Abstract
Multiple species within the basidiomycete genus Cryptococcus cause cryptococcal disease. These species are estimated to affect nearly a quarter of a million people leading to ∼180,000 mortalities, annually. Sexual reproduction, which can occur between haploid yeasts of the same or opposite mating type, is a potentially important contributor to pathogenesis as recombination can generate novel genotypes and transgressive phenotypes. However, our quantitative understanding of recombination in this clinically important yeast is limited. Here, we describe genome-wide estimates of recombination rates in Cryptococcus deneoformans and compare recombination between progeny from α–α unisexual and a–α bisexual crosses. We find that offspring from bisexual crosses have modestly higher average rates of recombination than those derived from unisexual crosses. Recombination hot and cold spots across the C. deneoformans genome are also identified and are associated with increased GC content. Finally, we observed regions genome-wide with allele frequencies deviating from the expected parental ratio. These findings and observations advance our quantitative understanding of the genetic events that occur during sexual reproduction in C. deneoformans, and the impact that different forms of sexual reproduction are likely to have on genetic diversity in this important fungal pathogen.
Keywords: unisexual reproduction, whole-genome sequencing, genome-wide recombination map, crossover hot spots, allele segregation distortion
ANNUALLY, cryptococcal disease is estimated to affect > 200,000 people worldwide, accounting for ∼15% of AIDS-related mortalities (Rajasingham et al. 2017). While Cryptococcus species are preferentially haploid (Hull et al. 2002) and propagate primarily asexually, sexual reproduction and recombination have been demonstrated in both the laboratory and environment (Kwon-Chung 1975, 1976; Hull et al. 2002; Litvintseva et al. 2003; Lin et al. 2007). The sexual cycle in Cryptococcus has clinical relevance as sexual reproduction produces spores that are readily aerosolized and inhaled by hosts, serving as infectious propagules (Giles et al. 2009; Velagapudi et al. 2009; Coelho et al. 2014). Furthermore, recombination during sex produces new genotypes, some of which may display novel phenotypes linked to virulence, such as the ability of offspring to grow at higher temperatures than that of their parental strains (Sun et al. 2014). Thus, quantitatively characterizing recombination in Cryptococcus is a key step to developing a better understanding of the genetics of virulence in this clade.
Cryptococcus deneoformans [formerly C. neoformans var. neoformans, serotype D; see Hagen et al. (2015), Kwon-Chung et al. (2017), and Hagen et al. (2017) for recent discussions of nomenclature] possesses a bipolar mating system with the mating-type locus (MAT) on chromosome 4. The MAT locus, which is > 100 kb in size and contains > 20 genes, is represented in two mating-type alleles, α and a (Heitman et al. 1999; Lengeler et al. 2002; Loftus et al. 2005; Sun and Heitman 2016). In the laboratory setting, sexual reproduction has been observed between haploid MATα and MATa strains (Kwon-Chung 1976; Hull et al. 2002; Nielsen et al. 2007; Xue et al. 2007; Sun et al. 2014; Gyawali et al. 2017). Diploid strains and signatures of recombination have been documented in environmental isolates, indicating that sexual reproduction also occurs in nature (Litvintseva et al. 2003; Campbell et al. 2005; Lin et al. 2007, 2009; Bui et al. 2008). However, an analysis of environmental and clinical isolates of Cryptococcus species revealed a bias in the distribution of the mating-type alleles, with the majority of C. deneoformans isolates analyzed possessing the MATα allele (Kwon-Chung and Bennett 1978). This observation called into question the frequency and importance of bisexual reproduction, and thus recombination, in the wild. Lin et al. (2009) provided an answer to this conundrum with the discovery that C. deneoformans is also capable of undergoing same-sex or unisexual matings between MATα strains (Lin et al. 2005, 2007, 2009).
Meiosis is an integral component of sexual reproduction (Page and Hawley 2003) that occurs in both unisexual and bisexual reproduction (Lin et al. 2005; Feretzaki and Heitman 2013). Within a basidium, meiosis produces nuclei that will undergo several rounds of mitosis to generate subsequent nuclei that are packaged into spores (Kwon-Chung 1980). These basidiospores then bud from the basidium in four long chains (Kwon-Chung 1980; Idnurm 2010). Dissection of basidiospore chains and analysis of their genotypes shows segregation of alleles consistent with one round of meiosis, and demonstrates that postmeiotic nuclei undergo mitosis and randomly assort into different spore chains (Kwon-Chung 1980; Idnurm 2010).
Various studies have examined recombination rates in Cryptococcus species, as well as other phenomena that occur during meiosis, such as crossover hot spots, gene conversions, and allele segregation distortion (Forche et al. 2000; Marra et al. 2004; Hsueh et al. 2006; Sun and Xu 2007; Sun et al. 2014; Sun and Heitman 2016). Genome-wide, our quantitative understanding of recombination is limited to a few studies of C. deneoformans crosses (Forche et al. 2000; Marra et al. 2004) and hybrid crosses between C. deneoformans and C. neoformans strains (Sun and Xu 2007). Current estimates of recombination rates for C. deneoformans are based on linkage maps constructed via a modest number of genetic markers, with estimates varying between 13.2 kb/cM (Marra et al. 2004) and 7.13 kb/cM (Sun et al. 2014).
Sun et al. (2014) described the first quantitative analysis of recombination resulting from unisexual reproduction in Cryptococcus. They generated several hundred progeny derived from α–α unisexual and a–α bisexual matings between C. deneoformans strain backgrounds XL280 (XL280a and XL280αSS) and 431α. Based on 42 markers along chromosome 4, they observed no significant difference in recombination rates between segregants derived from unisexual vs. bisexual reproduction. Across both sets of progeny from the unisexual and bisexual crosses, large regions along chromosome 4 were observed with biased or distorted allele frequencies, deviating from the expected 2:2 parental ratio. While gene conversions could explain some of these anomalies in allelic ratios, most of the conversion tracks were too large to have been generated solely by gene conversion, and the authors hypothesized that such regions were due to mitotic recombination-induced loss-of-heterozygosity prior to meiosis (Sun et al. 2014).
In the present study, we utilize progeny from Sun et al. (2014) and whole-genome sequencing data to quantitatively analyze differences in genome-wide recombination rates between progeny from unisexual and bisexual reproduction, to identify recombination hot and cold spots and to identify chromosomal regions that exhibit biased or distorted allele frequencies. We find genome-wide differences in the average rates of recombination between progeny from α–α unisexual and a–α bisexual crosses, with higher rates of crossovers in samples from a–α bisexual crosses. Recombination hot and cold spots are identified, with hot spots associated with higher than average GC content and cold spots clustering near centromeres. Centromeric cold spots are often flanked by areas of increased crossover activity. Finally, we show that regions with allele frequencies deviating from the expected 2:2 parental allele ratio are not unique to chromosome 4 and are seen genome-wide. The high-resolution characterization of patterns and rates of recombination that this study provides helps to advance our understanding of the processes that generate genetic diversity in this fungus, and will serve as a foundation for future investigations of the population and quantitative genetics of C. deneoformans and related Cryptococcus species.
Materials and Methods
Strains, laboratory crosses, and isolation
Progenitor strains:
As previously described (Sun et al. 2014), parental strains 431α, XL280αSS, and XL280a were used in α–α unisexual and a–α bisexual mating. Progenitor strain 431α is a natural C. deneoformans isolate with the MATα allele (Sun et al. 2012, 2014). XL280αSS is an XL280α strain with an inserted NAT resistance marker in the URA5 gene. Analysis of sequenced reads of progeny from the α–α unisexual crosses confirmed the insertion site of the NAT resistance marker within the URA5 gene of XL280αSS. XL280αSS is congenic to XL280a, with the exceptions of the URA5 gene, the NAT resistance marker, the MAT locus, and a partial duplication of the left arm of chromosome 10 (Zhai et al. 2013; Sun et al. 2014).
Laboratory crosses and isolation:
As described in Sun et al. (2014), parental strains were mixed then spotted onto V8 media (pH = 5) and incubated at room temperature, in the dark, for approximately 1 week until abundant hyphae, basidia, and basidiospore chains were visible under a microscope (Sun et al. 2014). For a–α bisexual crosses between strains XL280a and 431α, chains of basidiospores from individual basidia were transferred onto fresh YPD medium and individual basidiospores were separated using a fiber optic needle. In total, 261 basidiospores were isolated from 27 basidia (Sun et al. 2014).
From α–α unisexual crosses between XL280αSS (NATR ura5) and 431α (NATS URA5), sections at the edge of mating spots were extracted and suspended in PBS. This suspension was then diluted and spread onto synthetic defined-uracil plates to screen for URA5 progeny (Sun et al. 2014). These URA5 progeny were transferred to plates with YPD + Natamycin (NAT) media and further screened for NAT resistance. Finally, 156 (NATR URA5) progeny, representing crossover/fusion of the parental markers, were retained (Sun et al. 2014). Segregants were maintained in 35% glycerol frozen stocks (−80) and subcultured from freezer stock to YPD solid media for study.
Sequencing, aligning, variant calling, and filtering
From the α–α unisexual and a–α bisexual crosses conducted in Sun et al. (2014), 105 segregants (63 from α–α unisexual matings and 42 from a–α bisexual matings) were initially isolated for whole-genome sequencing. Sequencing was performed on the Illumina Hisequation 2500 platform in the Next Generation Sequencing Facility at the University of North Carolina, Chapel Hill. A paired-end library with ∼300 base inserts was constructed for each sample and libraries were multiplexed, running 24 samples per lane using 100-bp paired-end reads. Raw reads were aligned to an XL280 C. deneoformans reference genome (Zhai et al. 2013; Sun et al. 2014) using BWA (v0.7.10-r789, Li and Durbin 2009). Variant calling was carried out using The Genome Analysis Toolkit (v3.1-1, McKenna et al. 2010) and SAMtools (v1.2, Li 2011), resulting in 139,469 variable sites across the 105 segregants. These sites were scored as 0 or 1 if inherited from the XL280αSS (or XL280a) or 431α parental strains, respectively. Variable sites were filtered on read depth and quality. Across segregants, variable sites were required to have > 15× coverage, a quality score, normalized by read depth, of ≥ 20, and a minor allele frequency per site of ≥ 1%. Only sites with 100% call rate were used in analysis. Variant calls were further filtered to include only sites exhibiting biallelic single-nucleotide polymorphisms (SNPs), yielding a final total of 86,767 sites.
Segregant filtering
Read count data for each SNP site was used to screen each of the initial 105 segregants for gross aneuploidy of chromosomes. In total, six segregants were removed due to partial or complete aneuploidy. Aneuploidy of chromosome 1 was detected in three segregants, a duplication of the right arm of chromosome 7 in one segregant, and aneuploidy of chromosome 10 in two segregants. For all samples, pairwise genetic correlations were calculated to identify pairs of segregants that were genetically identical. These duplicates were removed from analysis to avoid biasing estimates of recombination by sampling a genotype more than once. In total, four pairs of segregants from the α–α unisexual crosses were identified as genetically identical. From each of the four pairs of segregants, one was removed from analysis. One segregant from the a–α bisexual cross, SSB593, showed no recombination across its genome and inheritance of all chromosomes, except chromosome 7, from the XL280a parental strain. This segregant was removed from further analysis. After passing these filtering criteria, 94 segregants, 55 from α–α unisexual crosses and 39 from a–α bisexual crosses, were retained for analysis.
Haplotype construction and filtering
For each sample, SNP data were used to estimate regions with consecutive SNPs inherited from one parent (i.e., haplotypes) between XL280a, XL280αSS, and 431α. A “minimum” run approach based on intermarker intervals was used to determine the size of haplotypes (Mancera et al. 2008). Briefly, for a set of SNPs within a haplotype with positions along a chromosome, the size of the haplotype in nucleotide bases or length of the intramarker interval is calculated as The intermarker interval is defined as the distance between two SNPs with opposing genotypes (Mancera et al. 2008). Let be the positions of two adjacent SNPs along a chromosome with opposing genotypes, then the distance in nucleotide bases between the two SNPs is calculated as For each sample, SNP data were used to construct haplotype blocks, where runs of contiguous SNPs with shared genotypes are grouped. For the results shown here, haplotypes were retained if the size of the haplotype or intramarker interval was ≥ 6 kb.
Crossover frequency estimation
Poisson regression:
Haplotype data for each segregant was used to calculate the number of crossovers. For any given segregant with n haplotypes there are crossovers. Across all segregants, a region 10 kb in size, centered on the URA5 locus (chromosome 7), was masked from analysis to account for the insertion of the NAT marker in the progenitor strain XL280αSS, used in the α–α unisexual crosses. Genome-wide recombination rates were estimated using Poisson regression, modeling the number of crossovers as a function of chromosome length with the mode of sexual reproduction as a covariate using the ‟glm” function implemented in R (version 3.4.1). Our analysis indicated no support for an interaction term between chromosome length and mode of sexual reproduction; we therefore fit a simple additive model of the form where is chromosome length and is an indicator variable for the cross type (0 = α–α unisexual and 1 = a–α bisexual crosses).
The model was estimated as: The model fit failed to reject the null hypothesis of a zero intercept term () but there was strong support to reject the null hypothesis of zero-valued and coefficients (P-values ).
Analysis of crossovers per chromosome:
For each chromosome, the number of crossovers was compared between segregants from the α–α unisexual and a–α bisexual crosses. A two-sided, Mann–Whitney U-test with an was utilized to test for significant differences in the average number of crossovers (per chromosome), along with the Holm–Sidak step-down method to correct for multiple testing (Holm 1979).
Recapitulation of previous results:
Across progeny from the α–α unisexual and a–α bisexual crosses, SNPs were selected to approximate original marker locations along chromosome 4 from Sun et al. (2014). This was done by reducing the set of SNPs to those with positions ≤ 1 kb away from the previously utilized marker locations (Sun et al. 2014). This yielded 42 and 17 SNPs for the segregants from α–α unisexual and a–α bisexual crosses, respectively, similar to the size of the original marker sets used by Sun et al. (2014). These data were then used to reconstruct haplotypes as described above and to calculate crossover events.
Crossover hot and cold spot discovery and analysis
Statistical association testing:
For each chromosome, contiguous bins of varying size were used to tile each chromosome from the edges of the centromeres out to the ends of the chromosome (centromeric regions were excluded from hot/cold spot analysis). After investigating the total detected number of hot and cold crossover spots as a function of bin size (from 0.5 to 100 kb), a bin size of 41.5 kb was chosen because it minimized the difference between the detected number of crossover hot and cold spots (Supplemental Material, Figure S9). The outermost 5′ and 3′ bins of each chromosome were constructed to have at least half of their width overlap the last two annotated SNPs on the respective end of that chromosome. Within each bin, the number of intermarker intervals in which a crossover was detected were counted. For each intermarker interval, crossovers shared by meiotic siblings were only counted once. For every bin, a Poisson model, with parameters established from genome-wide analysis of crossover frequencies of meiotic progeny from the a–α bisexual crosses, was utilized to compare the number of crossovers observed vs. the number expected given the bin size. A two-tailed test was used to search for statistically cold and hot crossover spots. A false discovery rate (FDR) approach (Benjamini and Yekutieli 2001) was used to define genome-wide, significantly hot or cold crossover spots, using an FDR cutoff of 0.05. An “artificial” hot spot on chromosome 7, resulting from the use of selectable markers to isolate recombinant progeny from the α–α unisexual crosses (Sun et al. 2014), was removed from the analysis.
Analysis of GC content:
For each intermarker interval, nucleotide sequences were obtained from the XL280 reference genome (Zhai et al. 2013). The GC content for all intermarker intervals was calculated and classified as hot, cold, or other according to whether the interval fell within a hot or cold region, as defined above. In total, there were 7558 hot intermarker interval sequences, 7369 cold spot intermarker interval sequences, and 68,051 intervals defined as other. The GC content for intermarker intervals within hot and cold spots was compared using a two-sided, Mann–Whitney U-test (). For the three groups of intermarker interval sequences, 95% C.I.s were calculated via permutation (sampling with replacement), taking the difference between the observed mean GC content and the sampled mean, 1000 times. From these deviations, the 2.5 and 97.5% percentiles of the permuted distribution were used as critical values.
Identification of motifs associated with crossover hot spot sequences:
To search for sequence motifs associated with hot spots, 100 random sequences from hot spot intermarker intervals in which there was a crossover where chosen, such that the lengths of sequences ranged between 100 and 10,000 bases and the sum of the sequences was < 60 kb. A complementary control set of 100 randomly chosen sequences was selected from other genomic regions using the same parameters. The hot and control sets of sequences where analyzed using the MEME software suite, version 4.12.0 (Bailey and Elkan 1994). Analysis in MEME was conducted using discriminative mode, with zero or one occurrence of a contributing motif site per sequence, searching for four motifs between 6 and 50 bases wide.
Analysis of allele distortion and bias
Segregants used in haplotype analysis:
From the a–α bisexual cross, 22 of the 39 segregants were grouped by basidium, representing five unique basidia. Basidia groups where chosen for analysis if they contained three or more segregants with unique genotypes. Of the five basidia groups, two consisted of three segregants, two with four segregants, and one basidium exhibited eight unique genotypes.
Analysis of regions with distorted allele frequencies:
The allele frequency of regions across segregants germinated from the same basidium was analyzed. Specifically, deviations from the expected 2:2 parental allele ratio where quantified. Regions were removed from consideration if only a single SNP supported the observation or if the size of the region was only one base in width. An ANOVA was used to examine average differences in sizes of regions with distorted allele frequencies across the genome. A log-linear model was used to investigate the average number of regions as a function of chromosome size ().
Analysis of allele bias:
Across all the 39 segregants from the a–α bisexual crosses, a binomial model was used to identify chromosomal regions with bias toward one parental allele. This model assumed equal likelihood for inheriting either of the parental alleles (). SNP sites were collapsed across the 39 segregants based on recombination breakpoints and common allele frequencies. This generated 944 sites to test in the binomial model. An FDR approach (FDR = 0.05) was used to correct for multiple comparisons. A similar procedure was used for testing for allele bias in the α–α unisexual cross.
Data availability
Raw sequence reads generated from samples utilized in this study are available on the National Center for Biotechnology Information’s sequence read archive under Bio-Project identification number PRJNA420966, with individual accession numbers SAMN08130857–SAMN08130963. The generated variant call file from the aligned sequenced reads are publicly available on the GitHub repository (https://github.com/magwenelab/crypto-recombination-paper). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6083315.
Results
High-density SNP data allows fine mapping of genome-wide crossovers
Whole-genome sequencing data were obtained for 55 segregants from α–α unisexual crosses between parental strains XL280αSS and 431α, and 39 segregants from a–α bisexual crosses between the parental strains XL280a and 431α (Sun et al. 2014). Variants were called for each segregant (see Materials and Methods) and 86,767 biallelic SNPs between the parental strains were used as genetic markers. Across the 19-Mb genome, comprised of 14 chromosomes, the median distance between consecutive SNPs (intermarker interval) was 87 bases with only 0.5% of the 86,753 intermarker intervals larger than 2 kb (Figure S1). SNP data were used to infer haplotypes and crossover events per segregant (Figure 1). In total, 3240 crossovers were detected.
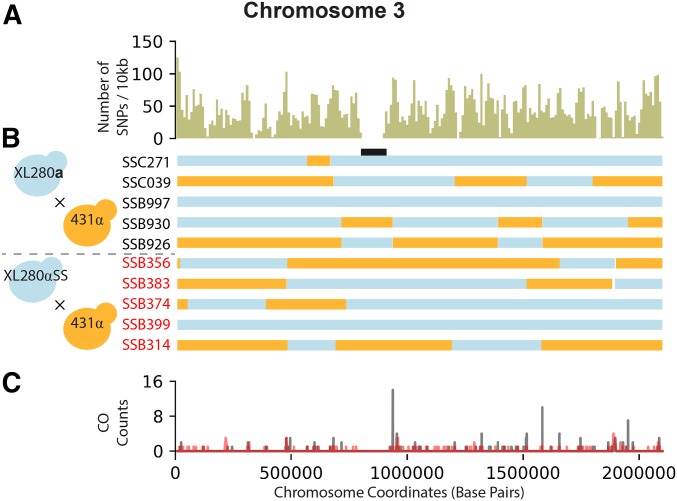
Figure 1.
SNP density, haplotypes, and crossover (CO) counts of chromosome 3. (A) The SNP density for chromosome 3 (length 2.1 Mb) across the progeny from the XL280a × 431α and XL280αSS × 431α crosses, calculated as the number of SNPs per 10 kb (total: 9779 SNPs). (B) Haplotypes, inferred from SNP data, are displayed as blue if inherited from XL280 (a or αSS) or orange if inherited from 431α for 10 segregants, five from the a–α bisexual (black) and five from the α–α unisexual (red) crosses. The position of the centromere is displayed in black. (C) CO counts (y-axis) along chromosome 3 (x-axis) for segregants from the a–α bisexual (black) and α–α unisexual (red) crosses. COs are detected by changes in genotype between two contiguous SNPs.
In each set of progeny from the α–α unisexual and a–α bisexual crosses, several segregants were identified as having at least one nonexchange chromosome. In 35 of 55 (64%) progeny from the α–α unisexual crosses and 19 of 39 (49%) progeny from the a–α bisexual crosses, at least one chromosome was nonrecombinant based on filtered SNP data and inferred haplotypes. There is no difference in the distributions of numbers of nonexchange chromosomes per segregant across the two cross types (Kolmogorov–Smirnov-test, P-value ). For these progeny, the median number of nonexchange chromosomes per segregant is between one and five. Smaller chromosomes are more likely to have zero crossovers. Of the 59 nonexchange chromosomes in the 35 progeny from the unisexual crosses, 32 (54%) have the parental XL280αSS genotype. However, in the 37 nonexchange chromosomes among the 19 progeny from the bisexual crosses, 29 (78%) have the XL280a parental copy.
Genome-wide recombination rates differ between unisexual and bisexual reproduction in C. deneoformans
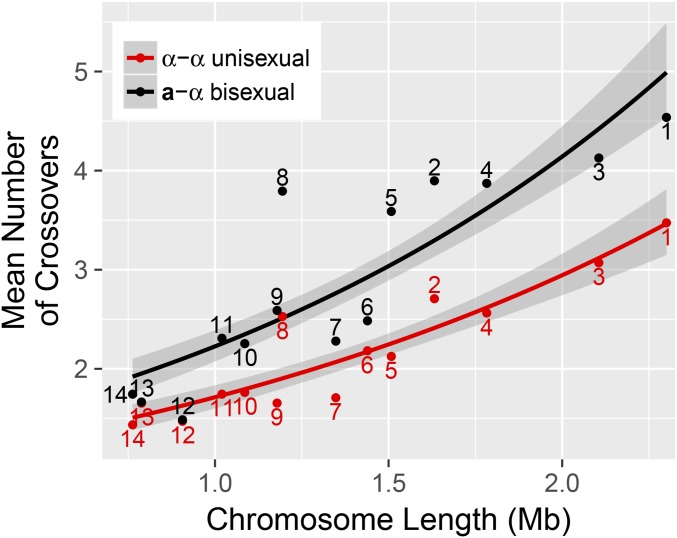
Genome-wide recombination rates were estimated using Poisson regression, modeling the number of crossovers as a function of chromosome length with the mode of sexual reproduction as a covariate (see Materials and Methods). This model predicts an obligatory ∼0.94 crossovers per chromosome for offspring from the unisexual crosses and ∼1.27 crossovers per chromosome for offspring from the bisexual cross. There is a significant difference in the expected number of crossovers between segregants from α–α unisexual and a–α bisexual crosses (P-value ). The expected number of crossovers is predicted to increase by a ratio of ∼1.79 per Mb increase in chromosome size (Figure 2). Based on the sum of the per chromosome average and the total genome length, we estimate an approximate physical–genetic distance of ∼6.33 kb/cM for the α–α unisexual crosses and ∼4.69 kb/cM for the a–α bisexual crosses.
Figure 2.
Unisexual vs. bisexual crossovers as a function of chromosome length. The average number of crossovers for progeny from the α–α unisexual (red) and a–α bisexual crosses (black) are shown per chromosome. Solid lines indicate the estimated Poisson regressions for the two cross types separately, relating the number of crossovers to chromosome lengths. Shaded regions are 95% C.I.s of the regression estimates. Numbers indicate chromosomes.
To explore this difference in greater detail, we compared recombination rates by chromosome for the two types of crosses. For chromosomes 1–5, 8, and 9 there are significant differences (Mann–Whitney U-test, FDR 0.042) in the average number of detected crossovers between the progeny from the α–α unisexual and a–α bisexual crosses. No significant difference in the average number of crossovers between the two cross types was detected on chromosomes 6, 7, and 10–14 (Figure S2).
Our finding that recombination rates differ in offspring generated via unisexual vs. bisexual reproduction contrasts with the conclusions of the earlier study of Sun et al. (2014), which utilized the same set of offspring. We reasoned that this difference was due to increases in the detected number of crossovers resulting from increased marker density. We confirmed this by using SNPs for reanalysis that were selected to best approximate marker locations from Sun et al. (2014) (see Materials and Methods). Using this smaller set of SNPs to reconstruct haplotypes and calculate crossover events recapitulated the findings of Sun et al. (2014) (Figure S3).
Analysis of crossover hot spots for segregants from α–α unisexual and a–α bisexual crosses in C. deneoformans
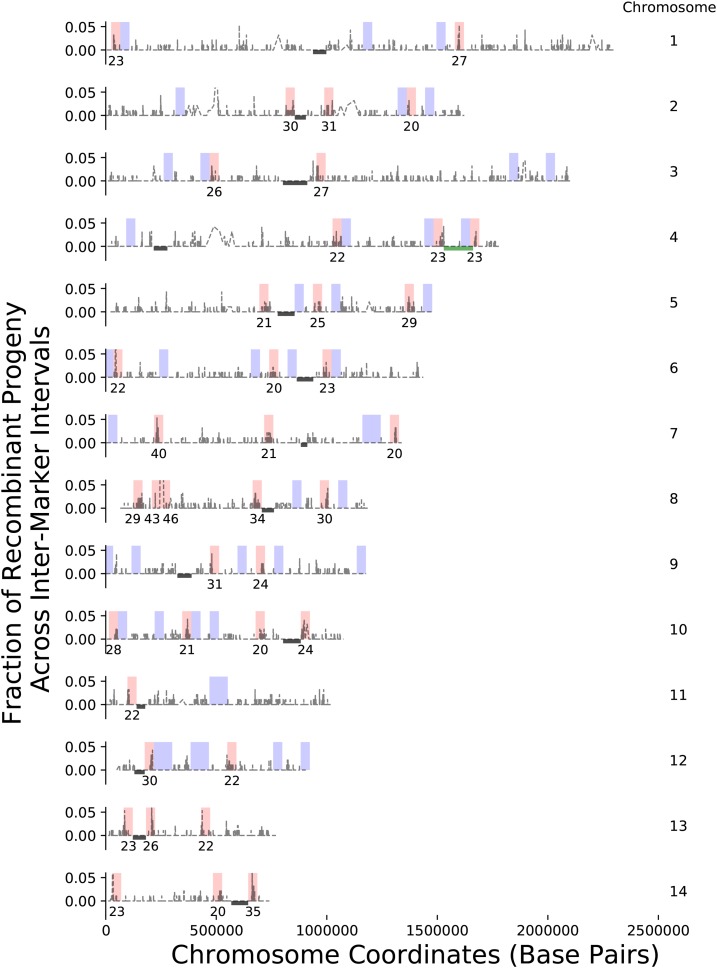
To identify regions of high- and low-crossover activity or crossover hot and cold spots (Petes 2001) along each chromosome, a binning approach was used. Bins of size 41.5 kb were tiled across each chromosome and the number of crossovers detected within each bin was counted. The bin size of 41.5 kb was chosen based on simulations, so as to minimize the difference in the total number of hot and cold spots (Figure S9). A Poisson model with this bin size and the expected genome-wide average crossover rate per segregant, as estimated from the observed data (see Materials and Methods), was used in two-tailed tests to examine each bin for significantly high- (hot) or low- (cold) crossover rates. An FDR procedure was used to establish genome-wide significance ( FDR 0.014) (Benjamini and Yekutieli 2001). This analysis revealed 39 hot spots, bins with 20 or more detected crossovers, and 44 cold spots, bins with zero detected crossovers (Figure 3). Along every chromosome, at least one crossover hot spot was identified and these regions were often found flanking or near centromeres.
Figure 3.
Genome-wide crossover hot and cold spots. In gray, the fraction of recombinant progeny between intermarker intervals (y-axis) for segregants from the α–α unisexual and a–α bisexual crosses along each of the 14 chromosomes (indicated by number to the right). Crossovers occur within an intermarker interval and are detected as a change in genotype between consecutive SNPs. Bins, 41.5 kb in size, were used to segment each chromosome. For bins identified as crossover hot spots (red), the number of crossovers detected is labeled underneath. All crossover cold spots (blue) have zero detected crossovers. Locations of centromeres and the MAT locus are displayed as black bars and a green bar, respectively. Note that the y-axis has been truncated in many instances to visualize crossovers along each chromosome.
Previous studies have demonstrated an association between recombination hot and cold spots and GC content (Sun et al. 2012; Sun and Heitman 2016). For 7558 intermarker interval sequences within the 39 hot spots, the mean GC content was 0.49 (95% C.I.: 0.489, 0.494), while the mean GC content for 7369 intermarker interval sequences contained within the 44 cold spots was 0.475 (95% C.I.: 0.473, 0.477). The mean GC content of hot spots differs significantly from the cold spots (Mann–Whitney U-test, P-value Figure S4). Both of these differ from the reported genome-wide average GC content (0.486) and the mean (0.483, 95% C.I.: 0.482, 0.484) of the other 68,051 intermarker interval sequences not associated with hot or cold spots (Sun et al. 2012). Of the 7558 intermarker interval sequences within identified hot spots, 584 detect a genotype change (i.e., the approximate sites of double-strand breaks), and of these intermarker interval sequences 64.4% overlapped with intergenic regions when compared to the annotated reference strain, JEC21 (Loftus et al. 2005).
From the set of 584 intermarker interval sequences associated with hot spots and in which a crossover occurs, 100 random sequences were analyzed using MEME to identify sequence motifs associated with crossover hot spots. These sequences were compared to a control set of sequences selected in a similar fashion from other genomic regions. A poly(G) motif that is 29 bases long was identified in all of the 100 hot spot-associated sequences (E-value Figure S5).
Allele bias and allele distortion seen in segregants generated via bisexual reproduction in C. deneoformans
Separately for the 55 segregants from the α–α unisexual crosses and 39 segregants from the a–α bisexual crosses, a binomial model was used to identify chromosomal regions with bias toward one parental allele, using a null model of equal likelihood of inheriting either of the parental alleles (P = 0.5). The allele frequencies across SNP sites in segregants from the α–α unisexual cross do not show evidence of bias toward either parental allele that reaches genome-wide significance. Across the 39 progeny from a–α bisexual crosses, five regions show evidence of biased allele inheritance toward the XL280a allele (FDR ). These regions are located on chromosomes 1, 2, 4, 6, and 12, with lengths of ∼364, 260, 303, 41, and 60 kb, respectively (Figure S6).
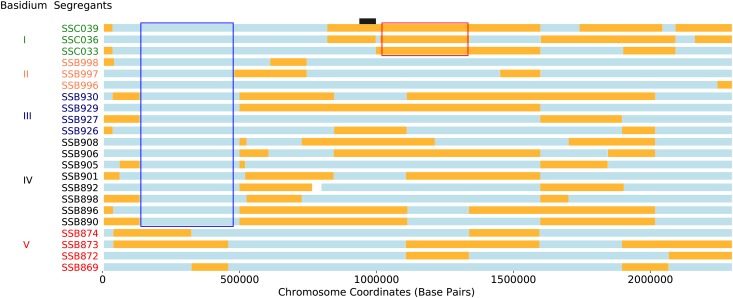
Allelic inheritance patterns within basidia were then examined for segregants from the a–α bisexual cross. Of the 39 progeny from the a–α bisexual crosses, 22 may be grouped by basidia of dissection. This grouping method generates five groups for analysis with three (), four (), and eight () segregants, all with unique genotypes (Figure 4). Using these segregants, 197 regions were identified across the genome with allelic ratios deviating from the expected parental ratio of 2:2 (allelic distortion). The size of these regions with allelic distortion ranged from a minimum of six bases to a maximum of 1.4 Mb (Figure S7A). The average size of regions exhibiting allelic distortion does not differ significantly across chromosomes (ANOVA, P-value = 0.092). The locations of regions exhibiting allelic distortion are often similar across basidia (Figure 4). Of the 197 allelic distortions, 83 were identified from basidia III, IV, and V, with allele ratios consistent with possible gene conversions.
Figure 4.
Allelic distortions along chromosome 1 in segregants from a–α bisexual crosses. Haplotypes (blue indicating inheritance from XL280a, orange from 431α) for 22 segregants from the a–α bisexual crosses, grouped by basidium of dissection (numbered I to V). Framed in red is a region, in a single basidium, exhibiting allelic distortion in the direction of 431α. Framed in blue is a region that exhibits allelic distortion (toward XL280a) across multiple basidia. This second region overlaps with a region of allelic bias, as determined from analysis of all progeny from the bisexual crosses. The position of the centromere is displayed as a black bar. Other regions with allelic distortion are present within this figure but have not been highlighted to preserve visual clarity.
Across the 197 regions with distorted parental allele frequencies, the direction of bias was examined. Along chromosome 12, 11 regions with distorted allele frequencies were identified, and 10 of these retain the XL280a allele. However, genome-wide, no evidence of consistent bias toward either parental genotype was observed (Figure S7B).
The average number of regions with distorted allele frequencies across the genome was established as a function of chromosome size for our 22 segregants, representing five unique basidia from the a–α bisexual crosses (Figure S8). A log-linear model provides evidence supporting a significant association between chromosome size and the average number of regions with distorted allele frequencies (P-value ).
Unique patterns of allele segregation
Two groups of segregants from the a–α bisexual crosses representing two unique basidia showed interesting patterns of allele segregation. The first group of samples dissected from one basidium was comprised of eight spores and analysis of their recombinant haplotypes indicates that all eight samples are genetically unique (for example see Figure 4, basidium IV). This observation deviates from the expected four unique gametes expected to result from meiosis (Kwon-Chung 1980; Page and Hawley 2003; Idnurm et al. 2005). The second basidium showing interesting allele segregation was composed of four segregants. These four samples are all recombinant and were previously thought to be genetically unique, as indicated by marker genotypes along chromosome 4 (Sun et al. 2014). However, our reanalysis indicates that two of the four segregants are nearly genetically identical; chromosome 4 is the only distinct chromosome differentiating the two samples, which are identical along the other 13 chromosomes, including a duplication of chromosome 10.
Discussion
C. deneoformans is capable of sexual reproduction between strains of the opposite and the same mating types. In this study, we document higher rates of recombination in offspring generated from bisexual crosses, which is surprising, given our previous results in Sun et al. (2014). Progeny from the bisexual cross are predicted to have a basal rate of ∼1.27 crossovers per chromosome vs. ∼0.94 crossovers per chromosome for progeny in the unisexual cross. For both sets of progeny, the number of crossovers is predicted to increase by a ratio of ∼1.79 per Mb increase in chromosome size. Of the 14 chromosomes in the C. deneoformans genome, seven show differences in the average number of crossovers per segregant when comparing samples from a–α bisexual and α–α unisexual crosses. Converting these crossover rates, we estimate an approximate physical–genetic distance of ∼6.33 and ∼4.69 kb/cM for the α–α unisexual and a–α bisexual crosses, respectively. These estimates are nearly three times lower than the estimated crossover rate of Saccharomyces cerevisiae (∼2 kb/cM; Cherry et al. 1997; Barton et al. 2008) and far higher than the crossover rates estimated for Drosophila melanogaster (∼100 kb/cM; Comeron et al. 2012), Arabidopsis thaliana (∼278 kb/cM; Salomé et al. 2012), and Homo sapiens (∼840 kb/cM; Kong et al. 2002).
Our results differ from previous estimates because they are based on information from the entire C. deneoformans genome and utilize > 200-fold higher density of markers than has been employed in any previous study of recombination in C. deneoformans (Forche et al. 2000; Marra et al. 2004; Sun et al. 2014). For example, relative to the earlier study of Sun et al. (2014), which utilized the same set of offspring, we detected differences in the average number of crossovers along chromosome 4 between progeny from α–α unisexual and a–α bisexual crosses. Our analysis shows that this difference is due to an increase in the detected number of crossovers resulting from greater marker density, which facilitates the detection of genotype changes previously masked by double-crossover events.
The regression model used to relate chromosome length and the number of crossovers predicts nearly one obligate crossover on average per chromosome for both sets of progeny from the α–α unisexual and a–α bisexual crosses (see Results). A significant number of segregants had chromosomes that had zero detected crossovers (nonexchange chromosomes), but analysis of segregants from basidia groups suggests that the standard model of crossover assurance holds (i.e., there is at least one crossover per homologous chromosome pair per meiosis; Ault and Nicklas 1989). The nonexchange chromosomes we observed may thus be due to Holiday junctions resolving into noncrossover events during chromosome disjunction or may reflect chromatids that were not involved in crossovers during meiosis.
The analysis of crossover hot and cold spots identified at least one crossover hot spot along each of the 14 chromosomes, and cold spots on every chromosome except chromosomes 13 and 14. Analyses based on a subset of the hot spot intermarker interval sequences, in which crossovers were detected, identified a poly(G) motif significantly enriched within these sequences. Furthermore, intermarker interval sequences within crossover hot spots have on average higher GC content, as documented in other studies of C. deneoformans as well as other fungi (Gerton et al. 2000; Petes 2001; Mancera et al. 2008; Marsolier-Kergoat and Yeramian 2009; Sun et al. 2012; Sun and Heitman 2016). Of the crossover hot spots, two were identified that flank the MAT locus, recapitulating the findings of several other studies (Marra et al. 2004; Hsueh et al. 2006; Sun et al. 2012; Sun and Heitman 2016). While recombination hot spots flank the MAT locus, the MAT locus itself contains a crossover cold spot, consistent with previous findings (Lengeler et al. 2002; Sun et al. 2014). Parallel to the pattern observed at the MAT locus, we noted a tendency for crossover hot spots to flank or surround centromeric regions and crossover cold spots. Some caution is required in interpreting the total number of hot and cold spots, and their precise locations. Due to the SNP and haplotype-filtering criteria we employed, some genomic regions such as centromeres and telomeres are excluded from analysis. Thus, we are unable to access recombination or gene conversion events that could have taken place within centromeric regions, as suggested in previous studies of Cryptococcus (Janbon et al. 2014; Sun et al. 2017) and other fungal species such as Candida albicans (Thakur and Sanyal 2013). The precise location of inferred hot and cold spots is also a function of the choice of bin widths and starting coordinates.
In addition to providing genome-wide information on crossover hot and cold spots, our analysis identified numerous regions that have allele ratios that deviate from the expected 2:2 parental ratio in progeny from the a–α bisexual crosses, consistent with and extending the findings of Sun et al. (2014) for chromosome 4. Some of the regions with deviant allele frequencies have 3:1 allele ratios, which would be consistent with gene conversion, but most of the regions of allelic distortion are quite large, nearing 100 kb. Thus, it is unlikely that gene conversions alone explain the observed loss-of-heterozygosity genome-wide, as conversion tracks from gene conversions are thought to be small, on the order of only a few kilobases, as observed in S. cerevisiae (Mancera et al. 2008). Alternate models that could explain the observed allelic distortions include mitotic recombination that takes place after nuclear fusion but prior to meiosis, or chromosomal mis-segregation that takes place during cell fusion prior to meiosis and the formation of a basidium, leading to loss of a parental genotype. Chromosomal breakage prior to meiosis followed by repair using the homologous chromosome could also lead to a loss of one of the parental alleles (Sun et al. 2014). It is also possible that combinations of alleles between the progenitor strains 431α and XL280a could have led to the creation of nonviable genotypes, thus resulting in observed biased allele inheritance in the sampled spores.
Of the segregants from the a–α bisexual crosses, two groups are worth discussing in detail. The first group is comprised of four segregants from a single basidium. All four segregants were previously described as unique based on marker genotypes along chromosome 4 (Sun et al. 2014). However, genome-wide analysis revealed that two of the segregants are genetically identical except for chromosome 4 and are aneuploid for chromosome 10. For this set of segregants, the patterns of allele segregation could be explained by chromosomal nondisjunction. During the formation of the basidium and during meiosis, chromosomal nondisjunction could have produced three nuclei, two with the correct ploidy of both chromosome 4 and 10 and one nucleus with two unique, recombinant copies of chromosome 4. Such patterns have been observed in hybrid crosses between C. neoformans and C. deneoformans (Vogan et al. 2013). During mitosis and basidiospore packaging, this aneuploid nucleus may have produced several copies of itself with varying arrangements of the genome, thus generating haplotypes that are genetically identical except for chromosome 4, as seen in two of these segregants. Another basidium from the a–α bisexual crosses that exhibited interesting patterns of allele segregation was a collection of eight segregants. Analysis of the haplotypes of these eight segregants indicates that all are genetically unique. In this instance, fusion between sister haploid nuclei could have taken place postmeiosis within the basidium, providing an opportunity for mitotic recombination to occur, and, through subsequent rounds of mitosis, produce more than four unique gametes (Vogan et al. 2013). Due to the nature of C. deneoformans and the methods of dissection, it is almost impossible to determine if crossover events occur during meiosis or mitosis.
Our analyses provide evidence of different rates of recombination in unisexual and bisexual crosses. What are the molecular mechanisms that could drive such differences in recombination and how might they have evolved? A strong candidate is mating type-specific regulation of meiotic genes. For example, in the budding yeast S. cerevisiae, the meiotic inhibitor Rme1 is regulated in a mating type-specific manner; repression of RME1 by the a1/α2 complex is required for the initiation of meiosis, thus restricting meiosis only to a/α diploids (Covitz et al. 1991; Covitz and Mitchell 1993). Similarly, mating type-specific regulation of DNA repair and mitotic recombination has been shown to occur in S. cerevisiae (Haber 2012). While there is no ortholog of Rme1 in Cryptococcus, mating type-specific transcriptional regulators such as SXI1α and SXI2a (Hull et al. 2005; Mead et al. 2015) may directly or indirectly regulate genes that are critical for recombination, such as DMC1 and SPO11 (Lin et al. 2005), leading to higher or lower crossover rates during sexual reproduction.
Given that bisexual mating is the most parsimonious hypothesis for the ancestral mode of sexual reproduction for the Cryptococcus clade (Hsueh et al. 2011; Heitman 2015), the evolutionary origin of unisexual reproduction in Cryptococcus species likely required rewiring of the gene networks that regulate meiosis. A key first step would be mutations that allowed for the initiation of meiosis in genotypes other than a/α-diploids. Once this basic rewiring had occurred, additional mating type-specific modifier mutations that induce quantitative effects on recombination rates or patterns of allelic inheritance could evolve, perhaps in a lineage-specific manner.
In this report, we have focused on a pair of crosses between two strains within a single species of the Cryptococcus complex, and the extent to which the patterns and rates of recombination we document here hold across all of the Cryptococcus species and lineages is as yet unknown. Like C. deneoformans, in the VNI and VNII lineages of C. neoformans, most isolates are of the MATα mating type (Kwon-Chung and Bennett 1978). Only in populations of the VNBI and VNBII lineages are MATa strains found with significant frequency (Litvintseva et al. 2003; Desjardins et al. 2017). This has led to the hypothesis that sexual reproduction in many C. neoformans lineages may be primarily unisexual (Fu et al. 2015). The differences in the rates of recombination we document here between a–α bisexual and α–α unisexual matings may contribute to differences in population recombination rates, even if a–α bisexual and α–α unisexual matings occur at similar frequencies. Consistent with this idea, the analysis of Desjardins et al. (2017) indicates that linkage disequilibrium decays at a relatively similar rate in both VNB lineages (bisexual) and the VNI lineage (unisexual). However, the primarily unisexual VNI lineage shows an overall higher rate of linkage disequilibrium. New high-resolution genomic data, both from crosses and from population studies (Desjardins et al. 2017; Rhodes et al. 2017), will help to clarify the relative contributions that sex, mitotic recombination (Vogan et al. 2013), hypermutation (Billmyre et al. 2017), and other mechanisms for generating genomic variation have on the origins and maintenance of genetic diversity within this clade of fungal pathogens.
Acknowledgments
We thank Debra Murray and Selcan Aydin for comments and feedback on the manuscript. This research was funded by a Research Training Grant from the University Program in Genetics and Genomics, Duke University, the Office of Biomedical Graduate Diversity’s Initiative for Maximizing Student Diversity program, BioCoRE, and National Institutes of Health grants R56 AI-123502, R01 AI-133654, and R37 AI-39115-20. The authors declare no conflicts of interest.
Author contributions: S.S. and J.H. conceived and designed the experiments. S.S. performed the experiments C.R., R.B.B., and P.M.M. analyzed the data. S.S., R.B.B., and J.H. contributed reagents and materials. C.R. and P.M.M. wrote the paper. C.R., S.S., R.B.B., J.H., and P.M.M edited the paper.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6083315.
Communicating editor: A. Mitchell
Literature Cited
- Ault J. G., Nicklas R. B., 1989. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma 98: 33–39. 10.1007/BF00293332 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Barton A. B., Pekosz M. R., Kurvathi R. S., Kaback D. B., 2008. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics 179: 1221–1235. 10.1534/genetics.107.083493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D., 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Billmyre R. B., Clancey S. A., Heitman J., 2017. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. Elife 6: e28802 10.7554/eLife.28802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T., Lin X., Malik R., Heitman J., Carter D., 2008. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, α mating type populations. Eukaryot. Cell 7: 1771–1780. 10.1128/EC.00097-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. T., Currie B. J., Krockenberger M., Malik R., Meyer W., et al. , 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell 4: 1403–1409. 10.1128/EC.4.8.1403-1409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M., Ball C., Weng S., Juvik G., Schmidt R., et al. , 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67. [PMC free article] [PubMed] [Google Scholar]
- Coelho C., Bocca A. L., Casadevall A., 2014. The tools for virulence of Cryptococcus neoformans. Adv. Appl. Microbiol. 87: 1–41. 10.1016/B978-0-12-800261-2.00001-3 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covitz P. A., Mitchell A. P., 1993. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 7: 1598–1608. 10.1101/gad.7.8.1598 [DOI] [PubMed] [Google Scholar]
- Covitz P. A., Herskowitz I., Mitchell A. P., 1991. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-α 2. Genes Dev. 5: 1982–1989. 10.1101/gad.5.11.1982 [DOI] [PubMed] [Google Scholar]
- Desjardins C. A., Giamberardino C., Sykes S. M., Yu C.-H., Tenor J. L., et al. , 2017. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 27: 1207–1219. 10.1101/gr.218727.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M., Heitman J., 2013. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet. 9: e1003688 10.1371/journal.pgen.1003688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Xu J., Vilgalys R., Mitchell T. G., 2000. Development and characterization of a genetic linkage map of Cryptococcus neoformans var. neoformans using amplified fragment length polymorphisms and other markers. Fungal Genet. Biol. 31: 189–203. 10.1006/fgbi.2000.1240 [DOI] [PubMed] [Google Scholar]
- Fu C., Sun S., Billmyre R. B., Roach K. C., Heitman J., 2015. Unisexual vs. bisexual mating in Cryptococcus neoformans: consequences and biological impacts. Fungal Genet. Biol. 78: 65–75. 10.1016/j.fgb.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O., et al. , 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. 10.1073/pnas.97.21.11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles S. S., Dagenais T. R. T., Botts M. R., Keller N. P., Hull C. M., 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77: 3491–3500. 10.1128/IAI.00334-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali R., Zhao Y., Lin J., Fan Y., Xu X., et al. , 2017. Pheromone independent unisexual development in Cryptococcus neoformans. PLoS Genet. 13: e1006772 10.1371/journal.pgen.1006772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F., Khayhan K., Theelen B., Kolecka A., Polacheck I., et al. , 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78: 16–48. 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Hagen F., Lumbsch H. T., Arsic Arsenijevic V., Badali H., Bertout S., et al. , 2017. Importance of resolving fungal nomenclature: the case of multiple pathogenic species in the Cryptococcus genus. MSphere 2: e00238–e00217. 10.1128/mSphere.00238-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., 2015. Evolution of sexual reproduction: a view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol. Rev. 29: 108–117. 10.1016/j.fbr.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Allen B., Alspaugh J. A., Kwon-Chung K. J., 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28: 1–5. 10.1006/fgbi.1999.1155 [DOI] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70. [Google Scholar]
- Hsueh Y.-P., Idnurm A., Heitman J., 2006. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2: e184 10.1371/journal.pgen.0020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh, Y.-P., B. Metin, K. Findley, M. Rodriguez-Carres, and J. Heitman, 2011 The mating-type locus of cryptococcus: evolution of gene clusters governing sex determination and sexual reproduction from the phylogenomic perspective, pp. 139–149 in Cryptococcus: From Human Pathogen to Model Yeast. American Society for Microbiology (ASM), Washington, DC. [Google Scholar]
- Hull C. M., Davidson R. C., Heitman J., 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16: 3046–3060. 10.1101/gad.1041402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C. M., Boily M.-J., Heitman J., 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4: 526–535. 10.1128/EC.4.3.526-535.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185: 153–163. 10.1534/genetics.109.113027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Bahn Y.-S., Nielsen K., Lin X., Fraser J. A., et al. , 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3: 753–764. 10.1038/nrmicro1245 [DOI] [PubMed] [Google Scholar]
- Janbon G., Ormerod K. L., Paulet D., Byrnes E. J., III, Yadav V., et al. , 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10: e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D. F., Sainz J., Jonsdottir G. M., Gudjonsson S. A., et al. , 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. 10.1038/ng917 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67: 1197–1200. 10.2307/3758842 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68: 943–946. [PubMed] [Google Scholar]
- Kwon-Chung K. J., 1980. Nuclear genotypes of spore chains in Filobasidiella neoformans (Cryptococcus neoformans). Mycologia 72: 418–422. 10.2307/3759266 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Bennett J. E., 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108: 337–340. 10.1093/oxfordjournals.aje.a112628 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Bennett J. E., Wickes B. L., Meyer W., Cuomo C. A., et al. , 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. MSphere 2: e00357–e00316. 10.1128/mSphere.00357-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler K. B., Fox D. S., Fraser J. A., Allen A., Forrester K., et al. , 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1: 704–718. 10.1128/EC.1.5.704-718.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Hull C. M., Heitman J., 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434: 1017–1021. 10.1038/nature03448 [DOI] [PubMed] [Google Scholar]
- Lin X., Litvintseva A. P., Nielsen K., Patel S., Floyd A., et al. , 2007. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3: 1975–1990. 10.1371/journal.pgen.0030186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Patel S., Litvintseva A. P., Floyd A., Mitchell T. G., et al. , 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5: e1000283 10.1371/journal.ppat.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva A. P., Marra R. E., Nielsen K., Heitman J., Vilgalys R., et al. , 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2: 1162–1168. 10.1128/EC.2.6.1162-1168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B. J., Fung E., Roncaglia P., Rowley D., Amedeo P., et al. , 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. 10.1126/science.1103773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra R. E., Huang J. C., Fung E., Nielsen K., Heitman J., et al. , 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167: 619–631. 10.1534/genetics.103.023408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier-Kergoat M.-C., Yeramian E., 2009. GC content and recombination: reassessing the causal effects for the Saccharomyces cerevisiae genome. Genetics 183: 31–38. 10.1534/genetics.109.105049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead M. E., Stanton B. C., Kruzel E. K., Hull C. M., 2015. Targets of the sex inducer homeodomain proteins are required for fungal development and virulence in Cryptococcus neoformans. Mol. Microbiol. 95: 804–818. 10.1111/mmi.12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., De Obaldia A. L., Heitman J., 2007. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot. Cell 6: 949–959. 10.1128/EC.00097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. 10.1126/science.1086605 [DOI] [PubMed] [Google Scholar]
- Petes T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. 10.1038/35072078 [DOI] [PubMed] [Google Scholar]
- Rajasingham R., Smith R. M., Park B. J., Jarvis J. N., Govender N. P., et al. , 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17: 873–881. 10.1016/S1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J., Desjardins C. A., Sykes S. M., Beale M. A., Vanhove M., et al. , 2017. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics 207: 327–346. 10.1534/genetics.117.203836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P., Bomblies K., Fitz J., Laitinen R., Warthmann N., et al. , 2012. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 108: 447–455. 10.1038/hdy.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Heitman J., 2016. Running hot and cold: recombination around and within mating-type loci of fungi and other eukaryotes, pp. 3–13 in Environmental and Microbial Relationships. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), Vol. IV. Springer-Verlag, New York: 10.1007/978-3-319-29532-9_1 [DOI] [Google Scholar]
- Sun S., Xu J., 2007. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics 177: 1475–1486. 10.1534/genetics.107.078923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Hsueh Y.-P., Heitman J., 2012. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 8: e1002810 10.1371/journal.pgen.1002810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Billmyre R. B., Mieczkowski P. A., Heitman J., 2014. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet. 10: e1004849 10.1371/journal.pgen.1004849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Yadav V., Billmyre R. B., Cuomo C. A., Nowrousian M., et al. , 2017. Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLoS Biol. 15: e2002527 10.1371/journal.pbio.2002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J., Sanyal K., 2013. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 23: 638–652. 10.1101/gr.141614.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi R., Hsueh Y.-P., Geunes-Boyer S., Wright J. R., Heitman J., 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77: 4345–4355. 10.1128/IAI.00542-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan A. A., Khankhet J., Xu J., 2013. Evidence for mitotic recombination within the basidia of a hybrid cross of Cryptococcus neoformans. PLoS One 8: e62790 10.1371/journal.pone.0062790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Tada Y., Dong X., Heitman J., 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1: 263–273. 10.1016/j.chom.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Zhai B., Zhu P., Foyle D., Upadhyay S., Idnurm A., et al. , 2013. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect. Immun. 81: 2626–2637. 10.1128/IAI.00259-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequence reads generated from samples utilized in this study are available on the National Center for Biotechnology Information’s sequence read archive under Bio-Project identification number PRJNA420966, with individual accession numbers SAMN08130857–SAMN08130963. The generated variant call file from the aligned sequenced reads are publicly available on the GitHub repository (https://github.com/magwenelab/crypto-recombination-paper). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6083315.