A root-knot nematode gene (MiIDL1) produces a peptide that mimics the Arabidopsis IDA signaling peptide that regulates cell separation events including abscission and lateral root emergence.
Keywords: Abscission, Arabidopsis, IDA, Meloidogyne incognita, MiIDL1, peptide signal, RNAi, root-knot nematode
Abstract
INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) is a signaling peptide that regulates cell separation in Arabidopsis including floral organ abscission and lateral root emergence. IDA is highly conserved in dicotyledonous flowering plant genomes. IDA-like sequences were also found in the genomic sequences of root-knot nematodes, Meloidogyne spp., which are globally deleterious pathogens of agriculturally important plants, but the role of these genes is unknown. Exogenous treatment of the Arabidopsis ida mutant with synthetic peptide identical to the M. incognita IDA-like 1 (MiIDL1) protein sequence minus its N-terminal signal peptide recovered both the abscission and root architecture defects. Constitutive expression of the full-length MiIDL1 open reading frame in the ida mutant substantially recovered the delayed floral organ abscission phenotype whereas transformants expressing a construct missing the MiIDL1 signal peptide retained the delayed abscission phenotype. Importantly, wild-type Arabidopsis plants harboring an MiIDL1-RNAi construct and infected with nematodes had approximately 40% fewer galls per root than control plants. Thus, the MiIDL1 gene produces a functional IDA mimic that appears to play a role in successful gall development on Arabidopsis roots.
Introduction
Abscission is the process by which organs (leaves, flowers, fruit, petals, stems, etc.) are shed from the parent plant (Roberts et al., 2002; Estornell et al., 2013). An innate part of abscission is cell separation, which is essential for additional developmental processes including organogenesis, emergence of lateral roots, pollen tube elongation through the pistil, seed germination, and pod and anther dehiscence (Roberts et al., 2002). A part of all cell separation events is the decomposition of the middle lamella that glues cells together and, very often, a loosening or restructuring of the primary cell wall (Roberts et al., 2002).
In 2003, Butenko et al. (2003) described an Arabidosis thaliana mutant inflorescence deficient in abscission (ida) that does not abscise (drop) its floral organs. The IDA gene encodes a small protein of 77 amino acids that includes an N-terminal signal peptide for secretion into the apoplast. IDA is a member of a small gene family in Arabidopsis that encodes proteins with a highly conserved proline-rich, C-terminal sequence of 12 amino acids. Since its nascent discovery, an understanding of the IDA signaling pathway and mode of action have progressed considerably (Cho et al., 2008; Stenvik et al., 2008b; Butenko and Simon, 2015; Vie et al., 2015; Meng et al., 2016; Santiago et al., 2016). The IDA protein is secreted into the apoplast where it is cleaved into a smaller peptide (Stenvik et al., 2008b; Butenko and Simon, 2015; Schardon et al., 2016). It was demonstrated that the previously identified, conserved 12 amino acid C-terminal sequence of IDA was sufficient to rescue the delayed abscission phenotype of ida, but a slightly longer 14 amino acid fragment of the C-terminus induced a higher percentage of abscission at a lower concentration (Schardon et al., 2016). The resultant mature IDA peptide binds to two redundant leucine-rich receptor-like kinases HAESA (HAE) and HAESA-like 2 (HSL2) localized in the plasma membrane (Cho et al., 2008; Stenvik et al., 2008a; Santiago et al., 2016). Recent studies demonstrate that another class of receptor-like kinases, SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK), facilitates binding of IDA to HAE and HSL2 (Meng et al., 2016; Santiago et al., 2016), which then relay the binding signal through a MAP kinase phosphorylation signaling pathway that ultimately activates a set of KNOX transcription factors that regulate a change in gene expression. It was proposed that the role of IDA in abscission is to regulate gene expression for cell wall modifying proteins, e.g. polygalacturonases (PGs), expansins (EXPs), cellulases, xyloglucan endotransglucosylase/hydrolase (XTH) and others (Butenko et al., 2003; Shi et al., 2011).
Because of the proposed role for IDA in regulating genes for cell wall modification, it was suggested that IDA might regulate other cell separation events in Arabidopsis (Butenko et al., 2003). Subsequently, IDA signaling was demonstrated to be involved in the emergence of lateral roots through the root cortex (Kumpf et al., 2013). In lateral root emergence, the HAE and HSL2 receptors each appear to play a role at a different stage of root emergence. Comparison of gene expression patterns for cell wall modifying proteins including PG LATERAL ROOT (PGLR), PG ABSCISSION ZONE ARABIDOPSIS THALIANA (PGAZAT/ADPG2), EXPANSIN 17 (EXP17) and XYLOGLUCAN TRANSFERASE 6 (XTR6/XTH23) in the wild-type, ida and the hae/hsl2 double mutant indicated that expression of these genes during lateral root emergence increased in an IDA signaling-dependent manner (Kumpf et al., 2013).
More recently, it was reported that IDA-like genes, AtIDL6 and AtIDL7, play a role in moderating stress and defense responses in Arabidopsis (Vie et al., 2015, 2017; Wang et al., 2017). It was demonstrated that AtIDL6 and AtIDL7 were strongly up-regulated by treatment with the bacterial flagellin elicitor, flg22, and infection with Pseudomonas syringae (Vie et al., 2015, 2017; Wang et al., 2017). An elevated expression of IDL6 during infection by P. syringae increased susceptibility to the pathogen (Wang et al., 2017). It was proposed that IDL6 increased the expression of poylgalacturonases that enhanced infection by P. syringae. Moreover, the authors noted an IDL6-dependent suppression of the PATHOGENESIS-RELATED PROTEIN-1 gene, PR1. Also, of interest was that the IDL6 response was at least partially dependent upon the presence of the HAE and HSL2 receptors. It was subsequently demonstrated that both IDL6 and IDL7 suppressed several stress- and defense-associated genes, and that IDL7 and, maybe IDL6, reduced stress-induced reactive oxygen species (ROS) signaling (Vie et al., 2017).
Genes encoding IDA-like (IDL) proteins are conserved in land plants (Tucker and Yang, 2012; Estornell et al., 2015; Stø et al., 2015; Vie et al., 2015). IDA-like genes have been found in dicots, monocots, and some gymnosperms (Stø et al., 2015; Vie et al., 2015). Of interest is that the conserved C-terminal domain of IDA-like proteins shares sequence similarity with the conserved C-terminus of CLAVATA3 (CLV3) (Stenvik et al., 2008b; Butenko and Simon, 2015) and PAMP-INDUCED PEPTIDES (PIP) (Vie et al., 2015). CLV3 is a small, secreted peptide that is transcribed and translated in the central zone of the shoot apical meristem where it is secreted and then processed in the apoplast (Barton, 2010). CLV3 binds to leucine-rich repeat receptor-like kinase proteins (LRR-RLK) CLAVATA1 and CLAVATA2 in the plasma membranes of cells immediately below the undifferentiated stem cells where CLV3 is synthesized (Barton, 2010). The role of CLV3 secretion and feedback regulation from affected cells is to limit cell divisions in the shoot apical meristem (Barton, 2010). It has been proposed that IDA may have evolved from the more primal CLV3 protein to fulfill a different role in regulating plant development (Butenko and Simon, 2015). Thus, although IDA and CLV3 (CLAVATA3/ESR-related or CLE) proteins share sequence similarity (Butenko and Simon, 2015), IDA and CLE bind to different LRR receptor-like proteins and are functionally distinct (Guo et al., 2010; Hou et al., 2014; Butenko and Simon, 2015; Santiago et al., 2016). PIP1 and PIP2, which are the progenitors for a larger gene family, were first identified as being up-regulated by the pathogen-associated molecular pattern (PAMP) bacterial flagellin (flg22) (Hou et al., 2014). Similar to CLV3, PIP1 and PIP2 are secreted into the apoplast where they are cleaved into smaller peptides that bind to the RECEPTOR-LIKE KINASE 7 (RLK7), which activates a plant immune response (Hou et al., 2014).
Root-knot and cyst nematodes are pandemic pathogens that parasitize the roots of many agriculturally important plant species (Williamson and Hussey, 1996; Jung and Wyss, 1999; Hewezi and Baum, 2013; Mitchum et al., 2013; Goverse and Smant, 2014; Hewezi, 2015). Cyst nematodes (Heterodera spp. and Globodera spp.) migrate through the root and attach to a pericycle or procambial cell in the vascular bundle to establish a feeding structure (syncytium) by secreting proteins (effectors) that alter root cell development and inhibit host defense responses (Hewezi and Baum, 2013; Goverse and Smant, 2014; Hewezi, 2015). The syncytium for cyst nematodes is formed by degrading the cell walls and membranes between existing root cells to form one large multinucleated cell that can eventually include as many as 200 host cells (Jung and Wyss, 1999). Root-knot nematodes (Meloidogyne spp.), on the other hand, migrate through the root tip to a parenchymatous cell in the still developing vascular bundle where they then become sessile and secrete effectors to establish their feeding structure (Hewezi and Baum, 2013; Mitchum et al., 2013; Goverse and Smant, 2014). The root-knot nematode forms its feeding structure by inducing cell division without cytokinesis to produce several large multinucleated giant cells (Jung and Wyss, 1999). The giant cells are surrounded by more dividing cells that form a protective gall around the nematode and giant cells (Jung and Wyss, 1999).
Numerous effector proteins have been identified in both cyst and root-knot nematodes. Of these effectors, it has been shown that a few have similarity to signaling peptides found in the plant (Gheysen and Mitchum, 2011; Mitchum et al., 2013; Goverse and Smant, 2014). For example, a CLV3-like (CLE) protein was one of the earliest effectors identified in the secretory esophageal glands of cyst nematodes (Heterodera spp.; Olsen and Skriver, 2003). Although it has been clearly demonstrated that the nematode CLE proteins play a role in establishing the syncytium (Hewezi, 2015) and that the nematode CLE peptide signals through a receptor complex that includes CLV1, CLV2 and RPK2 (Replogle et al., 2011; Replogle et al., 2013), the actual mechanism of action of the nematode CLE peptides that is needed for formation of the nematode feeding site is still being worked out (Guo et al., 2015, 2017). Interestingly, the CLE motif is also found in root-knot nematodes; however, it exists as multiple tandem repeats of CLE-like motifs within a single gene (Rutter et al., 2014), which was named MAP (Meloidogyne Avirulence Protein). The MAP protein is secreted from the nematode (Semblat et al., 2001; Vieira et al., 2011), but it is unknown if the larger MAP protein is processed into functional CLE peptides (Rutter et al., 2014); nevertheless, similar to the root-knot nematode, the potato cyst nematode (Globodera rostochiensis) also possesses CLE-like genes with multiple CLE motifs within the same gene and these gene products are correctly processed into functional CLE-like peptides that rescued the Arabidopsis CLV3 mutant, clv3-2 (Guo et al., 2011).
The proposed role for IDA in regulating gene expression for cell separation led us to hypothesize that IDA or an IDA-like peptide native to the plant (host) or nematode might play a role in dissolution and/or modification of the cell walls during formation of the syncytia or giant cells of cyst or root-knot nematodes, respectively. Quantitative PCR (qPCR) of RNA from soybean cyst nematode (SCN, Heterodera glycines)-infected soybean roots and southern root-knot nematode (Meloidogyne incognita)-infected tomato roots did not indicate a significant change in a plant IDA-like gene (Tucker and Yang, 2012, 2013) that was associated with the infection (unpublished results). Moreover, no IDA-like genes were found in the sequence data for cyst nematodes (Heterodera spp.); however, we did discover IDA-like genes in root-knot nematodes (Meloidogyne spp.; Tucker and Yang, 2013). M. incognita has two IDA-like genes, MiIDL1 and MiIDL2, and the open reading frames (ORFs) of both genes contain a sequence for an N-terminal secretion peptide and are expressed early in the infection and development of galls on tomato roots (Tucker and Yang, 2013). Although the C-terminus of the MiIDL1 and MiIDL2 translational products are similar to that of IDA and IDA-like genes found in plants, we had no proof that the nematode IDA-like genes produce a functional IDA peptide. Herein, we demonstrate that the MiIDL1 gene product can complement the ida mutant in Arabidopsis and thereby is capable of functioning as an IDA signal, which can bind to the receptor-like kinases such as HAE and HSL2 in Arabidopsis. In addition, we used an RNA interference (RNAi) approach to suppress MiIDL1 expression in the nematode (Dutta et al., 2014) and demonstrated that expression of the MiIDL1-RNAi construct in Arabidopsis roots significantly correlated with a reduction in the number of nematode infections (galls) that formed on the roots.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) seeds of Columbia (Col-0), C24, ida-2 (Col-0 background), ida-1 (C24 background) and the hae-3/hsl2-3 (Col-0 background) double mutant were surface sterilized by treatment with 20% (v/v) bleach for 2 min followed by 70% ethanol for 5 min and then rinsed several times with sterile distilled water. After stratification at 4 °C for 2–4 d, sterilized seeds were geminated on agar plates containing 0.8% (w/v) agar, half-strength Murashige and Skoog basal salt mixture (Murashige and Skoog, 1962), pH 5.7. Plants used for peptide assays were grown on agar plates with or without synthetic peptides as described. For other experiments, after seed germination on agar plates, plants were transferred to Promix BX potting soil (Griffin Greenhouse Supplies, Richmond, VA, USA). Both plants on agar and plants in soil were grown in a growth chamber set to 23 °C, 16 h light (90–150 μE m−2 s−1), 8 h dark and 50–60% humidity.
Retrieval of IDL and similar sequences and peptide alignment
The soybean (Glycine max) and tomato (Solanum lycopersicum) IDA-like sequences were those previously identified by a TBLASTN search of soybean and tomato genomic sequence data using the AtIDA (accession NP564941) ORF minus the predicted N-terminal signal peptide (Tucker and Yang, 2012). Additional genomes were later searched for IDA-like peptides using only the AtIDA EPIP domain (Stenvik et al., 2008b). Other genes encoding similar C-terminal protein sequences, CLV3, CLE-like, PIP, PIPL, MiMAP, and Mi16D10, were selected from the literature and downloaded from the National Center for Biotechnology Information (NCBI). Alignments were performed using T-Coffee in the MacVector software (version 15.1). A dendrogram was created using UPMGA, best-tree with distances uncorrected. These settings were selected to align sequences based simply on identity and chemically similar amino acids and not a predicted evolutionary relationship. When a conserved C-terminal domain was not predicted in the literature, the sequence was first aligned with predicted domains and the most similar C-terminal sequence used in subsequent alignments. The accession numbers for the nucleotide and protein sequences are listed in Supplementary Table S1 at JXB online. Where an accession number was not available, the nucleotide and protein sequence is included in Supplementary Table S1.
Peptide synthesis and mutant complementation assays
The peptides used in this study for complementation of the ida mutant were synthesized by Life Technologies (now Thermo Fisher Scientific, Waltham, MA, USA) at 90% purity (Table 1). Synthetic peptides were suspended in 1/10 volume of sterilized NaH2PO4 (0.125 M, pH 7.0) buffer and then transferred to 9/10 volume of ddH2O. For the floral organ abscission assay, Arabidopsis flowers were collected at anthesis from plants of wild-type (Col-0 and C24), ida-1 (C24), ida-2 (Col-0) and the hae/hsl2 double mutant, hae-3/hsl2-3 (Col-0) (abbreviated herein as hh33), and the pedicel of each flower was inserted into agar plates containing 10 μM of the corresponding peptides. To measure the percentage of abscission, individual flowers were touched with forceps and the numbers of flowers displaying petal separation were recorded after 48 h of treatment. Three to five biological replicates were performed for each peptide rescue assay and each assay included between 10 and 30 flowers. To assess the physiological response of Arabidopsis roots to the synthetic peptides, wild-type (Col-0) and ida-2 seeds were grown for 48 h on agar plates containing 0, 0.1, 0.5, and 1.0 μM of the MiIDL1p peptide and the root development recorded.
Table 1.
Sequence of synthetic peptides used in this study
| Peptide name | Sequence |
|---|---|
| MiIDL1p | IKGVPPNSGPSRRGNKVPGPGR |
| AtIDAp | FGYLPKGVPIPPSAPSKRHN |
| GmIDA1ap | FNFLPKGVPIPPSGPSKRHN |
| AtCLV3p | LRTVPSGPDPLHHH |
| Mi16D10p | GKKPSGPNPGGNN |
Nematode culture and infection assay
Root-knot nematodes, Meloidogyne incognita, were maintained on the roots of pepper (Capsicum annuum) in a greenhouse at the Soybean Genomics and Improvement Laboratory, United States Department of Agriculture, Beltsville, MD, USA. Meloidogyne incognita-infected roots were harvested from pepper plants 3–4 months after inoculation. To isolate second-stage juveniles (J2), roots with galls were collected and shaken in 15% bleach for 5 min to release the eggs, and the eggs isolated by successively passing the liquid through stainless steel sieves with pore sizes of 850, 150, and 25 μm (no. 20, no. 150, and no. 500 sieves from Newark Wire Cloth, Clifton, NJ, USA; Meyer et al., 2000). Eggs were collected from inside the no. 500 sieve and hatched in 100 ml of water with 50 μg ml−1 of ampicillin shaken at 28 °C. After 3–4 d hatching, J2 were isolated from the eggs using the no. 500 sieve. Arabidopsis transgenic seedlings were germinated on agar plates containing kanamycin 50 μg ml−1 for 10 d and transferred to 3.8 cm pots containing 90% sand 10% clay soil (Xue et al., 2013). After 1 week of recovery in soil, approximately 1000 freshly hatched J2 were added to each pot. All J2-infected plants were fertilized with liquid Knops medium (Sijmons et al., 1991) once a week for the first 2 weeks after transplanting. After 2, 3, 7, 14, 15, 35, 42, and 56 d post-inoculation (dpi), a sampling of infected roots of T3 transgenic plants was gently washed in tap water, blotted dry, and observed for a general assessment of infection. Based on our preliminary assessment of gall development and our ability to reproducibly count galls, a larger population of roots was harvested at 14, 35 and 42 dpi and the roots washed and stained using acid fuchsin as described by Bybd et al. (1983), which stains the nematodes red and enhances their detection. The number of stained galls per root was counted under a dissecting microscope.
RNA isolation and qPCR
Total RNA was extracted from roots of root knot nematode-infected wild-type (Col-0) and transgenic plants using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA). One microgram of total RNA was used for cDNA synthesis using Superscript-III™ Reverse Transcription System (Invitrogen), following the manufacturer’s instructions. The resultant cDNA was diluted 5-fold. Sequences of primers used for qPCR are listed in Supplementary Table S2. Gene-specific primers were used in a total volume of 15 μl consisting of 3 μl of diluted cDNA, 1.5 μl each of 1.0 μM gene-specific primers, 1.5 μl of ddH2O and 7.5 μl of Brilliant II SYBR Green QPCR Master Mix in an Mx3000P instrument (Stratagene, La Jolla, CA, USA). The qPCR results were normalized to either MiEF1b (Tucker and Yang, 2013) for the nematode genes or AtACT2 (Cartieaux et al., 2008) for the Arabidopsis genes and the transgenes β-glucuronidase (GUS), MiIDL1, or NPTII.
Constructs and transgenic Arabidopsis that overexpressed MiIDL1 and MiIDL1-RNAi
For overexpression of MiIDL1 in plants, the ORFs with and without predicted signal peptide sequences of MiIDL1 (Tucker and Yang, 2013) were PCR amplified from genomic DNA of root-knot nematode by primers as described in Supplementary Table S2. Amplified PCR products were digested with XhoI and XbaI and then cloned into pHANNIBAL (Wesley et al., 2001), which excised and replaced the PYRUVATE ORTHOPHOSPHATE DIKINASE (PDK) intron. The vector, pHANNIBAL, includes the CaMV 35S promoter for constitutive gene expression in plants. For root-knot nematode suppression assays, a 347 nt and 324 nt sense and antisense fragment of E. Coli GUS and root-knot nematode MiIDL1, respectively, were amplified using primers as described in Supplementary Table S2. The sense strand PCR product was introduced into the XhoI-EcoRI and antisense into XbaI–HindIII of pHANNIBAL on either side of the PDK intron, which generated the GUS and MiIDL1-RNAi (hairpin dsRNA) constructs. Both overexpression of MiIDL1 and of GUS and MiIDL1-RNAi in the pHANNIBAL vector were digested with NotI, which included the 35S promoter and the OCTOPINE SYNTHASE (OCS) terminator, and cloned into pART27 (Wesley et al., 2001). Plasmids pART27-MiIDL1 (full length ORF, named MG) and pART27-MiIDL1 (ORF minus signal peptide, named MS), pART27-GUS-RNAi (named DG), and pART27-MiIDL1-RNAi (named DM) were introduced into Agrobacterium tumefaciens GV3101 by electroporation, verified by PCR, and transformed into Arabidopsis ida-2 (Col-0) and wild-type Col-0 plants by floral spray transformation (Chung et al., 2000). Transgenic seeds were selected on half-strength Murashige and Skoog medium supplemented with kanamycin at 50 μg ml−1. Kanamycin-resistant plants were verified for expression of constructs and further used for the study as described.
Results
The nematode IDLs are more similar to the plant IDLs than to PIPs or CLEs
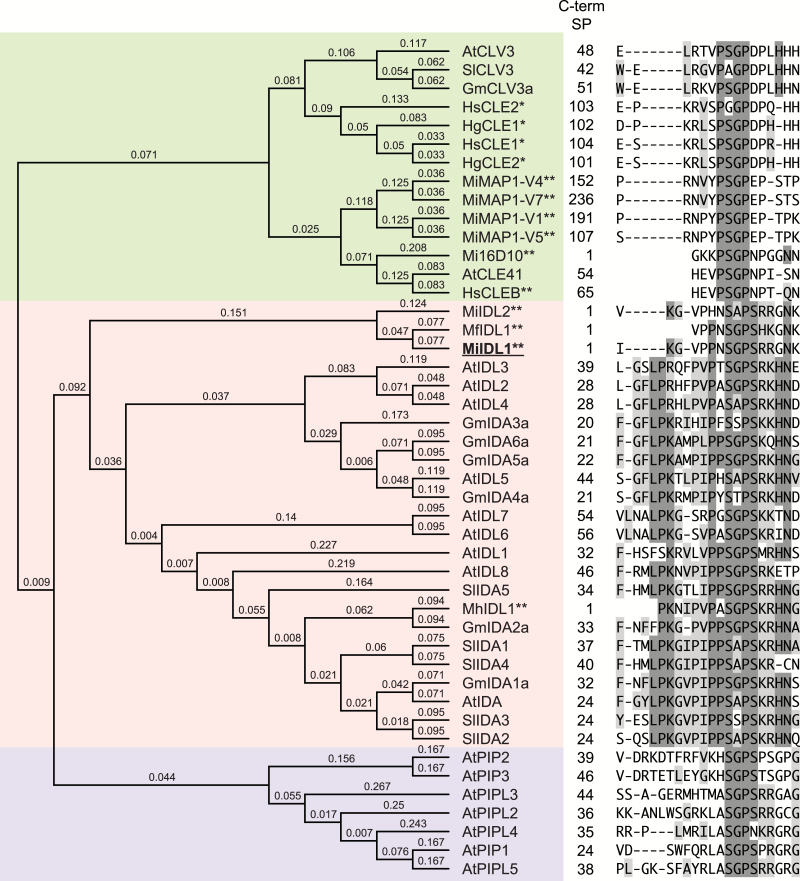
In an earlier study (Tucker and Yang, 2013), we concluded that MiIDL1 is more similar to the plant IDA and IDLs than CLE-like peptides. As more information has since been published regarding the phylogeny and sequence similarity among several different signaling peptides (Butenko and Simon, 2015; Stø et al., 2015; Vie et al., 2015) we performed a new, more extensive sequence alignment that included the active domains of these similar plant gene products: CLAVATA3/ESR-related (CLE), PAMP-INDUCED SECRETED PEPTIDE (PIP), and PIP-LIKE (PIPL) (Vie et al., 2015) (Fig. 1). In addition to the IDA-like proteins from the root-knot nematodes M. incognita, M. hapla, and M. floridensis, we included sequences for other root-knot and cyst nematode signaling peptides that share sequence similarity with IDL, CLE, and PIP peptides (Davis et al., 2008; Bobay et al., 2013). Only the most conserved C-terminal motif for each of these protein classes was used in the alignment; however, we included one extra amino acid after the conserved IDA EPIP domain (Stenvik et al., 2008b) to allow a potential alignment with the conserved domain in the PIP peptides (Fig. 1). In line with previous findings (Stø et al., 2015; Vie et al., 2015), we identified IDA-like genes in every dicot genome examined and several monocots, including banana, oil palm, and rice, and also Pinus taedea, a gymnosperm (results not shown). As previously observed with a much smaller set of proteins (Tucker and Yang, 2013), the root-knot nematode IDA-like genes clustered with the plant IDA-like genes and no other gene families (Fig. 1). Interestingly, the root-knot nematode 16D10 peptide, which shares similarity with IDLs and CLEs (Tucker and Yang, 2013) and is known to interact with a SCARECROW-like transcription factor (Huang et al., 2006b), clustered with the CLE-like peptides. The clustering of Mi16D10 with CLEs demonstrates how difficult it can be to predict protein function simply based on sequence alignments. Moreover, with more sequence data available for cyst nematodes, we did not find IDA-like genes in any genera of cyst nematodes including the potato cyst nematode, genus Globodera.
Fig. 1.
Dendrogram and alignment of conserved C-terminal amino acid sequences for a selection of representative plant and nematode genes with similarity to the AtIDA EPIP domain: Arabidopsis (Arabidopsis thaliana, At), soybean (Glycine max, Gm), tomato (Solanum lycopersicum, Sl), soybean cyst nematode (Heterodera glycines, Hg), sugar beet nematode (H. schachtii, Hs), southern root-knot nematode (Meloidogyne incognita, Mi), northern root-knot nematode (M. hapla, Mh) and peach root-knot nematode (M. floridensis). Sequence names marked with a single asterisk are from cyst nematodes (Hs and Hg), those with two asterisks are from root-knot nematodes, and the MiIDL1 peptide is highlighted in bold and underscored. The numbers under the heading of C-term SP indicate the amino acid position C-terminal from the end of the predicted signal peptide.
Exogenous application of synthetic MiIDL1 peptide rescues the Arabidopsis ida mutant
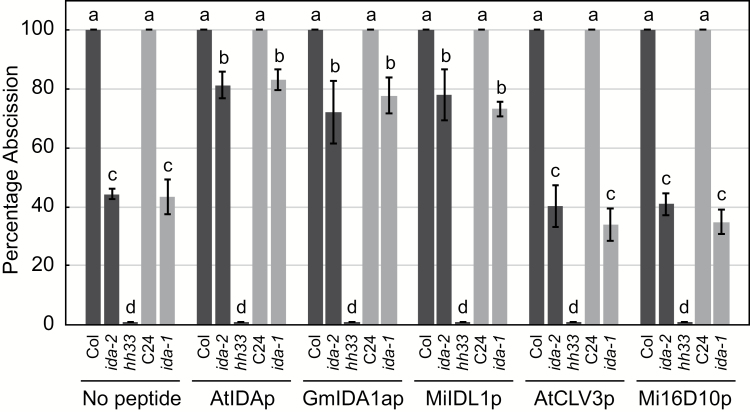
As a first step in determining if the nematode MiIDL1 gene product can function as an IDA mimic, we tested whether a synthetic MiIDL1 peptide could rescue the phenotypes of the Arabidopsis ida mutant. The MiIDL1 peptide we used encompassed the entire translation product of the MiIDL1 transcript starting at the end of the predicted N-terminal signal peptide (Tucker and Yang, 2013) (Table 1). For comparison, we also prepared synthetic peptides for the Arabidopsis AtIDA-EPIP (Stenvik et al., 2008b), soybean GmIDA1a-EPIP (Tucker and Yang, 2012), root-knot nematode Mi16D10 (Huang et al., 2006b), and the Arabidopsis AtCLV3 C-terminal motif (Olsen and Skriver, 2003) (Table 1). Each of these five peptides was used to treat ida mutant flowers to test for effects on petal abscission. Individual Arabidopsis flowers at anthesis were cut from wild-type (Columbia (Col-0) and C24 ecotypes) plants and the loss of function mutants ida-1 (C24), ida-2 (Col-0), and the hae-3/hsl2-3 (hh33) double mutant (Col-0) and the pedicel inserted into agar in Petri plates each containing 10 μM of peptide. In line with an earlier study (Stenvik et al., 2008b), 100% of the wild-type flower petals abscised after 48 h from both the Columbia and C24 ecotypes (Fig. 2). At 48 h, approximately 45% of the flowers abscised their petals in both the ida-2 and ida-1 mutants and none of the petals had abscised from the hh33 double mutant. Treatment with 10 μM of the MiIDL1p peptide caused approximately 80% of the petals to abscise, which was similar to the percent abscission for the Arabidopsis IDA peptide (AtIDAp) and the soybean GmIDA1ap peptide (Fig. 2). However, the 10 μM treatments with the AtCLV3p peptide and Mi16D10p, which share sequence similarity with IDA and IDLs, were similar to the non-treated (no peptide) ida mutants, i.e. approximately 35% abscission (Fig. 2). Thus, the synthetic nematode MiIDL1p peptide rescued the abscission phenotype of ida mutants much like the native AtIDAp peptide and none of the treatments affected the hh33 receptor mutant, suggesting that the MiIDL1p treatment is signaling via the same pathway as AtIDAp.
Fig. 2.
Percentage abscission of individual flowers cut from wild-type and mutant Arabidopsis plants and then treated in agar with 10 μM of the indicated peptide (listed in Table 1). Columbia (Col-0) and C24 are ecotypes of Arabidopsis that were used as controls. The mutant ida-1 is in the C24 background (lighter gray bars), whereas, the ida-2 and hh33 mutants are in the Columbia background (darker gray bars). Standard error bars indicate the variance of the means for between three and five experiments and each experiment included between 10 and 30 flowers for each treatment. Letters above standard-error bars indicate means that are statistically grouped using a Tukey–Kramer test in SAS.
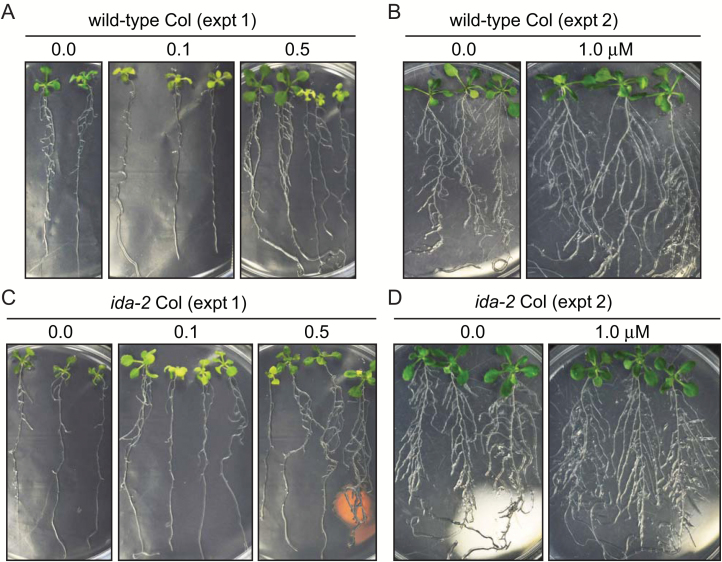
We next tested the effect of MiDL1p on root development, as the Arabidopsis ida mutant is known to have a lower density of lateral roots, i.e. a less branched root architecture than wild-type roots (Kumpf et al., 2013). We grew the wild-type and ida mutant seedlings on agar plates containing a series of concentrations of the synthetic MiIDL1p peptide and monitored the effect of the treatments on root development. A change in root architecture is a complex phenotype because growth of the above-ground parts (carbohydrate source tissue) affects the growth of roots (sink tissue) and, vice versa, an enhanced growth of roots (nutrient source) affects the growth of above-ground parts. Nonetheless, the ida mutant had a less branched root architecture (Fig. 3; Supplementary Fig. S1); moreover, when both the wild-type plants and the ida mutant plants were grown in the presence of different concentrations of MiIDL1p, there was a trend for the roots to be progressively more branched as the concentration of MiIDL1p increased from 0.0 to 1.0 μM (Fig. 3; Supplementary Fig. S1).
Fig. 3.
Arabidopsis wild-type Columbia (A, B) and ida-2 mutant seeds (C, D) were germinated and grown for 2 weeks in two separate experiments on agar containing 0.0, 0.1, and 0.5 (A, C) or 0.0 and 1.0 μM MiIDL1p peptide (B, D).
MiIDL1 complements the ida mutant phenotype in transgenic Arabidopsis
To further substantiate that MiIDL1 can function as an IDA mimic, we transformed the ida-2 mutant plants (Col-0) with a full-length MiIDL1 gene that included the predicted N-terminal signal peptide (Tucker and Yang, 2013), and an MiIDL1 construct in which the signal peptide was deleted. If a processed MiIDL1 peptide binds to the apoplast-exposed side of the HAE and/or HSL2 receptors in the plasma membrane, as would be expected for complementation of the abscission phenotype, a signal peptide would be required for translation on rough endoplasmic reticulum and secretion into the apoplast. Indeed, the transgenic ida-2 plants that included the full-length MiIDL1 gene (MG) displayed nearly wild-type floral organ abscission whereas the transgenic plants containing the construct lacking the signal peptide (MS) had a petal abscission phenotype very similar to the ida-2 plants, i.e. delayed or no petal abscission (Fig. 4). Nevertheless, a range of abscission rescue was observed in transgenic plants with the full-length construct. Included in Fig. 4 are two representative transgenic lines with the full-length MiIDL1 gene, MG4-8 and MG7-3. MG4-8 displayed a near complete recovery of wild-type abscission whereas MG7-3 displayed a partial recovery of abscission where most of the petals had abscised by position 11 but some of the stamens remained attached (Fig. 4). However, in the MS lines (minus the signal peptide) the floral organ abscission was very similar to the ida-2 mutant, i.e. the stamens, sepals, and petals remained attached beyond flower position 11 (Fig. 4).
Fig. 4.
Arabidopsis ida-2 mutant plants transformed with CaMV 35S promoter-driven full-length MiIDL1 open reading frame (ORF), MiIDL1 ORF minus the predicted signal peptide, or non-transformed wild-type and ida-2 mutant plants for comparison. Two transformation events (MG4-8 and MG7-3; MS2-4 and MS7-1) are included for each of the different constructs. The boxed insets are a 4-fold zoom of the indicated silique base.
RNAi suppression of the MiIDL1 gene in Arabidopsis reduces gall numbers and size
As an initial step to determine if the MiIDL1 gene plays a role in the infection process, we used a double-stranded, RNA interference approach (dsRNAi) to suppress expression of the MiIDL1 gene in the nematode. It has been demonstrated that expression of a nematode-targeted RNAi construct in the host plant can inhibit expression of genes in the parasitic nematode (Dutta et al., 2014). We prepared an MiIDL1-RNAi construct and a separate RNAi construct expressing a dsRNA for the E. coli β-glucuronidase (GUS) gene, which served as a control for these experiments. We reasoned that expression of a GUS-RNAi construct, which in our case would not specifically suppress the expression of any host or nematode gene but would produce a 300 bp dsRNA that would be processed into smaller RNA, would be a better control than transformation with an empty vector construct as is commonly done (Ibrahim et al., 2011; Youssef et al., 2013; Lischweski et al., 2015). Although the most conserved 45 nt region of the MiIDL1-RNAi has only 28–33% identity with the Arabidopsis IDA and IDLs and is unlikely to affect their expression, we still considered the possibility that expression of the MiIDL1-RNAi might inhibit the native Arabidopsis IDA. We therefore examined flower petal abscission in the MiIDL1-RNAi and GUS-RNAi transgenics. Both MiIDL1 and GUS-RNAi transgenic lines displayed a normal wild-type petal abscission phenotype and wild-type root architecture (Supplementary Fig. S2). No discernable phenotype different from the wild-type plants was observed for either the MiIDL1 or GUS-RNAi plants. Thus, expression of the MiIDL1-RNAi did not appear to significantly suppress expression of AtIDA or an AtIDL that might have manifested itself in growth or development.
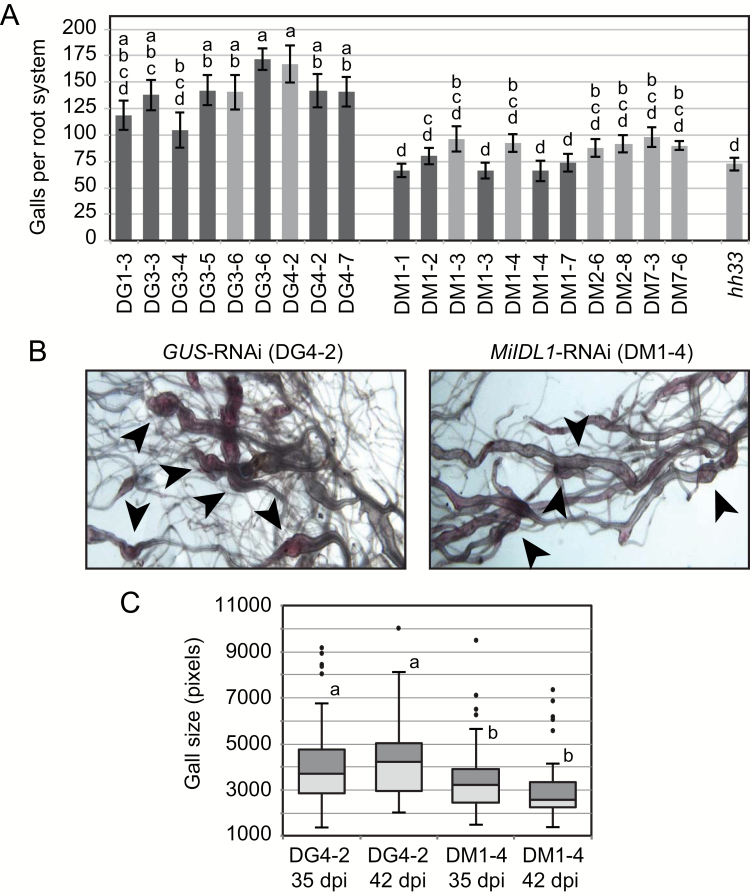
Multiple transgenic plants for both MiIDL1-RNAi and GUS-RNAi were infected with M. incognita and the roots examined at 2, 3, 7, 14, 15, 35, 42, and 56 d post-inoculation (dpi). To quantify the number of galls per root system, infected roots were harvested at 14, 35, and 42 dpi and stained with acid fuchsin to enhance detection of the nematodes (Bybd et al., 1983). At 14 dpi, when the galls were just beginning to form and be detectable under our conditions, we did not observe any clear reduction in gall number. At 35 and 42 dpi there were approximately 40% fewer galls on the MiIDL1-RNAi roots relative to GUS-RNAi roots (Fig. 5A). In addition, the galls that formed on the MiIDL1-RNAi plants appeared smaller than the galls on the GUS-RNAi plants (Fig. 5B, C).
Fig. 5.
Gall number and size on M. incognita infected roots of wild-type Arabidopsis transformed with GUS and MiIDL1-RNAi. (A) Mean number of galls per root system of 10–12 independently grown plants for multiple transgenic lines for GUS (DG) and MiIDL1 (DM) at 35 dpi (light gray bars) and 42 dpi (darker gray bars). (B) Photos showing the relative size of galls at 35 dpi on GUS and MiIDL1-RNAi plants. (C) Box chart of gall sizes (pixels counted using ImageJ software) from multiple photos of different plants from the lines indicated (the area of 50–100 galls was measured per transgenic line). In (A), DG and DM indicate GUS or MiIDL1-RNAi, respectively, and the first number after DG or DM indicate the transformation event from T0 plants and the second number the seed collection (line) from T1 plants. hh33 is the hae-3/hsl2-3 double mutant. Galls were counted on T3 plants. The standard error bars in (A) reflect gall counts for 9–14 plants for each line. Letters above or next to the bars in (A, C) indicate means that are statistically grouped using a Tukey–Kramer test in SAS. The P value (two-tailed t-test) for the significance of the difference between the means for the GUS lines compared with the MiDL1 lines in (A) is 4.1 × 10−7. Arrows in (B) point to representative galls. Points in (C) are outliers.
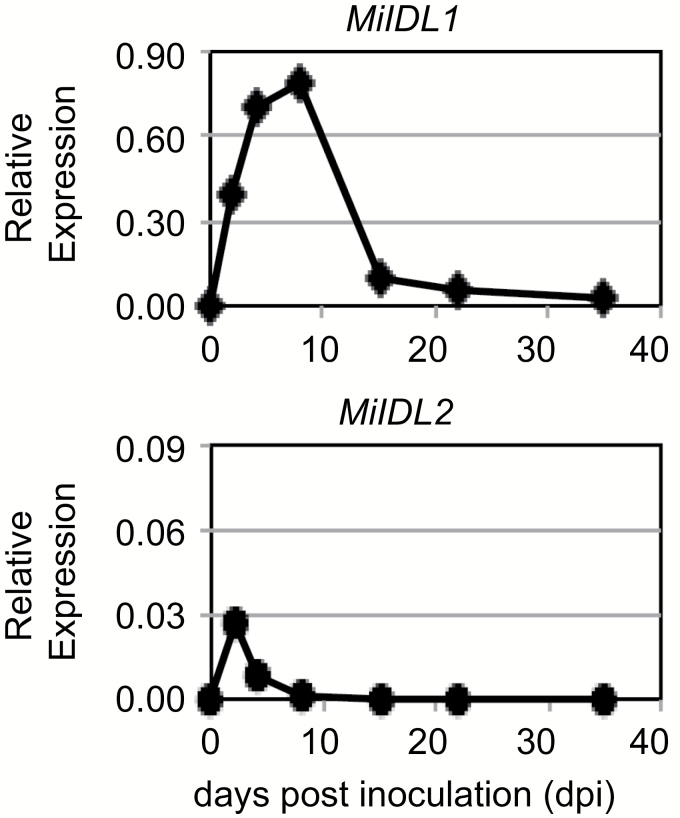
There are two IDA-like genes in M. incognita, MiIDL1 and MiIDL2 (Tucker and Yang, 2013). MiIDL1 and MiIDL2 are both expressed early after infection of wild-type Arabidopsis; however, MiIDL2 abundance is 10-fold less than MiIDL1 (Fig. 6). MiIDL2 is 90% identical to MiIDL1 and it is likely that the MiIDL1-RNAi would suppress both MiIDL1 and MiIDL2. Our focus for these early experiments was to establish a significant change in infection or gall development on MiIDL1-RNAi plants; nonetheless, total RNA was isolated from a sampling of MiIDL1 and GUS-RNAi-infected roots at 14 and 42 dpi. All the transgenic plants had high levels of the transcript for NPTII (the selection marker) and the GUS or MiDL1 transcripts in the respective transformed plants (Supplementary Fig. S3). We did not, however, see a significant reduction of MiIDL1 expression in the MiIDL1-RNAi lines at either 14 or 42 dpi; nonetheless, we did see an approximately 40% reduction in the constitutively expressed nematode elongation factor, MiEF1b, at 42 dpi, which supports the reduction in gall numbers that we observed at 42 dpi (Supplementary Fig. S3).
Fig. 6.
Gene expression profile for MiIDL1 (diamonds) and MiIDL2 (circles) normalized to the nematode elongation factor 1b (MiEF1b) in wild-type Arabidopsis infected with root-knot nematodes (M. incognita).
Discussion
INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) is highly conserved in plants. IDA and IDA-like peptides, however, have sequence similarity with CLE and PIP and PIP-like peptides but each has functionally distinct activities in the plant (Tucker and Yang, 2012; Vie et al., 2015). Moreover, although each of the plant IDA, CLE, and PIP peptides activates LRR-RLKs within the same class of RLKs (Class XI), they each bind to different RLKs (Hou et al., 2014; Tabata et al., 2014). The MiIDL1 peptide is more similar to plant IDA-like peptides than CLE or PIP peptides (Fig. 1), but sequence similarity over a short stretch of amino acids is not sufficient to prove that the peptide can function as an IDA-like signal. An example of this would be Mi16D10, which has sequence similarity with CLE peptides (Fig. 1) but binds to and alters the activity of a SCARECROW-like transcription factor (Huang et al., 2006b).
We have demonstrated herein that indeed the root-knot nematode MiIDL1 gene product can function as an IDA signal. Treatment of excised Arabidopsis ida mutant flowers with the nematode MiIDL1p peptide rescued the delayed abscission phenotype, as did the AtIDAp and GmIDA1ap peptides, whereas the AtCLV3p and Mi16D10p peptides did not rescue the ida mutant phenotype (Fig. 2). Moreover, transformation of the Arabidopsis ida-2 mutant with full-length and signal-peptide-attenuated MiIDL1 constructs indicates that not only can the MiIDL1 peptide function as an IDA signal but it also must be secreted into the apoplast to be functional (Fig. 4). Thus, the reversal of the ida mutant phenotype by MiIDL1 indicates that the MiIDL1 gene produces a protein that presumably can bind to the HAE/HSL2 receptors to propagate the signaling events necessary to complement the mutant phenotype. Interestingly, however, we did not observe an accumulation of a white substance reportedly rich in arabinogalactan at the abscission zone (AZ) in the MiIDL1-overexpressing transformants as was observed for the overexpression of the Arabidopsis IDA and IDLs in Arabidopsis (Stenvik et al., 2006, 2008b). The lack of a white secretion may be unique to MiIDL1 or simply reflects nuances in how IDA and IDA-like signals elicit a signaling pathway in Arabidopsis.
The Arabidopsis IDA precursor protein is secreted into the apoplast where it is processed into a smaller peptide (Stenvik et al., 2008b; Butenko and Simon, 2015; Schardon et al., 2016). The secreted IDA protein is predicted to be cleaved by subtilisin-like serine proteinases at both the N-terminus and C-terminus resulting in a 14 amino acid bioactive product (GVPIPPSAPSKRHN) (Schardon et al., 2016; Stührwohldt et al., 2017). When the N-terminal cleavage site was blocked with N-methylation of the Lys/Gly bond in a longer 23 amino acid peptide (SHIFGYLPK[m]GVPIPPSAPSKRHN), the methylated peptide was completely inactive whereas the non-methylated 23 amino acid peptide retained activity. This is interesting in regard to the prediction of the end of the signal peptide for MiIDL1. Previously, we predicted the most probable N-terminal cleavage site of the MiIDL1 for the signal peptide between amino acids 25 and 26 (Ser and Ile) (Tucker and Yang, 2013); however, a new version of SignalP (version 4.1) (Petersen et al., 2011) predicts two possible signal peptide cleavage sites, one at the Ser/Ile and another with slightly higher probability three amino acids further down between the Gly and Val (Table 1), which would remove the glycine at the N-terminus of the predicted bioactive IDA peptide (Schardon et al., 2016). The earlier Ser/Ile signal peptide cleavage site for MiIDL1 (Tucker and Yang, 2013) was used to prepare the MiIDL1p peptide (Table 1) that was used for rescue of the ida phenotype (Fig. 2). The MiIDL1p peptide clearly rescued the ida phenotype; however, we do not know if this peptide is proteolytically cleaved in the apoplast at the N-terminus or C-terminus to produce a shorter peptide in vivo. Also of interest is that, when the proline within the highly conserved active domain S(A/G)PS is hydroxylated, the peptide binds much more strongly to the HAE and HSL2 receptors (Butenko et al., 2014). The MiIDL1p peptide did not contain a hydroxyproline and typically it would be assumed that an exogenous application of a peptide would not be hydroxylated in vivo since proline hydroxylation normally occurs in the endoplasmic reticulum prior to secretion (Yuasa et al., 2005; Vlad et al., 2007). In addition, others have demonstrated that the ida mutant phenotype could be rescued with IDA-like peptides that do not contain a hydroxyproline modification (Stenvik et al., 2008b; Schardon et al., 2016). Interestingly, a recent report argues that the exogenously applied peptides might still be hydroxylated at proline by an extracellular enzyme (Stührwohldt et al., 2017).
To determine the importance of MiIDL1 expression to M. incognita pathogenesis, we transformed wild-type Arabidopsis with a 324 double-stranded RNAi construct of MiIDL1 that we predicted would inhibit MiIDL1 expression in the nematode (Dutta et al., 2014). There are, however, two IDA-like genes in M. incognita, MiIDL1 and MiIDL2; however, MiIDL2 is expressed at a much lower level, 10-fold less, than MiIDL1 (Fig. 6). Nevertheless, MiIDL2 is 90% identical to MiIDL1 and it is expected that the MiIDL1-RNAi would suppress both MiIDL1 and MiIDL2. In regard to phenotypes associated with the expression of the MiIDL1-RNAi, we did not observe a reduction in gall number at 14 dpi when galls were just beginning to form, but we did observe a significant 40% reduction in gall numbers at 35 and 42 dpi as well as notably smaller galls at these time points (Fig. 5). RNAi inhibition of the nematode gene requires ingestion by the nematode of the processed ~22 bp RNAi (Huang et al., 2006a). Meloidogyne incognita does not feed until it has established a rudimentary feeding structure (giant cells), which can be several days post-inoculation (Wyss and Grundler, 1992), and feeding stops when the J2 progressively molts into an adult female (Jung and Wyss, 1999). At 14 dpi MiIDL1 expression is already declining relative to a constitutively expressed MiEF1b gene but is near the peak of expression per nematode (i.e. normalized to expression of an Arabidopsis actin gene AtACT2) (Supplementary Fig. S3). Under our soil infection conditions, we did not see a significant reduction of MiIDL1 expression in the MiIDL1-RNAi lines at either 14 or 42 dpi, but did observe approximately 40% reduction in the constitutively expressed nematode elongation factor, MiEF1b, at 42 dpi, which reflects the 40% reduction in gall numbers on the MiIDL1-RNAi roots at 42 dpi (Supplementary Fig. S3). Because we did not observe any plant phenotype associated with the MiIDL1-RNAi or GUS-RNAi lines relative to wild-type Arabidopsis, it is reasonable to conclude that the MiIDL1-RNAi reduction in gall numbers is due to a suppression of the native MiIDL1/2 genes. The MiIDL mRNAs are low-abundance transcripts in the root RNA, which makes it difficult to quantify small changes in expression, and, moreover, the peak of expression of MiIDL1 may shift due to a change in the development of the nematode feeding structure as a result of the RNAi inhibition of MiIDL1; nonetheless, an extensive time course collection of RNA with more experimental replication may confirm a suppression of mRNA accumulation.
Our results indicated that gall numbers on hh33 roots were very similar to those observed for MiIDL1-RNAi roots, which was approximately 40% fewer than on wild-type Arabidopsis roots (Fig. 6A). This observation in the context of the complementation results supports a model where MiIDL1 functions through binding and activation of HAE, HSL2, or a similar HASEA-like receptor in the Arabidopsis roots. However, although our results strongly suggest that MiIDL1 is secreted from the nematode and is deposited directly into the apoplast or is somehow translocated to the apoplast, this remains to be confirmed experimentally.
As mentioned above, IDA was demonstrated to play an important role in Arabidopsis floral organ abscission and lateral root emergence and presumably the regulation of genes associated with cell wall modification that accompanies these developmental processes (Butenko et al., 2003; Kumpf et al., 2013). An up-regulation of gene expression for cell wall modifying EXPs and PGs is well established for root-knot nematode infection of Arabidopsis (Jammes et al., 2005; Cabrera et al., 2014). Thus, our original hypothesis for the role of the MiIDL1 peptide (Tucker and Yang, 2013) was that it might play a role in cell wall modifications associated with formation of the gall similar to that described for abscission and lateral root emergence (Butenko et al., 2003; Kumpf et al., 2013). However, recently it was reported that AtIDL6 and AtIDL7 play a role in suppressing defense and stress responses in the plant (Vie et al., 2015, 2017; Wang et al., 2017). AtIDL6 and AtIDL7 are rapidly up-regulated by PAMPs, e.g. flg22, and Pseudomonas syringae pv. tomato (Pst) DC3000 (Vie et al., 2015, 2017; Wang et al., 2017). In the particular case of P. syringae, up-regulation of AtIDL6 increased the number of bacterial infections (Wang et al., 2017). One explanation given for the enhanced infection was that the AtIDL6 peptide acted through HAE/HSL2 to up-regulate genes linked to cell wall modification, which included the polygalacturonase AtADPG2 (AtPGAZAT). They proposed that expression of these enzymes loosens the cell wall and aids the assembly of a type III secretion system that the bacteria use to inject virulence factors. Also of interest, Wang et al. (2017) suggested that expression of AtIDL6 directly or indirectly suppressed a salicylic acid (SA)-dependent defense response, which was indicated by an AtIDL6-dependent decrease in PATHOGENESIS-RELATED PROTEIN 1 (PR1) expression. A more recent study demonstrated that AtIDL7 and possibly AtIDL6 peptides were negative regulators of stress-induced ROS-signaling (Vie et al., 2017). Treatment of Arabidopsis leaves with either AtIDL6 or AtIDL7 peptide caused a significant decrease in several stress-related genes including ZINC FINGER PROTEIN (ZFP), WRKY, and genes encoding calcium-dependent proteins (Vie et al., 2017).
Suppression of the SA- and ROS-associated defense response or stress are interesting in regard to a potential role for MiIDL1 as elevation of an SA and ROS defense response has been implicated in root-knot nematode resistance (Branch et al., 2004; Goverse and Smant, 2014), and nematode migration and infection of roots elevates many defense- and stress-associated genes (Gheysen and Fenoll, 2002; Jammes et al., 2005; Alkharouf et al., 2006; Puthoff et al., 2007). Recent studies to characterize nematode elicitors have identified several that are involved in suppressing host defense and stress responses (Goverse and Smant, 2014; Hewezi, 2015). Moreover, it has been demonstrated that an extracellular network of LRR-RKs, which includes the HAESA-like receptors, is utilized to balance the response to extracellular signal (Smakowska-Luzan et al., 2018). Determining the mechanism of action for the IDA-like effectors in root-knot nematode infection awaits further research. Our original hypothesis was that MiIDL expression would directly affect gall development but it seems equally possible that MiIDL expression is involved in moderating stress and defense responses in the host; nonetheless, our findings here are significant in that we demonstrate that MiIDL1 can mimic IDA in Arabidopsis and that the MiIDL1 protein likely signals through HAE, HSL2, or another HAESA-like receptor.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Arabidopsis wild-type C24 and ida-1 mutant seeds were germinated and grown for 2 weeks in two separate experiments on agar containing 0.0, 0.1, and 0.5 or 0.0 and 1.0 μM MiIDL1p peptide.
Fig. S2. Arabidopsis seedlings of wild-type (Columbia) plants transformed independently with the GUS-RNAi and MiIDL1-RNAi constructs.
Fig. S3. Mean relative expression days post inoculation (dpi) of T-DNA and native MiIDL1 in GUS and MiIDL1-RNAi events and lines as included in Fig. 5A, and time course expression of MiIDL1 in inoculated wild-type Arabidopsis.
Table S1. Accession numbers or sequence for gene motifs included in Fig. 1.
Table S2. Primers for qPCR and construct preparation.
Funding
This work was supported by a US-Israel Binational Agricultural and Development Fund (BARD) US-4571-12C grant to MLT, Shimon Meir and Sonia Philosoph-Hadas, the United States Department of Agriculture (USDA Project 1245-21220-232-00D) and by the National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science, ICT & Future Planning) (NRF-2017R1A2B4010356) to JK.
Acknowledgements
We thank John Walker for the ida-2 and hae-3/hsl2-3 (hh33) double-mutant seeds, Melinka Butenko for ida-1 and wild-type C24 seeds, Eric Davis for sharing the Knop’s media protocol, Tarek Hewezi for the soil infection system protocol and Qijiang Song for statistical analysis. Special thanks to Shimon Meir and Sonia Philosoph-Hadas for suggestions and comments relating to this study.
References
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF. 2006. Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224, 838–852. [DOI] [PubMed] [Google Scholar]
- Barton MK. 2010. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Developmental Biology 341, 95–113. [DOI] [PubMed] [Google Scholar]
- Bobay BG, DiGennaro P, Scholl E, Imin N, Djordjevic MA, Mck Bird D. 2013. Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Letters 587, 3979–3985. [DOI] [PubMed] [Google Scholar]
- Branch C, Hwang CF, Navarre DA, Williamson VM. 2004. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Molecular Plant-Microbe Interactions 17, 351–356. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB. 2003. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. The Plant Cell 15, 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Simon R. 2015. Beyond the meristems: similarities in the CLAVATA3 and INFLORESCENCE DEFICIENT IN ABSCISSION peptide mediated signalling pathways. Journal of Experimental Botany 66, 5195–5203. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G. 2014. Tools and strategies to match peptide-ligand receptor pairs. The Plant Cell 26, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bybd DW, Kirkpatrick T, Barker KR. 1983. An improved technique for clearing and staining plant tissues for detection of nematodes. Journal of Nematology 15, 142–143. [PMC free article] [PubMed] [Google Scholar]
- Cabrera J, Díaz-Manzano FE, Sanchez M, et al. 2014. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytologist 203, 632–645. [DOI] [PubMed] [Google Scholar]
- Cartieaux F, Contesto C, Gallou A, Desbrosses G, Kopka J, Taconnat L, Renou JP, Touraine B. 2008. Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Molecular Plant-Microbe Interactions 21, 244–259. [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. 2008. Regulation of floral organ abscission in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105, 15629–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MH, Chen MK, Pan SM. 2000. Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Research 9, 471–476. [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Mitchum MG, Baum TJ. 2008. Parasitism proteins in nematode-plant interactions. Current Opinion in Plant Biology 11, 360–366. [DOI] [PubMed] [Google Scholar]
- Dutta TK, Banakar P, Rao U. 2014. The status of RNAi-based transgenic research in plant nematology. Frontiers in Microbiology 5, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell LH, Agustí J, Merelo P, Talón M, Tadeo FR. 2013. Elucidating mechanisms underlying organ abscission. Plant Science 199–200, 48–60. [DOI] [PubMed] [Google Scholar]
- Estornell LH, Wildhagen M, Pérez-Amador MA, Talón M, Tadeo FR, Butenko MA. 2015. The IDA peptide controls abscission in Arabidopsis and Citrus. Frontiers in Plant Science 6, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G, Fenoll C. 2002. Gene expression in nematode feeding sites. Annual Review of Phytopathology 40, 191–219. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. 2011. How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology 14, 415–421. [DOI] [PubMed] [Google Scholar]
- Goverse A, Smant G. 2014. The activation and suppression of plant innate immunity by parasitic nematodes. Annual Review of Phytopathology 52, 243–265. [DOI] [PubMed] [Google Scholar]
- Guo X, Chronis D, De La Torre CM, Smeda J, Wang X, Mitchum MG. 2015. Enhanced resistance to soybean cyst nematode Heterodera glycines in transgenic soybean by silencing putative CLE receptors. Plant Biotechnology Journal 13, 801–810. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang J, Gardner M, Fukuda H, Kondo Y, Etchells JP, Wang X, Mitchum MG. 2017. Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathogens 13, e1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark SE. 2010. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. The Plant Journal 63, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ni J, Denver R, Wang X, Clark SE. 2011. Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiology 157, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T. 2015. Cellular signaling pathways and posttranslational modifications mediated by nematode effector proteins. Plant Physiology 169, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ. 2013. Manipulation of plant cells by cyst and root-knot nematode effectors. Molecular Plant-Microbe Interactions 26, 9–16. [DOI] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W. 2014. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathogens 10, e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. 2006a Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences, USA 103, 14302–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Dong R, Allen R, Davis EL, Baum TJ, Hussey RS. 2006b A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Molecular Plant-Microbe Interactions 19, 463–470. [DOI] [PubMed] [Google Scholar]
- Ibrahim HM, Alkharouf NW, Meyer SL, Aly MA, Gamal El-Din Ael K, Hussein EH, Matthews BF. 2011. Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Experimental Parasitology 127, 90–99. [DOI] [PubMed] [Google Scholar]
- Jammes F, Lecomte P, de Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, Abad P, Favery B. 2005. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. The Plant Journal 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jung C, Wyss U. 1999. New approaches to control plant parasitic nematodes. Applied Microbiology and Biotechnology 51, 439–446. [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Peret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischweski S, Muchow A, Guthörl D, Hause B. 2015. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biology 15, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou J, Tang J, Li B, de Oliveira MVV, Chai J, He P, Shan L. 2016. Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Reports 14, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SLF, Massoud SI, Chitwood DJ, Roberts DP. 2000. Evaluation of Trichoderma virens and Burkholderia cepacia for antagonistic activity against root-knot nematode, Meloidogyne incognita. Nematology 2, 871–879. [Google Scholar]
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben M, Davis EL. 2013. Nematode effector proteins: an emerging paradigm of parasitism. New Phytologist 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Olsen AN, Skriver K. 2003. Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends in Plant Science 8, 55–57. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Puthoff DP, Ehrenfried ML, Vinyard BT, Tucker ML. 2007. GeneChip profiling of transcriptional responses to soybean cyst nematode, Heterodera glycines, colonization of soybean roots. Journal of Experimental Botany 58, 3407–3418. [DOI] [PubMed] [Google Scholar]
- Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, Sawa S, Davis EL, Wang X, Simon R, Mitchum MG. 2011. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. The Plant Journal 65, 430–440. [DOI] [PubMed] [Google Scholar]
- Replogle A, Wang J, Paolillo V, Smeda J, Kinoshita A, Durbak A, Tax FE, Wang X, Sawa S, Mitchum MG. 2013. Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Molecular Plant-Microbe Interactions 26, 87–96. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH. 2002. Abscission, dehiscence, and other cell separation processes. Annual Review of Plant Biology 53, 131–158. [DOI] [PubMed] [Google Scholar]
- Rutter WB, Hewezi T, Maier TR, Mitchum MG, Davis EL, Hussey RS, Baum TJ. 2014. Members of the Meloidogyne avirulence protein family contain multiple plant ligand-like motifs. Phytopathology 104, 879–885. [DOI] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M. 2016. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5, e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A. 2016. Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Semblat JP, Rosso MN, Hussey RS, Abad P, Castagnone-Sereno P. 2001. Molecular cloning of a cDNA encoding an amphid-secreted putative avirulence protein from the root-knot nematode Meloidogyne incognita. Molecular Plant-Microbe Interactions 14, 72–79. [DOI] [PubMed] [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA. 2011. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. The Plant Cell 23, 2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons PC, Grundler FMW, von Mende N, Burrows PR, Wyss U. 1991. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. The Plant Journal 1, 245–254. [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, et al. 2018. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Aalen RB. 2008a Identification of a putative receptor-ligand pair controlling cell separation in plants. Plant Signaling & Behavior 3, 1109–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. 2006. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. The Plant Cell 18, 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. 2008b The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. The Plant Cell 20, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stø IM, Orr RJ, Fooyontphanich K, Jin X, Knutsen JM, Fischer U, Tranbarger TJ, Nordal I, Aalen RB. 2015. Conservation of the abscission signaling peptide IDA during Angiosperm evolution: withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Frontiers in Plant Science 6, 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Hohl M, Schardon K, Stintzi A, Schaller A. 2017. Post-translational maturation of IDA, a peptide signal controlling floral organ abscission in Arabidopsis. Communicative & Integrative Biology 11, doi:10.1080/19420889.2017.1395119. [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Yang R. 2012. IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants 2012, pls035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ML, Yang R. 2013. A gene encoding a peptide with similarity to the plant IDA signaling peptide (AtIDA) is expressed most abundantly in the root-knot nematode (Meloidogyne incognita) soon after root infection. Experimental Parasitology 134, 165–170. [DOI] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Liu B, Winge P, Butenko MA, Hornslien KS, Kumpf R, Aalen RB, Bones AM, Brembu T. 2015. The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. Journal of Experimental Botany 66, 5351–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Winge P, Cattan E, Wrzaczek M, Kangasjärvi J, Miller G, Brembu T, Bones AM. 2017. The IDA-LIKE peptides IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis thaliana. Journal of Experimental Botany 68, 3557–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P, Danchin EG, Neveu C, et al. 2011. The plant apoplasm is an important recipient compartment for nematode secreted proteins. Journal of Experimental Botany 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Spano T, Vlad D, Bou Daher F, Ouelhadj A, Kalaitzis P. 2007. Arabidopsis prolyl 4-hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiologia Plantarum 130, 471–483. [Google Scholar]
- Wang X, Hou S, Wu Q, Lin M, Acharya BR, Wu D, Zhang W. 2017. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. The Plant Journal 89, 250–263. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Williamson VM, Hussey RS. 1996. Nematode pathogenesis and resistance in plants. The Plant Cell 8, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss U, Grundler FMW. 1992. Feeding behavior of sedentary plant parasitic nematodes. Netherlands Journal of Plant Pathology 98, 165–173. [Google Scholar]
- Xue B, Hamamouch N, Li C, Huang G, Hussey RS, Baum TJ, Davis EL. 2013. The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 103, 175–181. [DOI] [PubMed] [Google Scholar]
- Youssef RM, Kim KH, Haroon SA, Matthews BF. 2013. Post-transcriptional gene silencing of the gene encoding aldolase from soybean cyst nematode by transformed soybean roots. Experimental Parasitology 134, 266–274. [DOI] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. 2005. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. The Plant Journal 41, 81–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.