Abstract
Small molecule lipid-related metabolites are important components of fatty acid and steroid metabolism—two important contributors to human health. This study investigated the extent to which rare and common genetic variants spanning the human genome influence the lipid-related metabolome. Sequence data from 1552 European-Americans (EA) and 1872 African-Americans (AA) were analyzed to examine the impact of common and rare variants on the levels of 102 circulating lipid-related metabolites measured by a combination of chromatography and mass spectroscopy. We conducted single variant tests [minor allele frequency (MAF) > 5%, statistical significance P-value ≤ 2.45 × 10−10] and tests aggregating rare variants (MAF ≤ 5%) across multiple genomic motifs, such as coding regions and regulatory domains, and sliding windows. Multiethnic meta-analyses detected 53 lipid-related metabolites-locus pairs, which were inspected for evidence of consistent signal between the two ethnic groups. Thirty-eight lipid-related metabolite-genomic region associations were consistent across ethnicities, among which seven were novel. The regions contain genes that are related to metabolite transport (SLC10A1) and metabolism (SCD, FDX1, UGT2B15, and FADS2). Six of the seven novel findings lie in expression quantitative trait loci affecting the expression levels of 14 surrounding genes in multiple tissues. Imputed expression levels of 10 of the affected genes were associated with four corresponding lipid-related traits in at least one tissue. Our findings offer valuable insight into circulating lipid-related metabolite regulation in a multiethnic population.
Keywords: lipids, metabolome, rare variants, whole genome sequencing
LIPIDS are a diverse and omnipresent group of organic compounds that are not soluble in water (Smith 2006) and have complex structures due to a number of biochemical transformations they go through during biosynthesis (Fahy et al. 2011). Both lipids and lipid-related metabolites have multiple roles in fetal growth and development, and are important determinants of adult health and disease as a result of their contribution to cell membranes, energy metabolism, and the endocrine system (Kim et al. 2013; Calder 2015; El-Hattab and Scaglia 2015). We measured and analyzed 102 lipid-related metabolites, belonging to four groups: fatty acyls, glycerolipids, glycerophospholipids, and sterol lipids (Fahy et al. 2009). Fatty acids function to regulate cell membrane permeability, gene transcription, and signaling pathway regulation (Calder 2015). Glycerolipids are mainly used for energy storage (Reue and Brindley 2008). Sterol lipids include bile acids, which, together with their metabolites, participate in digestion and adsorption of lipid soluble nutrients (Maldonado-Valderrama et al. 2011), and have been shown to possess endocrine effects (Houten et al. 2006; Vítek and Haluzík 2016). Other sterol lipids, such as cortisol and androsterone, function as signaling molecules, while their precursor, cholesterol, forms cell membranes together with phospholipids (Bastiaanse et al. 1997). The measured metabolites analyzed here included 57 fatty acyls (represented by fatty esters, fatty acids, and their derivatives), four glycerolipids, 15 glycerophospholipids, 22 sterols, and their derivatives (including eight bile acids), as well as carnitine and deoxycarnitine because of their role in fatty acid transport (El-Hattab and Scaglia 2015), and inositol-1-phosphate and myo-inositol because of their role in multiple structural lipids (e.g., phosphatidylinositol) (Kim et al. 2013).
Many genes affecting various lipids and lipid-related traits have been identified previously by candidate gene studies (Niemsiri et al. 2015), genome-wide association studies (Pollin et al. 2008; Illig et al. 2010; Teslovich et al. 2010; Demirkan et al. 2012; Shin et al. 2014; Draisma et al. 2015), whole exome sequencing (WES) (Fouchier et al. 2014; Peloso et al. 2014; Yu et al. 2016b) and whole genome sequencing (WGS) (Long et al. 2017; Morrison et al. 2017) studies. WGS captures common and rare coding variation across the entirety of the genome, and has been used successfully to identify loci playing an important role in clinically recognized lipid traits, such as HDL-C levels (Morrison et al. 2013). For the current study, sequencing was performed on 1552 European-Americans (EA) and 1872 African-Americans (AA), to detect new genetic loci associated with the serum lipid-related metabolome. To our knowledge, this is the first multiethnic study using WGS and WES data to unravel pathways playing a role in the regulation of a broad spectrum of lipid-related metabolites.
Materials and Methods
Study populations and lipid metabolite measurements
A detailed description of the Atherosclerosis Risk in Communities (ARIC) study can be found elsewhere (The ARIC investigators 1989). Participants 45–64 years of age at the baseline examination were recruited from four communities (Forsyth County, North Carolina; Jackson, Mississippi; Minneapolis, Minnesota; and Washington County, Maryland). A total of 15,792 individuals, mostly of European and African ancestry, participated in the baseline examination in 1987–1989, with four follow-up visits in 1990–1992 (Visit 2), 1993–1995 (Visit 3), 1996–1998 (Visit 4) and 2011–2013 (Visit 5), with a sixth visit ongoing, which began in 2016. The current investigation was performed on 1850 AAs and 1330 EAs with WES data, and 1679 AAs and 1458 EAs with WGS data, all of whom had metabolomic measurements on 102 fasting serum lipid-related compounds. Both WES and WGS were available on a sample of 1657 AAs and 1236 EAs, while 193 AAs and 94 EAs had only WES data, and 22 AAs and 222 EAs had only WGS data. WES, WGS, or both were performed on a total sample of 1872 AAs and 1552 EAs.
Information on the method for baseline metabolites measurements is summarized in the Supplemental Methods. Briefly, 140 lipid-related metabolites were detected and quantified by Metabolon (Durham, NC), using an untargeted, gas chromatography–mass spectrometry and liquid chromatography–mass spectrometry (GC/LC-MS)-based metabolomic quantification protocol (Evans et al. 2009; Ohta et al. 2009). In 97 selected samples, lipid-related metabolites were measured twice (in 2010 and 2014). In order for measured metabolites to be included in this analysis, each compound had to have no more than 25% samples with missing values or values below the detection limit of the technology, and had to have the Pearson correlation coefficients between the 2010 and 2014 measurements ≥0.3 (Cohen 1988), with repeated measurements from 2014 used in the analyses. Therefore, the present study is based on an evaluation of 102 lipid-related metabolites.
Sequencing, variant calling, quality control, and annotation
All sequencing was done at the Baylor College of Medicine Human Genome Sequencing Center (HGSC). For WES, exomes were captured using the HGSC VCRome 2.1 reagent (Bainbridge et al. 2011) (42 Mb; NimbleGen) and all samples were paired-end sequenced using Illumina GAII or HiSeq instruments. For WGS, genomic DNA samples were made into Illumina paired-end libraries according to the manufacturer’s recommendation (Illumina Multiplexing_SamplePrep_Guide_1005361_D) and sequenced on a Hisequation 2000 (Illumina, San Diego, CA) in a pooled format to generate a minimum of 18 unique aligned giga-basepairs per sample. As previously reported, variant calling was completed using the Atlas2 (Challis et al. 2012) suite for WES, and goSNAP (https://sourceforge.net/p/gosnap/git/ci/master/tree/) for WGS (Yu et al. 2016a; de Vries et al. 2017). Detailed methods for the sequencing, variant calling and variant quality control for both WES and WGS are provided in the Supplemental Methods.
Whole exome variants were annotated using ANNOVAR (Wang et al. 2010) and dbNSFP v2.0 (Liu et al. 2013) according to the reference genome GRCh37 and National Center for Biotechnology Information RefSeq. Coding variants were annotated to a unique gene as well as the following categories used for inclusion in gene-based tests: splicing, stop-gain, stop-loss, nonsynonymous variants, and indels. WGS variation was annotated across the genome and functional domains using the Whole Genome Sequencing Annotation (WGSA) pipeline version 5 (Liu et al. 2016). Variants in eQTL were specified using GTEx annotation, V6p (GTEx Consortium 2015). All imputed gene expression levels had a q-value <0.05 (tissue-specific false-discovery rate), indicating a good fit to the model (Wang et al. 2016). Since the vast majority of tissue donors in GTEx were EAs (GTEx Consortium 2015), we limited this lipid-related metabolomics analysis of estimated gene expression to EAs.
Genotype-phenotype analyses
For each circulating lipid-related metabolite, observations outside of the 99% were winsorized, and the levels below the detectable limit of the assay were imputed to the lowest detected value in the whole sample. Prior to genetic analyses, each metabolite was investigated for its goodness-of-fit to normality. Suberate, azelate, sebacate, and undecanedioate were not transformed before analysis. Square root and negative square root transformations were applied to dodecanedioate and glycerol-3-phosphate levels, respectively. Other metabolites levels were natural log-transformed. Ethnic-specific analyses were performed for each lipid-related metabolite using additive genetic models, adjusting for age, sex, the first three principal components (PCs), batch and study site (as needed). Adjustment for estimated glomerular filtration rate was also performed (Levey et al. 2009), because some lipid-related traits were associated with this indicator of kidney function (Yu et al. 2014). Afterward, a transethnic meta-analysis was performed.
All analyses were carried out separately for common and rare variants. Single common variant meta-analysis (MAF > 5%) was performed using the inverse-variance-weighted fixed-effect method (Tang and Lin 2015) for individuals with WGS data. For rare variants, aggregate tests were conducted across functional motifs, because in either ethnic group any one variant was too rare to support single site analyses. Annotated functional motifs consisted of genes (coding regions), regulatory domains (including 3′UTR, 5′UTR, promoter and enhancer elements for each gene), and sliding windows (starting at position 0 bp for each chromosome, 4 kb in length, with a skip length of 2 kb) (Morrison et al. 2017). Gene-based analyses were the only analyses performed using the WES data, with splicing, stop-gain, stop-loss, nonsynonymous variants, and indels included in the aggregate tests. All aggregate tests contained only rare variants (MAF ≤ 5%). Meta-analyses of aggregate test results were performed using variants with pooled MAF ≤ 5%. Within each annotated functional motif, we carried out a burden test (Li and Leal 2008), which collapses variants into a single genetic score, using the T5 count method (https://cran.r-project.org/web/packages/seqMeta/vignettes/seqMeta.pdf). Burden tests have higher power than alternative sequence-kernel association tests if a large proportion of the rare variants in a region are causal, and have the same direction of the effect (Lee et al. 2012).
For each test, we used a two-step process to define statistical significance and transethnic consistency. In the first step, the result must be statistically significant in a meta-analysis across the two ethnic groups after Bonferroni correction for the number of statistical tests. For the single variant analysis, the P-value threshold was ≤ 2.45 × 10−10 [accounting for ∼2,000,000 independent variants (Pe’er et al. 2008) and 102 traits]; for the gene-based aggregate test the threshold was ≤ 3.04 × 10−8 (accounting for 16,117 genes and 102 metabolites); for the regulatory aggregate test, the threshold was ≤ 2.37 × 10−8 (accounting for 20,677 regulatory domains and 102 lipid compounds); for the sliding window aggregating analysis the threshold was ≤ 7.43 × 10−10 (accounting for 102 traits and 659,982 contiguous and nonoverlapping windows). Each aggregation unit had to have at least one rare variant in both AA and EA, with cumulative minor allele counts (cMAC) across the two ethnic groups ≥7 (Li et al. 2015). In the second step, statistically significant findings from the first step were considered consistent between ethnicities under the following conditions: (1) the effect estimate had consistent direction in both ethnic groups, and either (2) the P-value ≤ 1 × 10−5 in both ethnicities, or (3) the P-value reached an a priori definition of statistical significance used in the first step in any one ethnic group and was significant in the other group, after accounting for the number of significant findings in the first step. This value was 0.001, 0.006, 0.005, and 0.005 for the single locus (accounting for 48 findings), gene-based (accounting for nine findings), regulatory (accounting for 11 findings), and sliding window (accounting for 10 findings), respectively.
For significant functional motifs that were consistent between ethnic groups, analyses conditioning on the most significant rare variant were performed to identify whether the association with a leading rare variant could explain the detected association. The association between the genomic region and the lipid-related metabolite was considered to be novel if it was not reported by previous GWAS or sequence-based analysis, as verified with the GRASP search v2.0.0.0, GWAS catalog v1.0 and manual literature review. All analyses were carried out using the R seqMeta package (http://cran.r-project.org/web/packages/seqMeta/index.html, version 1.6.0).
PrediXcan was used to perform tissue-specific estimation of the genetically regulated gene expression levels of 14 unique genes, with the expression of each gene being estimated in up to 12 tissues with GTEx V6p (https://github.com/hakyimlab/PrediXcan downloaded on June 8, 2015) (Gamazon et al. 2015). PrediXcan analysis quantifies the association between the genetically regulated expression component of an individual’s gene expression level and the selected phenotype (Gamazon et al. 2015). Briefly, prediction models in PredictDB were built using data from multiple reference transcriptome studies, such as GTEx, GEUVADIS, DGN; 1000 Genomes imputed genotype data (including variants with MAF > 5% and imputation certainty >0.8) were used with the weights from PredictDB to impute gene expression levels for ARIC study participants (used variants are reported in Supplemental Material, Table S12). In EA participants, the association between the predicted gene expression and the lipid-related trait was performed using the general linear model (“glm”) in R, with lipid-related trait level as the outcome, and gene expression level, age, sex, the first three PCs and estimated glomerular filtration rate as covariates.
Data availability
Supporting data for the ARIC cohort is available via dbGaP study accession phs000280. The summary statistics for this paper are deposited into the dbGaP CHARGE Summary Results site (Rich et al. 2016) (dbGaP Study Accession: phs000930). Figure S1 shows proportion of single nucleotide variants by minor allele frequency for sequencing data. Figure S2 contains Manhattan plots and QQ-plots for the statistically significant traits with transethnic consistency. Figure S3 demonstrates distribution of rare variants in EAs and AAs. Figure S4 pictures predicted levels of FADS1 and FADS2 in skeletal muscle by rs174564 genotypes in EAs. Table S1 contains basic characteristics of the 102 lipid-related traits. Common variants significantly associated with lipid-related traits in EAs, AAs, or in meta-analysis can be found in Table S2, with sentinel common variant-region pairs significantly associated in meta-analysis available in Table S3. Association of predicted gene transcription levels with lipid-related phenotypes in EAs is presented in Table S4. Table S5 contains Whole Genome T5 results significantly associated in meta-analysis. Rare nonsynonymous exonic variants of SLC10A1-glycocholate are listed in Table S6. Table S7 contains regulatory domain T5 results significantly associated in meta-analysis. Rare variants and conditional analyses for the regulatory element of CYP3A43 for three traits are available in Tables S8 and S9, respectively. Table S10 has sliding windows T5 results significantly associated in meta-analysis, with rare variants of the significant and consistent across ethnicities sliding windows listed in Table S11. Table S12 contains variants used for gene transcription levels imputation in EAs. Institutional Review Board registration numbers: HSC-SPH-09-0490; HSC-SPH-09-0494. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6050099.
Results
Sample characteristics
The study sample size comprised 3424 individuals, 1872 AAs, and 1552 EAs. Individuals tended to be middle aged, with a greater proportion of females than males in both EAs and AAs. Characteristics of the study participants as well as the basic descriptive statistics for 102 lipid-related traits measured as part of the metabolomic profile for the sample of individuals having WGS data are summarized in Table S1. Among the study participants, there were 462,292 exonic variants, and 60,699,789 variants in the whole genomes. Figure S1, A–D shows the distribution of variants by frequency.
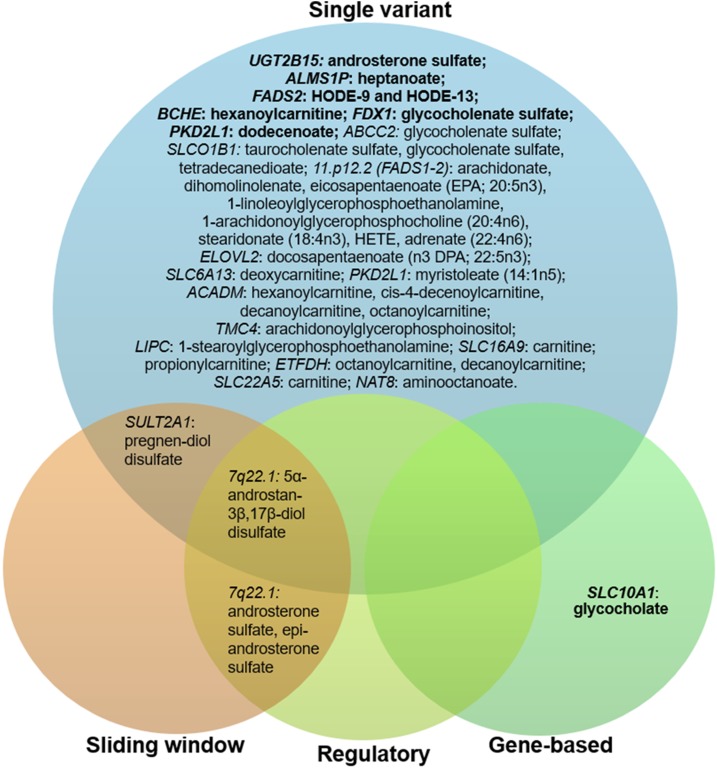
We used four analytical strategies to assess associations between the lipid-related traits and genomic variants: (1) single site common variant analysis; (2) gene-based rare variant analysis; (3) regulatory element rare variant analysis; and (4) a sliding window rare variant analysis. A total of 38 metabolite-region relationships were identified to be consistent across ethnicities with 31 associations reported previously and seven novel findings (Figure S2 contains Manhattan plots and QQ-plots for the seven lipid-related traits identified to be associated with novel regions for each of the ethnic groups and for the meta-analysis). The distributions of statistically significant lipid-related metabolite-region pairs among all analytical strategies are presented in Figure 1; four pairs were detected by more than one method. In the following paragraphs, we present the novel significant and consistent findings for each analytical strategy.
Figure 1.
An overview of identified consistent significant genetic association with lipid-related metabolites. Gene names in bold indicate novel findings, consistent between ethnicities; regular font indicate findings that were reported in previous studies.
Association with common variants
We conducted a whole genome survey of all common variants in AAs and EAs (MAF > 5%), followed by a multiethnic meta-analysis (Table S2). We report sentinel statistically significant lipid-related metabolite-genomic region pairs for common variants in Table S3. Among a total of 48 detected lipid-related metabolite-variant pairs, 40 pairs were reported previously. Six out of eight novel findings had evidence of consistency between the ethnic groups, and these novel consistent significant metabolite-variant pairs are shown in Table 1.
Table 1. Sentinel novel common SNP-metabolite pairs (MAF > 5%) significantly associated in meta-analyses (P ≤ 2.45 × 10−10), consistent between ethnic groups.
| Trait | Closest Gene | Chr:Position | SNP Name | R/Aa | P-Value | Beta | Location |
|---|---|---|---|---|---|---|---|
| Androsterone sulfate | UGT2B15 | 4:69491183 | rs13121671 | G/T | 8.91 × 10−15 | 0.19 | Intergenic |
| 5-Dodecenoate | PKD2L1 | 10:102075479 | rs603424 | G/A | 1.54 × 10−10 | −0.09 | Second intron |
| Glycocholenate sulfate | FDX1 | 11:110241999 | rs2051466 | C/A | 1.25 × 10−11 | 0.08 | Intergenic |
| Heptanoate | ALMS1P | 2:73883072 | rs13408433 | G/A | 4.28 × 10−12 | −0.04 | First intron |
| Hexanoyl-carnitine | BCHE | 3:165481945 | rs73165061 | G/A | 1.91 × 10−13 | 0.14 | Intergenic |
| 13-HODE + 9-HODE | FADS2 | 11:61588305 | rs174564 | A/G | 4.13 × 10−16 | 0.11 | Upstream |
Reference allele/Alternative allele.
All of the six novel findings consistent between the ethnicities lie in expression quantitative trait loci (eQTL) influencing the expression levels of 14 surrounding genes in multiple tissues (GTEx Consortium 2015) (Table S3). We imputed tissue-specific genetically regulated gene expression levels for each of the above genes (Table S3), and then investigated the relationship between each of the estimated gene expression levels and the corresponding lipid-related trait (Table S4). Out of 14 genes tested, 10 reached the predefined statistical significance threshold (P-value ≤ 0.00086, accounting for 58 tests corresponding to 58 gene-tissue pairs) in at least one tissue tested (Table 2). These 10 significant associations were found in four of the six lipid-related traits tested.
Table 2. Leading estimated gene transcription levels-metabolite pairs in EAs.
| Trait | Gene | Tissue | Beta | P-value |
|---|---|---|---|---|
| Androsterone sulfate | UGT2B15 | Transverse colon | 0.65 | 0.17 |
| Androsterone sulfate | UGT2B17 | Lung | 0.33 | 0.36 |
| 5-Dodecenoate | SEMA4G | Nerve tibial | 0.24 | 1.9 × 10−3 |
| 5-Dodecenoate | SCD | Adipose subcutaneous | 0.28 | 5.14 × 10−3 |
| Glycocholenate sulfate | FDX1 | Whole blood | 0.31 | 2.93 × 10−6 |
| Heptanoate | NAT8 | Skin sun exposed lower leg | −0.20 | 2.80 × 10−9 |
| Heptanoate | TPRKB | Artery tibial | 0.15 | 1.39 × 10−6 |
| Heptanoate | ALMS1 | Pancreas | −0.08 | 1.49 × 10−5 |
| Hexanoylcarnitine | BCHE | Heart atrial appendage | −0.75 | 9.57 × 10−6 |
| 13-HODE + 9-HODE | FADS1 | Cerebellum | −0.17 | 3.56 × 10−9 |
| 13-HODE + 9-HODE | FADS2 | Muscle skeletal | 0.34 | 2.85 × 10−7 |
| 13-HODE + 9-HODE | TMEM258 | Muscle skeletal | 0.75 | 1.12 × 10−8 |
| 13-HODE + 9-HODE | BEST1 | Heart atrial appendage | 0.33 | 2.29 × 10−5 |
| 13-HODE + 9-HODE | DAGLA | Cells transformed fibroblasts | −0.26 | 7.72 × 10−4 |
Gene-based results
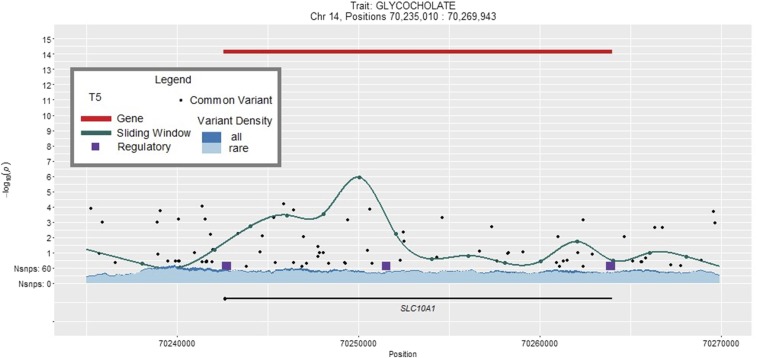
There were 16,117 genes (cMAC across the two ethnic groups ≥7) with at least one annotated variant (splicing, stop-gain, stop-loss, nonsynonymous variant, or an indel) in both AAs and EAs, and the distribution of variants ranged from 1 to 1798, with a median cMAC of 140 per gene (Figure S3A). Results of the aggregate gene-based tests that reached the predefined significance levels in the multiethnic meta-analysis (P-value ≤ 3.04 × 10−8) are presented in Table S5, and contained nine lipid-related metabolite-gene associations. Among four novel findings, one met the prespecified criteria for transethnic consistency. Aggregation of 29 rare coding variants in SLC10A1 was associated with 77% increase in glycocholate levels (P-valueMETA = 6.51 × 10−15) (Table S5), with 21 variants in AAs (cMAF = 5.6%), and 11 variants in EAs (cMAF = 0.5%). Overall, four variants in AAs and two variants in EAs had P-value ≤ 0.05 (Table S6), with no individual variants shared by the two ethnic groups. A graphical representation of the results of the four analytical strategies as applied to SLC10A1 is shown in Figure 2, where the low P-value for the gene-based analysis can be clearly seen.
Figure 2.
Results of meta-analyses of glycocholate levels and SLC10A1 by four methods.
Regulatory domain-based results
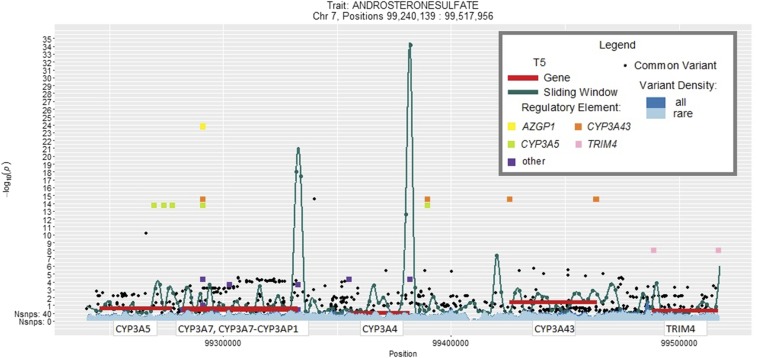
Among the 20,677 annotated regulatory domains (meta-analysis cMAC ≥7) that have at least one annotated variant in both AA and EA, we observed a distribution of 1–843 variants per domain and a median cMAC of 762 (Figure S3B). Statistically significant multiethnic meta-analysis burden test results (P-value ≤ 2.37 × 10−8) for the regulatory domains are shown in Table S7. Three significant consistent lipid-related metabolite-genetic region associations were observed involving steroid metabolites and regulatory domains for the gene CYP3A43 (Table 3). The aggregation test for this gene contained 5′UTR, 3′UTR and nearby enhancer elements (Table S8). All three significant sulfated steroids with transethnic consistency (androsterone sulfate, epiandrosterone sulfate, 5α-androstan-3β,17β-diol disulfate) are dehydroepiandrosterone metabolites (Labrie et al. 2005), which had a moderate-to-strong correlation among their levels (0.67 < r < 0.95). Figure 3 displays the detected region and the results for androsterone sulfate levels. There is a single variant (rs118168183) in an annotated enhancer located in the last intron of the CYP3A7-CYP3AP1 transcript that is acting as a lead or driver SNP for the burden tests (MAFEA = 0.023; P-valueEA ≤ 5 × 10−25 and MAFAA = 0.004; P-valueAA ≤ 2.95 × 10−4 for all three sulfated steroids) (Table S8). Conditioning on this lead SNP greatly attenuated the observed burden test P-values so that they were no longer significant (P > 0.05) (Table S9).
Table 3. Regulatory domains significantly associated (P ≤ 2.37 × 10−8) in meta-analysis and consistent between ethnic groups.
| Trait | Chr | Gene | cMaf | #Var | P-Value | Beta |
|---|---|---|---|---|---|---|
| 5α-androstan-3β,17β-diol disulfate | 7 | CYP3A43 | 0.052 | 8 | 1.06 × 10−12 | −0.371 |
| Androsterone sulfate | 7 | CYP3A43 | 0.052 | 8 | 3.33 × 10−15 | −0.388 |
| Epiandrosterone sulfate | 7 | CYP3A43 | 0.052 | 8 | 5.11 × 10−10 | −0.243 |
Figure 3.
Results of meta-analyses of androsterone sulfate levels and CYP3A43 by four methods.
Sliding window-based results
A total of 1,319,963 4-kb overlapping windows (meta-analysis cMAC ≥7) with at least one rare variant in both AA and EA, had a distribution of 1–749 variants per window (Figure S3C) with median cMAC of 1656. Multiethnic burden test meta-analyses revealed 10 statistically significant (P-value ≤ 7.43 × 10−10) lipid-related metabolite-region pairs, including two novel findings. Four region-metabolite pair associations were consistent between ethnicities (Figure 1 and Table S10). Aggregation of rare variants in a window, located downstream of SULT2A1 was associated with decreased pregnen-diol disulfate levels (P-valueMETA = 1.28 × 10−16) (Table S10). The window contains a total of 96 rare and low frequency variants, with 64 variants in AA, and 57 variants in EA (Table S11). Overall, 15 variants in AAs and 12 variants in EAs had a P-value ≤ 0.05, with seven variants belonging to a 3′UTR, and six variants—shared between the ethnicities. The most statistically significant variant among all the variants, rs296383, belongs to an eQTL increasing SULT2A1 expression in the adrenal gland (GTEx Consortium 2015).
Discussion
We performed a multiethnic study using WGS and WES data to identify loci influencing a broad spectrum of the lipid-related metabolome. Seven novel lipid-related metabolite-region significant signals with transethnic consistency were identified, with six associations from single variant tests and one association from gene-based tests. All six consistent significant single variant tests resided in the nonprotein-encoding region of the genome. There was one significant and consistent association influencing three sulfated steroids that was identified by analysis of annotated regulatory elements.
The analysis strategy to identify novel discoveries in this study is, by its very nature, conservative because it was defined a priori to include three criteria: (1) sufficient sequence variation in both ethnic groups, (2) statistical and nominal significance in both groups, and (3) consistency of the direction of effect. It is possible, indeed likely, that statistically significant findings in one ethnic group are real or true findings, but were not consistent according to the criterion set forth here because of lack of sufficient genetic variation in the other group. For example, although the association between glycochenodeoxycholate and SLC10A1 does not meet our criteria for consistency, this is likely due to the fact that the variant leading the association in AAs (rs61745930) is monomorphic in the sample of EAs. At the same time, we observe consistent direction of the effect of glycochenodeoxycholate-SLC10A1 pair between ethnicities and with the glycocholate-SLC10A1 association (declared as novel in this manuscript), and, as mentioned before, both glycochenodeoxycholate and glycocholate are biological substrates for SLC10A1 (Mita et al. 2005). Another reason for lack of consistency between the groups may be gene by environment interactions, and different environments between groups. Therefore, the significant and consistent locus-metabolite pairs reported here are likely to be true or real findings. Of the 38 significant and consistent region-metabolite pairs, seven were deemed novel by virtue of not being reported by previous GWAS or candidate gene studies. The biology of the significant, consistent, and novel findings are discussed below.
The results presented here improve our understanding of genetic influences on serum fatty acyl levels. Measured together, 9-hydroxyoctadecadienoic acid (9-HODE) and 13-HODE, the products of linoleic acid oxygenation by 12-lipoxygenase and 15-lipoxygenase, respectively (Cabral et al. 2014; Vangaveti et al. 2016), were significantly associated with a variant located in the first intron of the gene encoding fatty acid desaturase 2 (FADS2; rs174564, Table 1). Rs174564 is an eQTL associated with increased expression levels of FADS2, and decreased expression of FADS1 in multiple tissues (GTEx Consortium 2015). Both FADS1 and FADS2 are desaturases, involved in polyunsaturated fatty acids metabolism. Downregulation of this pathway may lead to activation of an alternative pathway of linoleic acid metabolism—such as conversions to various HODEs (Choque et al. 2014). This hypothesized mechanism may explain the observed association of rs174564 with increased levels of HODEs and is supported by data from Long et al. (2017) showing that the same variant is associated with structural derivatives of arachidonic and linoleic acids. Our analyses of predicted gene expression suggest that genetically regulated FADS1 expression level is associated with decreased HODEs levels, while FADS2 is associated with increased HODEs levels (Figure S4 and Table 2).
A common intronic variant (rs603424) in PKD2L1 was associated with decreased 5-dodecenoate levels, a conjugated base of the unsaturated fatty acid, dodecenoic acid (Feng and Cronan 2009). This variant previously was reported to be associated with decreased 5-dodecenoate levels (Long et al. 2017), and resides in an eQTL decreasing expression levels of the nearby gene SCD in adipose tissues (GTEx Consortium 2015). SCD encodes a stearoyl-CoA desaturase, a rate-limiting enzyme in the biosynthesis of unsaturated fatty acids from saturated fatty acids (Poudyal and Brown 2011).
A gene-based burden-test revealed an association between rare coding variants in a sodium-bile acid cotransporter SLC10A1 (Hallén et al. 2002), with increased levels of glycocholate (Table S5). SLC10A1 takes both glycocholate and glycochenodeoxycholate, with which a suggestive association was observed, as its substrates, as well as several other bile salts (Mita et al. 2005).
The results also provide insights into genetic regulation of sterol lipids. For example, a novel intergenic variant 58 kbp upstream of FDX1 was detected to be associated with increased levels of glycocholenate sulfate (Table S3). FDX1 participates in transport of electrons to mitochondrial cytochrome P450, which is involved in bile acid metabolism (Miller 2005). Rs2051466 lies in an eQTL, increasing the expression of FDX1 (GTEx Consortium 2015), and was recently reported to be suggestively associated with glycocholenate sulfate levels in a European population (Long et al. 2017).
The levels of serum steroid hormones were also investigated. Multiethnic meta-analysis revealed a novel intergenic variant 21 kbp downstream of UGT2B15 to be associated with increased androsterone sulfate levels. Rs13121671 lies in an eQTL decreasing the expression of UGT2B15 and/or UGT2B17 in several tissues (GTEx Consortium 2015). These genes encode UDP-glycosyltransferases, which catalyze the glucuronidation of endogenous androgens (Gauthier-Landry et al. 2015), thus affecting their clearance from the circulation (Yong et al. 2010). Decreased glucuronidation of androsterone may potentially lead to a compensatory increase in an alternative clearance mechanism, such as sulfation (Schulze et al. 2011).
A regulatory domain of CYP3A43 was associated with decreased levels of three sulfated steroids: androsterone sulfate, α-androstan-3-β-17-β-diol disulfate and epiandrosterone sulfate. It is located in a complex multigene region on chromosome 7, with several variants in surrounding genes previously reported to be associated with detected sulfated steroids (Shin et al. 2014). CYP3A43 is an oxidoreductase, that can act as testosterone 6β-hydroxylase (Domanski et al. 2001), leading to steroid hormone deactivation (Han et al. 2015). The association is driven primarily by a single variant (rs118168183) located in the last intron of CYP3A7-CYP3AP1. Rs118168183 belongs to an established enhancer and could potentially affect the transcription of several genes, including CYP3A43, PTCD1, CYP3A4, AZGP1, and CYP3A5 (Andersson et al. 2014).
The analysis strategy and the data presented here provide further insight and information about the benefits of WGS data relative to WES data for the analysis of common complex phenotypes that underlie the majority of morbidity and mortality in the US population. The tension becomes especially taught when taking into account the cost of WGS, which includes not only the sequencing itself, but also substantial data processing and data storage costs. Despite the vast majority of GWAS signals residing in noncoding regions of the genome, there are only a handful of examples of insight gained from the investigation of whole genome sequence for a common complex phenotype or disease (Fuchsberger et al. 2016; Morrison et al. 2017). There are even fewer examples where the mechanisms of the nongenic association are well-understood. In the data presented here all of the novel consistent between ethnicities common variants reside outside of protein encoding exons and are in eQTLs affecting the expression of the genes involved in crucial biological processes, such as inflammation and bile acids metabolism, in multiple tissues (GTEx Consortium 2015). These results underscore the importance of integrating WGS analyses with nongenic annotation such as ENCODE, FANTOM, and GTEx.
In summary, we identified seven novel consistent lipid-related metabolite-genomic region pairs. Discovered loci, identified by either single variant or aggregation tests, lie in or near the genes involved in transport (SLC10A1) and metabolism (SCD, FDX1, UGT2B15, UGT2B17, FADS1, and FADS2) of lipid-related metabolites. Most of the significant and consistent findings belonged to presumed regulatory regions near annotated protein-encoding genes, emphasizing the importance of investigating noncoding regions and applying versatile analytical approaches to improve our understanding of the genetic architecture of quantitative traits.
Acknowledgments
The authors thank the staff and participants of the Atherosclerosis Risk in Communities (ARIC) study for their important contributions. The ARIC study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Funding support for “Building on GWAS for NHLBI-diseases: the US CHARGE consortium” was provided by the National Institutes of Health (NIH) through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Metabolomics measurements were sponsored by the National Human Genome Research Institute (3U01HG004402-02S1). Sequencing was carried out at the Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 and R01HL086694).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6050099.
Communicating editor: E. Hauser
Literature Cited
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., et al. , 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge M. N., Wang M., Wu Y., Newsham I., Muzny D. M., et al. , 2011. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 12: R68 10.1186/gb-2011-12-7-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaanse E. M., Hold K. M., Van der Laarse A., 1997. The effect of membrane cholesterol content on ion transport processes in plasma membranes. Cardiovasc. Res. 33: 272–283. 10.1016/S0008-6363(96)00193-9 [DOI] [PubMed] [Google Scholar]
- Cabral M., Martin-Venegas R., Moreno J. J., 2014. Differential cell growth/apoptosis behavior of 13-hydroxyoctadecadienoic acid enantiomers in a colorectal cancer cell line. Am. J. Physiol. Gastrointest. Liver Physiol. 307: G664–G671. 10.1152/ajpgi.00064.2014 [DOI] [PubMed] [Google Scholar]
- Calder P. C., 2015. Functional roles of fatty acids and their effects on human health. JPEN J. Parenter. Enteral Nutr. 39(1 suppl.): 18S–32S. 10.1177/0148607115595980 [DOI] [PubMed] [Google Scholar]
- Challis D., Yu J., Evani U. S., Jackson A. R., Paithankar S., et al. , 2012. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 13: 8 10.1186/1471-2105-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choque B., Catheline D., Rioux V., Legrand P., 2014. Linoleic acid: between doubts and certainties. Biochimie 96: 14–21. 10.1016/j.biochi.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Cohen J., 1988. Statistical Power Analysis for the Behavioral Sciences. L. Erlbaum Associates, Hillsdale, NJ. [Google Scholar]
- Demirkan A., van Duijn C. M., Ugocsai P., Isaacs A., Pramstaller P. P., et al. , 2012. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 8: e1002490 10.1371/journal.pgen.1002490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries P. S., Yu B., Feofanova E. V., Metcalf G. A., Brown M. R., et al. , 2017. Whole-genome sequencing study of serum peptide levels: the Atherosclerosis Risk in Communities study. Hum. Mol. Genet. 26: 3442–3450. 10.1093/hmg/ddx266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski T. L., Finta C., Halpert J. R., Zaphiropoulos P. G., 2001. cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450. Mol. Pharmacol. 59: 386–392. 10.1124/mol.59.2.386 [DOI] [PubMed] [Google Scholar]
- Draisma H. H. M., Pool R., Kobl M., Jansen R., Petersen A. K., et al. , 2015. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 6: 7208 10.1038/ncomms8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab A. W., Scaglia F., 2015. Disorders of carnitine biosynthesis and transport. Mol. Genet. Metab. 116: 107–112. 10.1016/j.ymgme.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Evans A. M., DeHaven C. D., Barrett T., Mitchell M., Milgram E., 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81: 6656–6667. 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., et al. , 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50(Suppl.): S9–S14. 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E., Cotter D., Sud M., Subramaniam S., 2011. Lipid classification, structures and tools. Biochim. Biophys. Acta 1811: 637–647. 10.1016/j.bbalip.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Cronan J. E., 2009. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284: 29526–29535. 10.1074/jbc.M109.023440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier S. W., Dallinga-Thie G. M., Meijers J. C., Zelcer N., Kastelein J. J., et al. , 2014. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ. Res. 115: 552–555. 10.1161/CIRCRESAHA.115.304660 [DOI] [PubMed] [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., et al. , 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. 10.1038/nature18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E. R., Wheeler H. E., Shah K. P., Mozaffari S. V., Aquino-Michaels K., et al. , 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47: 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Landry L., Belanger A., Barbier O., 2015. Multiple roles for UDP-glucuronosyltransferase (UGT)2B15 and UGT2B17 enzymes in androgen metabolism and prostate cancer evolution. J. Steroid Biochem. Mol. Biol. 145: 187–192. 10.1016/j.jsbmb.2014.05.009 [DOI] [PubMed] [Google Scholar]
- GTEx Consortium , 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallén S., Mareninova O., Brändén M., Sachs G., 2002. Organization of the membrane domain of the human liver sodium/bile acid cotransporter. Biochemistry 41: 7253–7266. 10.1021/bi012152s [DOI] [PubMed] [Google Scholar]
- Han J. H., Lee Y. S., Kim H. J., Lee S. Y., Myung S. C., 2015. Association between cytochrome CYP17A1, CYP3A4, and CYP3A43 polymorphisms and prostate cancer risk and aggressiveness in a Korean study population. Asian J. Androl. 17: 285–291. 10.4103/1008-682X.133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten S. M., Watanabe M., Auwerx J., 2006. Endocrine functions of bile acids. EMBO J. 25: 1419–1425. 10.1038/sj.emboj.7601049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., et al. , 2010. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42: 137–141. 10.1038/ng.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Hernandez M. L., Balla T., 2013. Inositol lipid regulation of lipid transfer in specialized membrane domains. Trends Cell Biol. 23: 270–278. 10.1016/j.tcb.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie F., Luu-The V., Belanger A., Lin S. X., Simard J., et al. , 2005. Is dehydroepiandrosterone a hormone? J. Endocrinol. 187: 169–196. 10.1677/joe.1.06264 [DOI] [PubMed] [Google Scholar]
- Lee S., Wu M. C., Lin X., 2012. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 13: 762–775. 10.1093/biostatistics/kxs014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey A. S., Stevens L. A., Schmid C. H., Zhang Y. L., Castro A. F., III, et al. , 2009. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 150: 604–612. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. H., Morrison A. C., Kovar C., Cupples L. A., Brody J. A., et al. , 2015. Analysis of loss-of-function variants and 20 risk factor phenotypes in 8,554 individuals identifies loci influencing chronic disease. Nat. Genet. 47: 640–642. 10.1038/ng.3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321. 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jian X., Boerwinkle E., 2013. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34: E2393–E2402. 10.1002/humu.22376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., White S., Peng B., Johnson A. D., Brody J. A., et al. , 2016. WGSA: an annotation pipeline for human genome sequencing studies. J. Med. Genet. 53: 111–112. 10.1136/jmedgenet-2015-103423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T., Hicks M., Yu H.-C., Biggs W. H., Kirkness E. F., et al. , 2017. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49: 568–578. 10.1038/ng.3809 [DOI] [PubMed] [Google Scholar]
- Maldonado-Valderrama J., Wilde P., Macierzanka A., Mackie A., 2011. The role of bile salts in digestion. Adv. Colloid Interface Sci. 165: 36–46. 10.1016/j.cis.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Miller W. L., 2005. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology 146: 2544–2550. 10.1210/en.2005-0096 [DOI] [PubMed] [Google Scholar]
- Mita S., Suzuki H., Akita H., Stieger B., Meier P. J., et al. , 2005. Vectorial transport of bile salts across MDCK cells expressing both rat Na+-taurocholate cotransporting polypeptide and rat bile salt export pump. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G159–G167. 10.1152/ajpgi.00360.2003 [DOI] [PubMed] [Google Scholar]
- Morrison A. C., Voorman A., Johnson A. D., Liu X., Yu J., et al. , 2013. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat. Genet. 45: 899–901. 10.1038/ng.2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. C., Huang Z., Yu B., Metcalf G., Liu X., et al. , 2017. Practical approaches for whole-genome sequence analysis of heart- and blood-related traits. Am. J. Hum. Genet. 100: 205–215. 10.1016/j.ajhg.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemsiri V., Wang X., Pirim D., Radwan Z. H., Bunker C. H., et al. , 2015. Genetic contribution of SCARB1 variants to lipid traits in African Blacks: a candidate gene association study. BMC Med. Genet. 16: 106 10.1186/s12881-015-0250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Masutomi N., Tsutsui N., Sakairi T., Mitchell M., et al. , 2009. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol. Pathol. 37: 521–535. 10.1177/0192623309336152 [DOI] [PubMed] [Google Scholar]
- Pe'er I., Yelensky R., Altshuler D., Daly M. J., 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32: 381–385. 10.1002/gepi.20303 [DOI] [PubMed] [Google Scholar]
- Peloso G. M., Auer P. L., Bis J. C., Voorman A., Morrison A. C., et al. , 2014. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 94: 223–232. 10.1016/j.ajhg.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollin T. I., Damcott C. M., Shen H., Ott S. H., Shelton J., et al. , 2008. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science 322: 1702–1705. 10.1126/science.1161524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudyal H., Brown L., 2011. Stearoyl-CoA desaturase: a vital checkpoint in the development and progression of obesity. Endocr. Metab. Immune Disord. Drug Targets 11: 217–231. 10.2174/187153011796429826 [DOI] [PubMed] [Google Scholar]
- Reue K., Brindley D. N., 2008. Thematic review series: glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49: 2493–2503. 10.1194/jlr.R800019-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich S. S., Wang Z. Y., Sturcke A., Ziyabari L., Feolo M., et al. , 2016. Rapid evaluation of phenotypes, SNPs and results through the dbGaP CHARGE Summary Results site. Nat. Genet. 48: 702–703. 10.1038/ng.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. J., Thorngren J. O., Garle M., Ekstrom L., Rane A., 2011. Androgen sulfation in healthy UDP-glucuronosyl transferase 2B17 enzyme-deficient men. J. Clin. Endocrinol. Metab. 96: 3440–3447. 10.1210/jc.2011-0521 [DOI] [PubMed] [Google Scholar]
- Shin S. Y., Fauman E. B., Petersen A. K., Krumsiek J., Santos R., et al. , 2014. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46: 543–550. 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. D., 2006. Oxford Dictionary of Biochemistry and Molecular Biology. Oxford University Press, Oxford. [Google Scholar]
- Tang Z. Z., Lin D. Y., 2015. Meta-analysis for discovering rare-variant associations: statistical methods and software programs. Am. J. Hum. Genet. 97: 35–53. 10.1016/j.ajhg.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., et al. , 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ARIC investigators. 1989. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 129: 687–702. 10.1093/oxfordjournals.aje.a115184 [DOI] [PubMed] [Google Scholar]
- Vangaveti V. N., Jansen H., Kennedy R. L., Malabu U. H., 2016. Hydroxyoctadecadienoic acids: oxidised derivatives of linoleic acid and their role in inflammation associated with metabolic syndrome and cancer. Eur. J. Pharmacol. 785: 70–76. 10.1016/j.ejphar.2015.03.096 [DOI] [PubMed] [Google Scholar]
- Vítek L., Haluzí M.k 2016. The role of bile acids in metabolic regulation. J. Endocrinol. 228: R85–R96. 10.1530/JOE-15-0469 [DOI] [PubMed] [Google Scholar]
- Wang J., Gamazon E. R., Pierce B. L., Stranger B. E., Im H. K., et al. , 2016. Imputing gene expression in uncollected tissues within and beyond GTEx. Am. J. Hum. Genet. 98: 697–708. 10.1016/j.ajhg.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong M., Schwartz S. M., Atkinson C., Makar K. W., Thomas S. S., et al. , 2010. Associations between polymorphisms in glucuronidation and sulfation enzymes and mammographic breast density in premenopausal women in the United States. Cancer Epidemiol. Biomarkers Prev. 19: 537–546. 10.1158/1055-9965.EPI-09-0898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Zheng Y., Nettleton J. A., Alexander D., Coresh J., et al. , 2014. Serum metabolomic profiling and incident CKD among African Americans. Clin. J. Am. Soc. Nephrol. 9: 1410–1417. 10.2215/CJN.11971113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., de Vries P. S., Metcalf G. A., Wang Z., Feofanova E. V., et al. , 2016a Whole genome sequence analysis of serum amino acid levels. Genome Biol. 17: 237 10.1186/s13059-016-1106-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Li A. H., Metcalf G. A., Muzny D. M., Morrison A. C., et al. , 2016b Loss-of-function variants influence the human serum metabolome. Sci. Adv. 2: e1600800 10.1126/sciadv.1600800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supporting data for the ARIC cohort is available via dbGaP study accession phs000280. The summary statistics for this paper are deposited into the dbGaP CHARGE Summary Results site (Rich et al. 2016) (dbGaP Study Accession: phs000930). Figure S1 shows proportion of single nucleotide variants by minor allele frequency for sequencing data. Figure S2 contains Manhattan plots and QQ-plots for the statistically significant traits with transethnic consistency. Figure S3 demonstrates distribution of rare variants in EAs and AAs. Figure S4 pictures predicted levels of FADS1 and FADS2 in skeletal muscle by rs174564 genotypes in EAs. Table S1 contains basic characteristics of the 102 lipid-related traits. Common variants significantly associated with lipid-related traits in EAs, AAs, or in meta-analysis can be found in Table S2, with sentinel common variant-region pairs significantly associated in meta-analysis available in Table S3. Association of predicted gene transcription levels with lipid-related phenotypes in EAs is presented in Table S4. Table S5 contains Whole Genome T5 results significantly associated in meta-analysis. Rare nonsynonymous exonic variants of SLC10A1-glycocholate are listed in Table S6. Table S7 contains regulatory domain T5 results significantly associated in meta-analysis. Rare variants and conditional analyses for the regulatory element of CYP3A43 for three traits are available in Tables S8 and S9, respectively. Table S10 has sliding windows T5 results significantly associated in meta-analysis, with rare variants of the significant and consistent across ethnicities sliding windows listed in Table S11. Table S12 contains variants used for gene transcription levels imputation in EAs. Institutional Review Board registration numbers: HSC-SPH-09-0490; HSC-SPH-09-0494. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6050099.