch’s t-test, n = 8) (Figure 7A). These data indicate that MAK-2 functions upstream of both pp-1 and adv-1, while MAK-1 functions upstream of adv-1 independently of pp-1. To test the effect of misexpressing pp-1 in Δmak-2 cells, we used a Δmak-2 (Pccg1-pp-1-gfp) strain; attempts to obtain Δmak-2 (Ptef1-pp-1-v5) transformants were unsuccessful. The expression of Pccg1-pp-1-gfp complemented the germling communication, cell fusion, aerial hyphae, and growth rate phenotype of Δpp-1 cells, but not the sexual defects of the Δpp-1 mutant (Figure S3) (Leeder et al. 2013). In contrast to the restoration of adv-1 expression in Δpp-1 (Ptef1-pp-1-v5) germlings (Figure 4A) and in Δpp-1 (Ptef1-pp-1-gfp) germlings (Figure S3A), neither Δmak-2 (Pccg1-pp-1-gfp) nor Δmak-1(Ptef1-pp-1-v5) germlings showed increased expression of adv-1 as compared to Δmak-2 or Δmak-1 germlings, respectively (P = 0.73 and 0.06, respectively, Welch’s t-test, n = 8). These data indicate that MAK-1 and MAK-2 are necessary for PP-1-dependent transcription of adv-1.
We next assessed the phenotypes of each misexpression mutant. In contrast to Δpp-1 (Ptef1-adv-1-v5), Δpp-1 (Ptef1-pp-1-v5), or Δpp-1 (Pccg1-pp-1-gfp) cells, misexpression of Ptef1-adv-1-v5 or Pccg1-pp-1-gfp did not affect the phenotype of the Δmak-2 mutant (Figure 7, Figure S3, and Figure S6). The misexpression of Ptef1-pp-1-v5 also had no effect on the phenotype of the Δmak-1 mutant. However, the misexpression of Ptef1-adv-1-v5 significantly affected the growth phenotype of the Δmak-1 mutant (Figure 7, B–D). The introduction of Ptef1-adv-1-v5 into Δmak-1 cells was sufficient to fully rescue the growth rate defect of the Δmak-1 mutant (Figure 7C). The Δmak-1 (Ptef1-adv-1-v5) strain also produced significantly taller aerial hyphae than the Δmak-1 mutant itself (P = 3.6 × 10−8, Welch’s t-test, n = 6) (Figure 7B), and misexpression of Ptef1-adv-1-v5 was sufficient to rescue the compact, rosette-like colony morphology of the Δmak-1 mutant (Figure 7D). However, the introduction of Ptef1-adv-1-v5 was insufficient to rescue the communication and cell fusion defects of the Δmak-1 mutant (Figure 7, E and F), including protoperithecial formation and perithecial development (Figure S6). Together these data indicate that the growth defects of the Δmak-1 mutant can be explained by a simple lack of ADV-1. However, for cell-to-cell communication, cell fusion, and sexual reproduction, MAK-1 is clearly necessary, even in the presence of misexpressed adv-1.
Δpp-1, Δmak-2, and Δmak-1 cells are sensitive to cell wall stress, and misexpression of adv-1 or pp-1 increases resistance in sensitive strains
MAK-1 is the terminal MAP kinase in the cell wall integrity (CWI) pathway, which maintains the cell wall during growth and in response to stress (Park et al. 2008; Maddi et al. 2012). Previous studies demonstrated that the Δmak-1 mutant and other mutants in the CWI pathway are sensitive to the cell wall targeting drugs Caspofungin and Calcofluor White (Maddi et al. 2012). We reasoned that if Ptef1-adv-1-v5 was sufficient to rescue the growth defects of the Δmak-1 mutant, then misexpression of Ptef1-adv-1-v5 might also be sufficient to suppress sensitivity to cell wall stress reagents in Δmak-1 cells. Additionally, since our data suggested that the MAK-1 and MAK-2 pathways function upstream of both ADV-1 and PP-1, we hypothesized that Δadv-1, Δpp-1, and Δmak-2 cells may also be more sensitive to cell wall targeting drugs.
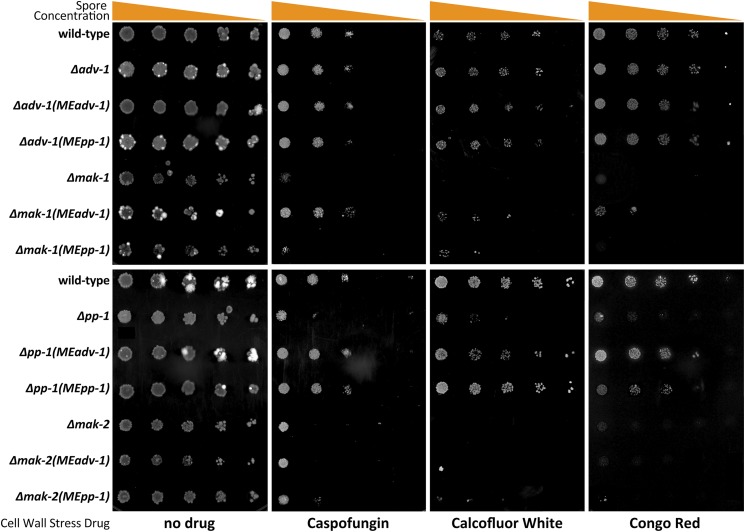
To test these hypotheses, we assessed growth on agar media containing one of three different cell wall stress drugs: the β-1,3-glucan synthase inhibitor Caspofungin, and two different anionic dyes that bind chitin and block chitin–glucan cross-linking, Calcofluor White and Congo Red. Wild-type and Δadv-1 cells were mildly sensitive to all three drugs, while Δpp-1, Δmak-1, and Δmak-2 mutants were almost completely unable to grow on all three drugs (Figure 8). Similar to our other growth phenotype data, both the Δpp-1 (Ptef1-pp-1-v5) and Δpp-1 (Ptef1-adv-1-v5) strains showed wild-type-like resistance to all three drugs. Additionally, the misexpression of Ptef1-adv-1-v5 in Δmak-1 cells increased its resistance to all three drugs, although this effect was more pronounced on Caspofungin than on Calcofluor White or Congo Red. The misexpression of Ptef1-pp-1-v5 in Δmak-1 cells modestly increased resistance on Calcofluor White only. Similarly, the misexpression of Pccg1-pp-1-gfp in Δmak-2 cells modestly increased its resistance to all three drugs, while misexpression of Ptef1-adv-1-v5 in Δmak-2 cells did not affect resistance. As expected, the introduction of Ptef1-pp-1-v5 and Ptef1-pp-1-gfp into the Δpp-1 mutant was sufficient to complement the growth defects of the Δpp-1 mutant on all three drugs (Figure S7). These data indicate that adv-1 and pp-1 function downstream of both MAK-2 and MAK-1 to regulate cell wall stress-responsive gene expression.
Figure 8.
Misexpression of adv-1 restores resistance to cell wall stress agents in Δpp-1 and Δmak-1 cells. A 1:5 serial dilution from ∼5000 spores per spot to ∼8 spores per spot was performed on Δpp-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)), Δpp-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)), Δmak-2 (Ptef1-adv-1-v5; his-3 (MEadv-1)), Δmak-2 (Pccg1-pp-1-gfp; his-3 (MEpp-1)), Δadv-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)), Δadv-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)), Δmak-1 (Ptef1-adv-1-v5; his-3 (MEadv-1)), and Δmak-1 (Ptef1-pp-1-v5; his-3 (MEpp-1)) cells compared to Δpp-1, Δadv-1, Δmak-1, Δmak-2, and wild-type cells. All agar media contains VMM and FGS to force colonial growth. Plates were incubated at 30° for 5 days. Drug concentrations: 1.3 μg/ml Caspofungin, 1.5 mg/ml Calcofluor White, and 1 mg/ml Congo Red.
Discussion
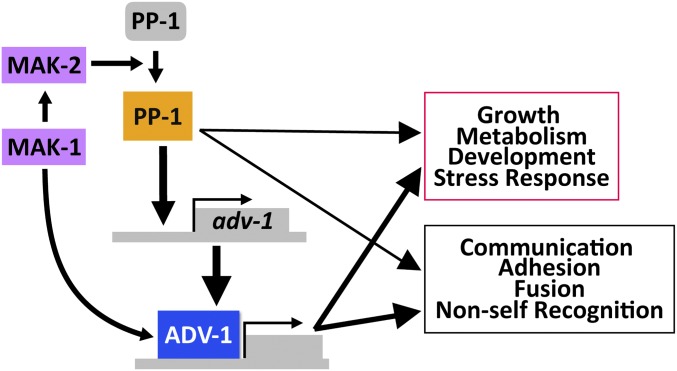
Our data supports a model for integrated phosphorylation and transcriptional regulation of genes involved in communication, fusion, growth, development, and cell wall stress response by a network of two MAPK pathways and two transcription factors (Figure 9). The MAK-2 pathway primarily functions upstream of PP-1, while the MAK-1/CWI pathway functions upstream of ADV-1. However, PP-1 also directly regulates transcription of adv-1, and there are several points of phosphorylation-mediated cross-talk between the MAK-1 and MAK-2 pathways (Maerz et al. 2008; Dettmann et al. 2012, 2013; Maddi et al. 2012; Leeder et al. 2013; Fu et al. 2014). Additionally, the catalytic activity of MAK-2 is essential for its function (Leeder et al. 2013; Jonkers et al. 2014). There are seven MAK-2-dependent phosphorylation sites on PP-1 (Jonkers et al. 2014), and the catalytic activity of MAK-2 is required for expression of many PP-1 (and ADV-1) regulated genes (Leeder et al. 2013). Our qRT-PCR data demonstrated that both MAK-1 and MAK-2 were required for PP-1-dependent transcription of adv-1 (Figure 7A). Our data also indicate that MAK-1 influences adv-1-dependent transcription both directly and indirectly via MAK-2 and PP-1. These data combined with the observation that phosphorylation of MAK-2 is reduced in the Δmak-1 mutant (Maerz et al. 2008; Dettmann et al. 2012) indicate that MAK-1 is required for full activation of MAK-2, and that MAK-2 is essential for activating or derepressing PP-1, likely via phosphorylation. Thus, in the absence of fully activated MAK-2, misexpression of pp-1 alone is not sufficient to trigger transcription of adv-1. Additionally, our observations that MAK-2 is essential for optimal growth rate and resistance to cell wall stress despite misexpression of pp-1 or adv-1 (Figure 7 and Figure 8) suggests that the catalytic activity of MAK-2 may also be important for post-transcriptional regulation of genes important for growth and cell wall stress response.
Figure 9.
Model for transcriptional regulation by MAK-1, MAK-2, PP-1, and ADV-1. MAK-1 activates both MAK-2- and ADV-1-dependent transcription. MAK-2 activates or derepresses PP-1, which is necessary for transcription of adv-1 and other downstream genes (inactivated PP-1 is gray, activated PP-1 is orange). PP-1 directly binds and regulates transcription of adv-1; ADV-1 is the direct transcriptional activator of many of the genes required for cell-to-cell communication, cell fusion, growth, development, and metabolism. Additionally, PP-1 and ADV-1 are important for mediating the cell wall stress response downstream of both MAK-1 and MAK-2. While our data indicates that ADV-1 is the primary regulatory for many downstream genes, PP-1 also contributes to the transcription of some of these genes independently of adv-1. Downstream gene groups are boxed with colors (magenta or black) that match the colors detailing the same groups in Figure 2.
The proteins that are dependent on MAK-2 for phosphorylation (either directly or indirectly) are enriched for proteins involved in growth, cell cycle progression, development, signaling, and metabolism (Jonkers et al. 2014). There are 40 genes that are both phosphorylated in a MAK-2-dependent manner (Jonkers et al. 2014) and regulated by ADV-1 or PP-1 [1 < log2 fold change < −1, P < 0.01, both our data and data from Dekhang et al. (2017)] (Table S3). These 40 genes include the CWI pathway MAPKK mek-1, two hypothetical proteins required for germling communication and fusion (ham-9 and ham-11), one gene that modulates long-distance nonself recognition (doc-2), and several genes broadly involved in growth and metabolism. These genes, in addition to the other communication genes that are regulated by ADV-1 and PP-1 may represent a positive feedback loop, in which pathway activation leads to reinforcement. This is particularly true where MEK-1 phosphorylates MAK-1, and then MAK-1 activates transcription of adv-1, which in turn activates transcription of MEK-1.
In addition to the communication and fusion genes, there are three calcium signaling genes that are both phosphorylated by MAK-2 (Jonkers et al. 2014) and regulated by ADV-1 and PP-1. These genes, a calcineurin binding protein (NCU01504), a calmodulin-dependent kinase (NCU09212), and a predicted Ca2+/cation channel (NCU08283) are particularly interesting because calcium is required for polarized growth, cell fusion, and stress response (Silverman-Gavrila 2003; Palma-Guerrero et al. 2014, 2015; Virgilio et al. 2017). In other systems, the integration of calcium signaling pathways with MAPK pathways has been well documented. For example, in the dicot plant Nicotiana benthamiana, ethylene mediates cross-talk between a hormone-responsive MAPK pathway and a calcium signaling pathway (Ludwig et al. 2005).
Once activated, the CWI and MAK-2 pathways engage in cross-talk that modulates their responses. The signal(s) and receptor(s) involved in germling communication remain elusive, but it is likely that an unidentified receptor leads to activation of the MAK-2 pathway. An individual receptor may activate both CWI and MAK-2 pathways, similar to how receptor-associated Ras-GTPase activates both the ERK pathway (mak-2 ortholog) and the PI3K-TORC1 MAPK pathway to regulate cell survival, proliferation, and metabolism in mammalian cells (Mendoza et al. 2011). Alternatively, there could be more than one receptor such that some receptors lead to activation of the MAK-2 pathway, while other receptors lead to activation of the CWI pathway. The cell wall sensors HAM-7 and WSC-1 both function in the CWI pathway and are required for phosphorylation of MAK-1, and to a lesser extent MAK-2 (Maddi et al. 2012). However, ham-7 is essential for germling fusion while wsc-1 is dispensable, indicating that these two inputs are differentially interpreted through the CWI pathway. The current model suggests that the MAK-2 pathway is primarily activated or repressed by the sensing of extracellular self or nonself signals, similar to the orthologous pheromone sensing pathway in yeast (Heller et al. 2016). Once activated, the MAK-2 pathway works with the CWI pathway to coordinate growth and prepare for cell fusion by regulating expression of genes involved in adhesion, cell wall remodeling, membrane merging, and postfusion nonself recognition. Cross-talk reinforces signaling between these pathways when the integrity of the cell wall is maintained and an extracellular self signal is present. However, if cell wall integrity is compromised or if extracellular signaling is dampened, then signaling through both the CWI pathway and the MAK-2 is adjusted to confer the correct response to cell wall stress or termination of communication.
MAPKs are evolutionarily conserved central regulators for a broad range of cellular processes (Caffrey et al. 1999; Xu et al. 2017). Cross-talk between MAPK pathways allows for efficient integration of multiple inputs into multiple outputs while increasing the robustness and plasticity of a signaling network (Jordan et al. 2000; Barabási and Oltvai 2004; Komarova et al. 2005). In S. cerevisiae, the CWI pathway and the osmotic stress (HOG) MAPK pathway respond to environmental stress by integrating input from several different cell-surface sensors via cross-activation and downstream cross-talk, which results in a transcriptional response controlled by several different transcription factors (Rodríguez-Peña et al. 2010). Similarly, flowering plants such as A. thaliana and Oryza sativa have complex MAPK pathways that are characterized by having several modular MAPKKKs and MAPKs for each individual MAPKK (Fujita et al. 2006; Chardin et al. 2017).
In N. crassa and closely related S. macrospora, the Striatin-Interacting protein Phosphatase and Kinase (STRIPAK) complex regulates MAPK signaling and mediates some cross-talk between the CWI and MAK-2 pathways. The STRIPAK complex localizes to the nuclear envelope, and mutants in the STRIPAK complex have a similar phenotype to mutants in the CWI or MAK-2 pathways (Simonin et al. 2010; Bernhards and Pöggeler 2011; Fu et al. 2011; Dettmann et al. 2013; Nordzieke et al. 2015; Beier et al. 2016; Kück et al. 2016). Both MAK-1 and MAK-2 have been observed in the nucleus, and the amount of MAK-1 inside the nucleus is dependent on MAK-2 phosphorylating MOB-3, which is a core component of the STRIPAK complex. Furthermore, phosphorylation of MAK-1 is reduced in both the Δmob-3 mutant and another STRIPAK complex mutant, Δham-3 (Dettmann et al. 2012, 2013). Phosphorylated MOB-3 is also broadly required for fruiting body development, but not chemotropic interactions or STRIPAK complex assembly at the nuclear envelope (Dettmann et al. 2013). These data illustrate another potential positive feedback loop in which MAK-1 is required for full phosphorylation of MAK-2, and then MAK-2 phosphorylates MOB-3, which is required for both full phosphorylation of MAK-1 and entry of MAK-1 into the nucleus. Furthermore, the STRIPAK complex clearly mediates CWI-MAK-2 pathway cross-talk and modulates the output of both pathways. Future experiments will dissect the interconnected signaling network of the STRIPAK complex, the CWI pathway, and the MAK-2 pathway. Additionally, further characterization of cell-to-cell communication proteins will reveal novel and potentially conserved features of cell-to-cell communication, cell wall dissolution, and membrane merger across the fungal kingdom.
Acknowledgments
This work was funded by a grant from the National Science Foundation to N.L.G. (MCB1412411). M.S.F. was supported by National Institutes of Health (NIH) Genetics training grant #5T32GM007127-40. V.W.W. was supported by NIH Genetics training grant #5T32GM007127-39. We acknowledge the use of deletion strains generated by grant P01 GM068087 “Functional Analysis of a Model Filamentous Fungus,” which are publicly available at the FGSC. The RNA-seq library preparation and sequencing was carried out at the DNA Technologies and Expression Analysis Cores at the University of California, Davis Genome Center, supported by NIH Shared Instrumentation grant 1S10OD010786-01. DAP-seq work was conducted by the US Department of Energy Joint Genome Institute, a Department of Energy Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6146681.
Communicating editor: M. Freitag
Literature Cited
- Barabási A. L., Oltvai Z. N., 2004. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- Bastiaans E., Debets A. J., Aanen D. K., 2015. Experimental demonstration of the benefits of somatic fusion and the consequences for allorecognition. Evolution 69: 1091–1099. 10.1111/evo.12626 [DOI] [PubMed] [Google Scholar]
- Beier A., Teichert I., Krisp C., Wolters D. A., Kück U., 2016. Catalytic subunit 1 of protein phosphatase 2A is a subunit of the STRIPAK complex and governs fungal sexual development. MBio 7: e00870–16 10.1128/mBio.00870-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. D., Beremand P., Thomas T. L., Bell-Pedersen D., 2013. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryot. Cell 12: 59–69. 10.1128/EC.00207-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhards Y., Pöggeler S., 2011. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr. Genet. 57: 133–149. 10.1007/s00294-010-0333-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E., Kim H.-J. N., Stone D. E., 2007. The pheromone-induced nuclear accumulation of the Fus3 MAPK in yeast depends on its phosphorylation state and on Dig1 and Dig2. BMC Cell Biol. 8: 44 10.1186/1471-2121-8-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey D. R., O’neill L. A., Shields D. C., 1999. The evolution of the MAP kinase pathways: coduplication of interacting proteins leads to new signaling cascades. J. Mol. Evol. 49: 567–582. 10.1007/PL00006578 [DOI] [PubMed] [Google Scholar]
- Chardin C., Schenk S. T., Hirt H., Colcombet J., Krapp A., 2017. Review: mitogen-activated protein kinases in nutritional signaling in Arabidopsis. Plant Sci. 260: 101–108. 10.1016/j.plantsci.2017.04.006 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 (erratum: Proc. Natl. Acad. Sci. USA 103: 16614) 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekhang R., Wu C., Smith K. M., Lamb T. M., Peterson M., et al. , 2017. The Neurospora transcription factor ADV-1 transduces light signals and temporal information to control rhythmic expression of genes involved in cell fusion. G3 (Bethesda) 7: 129–142. 10.1534/g3.116.034298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmann A., Illgen J., März S., Schürg T., Fleissner A., et al. , 2012. The NDR kinase scaffold HYM1/MO25 is essential for MAK2 MAP kinase signaling in Neurospora crassa. PLoS Genet. 8: e1002950 10.1371/journal.pgen.1002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmann A., Heilig Y., Ludwig S., Schmitt K., Illgen J., et al. , 2013. HAM-2 and HAM-3 are central for the assembly of the Neurospora STRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol. Microbiol. 90: 796–812. 10.1111/mmi.12399 [DOI] [PubMed] [Google Scholar]
- Dettmann A., Heilig Y., Valerius O., Ludwig S., Seiler S., 2014. Fungal communication requires the MAK-2 pathway elements STE-20 and RAS-2, the NRC-1 adapter STE-50 and the MAP kinase scaffold HAM-5. PLoS Genet. 10: e1004762 10.1371/journal.pgen.1004762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Festa R. A., Chen Y.-L., Espart A., Palacios Ò., et al. , 2013. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13: 265–276. 10.1016/j.chom.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh I., Nowrousian M., Kück U., 2010. Sordaria macrospora, a model organism to study fungal cellular development. Eur. J. Cell Biol. 89: 864–872. 10.1016/j.ejcb.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Errede B., Ammerer G., 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3: 1349–1361. 10.1101/gad.3.9.1349 [DOI] [PubMed] [Google Scholar]
- Fleissner A., Diamond S., Glass N. L., 2008. The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics 181: 497–510. 10.1534/genetics.108.096149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A., Leeder A. C., Roca M. G., Read N. D., Glass N. L., 2009. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. USA 106: 19387–19392. 10.1073/pnas.0907039106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleißner A., Herzog S., 2016. Signal exchange and integration during self-fusion in filamentous fungi. Semin. Cell Dev. Biol. 57: 76–83. 10.1016/j.semcdb.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U., Read N. D., 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41: 897–910. 10.1016/j.fgb.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Fu C., Iyer P., Herkal A., Abdullah J., Stout A., et al. , 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10: 1100–1109. 10.1128/EC.05003-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Ao J., Dettmann A., Seiler S., Free S. J., 2014. Characterization of the Neurospora crassa cell fusion proteins, HAM-6, HAM-7, HAM-8, HAM-9, HAM-10, AMPH-1 and WHI-2. PLoS One 9: e107773 10.1371/journal.pone.0107773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., et al. , 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442. 10.1016/j.pbi.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Garnjobst L., Wilson J. F., 1952. Heterocaryosis and protoplasmic incompatibility in Neurospora crassa. Proc. Natl. Acad. Sci. USA 42: 613–618. 10.1073/pnas.42.9.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12: 135–141. 10.1016/j.tim.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Kaneko I., 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 2: 1–8. 10.1128/EC.2.1.1-8.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Vollmer S., Staben C., Grotelueschen J., Metzenberg R. L., et al. , 1988. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241: 570–573. 10.1126/science.2840740 [DOI] [PubMed] [Google Scholar]
- Han K. H., Kim J. H., Moon H., Kim S., Lee S. S., et al. , 2008. The Aspergillus nidulans esdC (early sexual development) gene is necessary for sexual development and is controlled by veA and a heterotrimeric G protein. Fungal Genet. Biol. 45: 310–318. 10.1016/j.fgb.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Harner M. E., Unger A. K., Izawa T., Walther D. M., Özbalci C., et al. , 2014. Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife 3: e01684 10.7554/eLife.01684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Zhao J., Rosenfield G., Kowbel D. J., Gladieux P., et al. , 2016. Characterization of greenbeard genes involved in long-distance kind discrimination in a microbial eukaryote. PLoS Biol. 14: e1002431 10.1371/journal.pbio.1002431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Clavé C., Gladieux P., Saupe S. J., Glass N. L., 2018. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. USA 115: E2292–E2301. 10.1073/pnas.1719705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Galván M., Cano-Domínguez N., Robledo-Briones A., Castro-Longoria E., Lichius A., et al. , 2015. The tetraspanin PLS-1 is required for NOX-1 and NOX-2 functions in cell fusion, cell growth and differentiation in the fungus Neurospora crassa, pp. 66 in Fungal Genetics Reports, 61. New Prairie Press, Pacific Grove, CA. [Google Scholar]
- Hickey P. C., Jacobson D. J., Read N. D., Glass N. L., 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37: 109–119. 10.1016/S1087-1845(02)00035-X [DOI] [PubMed] [Google Scholar]
- Jonkers W., Leeder A. C., Ansong C., Wang Y., Yang F., et al. , 2014. HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLoS Genet. 10: e1004783 10.1371/journal.pgen.1004783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J. D., Landau E. M., Iyengar R., 2000. Signaling networks: the origins of cellular multitasking. Cell 103: 193–200. 10.1016/S0092-8674(00)00112-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko I., Dementhon K., Xiang Q., Glass N. L., 2006. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172: 1545–1555. 10.1534/genetics.105.051490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Borkovich K. A., 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52: 1781–1798. 10.1111/j.1365-2958.2004.04096.x [DOI] [PubMed] [Google Scholar]
- Kim H., Borkovich K. A., 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5: 544–554. 10.1128/EC.5.3.544-554.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N. L., Zou X., Nie Q., Bardwell L., 2005. A theoretical framework for specificity in cell signaling. Mol. Syst. Biol. 1: E1–E5. 10.1038/msb4100031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U., Beier A. M., Teichert I., 2016. The composition and function of the striatin-interacting phosphatases and kinases (STRIPAK) complex in fungi. Fungal Genet. Biol. 90: 31–38. 10.1016/j.fgb.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Lalucque H., Malagnac F., Green K., Gautier V., Grognet P., et al. , 2016. IDC2 and IDC3, two genes involved in cell non-autonomous signaling of fruiting body development in the model fungus Podospora anserina. Dev. Biol. 421: 126–138. 10.1016/j.ydbio.2016.12.016 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeder A. C., Jonkers W., Li J., Glass N. L., 2013. Early colony establishment in Neurospora crassa requires a MAP kinase regulatory network. Genetics 195: 883–898. 10.1534/genetics.113.156984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170: 1091–1104. 10.1534/genetics.104.036772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A. A., Saitoh H., Felix G., Freymark G., Miersch O., et al. , 2005. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102: 10736–10741. 10.1073/pnas.0502954102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddi A., Dettman A., Fu C., Seiler S., Free S. J., 2012. WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa. PLoS One 7: e42374 10.1371/journal.pone.0042374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz S., Ziv C., Vogt N., Helmstaedt K., Cohen N., et al. , 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179: 1313–1325. 10.1534/genetics.108.089425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masloff S., Pöggeler S., Kück U., 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masloff S., Jacobsen S., Pöggeler S., Kück U., 2002. Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 36: 107–116. 10.1016/S1087-1845(02)00010-5 [DOI] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K., 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M. C., Er E. E., Blenis J., 2011. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36: 320–328. 10.1016/j.tibs.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Dudin O., Martin S. G., 2013. Mate and fuse: how yeast cells do it. Open Biol. 3: 130008 10.1098/rsob.130008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordzieke S., Zobel T., Fränzel B., Wolters D. A., Kück U., et al. , 2015. A fungal sarcolemmal membrane-associated protein (SLMAP) homolog plays a fundamental role in development and localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria. Eukaryot. Cell 14: 345–358. 10.1128/EC.00241-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Frank S., Koers S., Strauch P., Weitner T., et al. , 2007. The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 64: 923–937. 10.1111/j.1365-2958.2007.05694.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R. C., Huang S. C., Song L., Lewsey M. G., Bartlett A., et al. , 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 (erratum: Cell 166: 1598) 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J., Hall C. R., Kowbel D., Welch J., Taylor J. W., et al. , 2013. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 9: e1003669 10.1371/journal.pgen.1003669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J., Leeder A. C., Welch J., Glass N. L., 2014. Identification and characterization of LFD1, a novel protein involved in membrane merger during cell fusion in Neurospora crassa. Mol. Microbiol. 92: 164–182. 10.1111/mmi.12545 [DOI] [PubMed] [Google Scholar]
- Palma-Guerrero J., Zhao J., Gonçalves A. P., Starr T. L., Glass N. L., 2015. Identification and characterization of LFD-2, a predicted fringe protein required for membrane integrity during cell fusion in Neurospora crassa. Eukaryot. Cell 14: 265–277. 10.1128/EC.00233-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A., Maheshwari R., 1994. A simple method of obtaining pure microconidia in Neurospora crassa. Fungal Genet. Newsl. 41: 64–65. [Google Scholar]
- Park G., Pan S., Borkovich K. A., 2008. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot. Cell 7: 2113–2122. 10.1128/EC.00466-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B., 2003. Communication is the key. Cell Commun. Signal. 1: 3 10.1186/1478-811X-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read N. D., Goryachev A. B., Lichius A., 2012. The mechanistic basis of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biol. Rev. 26: 1–11. 10.1016/j.fbr.2012.02.003 [DOI] [Google Scholar]
- Richard F., Glass N. L., Pringle A., 2012. Cooperation among germinating spores facilitates the growth of the fungus, Neurospora crassa. Biol. Lett. 8: 419–422. 10.1098/rsbl.2011.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J. L., Pachter L., 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12: R22 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., García R., Nombela C., Arroyo J., 2010. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast 27: 495–502. 10.1002/yea.1792 [DOI] [PubMed] [Google Scholar]
- Saupe S. J., 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64: 489–502. 10.1128/MMBR.64.3.489-502.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman-Gavrila L. B., 2003. Calcium gradient dependence of Neurospora crassa hyphal growth. Microbiology 149: 2475–2485. 10.1099/mic.0.26302-0 [DOI] [PubMed] [Google Scholar]
- Simonin A., Palma-Guerrero J., Fricker M., Glass N. L., 2012. Physiological significance of network organization in fungi. Eukaryot. Cell 11: 1345–1352. 10.1128/EC.00213-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin A. R., Rasmussen C. G., Yang M., Glass N. L., 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47: 855–868. 10.1016/j.fgb.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Smith K. M., Sancar G., Dekhang R., Sullivan C. M., Li S., et al. , 2010. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora white collar complex. Eukaryot. Cell 9: 1549–1556. 10.1128/EC.00154-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert I., Steffens E. K., Schnaß N., Fränzel B., Krisp C., et al. , 2014. PRO40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase C. PLoS Genet. 10: e1004582 10.1371/journal.pgen.1004582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio S., Cupertino F. B., Ambrosio D. L., Bertolini M. C., 2017. Regulation of the reserve carbohydrate metabolism by alkaline pH and calcium in Neurospora crassa reveals a possible cross-regulation of both signaling pathways. BMC Genomics 18: 457 10.1186/s12864-017-3832-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora (Medium N). Microb. Genet. Bull. 13: 42–43. [Google Scholar]
- Weichert M., Lichius A., Priegnitz B.-E., Brandt U., Gottschalk J., et al. , 2016. Accumulation of specific sterol precursors targets a MAP kinase cascade mediating cell–cell recognition and fusion. Proc. Natl. Acad. Sci. USA 113: 11877–11882. 10.1073/pnas.1610527113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M. T., Yang A., Albu M., Cote A. G., Montenegro-Montero A., et al. , 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. 10.1002/j.1537-2197.1947.tb13032.x [DOI] [Google Scholar]
- Xiong Y., Coradetti S. T., Li X., Gritsenko M. A., Clauss T., et al. , 2014. The proteome and phosphoproteome of Neurospora crassa in response to cellulose, sucrose and carbon starvation. Fungal Genet. Biol. 72: 21–33. 10.1016/j.fgb.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Liu R., Zhang Q., Chen X., Qian Y., et al. , 2017. The diversification of evolutionarily conserved MAPK cascades correlates with the evolution of fungal species and development of lifestyles. Genome Biol. Evol. 9: 311–322. 10.1093/gbe/evw051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Some strains are also publicly available through the FGSC. DAP-seq raw data are available at the NCBI Sequence Read Archive with accession number (SRP133627). RNA-seq raw data (.fastq) is available at the NCBI Sequence Read Archive, accession number SRP133239. Differential expression analysis outputs are included with this publication as Supplemental Material, File S1. File S2 includes detailed information about the 155 differentially expressed genes that we focused on. Results from DAP-seq data analysis are in File S3. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6146681.