Genomic regions that regulate multiple traits, called supergenes, have been found in a range of organisms, including threespine stickleback fish. Here, Erickson et al. show that closely linked but genetically separable changes in ...
Keywords: QTL, fine mapping, skeletal evolution, genome editing, supergene
Abstract
In nature, multiple adaptive phenotypes often coevolve and can be controlled by tightly linked genetic loci known as supergenes. Dissecting the genetic basis of these linked phenotypes is a major challenge in evolutionary genetics. Multiple freshwater populations of threespine stickleback fish (Gasterosteus aculeatus) have convergently evolved two constructive craniofacial traits, longer branchial bones and increased pharyngeal tooth number, likely as adaptations to dietary differences between marine and freshwater environments. Prior QTL mapping showed that both traits are partially controlled by overlapping genomic regions on chromosome 21 and that a regulatory change in Bmp6 likely underlies the tooth number QTL. Here, we mapped the branchial bone length QTL to a 155 kb, eight-gene interval tightly linked to, but excluding the coding regions of Bmp6 and containing the candidate gene Tfap2a. Further recombinant mapping revealed this bone length QTL is separable into at least two loci. During embryonic and larval development, Tfap2a was expressed in the branchial bone primordia, where allele specific expression assays revealed the freshwater allele of Tfap2a was expressed at lower levels relative to the marine allele in hybrid fish. Induced loss-of-function mutations in Tfap2a revealed an essential role in stickleback craniofacial development and show that bone length is sensitive to Tfap2a dosage in heterozygotes. Combined, these results suggest that closely linked but genetically separable changes in Bmp6 and Tfap2a contribute to a supergene underlying evolved skeletal gain in multiple freshwater stickleback populations.
INTRASPECIFIC morphological variation offers the opportunity to dissect the genetic changes that underlie evolution, including adaptation to novel environments. Genetic mapping studies of naturally varying traits can reveal the genomic regions that contribute to evolutionary changes. When multiple phenotypes are controlled by the same region of a genome, a single pleiotropic locus could affect multiple traits, or separate but closely linked genes could affect each trait independently. Clustering of two or more QTL into supergenes is a commonly described feature of evolution (Schwander et al. 2014; Thompson and Jiggins 2014), likely because close linkage of loci allows advantageous combinations of traits to be inherited together. Supergenes have been shown to control pigmentation patterns in locusts (Nabours 1933); mimetic patterns in butterflies (Joron et al. 2011; Kunte et al. 2014); color and patterning in snails (Murray and Clarke 1976a,b); social behavior in ants (Wang et al. 2013; Pracana et al. 2017); breeding behavior, morphology, and sperm traits in birds (Thomas et al. 2008; Küpper et al. 2016; Lamichhaney et al. 2016; Tuttle et al. 2016; Kim et al. 2017); and life history, morphology, and pollination syndromes in plants (Mather 1950; Lowry and Willis 2010; Hermann et al. 2013). The supergenes that have been molecularly characterized to date are often controlled by chromosomal inversions with large phenotypic effects. In Heliconius butterflies, a supergene in one species was shown to be an inversion containing tightly linked loci that can recombine in other species (Joron et al. 2006, 2011; Nadeau et al. 2016). Determining whether clustering of QTL controlling different traits is caused by close linkage or pleiotropy requires careful genetic dissection of the genomic intervals of interest.
While QTL for different traits can cluster in the genome, multiple genetic changes affecting a single phenotype can also be clustered within a QTL. Whether a given QTL typically represents an individual locus, or alternatively, multiple tightly linked loci, remains a largely open question in quantitative genetics. For example, QTL mapping in maize × teosinte crosses has revealed that some large-effect QTL can fractionate (Studer and Doebley 2011), including a QTL that was fractionated into up to five distinct tightly linked loci (Lemmon and Doebley 2014). In contrast, other maize × teosinte QTL contain single causative loci (Wang et al. 2005; Hung et al. 2012; Wills et al. 2013). In mice, one QTL controlling pigmentation maps to a single coding mutation in Mc1r (Steiner et al. 2007), but a second QTL maps to multiple smaller effect mutations affecting different aspects of Agouti expression (Linnen et al. 2013). Interspecific differences in Drosophila pigmentation map, in part, to a single regulatory element of tan (Jeong et al. 2008) and multiple point mutations in a single enhancer of ebony (Rebeiz et al. 2009), but differences in Drosophila trichome patterning are caused by changes in regulatory elements of the shavenbaby gene that are spread over 50 kb (McGregor et al. 2007; Frankel et al. 2011).
Testing whether the same or different genetic changes underlie the evolution of similar phenotypes in multiple lineages may shed light on the predictability and repeatability of evolution (Stern and Orgogozo 2008; Stern 2013; Rosenblum et al. 2014). Both theoretical and empirical studies indicate that parallel genetic evolution can occur quite often (Orr 2005; Conte et al. 2012). However, given the relatively small number of cases where convergently evolved phenotypes have been associated with specific genes, the extent of genetic parallelism remains largely unknown, especially for quantitative traits.
The threespine stickleback fish has convergently evolved countless freshwater forms from ancestral marine populations and has emerged as a powerful model system for studying both the genetic basis and repeatability of morphological evolution (Peichel and Marques 2017). In freshwater environments, sticklebacks repeatedly evolve a suite of craniofacial and other morphological adaptations to cope with differences in diet, predation, and other environmental variables (Bell and Foster 1994). Individuals from different populations are easily intercrossed to produce large clutches in the laboratory (Peichel et al. 2001), the stickleback genome is well assembled and annotated (Jones et al. 2012; Glazer et al. 2015), and reverse genetic techniques are available (Erickson et al. 2016a), facilitating both genetic and genomic dissection of the molecular basis of evolved traits.
Marine and freshwater sticklebacks occupy different trophic niches: while marine fish feed on small planktonic prey, freshwater fish typically consume diets of larger macroinvertebrates (Kislalioglu and Gibson 1977; Gross and Anderson 1984). Sticklebacks process food primarily in the throat, with the branchial skeleton and pharyngeal jaw used to chew and crush food en route to the gut (McGee and Wainwright 2013; McGee et al. 2013). The branchial skeleton forms in the posterior-most five pharyngeal arches and consists of segmental homologs of the upper and lower jaw. The branchial skeleton is composed of five bilateral pairs of ventral bones (ceratobranchials), four bilateral pairs of dorsal bones (epibranchials), and three bilateral pairs of tooth plates (Anker 1974; see Supplemental Material, Figure S1 for anatomy). We described increases in both pharyngeal tooth number and branchial bone length as repeatable and heritable features of freshwater adaptation (Cleves et al. 2014; Erickson et al. 2014; Ellis et al. 2015). We hypothesize that these increases permit freshwater fish to eat larger prey items via a larger pharyngeal cavity and greater chewing capacity. The first epibranchial (EB1) bone, a serial homolog of the upper jaw, serves as a critical lever for the mastication motion of the pharyngeal jaw (Wainwright 2006) and is the most proportionally elongated branchial bone in freshwater sticklebacks (Erickson et al. 2014). The branchial bones are endochondral bones that form from cartilage templates during late embryonic development (Haines 1934), much like mammalian long bones (Haines 1942). Changes to both the early patterning of cartilage and the relative growth of bones contribute to stickleback branchial bone length differences (Erickson et al. 2014) and to skeletal evolution in other systems (Farnum et al. 2008a,b; Sanger et al. 2011, 2012).
Previous genetic mapping of evolved stickleback skeletal variation has identified >100 QTL controlling a variety of traits, including pharyngeal tooth number and branchial bone length, in the benthic population from Paxton Lake, British Columbia (PAXB). These trophic traits are highly polygenic, but the QTL controlling skeletal adaptation are significantly clustered into supergenes on three chromosomes, including chromosome 21 (Miller et al. 2014). Although chromosome 21 contains an inversion that typically differs between marine and freshwater populations (Jones et al. 2012), the PAXB pharyngeal tooth number QTL was fine-mapped to a genomic region over 1 Mb outside this inversion, to an intronic enhancer of the gene Bone morphogenetic protein 6 (Bmp6) (Cleves et al. 2014; P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data 2018). Additional work mapped branchial bone length QTL to peaks near Bmp6 on chromosome 21 in both PAXB and a stream population from Fishtrap Creek, Washington (FTC) (Erickson et al. 2014). BMP ligands are critical for both tooth and bone development (Balic and Thesleff 2015; Salazar et al. 2016), so Bmp6 is an excellent candidate gene for both the tooth number and branchial bone length QTL. However, a second excellent candidate gene for the branchial bone length QTL, Tfap2a, a known regulator of pharyngeal skeletal development (Schorle et al. 1996; Knight et al. 2004; Milunsky et al. 2008; Tekin et al. 2009; Van Otterloo et al. 2018), is tightly linked to Bmp6. We sought to further elucidate the genetic basis of the increased bone length QTL in both the FTC and PAXB freshwater populations to answer three questions: (1) Are the bone length and tooth number QTL genetically separable? (2) What is the developmental genetic basis of the bone length QTL? and (3) Does convergent evolution of branchial bone gain in two freshwater populations have a similar genetic basis?
Materials and Methods
Animal statement
All animal work was approved by University of California, Berkeley (animal protocol #R330). Fish were reared as previously described (Erickson et al. 2014).
Recombinant mapping and statistical analysis
Fish segregating for the chromosome 21 EB1 length QTL from the PAXB × Little Campbell Marine (LITC) and FTC × LITC crosses (Erickson et al. 2014) were propagated and genotyped with markers Stn487 (Cleves et al. 2014), PAE309, PAE323, and/or PAE349 (Table S1) to ensure that marine and freshwater alleles of the QTL interval were passed on to each generation and to look for recombination events within the QTL interval. To test the phenotypic effects of recombinant chromosomes, recombinant fish were crossed to related fish that were either heterozygous for the QTL or homozygous for the marine chromosome within the QTL interval. Offspring were grown to 25–28 days postfertilization (dpf) (10–12 mm, FTC cross) or to roughly 80 dpf (∼20 mm, PAXB cross). These fish were fixed, stained with Alizarin red, cleared, dissected, and photographed, and EB1 bone length was measured as previously described, using ImageJ (Erickson et al. 2014; Miller et al. 2014; Ellis and Miller 2016). EB1 length was measured on both the left and right side and averaged. Bone length was corrected for fish standard length in each clutch separately and residuals of bone length or back-transformed residuals were used for all analyses. Pharyngeal tooth number was counted as described (Cleves et al. 2014).
For recombinant fish crossed to heterozygotes, the R package lmtest was used to perform a likelihood ratio test (lrtest) to determine whether the recombinant chromosome behaved as marine or freshwater. The recombinant chromosome (R) was coded as either (a) a marine (M) or (b) a freshwater (F) chromosome. For (a), the MR and FR genotypes were replaced with MM and MF, respectively, and for (b), they were replaced with MF and FF. The genotypes were then coded numerically with MM = 0, MF = 1, and FF = 2. The lrtest function was used to compare two nested linear models of bone length: the model (bone length ∼a + b) was compared to the model (bone length ∼a) and to the model (bone length ∼b). A significant difference between the model containing both possible genotypes and a model containing one possible genotype indicated that the addition of the second genotype significantly improved the fit of the model, and therefore that the second genotype best represented the effect of the recombinant chromosome. For example, a significant P–value in the likelihood ratio test comparing (bone length ∼a + b) to (bone length ∼a) would indicate that the addition of b significantly improved the model and therefore that R behaves as F. For recombinant chromosomes crossed to homozygous marine fish (MM), a t-test was used to determine whether the recombinant chromosome behaved like a freshwater chromosome (i.e., there was a significant increase in bone length between the MM and MR genotypic classes).
Analysis of chromosome 21 QTL in previously published crosses
Genotype data from Glazer et al. (2015) and Erickson et al. (2016b) were used to test for the presence of a bone length QTL at the genotype-by-sequencing (GBS) marker containing Tfap2a (binned marker 16_9) in the LITC × FTC (a different cross than the FTC cross presented here) and LITC × Enos Benthic (ENOB) crosses. Adult bone lengths were measured for 210 fish from the three largest families in the Glazer et al. (2015) study as described above, and processed to correct for size and sex as described in Erickson et al. (2016b). Processed adult bone lengths for the ENOB × LITC cross were used as reported in Erickson et al. (2016b).
Genome resequencing and Tfap2a genotyping
A synonymous polymorphism in Tfap2a was initially identified by examining the genome sequences of the grandparents of a previously studied LITC × FTC cross (a different cross than the one studied here; Glazer et al. 2015) and a PAXB × LITC cross (the same cross as the one studied here; P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data). This mutation (see Figure S2 for location) disrupts an AvaI cut site and was genotyped by amplifying with PAE414/416 (Table S1) and digesting the PCR product with AvaI; presence of an uncut band indicated presence of the freshwater alternate allele.
Allele-specific expression assay for Tfap2a
We modified a GBS protocol (Elshire et al. 2011; Glazer et al. 2015) to assay allele-specific expression (ASE) in RT-PCR products amplified from complementary DNA (cDNA) relative to genomic DNA (gDNA) PCR controls. PAXB × LITC fish segregating marine and freshwater alleles of the QTL interval (F9 generation) were intercrossed, their F10 larval offspring were euthanized with MS-222, and the branchial skeleton or EB1 was immediately dissected on ice and stored in 500 μl TRI reagent (Ambion) at −80°. Tissue was collected from five time points: 9, 13, 17, 22, and 35 dpf. At 9 and 13 dpf, the entire branchial skeleton was collected. At 17 and 22 dpf, the dorsal portion of the branchial skeleton was collected, and at 35 dpf individual EB1 bones were isolated. The remaining tissue was stored in ethanol. DNA was isolated from the excess tissue by digesting overnight with Proteinase K in lysis buffer at 55° and then performing a phenol-chloroform extraction followed by ethanol precipitation (Green and Sambrook 2012). DNA was genotyped with indel marker PAE311/312 (Table S1) to identify heterozygous fish. RNA was extracted from heterozygous fish following the TRI reagent manufacturer’s protocol, with the final RNA pellet resuspended in 20 μl RNase free water. For each time point, 24 fish were collected, yielding ∼12 heterozygotes.
To prepare cDNA, 4.5 μl RNA was treated with 0.5 μl amplification grade DNase I (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. cDNA was synthesized with Superscript III (Invitrogen), using random hexamers according to the manufacturer’s protocol but halving all volumes to produce a final volume of 10 μl cDNA. One microliter of cDNA or 50 ng of gDNA from each fish was used as template in separate Phusion PCR reactions with primers PAE414 and PAE416 (Table S1), designed to amplify the synonymous SNP in Tfap2a at position chrXXI:4,265,995, which was shared in both the PAXB and FTC cross parents. The 5′ end of each primer contained an ApeKI cut site so that the product could be ligated into the GBS adapters. A titration control was performed by mixing marine and freshwater DNA in fixed ratios. Technical replicates were performed by (1) amplifying the same gDNA twice, (2) preparing cDNA from the same RNA sample twice, and (3) amplifying from the same cDNA sample twice. All PCR products were purified in a 96-well format by the University of California, Berkeley DNA Sequencing Facility and quantified using a PicoGreen assay (ThermoFisher) on a BioTek Flx800 plate reader.
Up to 25 ng of each purified PCR product was used in the GBS library preparation as previously described (Elshire et al. 2011; Glazer et al. 2015). Briefly, the PCR products were combined with 1.8 ng of barcoded adapters and digested with ApeKI (New England BioLabs, Ipswich, MA) for 2 hr at 75°. The adapters were ligated to the PCR products with T4 DNA Ligase (New England BioLabs) for 1 hr at 22°. Then, 5 μl of each ligation product was pooled for a PCR purification (Qiagen, Valencia, CA). Sequencing adapters were added with 10 cycles of PCR using Taq 2× master mix (New England BioLabs). The PCR product was cleaned up and size-selected twice with 1.5 vol of Sera-Mag beads prepared as previously described (Rohland and Reich 2012). Six libraries were pooled equally into one Illumina HiSequation 2000 lane and sequenced on 50 bp single-end rapid run mode at the University of California, Berkeley Vincent J. Coates Genomic Sequencing Laboratory. Indexed and barcoded sequences were demultiplexed and aligned to the expected PCR product sequence to count marine and freshwater alleles at the polymorphic site with the custom script nuc_by_pos.py (available at https://github.com/trahsemaj/TFAP2). Samples receiving <300 aligned reads were removed from the analysis (n = 4), as preliminary experiments suggested that at least 300 reads would be required to obtain reliable 1:1 allelic ratios for gDNA PCR products. Outliers >3 SD from the mean were also removed (n = 2). The gDNA and cDNA freshwater/marine ratios were compared at each time point to test for ASE with a Mann–Whitney U-test using the wilcox.test function in R.
Genome editing of Tfap2a
TALENs were generated to target the second exon of Tfap2a (see Table S2 for RVD design and Figure S2 for binding sites) following established protocols (Cermak et al. 2011; Doyle et al. 2012) and injected into one-cell FTC and LITC stickleback embryos as previously described (Erickson et al. 2015, 2016a). A subset of injected embryos (n = 10–12) were screened for mutations by amplifying with primers PAE379 and PAE381 (Table S1) in a standard Phusion (New England BioLabs) reaction, digesting the PCR product with PvuII, and running the digested product on a 1% agarose gel. Undigested product of ∼297 bp indicated molecular lesions that disrupted a PvuII sequence at the expected DNA cleavage site. Clutches carrying lesions were raised to adulthood and outcrossed to wild-type fish. Lesions were sequenced by extracting the undigested PCR band from an agarose gel followed by Sanger sequencing (see Table S3 for lesions studied). F1 fish carrying lesions were propagated for further analysis.
TALEN phenotyping
Tfap2a+/− heterozygotes were intercrossed to produce clutches containing homozygous mutants. These larval fish were euthanized immediately after hatching (9–10 dpf), fixed overnight in 4% paraformaldehyde in 1× PBS, stained with Alcian blue, digested with trypsin, mounted in glycerol (Kimmel et al. 1998), and imaged on a Leica DM2500 compound microscope. For bone length phenotyping, heterozygous fish were outcrossed to wild-type laboratory-reared individuals. In the FTC background, a total of 144 fish were studied from three families carrying a 16 bp insertion, an 8 bp deletion, and a 10 bp deletion. In LITC, two families totaling 96 fish carrying a 13 bp deletion were studied. Fish were raised to 25–28 dpf and genotyped and phenotyped as described above. Bone lengths were not back-transformed in this analysis due to different standard length ranges and different y-intercepts of the bone length ∼ standard length regression for fish in different families.
In situ hybridization
In situ hybridization for Tfap2a was performed on whole embryos as previously described (Cleves et al. 2014).
Data and reagent availability
Raw data used in this study including genotypes, phenotypes, and ASE counts are available in File S1. Supplemental material is available at Figshare: https://doi.org/10.25386/genetics.6024338. All reagents used are available upon request. Custom Python script used for ASE is available at https://github.com/trahsemaj/TFAP2.
Results
Fine mapping the chromosome 21 bone length QTL
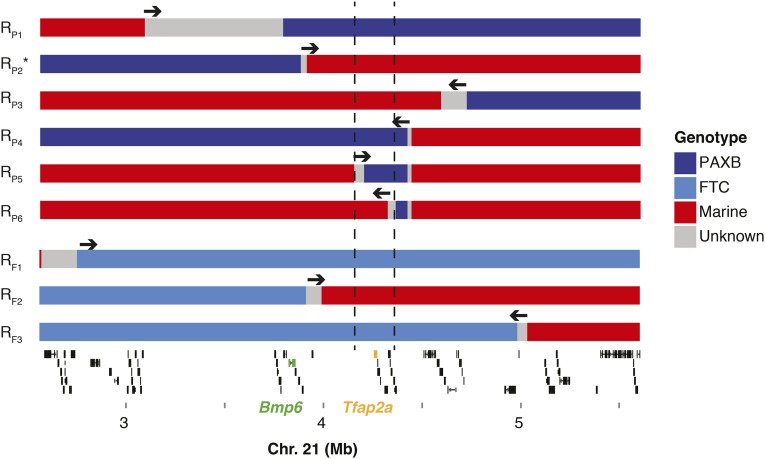
We tested whether bone length and tooth number were controlled by the same genomic region in two freshwater × marine crosses known to carry the bone length QTL (Erickson et al. 2014): PAXB and FTC, both crossed to LITC. We analyzed a series of recombinant chromosomes derived from later generations of the original crosses. For each recombinant chromosome, we used likelihood ratio tests to ask whether the recombinant chromosome produced a phenotypic effect significantly more similar to a marine or freshwater chromosome. If the chromosome behaved like a freshwater chromosome (significantly increased branchial bone length relative to a marine chromosome or did not differ from a freshwater chromosome), we concluded that the QTL was contained within the freshwater portion of the recombinant chromosome (blue in Figure 1). If the chromosome behaved like a marine chromosome (did not increase branchial bone length relative to a marine chromosome or caused significantly shorter bone length when compared to a freshwater chromosome), we concluded that the QTL was contained within the marine portion (red in Figure 1).
Figure 1.
Recombinant mapping of the chromosome 21 bone length QTL in two crosses. Recombinant chromosomes were identified in later generations (>F4) of the original F2 crosses described in Erickson et al. (2014). Fish with recombinant chromosomes were crossed to known genotype relatives descended from the same cross. Colors indicate the marine/freshwater identity along each recombinant chromosome; regions of unknown genotype between confirmed breakpoint genotypes are colored gray (see key). Arrows above each chromosome indicate the direction of the bone length QTL relative to the recombination break point; see Table 1 for full statistical analysis of each cross. Note that recombinant chromosomes RP5 and RP6 in the PAXB cross are both double recombinants derived from chromosome RP4. The asterisk on RP2 indicates that this chromosome also carries a tooth number QTL (P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data). The vertical dashed lines indicate the boundaries of the fine-mapped region in the PAXB × LITC cross. An Ensembl-based gene prediction track from the stickleback genome (chrXXI: 2.56–5.60 Mb) is shown below (Jones et al. 2012). Thick lines indicate predicted exons and thin lines indicate introns. Bmp6 and Tfap2a are highlighted in green and yellow, respectively.
Using this approach, we identified recombinant chromosomes in each cross with recombination events located within ∼2 Mb of Bmp6. In the PAXB cross, we identified one recombinant chromosome that separated the Bmp6 locus from the region controlling bone length (Figure 1, chromosome RP2). This particular recombinant chromosome significantly increases pharyngeal tooth number (P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data), but did not significantly affect the length of the EB1 bone (Table 1). Therefore, bone length and tooth number are controlled by different genetic loci on chromosome 21 in the PAXB × LITC cross. In the FTC cross, we also found a chromosome that carried a freshwater allele of Bmp6 but did not affect bone length (Figure 1, chromosome RF2 and Table 1). We found that this recombinant chromosome increased pharyngeal tooth number relative to a marine chromosome (Figure S3), suggesting that the FTC population also harbors a tooth QTL in a portion of the chromosome containing Bmp6, and like in the PAXB population, the regions controlling tooth number and bone length are genetically separable. The coding sequence of Bmp6 is excluded from the bone length QTL interval in both crosses.
Table 1. Statistical analysis of recombinant chromosomes.
| Name | Cross | Design | Generation | N | Clutches | Geno 1 | Geno 2 | Geno 3 | Geno 4 | LR test (M) | LR test (F) | t-test | Conclusion (Mb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RP1 | PAXB × LITC | MR × MM | F7 | 125 | 3 | MM (−4.7 ± 21.2) | MR (5.6 ± 21.1) | — | — | — | — | 0.008 | right of 3.09 |
| RP2 | PAXB × LITC | MR × MM | F7 | 96 | 2 | MM (1.5 ± 15.8) | MR (−1.2 ± 17.7) | — | — | — | — | 0.43 | right of 3.88 |
| RP3 | PAXB × LITC | MR × MF | F6 | 192 | 2 | MM (−3.4 ± 27.8) | MR (−10.4 ± 25) | MF (2.6 ± 22.2) | FR (9.3 ± 29.0) | 0.04 | 0.99 | — | left of 4.89 |
| RP4 | PAXB × LITC | MR × MF | F6 | 171 | 2 | MM (−10.4 ± 20.1) | MR (3.1 ± 30.9) | MF (0.4 ± 23.4) | FR (8.6 ± 25.2) | 0.59 | 0.005 | — | left of 4.4 |
| RP5 | PAXB × LITC | RP4RP5 × MM | F8 | 62 | 1 | MRP5 (4.6 ± 23.9) | MRP4 (−4.4 ± 32.5) | — | — | — | — | 0.22 | between 4.15–4.44 |
| RP6 | PAXB × LITC | MR × MM | F9 | 78 | 1 | MM (4.2 ± 28.2) | MR (1.9 ± 28.0) | — | — | — | — | 0.72 | excludes 4.32–4.44 |
| RF1 | FTC × LITC | FR × MF | F8 | 96 | 2 | MR (−7.4 ± 9.7) | MF (−5.5 ± 11.4) | FR (5.6 ± 8.9) | FF (9.3 ± 10.3) | 0.22 | 0.0002 | — | right of 2.57 |
| RF2 | FTC × LITC | MR × MM | F8 | 76 | 2 | MM (−2.0 ± 11.8) | MR (1.0 ± 10.0) | — | — | — | 0.25 | right of 3.91 | |
| RF3 | FTC × LITC | FR × MF | F8 | 63 | 1 | MR (−11.8 ± 10.3) | MR (1.7 ± 13.3) | MF (1.8 ± 12.9) | FR (11.3 ± 13.5) | 0.93 | 0.0004 | — | left of 5.03 |
Each row corresponds to one chromosome in Figure 1 (in order from top to bottom). All bone length measurements were size-corrected within individual clutches and then pooled. Reported values for each genotypic class are mean residual bone lengths (in micrometer) ± SD. P-values for linear model likelihood ratio (LR) tests or t-tests are reported when appropriate and bolded when significant; see Materials and Methods for details on the statistical tests. Arrows in Figure 1 indicate the conclusion for each chromosome. M, marine; F, freshwater; R, recombinant.
With subsequent fine mapping, we narrowed the QTL interval in the PAXB cross (Figure 1 and Table 1). Notably, a double recombinant chromosome in the PAXB cross (chromosome RP5) shows no phenotypic difference from the original recombinant chromosome (RP4), which had a strong effect on bone length (Table 1), suggesting that the freshwater alleles within the double recombinant region are sufficient to increase bone length. A second double recombinant sharing the same 3′ breakpoint (recombinant RP6) did not differ from a marine chromosome, suggesting that this smaller freshwater portion of the chromosome does not carry the bone length QTL. Combined, these two results map the bone length QTL to a 155 kb region (chrXXI:4,200,364–4,355,895) in the PAXB cross, containing eight Ensembl-predicted genes (Tfap2a, Tmem14b, Mak, Plcxd2, Phldb2, Tmem56, ENSGACG00000002373, and Bco1; Jones et al. 2012. Seven of these eight genes have no described roles in mouse skeletal development (Smith et al. 2014). However, one gene, Transcription Factor Activating Protein 2 alpha (Tfap2a), is an outstanding candidate for craniofacial evolution because it has roles in patterning the craniofacial skeleton in humans, mice, and zebrafish (Schorle et al. 1996; Knight et al. 2004; Milunsky et al. 2008), and in regulating the growth and maturation of mammalian chondrocytes (Wenke and Bosserhoff 2010).
Fine mapping the bone length QTL in the FTC cross supported a larger ∼1.1 Mb genomic interval (chrXXI: 3,906,104–5,003,790 bp; Figure 1 and Table 1), also containing Tfap2a. The entire PAXB fine-mapped interval is contained within the FTC fine-mapped interval. Therefore, together these data support a model of a shared genomic basis for branchial bone length gain in these two independently derived freshwater populations.
Fractionation of the QTL in the FTC cross
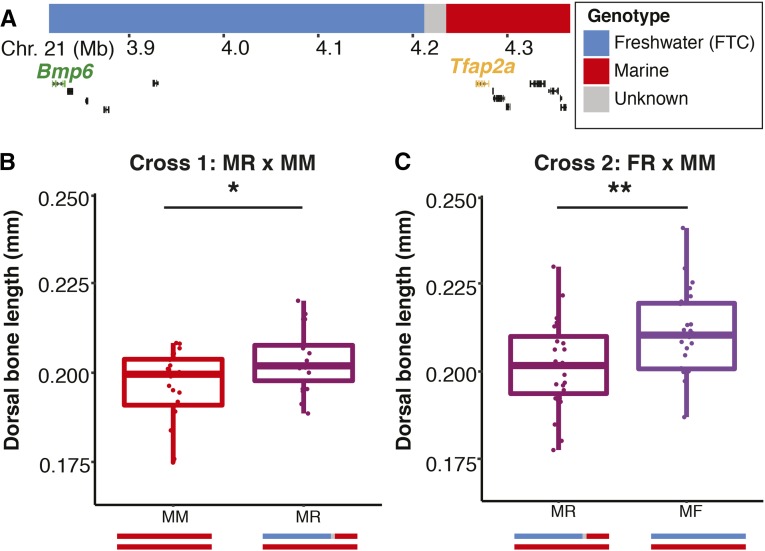
For one FTC recombinant chromosome, we found evidence of fractionation of the bone length QTL. The breakpoint of this recombinant was within a large (∼200 kb) gene desert between Bmp6 and Tfap2a (Figure 2A). We compared the effect of this recombinant chromosome to both marine and freshwater chromosomes that were nonrecombinant within the QTL region. We found that this recombinant chromosome produced significantly longer bones than a marine chromosome (Figure 2B); however, it also produced significantly shorter bones than a freshwater chromosome (Figure 2C). These data suggest the recombinant chromosome has effects intermediate between fully marine and freshwater alleles, and the freshwater portion of this chromosome does not recapitulate the full QTL effect.
Figure 2.
Fractionation of the bone length QTL in the FTC × LITC cross. A single recombinant chromosome with breakpoint between 4,210,854 and 4,231,226 bp (A) was identified and then bred to produce fish with genotypes MR and FR (M, marine; F, freshwater; R, recombinant). Note this chromosome is distinct from those shown in Figure 1. The Ensembl gene predictions for this region (chrXXI: 3.85–4.35 Mb) are shown below with Bmp6 and Tfap2a highlighted (Jones et al. 2012). Each genotype was crossed to an MM fish (homozygous for marine genotypes on chromosome 21) and epibranchial one bone length (size-corrected and back transformed to a 9 mm fish) was compared between genotypes in each cross. The recombinant chromosome diagrammed in A results in bones significantly longer than those from an M chromosome (B) (t-test: d.f. = 38.98, t = −2.51, P = 0.017) but significantly shorter than those from an F chromosome (C) (t-test: d.f. = 49.13, t = 2.75, P = 0.008). Schematics depicting the genotypes of the offspring are illustrated below the x-axes in B and C. * P < 0.05; ** P < 0.001.
Variable presence of the chromosome 21 bone length QTL in freshwater populations
We previously reported a relatively weak EB1 bone length QTL at the far end of chromosome 21 (41.7 cM away from Tfap2a) in a LITC × ENOB cross (Erickson et al. 2016b), but in this cross there is no association between Tfap2a genotype and EB1 length (Figure S4A). Additionally, a LITC × FTC cross with different grandparents than those used in the present study (Glazer et al. 2014, 2015) also shows no evidence for a bone length QTL near Tfap2a (Figure S4B). Therefore, while this bone length QTL was identified in two populations, it appears to not be present in all freshwater populations nor fixed in the FTC population. These cases where this bone length QTL was not detected could reflect differences in genetic background that mask the penetrance of the QTL, or alternatively, a true absence of the mutation(s) underlying the QTL.
The chromosome 21 QTL affects bone length at stages soon after ossification
Our previous findings that cartilage template size was larger and bone growth rate was accelerated in the FTC population (Erickson et al. 2014) raised the question of whether the chromosome 21 QTL controlled cartilage development, bone development, or both. To address this question, we raised F5 fish from the FTC × LITC cross to 13 days dpf (a stage when the EB1 cartilage template has formed but not ossified) and 20 dpf (a stage soon after EB1 ossification) to test for an effect of the chromosome 21 QTL on cartilage and bone length. The QTL did not have a significant effect on EB1 cartilage length in a heterozygous × marine backcross at 13 dpf, but had a strong effect on bone length by 20 dpf (Figure S5). Therefore, the chromosome 21 QTL likely has an effect on either initial bone ossification or bone elongation, but does not appear to have a substantial effect on EB1 cartilage template length.
Expression of Tfap2a in developing branchial skeletons
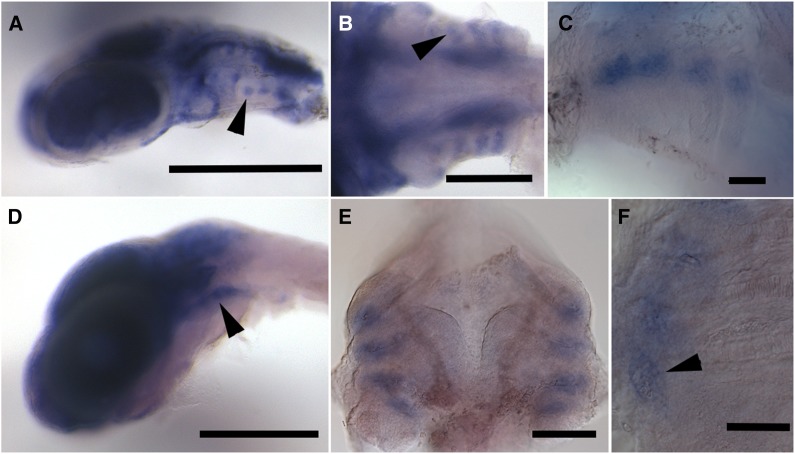
We previously showed that Tfap2a is expressed in undifferentiated mesenchymal cells in the dorsal branchial skeletal primordia prior to epibranchial chondrification, but not in developing teeth (Cleves et al. 2014). To further characterize the temporal emergence of this dorsal branchial expression domain, we assayed Tfap2a expression at earlier stages in development. At 4 dpf, Tfap2a was broadly expressed in postmigratory cranial neural crest in the posterior pharyngeal arches (Figure 3, A and B). By 5 dpf, expression was detected in postmigratory cranial neural crest in the dorsal pharyngeal arches (Figure 3C). At 7 dpf, as previously reported, expression was detected in undifferentiated mesenchymal cells in the dorsal branchial arches, in precisely the future locations of the epibranchial cartilages (Figure 3, D–F). Thus, Tfap2a expression during craniofacial development is found in the right place and right time to regulate branchial bone development, further supporting Tfap2a as contributing to the branchial bone length QTL. We detected no obvious qualitative differences in expression patterns or levels between marine and freshwater embryos.
Figure 3.
In situ hybridization for Tfap2a. (A) Lateral view of a 4 dpf embryonic head. Tfap2a expression in dorsal pharyngeal arch postmigratory cranial neural crest is indicated with an arrowhead. (B) Ventral view of a 5 dpf embryo; arrowhead indicates pharyngeal arch expression. (C) Ventrolateral view of a 5 dpf embryo; Tfap2a expression is visible at the dorsal ends of the pharyngeal pouches. (D) Lateral view of 7 dpf embryo. Tfap2a expression remains in the dorsal pharyngeal arches (arrowhead). (E) Dorsal view of a dissected 7 dpf branchial skeleton. (F) Higher magnification of dorsal view of 7 dpf branchial skeleton. Arrowhead indicates expression in condensing epibranchial mesenchymal cells. Scale bars: A and D = 500 µm; B = 200 µm; C, E, and F = 100 µm. A–D show the anterior on the left; E and F show the anterior at the top.
Evolved changes in the cis-regulation of Tfap2a
We first tested for coding changes in Tfap2a with genome resequencing data of a FTC grandparent from a previous study (Glazer et al. 2015) and the PAXB grandparent used in this study (P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data). We identified a silent mutation shared between PAXB and FTC and an additional silent mutation in PAXB, but no nonsynonymous changes. Since no predicted protein coding changes exist in either freshwater population, we next tested for cis-regulatory differences in Tfap2a.
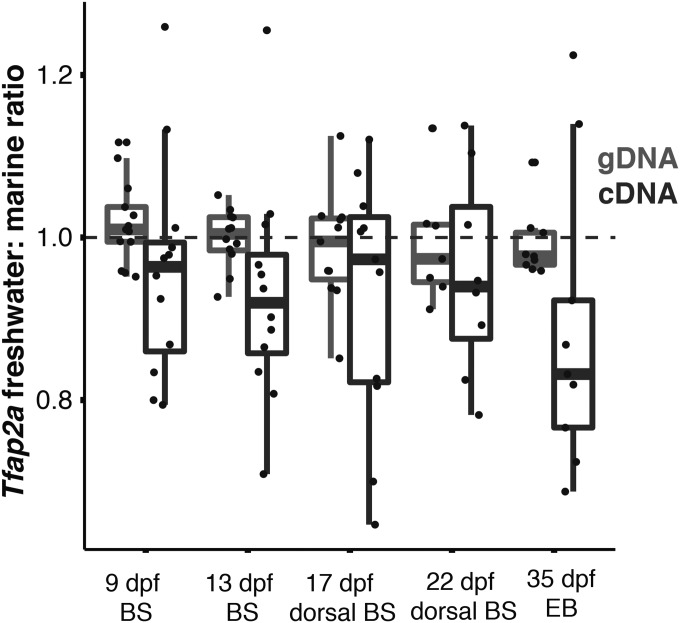
We compared the ratios of expressed marine (LITC) and freshwater (PAXB) alleles of Tfap2a in developing branchial skeletal tissue of marine × freshwater hybrid fish to look for allele-specific expression (ASE) (Cowles et al. 2002; Yan et al. 2002; Wittkopp et al. 2004). We adapted a barcoded next-generation sequencing assay originally developed for GBS (Elshire et al. 2011; Glazer et al. 2015) and verified that this assay is capable of detecting a range of allelic ratios despite some inherent variability (Figure S6). We tested a series of developmental stages of PAXB × LITC F9 hybrids for ASE of Tfap2a. The earliest stage collected for ASE was hatching (9 dpf), when the branchial skeleton could be accurately dissected. We collected additional time points at 13 and 17 dpf (immediately before ossification of EB1), 22 dpf (immediately following ossification of EB1), and 35 dpf (during growth of EB1). We compared the freshwater/marine allelic ratio of RT-PCR products amplified from cDNA to that of PCR products amplified from gDNA controls (which should have a ratio of 1:1 in heterozygous fish). While the observed allelic ratios have a high variance and only one developmental stage showed a significant difference between the gDNA ratio and the cDNA ratio, at every stage the median expression of the freshwater allele was lower than the marine allele (Figure 4). When the data from all five stages were pooled, the measured freshwater/marine allelic ratio was significantly reduced in cDNA samples relative to gDNA controls (P = 0.001, Mann–Whitney U, Figure 4). These findings reveal that Tfap2a has evolved cis-regulatory changes and further suggest that Tfap2a might have an inhibitory role in bone development, as reduction of expression of the freshwater allele of Tfap2a is associated with longer bones.
Figure 4.
Allele-specific expression of Tfap2a. A SNP within exon 2 of Tfap2a was amplified by PCR from genomic DNA (gDNA) and by RT-PCR from cDNA collected from ∼12 individuals from the PAXB × LITC cross at each of five stages: 9 and 13 dpf branchial skeleton (BS), 17 and 22 dpf dorsal BS, and 35 dpf EB1 tissue. The freshwater/marine allele ratio was calculated for each individual, and the gDNA ratio was compared to the cDNA ratio for each time point by Mann–Whitney U-test. While only one developmental stage was significant (9 dpf: W = 43, P = 0.06; 13 dpf: W = 35, P = 0.03; 17 dpf: W = 50, P = 0.51; 22 dpf: W = 21, P = 0.46; 35 dpf: W = 18 P = 0.05), collectively the reduced expression of the freshwater allele across the entire data set was highly significant (W = 851, P = 0.001).
Tfap2a dosage affects branchial bone length and craniofacial development
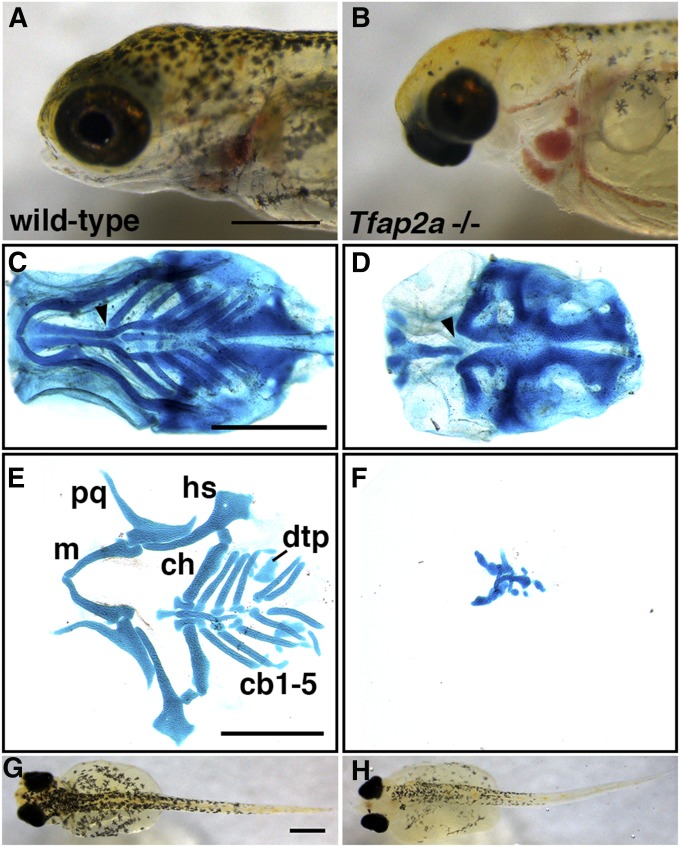
Based on the results of the fine mapping and ASE assays, we hypothesized that manipulation of Tfap2a levels could affect branchial bone length. We used TALENs to induce predicted loss-of-function mutations (see Table S3) in Tfap2a in the FTC and LITC genetic backgrounds. We intercrossed stable heterozygous mutants to produce trans-heterozygotes. As expected based upon the zebrafish Tfap2a mutant phenotype (Holzschuh et al. 2003; Knight et al. 2003), homozygous mutants with a severe lethal craniofacial phenotype (Figure 5, A and B) were obtained at the expected 25% proportion in each of nine crosses carrying various combinations of the alleles in Table S3. Presence of this phenotype was perfectly concordant with homozygous lesions indicated by loss of a PvuII cut site (see Materials and Methods). Homozygous mutant fish were almost entirely lacking a pharyngeal skeleton, while in contrast, the mesodermally derived posterior neurocranium was present and well-formed (Figure 5, C and D). The remnants of the pharyngeal skeletal elements were severely malformed, often asymmetric, and not identifiable (Figure 5, E and F). The branchial skeleton was particularly hypoplastic, and no identifiable ceratobranchial or epibranchial cartilages were present. The craniofacial defects were seen in homozygous mutants in both FTC and LITC genetic backgrounds. Consistent with the known roles of Tfap2a in controlling neural crest development and migration (Knight et al. 2003, 2004), we also observed severe pigmentation defects in homozygous mutants, including reduced melanophore numbers (Figure 5, G and H). Like the zebrafish lockjaw mutant (Knight et al. 2004), xanthophores appeared unaffected and iridophores were reduced in homozygous mutant fish (Figure S7).
Figure 5.
Induced homozygous mutations in Tfap2a result in severe craniofacial defects and reduced pigmentation. Wild-type FTC freshwater (A, C, E, and G) and homozygous Tfap2a mutant sibling fish (B, D, F, and H). (A and B) Relative to wild-type (A), mutants (B) have severely hypoplastic craniofacial tissue that results in an inability to feed. (C and D) Ventral view of the heads of Alcian blue stained fish reveal defects to the anterior neurocranium in mutant fish (arrowheads). (E and F) Flat-mounted, Alcian blue-stained pharyngeal arch cartilage elements. The remnant cartilages in the mutants are unidentifiable. (G and H) Dorsal views of 9 dpf larvae showing reduced melanophore pigmentation in Tfap2a mutants. While FTC fish are shown here, no obvious differences were observed between FTC−/− and LITC−/− fish. Anterior is to the left for all images; scale bar = 500 μm. cb, ceratobranchial; ch, ceratohyal; dtp, dorsal tooth plate; hs, hyosymplectic; m, Meckel’s cartilage (lower jaw); pq, palatoquadrate.
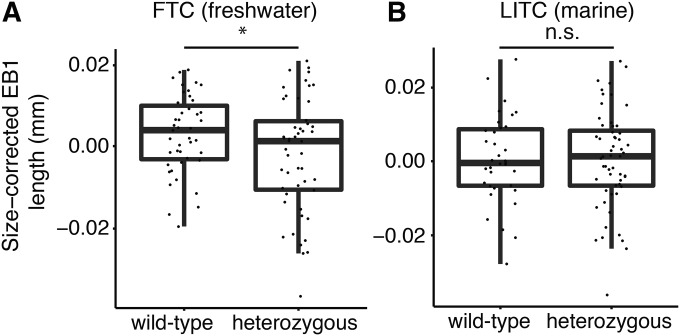
Because we could not study branchial bone length in homozygous mutants, we outcrossed heterozygotes to wild-type fish to compare bone length in wild-type and heterozygous siblings. We grew fish to ∼4 weeks postfertilization (∼10 mm standard length, a stage just after the effect of the chromosome 21 branchial bone length QTL could first be detected in the FTC × LITC cross) (Erickson et al. 2014). For the Tfap2a mutations in the FTC background, we found that EB1 length was slightly but significantly decreased in heterozygous fish (Figure 6A), while surprisingly, the third, fourth, and fifth ceratobranchials (CB3, CB4, and CB5) were slightly but significantly increased (Figure S8). However, in the LITC background, none of the branchial bones measured (CB1–5 and EB1) were significantly different between wild-type and heterozygous fish (Figure 6B and Figure S8). Although our allele specific expression data predict that reducing Tfap2a function might lead to an increase in branchial bone length, our functional data suggest that halving the dosage of Tfap2a on a freshwater genetic background can both increase (for ventral posterior ceratobranchials) and decrease (for dorsal EB1) branchial bone length.
Figure 6.
Heterozygous loss of Tfap2a produces slightly but significantly shorter EB1 bone length in freshwater, but not marine fish. (A) FTC freshwater fish (25–28 dpf) heterozygous for predicted Tfap2a loss-of-function mutations have significantly shorter first epibranchial (EB1) length relative to wild-type siblings (t-test: t = 1.99, d.f. = 87.1, P = 0.049, asterisk). (B) LITC marine heterozygous Tfap2a mutant fish have no significant difference in EB1 length compared to wild-type siblings. Additional bone length phenotypes are shown in Figure S8. There was no difference in standard length between +/+ and +/− fish in either population (P > 0.5). EB1 length is size-corrected for standard length; residuals are shown.
Discussion
Genetic dissection of a supergene involved in parallel evolution
Many putatively pleiotropic QTL affecting traits of evolutionary, economic, and biomedical interest have been discovered (Feitosa et al. 2006; Hall et al. 2006; Ookawa et al. 2010; Stearns 2010; Saatchi et al. 2014), but determining whether the underlying causative loci are the same or different remains a challenge (Wagner and Zhang 2011; Paaby and Rockman 2013). Using recombinant mapping in two genetic crosses, we show that two separate but tightly linked genes, Bmp6 and Tfap2a, form a supergene that increases both the length of bones and the number of teeth in the stickleback branchial skeleton in two freshwater populations. Clustering of adaptive loci is predicted by population genetic theory when migration occurs between differentially adapted populations (Yeaman 2013), such as occurs between marine and freshwater adapted sticklebacks (Schluter and Conte 2009; Bell and Aguirre 2013). Migration could maintain close genetic linkage of two alleles affecting trophic morphology, which in turn could facilitate rapid adaptation to freshwater diets upon colonization of new habitats. After the causative mutation(s) for both QTL have been determined, testing for linkage disequilibrium of these alleles in wild marine and estuarine populations could test this hypothesis.
While no study has directly tested whether tooth number or bone length are under selection in freshwater stickleback populations, the causative mutations (once found) could be tested for genomic signatures of selection. It is possible that the mutations underlying only one or neither of these phenotypes are under selection, and that they are instead hitchhiking with other adaptive mutations. The bone length QTL is either not fixed in the FTC population or is dependent upon interactions at other loci, and the ENOB population has a tooth number QTL on chromosome 21 (Erickson et al. 2016b) but not a bone length QTL, suggesting that the two elements of this supergene are genetically separable and do not always covary in the wild. The alleles may be maintained at low frequency in marine populations, allowing for repeatable genetic evolution upon freshwater colonization. Alternatively, genetic and developmental constraints may have led to the independent evolution of these craniofacial QTL in multiple populations. The presence and absence of this bone length QTL could be leveraged in future studies to identify polymorphisms associated with the QTL.
In addition to separating the loci controlling bone length and tooth number, we also separated the bone length QTL into at least two loci in the FTC freshwater population by identifying a single recombinant chromosome with effects that are significantly different from both marine and freshwater alleles. These data suggest that a portion of the phenotypic effect maps nearer to the coding sequence of Tfap2a, and a portion of the phenotypic effect is attributable to a large non-coding region between Tfap2a and Bmp6. QTL fractionation has also been observed in a variety of other animal and plant systems (Mackay 2004; Yalcin et al. 2004; Willis-Owen and Flint 2006; Studer and Doebley 2011; Johnson et al. 2012; Linnen et al. 2013), suggesting QTL for complex traits might often represent multiple mutations, especially when the underlying gene has many roles in development, such as Tfap2a.
Tfap2a as a candidate gene for craniofacial evolution
Our fine mapping and functional results support Tfap2a as a candidate for contributing to craniofacial evolution in sticklebacks. Tfap2a was initially identified as an activating protein that binds the SV40 viral enhancer to promote transcription (Williams et al. 1988) and has roles in both activating and inhibiting gene expression (Mitchell et al. 1987; Williams and Tjian 1991; Gaubatz et al. 1995; Pfisterer et al. 2002; Eckert et al. 2005). During vertebrate embryogenesis, Tfap2a is expressed in a variety of tissues in both mice and zebrafish (Mitchell et al. 1991; Thisse et al. 2001), including the neural crest, an embryonic migratory cell population that gives rise to a variety of tissues, including the pharyngeal skeleton and pigment cells (Bronner and LeDouarin 2012; Mayor and Theveneau 2013). Further studies found a critical role for Tfap2a as a master regulator of neural crest development (de Crozé et al. 2011; Rada-Iglesias et al. 2012). Mice and zebrafish with loss-of-function mutations have severe craniofacial defects (Schorle et al. 1996; Zhang et al. 1996; Holzschuh et al. 2003; Knight et al. 2003, 2004) likely due to Tfap2a’s role in the pharyngeal ectoderm (Knight et al. 2005). Conditional knockout of Tfap2a in the frontonasal prominence of mice results in a failure of craniofacial outgrowth during postnatal development (Nelson and Williams 2004), while conditional knockout of Tfap2a/b in cranial neural crest cells results in a loss of the jaw hinge and other craniofacial phenotypes (Van Otterloo et al. 2018). In humans, familial mutations in Tfap2a cause branchio-oculo-facial syndrome (Milunsky et al. 2008; Stoetzel et al. 2009; Tekin et al. 2009). Thus, Tfap2a plays complex roles at multiple stages of craniofacial development in vertebrates, consistent with the possibility of cis-regulatory changes contributing to evolved branchial bone length differences in freshwater sticklebacks.
Evolved differences in neural crest cell regulation and patterning are hypothesized to underlie craniofacial evolution in primates, birds, and cichlids, as well as the domestication of a variety of mammals (Fish et al. 2014; Powder et al. 2014; Wilkins et al. 2014; Prescott et al. 2015). Therefore, Tfap2a might affect stickleback branchial bone length by altering the early specification and patterning of cranial neural crest cells that eventually form the pharyngeal skeleton. Consistent with this possibility, we observed tissue-restricted expression of Tfap2a in dorsal postmigratory cranial neural crest in developing pharyngeal arches, as well as later in development in the primordia of stickleback branchial bones. Tfap2a might regulate dorsal branchial skeleton morphology by modifying the dorsal–ventral partitioning of cells within the pharyngeal arches. The cis-regulatory differences in freshwater Tfap2a expression might reflect spatial and/or quantitative changes in Tfap2a expression, which could be tested once Tfap2a enhancers of craniofacial expression are determined. While conserved regulatory elements of Tfap2a have been described in mice (Zhang and Williams 2003; Donner and Williams 2006; Feng et al. 2008), an enhancer driving pharyngeal arch neural crest has not been reported. Future genomic sequence comparisons and transgenic assays could identify additional enhancers driving expression in pharyngeal arch neural crest (including the precursors of the EB1 studied here). In addition to possible changes in pharyngeal arch neural crest regulatory elements, it is also possible that the synonymous change identified here contributes to or even fully explains the observed allelic expression differences through differences in mRNA processing or stability, both of which could also contribute to the observed allele-specific difference in transcript abundance.
In addition to its roles in craniofacial development, Tfap2a also regulates the differentiation and maintenance of cell types important for skeletal development. Tfap2a mutant mice and chimeric mice have partially penetrant absence of zeugopods and polydactyly (Schorle et al. 1996; Zhang et al. 1996; Nottoli et al. 1998). During mammalian long bone development, analogous to fish branchial bone development (Haines 1942), an initial cartilage template for the bone is formed. Osteoblasts then secrete bone matrix surrounding this template, but a region of cartilage cells (chondrocytes) is left behind at either end as a growth plate for further bone elongation (Kronenberg 2003; Hall 2005; Karsenty et al. 2009). TFAP2A protein is expressed in the growth plate of mouse long bones (Davies et al. 2002), is thought to regulate a variety of genes related to chondrocyte development (Xie et al. 1998; Tuli et al. 2002), and is a negative regulator of chondrocyte differentiation in vitro (Huang et al. 2004). Therefore, a downregulation or reduced function of Tfap2a in freshwater fish might cause an increase in chondrocyte differentiation, resulting in greater long bone growth. The allele specific expression of Tfap2a seen during early bone development, combined with the manifestation of the bone length QTL observed in juveniles, suggests that some combination of Tfap2a’s roles in neural crest development as well as chondrocyte differentiation might underlie the evolved increases in bone length. Furthermore, the fractionation of the QTL in the FTC cross suggests that at least two regions surrounding Tfap2a, and possibly two developmental functions, contribute to the evolved phenotypes.
Surprisingly, the effect of Tfap2a mutation was more severe in sticklebacks than in zebrafish. While zebrafish lockjaw mutants still have identifiable pharyngeal cartilage elements in the jaw and branchial skeleton (Knight et al. 2003), stickleback larvae homozygous for Tfap2a mutations had only a few severely hypoplastic and unidentifiable pharyngeal skeletal elements, as well as defects in the anterior neurocranium. The remaining chondrocytes appear to be well developed in mutant fish, so the primary craniofacial defect appears to be pharyngeal arch patterning, not cartilage differentiation. Therefore, compared to zebrafish, sticklebacks may have fewer redundant functions of other genes with Tfap2a during craniofacial development.
We found that while induced mutations in Tfap2a caused lethal craniofacial defects, heterozygous loss of Tfap2a was sufficient to alter branchial bone length in the FTC freshwater background. Based on our ASE data, we predict that freshwater fish likely have lower levels of Tfap2a, so they may be more sensitive to reductions in Tfap2a function than marine fish. FTC fish heterozygous for mutations in Tfap2a had slightly but significantly shorter dorsal (EB1) bones, but significantly longer ventral (CB5) bones. These opposite results suggest that Tfap2a could play some role in allocating cells to dorsal and ventral pharyngeal elements, but also suggest that branchial skeleton morphology can be sensitive to Tfap2a dosage in a genetic background-specific manner, perhaps similar to the quantitative craniofacial phenotypes reported in heterozygous Tfap2a mutant mice (Green et al. 2015). The predicted strong loss-of-function mutations generated by TALENs likely do not fully recapitulate cis-regulatory mutations in Tfap2a that occur in nature, which may affect the timing, location, and/or level of gene expression.
Cis-regulatory changes and morphological evolution
Evolution of cis-regulatory modules is thought to be an important general feature of morphological evolution (Stern 2000; Carroll 2008). Because developmental regulatory genes are often expressed and required in a wide variety of tissues at time points throughout development, changes to their regulation, rather than changes to their coding sequences, may prevent deleterious pleiotropic effects. Both Tfap2a and Bmp6 are expressed dynamically in multiple tissues during development (Cleves et al. 2014; Erickson et al. 2015 and this study) and can have deleterious and lethal loss-of-function phenotypes (P. Cleves, J. Hart, R. Agoglia, M. Jimenez, P. Erickson, L. Gai, and C. Miller, unpublished data, and this study). Here, we provide evidence that a subtle regulatory difference in Tfap2a is associated with evolved skeletal changes related to dietary adaptation, but we cannot exclude the possibility that a long distance regulatory element of another gene outside the eight-gene interval (including Bmp6) could underlie at least part of the QTL effect (Montavon et al. 2011; Marinić et al. 2013; Smallwood and Ren 2013; Anderson and Hill 2014). However, the paralogous tightly linked Tfap2c and Bmp7 genes in mice have been found to be in distinct topologically associating domains, suggesting that their regulatory elements are largely distinct (Tsujimura et al. 2015).
In the stickleback genome (Jones et al. 2012; Glazer et al. 2015), Tfap2a and Bmp6 are separated by a large gene desert that is replete with conserved noncoding sequence. These many putative regulatory elements might provide fodder for cis-regulatory changes controlling evolved morphological differences. Regulatory changes to other developmental signaling molecules have also been implicated in stickleback evolution (Colosimo et al. 2005; Miller et al. 2007; Chan et al. 2010; Indjeian et al. 2016; Ishikawa et al. 2017), suggesting that changes to gene regulation may be used to fine-tune developmental processes to produce novel and ecologically beneficial phenotypes. Interestingly, our studies indicate evolved increases in both branchial bone length and pharyngeal tooth number in freshwater stickleback are associated with cis-regulatory reductions in expression (Cleves et al. 2014). Combined, these findings suggest that evolved gain traits may make “more from less”: constructive morphological phenotypes may commonly evolve from mutations that reduce function and/or expression of genes that repress tissue growth (e.g., inhibit tooth replacement or repress chondrocyte differentiation).
Acknowledgments
We thank Monica Jimenez for assistance in designing the Tfap2a TALENs, Alyson Smith for assistance in cartilage phenotyping, and Andrew Glazer for identifying polymorphic indels and for helpful discussions on the ASE assay development. This work was funded in part by National Institutes of Health (NIH) grant R01 #DE021475 (C.T.M.), NIH predoctoral training grants #5T32GM007127 (P.A.E.) and # 5T32HG000047-15 (J.C.H.), and the National Science Foundation Graduate Research Fellowship (P.A.C.). Experiments in this study used the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, supported by NIH S10 instrumentation grants S10RR029668 and S10RR027303.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6024338.
Communicating editor: K. Peichel
Literature Cited
- Anderson E., Hill R. E., 2014. Long range regulation of the sonic hedgehog gene. Curr. Opin. Genet. Dev. 27: 54–59. 10.1016/j.gde.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Anker G. C., 1974. Morphology and kinetics of the head of the stickleback, Gasterosteus aculeatus. Trans. Zool. Soc. London 32: 311–416. [Google Scholar]
- Balic A., Thesleff I., 2015. Tissue interactions regulating tooth development and renewal, pp. 157–186 in Current Topics in Developmental Biology, Craniofacial Development, edited by Chai Y. Academic Press, New York: 10.1016/bs.ctdb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Bell M. A., Aguirre W. E., 2013. Contemporary evolution, allelic recycling, and adaptive radiation of the threespine stickleback. Evol. Ecol. Res. 15: 377–411. [Google Scholar]
- Bell M. A., Foster S. A., 1994. The Evolutionary Biology of the Threespine Stickleback. Oxford University Press, Oxford, UK. [Google Scholar]
- Bronner M. E., LeDouarin N. M., 2012. Development and evolution of the neural crest: an overview. Dev. Biol. 366: 2–9. 10.1016/j.ydbio.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-eevo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. F., Marks M. E., Jones F. C., Villarreal G., Shapiro M. D., et al. , 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. 10.1126/science.1182213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves P. A., Ellis N. A., Jimenez M. T., Nunez S. M., Schluter D., et al. , 2014. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc. Natl. Acad. Sci. USA 111: 13912–13917. 10.1073/pnas.1407567111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G., Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. 10.1126/science.1107239 [DOI] [PubMed] [Google Scholar]
- Conte G. L., Arnegard M. E., Peichel C. L., Schluter D., 2012. The probability of genetic parallelism and convergence in natural populations. Proc. R. Soc. Lond. B Biol. Sci. 279: 5039–5047. 10.1098/rspb.2012.2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Hirschhorn J. N., Altshuler D., Lander E. S., 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. 10.1038/ng992 [DOI] [PubMed] [Google Scholar]
- Davies S. R., Sakano S., Zhu Y., Sandell L. J., 2002. Distribution of the transcription factors Sox9, AP-2, and [delta]EF1 in adult murine articular and meniscal cartilage and growth plate. J. Histochem. Cytochem. 50: 1059–1065. 10.1177/002215540205000808 [DOI] [PubMed] [Google Scholar]
- de Crozé N., Maczkowiak F., Monsoro-Burq A. H., 2011. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc. Natl. Acad. Sci. USA 108: 155–160. 10.1073/pnas.1010740107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A. L., Williams T., 2006. Frontal nasal prominence expression driven by Tcfap2a relies on a conserved binding site for STAT proteins. Dev. Dyn. 235: 1358–1370. 10.1002/dvdy.20722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle E. L., Booher N. J., Standage D. S., Voytas D. F., Brendel V. P., et al. , 2012. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40: W117–W122. 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D., Buhl S., Weber S., Jäger R., Schorle H., 2005. The AP-2 family of transcription factors. Genome Biol. 6: 246 10.1186/gb-2005-6-13-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N. A., Miller C. T., 2016. Dissection and flat-mounting of the threespine stickleback branchial skeleton. J. Vis. Exp. (111): e54056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N. A., Glazer A. M., Donde N. N., Cleves P. A., Agoglia R. M., et al. , 2015. Distinct developmental and genetic mechanisms underlie convergently evolved tooth gain in sticklebacks. Development 142: 2442–2451. 10.1242/dev.124248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Glazer A. M., Cleves P. A., Smith A. S., Miller C. T., 2014. Two developmentally temporal quantitative trait loci underlie convergent evolution of increased branchial bone length in sticklebacks. Proc. R. Soc. Lond. B Biol. Sci. 281: 20140822 10.1098/rspb.2014.0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Cleves P. A., Ellis N. A., Schwalbach K. T., Hart J. C., et al. , 2015. A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev. Biol. 401: 310–323. 10.1016/j.ydbio.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Ellis N. A., Miller C. T., 2016a Microinjection for transgenesis and genome editing in threespine sticklebacks. J. Vis. Exp. (111): e54055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Glazer A. M., Killingbeck E. E., Agoglia R. M., Baek J., et al. , 2016b Partially repeatable genetic basis of benthic adaptation in threespine sticklebacks. Evolution 70: 887–902. 10.1111/evo.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnum C. E., Tinsley M., Hermanson J. W., 2008a Postnatal bone elongation of the manus vs. pes: analysis of the chondrocytic differentiation cascade in Mus musculus and Eptesicus fuscus. Cells Tissues Organs 187: 48–58. 10.1159/000109963 [DOI] [PubMed] [Google Scholar]
- Farnum C. E., Tinsley M., Hermanson J. W., 2008b Forelimb vs. hindlimb skeletal development in the big brown bat, Eptesicus fuscus: functional divergence is reflected in chondrocytic performance in autopodial growth plates. Cells Tissues Organs 187: 35–47. 10.1159/000109962 [DOI] [PubMed] [Google Scholar]
- Feitosa M. F., Rice T., North K. E., Kraja A., Rankinen T., et al. , 2006. Pleiotropic QTL on chromosome 19q13 for triglycerides and adiposity: the HERITAGE family study. Atherosclerosis 185: 426–432. 10.1016/j.atherosclerosis.2005.06.023 [DOI] [PubMed] [Google Scholar]
- Feng W., Huang J., Zhang J., Williams T., 2008. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Mol. Cell. Biol. 28: 315–325. 10.1128/MCB.01168-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J. L., Sklar R. S., Woronowicz K. C., Schneider R. A., 2014. Multiple developmental mechanisms regulate species-specific jaw size. Development 141: 674–684. 10.1242/dev.100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N., Erezyilmaz D. F., McGregor A. P., Wang S., Payre F., et al. , 2011. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603. 10.1038/nature10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S., Imhof A., Dosch R., Werner O., Mitchell P., et al. , 1995. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 14: 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A. M., Cleves P. A., Erickson P. A., Lam A. Y., Miller C. T., 2014. Parallel developmental genetic features underlie stickleback gill raker evolution. Evodevo 5: 19 10.1186/2041-9139-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A. M., Killingbeck E. E., Mitros T., Rokhsar D. S., Miller C. T., 2015. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3 5: 1463–1472. 10.1534/g3.115.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Sambrook J., 2012. Molecular Cloning : A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Green R. M., Feng W., Phang T., Fish J. L., Li H., et al. , 2015. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis. Model. Mech. 8: 31–43. 10.1242/dmm.017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H. P., Anderson J. M., 1984. Geographic variation in the gillrakers and diet of European threespine sticklebacks, Gasterosteus aculeatus. Copeia 1984: 87–97. 10.2307/1445038 [DOI] [Google Scholar]
- Haines R. W., 1934. Epiphyseal growth in the branchial skeleton of fishes. Q. J. Microsc. Sci. 77: 77–97. [Google Scholar]
- Haines R. W., 1942. The evolution of epiphyses and of endochondral bone. Biol. Rev. Camb. Philos. Soc. 17: 267–292. 10.1111/j.1469-185X.1942.tb00440.x [DOI] [Google Scholar]
- Hall B. K., 2005. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology. Academic Press, London. [Google Scholar]
- Hall M. C., Basten C. J., Willis J. H., 2006. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics 172: 1829–1844. 10.1534/genetics.105.051227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann K., Klahre U., Moser M., Sheehan H., Mandel T., et al. , 2013. Tight genetic linkage of prezygotic barrier loci creates a multifunctional speciation island in Petunia. Curr. Biol. 23: 873–877. 10.1016/j.cub.2013.03.069 [DOI] [PubMed] [Google Scholar]
- Holzschuh J., Barrallo-Gimeno A., Ettl A.-K., Durr K., Knapik E. W., et al. , 2003. Noradrenergic neurons in the zebrafish hindbrain are induced by retinoic acid and require tfap2a for expression of the neurotransmitter phenotype. Development 130: 5741–5754. 10.1242/dev.00816 [DOI] [PubMed] [Google Scholar]
- Huang Z., Xu H., Sandell L., 2004. Negative regulation of chondrocyte differentiation by transcription factor AP-2alpha. J. Bone Miner. Res. 19: 245–255. 10.1359/jbmr.2004.19.2.245 [DOI] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L. M., Tian F., Bradbury P. J., Chen C., et al. , 2012. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109: E1913–E1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V. B., Kingman G. A., Jones F. C., Guenther C. A., Grimwood J., et al. , 2016. Evolving new skeletal traits by cis-regulatory changes in Bone Morphogenetic Proteins. Cell 164: 45–56. 10.1016/j.cell.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A., Kusakabe M., Yoshida K., Ravinet M., Makino T., et al. , 2017. Different contributions of local- and distant-regulatory changes to transcriptome divergence between stickleback ecotypes. Evolution 71: 565–581. 10.1111/evo.13175 [DOI] [PubMed] [Google Scholar]
- Jeong S., Rebeiz M., Andolfatto P., Werner T., True J., et al. , 2008. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132: 783–793. 10.1016/j.cell.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Johnson E. B., Haggard J. E., St.Clair D. A., 2012. Fractionation, stability, and isolate-specificity of QTL for resistance to Phytophthora infestans in cultivated tomato (Solanum lycopersicum). G3 2: 1145–1159. 10.1534/g3.112.003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M., Papa R., Beltrán M., Chamberlain N., Mavárez J., et al. , 2006. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 4: e303 10.1371/journal.pbio.0040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M., Frezal L., Jones R. T., Chamberlain N. L., Lee S. F., et al. , 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H. M., Settembre C., 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25: 629–648. 10.1146/annurev.cellbio.042308.113308 [DOI] [PubMed] [Google Scholar]
- Kim K.-W., Bennison C., Hemmings N., Brookes L., Hurley L. L., et al. , 2017. A sex-linked supergene controls sperm morphology and swimming speed in a songbird. Nat. Ecol. Evol. 1: 1168–1176. 10.1038/s41559-017-0235-2 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Miller C. T., Kruze G., Ullmann B., BreMiller R. A., et al. , 1998. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev. Biol. 203: 245–263. 10.1006/dbio.1998.9016 [DOI] [PubMed] [Google Scholar]
- Kislalioglu M., Gibson R. N., 1977. The feeding relationship of shallow water fishes in a Scottish sea loch. J. Fish Biol. 11: 257–266. 10.1111/j.1095-8649.1977.tb04118.x [DOI] [Google Scholar]
- Knight R. D., Nair S., Nelson S. S., Afshar A., Javidan Y., et al. , 2003. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130: 5755–5768. 10.1242/dev.00575 [DOI] [PubMed] [Google Scholar]
- Knight R. D., Javidan Y., Nelson S., Zhang T., Schilling T., 2004. Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev. Dyn. 229: 87–98. 10.1002/dvdy.10494 [DOI] [PubMed] [Google Scholar]
- Knight R. D., Javidan Y., Zhang T., Nelson S., Schilling T. F., 2005. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development 132: 3127–3138. 10.1242/dev.01879 [DOI] [PubMed] [Google Scholar]
- Kronenberg H. M., 2003. Developmental regulation of the growth plate. Nature 423: 332–336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Kunte K., Zhang W., Tenger-Trolander A., Palmer D. H., Martin A., et al. , 2014. Doublesex is a mimicry supergene. Nature 507: 229–232. 10.1038/nature13112 [DOI] [PubMed] [Google Scholar]
- Küpper C., Stocks M., Risse J. E., dos Remedios N., Farrell L. L., et al. , 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48: 79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S., Fan G., Widemo F., Gunnarsson U., Thalmann D. S., et al. , 2016. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 48: 84–88. 10.1038/ng.3430 [DOI] [PubMed] [Google Scholar]
- Lemmon Z. H., Doebley J. F., 2014. Genetic dissection of a genomic region with pleiotropic effects on domestication traits in maize reveals multiple linked QTL. Genetics 198: 345–353. 10.1534/genetics.114.165845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen C. R., Poh Y.-P., Peterson B. K., Barrett R. D. H., Larson J. G., et al. , 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339: 1312–1316. 10.1126/science.1233213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500 (erratum: PLoS Biol. 10 (2012)). 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14: 253–257. 10.1016/j.gde.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Marinić M., Aktas T., Ruf S., Spitz F., 2013. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell 24: 530–542. 10.1016/j.devcel.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Mather K., 1950. The genetical architecture of heterostyly in Primula sinensis. Evolution 4: 340–352. 10.1111/j.1558-5646.1950.tb01404.x [DOI] [Google Scholar]
- Mayor R., Theveneau E., 2013. The neural crest. Development 140: 2247–2251. 10.1242/dev.091751 [DOI] [PubMed] [Google Scholar]
- McGee M. D., Wainwright P. C., 2013. Convergent evolution as a generator of phenotypic diversity in threespine stickleback. Evolution 67: 1204–1208. 10.1111/j.1558-5646.2012.01839.x [DOI] [PubMed] [Google Scholar]
- McGee M. D., Schluter D., Wainwright P. C., 2013. Functional basis of ecological divergence in sympatric stickleback. BMC Evol. Biol. 13: 277 10.1186/1471-2148-13-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A. P., Orgogozo V., Delon I., Zanet J., Srinivasan D. G., et al. , 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448: 587–590. 10.1038/nature05988 [DOI] [PubMed] [Google Scholar]
- Miller C. T., Beleza S., Pollen A. A., Schluter D., Kittles R. A., et al. , 2007. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131: 1179–1189. 10.1016/j.cell.2007.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Glazer A. M., Summers B. R., Blackman B. K., Norman A. R., et al. , 2014. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics 197: 405–420. 10.1534/genetics.114.162420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky J. M., Maher T. A., Zhao G., Roberts A. E., Stalker H. J., et al. , 2008. TFAP2A mutations result in branchio-oculo-facial syndrome. Am. J. Hum. Genet. 82: 1171–1177. 10.1016/j.ajhg.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R., 1987. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell 50: 847–861. 10.1016/0092-8674(87)90512-5 [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Timmons P. M., Hébert J. M., Rigby P. W., Tjian R., 1991. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 5: 105–119. 10.1101/gad.5.1.105 [DOI] [PubMed] [Google Scholar]
- Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., et al. , 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Murray J., Clarke B., 1976a Supergenes in polymorphic land snails. II. Partula suturalis. Heredity 37: 271–282. 10.1038/hdy.1976.87 [DOI] [PubMed] [Google Scholar]
- Murray J., Clarke B., 1976b Supergenes in polymorphic land snails. I. Partula taeniata. Heredity 37: 253–269. 10.1038/hdy.1976.86 [DOI] [PubMed] [Google Scholar]
- Nabours R. K., 1933. Inheritance of color patterns in the grouse locust Acrydium arenosum burmeister (Tettigidae). Genetics 18: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Pardo-Diaz C., Whibley A., Supple M. A., Saenko S. V., et al. , 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534: 106–110. 10.1038/nature17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. K., Williams T., 2004. Frontonasal process-specific disruption of AP-2alpha results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Dev. Biol. 267: 72–92. 10.1016/j.ydbio.2003.10.033 [DOI] [PubMed] [Google Scholar]
- Nottoli T., Hagopian-Donaldson S., Zhang J., Perkins A., Williams T., 1998. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc. Natl. Acad. Sci. USA 95: 13714–13719. 10.1073/pnas.95.23.13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookawa T., Hobo T., Yano M., Murata K., Ando T., et al. , 2010. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1: 132 10.1038/ncomms1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2005. The probability of parallel evolution. Evolution 59: 216–220. 10.1111/j.0014-3820.2005.tb00907.x [DOI] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2013. The many faces of pleiotropy. Trends Genet. 29: 66–73. 10.1016/j.tig.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Marques D. A., 2017. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20150486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Nereng K. S., Ohgi K. A., Cole B. L. E., Colosimo P. F., et al. , 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414: 901–905. 10.1038/414901a [DOI] [PubMed] [Google Scholar]
- Pfisterer P., Ehlermann J., Hegen M., Schorle H., 2002. A subtractive gene expression screen suggests a role of transcription factor AP-2α in control of proliferation and differentiation. J. Biol. Chem. 277: 6637–6644. 10.1074/jbc.M108578200 [DOI] [PubMed] [Google Scholar]
- Powder K. E., Cousin H., McLinden G. P., Albertson R. C., 2014. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol. Biol. Evol. 31: 3113–3124. 10.1093/molbev/msu267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracana R., Priyam A., Levantis I., Nichols R. A., Wurm Y., 2017. The fire ant social chromosome supergene variant Sb shows low diversity but high divergence from SB. Mol. Ecol. 26: 2864–2879. 10.1111/mec.14054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott S. L., Srinivasan R., Marchetto M. C., Grishina I., Narvaiza I., et al. , 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163: 68–83. 10.1016/j.cell.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S. A., Swigut T., et al. , 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11: 633–648. 10.1016/j.stem.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M., Pool J. E., Kassner V. A., Aquadro C. F., Carroll S. B., 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326: 1663–1667. 10.1126/science.1178357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N., Reich D., 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22: 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum E. B., Parent C. E., Brandt E. E., 2014. The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 45: 203–226. 10.1146/annurev-ecolsys-120213-091851 [DOI] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., Garrick D. J., 2014. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics 15: 442 10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar V. S., Gamer L. W., Rosen V., 2016. BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 12: 203–221. 10.1038/nrendo.2016.12 [DOI] [PubMed] [Google Scholar]
- Sanger T. J., Norgard E. A., Pletscher L. S., Bevilacqua M., Brooks V. R., et al. , 2011. Developmental and genetic origins of murine long bone length variation. J. Exp. Zoolog. B Mol. Dev. Evol. 316B: 146–161. 10.1002/jez.b.21388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger T. J., Revell L. J., Gibson-Brown J. J., Losos J. B., 2012. Repeated modification of early limb morphogenesis programmes underlies the convergence of relative limb length in Anolis lizards. Proc. Biol. Sci. 279: 739–748. 10.1098/rspb.2011.0840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., Conte G. L., 2009. Genetics and ecological speciation. Proc. Natl. Acad. Sci. USA 106(Suppl. 1): 9955–9962. 10.1073/pnas.0901264106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H., Meier P., Buchert M., Jaenisch R., Mitchell P. J., 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381: 235–238. 10.1038/381235a0 [DOI] [PubMed] [Google Scholar]
- Schwander T., Libbrecht R., Keller L., 2014. Supergenes and complex phenotypes. Curr. Biol. 24: R288–R294. 10.1016/j.cub.2014.01.056 [DOI] [PubMed] [Google Scholar]
- Smallwood A., Ren B., 2013. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 25: 387–394. 10.1016/j.ceb.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M., Finger J. H., Hayamizu T. F., McCright I. J., Xu J., et al. , 2014. The mouse Gene Expression Database (GXD): 2014 update. Nucleic Acids Res. 42: D818–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns F. W., 2010. One hundred years of pleiotropy: a retrospective. Genetics 186: 767–773. 10.1534/genetics.110.122549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C. C., Weber J. N., Hoekstra H. E., 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5: e219 (erratum: PLoS Biol. 6: e36). 10.1371/journal.pbio.0050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., 2000. Perspective: evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. 10.1111/j.0014-3820.2000.tb00544.x [DOI] [PubMed] [Google Scholar]
- Stern D. L., 2013. The genetic causes of convergent evolution. Nat. Rev. Genet. 14: 751–764. 10.1038/nrg3483 [DOI] [PubMed] [Google Scholar]
- Stern D. L., Orgogozo V., 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62: 2155–2177. 10.1111/j.1558-5646.2008.00450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoetzel C., Riehm S., Bennouna Greene V., Pelletier V., Vigneron J., et al. , 2009. Confirmation of TFAP2A gene involvement in branchio-oculo-facial syndrome (BOFS) and report of temporal bone anomalies. Am. J. Med. Genet. A. 149A: 2141–2146. 10.1002/ajmg.a.33015 [DOI] [PubMed] [Google Scholar]
- Studer A. J., Doebley J. F., 2011. Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 188: 673–681. 10.1534/genetics.111.126508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin M., Sırmacı A., Yüksel-Konuk B., Fitoz S., Sennaroğlu L., 2009. A complex TFAP2A allele is associated with branchio-oculo-facial syndrome and inner ear malformation in a deaf child. Am. J. Med. Genet. A. 149A: 427–430. 10.1002/ajmg.a.32619 [DOI] [PubMed] [Google Scholar]
- Thisse, B., S. Pflumio, M. Furthauer, B. Loppin, V. Heyer et al., 2001 Expression of the Zebrafish Genome During Embryogenesis ZFIN Direct Data Submission. Available at: https://zfin.org/ZDB-PUB-010810-1. [Google Scholar]
- Thomas J. W., Cáceres M., Lowman J. J., Morehouse C. B., Short M. E., et al. , 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179: 1455–1468. 10.1534/genetics.108.088229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. J., Jiggins C. D., 2014. Supergenes and their role in evolution. Heredity 113: 1–8. 10.1038/hdy.2014.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura T., Klein F. A., Langenfeld K., Glaser J., Huber W., et al. , 2015. A discrete transition zone organizes the topological and regulatory autonomy of the adjacent Tfap2c and Bmp7 genes. PLoS Genet. 11: e1004897 10.1371/journal.pgen.1004897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuli R., Seghatoleslami M. R., Tuli S., Howard M. S., Danielson K. G., et al. , 2002. p38 MAP kinase regulation of AP-2 binding in TGF-β1-stimulated chondrogenesis of human trabecular bone-derived cells. Ann. N. Y. Acad. Sci. 961: 172–177. 10.1111/j.1749-6632.2002.tb03077.x [DOI] [PubMed] [Google Scholar]
- Tuttle E. M., Bergland A. O., Korody M. L., Brewer M. S., Newhouse D. J., et al. , 2016. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26: 344–350. 10.1016/j.cub.2015.11.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Otterloo E., Li H., Jones K. L., Williams T., 2018. AP-2α and AP-2β cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 145: dev157438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J., 2011. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213. 10.1038/nrg2949 [DOI] [PubMed] [Google Scholar]
- Wainwright P. C., 2006. Functional morphology of the pharyngeal jaw apparatus, pp. 77–101 in Fish Physiology: Fish Biomechanics. Academic Press, San Diego. [Google Scholar]
- Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y., et al. , 2005. The origin of the naked grains of maize. Nature 436: 714–719. 10.1038/nature03863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wurm Y., Nipitwattanaphon M., Riba-Grognuz O., Huang Y.-C., et al. , 2013. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493: 664–668. 10.1038/nature11832 [DOI] [PubMed] [Google Scholar]
- Wenke A.-K., Bosserhoff A. K., 2010. Roles of AP-2 transcription factors in the regulation of cartilage and skeletal development. FEBS J. 277: 894–902. 10.1111/j.1742-4658.2009.07509.x [DOI] [PubMed] [Google Scholar]
- Wilkins A. S., Wrangham R. W., Fitch W. T., 2014. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197: 795–808. 10.1534/genetics.114.165423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Tjian R., 1991. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 5: 670–682. 10.1101/gad.5.4.670 [DOI] [PubMed] [Google Scholar]
- Williams T., Admon A., Lüscher B., Tjian R., 1988. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 2: 1557–1569. 10.1101/gad.2.12a.1557 [DOI] [PubMed] [Google Scholar]
- Wills D. M., Whipple C. J., Takuno S., Kursel L. E., Shannon L. M., et al. , 2013. From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9: e1003604 10.1371/journal.pgen.1003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Owen S. A. G., Flint J., 2006. The genetic basis of emotional behaviour in mice. Eur. J. Hum. Genet. 14: 721–728. 10.1038/sj.ejhg.5201569 [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- Xie W. F., Kondo S., Sandell L. J., 1998. Regulation of the mouse cartilage-derived retinoic acid-sensitive protein gene by the transcription factor AP-2. J. Biol. Chem. 273: 5026–5032. 10.1074/jbc.273.9.5026 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Willis-Owen S. A. G., Fullerton J., Meesaq A., Deacon R. M., et al. , 2004. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat. Genet. 36: 1197–1202. 10.1038/ng1450 [DOI] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V. E., Vogelstein B., Kinzler K. W., 2002. Allelic variation in human gene expression. Science 297: 1143 10.1126/science.1072545 [DOI] [PubMed] [Google Scholar]
- Yeaman S., 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc. Natl. Acad. Sci. USA 110: E1743–E1751. 10.1073/pnas.1219381110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Williams T., 2003. Identification and regulation of tissue-specific cis-acting elements associated with the human AP-2α gene. Dev. Dyn. 228: 194–207. 10.1002/dvdy.10365 [DOI] [PubMed] [Google Scholar]
- Zhang J., Hagopian-Donaldson S., Serbedzija G., Elsemore J., Plehn-Dujowich D., et al. , 1996. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381: 238–241. 10.1038/381238a0 [DOI] [PubMed] [Google Scholar]