New mutations are rare, which makes their discovery laborious and time-consuming. Arthur and Bennetzen describe an approach for enriching recent mutations that relies only on a reference genome sequence and resequencing data for other...
Keywords: de novo mutation, indels, mutation, mutation enrichment, transition, transversion
Abstract
Genome comparisons provide information on the nature of genetic change, but such comparisons are challenged to differentiate the importance of the actual sequence change processes relative to the role of selection. This problem can be overcome by identifying changes that have not yet had the time to undergo millions of years of natural selection. We describe a strategy to discover accession-specific changes in the rice genome using an abundant resource routinely provided for many genome analyses, resequencing data. The sequence of the fully sequenced rice genome from variety Nipponbare was compared to the pooled (∼114×) resequencing data from 126 japonica rice accessions to discover “Nipponbare-specific” sequences. Analyzing nonrepetitive sequences, 8504 “candidate” Nipponbare-specific changes were detected, of which around two-thirds are true novel sequence changes and the rest are predicted genome sequencing errors. Base substitutions outnumbered indels in this data set by > 28:1, with ∼8:5 bias toward transversions over transitions, and no transposable element insertions or excisions were observed. These results indicate that the strategy employed is effective for finding recent sequence changes, sequencing errors, and rare alleles in any organism that has both a reference genome sequence and a wealth of resequencing data.
ONE of the primary questions in biology is the origin of genetic change. Evolutionary biologists routinely use comparative genomic analyses to identify the changes that differentiate individuals within a species or between species. However, these methods uncover variations that are the outcomes of multiple phenomena, including rates and natures of de novo mutation, natural selection acting on these changes, and transmission issues associated with mating strategies, population sizes, and geographical distributions. Mutation may arise due to spontaneous or environmentally driven base modification, errors during DNA replication, inaccurate DNA repair, transposon insertion/deletion, or chromosome breakage (Burrus and Waldor 2004; Aminetzach et al. 2005). Multiple DNA repair mechanisms work in concert to minimize change, such that the tens of thousands of DNA changes generated every cell generation still yield mutation rates of only to per base per organismal generation (Vazquez et al. 2000; Ossowski et al. 2010; Roach et al. 2010; Lee et al. 2012). For instance, in the model plant Arabidopsis, with just over 108 bp in its nuclear genome, around one de novo mutation is expected to be transmitted in a single plant generation (Ossowski et al. 2010).
The rate at which mutations occur can vary within or between species, and taxa also differ in the relative frequencies of types of mutation, although point mutations (both substitutions and tiny indels) are routinely far more common than larger indels (Drake et al. 1998; Vazquez et al. 2000; Ossowski et al. 2010; Roach et al. 2010; Walser and Furano 2010; Lee et al. 2012). Within genomes, genic regions exhibit a lower number of accumulated mutations, at least partly due to the fact that coding sequences are usually subject to purifying selection (Drake et al. 1998). Regions in genomes that are methylated, such as CG dinucleotides in many animals and all plants, also display a higher point mutation rate because 5-methyl cytosine deaminates at a higher rate than unmethylated cytosine, leading to frequent cytosine-to-thymidine transitions (Ma and Bennetzen 2004; Walser and Furano 2010; Wang et al. 2012). Taxa with active transposable elements (TEs) can accumulate dozens of de novo insertion mutations per generation, while sister lineages with quiescent TEs can go thousands, perhaps millions, of years without any new TE insertions (International Rice Genome Sequencing Project 2005; Huang et al. 2012; Kawahara et al. 2013).
While most mutation analysis studies have focused on changes that have accumulated over evolutionary time, few studies have investigated de novo change because of the cost and temporal demands of such investigations. Estimation of the spontaneous mutation rate in Escherichia coli was de novo changes per genome per generation, with point mutations outnumbering indels by > 9:1 (Lee et al. 2012). In Schizosaccharomyces pombe, the rate of point mutations was bases/generation (Behringer and Hall 2016). In humans, sperm DNA sequencing was utilized to investigate de novo DNA change and predicted a mutation rate of mutations/base/generation (Wang et al. 2012). In the plant kingdom, Ossowski and colleagues conducted a mutation-accumulation study in Arabidopsis thaliana that discovered 99 new base substitutions and 17 indels that had accumulated in 5 lineages within 30 generations, yielding an overall mutation rate of for point mutations/base/generation and for insertions and deletions/base/generation, respectively (Ossowski et al. 2010).
Rather than spend several plant generations creating mutation-accumulation lines (Drake et al. 1998; Vazquez et al. 2000; Ossowski et al. 2010; Roach et al. 2010; Lee et al. 2012) and then demanding deep full-genome sequencing to identify/confirm any de novo mutations, we have chosen to utilize currently available genome data to enrich for de novo mutations without any investment of plant growth time or sequencing expense. We have chosen to undertake this initial study with Nipponbare, the rice cultivar that was the target of the first high-quality reference genome sequence for Oryza sativa (International Rice Genome Sequencing Project 2005; Kawahara et al. 2013). Because Nipponbare is a unique cultivar, it has accumulated de novo mutations in the generations that it has been separate from any other rice germplasm. Recent genome resequencing studies have investigated a great deal of the germplasm of domesticated rice, providing the raw material for genome comparisons (Huang et al. 2012). In this study, we present a novel protocol whereby resequencing data can be used in tandem with a reference genome to analyze de novo genomic instability, and we herein identify and confirm thousands of recent mutations in the Nipponbare lineage of O. sativa ssp. japonica.
Materials and Methods
DNA isolation, PCR, and sequencing
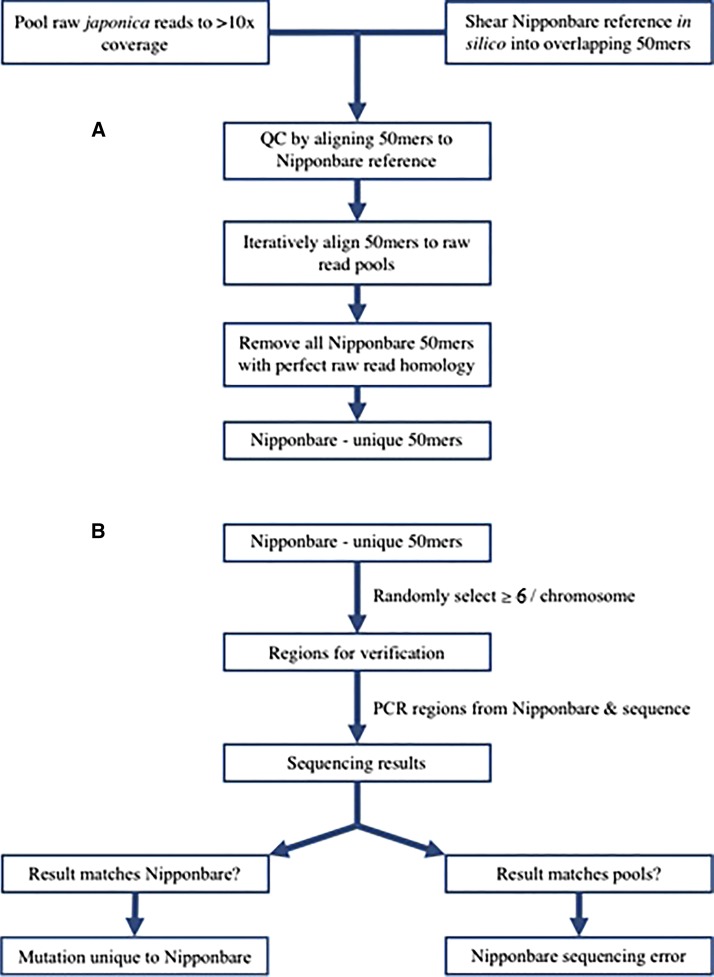
Seeds from O. sativa ssp. japonica cultivar Nipponbare (accession GSOR100) were provided by the US Department of Agriculture. PCR investigation of candidate Nipponbare-specific alleles from the japonica “pools” comparison used seed from the original 2005 distribution of GSOR100. Genomic DNA was prepared from pools of multiple plants. DNA isolation, PCR primer design, and PCR amplification were performed as in Vaughn et al. (2014). Primers were designed with the software Primer3 by using 500-bp flanking regions surrounding the candidate 50-bp oligomer (50mer), with the target PCR product size being 100–200 bp. PCR product purification was carried out as described by the manufacturer with Macrogen kits (Macrogen Corporation, Rockville, MD), and samples were directly sequenced by Sanger technology, then read on an ABI 3730 machine. Inspection of the resulting PCR fragment sequences was conducted manually and via the usage of BLASTn (Camacho et al. 2009). As in Figure 1, if the sequencing result matched Nipponbare genomic data, it was considered a Nipponbare-specific mutation and if it matched the japonica pools, it was considered a Nipponbare-specific sequencing error.
Figure 1.
Flow chart depicting the steps taken in finding recent mutations in Nipponbare via comparison to pools of other japonica rice accessions. (A) Candidate Nipponbare-specific 50-bp oligomer (50mer) discovery. First, sequence data from japonica accessions were pooled to ∼10× coverage per pool and Nipponbare was sheared in silico to create overlapping 50mers. Next, iterative alignments were conducted between the Nipponbare 50mers and the japonica pools, and all Nipponbare 50mers with perfect homology to the japonica pools were removed from consideration, yielding 50mers unique to the Nipponbare line. (B) Resolution of candidate Nipponbare-specific 50mers between sequencing error and de novo mutation possibilities. Random selection of at least six Nipponbare-specific 50mers per chromosome was conducted to identify potential changes across the genome. Polymerase chain reaction (PCR) and Sanger sequencing of PCR products were performed on the selected Nipponbare 50mers, followed by classification of either a mutation unique to Nipponbare or a Nipponbare sequencing error.
Sequence acquisition, alignment, and comparison
The reference genome of Nipponbare, version IRGSP 1.0, used as the basis for this study, was downloaded from the International Rice Genome Sequencing Project 1.0 website (http://rapdb.dna.affrc.go.jp/download/irgsp1.html) on December 4, 2014 (Kawahara et al. 2013). O. sativa ssp. japonica pools of raw reads were obtained on December 5, 2014 from EBI (http://www.ebi.ac.uk/ena/data/view/ERP000106) and were generated by the Bin Han group (Huang et al. 2012). Of the 131 japonica accessions available (Huang et al. 2012), 126 accessions with good quality sequence data were chosen to provide a combined ∼113.8× coverage.
Nipponbare was sheared in silico into 50mers (Figure 1) via a custom PERL script. A quality control step was conducted by aligning the 50mers back to the Nipponbare reference genome assembly to ensure 50mer accuracy. A 100% homology between all 50mers and the assembled Nipponbare genome was observed. The overlapping 50mers were iteratively aligned to raw japonica reads in pools of ∼10× coverage, each sorted by geographical location (Huang et al. 2012), using Bowtie2 (Langmead and Salzberg 2012) to conduct alignments under the “very sensitive parameters” setting (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml). The 50mers that mapped back to japonica pools with perfect homology were removed from further analysis using the SAMTools suite (Li et al. 2009). Hence, any Nipponbare 50mers that did not perfectly align with even one resequencing raw read using Bowtie2 were the oligos that were considered to be Nipponbare-specific. Target candidate Nipponbare-specific 50mers (Figure 1) were then chosen and their genomic coordinates extracted using BLASTn+ (Camacho et al. 2009) against the full Nipponbare genome.
Several apparent Nipponbare-specific 50mers were chosen at random across all chromosomes for verification via PCR from Nipponbare genomic DNA, with at least six per chromosome, out of the total of 8504 candidate 50mers. When the Nipponbare PCR fragments were found to be identical in sequence to the candidate Nipponbare-specific 50mer, then this confirmed that the novel sequence was actually a product of Nipponbare-specific mutation during the descent of this cultivar. When the Nipponbare PCR fragments were found to differ in sequence from the candidate Nipponbare-specific sequence, then this was concluded to be caused by an error in the Nipponbare IRGSP 1.0 sequence assembly.
The true Nipponbare-specific 50mers that were confirmed by PCR were then compared to the most homologous sequences in the japonica pools. Sequences chosen as the best japonica read candidate by BowTie2 had ≥ 90% consensus among all japonica reads (Supplemental Material, Table S2), and the > 90% nucleotide was considered the ancestral nucleotide to the Nipponbare-specific change. Thus, the nature of sequence change from the japonica pool sequence to the Nipponbare-specific sequence was extracted from the alignment BAM files created by Bowtie2 through the use of a custom Python script, which recorded the types and number of changes and positions as they occurred on the Nipponbare-specific 50mers.
To search for large indels like those expected from transposable element insertion or excision, we used the Bowtie2 alignment results and SAMtools to search for any Nipponbare-specific 50mers that had ≥ 70% identity to their best hit in the japonica pools. The result would be expected for 2–4 50mers for an insertion (which would create two novel junction sites) and for 1–2 50mers for an excision (which would create one novel junction site). No such low-homology best hits were found with any of the candidate Nipponbare-specific 50mers, indicating a complete absence of large indels that were Nipponbare-specific.
Sequence comparisons performed to calculate changes per kilobase in coding sequences (CDS)/UTR/introns between Nipponbare and O. glaberrima were carried out using BLAST. The data sources for O. glaberrima were from Camacho et al. (2009) and Zhang et al. (2014).
Data availability statement
Unless otherwise specified, UNIX shell, PERL, and Python scripts were written to execute the above analyses. No other external libraries were used. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6151094.
Results
De novo mutation discovery
The Nipponbare genome sequence was generated as part of an international consortium effort to sequence O. sativa ssp. japonica at the highest resolution and quality possible (International Rice Genome Sequencing Project 2005). It has since been updated by correction of genomic sequencing errors, the extension of previous gap sequences (N chains), and the correction/expansion of annotation (Kawahara et al. 2013). Nipponbare is currently one of the best annotated and highest sequence quality genome assemblies in the plant genomics world, and as such provides an excellent resource for many types of evolutionary, genetic, and molecular studies.
To discover the nature and relative frequencies of different types of de novo mutations in rice, we selected Nipponbare as the target genome. The basic concept is that Nipponbare has had a unique breeding history (as has any unique cultivar or lineage within any species), and that any de novo mutations during the unique descent of Nipponbare would not be shared with any other rice variety. Hence, Nipponbare IRGSP 1.0 was broken in silico into 50mers that cover the entire genome twofold because they overlap by 25 bp. Because Nipponbare is a japonica cultivar, our goal was to compare it to all other japonica cultivars. The Nipponbare 50mers were thus compared to pools of shotgun sequence data from the resequencing of closely related rice cultivars, all from subspecies japonica (Huang et al. 2012). Any 50mers that had an exact match with any read (even a single read) from the shotgun resequencing data were judged to be not specific to the Nipponbare lineage (and, thus, probably ancestral), and were then removed from further analysis. The Nipponbare-specific 50mers that remained could then be investigated to see if they were due to sequencing errors in the original Nipponbare assembly or were truly unique to the Nipponbare cultivar. The usage of overlapping 50mers allowed precise positioning and confirmation of any Nipponbare-specific mutation, because any change (even a single-base pair indel or substitution) should affect at least two overlapping 50mers. Any category of sequence change can be detected by this strategy, including base substitutions and small indels (each of which would create novel oligo sequences) and larger indels like TE insertions or excisions, which create novel junction sites.
Properties of the japonica data pools
Many O. sativa ssp. japonica cultivars were resequenced at low redundancy in efforts to study the domestication of Asian rice (Huang et al. 2012). Out of the publicly available data from this publication, we chose 126 accessions with good quality sequence data to provide a combined ∼113.8× coverage. The sequences generated were between 0.5× and 2× coverage for each japonica accesson, and were generated via the Illumina Genome Analyzer llx platform as paired end reads with an average size of ∼73 bp (Huang et al. 2012).
Workflow
Figure 1 depicts the workflow for this project to identify and confirm Nipponbare-specific sequence changes. Overlapping 50mers from the Nipponbare reference genome were created by a custom PERL script using the repeat-masked Nipponbare genome. Repeat masking removed 5,756,938 of the original 14,929,820 overlapping 50mers (∼38%) from our analysis. Then, comparisons were performed of the Nipponbare 50mers to the japonica line resequencing data. The japonica reads from Huang et al. (2012) were combined into pools that had a total coverage of ∼10× per pool, resulting in 11 total pools. The Nipponbare 50mers were then iteratively aligned to each pool using Bowtie2, bringing the resulting total alignment coverage to ∼113.8× across all 126 accessions in the data pools. Any 50mers that mapped to the japonica pools with perfect homology (50/50 bp match) were removed, leaving only those 50mers that showed a possible difference between Nipponbare and other japonica lines to be analyzed.
The genomic coordinates of the candidate missing 50mers were extracted, and some candidates from each chromosome were chosen at random to be verified using PCR and sequencing. The PCR reactions were carried out with template data from Nipponbare and the resulting PCR products were analyzed by direct Sanger sequence analysis of excised PCR bands. If the 50mer that was amplified and sequenced matched the original Nipponbare genomic sequence, the 50mer was considered to be a Nipponbare-specific mutation. If the 50mer sequence did not match Nipponbare, it was considered to be an error in the Nipponbare sequencing project assembly.
Nipponbare-specific 50mers
We created 14,929,820 starting 50mers because each 50mer has an overlapping 50mer at 25-bp intervals in the ∼373-Mb Nipponbare genome assembly (Kawahara et al. 2013). The candidates were 50mers that were not identical to any raw sequence in the 11 japonica pools, and thus not removed from our analysis. Our computational analysis comparing the DNA sequences of the Nipponbare and japonica pools yielded 17,008 50mers of interest out of the ∼14.9 million starting 50mers. Because each of these 17,008 were covering each sequence novelty twofold, due to the overlap, this led to two candidate 50mers representing the same sequence novelty region. Hence, these results indicated 8504 novel sequence candidate sites. These candidates could represent novel recent mutations unique to the Nipponbare lineage or errors in the Nipponbare assembly (Figure 1), so 250 of them were investigated via PCR and sequencing.
Novel 50mer PCR sequencing analysis
Primers were designed from regions flanking the candidate de novo sequence sites to generate predicted amplification products of 100–200 bp. These primer pairs yielded amplification products ∼83% (207/250) of the time. Amplification products were subjected to direct Sanger sequence analysis of the PCR product excised from an agarose gel. Useful sequences were found in ∼94% (194/207) of these sequencing attempts.
Confirmation sites were initially selected to provide comparable numbers per chromosome, but otherwise randomly selected across each chromosome. However, the high frequency of sequencing errors on some chromosomes (Table 1) led to the selection of additional sites for amplification from those error-rich chromosomes, so that at least four confirmed Nipponbare-specific sequence changes were found on each chromosome.
Table 1. Distribution of verified Nipponbare-specific sequences and Nipponbare sequencing errors organized by chromosome across the Nipponbare genome.
| Chromosome | Number of 50mers analyzed | Number of de novo changes verified | Number of Nipponbare sequencing errors |
|---|---|---|---|
| 1 | 8 | 6 | 2 |
| 2 | 9 | 7 | 2 |
| 3 | 13 | 9 | 4 |
| 4 | 61 | 4 | 57 |
| 5 | 17 | 11 | 6 |
| 6 | 9 | 7 | 2 |
| 7 | 6 | 5 | 1 |
| 8 | 13 | 11 | 2 |
| 9 | 10 | 5 | 5 |
| 10 | 20 | 11 | 9 |
| 11 | 11 | 9 | 2 |
| 12 | 17 | 8 | 9 |
| All | 194 | 93 (148) | 101 (141) |
The numbers in brackets indicate the number of actual sequence changes within these 194 analyzed sequences. 50mer, 50-bp oligomer.
Overall, there were 93 candidate 50mers with confirmed variants and 101 with confirmed errors (Table 1). On most chromosomes, the number of confirmed de novo alleles was equal to or greater than the number of detected sequencing errors. The exception was chromosome 4, where the ratio of de novo alleles to sequencing errors was 4/57. Not including chromosome 4, 89 sequence changes were true de novo alleles and 44 were sequencing errors, suggesting that about two-thirds of our candidates are actually de novo mutations that are Nipponbare-specific. Our analysis indicated that the nonrepetitive portion of the IRGSP 1.0 Nipponbare genome sequence includes a predicted ∼7400 sequencing errors. Because our analysis investigated all of the nonrepetitive parts of the rice genome (> 250 Mb) and 7400/250,000,000 is < 0.01%, then the overall Nipponbare genome sequencing accuracy is > 99.99%.
De novo sequence changes
Of the 93 50mers confirmed with Nipponbare-specific sequences, many had more than one changed nucleotide compared to a specific 50mer. The specific nucleotide changes were determined by comparing the novel 50mer sequence to the consensus sequence of the japonica pools. In most cases, the japonica pools had ≥ 90% agreement on the consensus sequence, with the exceptions presumably due to actual allelic variation among pool lineages or to sequencing errors in the data generation. Overall, there was a higher number of transversions compared to transitions, and indels were quite rare (Table 2). All of the indels were tiny, involving only 1 or 2 bp.
Table 2. Summary of the nature of the Nipponbare-specific variants discovered in the Nipponbare genome, organized by chromosome.
| Chromosome | Number of 50mers analyzed | Transitions | Transversions | Insertions | Deletions |
|---|---|---|---|---|---|
| 1 | 6 | 5 | 5 | 0 | 0 |
| 2 | 7 | 2 | 8 | 0 | 0 |
| 3 | 9 | 5 | 11 | 0 | 0 |
| 4 | 4 | 1 | 2 | 2 | 0 |
| 5 | 11 | 6 | 10 | 0 | 1 |
| 6 | 7 | 6 | 5 | 0 | 0 |
| 7 | 5 | 2 | 4 | 0 | 0 |
| 8 | 11 | 7 | 6 | 0 | 0 |
| 9 | 5 | 2 | 8 | 0 | 0 |
| 10 | 11 | 6 | 14 | 1 | 0 |
| 11 | 9 | 6 | 6 | 1 | 0 |
| 12 | 8 | 9 | 7 | 0 | 0 |
| Total | 93 | 57 | 86 | 4 | 1 |
The numbers of observed indels, transitions, and transversions are denoted per chromosome. 50mer, 50-bp oligomer.
A comprehensive analysis of the entire data set of 8504 novel oligos was undertaken to see if any had < 70% identity in their best hit within the japonica pools. This would be expected if any of the novel oligos were created by the insertion or deletion of a large (> 20 bp) fragment of DNA, for instance due to TE activity. No such case was found, indicating that TE insertion and deletion activity had been zero during the unique descent/creation of the cultivar Nipponbare.
Of the confirmed 56 sequence changes associated with genes, 26 were from CDS, 17 were in introns, and 13 were from either 5′- or 3′-UTRs. The 26 CDS changes were found to have a dN/dS ratio of 1.54 using the PAML package (Yang 1997). Overall, the dN/dS ratios for CDS changes when comparing different Orzya species has been found to be 0.28–0.47 (Zhang et al. 2014), in agreement with the fact that most genes are under strong purifying selection. The much higher ratio observed in the de novo 50mer data set is compatible with a 1:1 ratio indicative of random drift, as would be expected for de novo mutations that have not yet undergone long periods of selection.
We randomly chose and compared 10 10-kb windows of orthologous genes and flanking regions from Nipponbare and its close relative O. glaberrima. In these comparisons, we found 31 sequence differences in 15.6 kb of CDS (2/kb), 27 in 5 kb of UTR (5/kb), and 188 in 25.9 kb of introns (7/kb) (Table S1). Hence, the frequencies of sequence variation per kilobase in introns were much higher than other gene components in the comparison of rice with O. glaberrima. In contrast, the 26 in 31 kb (CDS), 13 in 19.6 kb (UTRs), and 17 in 57.2 kb (introns) in the de novo 50mer data set were closer to a 1:1:1 ratio per kilobase, as expected of a de novo mutation data set.
Clustered sequence variation in the novel 50mers
Our data indicate that a single mutation in a given 50-bp region is the most common observed event (Table 3). With the sequence difference frequency observed (one difference per 28,702 bp), the results demonstrate that significantly (P-value < 0.0001, Fisher’s exact test) more multiple changes per 50-bp regions were observed than predicted by a fully independent mutation model. The confirmation analyses by PCR indicated that multiple sequence differences per 50mer were about equally likely in both the sequencing error category and the verified sequence change category (Table 1), so around two-thirds of these observed results indicate true clusters of mutation.
Table 3. Number of sequence differences per 50mer, based on alignment.
| Number (N) of changes | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Predicted number of 50mers with N | 13,004 | 37 | 0 | 0 | 0 | 0 | 0 | 0 |
| changes for an independent model | ||||||||
| Observed number of 50mers | 5,979 | 1,277 | 712 | 365 | 158 | 8 | 3 | 2 |
50mer, 50-bp oligomer.
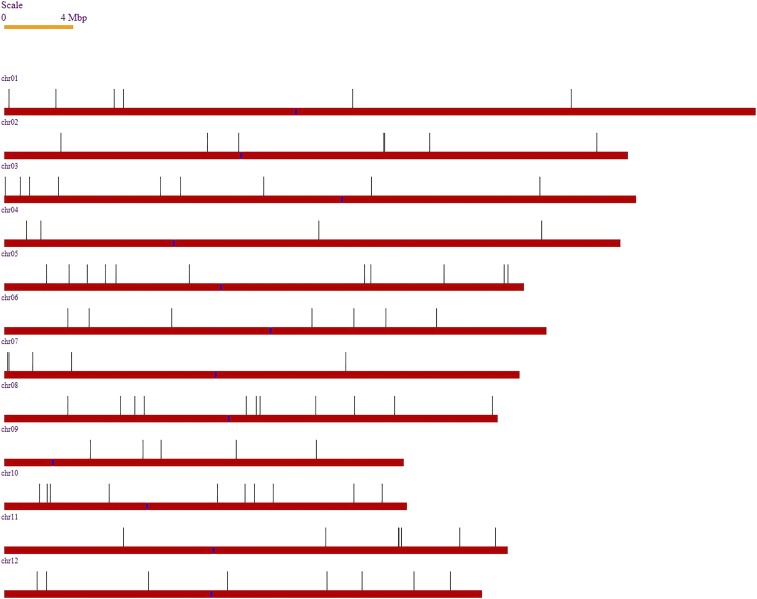
The chromosomal locations of all candidate de novo 50mers were plotted (Figure S1) and also of all confirmed de novo sequence changes (Figure 2). Sequence variants were distributed across all chromosomes, with no major clusters of de novo or candidate de novo sequences obvious, although chromosome 4 exhibited a much higher number of candidate novel 50mers because of the higher rate of sequencing errors (Table 1). The majority of the novel 50mers (67%) were mapped to areas in the Nipponbare genome that do not contain annotated genes.
Figure 2.
The distribution of verified mutations across all chromosomes of Nipponbare. Centromeres are highlighted on each chromosome in blue, and each vertical line corresponds to a confirmed Nipponbare-specific sequence variant.
Discussion
Discovery of recent genome sequence variation
The abundance of sequencing and resequencing data for multiple organisms presents a novel opportunity for the study of sequence variation. Commonly, these studies use variation across individuals and populations within a species to investigate population histories, especially with respect to geographic origin and population dispersal (Roach et al. 2010; Huang et al. 2012; Zhang et al. 2014). However, this wealth of data could also be mined to study the molecular nature and patterns in sequence change by comparing high-quality reference genomes to the resequencing data.
All individuals, even biological clones, will differ somewhat in genome sequence from their closest relatives because of the vagaries of mutation (somatic and germinal), segregation, and selection. The heritable differences that make each individual unique are an outcome of these genetic alterations. In crop improvement, a subject that inspired Darwin’s discovery and elaboration of natural selection (Darwin 1868), each developed variety has novel genetic characteristics that make it particularly productive in a specific agricultural environment. Most of the sequence variations that make each variety unique are presumed to be derived from the segregation of preexisting variants dispersed in the germplasm pool, but some variation is also generated during the crop breeding process. Hence, the discovery of alleles unique to a single developed variety is expected to enrich for de novo mutations among the variety-specific alleles.
Most molecular evolution studies investigate the current status of allelic variation across individuals within a closely related set of taxa, for instance members of the same species. The observed changes are a combined outcome of de novo mutation type and rate, plus population history, plus selection. Even with large studies that provide some knowledge of the population histories, it is still difficult or impossible to determine how much of the genetic change is due to these three contributing factors. One way to begin to sort this out is to investigate the natures and rates of de novo change per se. However, because mutation is so rare, the expense and time demands of such studies are so great that relatively few have been performed (Vazquez et al. 2000; Ossowski et al. 2010; Behringer and Hall 2016). We herein develop and describe an inexpensive and rapid alternative to methods such as mutation accumulation studies for discovering de novo mutations.
A novel strategy for the discovery of lineage-specific de novo sequence change
Our strategy utilizes a high-quality reference genome sequence for the targeted organism, and then compares that sequence to all other genome sequence data that are available for that organism. The quality of the results depends on the depth of that additional sequence data and the degree to which the other sequenced genomes are closely related to the targeted (reference sequence) genome. If the other genome sequences are deep and closely related, then the only novel sequences in the targeted reference genome will be ones that arose during the descent of the reference genome sequence lineage. Hence, this is the equivalent of a mutation enrichment study, but with all of the line progression having been done either by breeders (for a crop or domesticated animal) or by the natural process of lineage descent.
We decided to test this strategy on rice (O. sativa). The initial strategy for sequencing Nipponbare was a careful clone-by-clone analysis of rice bacterial artificial chromosome clones, with different laboratories taking responsibility for different chromosomes or chromosome segments. The Nipponbare rice genome has proven to be one of the best quality plant genome sequences (International Rice Genome Sequencing Project 2005). The sequence has also been much improved over the past 15 years (Kawahara et al. 2013). However, to our knowledge, no studies have been conducted to identify the recent genome sequence changes in Nipponbare rice. By dividing the Nipponbare genome in silico into overlapping 50mers, we were able to directly compare the Nipponbare 50mers to ∼114-fold depth data for O. sativa ssp. japonica resequencing accessions (Huang et al. 2012). This analysis uncovered 8504 candidate Nipponbare-specific regions of sequence change, of which a predicted ∼2800 are false positives derived from sequencing errors in the Nipponbare reference genome IRGSP 1.0, while the other ∼5700 are true Nipponbare-specific sequence-change regions.
The primary advantages of this strategy are the immediacy of the analysis (with all data only a database download away), zero cost for initial data generation and low costs for result confirmation, and the ability to find thousands of de novo sequence changes compared to the handful available from traditional mutation enrichment studies. Moreover, this technology can apply to any organism, even those with exceedingly long generation times (like many conifers) or very large genome sizes that make accurate full genome sequencing cost-prohibitive. In addition, this approach can lead to the identification of the errors in a reference genome sequence. However, there are some limitations. First, certain de novo changes will be missed. Any nucleotide variation that arose in Nipponbare, but was already present as a sequence polymorphism in one of the pooled japonica cultivars, will be missed as a sequence change. This should be a minor source of false negatives for most sequences, because these Nipponbare pools were near 100% identical at most nucleotide positions in the analyzed data, indicating a low level of standing polymorphism in the ancestral japonica germplasm. However, this is not true for repeats, where multiple paralogous variants are already observed, particularly with such hypervariable entities as simple sequence repeats (SSRs). Hence, we did not investigate repeats, and acknowledge that our data misses all sequencing errors and Nipponbare-originated changes that are within repeat sequences. Because of this category of missed changes, it is not possible to calculate sequence change rates with this approach. Second, it is possible that the large pool of assembled japonica varieties still did not contain some of the germplasm found in the ancestors of Nipponbare. Hence, some Nipponbare-specific 50mers would not be caused by de novo change during Nipponbare descent but by transmission of ancestral alleles not found in the japonica pools. If this were a major problem, then we would expect to see larger clusters of apparent Nipponbare-specific alleles caused by linkage drag on transmitted chromosomal segments, but such clusters were not observed. Hence, we feel that the great majority of our confirmed Nipponbare-specific sequences are the result of de novo mutation during the breeding of the Nipponbare variety.
Two additional lines of evidence support the conclusion that we have discovered novel Nipponbare alleles generated by recent de novo mutation. If many of the verified de novo changes that we observed were actually standing variation, then we would expect that those within genes would show strong evidence of purifying selection, both with a dN/dS ratio of < 1 and an overrepresentation in introns and UTRs relative to exons. However, our verified mutations were fairly evenly distributed across the various gene components and exhibited a 1.54 dN/dS ratio, both results expected of de novo variation.
Nipponbare-specific genome sequences
Comparison of japonica pools and Nipponbare revealed 8504 Nipponbare-specific chromosome sites, of which subsequent confirmation analysis indicated that around one-third were not actually Nipponbare-specific but were rather errors in the reference genome sequence. Of 101 confirmed sequencing errors, 76 contained precisely one error and the remaining 25 contained two to four sequencing errors per 50mer. Hence, the overall sequence accuracy of the Nipponbare reference genome appears to be excellent. The few clusters of candidate de novo 50mers in our analysis are from chromosomes with the highest sequence error rates, so we expect that those clusters may be caused by genomic regions that were particularly difficult to sequence, thus leading to small chromosome segments with lower quality overall.
Although some base substitutions should be missed by our analysis, primarily because they are identical to some standing variation in the japonica germplasm, our approach to search for best-hit 50mers that were < 70% identical to any japonica pool sequence should find 100% of the indels that are larger than 30 bp. Because of the vast number of different ways any indel covering a specific nucleotide position on any chromosome can have different indel end points, it is unlikely that any standing variation would be identical to any de novo allelic variation, except precise TE excision [which is a rare phenomenon, even for well-studied plant TEs (Rinehart et al. 1997; Bennetzen 2007)]. Hence, the fact that we discovered zero cases of TE excision, insertion, or other large indel variation indicates a very quiescent genome during the breeding of Nipponbare.
Although most angiosperm genome analysis shows a great deal of TE activity in the last few million years in all lineages examined (Rinehart et al. 1997; Ma and Bennetzen 2004), including rice (Rinehart et al. 1997; Huang et al. 2012; Kawahara et al. 2013), these studies rarely have the power to differentiate events that occurred a million years ago from one that happened in the last 1000 years. This surprising absence of novel large indels in the Nipponbare lineage suggests that none of Nipponbare’s improvement is associated with TE-induced genetic change, but is primarily an outcome of the improved combination of standing genetic variation.
Regarding the nature of de novo mutations, we confirmed 93 sites that contained Nipponbare-specific mutations. The investigated 50mers were chosen to represent each chromosome, and their locations on each chromosome were random in their selection. Confirmed de novo variants were not clustered on any chromosome. Of the 93 confirmed sites, 64 contained exactly one mutation and 29 contained more than one mutation.
A total of 57 transitions, 86 transversions, 4 insertions, and 1 deletion were confirmed in Nipponbare. A higher frequency of substitutions compared to indels was also found in Drosophila melanogaster, A. thaliana, and human (Vazquez et al. 2000; Ossowski et al. 2010; Wang et al. 2012). However, the substitution/indel ratio in our analysis of over 28/1 (143/5) is high compared to that seen in these other systems, such as 732/60 in Drosophila and 99/17 in A. thaliana (Ossowski et al. 2010; Schrider et al. 2013). These differences may be a result of different relative frequencies of mutation types (e.g., chromosome breaks vs. cross-linking vs. deamination vs. base modification) in each lineage, or differences in the relative efficiencies of different repair processes. Studies in plants have shown that even closely related species may differ dramatically in their rate of failure in certain types of DNA repair, and that this surprising “repair-efficiency variation” might have some selective value in creating different levels of genetic novelty in lineages that differ in their need to adapt genetically to changing environments (Vitte and Bennetzen 2006; Bennetzen and Wang 2014).
Although thousands of confirmed mutations across a genome provide a wealth of data, these are still rare events that should not be found in multiple types in a single 50-bp region if they were fully independent. Hence, we believe that some mutation processes in the rice genome act on stretches of nearby DNA rather than on single nucleotide positions. Clusters of genomic changes are typically due to repeat-rich regions being more prone to polymerase slippage during repair (Drake et al. 1998), the higher spontaneous deaminations in methylated DNA regions leading to nearby additional mutations because of repeated short-patch repair (Walser and Furano 2010), indels causing frameshifts that are associated with an increased frequency of point mutations around the indel (Tian et al. 2008), or double-strand breaks causing multiple new mutations and becoming “induced” hotspots (Shee et al. 2012). Many of these clusters are thus likely to be an outcome of repair using less-accurate DNA polymerases (Colis et al. 2008; Waters et al. 2009) that are called into action when DNA damage in a region is not easily repaired. Unfortunately, our results on these multiple-change regions are too sparse to provide any insight into the relative importance of these clustered mutation processes in rice, but the large data set of candidate sequence changes that we have detected should provide sufficient grist for this mill.
We are particularly excited to see our strategy for lineage-specific mutations applied to more species, especially in those animals and plants with excellent reference genome sequences and a wealth of resequencing data. This technique can be applied to any organism, so future studies could be designed for other plants, animals, and unicellular organisms. With the huge numbers of sequence changes that this technology can inexpensively provide, robust conclusions on the types of sequence change can be made in interspecies comparisons. Moreover, with identification of the nature and relative frequencies of de novo change, then the observed variation between lineages can help to indicate which changes inside and outside of genes are preferentially retained over evolutionary time.
Acknowledgments
This research was funded by the endowment associated with the Norman and Doris Giles Professorship to J.L.B. We thank S. Chaluvadi for his generous assistance in PCR confirmation experiments.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6151094.
Communicating editor: J. Birchler
Literature Cited
- Aminetzach Y. T., Macpherson J. M., Petrov D. A., 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309: 764–767. 10.1126/science.1112699 [DOI] [PubMed] [Google Scholar]
- Behringer M. G., Hall D. W., 2016. The repeatability of genome-wide mutation rate and spectrum estimates. Curr. Genet. 62: 507–512. 10.1007/s00294-016-0573-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., 2007. Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10: 176–181. 10.1016/j.pbi.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Wang H., 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 65: 505–530. 10.1146/annurev-arplant-050213-035811 [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K., 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155: 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colis L. C., Raychaudhury P., Basu A. K., 2008. Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry 47: 8070–8079. 10.1021/bi800529f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. R., 1868. . John Murray, London. [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., Crow J. F., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z., Wang A., et al. , 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project , 2005. The map-based sequence of the rice genome. Nature 436: 793–800. 10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., McCombie W. R., et al. , 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 6: 1–10. 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: 2774–2783. 10.1073/pnas.1210309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J. L., 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. 10.1073/pnas.0403715101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeburger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart T. A., Dean C., Weil C. F., 1997. Comparative analysis of non-random DNA repair following Ac transposition excision in maize and Arabidopsis. Plant J. 12: 1419–1427. [DOI] [PubMed] [Google Scholar]
- Roach J. C., Glusman G., Smit A. F., Huff C. D., Hubley R., et al. , 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328: 636–639. 10.1126/science.1186802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. 10.1534/genetics.113.151670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Gibson J. L., Rosenberg S. M., 2012. Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep. 2: 714–721. 10.1016/j.celrep.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang Q., Zhang P., Araki H., Yang S., et al. , 2008. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455: 105–108. 10.1038/nature07175 [DOI] [PubMed] [Google Scholar]
- Vaughn J. N., Chaluvadi S. R., Tushar L. R., Bennetzen J. L., 2014. Whole plastome sequences from five ginger species facilitate marker development and define limits to barcode methodology. PLoS One 9: e108581 10.1371/journal.pone.0108581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J. F., Perez T., Albornoz J., Dominguez A., 2000. Estimation of the mutation rates in Drosophila melanogaster. Genet. Res. 76: 323–326. [DOI] [PubMed] [Google Scholar]
- Vitte C., Bennetzen J. L., 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103: 17638–17643. 10.1073/pnas.0605618103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser J. C., Furano A., 2010. The mutational spectrum of non-CpG dna varies with CpG content. Genome Res. 20: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Fan H. C., Behr B., Quake S. R., 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. 10.1016/j.cell.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Minesinger B. K., Wiltrout M. E., D’Souza S., Woodruff R. V., et al. , 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73: 134–154. 10.1128/MMBR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhu T., Xia E. H., Shi C., Liu Y. L., et al. , 2014. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 111: 4954–4962. 10.1073/pnas.1418307111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Unless otherwise specified, UNIX shell, PERL, and Python scripts were written to execute the above analyses. No other external libraries were used. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6151094.