Histone H3.3, a major variant of canonical histone H3, is highly conserved and essential for viability or fertility in most lineages. In Caenorhabditis elegans, H3.3 is expressed from five genes. Here, Delaney...
Keywords: Histone variants, H3.3, germ line, C. elegans, stress response, HIRA
Abstract
Replication-independent variant histones replace canonical histones in nucleosomes and act as important regulators of chromatin function. H3.3 is a major variant of histone H3 that is remarkably conserved across taxa and is distinguished from canonical H3 by just four key amino acids. Most genomes contain two or more genes expressing H3.3, and complete loss of the protein usually causes sterility or embryonic lethality. Here, we investigate the developmental expression patterns of the five Caenorhabditis elegans H3.3 homologs and identify two previously uncharacterized homologs to be restricted to the germ line. Despite these specific expression patterns, we find that neither loss of individual H3.3 homologs nor the knockout of all five H3.3-coding genes causes sterility or lethality. However, we demonstrate an essential role for the conserved histone chaperone HIRA in the nucleosomal loading of all H3.3 variants. This requirement can be bypassed by mutation of the H3.3-specific residues to those found in H3. While even removal of all H3.3 homologs does not result in lethality, it leads to reduced fertility and viability in response to high-temperature stress. Thus, our results show that H3.3 is nonessential in C. elegans but is critical for ensuring adequate response to stress.
HISTONES are highly conserved proteins that assemble into heterotypic octamers to organize DNA into nucleosomes. Besides the replication-dependent “canonical” histones that organize newly synthesized DNA during S-phase, most lineages have evolved histone variants that are expressed independently of DNA replication and can replace the canonical histones in nucleosomes to carry out specialized functions. H3.3 is a major variant of histone H3 that is highly conserved among eukaryotes, with 94–97% amino acid sequence identity with canonical H3 (Supplemental Material, Figure S1A). Four key amino acid positions specify H3.3. Three of these residues (amino acids 87, 89, and 90) mediate recognition by the H3.3-specific histone chaperones HIRA and DAXX (Filipescu et al. 2013; Ricketts and Marmorstein 2017), and amino acid 31 can be modified and can influence the methylation of lysine 27 (Hake et al. 2005; Jacob et al. 2014). Genomic studies have revealed that H3.3 is incorporated into transcriptionally active regions as well as centromeric and telomeric regions (Henikoff and Smith 2015; Buschbeck and Hake 2017). Incorporation at transcriptionally active regions depends on HIRA, while incorporation at heterochromatic regions is mediated by ATRX and DAXX (Filipescu et al. 2013; Mattiroli et al. 2015; Dyer et al. 2017).
H3.3 has been implicated in a variety of biological processes: it is important for embryonic stem cell differentiation (Banaszynski et al. 2013), epigenetic reprogramming following somatic cell nuclear transfer (Jullien et al. 2012; Wen et al. 2014a,b), neuron plasticity (Maze et al. 2015), the DNA damage response (Adam et al. 2013; Frey et al. 2014), and centromere maintenance (Dunleavy et al. 2011). Moreover, specific H3.3 mutations have been identified as recurrent somatic driver mutations in gliomas and a subset of skeletal neoplasms (Schwartzentruber et al. 2012; Sturm et al. 2012; Wu et al. 2012; Behjati et al. 2013).
The importance of H3.3 is manifested in developmental defects upon loss or depletion of the protein in most organisms. Flies that lack both H3.3 genes (His3.3A and His3.3B) have reduced viability and individuals that survive to adulthood are completely sterile in both sexes (Hödl and Basler 2009; Sakai et al. 2009). H3.3 is also essential for germ line development in mammals, where it is required for the remodeling of both maternal and paternal gametes (Santenard et al. 2010; Akiyama et al. 2011). Deletion of both H3.3 genes (H3f3a and H3f3b) in mice results in early primary oocyte death. Moreover, deletion of H3f3b alone leads to developmental deficiencies and death at birth, and heterozygotes are male sterile. Deletion of H3f3a alone results in postnatal mortality, slow growth, and reduced male fertility (Couldrey et al. 1999; Tang et al. 2015). While the majority of nucleosomes are replaced by protamines during mammalian spermatogenesis, some H3.3 nucleosomes are retained and may transmit epigenetic information to the zygote (Erkek et al. 2013; Yuen et al. 2014). After fertilization, knockdown of H3.3 leads to overcondensation and missegregation of chromosomes, and developmental arrest at the morula stage in mice (Lin et al. 2013). Similarly, H3.3 depletion in Xenopus results in late gastrulation defects (Szenker et al. 2012). In Arabidopsis, removal of three H3.3 genes (HTR4, HTR5, and HTR8) causes defects in male gametogenesis and results in embryonic lethality (Wollmann et al. 2017).
In Caenorhabditis elegans, developmental analysis of H3.3 mutants is still lacking. Genome sequence analysis has revealed that C. elegans contains five H3.3 homologs (his-69, his-70, his-71, his-72, and his-74). Of those five genes, only the two with closest homology to human H3.3 (his-71 and his-72) have been previously studied (Ooi et al. 2006, 2010; Piazzesi et al. 2016). By introducing additional tagged copies of his-71 and his-72 into the genome, these proteins were shown to be localized to the nuclei of nearly every somatic cell, with his-72 also being expressed in the germ line (Ooi et al. 2006). HIS-72 is mainly incorporated into regions of the genome that are associated with active transcription (Ooi et al. 2010). Surprisingly, deletion of his-71 or his-72 did not result in any detectable phenotype, which was attributed to redundancy of these genes (Ooi et al. 2006). However, deletion of his-71 and his-72 affects worm life span when combined with additional mutations in the insulin pathway (Piazzesi et al. 2016).
In this study, we created deletion alleles of the uncharacterized H3.3 homologs and generated gfp fusions at the endogenous loci of all H3.3 genes to better understand the role of this histone variant in C. elegans. Our analysis demonstrates that four of the five H3.3 genes are expressed at detectable levels, and that the expression patterns range from ubiquitous to germ line-specific. Chromatin association of all H3.3 homologs depends on the conserved histone chaperone HIRA-1. Mutation of all five H3.3 homologs revealed that, surprisingly, H3.3 is nonessential in C. elegans, but is required for adequate response to stress at high temperatures.
Materials and Methods
Nematode strains
C. elegans strains were maintained using standard conditions at 20° unless otherwise noted. N2 (Bristol strain) was used as a wild-type strain and all of the genetic modifications were performed in this background unless otherwise indicated. A list of strains used in this study, including the numbers of alleles generated, is given in Table S1. A summary of the deletions and mutations is given in Figure S2.
All insertions and deletions were generated using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-9 technology as described in Arribere et al. (2014). Hira-1, his-69, and his-70 were deleted by removing the complete coding sequences, while his-74 knockout was generated by introducing a frameshift and premature STOP codon (Figure S2, A and B). Gfp fusions were generated by insertion at the 5′ end of each of the H3.3 genes. To change the chaperone specificity for HIS-72::GFP, we mutated the HIRA-recognition AAIG sequence to the canonical H3-specific SAVM sequence (Figure S2C). Single-guide RNAs and repair templates, as well as PCR primers used to detect and sequence the mutations, are listed in Table S2.
For crosses of H3.3::GFP males to feminized hermaphrodites, fem-2(b245) hermaphrodites were shifted from 15 to 25° as L4 larvae. Their adult progenies, which did not produce any sperm, were crossed with H3.3::GFP males at 25°. After 12 hr of crossing, hermaphrodites were dissected to release the embryos. Embryos were mounted on 2% agarose pads and immediately imaged to analyze the dynamics of paternally contributed H3.3::GFP.
Microscopy
For live-cell and DIC imaging, worms were anesthetized with levamisole, mounted on 2% agarose pads, and imaged on a Leica DM5000 B microscope. For confocal imaging, gonads were dissected in anesthetizing buffer (50 mM sucrose; 75 mM HEPES, pH 6.5; 60 mM NaCl; 5 mM KCl; 2 mM MgCl2; 10 mM EGTA, pH 7.5; and 1% NaN3 in PBS). Upon dissection, an equal amount of 5% paraformaldehyde (PFA; Alfa Aesar) in the same buffer was added and incubated for 5 min. Samples were incubated with 0.1% Trition X-100 in PBS for 5 min. Samples were washed with PBS three times for 5 min, stained with DAPI, and mounted with VECTASHIELD Antifade Mounting Medium. Images were obtained with a Leica SP8 confocal microscope. Pictures shown are merged 0.2–0.4 µm Z-sections of the entire nuclei, except for images showing whole gonads, for which 0.8–1 µm Z-sections were taken. Images were obtained with exposure times adjusted for each H3.3::GFP fusion to capture the specific expression patterns, unless otherwise noted, and processed in ImageJ. Images for Figure S6 were obtained from the same slides using the same settings to reflect differences in GFP expression levels.
Staining
For H3K4me3 staining, gonads were prepared as described above. Slides were washed with PBS three times for 5 min and incubated with an H3K4me3 antibody (ab8580, concentration 1:10,000; Abcam) overnight at 4°. Slides were washed with PBS three times for 5 min and incubated with a Cy3-conjugated secondary antibody (711-165-152, concentration 1:700; Jackson Immunoresearch) for 1.5 hr at room temperature. Samples were washed with PBS three times for 5 min, stained with DAPI, and mounted with VECTASHIELD Antifade Mounting Medium.
Detection of apoptotic cells in adult gonads was performed as previously described (Papaluca and Ramotar 2016). Briefly, young adults were incubated in the dark for 2 hr at 20° on OP50-seeded NGM plates containing 1 ml of 50 μg/μl of acridine orange DNA dye (Sigma [Sigma Chemical], St. Louis, MO). Animals were subsequently transferred to fresh OP50-seeded NGM plates and incubated for 2 hr. Acridine orange-positive apoptotic cells were detected and scored by live-cell imaging.
Brood size assessment
To count brood sizes, 20 L4 worms from cultures maintained at 20 or 25° for several generations were singled onto individual plates. The worms were transferred to new plates every 24 hr for 4 days. No egg-laying was observed on subsequent days. To assess embryonic lethality, laid embryos were counted, and hatched worms were recounted the next day. When the F1 progeny reached adulthood, the worms on each plate were counted manually. The sum of the F1 progeny on each of the four plates was used as the brood size. P-values were calculated using the “t.test” function in R.
Heat shock experiments
Worm cultures were maintained at 20 or 25° for several generations. For each experiment, ∼100–200 young adult worms were exposed to a heat shock of 37° for 100 min and then recovered at the maintenance temperature. Preconditioning prior to the heat shock was carried out at 30° for 3.5 hr. The percentage of surviving adult worms was counted after 24 hr. Sample sizes are given in the text. P-values were calculated using the t.test function in R.
RNA sequencing
RNA was isolated from N2 and H3.3 null mutant mixed-stage embryos obtained by bleaching a synchronized adult population grown at 20°, and from synchronized L1 larvae hatched in M9 in an absence of food at 20°. RNA was isolated using TRIzol, with two biological replicates for each sample. Total RNA was quantified with a Qubit fluorimeter (Life Technologies), and RNA integrity assessed with a Bioanalyzer (Agilent Technologies). A TruSeq stranded mRNA kit from Illumina was used for library preparation, with 1000 ng of total RNA as input. Library cDNA concentration and quality was assessed with the Qubit and Tapestation using a DNA high-sensitivity chip (Agilent Technologies). Pools of eight libraries were loaded at 8.5 pM for clustering on a single-read Illumina flow cell. Reads of 50 bases were generated using TruSeq SBS HS v3 chemistry on an Illumina HiSequation 2500 sequencer. The reads (length = 50 bp) were mapped with the TopHat v2.0.13 (default parameters) software to the C. elegans reference genome (WBcel235). Read counts representing the total number of reads aligning to each genomic feature were produced from aligned reads by the Python software htseq-count (–mode = union, with HTSeq v.0.6.1) with the reference .gtf file. Correlation plots of biological replicates are shown in Figure S3. The normalization and differential expression analysis between N2 and H3.3 null mutant samples was performed with the R/Bioconductor package edgeR v.3.10.5 for the genes annotated in the reference genome. Briefly, very low-expressed genes were filtered out and the data were normalized according to the library size. Differentially expressed genes were estimated using a General Linear Model approach, negative binomial distribution, and a quasi-likelihood F-test. Genes with a P-value < 0.05 and fold change > 1 were considered significant. Raw and normalized counts, as well as statistical measures, are provided in Tables S3 and S4.
Gene ontology (GO) enrichment analysis was done using the PANTHER Overrepresentation Test (release 20170413) with the GO Ontology database (release 2017-08-14, http://www.geneontology.org/). We tested for enrichment of GO biological process with all C. elegans genes in the database as a reference list.
Quantitative PCR
Quantitative RT-PCR was executed in four biological replicates following the protocol from Ly et al. (2015) with minor adaptations. N2 and H3.3 null mutant worms were exposed to a heat shock at 37° for 1 hr and recovered at 20°. After 0, 30, 60, and 120 min, five adults worms of each strain were lysed in 5 µl of lysis buffer [50 mM KCl, 10 mM Tris (pH 8.3), 2.5 mM MgCl2, 0.45% NP-40, and 0.45% Tween-20] with 1 mg/ml proteinase K. Next, 1 µl of the lysate was used for cDNA synthesis using the Maxima H Minus cDNA synthesis mix (Thermo Fisher) with random primers, following the manufacturer’s protocol. cDNA was diluted 100× and used to perform quantitative PCR using LightCycler 480 SYBR Green (Roche) and a LightCycler 480 instrument. Primers used to detect act-1, hsp-16.2, and hsp-70 are listed in Table S2. The Hsp-70 and hsp-16.2 signals were normalized with the act-1 signal by ΔΔCt analysis. To obtain relative expression levels, the maximum signal in each biological replicate was set to 1.
Data availability
Strains and plasmids are available upon request. The raw RNA sequencing (RNA-seq) data has been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE106889. Supplemental files are available at FigShare. Figure S1 shows an analysis of C. elegans H3.3 homologs. Figure S2 contains a summary of deletion and mutation alleles. Figure S3 contains correlation plots of biological replicates for the RNA-seq experiments. Figure S4 shows H3.3 homolog expression during embryogenesis. Figure S5 shows H3.3 homolog expression in the hermaphrodite and male germ line. Figure S6 shows a comparison of expression levels of H3.3 homologs. Figure S7 shows that H3.3 proteins are depleted from chromosome X. Figure S8 shows an alignment of C. elegans HIRA-1 with homologs from other species. Figure S9 shows the morphological defects and reduced brood size of hira-1 mutant worms. Figure S10 shows that HIS-71 and HIS-72 are detectable in the postembryonic somatic tissue even upon hira-1 deletion. Figure S11 shows embryonic lethality and apoptosis in H3.3 null mutant worms. Figure S12 shows the identity and expression of additional H3 homologs in C. elegans. Table S1 lists the C. elegans strains used in this study. Table S2 lists the reagents used for allele generation by CRISPR/Cas9 and quantitative PCR. Table S3 contains the RNA-seq results for embryos. Table S4 contains the RNA-seq results for L1 larvae. Table S5 lists all significantly enriched GO terms. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6106448.
Results
C. elegans H3.3 homologs are differentially expressed
Histone H3.3 is distinguished from canonical histone H3 by amino acid changes of alanine 31, serine 87, valine 89, and methionine 90. Moreover, expression of H3.3 genes is uncoupled from DNA replication. Basic local alignment search tool (BLAST) searches identified five genes with homology to human H3.3 in the C. elegans genome: his-69, his-70, his-71, his-72, and his-74 (Figure S1). Previous studies have identified his-71 and his-72 as the main C. elegans homologs of H3.3, based on close sequence homology (99% amino acid sequence identity) with human H3.3 (Ooi et al. 2006; Piazzesi et al. 2016). HIS-69, HIS-70, and HIS-74 have more diverged amino acid sequences, with 89, 79, and 93% sequence identity with human H3.3, respectively. However, all five proteins show the hallmarks of histone H3.3 variants, with amino acid substitutions at positions 31, 87, 89, and 90 (Figure S1). None of the five genes are present in a histone cluster, or are paired and divergently transcribed with an H4 gene, as is typical for replication-dependent H3 genes (Pettitt et al. 2002). Moreover, the 3′-UTRs of the his-70, his-71, his-72, and his-74 genes lack the hairpin sequence characteristic for replication-dependent histones (Pettitt et al. 2002). Based on these criteria, we consider his-69, his-70, his-71, his-72, and his-74 to be genes expressing H3.3 variants.
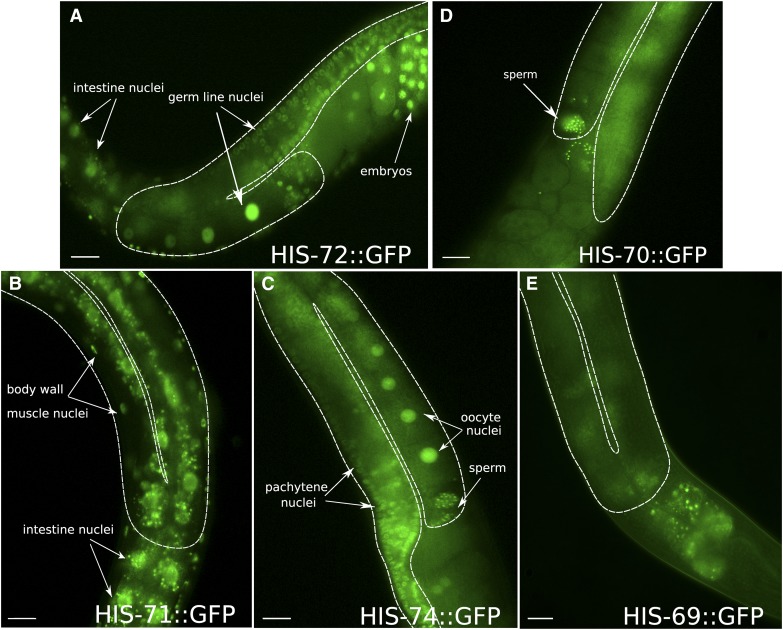
To gain insight into potential redundancies between the H3.3 genes in C. elegans, we created N-terminal gfp fusions for all five H3.3 genes by inserting gfp at the endogenous loci using CRISPR/Cas9 and analyzed the resulting expression patterns. As expected for histone proteins, the GFP signals of the fusion proteins are restricted to nuclei in all cells. The expression patterns of the endogenous his-71 and his-72 gfp fusions match those previously observed using transgenic low-copy arrays, with minor differences (Ooi et al. 2006). His-72 is expressed ubiquitously in every cell, both in the soma and germ line at every stage of development in both males and hermaphrodites (Figure 1A, Figure S4A, and Figure S5). Expression appears highest in embryos compared to other developmental stages based on GFP signal, consistent with RNA-seq data (Figure S1B). However, the HIS-72 signal remaines detectable in all cells throughout development. His-71 is expressed only in somatic cells and is undetectable in the germ line. (Figure 1B). His-71 expression is also absent in early embryonic development and only becomes detectable after gastrulation (Figure S4B). At postembryonic stages, somatic expression of his-71 resembles that of his-72, but showes slightly lower nuclear GFP levels compared to his-72 (Figure S6, A and B).
Figure 1.
H3.3 homologs are differentially expressed. Signals of GFP fusions with H3.3 homologs in representative parts of adult hermaphrodites are shown. (A) HIS-72 is expressed ubiquitously in both the soma and germ line. (B) HIS-71 is expressed in somatic cells, but absent from the germ line. (C) HIS-74 expression is restricted to the germ line at all stages of germ line development, including mature oocytes and sperm. (D) HIS-70 is restricted to the male germ line and only detectable in mature sperm in adult hermaphrodites. (E) HIS-69 is not detectable in any cells. The germ line is outlined by a dashed white line. Note the autofluorescence of the intestine in all panels. Bars represent 20 µm.
Strikingly, analysis of the previously uncharacterized H3.3 homologs revealed that his-70 and his-74 show germ line-restricted expression patterns. His-74 expression is restricted to the germ line at all stages of development in both males and hermaphrodites (Figure 1C and Figure S5). HIS-74::GFP is only faintly visible in early embryos, and robustly reappears in the primordial germ cells (PGCs; the germ line precursor cells that will give rise to the entire germ line of the worm) and faintly in some cells surrounding the PGCs (Figure S4C). Expression of his-70 is restricted to the germ line during spermatogenesis and to mature sperm in both males and hermaphrodites, and is absent from all other tissues and developmental stages (Figure 1D, Figure S4D, and Figure S5). His-69 expression is not detected, consistent with barely detectable transcript levels in RNA-seq experiments (Figure 1E and Figure S1B).
Taken together, we found that the H3.3 homologs have distinct, developmentally regulated expression patterns. The expression patterns observed for each GFP fusion closely match RNA expression levels at the different developmental stages (Figure S1B). As previously shown, HIS-72 is expressed ubiquitously in every cell, while HIS-71 is restricted to the soma (Ooi et al. 2006). Remarkably, the two additional H3.3 variants described here, HIS-74 and HIS-70, are restricted to the germ line. HIS-74 is found in both female and male germ cells, while HIS-70 is specific for the male germ line.
Germ line dynamics of H3.3 proteins
In the adult hermaphrodite C. elegans germ line, gametes are produced in an assembly line-like process from the distal stem cell niche to the proximal mature oocytes (Hirsh et al. 1976). This arrangement offers the possibility to image germ cell chromatin during mitosis in the distal region and at different stages of meiosis in the proximal region. Moreover, the condensed chromatin of the different meiotic stages allows for a more detailed analysis of how the different H3.3 homologs are associated with chromatin.
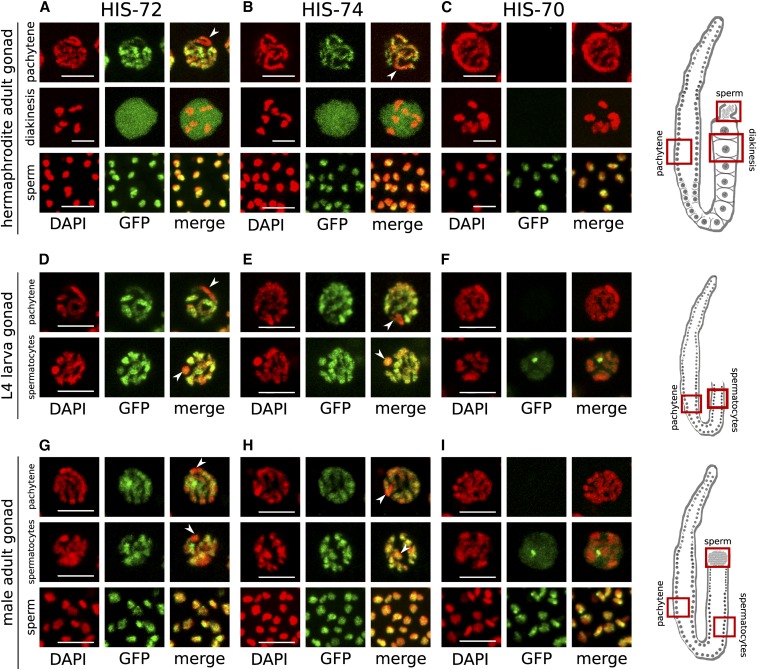
HIS-72 and HIS-74 display indistinguishable expression patterns in adult hermaphrodite germ lines, although HIS-72 levels are higher based on GFP signal (Figure 2, A and B, Figure S5, A and B, and Figure S6, C and D). Both proteins are visible on the condensed chromatin in the mitotic, transition, and pachytene zones. The signal of both proteins is depleted from one region of the chromatin in pachytene nuclei, as previously noted for HIS-72 (Figure 2, A and B, top panels) (Ooi et al. 2006). This depleted region was previously identified as the X chromosome, based on its anticorrelation with active histone modifications such as H3K4me3 and the correlation with repressive histone modifications such as H3K9me3, as the X chromosome is largely silenced in germ cells (Schaner and Kelly 2006; Ooi et al. 2010). We confirmed that the region depleted for both HIS-72 and HIS-74 also lacks H3K4me3 in pachytene nuclei of adult hermaphrodites as well as in L4 hermaphrodites and adult males, indicating that both proteins are depleted from the X chromosomes in these cells (Figure S7). During diakinesis, both HIS-72 and HIS-74 remain chromatin associated, but there are also high levels of the nonchromatin-associated protein in the nucleoplasm, obscuring the chromosomal GFP signal, as previously shown for HIS-72 (Ooi et al. 2006) (Figure 2, A and B, middle panels). The similarities of expression patterns and chromatin association suggest that the broad chromosomal incorporation in germ cells is similar for both proteins.
Figure 2.
HIS-72 and HIS-74 are expressed in all germ cells, whereas HIS-70 is male germ line-specific. (A–C) Hermaphrodite adult stage. HIS-72 (A) and HIS-74 (B) expression shown in pachytene, oocytes, and sperm. In pachytene nuclei, the GFP signal is chromatin associated, but appears depleted from parts of the chromatin (arrows). In oocytes, strong nucleoplasmic signals obscure the chromatin-associated signal. (C) HIS-70 is only detectable in sperm. (D–F) Hermaphrodite L4 larval stage. HIS-72 (D) and HIS-74 (E) expression is present in all germ cells, including pachytene and spermatocyte stages. (F) HIS-70 is only detectable in spermatocytes, but absent from pachytene. (G–I) Male adult stage. HIS-72 (G) and HIS-74 (H) expression is visible in all germ cells, including pachytene stage and in spermatocytes and sperm. (I) HIS-70 is only detectable in spermatocytes and sperm. Bars represent 5 µm. Cartoon images of germ lines highlight the regions shown in (A–I).
HIS-70 deviates from the germ line distribution observed for HIS-72 and HIS-74. Although it is also restricted to the germ line, the HIS-70 signal is only detectable in sperm in adult hermaphrodites, suggesting that it is specific to the male germ line (Figure 2C and Figure S5C). At the adult stage, spermatogenesis has been completed (while oogenesis is still ongoing). To assess the distribution of the germ line-expressed H3.3 homologs during hermaphrodite spermatogenesis, we analyzed the GFP expression in the gonads of L4 larvae. As observed in adults, HIS-72 and HIS-74 are localized to chromatin in all germ cells (Figure 2, D and E and Figure S5, D and E). In contrast, HIS-70 is only expressed in cells undergoing spermatogenesis (Figure 2F and Figure S5F), but is absent from mitotic and pachytene regions. Moreover, the nuclear distribution of HIS-70 appears different from those of HIS-72 and HIS-74, as the GFP signal seems to be more dispersed in the nucleoplasm and only faintly associated with the chromatin, with one distinct focus that is not clearly associated with chromatin and is distinct from the X chromosome (Figure 2F and Figure S7H). Although we could not identify the specific nuclear compartment, other factors (e.g., TOP-1) form similar foci in the male germ line, which have been identified as nucleoli (Chu et al. 2006), and it is therefore tempting to speculate that HIS-70 accumulates at the nucleoli as well. In males, spermatogenesis is also initiated at the L4 stage, but persists through adulthood. The differences in germ line expression between HIS-72, HIS-74, and HIS-70 observed in hermaphrodites are also apparent during spermatogenesis in the adult male germ line, where HIS-72 and HIS-74 are visible in all germ cells, but where expression of HIS-70 is absent from mitotic and pachytene regions and is only detected in spermatocytes and spermatids (Figure 2, G–I and Figure S5, G–I). Interestingly, spermatocytes in both adult male and L4 hermaphrodite germ lines, which correspond to diakinesis I-stage cells, lack the strong nucleoplasmic HIS-72 and HIS-74 signal observed in diakinesis I oocytes (Figure 2, D–I).
Dynamics of H3.3 proteins in early embryos
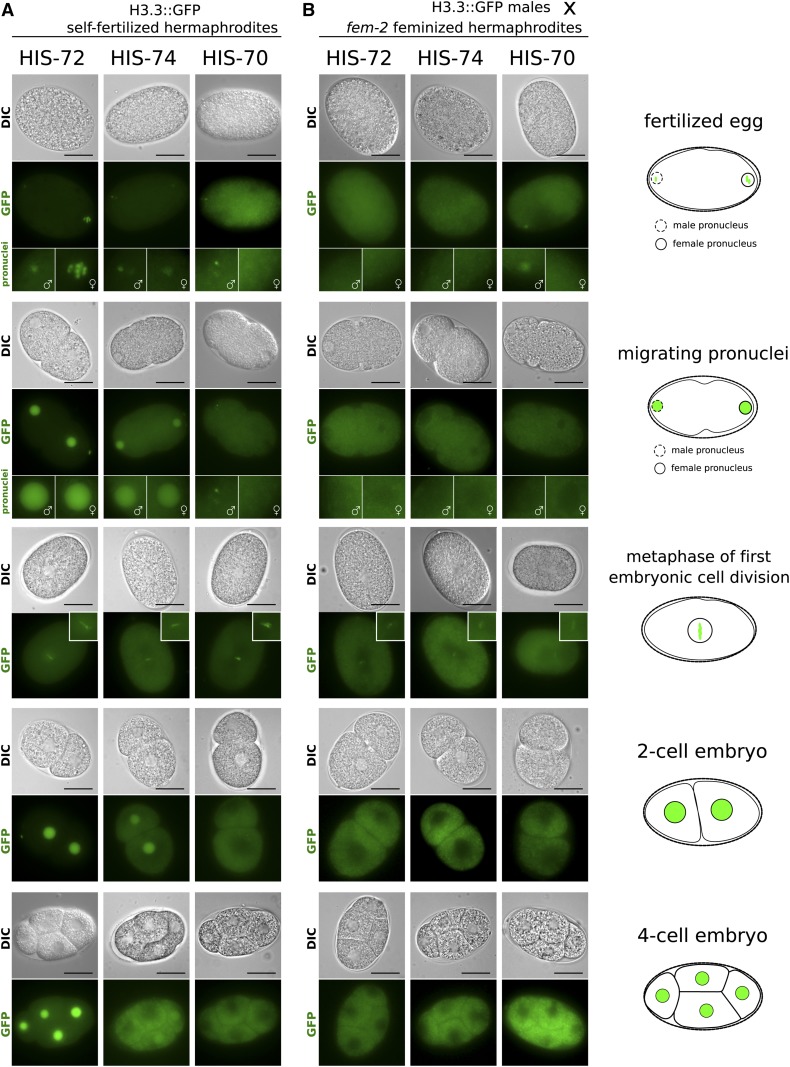
Post-translational histone modifications and histone variants can transmit epigenetic information from one generation to the next, both through the paternal and maternal gametes. Both HIS-72 and HIS-74 accumulate to high levels in the nucleoplasm of oocytes (Figure 2, A and B). It was previously shown that HIS-72 is provided only maternally, and that the presence of HIS-72::GFP in the male pronucleus is a result of chromatin incorporation of the histone pool present in the oocyte (Ooi et al. 2006). Consistent with these findings, we found that upon fertilization, both HIS-72::GFP and HIS-74::GFP signals are detectable in both female and male pronuclei prior to pronuclear fusion, and are visible at the metaphase plate during the first mitotic division in the embryo (Figure 3A). HIS-72 remains present at high levels in every cell in the embryo throughout development after the first division (Figure 3A and Figure S4A). However, HIS-74 levels appear to be diluted by subsequent cell divisions. The HIS-74::GFP signal is still present in the nucleoplasm of the two-cell embryo, becomes faintly visible in one or two cells at the four-cell stage (Figure 3A), and robustly reappears in the PGCs and faintly in some cells surrounding the PGCs (Figure S4C). Postembryonically, both HIS-72 and HIS-74 are detectable in all germ cells at all larval and adult stages (Figure 2 and Figure S5), with HIS-72 also being present in all somatic cells (Figure 1A and Figure S6, A and B). HIS-70::GFP is not detectable in mature oocytes and likely only arrives in the embryo through sperm (Figure 2C). It is present only in the male pronucleus immediately after fertilization and can be detected at metaphase of the first embryonic cell division, before it becomes undetectable (Figure 3A).
Figure 3.
Dynamics of H3.3 proteins in early embryos. Representative live-cell images of early stages in embryonic development are shown. Stages include eggs after fertilization, migrating pronuclei, the metaphase of first cell division, the two-cell embryo, and the four-cell embryo, as outlined by cartoons on the right. (A) Eggs of self-fertilizing hermaphrodites carrying H3.3::GFP fusions. HIS-72 (left panels) is present in both male and female pronuclei upon fertilization and remains chromatin associated during metaphase of the first mitotic division. It is subsequently present in all cells. HIS-74 (center panels) is present in both male and female pronuclei upon fertilization and remains chromatin associated during metaphase of the first mitotic division. It is present in both nuclei at the two-cell stage, but subsequently becomes diluted. It is sometimes still faintly visible in two nuclei at the four-cell stage and then becomes undetectable. HIS-70 (right panels) is faintly detectable in the male but not the female pronucleus, remains chromatin associated during the first mitotic division, and subsequently becomes undetectable. (B) Eggs of feminized fem-2 hermaphrodites fertilized by males carrying H3.3::GFP fusions. HIS-72 and HIS-74 are not detectable in the pronuclei, while HIS-70 is visible in the male pronucleus. All three GFP fusions appear faintly chromatin associated during the first mitotic division and become undetectable after that. In GFP images representing fertilized eggs and migrating pronuclei, female pronuclei are highlighted by enlarged images with female symbols and male pronuclei are highlighted by enlarged images with male symbols. Bars represent 20 µm.
The presence of HIS-72, HIS-74, and HIS-70 in sperm suggested that these histone variants might also be transmitted through the male germ line. To examine this possibility, we crossed males carrying HIS-70, HIS-72, and HIS-74 GFP fusions with feminized hermaphrodites that are unable to produce functional sperm (Figure 3B). Interestingly, we were only able to detect a HIS-70::GFP signal, but not a HIS-72::GFP or HIS-74::GFP signal, in the male pronuclei, suggesting that the latter two proteins are removed from paternal chromatin upon fertilization, consistent with previous finings (Ooi et al. 2006). However, we could observe weak signals in the metaphase plates of first divisions for all three GFP fusions, indicating that HIS-70, HIS-72, and HIS-74 are transmitted to the embryo paternally at low levels (Figure 3B). The signal becomes undetectable at the two- and four-cell stages.
Our results indicate that the GFP signal detected in embryos at early stages is mainly maternally provided, and at later stages is derived from embryonic expression of the fusion proteins.
Chromatin association of all H3.3 homologs depends on the H3.3-specific chaperone HIRA-1
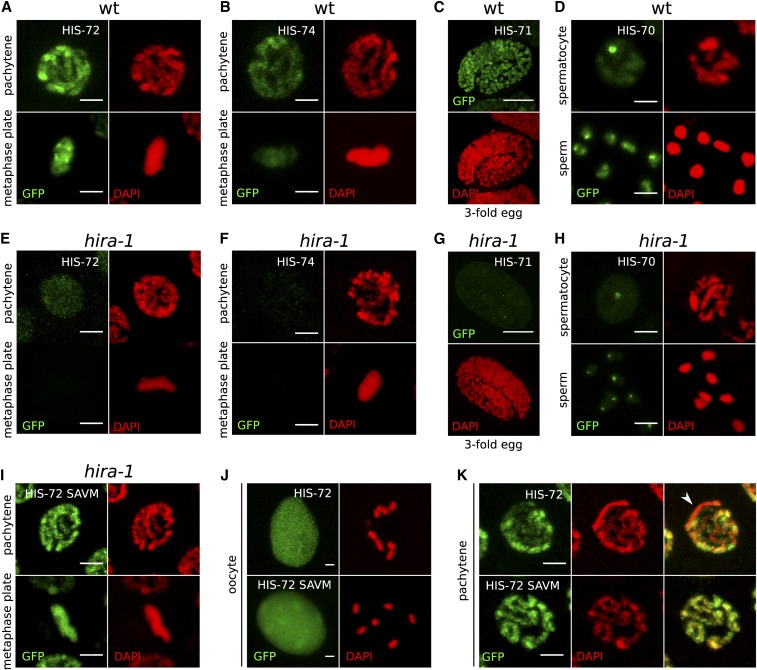
In higher eukaryotes, chromatin loading of H3.3 depends on the highly conserved histone chaperone HIRA or on the ATRX/DAXX complex (Filipescu et al. 2013). These chaperone complexes facilitate H3.3 incorporation into different chromatin regions; HIRA is responsible for incorporating the histones into actively transcribed chromatin like active gene promoters and gene bodies, while DAXX mediates the heterochromatic incorporation into telomeres and pericentric regions (Filipescu et al. 2013; Mattiroli et al. 2015; Dyer et al. 2017). The specificity of both ATRX/DAXX and HIRA to H3.3 was shown to be mediated by a conserved sequence motif in H3.3 (AAIG) that differs from canonical H3 (SAVM) (Figure S1A) (Lewis et al. 2010; Elsässer et al. 2012; Liu et al. 2012; Ricketts et al. 2015). This AAIG motif is found in HIS-71, HIS-72, and HIS-74, while HIS-70 contains a slight variation of the motif (AAIQ) (Figure S1A). However, there is no identifiable DAXX homolog in C. elegans. BLAST searches identified K10D2.1, which we named hira-1 for HIstone cell cycle RegulAtor homolog 1, as the only C. elegans homolog of vertebrate, insect, and plant HIRA. HIRA-1 shows significant conservation with HIRA from other lineages in multiple sequence alignments, particularly within annotated domains (Figure S8). To test if the chromatin association of all H3.3 homologs observable during mitosis and meiosis (Figure 4, A–D) depends on HIRA-1, we generated a hira-1 deletion allele using CRISPR/Cas9. The hira-1 deletion strain is viable, but shows morphological defects, including protruding vulvas, that are not observed in the H3.3 null mutant strain discussed below (Figure S9A). The hira-1 deletion strain also shows reduced brood sizes at higher temperatures (median at 20° = 56; median at 25° = 7; P = 1.3e−5; and N = 20), similar to the H3.3 null mutant strain discussed below (Figure S9B). However, the brood sizes are consistently lower at all temperatures compared to wild-type or H3.3 null mutant worms, presumably due to the morphological or developmental defects that may be independent of the role of HIRA-1 in H3.3 loading.
Figure 4.
Chromatin association of all H3.3 homologs depends on HIRA-1. (A–D) Chromatin association of H3.3 homologs in a wild-type (wt) background. (A) HIS-72 in pachytene and mitotic metaphase nuclei, (B) HIS-74 in pachytene and mitotic metaphase nuclei, (C) HIS-71 in threefold embryo, and (D) HIS-70 in spermatocyte nucleus and mature sperm. (E–H) H3.3 homolog signal is lost in hira-1 deletion background at the same stages as in (A–D). (E) HIS-72, (F) HIS-74, (G) HIS-71, and (H) HIS-70. Residual signal remains at HIS-70 foci. See text for details. (I–K) Chromatin localization of HIS-72 in hira-1 deletion background is restored upon mutation of the H3.3-specific motif in HIS-72 to the canonical H3-specific motif (AAIG to SAVM). (I) HIS-72 SAVM in pachytene and mitotic metaphase nuclei. (J and K) Chromatin association of HIS-72 (top panels) and HIS-72 SAVM (bottom panels) appears different. HIS-72 SAVM is more strongly chromatin associated in oocyte nuclei, with chromosomes being visible despite the strong nucleoplasmic background (J), and there is no obvious depletion of GFP signal in any part of the chromatin (K). Bars represent 2 µm in all panels except (C and G), where they represent 10 µm.
We crossed the GFP fusions of the H3.3 homologs to the hira-1 deletion strain and examined the distribution of the GFP-tagged proteins. Upon hira-1 deletion, the nuclear GFP signal is generally reduced or absent for all four H3.3 homologs (Figure 4, E–H). In germ line mitotic and meiotic nuclei with condensed chromosomes, no chromatin association was observed. These results demonstrate that loading of all four H3.3 homologs depends on this H3.3-specific chaperone HIRA-1 (Figure 4, E–H). Interestingly, distinct HIS-70 foci observed in spermatocytes and sperm persist in the hira-1 mutant (Figure 4, D and H). Low levels of HIS-72::GFP and very low levels of HIS-71::GFP signals can also be detected in somatic cells in the hira-1 mutant, presumably due to higher levels of his-72 expression compared to the other H3.3 genes (Figure S10).
To exclude the possibility that loss of HIRA-1 interfered with H3.3 expression rather than H3.3 loading, we mutagenized the HIRA recognition motif (AAIG) of HIS-72 to the sequence of canonical histone H3 (SAVM) in the endogenous gfp-tagged gene. The mutated HIS-72 SAVM protein showes chromatin association even in the absence of HIRA-1 (Figure 4I). This result shows that chromatin association, but not expression of HIS-72, depends on HIRA-1, and that HIS-72 with an SAVM motif is loaded onto chromatin by a different histone chaperone, possibly the canonical H3-specific chaperone CAF-1 (Nakano et al. 2011). In oocytes, where a large part of HIS-72 is nonchromatin associated, the GFP signal on chromosomes appears increased upon mutation to the SAVM motif (Figure 4J). Similarly, the depletion of HIS-72 from the X chromosome in pachytene nuclei is no longer observed in the strains carrying the SAVM motif (Figure 4K and Figure S7A). These observations imply that HIS-72 SAVM has a genomic distribution and loading dynamics similar to that of canonical H3.
Taken together, our results demonstrate that chromatin association of HIS-70, HIS-71, HIS-72, and HIS-74 depends on the H3.3-specific chaperone HIRA-1, and that this dependency can be bypassed by replacing the H3.3-specific AAIG motif with the H3 motif SAVM.
H3.3 is not essential in C. elegans
The germ line expression and incorporation into mature sperm of HIS-70, HIS-72, and HIS-74 appeared consistent with the severe fertility defects upon loss of H3.3 observed in other organisms (summarized above). However, worms carrying a deletion of the ubiquitously expressed his-72 gene, either alone or in combination with the somatically expressed his-71, show no fertility defects [Ooi et al. (2006), Piazzesi et al. (2016), and our own results]. His-69 and his-70 are localized in tandem on chromosome III, and we created a full deletion of the locus using the CRISPR/Cas9 system. Worms carrying this deletion appear superficially wild-type and do not show any fertility defects, neither in hermaphrodites nor males. The only available allele of his-74 (ok1219, embryonic lethal) also affects the flanking ccch-3 gene. Therefore, we created an early stop and frameshift mutant of his-74. The strain carrying this allele is viable, with no detectable phenotype. To assess the redundancy of the different H3.3 genes, we combined the mutants of his-69, his-70, his-71, his-72, and his-74 to create a strain that lacks all H3.3 homologs (H3.3 null mutant). Surprisingly, this strain looks superficially wild-type, lacking the lethality or complete sterility observed in other organisms. H3.3 null mutant animals shows no developmental delays and hermaphrodites appear to have normal brood sizes. We concluded that, in contrast to other organisms, H3.3 is not essential in C. elegans.
H3.3 modulates the response to stress at high temperatures
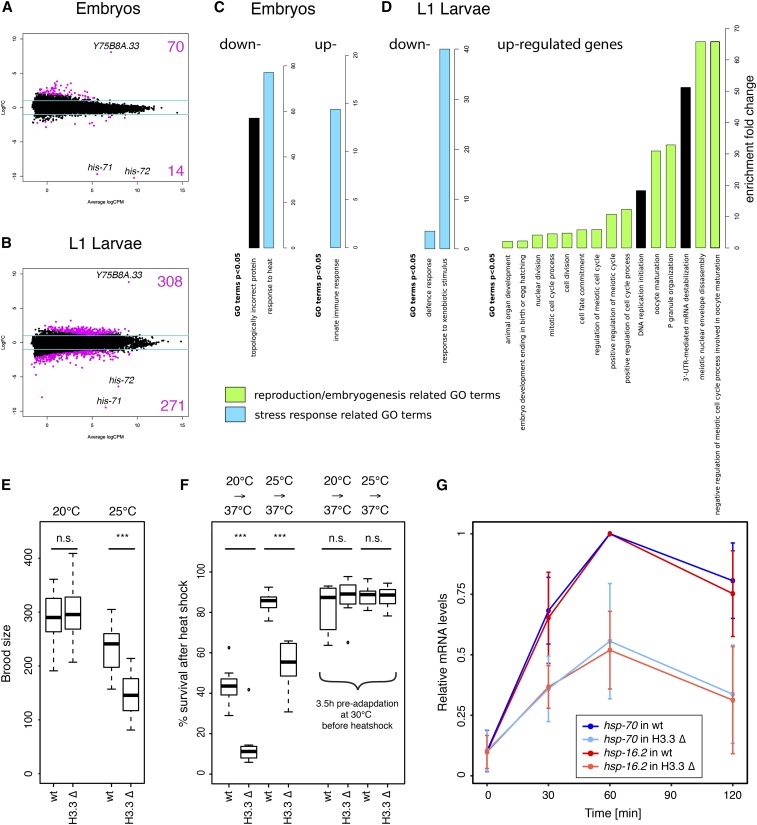
To gain insight into the function of H3.3 in C. elegans, we compared gene expression in wild-type and H3.3 null mutant animals at embryonic and L1 larval stages by RNA-seq. We identified 84 genes in embryos and 579 genes in L1 larvae with significantly changed expression levels (Tables S3 and S4). His-71 and his-72, as well as the gene adjacent to his-72 (Y75B8A.33), were by far the most affected in both data sets (Figure 5, A and B and Tables S3 and S4). Deletion alleles of his-71 and his-72 remove most of the coding sequences of the genes (Figure S2A), thus removing the mRNAs from the H3.3 null mutant data set. These two genes therefore appear to be the most depleted and serve as positive controls. Y75B8A.33, the gene adjacent to his-72, is only expressed at residual levels in wild-type worms, but most likely becomes fused to a his-72 regulatory element upon his-72 deletion, causing its upregulation. Changes in all other genes were surprisingly moderate, and it was difficult to select individual genes for follow-up studies. To identify pathways altered upon H3.3 deletion, we performed GO enrichment analysis separately for up- and downregulated genes at both developmental stages. Enriched GO terms with P-values < 0.05 are shown in Figure 5, C and D. For simplicity, we only show the most general subclass of each GO term, and full lists of significant GO terms are shown in Table S5. Consistent with the germ line expression of three of the five H3.3 homologs, we found a large number of GO terms related to reproduction or embryogenesis. The second major class of GO terms was related to immunity or stress response.
Figure 5.
H3.3 modulates fertility and temperature stress response. (A and B) MA plots of RNA sequencing results showing genes up- and downregulated in H3.3 null mutant (H3.3 ∆) worms compared to wild-type (wt) worms. (A) Embryos. (B) L1 larvae. Log2 fold changes (FC) are plotted against log2 counts per million (CPM). The values are averages from two biological replicates. The blue lines show the FC ± 2 cutoff. The most significant genes—his-71, his-72, and Y75B8A.33—are highlighted. (C and D) Gene ontology (GO) enrichment analysis for genes differentially expressed in H3.3 ∆ animals compared to wt animals at embryonic (C) and L1 larval stages (D). Enrichment fold change of significant GO terms is plotted separately for up- and downregulated genes. (E) Brood size analysis. Wt and H3.3 ∆ strain brood sizes at 20 and 25°. Brood sizes of individual worms were counted over a period of 4 days after the first egg was laid (N = 20). *** indicates P ≤ 0.001. n.s., not significant. (F) Resistance to heat shock. Percentage of surviving wt and H3.3 ∆ worms after 100 min heat shock at 37°, without and with 3.5 hr adaptation at 30° (N = 8). *** indicates P ≤ 0.001. n.s., not significant. Worms were either maintained at 20 or 25° prior to heat shock. (G) Relative mRNA levels of hsp-70 (blue) and hsp-16.2 (red) upon heat shock in wt (dark colors) and H3.3 ∆ (light colors) worms. Expression levels were determined by quantitative PCR, and the maximum expression levels set to 1.
Since both expression patterns and GO term analysis pointed to a role of H3.3 in reproduction and embryogenesis, we more thoroughly assessed the role of H3.3 in fertility. We counted the brood sizes of individual wild-type and H3.3 null worms after growing the worm cultures at different temperatures for several generations (Figure 5E). At 20°, the brood sizes of wild-type and H3.3 null worms were almost identical (wild-type median 290; H3.3 null mutant median 295.5; P = 0.7; and N = 20). However, at 25° we detected a significant reduction of the brood size for the H3.3 null mutant compared to wild-type worms (wild-type median 241; H3.3 null mutant median 145.5; P = 2.6e−15; and N = 20). Embryonic viability was decreased by < 10% in the H3.3 null mutant strain at 25° (wild-type 96%; H3.3 null mutant 88%; P = 2.1e−5; and N = 20), thus only partly explaining the reduction in brood size (Figure S11A). We did not observe an increase in the number of apoptotic cells in the germ line of the H3.3 null mutant strain compared to wild-type (Figure S11, B and C). These experiments indicate that, while the H3.3 null phenotype is unexpectedly mild, H3.3 functions in the germ line to modulate fertility at least at elevated temperatures.
The observed reduction in brood size of H3.3 null worms at 25° suggested a role of H3.3 in response to elevated temperatures. Additionally, GO terms related to stress response were enriched in our RNA-seq analysis. To test if H3.3 was involved in the response to high-temperature stress, we exposed populations of wild-type and H3.3 null animals grown at 20 or 25° to a 37° heat shock for 100 min, followed by recovery for 24 hr (Figure 5F). We found that H3.3 null mutants were much less able to respond to the heat shock, with significantly increased mortality compared to wild-type (P = 5.0e−4, N = 7 for 20° and P = 8.0e−5, N = 8 for 25°). Mortality was higher for worms grown at 20° than for worms grown at 25°, suggesting that the temperature difference between growth temperature and stress temperature influences the chances of survival. Therefore, we preadapted the worms at 30° for 3.5 hr prior to the heat shock. This pretreatment significantly increased the survival of worms grown at 20° and abolished the difference between wild-type and H3.3 null mutant worms (P = 0.28, N = 9 for 20° and P = 0.81, N = 9 for 25°) (Figure 5F). These results show that H3.3 is required for the response to the rapid change of temperature rather than for survival at higher temperature.
Previous studies have shown that H3.3 accumulates at hsp genes upon heat shock in Drosophila and human cell cultures (Schwartz and Ahmad 2005; Kim et al. 2011). Therefore, we hypothesized that expression of the heat shock genes in response to high-temperature stress could be impaired in C. elegans H3.3 null mutant worms. We analyzed the induced expression of hsp-16.2 and hsp-70 in adult wild-type and H3.3 null mutant worms upon heat shock by quantitative PCR (Figure S11D). We found that the maximum expression levels of both hsp-16.2 and hsp-70 were reduced by ∼50% in adult H3.3 null mutant worms (Figure 5G).
Taken together, the results of our H3.3 mutant analysis suggest that while H3.3 is nonessential in C. elegans, it is important for the response to stress at higher temperatures in both the soma and the germ line.
Discussion
In this study, we analyzed the developmental expression patterns of all H3.3 homologs in C. elegans using gfp fusions at the endogenous loci. We found that his-69 is not expressed at detectable levels and that the remaining four homologs have distinct, but partially overlapping, expression patterns. Only his-72 is expressed ubiquitously, while his-71 is soma-specific, and his-70 and his-74 are germ line-specific. His-72 and his-74 are expressed in all germ cells, and both proteins are depleted from the X chromosomes, which may indicate potential redundancy of the two proteins in these cell types. The expression pattern of his-70 is clearly distinct, as it is restricted to the male germ line, and the protein is less clearly associated with chromatin, suggesting a different genomic distribution (Figure 2).
Interestingly, all three germ line-expressed H3.3 homologs are present in mature sperm. In most organisms, canonical histone proteins in sperm are replaced by smaller structural proteins called sperm nuclear basic proteins, such as protamines (Ausió 1999). However, there are a number of reports demonstrating that histone variants are also retained in sperm chromatin (van Roijen et al. 1998; Loppin et al. 2001; Govin et al. 2007; Ishibashi et al. 2010). In C. elegans, the H2A variant HTAS-1 was previously reported to be specific for the worm male germ line (Chu et al. 2006). Male germ line-specific histone H3 variants have been described in different taxa, e.g., H3t in vertebrates, H3.5 in hominids, or AT1G19890 in Arabidopsis (Trostle-Weige et al. 1984; Okada et al. 2005; Schenk et al. 2011). Here, we find that C. elegans HIS-70 is male germ line-specific as well, the first such H3 variant described in nematodes, and we show that HIS-70 is restricted to spermatocytes and sperm (Figure 2 and Figure S5). The function of these male germ line-specific histone variants is not fully understood, but it is tempting to speculate that they carry epigenetic information about gene expression to the next generation.
We were able to observe that all three H3.3 variants present in sperm can be paternally provided into the embryo, and appear to be chromatin associated at the first mitotic division of the developing embryo (Figure 3B). This signal is normally obscured by maternally provided HIS-72 and HIS-74, which is incorporated into paternal chromatin shortly after fertilization and prior to pronuclear fusion, but becomes visible when the H3.3::GFP fusions are only transmitted through the paternal germ line by sexual crossing of H3.3::GFP-carrying males to nontagged feminized hermaphrodites. These findings indicate that the H3.3 dynamics in nematodes differ slightly from mammalian systems, where paternally provided H3.3 is removed from the zygote with the second polar body and replaced by maternal copies of the histone (Kong et al. 2018). Moreover, it has been shown that epigenetic patterns established during worm spermatogenesis, including the ones associated with H3 and H3.3, are retained in the early embryo in C. elegans (Arico et al. 2011). However, despite the presence of HIS-70, HIS-72, and HIS-74 in the male germ line and their persistence into the one-cell embryo, we were unable to detect any abnormalities in sperm function in the H3.3 null mutant worms. H3.3 null mutant males mate with wild-type efficiency and produce healthy offspring. Given the role of H3.3 in the stress response that we describe here, it is possible that loss of H3.3 affects male fertility or the transmission of epigenetic information under nonstandard conditions that remain to be further investigated.
Given the differences in expression patterns in both soma and the germ line, we tested if the H3.3 homologs depended on the same histone chaperone for chromatin association. Since there is no identifiable homolog of DAXX in the C. elegans genome, we focused our analysis on the HIRA homolog. Deletion of hira-1 did not result in lethality in C. elegans, allowing us to demonstrate that chromatin association of HIS-70, HIS-71, HIS-72, and HIS-74 depends on this histone chaperone. No chromatin association of H3.3 was observed in hira-1 deletion mutants in germ line cells, where chromatin is condensed and chromatin association can be assessed (Figure 4, E–H), consistent with the absence of DAXX in C. elegans (Figure 4). DAXX loads H3.3 into heterochromatic regions in higher eukaryotes (Filipescu et al. 2013; Mattiroli et al. 2015; Dyer et al. 2017), implying that H3.3 is not targeted to these regions in C. elegans. This is consistent with our observations that the X chromosome, which is associated with heterochromatic histone modifications in the germ line, does not show a significant H3.3::GFP signal in the wild-type background (Figure 2 and Figure S7). Additionally, H3.3 has been found to be mostly associated with transcribed regions in chromatin immunoprecipitation experiments (Ooi et al. 2010). The conserved AAIG motif of H3.3 is thought to confer H3.3 specificity to both HIRA and DAXX in human cells (Lewis et al. 2010; Ricketts et al. 2015). Structural studies have shown that H3.3 Ala87 and Gly90 are principal determinants of the specific interaction of H3.3 with DAXX in humans (Elsässer et al. 2012; Liu et al. 2012). An absence of C. elegans DAXX might lower the constraint on the AAIG motif, which could explain the substitution of Gly90 to Gln90 observed in HIS-70.
We found that the reliance on HIRA-1 for HIS-72 loading can be bypassed by changing the AAIG motif to the SAVM motif of H3 (Figure 4, I–K). This substitution resulted in chromatin association even in absence of HIRA-1. Moreover, HIS-72 SAVM distribution within the nucleus appeared changed, as the depletion from the X chromosome in pachytene was no longer evident (Figure 4K and Figure S7A). These results imply that the histone H3 chaperone CAF-1 recognizes and loads HIS-72 SAVM in a distribution that resembles that of canonical histone H3.
Despite the essential function of H3.3 in other organisms, we found that complete knockout of all genes encoding for H3.3 homologs does not lead to strong developmental phenotypes in C. elegans. The reasons for this discrepancy are unclear. It is unlikely that other H3 variants contribute to the compensation of H3.3 upon H3.3 deletion. In addition to the five histone H3.3 genes and 15 histone H3 genes, four additional histone H3 variants are found in the C. elegans genome: HCP-3 is the functional CENP-A homolog (Buchwitz et al. 1999; Gassmann et al. 2012; Steiner and Henikoff 2014), CPAR-1 is a CENP-A homolog expressed in the germ line and early embryos without known function at the centromere (Gassmann et al. 2012; Monen et al. 2015), F20D6.9 clusters with the CENP-A homologs in sequence alignments, and HIS-73 has a highly diverged N-terminal tail, but lacks the characteristic residues of H3.3 and is likely expressed in a replication-dependent manner (Pettitt et al. 2002) (Figure S12, A and B). Moreover, both F20D6.9 and his-73 are only expressed at marginal levels compared to the other H3 variants, based on RNA-seq data (Boeck et al. 2016) (Figure S12C).
The most likely explanation for the viability of the H3.3 null worms is that histone H3 is present in sufficient levels to compensate for the loss of H3.3. There is evidence for such compensation in Drosophila, where deletion of H3.3 leads to broader H3 expression during the cell cycle (Sakai et al. 2009). The precise cell cycle expression timing of the 15 H3 genes in C. elegans is not well established, and it is possible that some of the canonical H3 genes are sufficiently expressed outside S-phase to make H3.3 dispensable. These findings imply that while both H3 and H3.3 and their chaperones are remarkably conserved, the dynamics of expression and chromatin incorporation of these histones may vary between taxa. Consistent with the viability of the H3.3 null worms, we found that the strains carrying the hira-1 deletion were viable. However, we observed some morphological defects in adults, including protruding vulvas, which were not present in the H3.3 null mutants, suggesting that HIRA may have functions beyond H3.3 loading. Interestingly, similar defects were observed upon deletion of the C. elegans ATRX homolog xnp-1 (Cardoso et al. 2005).
GO term analysis of genes misregulated in H3.3 null mutant animals compared to wild-type suggested a role of H3.3 in fertility and embryogenesis, as well as in immunity and stress response. Indeed, we found that H3.3 null mutants had smaller brood sizes than wild-type worms, specifically when grown at higher temperatures (Figure 5E). We also showed that H3.3 null animals had reduced ability to survive a heat shock, possibly due to defects in heat shock gene expression (Figure 5, F and G), consistent with the finding that H3.3 stimulates HSP70 transcription in mammalian cells (Kim et al. 2011). Heat shock can result in transgenerational expression changes of heterochromatic elements in C. elegans (Klosin et al. 2017). Given our observation that H3.3 is involved in changes of gene expression upon heat shock, and that at least some H3.3 proteins are transmitted from the germ line to the early embryos, it is possible that H3.3 contributes to this transgenerational inheritance.
Temperature stress may not be the only condition affected by the loss of H3.3 in C. elegans. A previous study reported that his-71, his-72 double mutant worms were more sensitive to oxidative stress induced by paraquat (Piazzesi et al. 2016), suggesting that the H3.3 involvement in response to stress is more general. Moreover, H3.3 is required for DNA damage repair caused by UV stress in human cells (Adam et al. 2013). Despite our finding that H3.3 is required for the response to heat stress, the H3.3 null phenotypes that we found were surprisingly subtle, especially in light of the essential roles of H3.3 in other organisms. More context-dependent experiments will be required to uncover additional functions of H3.3 and to explain the significance of the specific expression patterns of the different C. elegans H3.3 homologs.
Acknowledgments
We thank the Genomics Platform and the Bioimaging Center of the University of Geneva for providing imaging and sequencing services, and Ramesh Pillai for advice on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). The work was funded by the Swiss National Science Foundation (grant #31003A_156774), the Republic and Canton of Geneva, and an Institute of Genetics and Genomics in Geneva (iGE3) PhD Students’ Award to K.D.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6106448.
Communicating editor: Greenstein D.
Literature Cited
- Adam S., Polo S. E., Almouzni G., 2013. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106. 10.1016/j.cell.2013.08.029 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Suzuki O., Matsuda J., Aoki F., 2011. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 7: e1002279 10.1371/journal.pgen.1002279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico J. K., Katz D. J., van der Vlag J., Kelly W. G., 2011. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 7: e1001391 10.1371/journal.pgen.1001391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausió J., 1999. Histone H1 and evolution of sperm nuclear basic proteins. J. Biol. Chem. 274: 31115–31118. 10.1074/jbc.274.44.31115 [DOI] [PubMed] [Google Scholar]
- Banaszynski L. A., Wen D., Dewell S., Whitcomb S. J., Lin M., et al. , 2013. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155: 107–120. 10.1016/j.cell.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S., Tarpey P. S., Presneau N., Scheipl S., Pillay N., et al. , 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45: 1479–1482 (erratum: Nat. Genet. 46: 316) 10.1038/ng.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck M. E., Huynh C., Gevirtzman L., Thompson O. A., Wang G., et al. , 2016. The time-resolved transcriptome of C. elegans. Genome Res. 26: 1441–1450. 10.1101/gr.202663.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz B. J., Ahmad K., Moore L. L., Roth M. B., Henikoff S., 1999. A histone-H3-like protein in C. elegans. Nature 401: 547–548. 10.1038/44062 [DOI] [PubMed] [Google Scholar]
- Buschbeck M., Hake S. B., 2017. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18: 299–314. 10.1038/nrm.2016.166 [DOI] [PubMed] [Google Scholar]
- Cardoso C., Couillault C., Mignon-Ravix C., Millet A., Ewbank J. J., et al. , 2005. XNP-1/ATR-X acts with RB, HP1 and the NuRD complex during larval development in C. elegans. Dev. Biol. 278: 49–59. 10.1016/j.ydbio.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Chu D. S., Liu H., Nix P., Wu T. F., Ralston E. J., et al. , 2006. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443: 101–105. 10.1038/nature05050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C., Carlton M. B., Nolan P. M., Colledge W. H., Evans M. J., 1999. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 8: 2489–2495. 10.1093/hmg/8.13.2489 [DOI] [PubMed] [Google Scholar]
- Dunleavy E. M., Almouzni G., Karpen G. H., 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2: 146–157. 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer M. A., Qadeer Z. A., Valle-Garcia D., Bernstein E., 2017. ATRX and DAXX: mechanisms and mutations. Cold Spring Harb. Perspect. Med. 7: a026567 10.1101/cshperspect.a026567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer S. J., Huang H., Lewis P. W., Chin J. W., Allis C. D., et al. , 2012. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 491: 560–565. 10.1038/nature11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S., Hisano M., Liang C. Y., Gill M., Murr R., et al. , 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 20: 868–875 (erratum: Nat. Struct. Mol. Biol. 20: 1236) 10.1038/nsmb.2599 [DOI] [PubMed] [Google Scholar]
- Filipescu D., Szenker E., Almouzni G., 2013. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 29: 630–640. 10.1016/j.tig.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Frey A., Listovsky T., Guilbaud G., Sarkies P., Sale J. E., 2014. Histone H3.3 is required to maintain replication fork progression after UV damage. Curr. Biol. 24: 2195–2201. 10.1016/j.cub.2014.07.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Rechtsteiner A., Yuen K. W., Muroyama A., Egelhofer T., et al. , 2012. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484: 534–537. 10.1038/nature10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., et al. , 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176: 283–294. 10.1083/jcb.200604141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S. B., Garcia B. A., Kauer M., Baker S. P., Shabanowitz J., et al. , 2005. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. USA 102: 6344–6349. 10.1073/pnas.0502413102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Smith M. M., 2015. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7: a019364 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. 10.1016/0012-1606(76)90267-0 [DOI] [PubMed] [Google Scholar]
- Hödl M., Basler K., 2009. Transcription in the absence of histone H3.3. Curr. Biol. 19: 1221–1226. 10.1016/j.cub.2009.05.048 [DOI] [PubMed] [Google Scholar]
- Ishibashi T., Li A., Eirin-Lopez J. M., Zhao M., Missiaen K., et al. , 2010. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 38: 1780–1789. 10.1093/nar/gkp1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Bergamin E., Donoghue M. T., Mongeon V., LeBlanc C., et al. , 2014. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253. 10.1126/science.1248357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J., Astrand C., Szenker E., Garrett N., Almouzni G., et al. , 2012. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 5: 17 10.1186/1756-8935-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Heo K., Choi J., Kim K., An W., 2011. Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1gamma. Nucleic Acids Res. 39: 8329–8341. 10.1093/nar/gkr529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B., 2017. Transgenerational transmission of environmental information in C. elegans. Science 356: 320–323. 10.1126/science.aah6412 [DOI] [PubMed] [Google Scholar]
- Kong Q., Banaszynski L. A., Geng F., Zhang X., Zhang J., et al. , 2018. Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J. Biol. Chem. 293: 3829–3838. 10.1074/jbc.RA117.001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D., 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107: 14075–14080. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. J., Conti M., Ramalho-Santos M., 2013. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140: 3624–3634. 10.1242/dev.095513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. P., Xiong C., Wang M., Yu Z., Yang N., et al. , 2012. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 19: 1287–1292. 10.1038/nsmb.2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Berger F., Couble P., 2001. The Drosophila maternal gene sesame is required for sperm chromatin remodeling at fertilization. Chromosoma 110: 430–440. 10.1007/s004120100161 [DOI] [PubMed] [Google Scholar]
- Ly K., Reid S. J., Snell R. G., 2015. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX 2: 59–63. 10.1016/j.mex.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F., D’Arcy S., Luger K., 2015. The right place at the right time: chaperoning core histone variants. EMBO Rep. 16: 1454–1466. 10.15252/embr.201540840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I., Wenderski W., Noh K. M., Bagot R. C., Tzavaras N., et al. , 2015. Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87: 77–94. 10.1016/j.neuron.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J., Hattersley N., Muroyama A., Stevens D., Oegema K., et al. , 2015. Separase cleaves the N-tail of the CENP-A related protein CPAR-1 at the meiosis I metaphase-anaphase transition in C. elegans. PLoS One 10: e0125382 10.1371/journal.pone.0125382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S., Stillman B., Horvitz H. R., 2011. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell 147: 1525–1536. 10.1016/j.cell.2011.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Endo M., Singh M. B., Bhalla P. L., 2005. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44: 557–568. 10.1111/j.1365-313X.2005.02554.x [DOI] [PubMed] [Google Scholar]
- Ooi S. L., Priess J. R., Henikoff S., 2006. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2: e97 10.1371/journal.pgen.0020097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S. L., Henikoff J. G., Henikoff S., 2010. A native chromatin purification system for epigenomic profiling in Caenorhabditis elegans. Nucleic Acids Res. 38: e26 10.1093/nar/gkp1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaluca A., Ramotar D., 2016. A novel approach using C. elegans DNA damage-induced apoptosis to characterize the dynamics of uptake transporters for therapeutic drug discoveries. Sci. Rep. 6: 36026 10.1038/srep36026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J., Crombie C., Schumperli D., Muller B., 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 115: 857–866. [DOI] [PubMed] [Google Scholar]
- Piazzesi A., Papic D., Bertan F., Salomoni P., Nicotera P., et al. , 2016. Replication-independent histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs. Cell Rep. 17: 987–996. 10.1016/j.celrep.2016.09.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts M. D., Marmorstein R., 2017. A molecular prospective for HIRA complex assembly and H3.3-specific histone chaperone function. J. Mol. Biol. 429: 1924–1933. 10.1016/j.jmb.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts M. D., Frederick B., Hoff H., Tang Y., Schultz D. C., et al. , 2015. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6: 7711 10.1038/ncomms8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Schwartz B. E., Goldstein S., Ahmad K., 2009. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19: 1816–1820. 10.1016/j.cub.2009.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A., Ziegler-Birling C., Koch M., Tora L., Bannister A. J., et al. , 2010. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 12: 853–862. 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Kelly W. G., 2006. Germline chromatin (January 24, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.73.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk R., Jenke A., Zilbauer M., Wirth S., Postberg J., 2011. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120: 275–285. 10.1007/s00412-011-0310-4 [DOI] [PubMed] [Google Scholar]
- Schwartz B. E., Ahmad K., 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19: 804–814. 10.1101/gad.1259805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., et al. , 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Steiner F. A., Henikoff S., 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife 3: e02025 10.7554/eLife.02025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D., Witt H., Hovestadt V., Khuong-Quang D. A., Jones D. T., et al. , 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437. 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- Szenker E., Lacoste N., Almouzni G., 2012. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 1: 730–740. 10.1016/j.celrep.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Tang M. C., Jacobs S. A., Mattiske D. M., Soh Y. M., Graham A. N., et al. , 2015. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 11: e1004964 10.1371/journal.pgen.1004964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trostle-Weige P. K., Meistrich M. L., Brock W. A., Nishioka K., 1984. Isolation and characterization of TH3, a germ cell-specific variant of histone 3 in rat testis. J. Biol. Chem. 259: 8769–8776. [PubMed] [Google Scholar]
- van Roijen H. J., Ooms M. P., Spaargaren M. C., Baarends W. M., Weber R. F., et al. , 1998. Immunoexpression of testis-specific histone 2B in human spermatozoa and testis tissue. Hum. Reprod. 13: 1559–1566. 10.1093/humrep/13.6.1559 [DOI] [PubMed] [Google Scholar]
- Wen D., Banaszynski L. A., Liu Y., Geng F., Noh K. M., et al. , 2014a Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. USA 111: 7325–7330. 10.1073/pnas.1406389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D., Banaszynski L. A., Rosenwaks Z., Allis C. D., Rafii S., 2014b H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus 5: 369–375. 10.4161/nucl.36231 [DOI] [PubMed] [Google Scholar]
- Wollmann H., Stroud H., Yelagandula R., Tarutani Y., Jiang D., et al. , 2017. The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18: 94 10.1186/s13059-017-1221-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., et al. , 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44: 251–253. 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen B. T., Bush K. M., Barrilleaux B. L., Cotterman R., Knoepfler P. S., 2014. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 141: 3483–3494. 10.1242/dev.106450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The raw RNA sequencing (RNA-seq) data has been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE106889. Supplemental files are available at FigShare. Figure S1 shows an analysis of C. elegans H3.3 homologs. Figure S2 contains a summary of deletion and mutation alleles. Figure S3 contains correlation plots of biological replicates for the RNA-seq experiments. Figure S4 shows H3.3 homolog expression during embryogenesis. Figure S5 shows H3.3 homolog expression in the hermaphrodite and male germ line. Figure S6 shows a comparison of expression levels of H3.3 homologs. Figure S7 shows that H3.3 proteins are depleted from chromosome X. Figure S8 shows an alignment of C. elegans HIRA-1 with homologs from other species. Figure S9 shows the morphological defects and reduced brood size of hira-1 mutant worms. Figure S10 shows that HIS-71 and HIS-72 are detectable in the postembryonic somatic tissue even upon hira-1 deletion. Figure S11 shows embryonic lethality and apoptosis in H3.3 null mutant worms. Figure S12 shows the identity and expression of additional H3 homologs in C. elegans. Table S1 lists the C. elegans strains used in this study. Table S2 lists the reagents used for allele generation by CRISPR/Cas9 and quantitative PCR. Table S3 contains the RNA-seq results for embryos. Table S4 contains the RNA-seq results for L1 larvae. Table S5 lists all significantly enriched GO terms. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6106448.