Abstract
Summary
Here we report on an R package for genome-wide association studies of orthologous genes in bacteria. Before using the software, orthologs from bacterial genomes or metagenomes are defined using local or online implementations of OrthoMCL. These presence–absence patterns are statistically associated with variation in user-collected phenotypes using the Mono-Associated GNotobiotic Animals Metagenome-Wide Association R package (MAGNAMWAR). Genotype-phenotype associations can be performed with several different statistical tests based on the type and distribution of the data.
Availability and implementation
MAGNAMWAR is available on CRAN.
1 Introduction
Bacterial genome-wide association (BGWA) studies are increasingly viewed as a useful alternative to traditional genetic screens, in part because they can accurately predict genetic variants associated with phenotype from a smaller set of measures (Chen and Shapiro, 2015; Power et al., 2017). In a traditional BGWA, genetic variants such as SNPs, kmers, or genes, are associated with an organism’s phenotype (Bayjanov et al., 2013; Lees et al., 2016; Lippert et al., 2011). A variation of BGWA includes metagenome-wide association (MGWA), where bacterial genetic variants are associated with a host organism’s phenotype. Genetic variants of coarse resolution, including genes (Brynildsrud et al., 2016; Chaston, 2014) or bacterial strains (Qin et al., 2012) must usually be used in MGWA because the taxa are often phylogenetically distant. Since most current software for BGWA is SNP-based, we present MAGNAMWAR, R software for gene-level BGWA. MAGNAMWAR performs associations based on gene presence–absence patterns and provides greater statistical flexibility than existing Chi-square test approaches, making it ideally suited for MGWA and ortholog-level BGWA.
MAGNAMWAR formally implements an analysis that previously identified bacterial genes associated with variation in Drosophila melanogaster phenotypes (Chaston et al., 2014). Previously, a single fruit fly line was individually associated with each of 42 genome-sequenced bacterial strains. Separately, the phenotype of flies bearing the different bacterial strains were measured and gene presence–absence patterns in the bacterial strain panel were defined by OrthoMCL (Li et al., 2003). The genotype-phenotype relationship for each gene was statistically defined and roles for bacterial genes that were predicted to influence fruit fly traits were confirmed by mutant analysis. Here we reanalyze the data for fly triglyceride content to show how MAGNAMWAR simplifies the pre-formatting and analysis steps, and the graphical presentation of the data. MAGNAMWAR also incorporates bacterial population structure into analyses and permits the use of additional statistical tests.
2 Implementation
MAGNAMWAR functions require user-created datasets. Core functionality requires a file that defines the orthologous gene (OG) sets and a file containing the phenotype measurements and metadata for the statistical models (Fig. 1A). Optional functions require additional datasets. Detailed instructions are provided in the documentation, including an R vignette.
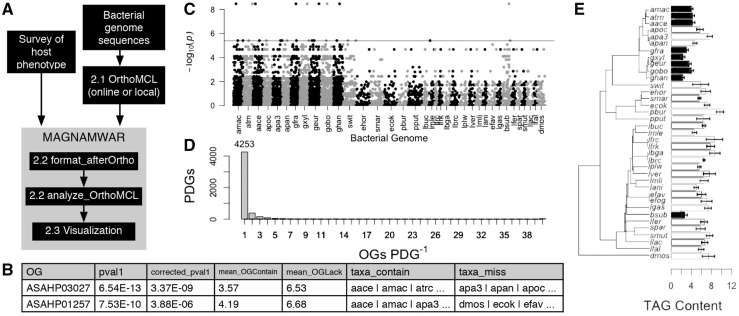
Fig. 1.
(A) MAGNAMWAR workflow. (B) Sample rows from matrix produced by AnalyzeOrthoMCL. (C) Manhattan plot with the different bacterial taxa along the X-axis and –log10P-value on the Y-axis. (D) PDG versus OG plot which shows the frequencies of unique OG distributions according to OrthoMCL clustering. (E) PDG plot of the mean effect of different bacteria on D.melanogaster triacylglyceride (TAG) content. Shading corresponds to the OG ASAHP03027 (black: gene present; white: gene absent). Data in B–E are from Chaston et al. (2014)
2.1 Pre-analysis/gene clustering
First, OGs must be called from the protein-coding sequences. Gene clustering is accomplished by providing OrthoMCL (Li et al., 2003), available locally or online, with custom-formatted amino acid FASTA files for each bacterial taxon or metagenome. For each file, features in MAGNAMWAR format the FASTA headers of each sequence, ensure all protein IDs are unique, and combine all target files into a single output named ‘MCLformatted_all.fasta,’ which is the input for the OrthoMCL clustering software.
2.2 Statistical analyses
In the second step the BGWA is performed. The data are analyzed by specifying the phenotype file, the gene presence–absence file and the type of model to be used in the statistical association. Different statistical tests can be performed, including mixed and survival models, and Wilcoxon tests. Bacterial population structure is automatically calculated as principal components, which can be included as covariates in models by specifying the percentage of variation or number of principal components to include. Most analyses are single-threaded, but the survival analysis can be run on multiple threads. The analysis produces an R matrix containing the gene cluster identifier, P-values, effect size and presence/absence pattern for each gene (Fig. 1B).
2.3 Post-statistical functions and visualization
Optional functions further assist the user in examining the data. OG functions can be assigned by appending to the main statistical results a protein annotation and sequence selected randomly from among proteins assigned to an OG. Visualization options include a modified Manhattan plot, where the bacterial gene content is ordered by species along the X-axis according to each protein’s numerical annotation (e.g. Ecol_0001), against –log10P (Fig. 1C). QQ-Plots present the relationship between expected (X-axis) and observed (Y-axis) P-values. A PDG versus OG plot reports the number of OGs with the same phylogenetic distribution group (PDG) (i.e. presence–absence pattern) (Fig. 1D). Single PDG plots display the mean and standard error of phenotypes by treatment, color-coded according to gene presence–absence with an optionally-attached phylogenetic tree (Fig. 1E).
3 Discussion
MAGNAMWAR enables BGWA to predict bacterial genes that influence organismal traits. Originally developed to identify bacterial determinants of fruit fly nutrition, this pipeline can be extended to define the genetic relationship between genome-sequenced bacteria or metagenomes and any organismal phenotype (bacterium, associated plant or animal). Because the statistical implementations are flexible, MAGNAMWAR can appropriately analyze a variety of datasets. The package is freely distributed (MIT license).
Acknowledgements
We thank N. Porter, C. Carroll and M. Koyle for testing the software and two anonymous reviewers for feedback that improved the software and the manuscript.
Funding
The work was supported by Foundation for the National Institutes of Health (FNIH) grant number R01GM095372 to AED and 1F32GM099374-01 to PDN; startup funds and a ME grant from Brigham Young University (BYU) to JMC; and a BYU ORCA grant to CES to promote undergraduate research.
Conflict of Interest: none declared.
References
- Bayjanov J.R. et al. (2013) Genotype-phenotype matching analysis of 38 Lactococcus lactis strains using random forest methods. BMC Microbiol., 13, 68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O. et al. (2016) Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol., 17, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J.M. et al. (2014) Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio, 5, e01631–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.E., Shapiro B.J. (2015) The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol., 25, 17–24. [DOI] [PubMed] [Google Scholar]
- Lees J.A. et al. (2016) Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat. Commun., 7, 12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res., 13, 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C. et al. (2011) FaST linear mixed models for genome-wide association studies. Nat. Methods, 8, 833–835. [DOI] [PubMed] [Google Scholar]
- Power R.A. et al. (2017) Microbial genome-wide association studies: lessons from human GWAS. Nat. Rev. Genet., 18, 41–50. [DOI] [PubMed] [Google Scholar]
- Qin J. et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature, 490, 55–60. [DOI] [PubMed] [Google Scholar]