A tomato GRAS transcription factor, designated as SlFSR (fruit shelf-life regulator), controls fruit shelf-life by regulating the expression of cell wall modification-related genes and metabolism of pectin and cellulose.
Keywords: Cell wall metabolism, GRAS transcription factor, overexpression, RNAi, shelf-life, SlFSR, tomato
Abstract
Fruit ripening represents a process that changes flavor and appearance and also a process that dramatically increases fruit softening. Fruit softening and textural variations mainly result from disruptions to the cell walls of the fruit throughout ripening, but the exact mechanisms and specific modifications of the cell wall remain unclear. Plant-specific GRAS proteins play a critical role in development and growth. To date, few GRAS genes have been functionally categorized in tomato. The expression of a novel GRAS gene described in this study and designated as SlFSR (fruit shelf-life regulator) specifically increased during fruit ripening, but was significantly decreased in the tomato mutant rin (ripening inhibitor). RNAi repression of SlFSR resulted in reduced expression of multiple cell wall modification-related genes, decreased the activities of PG (polygalacturonase), TBG (tomato β-galactosidase), CEL (cellulase), and XYL (β-D-xylosidase), and significantly prolonged fruit shelf-life. Furthermore, overexpression of SlFSR in mutant rin gave rise to up-regulated expression of multiple cell wall modification-related genes, such as PG, TBG4, CEL2, XYL1, PL, PE, MAN1, EXP1, and XTH5, and significantly shortened the fruit shelf-life. These findings reveal some of the genetic mechanisms underlying fruit cell wall metabolism and suggest that the SlFSR gene is another potential biotechnological target for the control of tomato fruit shelf-life.
Introduction
Fruits contain essential key nutrients of the diet of humans and many animals. Fruit ripening is a stepwise growth process that involves complex changes in physiological and metabolic processes such as fruit softening, carotenoid accumulation, chlorophyll degradation, and flavor biosynthesis. All these changes lead to the fruit developing the required quality for consumption, but the shelf-life of fruit is determined by how long these required features last. The main cause of fruit rotting is the extent of softening. The cost of fruit is also dependent on the extent of softening because it has a direct effect on palatability, shelf-life, resistance to post-harvest pathogen infection, transportation, storage, and consumer acceptability (Brummell and Harpster, 2001; Meli et al., 2010). Tomato belongs to the group of soft fruits characterized by a rapid and high loss of firm texture during the ripening process but is one of the most commonly used and versatile fruits in terms of its nutritional and commercial value. In addition, tomato has long served as an excellent model of fruit ripening and softening in research, primarily due to its small genome, efficient transient and stable transformation, short life cycle, well-characterized ripening mutants, rich genomic resources, and commercial importance (Moore et al., 2002; Giovannoni, 2007; Sato et al., 2012; Zhu et al., 2014).
Physiologically, tomato is a typical climacteric fruit and its ripening is determined by proper softening, which is caused by short-term ethylene biosynthesis and higher respiration (Adams-Phillips et al., 2004). Studies that have identified various mechanisms that control fruit ripening have greatly benefited from the availability of numerous ripening-deficient mutants, which have been very valuable in exploring the roles of cell wall-modifying proteins and changes in cell wall during softening. These mutants include rin (Vrebalov et al., 2002), Nr (never ripe) (Wilkinson et al., 1995), nor (non-ripening) (Giovannoni, 2007), and Cnr (Colorless non-ripening) (Orfila et al., 2001).
Fruits of rin and nor mutants do not show increases in lycopene and carotenes, and soften very gradually. Studies of ripening control mechanisms have shown that the RIN and NOR genes both act upstream of ethylene, and regulate both ethylene and non-ethylene-controlled functions (Tigchelaar et al., 1978; Moore et al., 2002). RIN encodes a MADS-box transcription factor, which is considered as a pivotal regulator of tomato fruit ripening (Vrebalov et al., 2002). However, both rin and nor fruits are unpalatable and have poor quality, which limits their commercial value. Therefore, genetic studies involving the transfer of genes to regulate specific genes involved in fruit softening are one of the major areas of biological research. This approach can also potentially reduce the level of fruit softening while permitting the accumulation of the necessary components of normal ripening (i.e. sugars, pigments, volatiles, and organic acids), increase shelf-life, and decrease spoilage rate. Recently, the suppression of cell wall modification-related genes has been used to reduce the softening of fruit in transgenic tomato (Han et al., 2016; Uluisik et al., 2016; Yang et al., 2017). However, these studies have had very little success. Disruption of the cell wall is mainly responsible for fruit softening and textural variations throughout ripening, but the exact mechanisms and particular functions of cell wall modifications during fruit ripening are still poorly understood. In addition, the improvements to fruit shelf-life accomplished to date have not been sufficient, thus the identification of more targets is required.
The GRAS proteins are a recently identified plant-specific family of putative transcription factors, whose name derives from the three initially identified members, GAI (gibberellic acid insensitive), RGA (repressor of GAI), and SCR (scarecrow) (Pysh et al., 1999). Typically, GRAS proteins are composed of 400–770 amino acid residues, and the GRAS domain contains five conserved motifs, including LHRI, VHIID, LHRII, PFYRE, and SAW (Bolle, 2004). To date, genes encoding GRAS proteins have been studied primarily in the model plants Arabidopsis and rice, in which 34 and 60 putative GRAS members have been identified, respectively (Liu and Widmer, 2014). Plant molecular genetics studies have shown that GRAS genes play various critical roles in growth and development, such as in root development, phytohormones, light signaling pathways, and transcriptional regulation in response to biotic and abiotic stress (Bolle, 2004; Smit et al., 2005; Ma et al., 2010; Sun et al., 2012; Liu and Widmer, 2014); for example, AtSCL3 (a member of the AtSCL3 subfamily) in assimilating several signals in root cell elongation of Arabidopsis (Heo et al., 2011), and DLT (a member of the DLT subfamily) in brassinosteroid signaling of rice (Tong et al., 2009). In addition, it has been reported that the overexpression of the Populus euphratica gene PeSCL7 (AtSCL4/7 subfamily) in transgenic Arabidopsis boosts drought and salt tolerance (Ma et al., 2010). Recently, a GRAS protein known as RAM1 has been considered essential for infection by arbuscular mycorrhizal fungi (Gobbato et al., 2012). Although the GRAS proteins are encoded by a large gene family and have been studied for several years, currently we have only an incomplete understanding of many of their features, and the specific biological functions of most members remain unclear.
Few GRAS family members have been functionally categorized in tomato, and the contribution of GRAS proteins to fruit ripening and/or softening has not been reported to date. Previously, 17 putative tomato GRAS genes were identified by Mayrose et al. (2006). Pseudomonas syringae pv. tomato was used to up-regulate six SlGRAS transcripts and the transcripts of eight SlGRAS genes increased in response to mechanical stress. Suppression of SlGRAS6 impaired tomato resistance to P. syringae pv. tomato. The first characterized tomato GRAS gene, Ls (Lateral suppressor), is obligatory for the initiation of axillary meristems (Mayrose et al., 2006). A gibberellin (GA)-constitutive-response tomato mutant pro (procera) carries a point mutation in the GRAS region of the gene encoding SlDELLA, a repressor in the GA signaling pathway, which was shown to function in the control of flower morphology, cell division, expansion, and the auxin-signaling pathway throughout fruit set and growth (Brummell and Harpster, 2001; Carrera et al., 2012). Solyc07g052960 was reported by Fei et al. (2004) to be the direct target of RIN, and it was revealed to be a ripening-specific GRAS gene. More recently, Solyc07g052960 was also identified as a direct target of RIN and subjected to qChip–PCR, which showed a high degree of RIN dependence, the highest FCWT value (683.5), and the highest ECS value (100.6) among the gene category associated with transcription factors (Fujisawa et al., 2013; Fujisawa et al., 2012). In this study, we explored the function of this gene, named as SlFSR (fruit shelf-life regulator), which was isolated from tomato fruit by a cDNA clone, and whose mRNA specifically accumulates in ripening fruits. RNAi repression of SlFSR was accomplished to further examine its role in tomato. In SlFSR-RNAi fruits, decreased expression of multiple cell wall modification-related genes, reduced PG (polygalacturonase), TBG (tomato β-galactosidase), CEL (cellulase), and XYL (β-D-xylosidase) activities, and significantly enhanced shelf-life were detected. A SlFSR-overexpressing rin mutant was also generated, in which the overexpression of SlFSR resulted in the up-regulation of multiple cell wall modification-related genes, including PG, TBG4, CEL2, XYL1, pectate lyase (PL), pectinesterase (PE), mannosidase (MAN1), xyloglucan endotransglucosylase/hydrolase (XTH5), and expansin 1 (EXP1), and significantly shortened fruit shelf-life. These findings suggest that SlFSR plays an essential role in fruit post-harvest storage, and its underlying molecular mechanisms involved in fruit cell wall modification are discussed. Our results also indicate another possible biotechnological approach to extend fruit shelf-life, in addition to altering ethylene biosynthesis and cell wall metabolism.
Materials and methods
Promoter analysis of SlFSR in tomato
To study the putative cis-elements in the promoter region of the SlFSR gene, the promoter sequence (2 kb region upstream of the 5ʹ end of the predicted open reading frame) of SlFSR was extracted from the SGN catalog (https://solgenomics.net/; accessed 20 October 2017) and searched against the promoter database PLACE (http://www.dna.affrc.go.jp/PLACE/index.html; accessed 20 October 2017) (Higo et al., 1999).
Plant materials and growth conditions
The wild-type (WT) tomato Solanum lycopersicum Mill. cv. Ailsa Craig, rin and Nr mutants, SlFSR-RNAi, and SlFSR-overexpressing transgenic lines were grown in a greenhouse under the following conditions: 16 h day (27 °C) and 8 h night (19 °C), at 80% relative humidity; plants were irrigated regularly. For tissue-specific expression of SlFSR, leaves, flowers, sepals, roots, and fruits at various stages of development were gathered. Flowers were sampled at anthesis. Fruit development was denoted as days post anthesis (DPA). Fruits at 20 DPA were defined as immature green (IMG). Fruits at 35 DPA were defined as mature green (MG) and considered as full fruit growth but with no clear ripe fruit color evident. Breaker (B) fruit was recorded as fruit with the first appearance of orange color. The following ripening periods were distinguished as B+4 (4 days after breaker) and B+7 (7 days after breaker). WT and rin lines were used to produce SlFSR-RNAi and SlFSR-overexpressing transgenic lines, respectively. Fruits from the Nr and rin mutants were harvested at IMG, MG, B, B+4, and B+7 stages when they showed equivalent characteristics to those defined in WT tomato. All samples were immediately transferred to liquid nitrogen and stored at –80 °C until required.
Construction of RNAi and overexpression vectors and plant transformation
The SlFSR RNAi and overexpression constructs were made using the pBIN19 and pBI121 vectors, respectively, as described previously (Dong et al., 2013; Xie et al., 2014). The detailed method was as follows: for the RNAi vector construction, a 718 bp fragment of DNA was amplified with SlFSR-RNAi-F/R primers (see Supplementary Table S1 at JXB online) which had been joined with KpnI/ClaI and XhoI/XbaI restriction sites at the 5ʹ end. The amplified products were digested with the restriction enzymes ClaI/XbaI and KpnI/XhoI and linked to the plasmid pHANNIBAL using the same restriction enzymes. The double-stranded RNAi expression unit was digested with the restriction enzymes SacI/SpeI and inserted into the plant binary vector PBIN19 via SacI/XbaI restriction sites to form the RNAi vector. For construction of the overexpression construction, the full-length cDNA of SlFSR was amplified with SlFSR-over-F/R primers to which XbaI/SacI restriction sites were inserted at the 5ʹ end (Supplementary Table S1). The amplified products were digested with XbaI/SacI and linked to the plant binary vector pBI121 at XbaI/SacI restriction sites. Finally, the RNAi vector was transformed into WT tomato and the overexpression vector was transformed into the tomato mutant rin through the freeze-thaw method, using Agrobacterium tumefaciens strain LBA4404 (An, 1987). Transgenic lines were selected on the basis of kanamycin (50 mg l−1) resistance. Genomic DNA of the WT and transgenic lines was isolated using a kit (Invitrogen, Shanghai, China) and the presence of T-DNA was confirmed by PCR using NPTII-F/R primers (Supplementary Table S1).
Total RNA extraction and qRT–PCR analysis
Total RNA was extracted from various samples using Trizol reagent (Invitrogen, Shanghai, China). First-strand cDNA was synthesized using a kit (Promega, Beijing, China). Quantitative reverse-transcription–PCR (qRT–PCR) was performed by using a CFX96™ Real-Time System (Bio-Rad, USA). The reaction mixture consisted of 5 μl enzyme solution (2×GoTaq® qPCR Master Mix, Promega, Beijing, China), 1 μl cDNA, 0.5 μl primer pairs (10 mM), and 3.5 μl distilled water. The reaction conditions were 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and Tm (the most suitable temperature for each gene) for 45 s, followed by a melting curve analysis. The CAC gene of tomato was used as an internal control for expression analysis (Expósito-Rodríguez et al., 2008; Nicot et al., 2005), and the 2–ΔΔCT method was used for the analysis of relative expression levels (Livak and Schmittgen, 2001). A no-template control was also included in each gene study. All qRT–PCRs were performed in three replicates. The primers used for each gene are listed in Supplementary Table S1; a standard curve was performed for each pair of specific primers.
Enzyme determination assays
For all enzyme determination assays, 0.1 g of fresh pericarp at the B+4 stage was ground in an ice water bath. The activity of PG, TBG, CEL, and XYL in rin, SlFSR transgenic lines, and WT tomato was analyzed using a kit (Komin Suzhou, China) according to the manufacturer’s instructions. Three individual fruits were sampled from each line and the assays were done in triplicate.
Metabolite analysis
For analysis of the quantity of pectin, 3 mg of pericarp at the B+4 stage was ground in liquid nitrogen. Total pectin, water-soluble pectin, cellulose, and hemicelluose were analyzed using a kit (Komin Suzhou, China) according to the manufacturer’s instructions. The levels of soluble sugar in the fruit were determined exactly as described previously (Fernie et al., 2001). Malic acid and citric acid contents were measured as described by Nunes-Nesi et al. (2007). Three independent fruits at the B+4 stage were sampled and the assays were performed in triplicate.
Ethylene measurement
The rin mutant, WT, and transgenic lines were harvested at the B, B+4, and B+7 stages and kept at room temperature for 3 h to reduce the influence of wound-induced ethylene produced in response to harvesting of the fruits. The fruits were weighed and then placed in 235 ml glass jars sealed with a plastic membrane and stored for 24 h at room temperature(Zhu et al., 2014). The ethylene concentration in a 1 ml sample of headspace gas from each glass jar was measured by using the method of Chung et al. (2010).
Pigment extraction
Carotenoids were extracted from a 5 mm wide rectangular strip of freeze-dried pericarp, sampled from around the equator of fruits, according to an improved protocol described by Forth and Pyke (2006). Each sample (of known weight) was ground into a powder in liquid nitrogen and then placed into a 2 ml tube. Pigments were extracted by the addition of hexane:acetone (6:4, v/v). The sample was then centrifuged at 2500 g for 5 min and the supernatant was placed in a new tube after centrifugation. The sediment was repeatedly extracted with hexane:acetone (6: 4, v/v) until it was colorless. The absorbance of the supernatant was immediately measured. The total carotenoids content was quantified using the equation: total carotenoids (mg ml–1)=4×(OD450)×10 ml/1 g. All experiments were repeated for individual samples at least three times.
Water loss measurements
Nine fruits from WT tomato and each of the SlFSR-RNAi lines were collected at the B+4 stage, and nine fruits from rin and each of the overexpressing lines were harvested at the B stage. Fruits of WT and SlFSR-RNAi lines were kept at room temperature (23–25 °C with 55–60% relative humidity) for 2 months after harvest; fruits of rin and SlFSR-overexpressing lines were stored at room temperature for 3 months after harvest. Water loss per unit fruit weight was calculated after recording the weight decrease over time. The weight loss of WT and SlFSR-RNAi fruits was measured at 0, 7, 10, 13, 16,19, 22, 25, 28, 31, 34, 37, and 40 days.
Storage assays of tomato fruits
Fruits of WT and SlFSR-RNAi lines were harvested at the B+4 stage, and fruits of rin and SlFSR-overexpressing lines were harvested at the B stage. All the fruits were disinfected with 10% bleach for 10 min, followed by rinsing with sterilized water and air-drying. The fruits of WT and SlFSR-RNAi lines were stored at room temperature for 2 months; fruits of rin and SlFSR-overexpressing lines were stored at room temperature for 3 months. The chromatic softening and collapse of the fruits were evaluated by taking photographs at the beginning (day 7 after harvesting) and end (day 90 after harvesting) of the storage period.
Microscopic observations
Approximately 2 cm of pericarp was collected from fruits stores for 2 months (WT and RNAi lines) or 3 months (rin and overexpressing lines). Samples were immediately fixed in FAA liquid (70% ethanol, acetic acid, and formaldehyde mixed 18:1:1 v/v) and subsequently dehydrated, wax embedded, sectioned, dewaxed, and stained with safranin and fast green. All observations were made under a light microscope (Olympus IX71, Japan) and photographed. Three replicates were performed for each sample.
Statistical analysis
Data were subjected to analysis of variance with SPSS Statistics 18.0. Differential expression levels were considered to be statistically significant when exceeding the Dunnett’s test critical value at the P<0.05 level. The difference was defined as ‘repressed’, ‘induced’, or ‘different’ only if such differences met the above standard.
Results
Expression profiles of SlFSR in WT tomato
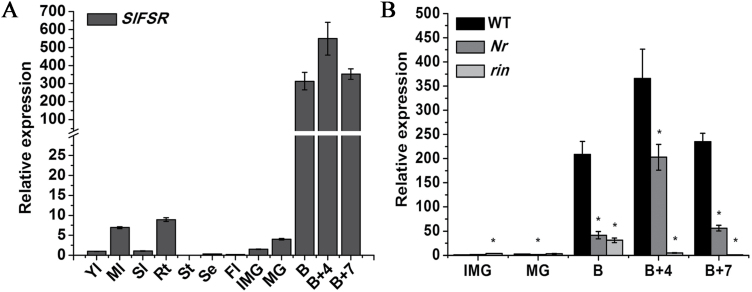
The expression profiles of the SlFSR gene in different tissues of WT tomato were detected by qRT–PCR. SlFSR mRNA was predominantly expressed in the fruit during the ripening stages (B, B+4, and B+7), but little or no expression was observed in all other tissues (Fig. 1A). The B stage sees the first ripening-related changes in tomato fruit due to climacteric changes in ethylene production, cell wall disruption, and synthesis of lycopene, followed by an obvious increase in the expression level of cell wall hydrolases (Fischer and Bennett, 1991; Giovannoni, 2004). Consequently, higher expression of SlFSR at the ripening stage indicates its role in tomato fruit ripening and softening.
Fig. 1.
(A) Expression of SlFSR in different tissues of WT tomato. (B) Expression of SlFSR in WT and ripening mutant fruits. Total RNA from rin and Nr fruits at the IMG, MG, B, B+4, and B+7 stages equivalent to WT tomato was subjected to qRT–PCR analysis. B, Breaker stage; B+4, 4 days after breaker stage; B+7, 7 days after breaker stage; Fl, flower; IMG, immature green; MG, mature green; Ml, mature leaf; Rt, root; Se, sepal; Sl, senescent leaf; St, stem; Yl, young leaf. Data are the mean ±SE of three independent experiments. Significant differences (P<0.05) are denoted by asterisks.
Expression of SlFSR is inhibited in tomato ripening mutants and regulated by ethylene
The level of SlFSR expression increased mainly in the ripening stage of tomato fruit; this led us to examine its expression in the ripening-impaired mutants rin (in which higher ethylene is not produced and ripening activities are affected) and Nr (which is unresponsive to ethylene). As in WT fruits, almost undetectable SlFSR gene transcription was observed in IMG and MG fruits of rin and Nr mutants (Fig. 1B). SlFSR expression was significantly down-regulated in both rin and Nr (especially in rin) (Fig. 1B), indicating that SlFSR expression is obstructed by both the RIN and Nr mutations. The reduced expression of SlFSR strongly indicates its contribution to fruit ripening and induction by ethylene. Indeed, a putative ethylene responsive element was found in the promoter sequence of SlFSR (Supplementary Fig S1). These observations indicate the relationship between ethylene and SlFSR expression, and the action of ripening regulators downstream of SlFSR.
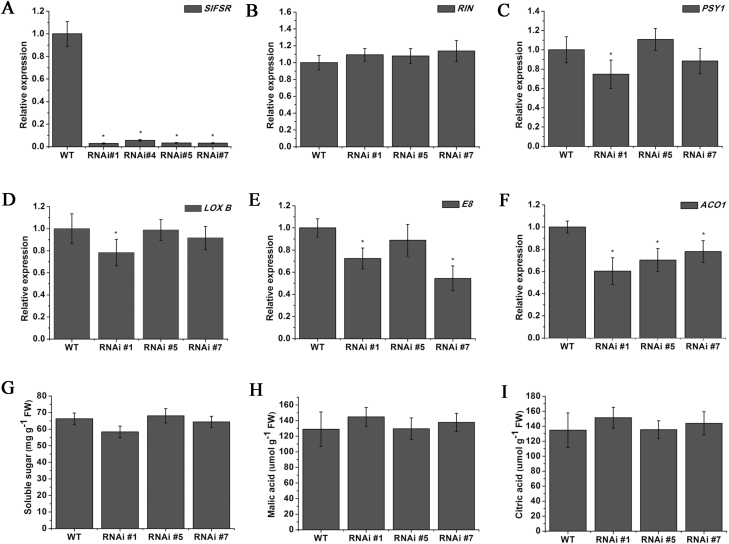
SlFSR-RNAi fruits go through normal climacteric ripening and color development
To further study the role of the SlFSR gene, several independent RNAi silencing lines were obtained. The accumulation of SlFSR transcript was greatly silenced, to approximately 2–5% of control levels at the B+4 stage, in the RNAi lines (Fig. 2A). Curiously, regardless of the specific accumulation of SlFSR in ripening fruits, the significantly silencing of SlFSR had no apparent effect on the tomato fruit ripening phenotype (data not shown). It is known that the production, perception, and transfer of ethylene signals are required for complete fruit ripening (Alexander and Grierson, 2002), and thus the expression levels of ripening-related genes were assessed in the SlFSR-RNAi lines. The expression of PHYTOENE SYNTHETASE1 (PSY1) (Fray and Grierson, 1993), RIN (Vrebalov et al., 2002), and TomloxB (Griffiths et al., 1999) were almost unchanged in SlFSR-RNAi lines relative to expression in WT tomato, but expression of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO1) (Barry and Giovannoni, 2007) and E8 (Lincoln et al., 1987) was significantly reduced in SlFSR-RNAi fruits (Fig. 2B–F). This slight reduction apparently did not affect the phenotype of SlFSR fruits. Moreover, some compounds characteristic of flavor, such as sugar, malic acid, and citric acid were measured in SlFSR-RNAi fruit, and no significant difference was found in the contents of each of them relative to WT fruit (Fig. 2G–I).
Fig. 2.
Silencing of SlFSR in WT tomato causes no obvious phenotypic changes. Relative expression profiles of SlFSR (A) and ethylene- and ripening-related genes (B–F) in WT and SlFSR-RNAi fruits at B+4 stage. (G–I) Analysis of flavor compound contents in WT and SlFSR-RNAi fruits at B+4 stage. The expression data for WT plants were normalized to a value of 1. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between WT and RNAi lines.
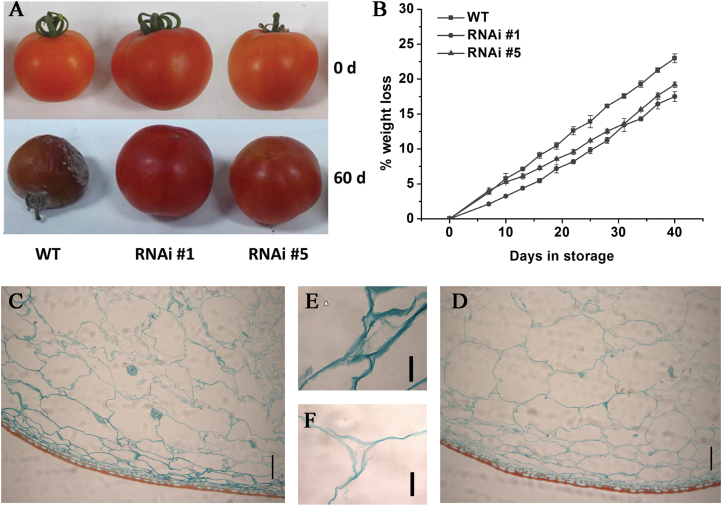
Silencing of SlFSR greatly extends tomato fruits shelf-life
To assess the shelf-life of SlFSR-RNAi fruits, storage tests were performed using B+4 fruits at room temperature. After 2 months of storage, WT tomato fruits were completely collapsed and severely infected, while SlFSR-RNAi fruits showed delayed signs of deterioration and no visible infection was observed (Fig. 3A). In addition, the RNAi lines showed obviously lower weight loss (Fig. 3B). Microscopic examination of pericarp revealed ruptured cells with an irregular shape in WT fruits after 40 days of storage (Fig. 3C), while the cells of the RNAi lines were comparatively round shaped and normal in appearance (Fig. 3D). In addition, the cell wall of WT fruits was degraded, while at this point the SlFSR-RNAi fruits showed a normal cell wall structure (Fig. 3E, F). These results indicate that silencing SlFSR in tomato is sufficient to change the post-harvest ripening process and greatly prolong fruit shelf-life.
Fig. 3.
Silencing SlFSR alters cell wall components and increases shelf-life of tomato fruit. (A) WT and SlFSR-RNAi fruits harvested at B+7 stage were stored at room temperature for 60 days. (B) Weight loss of WT and SlFSR-RNAi fruits during storage. (C, E) Microscopic observations of WT tomato fruit stored for 40 days. (D, F) Microscopic observations of SlFSR-RNAi tomato fruit stored for 40 days. (C, D) Bar=50 μm; (E, F) Bar=25 μm.
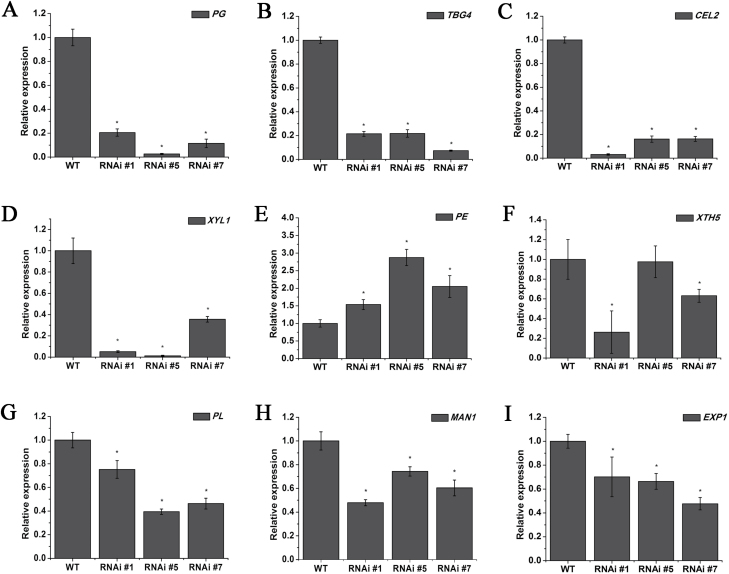
Expression profiles of cell wall modification-related genes in SlFSR-RNAi fruits
Fruit ripening is associated with cell wall modifications (Orfila et al., 2002). Tomato cell wall modification involves processes including depolymerization and solubilization of pectins and hemicellulosic polysaccharides (Brummell, 2006). This process is stimulated by various cell wall modifying enzymes and proteins, including PG, CEL, TBG, XYL, XTH, and PE, and other cell wall loosening proteins, such as EXP (Brummell and Harpster, 2001). To investigate whether the expression of cell wall modification-related genes differs between SlFSR-RNAi and WT fruits, transcripts of PE (Phan et al., 2007), PG (Giovannoni et al., 1989), CEL2 (Lashbrook et al., 1994), XYL1(Buanafina et al., 2015), XTH5 (Miedes and Lorences, 2009), TBG4 (Smith et al., 2002), MAN1 (Meli et al., 2010), PL (Uluisik et al., 2016), and EXP1 (Brummell et al., 1999b) were detected in B+4 fruits and quantified relative to expression in WT fruits (Fig. 4A–I). The expression levels of all genes except PE were down-regulated; notably, PG, TBG4, CEL2, and XYL1 were down-regulated by more than 80% (Fig. 4A–D). These results suggest that silencing of SlFSR may affect tomato cell wall modification.
Fig. 4.
(A–I) Relative expression profiles of cell wall metabolism genes in the pericarp of WT and SlFSR-RNAi tomato fruits. The expression data for WT plants were normalized to a value of 1. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between WT and RNAi lines.
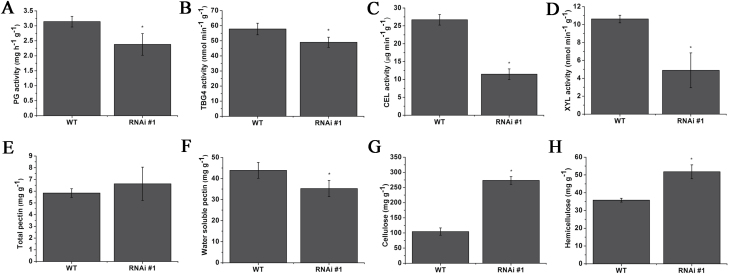
The significant inhibition of the expression of cell wall modification-related genes in SlFSR-RNAi fruits suggests that the activity of relevant enzymes is also reduced in these lines. Similar to the reduced PG, TBG4, CEL2, and XYL1 expression that was observed at the mRNA level (see above), reduced activity of PG, TBG, CEL, and XYL at the protein level was observed in SlFSR-RNAi fruits, relative to WT fruits, at the B+4 stage (Fig. 5A–D). The total pectin content in SlFSR-RNAi fruits at the B+4 stage was not significantly different from that in WT fruits (Fig. 5E). However, water-soluble pectin was lower in SlFSR-RNAi fruits (Fig. 5F). Moreover, the cellulose and hemicellulose contents in SlFSR-RNAi fruits at the B+4 stage were higher than their respective concentrations in WT fruits (Fig. 5G, H). These results suggest that down-regulation of cell wall modification-related genes and changes in the activity of cell wall modification-related enzymes and cell wall components in SlFSR-RNAi fruits contribute to the prolonged shelf-life observed for RNAi fruits.
Fig. 5.
(A–D) Activities of PG, TBG, CEL, and XYL in WT and SlFSR-RNAi fruits at the B+4 stage. (E–H) Contents of (E) total pectin, (F) water-soluble pectin, (G) cellulose and (H) hemicellulose in SlFSR-RNAi and WT fruits at the B+4 stage. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between WT and RNAi lines.
Overexpression of SlFSR cannot restore the course of fruit ripening in the rin mutant
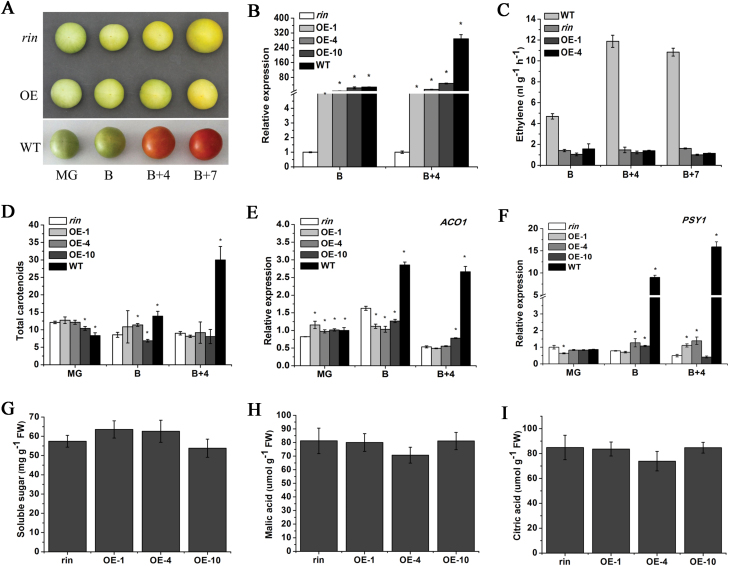
To explore the function of SlFSR in fruit ripening and color development in more depth, a SlFSR-overexpression vector was constructed and transformed into the tomato mutant rin. Transgenic rin lines (OE-1, OE-4, and OE-10) were generated; SlFSR mRNA accumulated to a higher level in these lines than in rin; expression in the transgenic lines was similar to that of WT at the B stage (Fig. 6B). However, there was no difference in fruit color between the transgenic rin lines and rin (Fig. 6A). In addition, the SlFSR-overexpressing lines did not exhibit obvious differences in ethylene production from B to B+7 stage, like rin; in contrast, WT fruits showed a rapid and considerable increase in ethylene production at the B+4 stage (Fig. 6C). Moreover, overexpression of SlFSR resulted in little change in carotenoid accumulation compared with rin, whereas a significant increase was observed in WT fruits at the B and B+4 stages (Fig. 6D). Similarly, overexpression of SlFSR did not activate the expression of ACO1 and PSY1, while a dramatic increase in transcripts of ACO1 and PSY1 was observed in WT fruits at the B and B+4 stages (Fig. 6E, F). Nevertheless, some flavor compounds, such as sugar, malic acid, and citric acid, showed no significant change in SlFSR-overexpressing transgenic lines compared with rin (Fig. 6G–I).These results indicate that the course of fruit ripening of the mutant rin cannot be restored to a WT-like phenotype by the overexpression of SlFSR.
Fig. 6.
Overexpression of SlFSR in rin leads to no obvious phenotypic changes. (A) Color of rin, SlFSR-overexpressing transgenic rin (OE), and WT fruits at the MG, B, B+4, and B+7 stages. (B) Relative levels of SlFSR mRNA in rin (control), WT, and SlFSR-overexpressing fruit at the B and B+4 stages. The expression data for rin fruits were normalized to a value of 1. (C–F) Ethylene production (C), accumulation of carotenoids (D), expression of ACO1 (E), and expression of PSY1 (F) in rin, WT and SlFSR-overexpressing transgenic rin fruits. (G–I) Analysis of flavor compounds in WT, rin and SlFSR-overexpressing transgenic rin fruits. The expression data for rin fruits were normalized to a value of 1. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between rin and the other lines.
Overexpression of SlFSR shortens the shelf-life of transgenic rin fruits
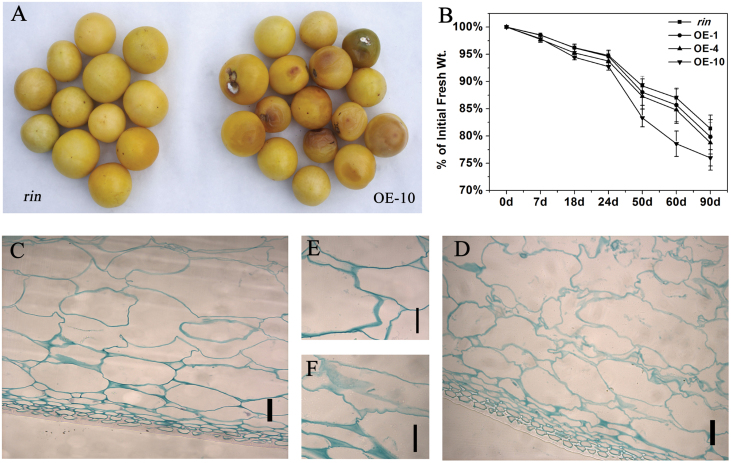
To investigate the effects of SlFSR overexpression on fruit shelf-life, fruits of the mutant rin and SlFSR-overexpressing transgenic rin were harvested at the breaker stage and stored at room temperature. Despite the lack of discernible differences in ripening and color development between SlFSR-overexpressing lines and rin fruits (see (Fig. 6A), the overexpressing lines exhibited a shorter fruit shelf-life than rin. SlFSR-overexpressing lines showed visible signs of rot and deterioration 3 months after harvest; by contrast, no obvious signs of deterioration or rot were observed in rin fruit stored under the same conditions for 3 months (Fig. 7A). The fresh weight of the fruits was also measured during post-harvest storage. SlFSR-overexpressing lines showed a significantly larger decrease in fruit fresh weight than rin (Fig. 7B). Microscopic examination of pericarp samples taken after 90 days of post-harvest storage revealed ruptured cells with irregular shape in the SlFSR-overexpressing lines (Fig. 7C), while in rin the cells were comparatively round shaped and normal in appearance (Fig. 7D). In addition, the cell walls of the fruit of SlFSR-overexpressing lines were degraded, whereas rin fruit showed a normal cell wall structure (Fig. 7E, F). These results suggest that the overexpression of SlFSR in rin significantly shortens fruit shelf-life.
Fig. 7.
Overexpression of SlFSR alters tomato fruit cell wall composition and shortens the shelf-life. (A) Appearance of rin and SlFSR-overexpression transgenic rin fruits harvested at B+4 and stored at room temperature for 90 days. (B) Fresh weight loss of rin and SlFSR-overexpressing fruits during storage. (C, E) Microscopic observations of rin tomato fruit stored for 60 days. (D, F) Microscopic observations of SlFSR-overexpressing tomato fruit stored for 60 days. (C, D) Bar=50 μm; (E, F) Bar=25 μm.
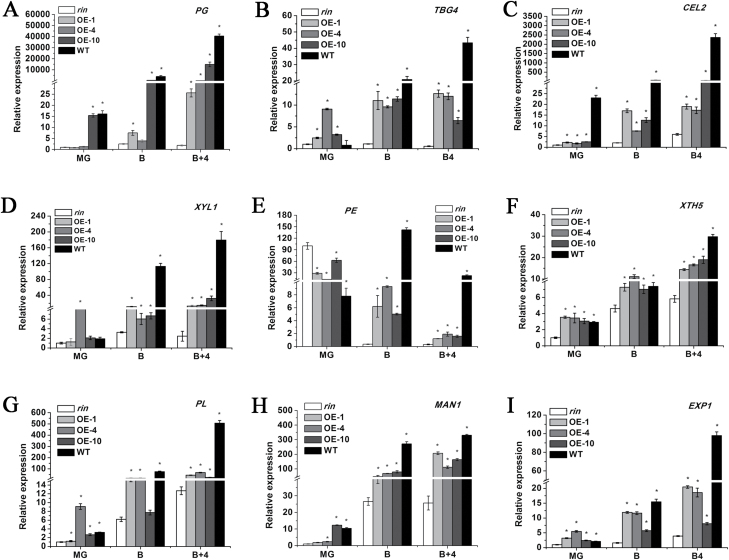
Expression profiles of cell wall modification-related genes in SlFSR-overexpressing fruits
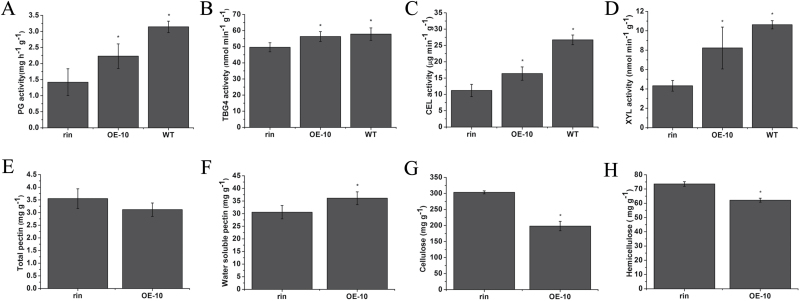
In order to further ascertain the molecular mechanisms of the shortened fruit shelf-life in SlFSR-overexpressing tomato lines, the expression profiles of cell wall modification-related genes, including PG, PE, TBG4, CEL2, XYL1, XTH5, EXP1, MAN1, and PL, were examined in SlFSR-overexpressing, rin, and WT fruits (Fig. 8A–I). The transcript levels of all these genes were up-regulated in SlFSR-overexpressing transgenic rin fruits compared with rin fruits at the B and B+4 stages, but did not reach the levels observed in WT fruits. The activity of the protein products of some of these genes was assessed: PG, TBG, CEL, and XYL showed a significant increase in activity in WT and SlFSR-overexpressing fruits, relative to the activity in rin fruits, at the B+4 stage (Fig. 9A–D). The total pectin content in SlFSR-overexpressing fruits at the B+4 stage was slightly (although not significantly) lower than that in rin (Fig. 9E). By contrast, the water-soluble pectin content was higher in SlFSR-overexpressing fruit than in rin (Fig. 9F), suggesting that the down-regulation of SlFSR promotes pectin degradation. Moreover, the cellulose and hemicellulose contents in SlFSR-overexpressing fruits at the B+4 stage were significantly lower than in rin (Fig. 9G, H). These results indicate that the expression of cell wall modification-related genes is positively regulated by SlFSR, and that the overexpression of SlFSR in rin indeed accelerates the degradation of the fruit cell wall.
Fig. 8.
(A–I) Relative expression profiles of cell wall metabolism genes in the pericarp of rin, WT, and SlFSR-overexpressing transgenic rin tomato fruits. The expression data for rin fruits were normalized to a value of 1. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between rin and the other lines.
Fig. 9.
(A–D) Activities of PG, TBG, CEL, and XYL in the pericarp of rin and SlFSR-overexpressing transgenic rin tomato fruits at the B+4 stage. (E–H) Contents of (E) total pectin, (F) water-soluble pectin, (G) cellulose, and (H) hemicellulose in SlFSR-overexpressing and rin fruits at the B+4 stage. Each value represents the mean ±SE of three replicates. Asterisks indicate significant differences (P<0.05) between rin and SlFSR-overexpressing lines.
Discussion
Tomato fruits are rich in vitamins, fiber, minerals, and antioxidants, which are key components of human nutrition. In tomato production, great losses often occur as a result of over-softening and subsequent fungal infections during post-harvest transportation and storage, and post-harvest loss is one of the major problems in tomato production. To date, numerous genes have been reported to control tomato fruit growth, ripening ,and softening (Brummell and Harpster, 2001; Karlova et al., 2014).
Plant-specific GRAS proteins play critical and diverse roles in growth and development (Sun et al., 2012). Here, we report on a new GRAS gene, designated as SlFSR (fruit shelf-life regulator), whose mRNA specifically accumulates in ripening fruits, implying its potential role in tomato fruit ripening and/or softening (Fig. 1). Interestingly, silencing SlFSR in tomato greatly prolonged the shelf-life and reduced cell degradation of fruits, as confirmed by the decreased expression of multiple cell wall modification-related genes and reduced PG, TBG, CEL, and XYL activities, but did not exert a significant influence on the normal fruit ripening phenotype (Figs 2–5). In addition, transgenic SlFSR-overexpressing lines exhibited a similar inhibited ripening process to that of the rin mutant and had comparable levels of ethylene and carotenoids production to rin (Fig. 6), suggesting that the overexpression of SlFSR is unable to recover the impaired ripening phenotype of rin. Essentially, overexpression of SlFSR in the rin background significantly reduced the shelf-life and increased the rate of water loss in stored fruits by up-regulating the expression of multiple cell wall modification-related genes (Figs 6–8); increased activities of PG, TBG, CEL, and XYL were detected in SlFSR-overexpressing fruits (Fig. 9).
It has been well documented that cell wall modification-related proteins, including PG, TBG4, CEL2, XYL1, PE, XTH5, PL, MAN1, and EXP1, function in cell wall disruption, and are generally considered as key factors in the changes that occur to the primary cell wall during fruit ripening (Owino et al., 2005; Brummell, 2006; Vicente et al., 2007). For instance, PG is involved in polyuronide solubilization and depolymerization during ripening, but is not necessary or sufficient for tomato fruit ripening. However, resistance to post-harvest pathogens cracking and shelf-life were improved in PG-RNAi lines (Kramer et al., 1992; Hadfield and Bennett, 1998). CELs are involved in fruit ripening, tissue abscission, cell extension and differentiation (Flors et al., 2007). Greatly increased levels of CEL2 mRNA were found at the onset of ripening, but the suppression of CEL2 did not affect the changes in fruit softening (Brummell et al., 1999a). Subsequently, Flors et al. (2007) found that lack of both CEL1 and CEL2 decreases susceptibility to Botrytis cinerea infection in tomato. XYL is involved in cell wall degradation, via participation in the breakdown of xylans (Buanafina et al., 2015). XTHs are believed to be related to the maintenance of the structural integrity of the cell wall, and XTH5 is evidently related to fruit ripening (Miedes and Lorences, 2009). PE is involved in pectin depolymerization and affects tissue integrity in over-ripe fruit (Brummell and Harpster, 2001). Silencing PL at the mRNA level improved tomato shelf-life; this improvement was caused by differences in the levels of total pectin and soluble pectin (Uluisik et al., 2016). MAN is involved in fruit shelf-life, without any negative effect on vegetative growth, fruit development, days to maturity, seed production, and yield (Meli et al., 2010). β-Galactosidase, encoded by TBG4, plays a role in the hydrolysis of galactan side-chains of pectic polysaccharides. Down-regulation of TBG4 results in significantly greater fruit firmness compared with WT fruits (Smith et al., 2002). The suppression and overexpression of EXP1 mRNA and protein accumulation caused changes in fruit softening during ripening, and multiple changes in cell wall polysaccharide metabolism (Brummell et al., 1999b). Therefore, given that these cell wall modification-related genes/proteins play important roles in fruit softening, shelf-life, and resistance to post-harvest pathogens, their reduced expression/activities may extend the shelf-life of SlFSR-RNAi fruits. In contrast, their increased expression/activities may result in the shortened shelf-life of SlFSR-overexpressing fruits. Taken together, these results suggest that the SlFSR transcription factor may participate in the modulation of tomato cell wall metabolism and may affect fruit shelf-life by regulating the expression of genes related to cell wall modification.
Although changes in the expression level of SlFSR could significantly influence the shelf-life of tomato fruits, the ripening phenotype of the SlFSR-RNAi lines and SlFSR-overexpressing transgenic rin fruits showed no significant changes. Generally, ethylene plays a critical role during fruit ripening and softening in climacteric fruits (Hiwasa et al., 2003; Ergun et al., 2005; Nishiyama et al., 2007); however, it is not the only key regulator of fruit ripening. RIN is believed to also act as a key ripening regulator by acting upstream of both ethylene-dependent and ethylene-independent pathways (Vrebalov et al., 2002). The rin mutation has been investigated extensively in studies to identify genes associated with the ripening process. RIN-targeted genes participate in a range of fruit ripening-associated metabolic and regulatory mechanisms, including cell wall metabolism and ethylene signaling, suggesting that RIN controls fruit ethylene production and softening through the transcriptional regulation of ethylene biosynthesis genes and cell wall-modifying genes during ripening (Fujisawa et al., 2011). To date, 342 genes positively regulated by RIN and 473 genes negatively regulated by RIN have been identified (Fujisawa et al., 2012). Moreover, 241 genes that are direct targets of RIN have been identified (Fujisawa et al., 2013). Most of the positively regulated genes contained possible RIN-binding (CArG-box motif) sequences [C(C/T)(A/T)6(A/G)G] in their promoters (Ito et al., 2008). Subsequently, a potential binding site for RIN was found in the promoter region of the SlFSR gene. The SlFSR gene promoter has three typical CArG-box sequences [C(A/T)8G] and one intermediate CArG-box sequence [CC(A/T)6AG] (Supplementary Fig S1), suggesting that that SlFSR expression can be regulated by RIN. Moreover, the expression level of SlFSR was down-regulated in the rin mutant compared with WT. Therefore, RIN may directly regulate the expression of SlFSR, providing an important clue to elucidate the complicated transcriptional cascade for tomato cell wall modification.
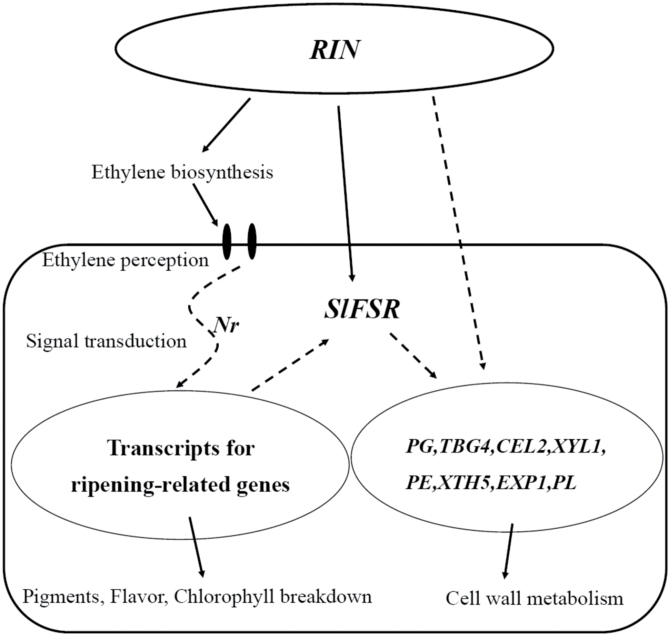
In conclusion, we have identified an important fruit shelf-life regulator, SlFSR. The results obtained from our experiments with both SlFSR-RNAi and SlFSR-overexpressing fruits enable us to conclude that there is a link between SlFSR and fruit shelf-life in tomato. We have attempted to summarize our results in a model to explain the potential role of SlFSR in regulating tomato fruit cell wall metabolism (Fig. 10). In brief, our results provide a valuable opportunity to deepen understanding of the genetic mechanism underlying this significant agronomic trait and to facilitate molecular breeding in tomato. Recognition of the role of SlFSR in post-harvest storage may be conducive to the design and development of approaches to limit losses during fruit storage, handling. and delivery.
Fig. 10.
Proposed model depicting the regulation of the SlFSR gene and its function in tomato fruit shelf-life.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Specific primer sequences used in this study.
Fig. S1. Putative cis-elements enriched in the promoter of the SlFSR gene.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant numbers 30600044 and 31572129), and the Natural Science Foundation of Chongqing of China (grant number cstc2015jcyjA80026).
Author contributions
LZ, LR, and AL performed the experiments and data analysis; LZ and MZ wrote the manuscript; GC and ZH conceived and directed the project and improved the manuscript.
References
- Adams-Phillips L, Barry C, Giovannoni J. 2004. Signal transduction systems regulating fruit ripening. Trends in Plant Science 9, 331–338. [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D. 2002. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. Journal of Experimental Botany 53, 2039–2055. [DOI] [PubMed] [Google Scholar]
- An G. 1987. Binary ti vectors for plant transformation and promoter analysis. Methods in Enzymology 153, 292–305. [Google Scholar]
- Barry CS, Giovannoni JJ. 2007. Ethylene and fruit ripening. Journal of Plant Growth Regulation 26, 143–159. [Google Scholar]
- Bolle C. 2004. The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692. [DOI] [PubMed] [Google Scholar]
- Brummell DA. 2006. Cell wall disassembly in ripening fruit. Functional Plant Biology 33, 103–119. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Hall BD, Bennett AB. 1999a. Antisense suppression of tomato endo-1,4-β-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Molecular Biology 40, 615–622. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH. 2001. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Molecular Biology 47, 311–340. [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. 1999b. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. The Plant Cell 11, 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buanafina MM, Dalton S, Langdon T, Timms-Taravella E, Shearer EA, Morris P. 2015. Functional co-expression of a fungal ferulic acid esterase and a β-1,4 endoxylanase in Festuca arundinacea (tall fescue) modifies post-harvest cell wall deconstruction. Planta 242, 97–111. [DOI] [PubMed] [Google Scholar]
- Carrera E, Ruiz-Rivero O, Peres LE, Atares A, Garcia-Martinez JL. 2012. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiology 160, 1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. The Plant Journal 64, 936–947. [DOI] [PubMed] [Google Scholar]
- Dong T, Hu Z, Deng L, Wang Y, Zhu M, Zhang J, Chen G. 2013. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiology 163, 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun M, Jeong JW, Huber DJ, Cantliffe DJ. 2005. Suppression of ripening and softening of ‘Galia’ melons by 1-methylcyclopropene applied at preripe or ripe stages of development. Hortscience 40, 170–175. [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. 2008. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology 8, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ. 2004. Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. The Plant Journal 40, 47–59. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ. 2001. Fructose 2,6-bisphosphate activates pyrophosphate: fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212, 250–263. [DOI] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB. 1991. Role of cell wall hydrolases in fruit ripening. Annual Review of Plant Biology 42, 675–703. [Google Scholar]
- Flors V, Leyva M de la O, Vicedo B, Finiti I, Real MD, García-Agustín P, Bennett AB, González-Bosch C. 2007. Absence of the endo-β-1,4-glucanases Cel1 and Cel2 reduces susceptibility to Botrytis cinerea in tomato. The Plant Journal 52, 1027–1040. [DOI] [PubMed] [Google Scholar]
- Forth D, Pyke KA. 2006. The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. Journal of Experimental Botany 57, 1971–1979. [DOI] [PubMed] [Google Scholar]
- Fray RG, Grierson D. 1993. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Molecular Biology 22, 589–602. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y. 2011. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biology 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y. 2013. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. The Plant Cell 25, 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, Ito Y. 2012. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235, 1107–1122. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. The Plant Cell 16(Suppl), S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. 2007. Fruit ripening mutants yield insights into ripening control. Current Opinion in Plant Biology 10, 283–289. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ, DellaPenna D, Bennett AB, Fischer RL. 1989. Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. The Plant Cell 1, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, et al. . 2012. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Current Biology 22, 2236–2241. [DOI] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis AG, Grierson D. 1999. Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. Journal of Experimental Botany 50, 793–798. [Google Scholar]
- Hadfield KA, Bennett AB. 1998. Polygalacturonases: many genes in search of a function. Plant Physiology 117, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Ban Q, Li H, Hou Y, Jin M, Han S, Rao J. 2016. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Scientific Reports 6, 39155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J-O, Chang KS, Kim IA, Lee M-H, Lee SA, Song S-K, Lee MM, Lim J. 2011. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proceedings of the National Academy of Sciences, USA 108, 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwasa K, Kinugasa Y, Amano S, Hashimoto A, Nakano R, Inaba A, Kubo Y. 2003. Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. Journal of Experimental Botany 54, 771–779. [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. 2008. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. The Plant Journal 55, 212–223. [DOI] [PubMed] [Google Scholar]
- Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. 2014. Transcriptional control of fleshy fruit development and ripening. Journal of Experimental Botany 65, 4527–4541. [DOI] [PubMed] [Google Scholar]
- Kramer M, Sanders R, Bolkan H, Waters C, Sheeny RE, Hiatt WR. 1992. Postharvest evaluation of transgenic tomatoes with reduced levels of polygalacturonase: processing, firmness and disease resistance. Postharvest Biology & Technology 1, 241–255. [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. 1994. Two divergent endo-beta-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. The Plant Cell 6, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. 1987. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proceedings of the National Academy of Sciences, USA 84, 2793–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Widmer A. 2014. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice. Plant Molecular Biology Reporter 32, 1129–1145. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma HS, Liang D, Shuai P, Xia XL, Yin WL. 2010. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. Journal of Experimental Botany 61, 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose M, Ekengren SK, Melech-Bonfil S, Martin GB, Sessa G. 2006. A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Molecular Plant Pathology 7, 593–604. [DOI] [PubMed] [Google Scholar]
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A. 2010. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proceedings of the National Academy of Sciences, USA 107, 2413–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Lorences EP. 2009. Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. Journal of Plant Physiology 166, 489–498. [DOI] [PubMed] [Google Scholar]
- Moore S, Vrebalov J, Payton P, Giovannoni J. 2002. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. Journal of Experimental Botany 53, 2023–2030. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Guis M, Rose JK, et al. . 2007. Ethylene regulation of fruit softening and cell wall disassembly in Charentais melon. Journal of Experimental Botany 58, 1281–1290. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, et al. . 2007. Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. The Plant Journal 50, 1093–1106. [DOI] [PubMed] [Google Scholar]
- Orfila C, Huisman MM, Willats WG, van Alebeek GJ, Schols HA, Seymour GB, Knox JP. 2002. Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening. Planta 215, 440–447. [DOI] [PubMed] [Google Scholar]
- Orfila C, Seymour GB, Willats WG, Huxham IM, Jarvis MC, Dover CJ, Thompson AJ, Knox JP. 2001. Altered middle lamella homogalacturonan and disrupted deposition of (1–>5)-α-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiology 126, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owino WO, Ambuko JL, Mathooko FM. 2005. Molecular basis of cell wall degradation during fruit ripening and senescence. Stewart Postharvest Review 1, 1–10. [Google Scholar]
- Phan TD, Bo W, West G, Lycett GW, Tucker GA. 2007. Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiology 144, 1960–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. 1999. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. The Plant Journal 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, et al. . 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. 2005. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. [DOI] [PubMed] [Google Scholar]
- Smith DL, Abbott JA, Gross KC. 2002. Down-regulation of tomato β-galactosidase 4 results in decreased fruit softening. Plant Physiology 129, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jones WT, Rikkerink EH. 2012. GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. The Biochemical Journal 442, 1–12. [DOI] [PubMed] [Google Scholar]
- Tigchelaar EC, Mcglasson WB, Buescher RW. 1978. Genetic regulation of tomato fruit ripening. Hortscience 13, 508–513. [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C. 2009. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. The Plant Journal 58, 803–816. [DOI] [PubMed] [Google Scholar]
- Uluisik S, Chapman NH, Smith R, et al. . 2016. Corrigendum: Genetic improvement of tomato by targeted control of fruit softening. Nature Biotechnology 34, 1072. [DOI] [PubMed] [Google Scholar]
- Vicente AR, Saladie M, Rose JKC, Labavitch JM. 2007. The linkage between cell wall metabolism and fruit softening: looking to the future. Journal of the Science of Food and Agriculture 87, 1435–1448. [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. 2002. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. 1995. An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270, 1807–1809. [DOI] [PubMed] [Google Scholar]
- Xie QL, Hu ZL, Zhu ZG, Dong TT, Zhao ZP, Cui BL, Chen GP. 2014. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Scientific Reports 4, 4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang W, Xiong F, Xian Z, Su D, Ren M, Li Z. 2017. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnology Journal 15, 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z. 2014. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant & Cell Physiology 55, 119–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.