See Huang and Gitler (doi:10.1093/brain/awy112) for a scientific commentary on this article.
Small molecule drugs that can reduce levels of the mutant huntingtin protein (mHTT) are sought for the treatment of Huntington’s disease. Song et al. demonstrate that deleting Gpr52, or inhibiting Gpr52 protein function with a novel small molecule antagonist, reduces mHTT levels and rescues Huntington’s disease-associated phenotypes in cellular and mouse models.
Keywords: GPCR, target validation, polyQ, neurodegeneration, antagonists
Abstract
See Huang and Gitler (doi:10.1093/brain/awy112) for a scientific commentary on this article.
Lowering the levels of disease-causing proteins is an attractive treatment strategy for neurodegenerative disorders, among which Huntington’s disease is an appealing disease for testing this strategy because of its monogenetic nature. Huntington’s disease is mainly caused by cytotoxicity of the mutant HTT protein with an expanded polyglutamine repeat tract. Lowering the soluble mutant HTT may reduce its downstream toxicity and provide potential treatment for Huntington’s disease. This is hard to achieve by small-molecule compound drugs because of a lack of effective targets. Here we demonstrate Gpr52, an orphan G protein-coupled receptor, as a potential Huntington’s disease drug target. Knocking-out Gpr52 significantly reduces mutant HTT levels in the striatum and rescues Huntington’s disease-associated behavioural phenotypes in a knock-in Huntington’s disease mouse model expressing endogenous mutant Htt. Importantly, a novel Gpr52 antagonist E7 reduces mutant HTT levels and rescues Huntington’s disease-associated phenotypes in cellular and mouse models. Our study provides an entry point for Huntington’s disease drug discovery by targeting Gpr52.
Introduction
Neurodegenerative disorders, characterized by progressive loss of neurons in the CNS, influence millions of people in the aged population. Treatment of such diseases has been extremely challenging, and there is currently no disease progression-modifying treatment. A common hallmark for these diseases is the accumulation of misfolded and aggregation-prone proteins, and lowering their levels is considered as an appealing therapeutic strategy (Soto, 2003). Among different neurodegenerative disorders, the monogenetic disorder Huntington’s disease provides appealing genetic models to study the disease-protein lowering strategy, because the genetics and the disease-causing protein is clear. Huntington’s disease is caused by the mutation of the HTT gene encoding the mutant HTT protein (mHTT) with expanded polyglutamine tract (polyQ) (The Huntington’s Disease Collaborative Research Group, 1993). Lowering the mHTT level ameliorates mHTT toxicity in multiple models. In a transgenic Huntington’s disease mouse model expressing inducible mHTT N-terminal fragments, turning off the transgene reversed neuropathology and motor deficits (Yamamoto et al., 2000). Delivery of short hairpin RNAs (shRNAs), small interference RNAs (siRNAs) or antisense oligonucleotides reducing mHTT attenuate neuropathology and disease-related phenotypes in several mouse models (Harper et al., 2005; Rodriguez-Lebron et al., 2005; DiFiglia et al., 2007; Kordasiewicz et al., 2012). CRISPR/Cas9-mediated genome editing of mHTT ameliorated Huntington’s disease neurotoxicity (Yang et al., 2017). Several screening studies revealed that genetic modifiers of mHTT levels rescued Huntington’s disease-associated phenotypes (Zhang et al., 2010; Baldo et al., 2012; Lu et al., 2013). A clinical trial using a non-allele specific antisense oligonucleotide to lower HTT levels has also been launched for Huntington’s disease treatment (https://www.clinicaltrials.gov/ct2/show/NCT02519036). In most studies, both mHTT and the wild-type HTT were lowered, and beneficial effects were still observed.
Meanwhile, delivering antisense oligonucleotides, siRNA/shRNAs or genome editing reagents into patient’s brains is challenging and expensive, and thus small molecule drugs that reduce HTT levels are highly desired. This is extremely challenging because of a lack of reliable drug targets. G protein-coupled receptors (GPCRs) constitute the largest drug target family of FDA-approved drugs (Overington et al., 2006). They are located on the plasma membrane and modulated by endogenous extracellular molecules, making them ideal targets for small molecule compounds.
We have previously identified an orphan GPCR, Gpr52, as a striatal-enriched modulator of soluble mHTT levels ex vivo and in vivo (Yao et al., 2015). Knocking-down Gpr52 or loss of function mutation of Gpr52 rescues Huntington’s disease-associated phenotypes in Huntington’s disease fly models and patient induced pluripotent stem cell (iPSC)-derived neurons (Yao et al., 2015). To investigate its therapeutic potential as a Huntington’s disease drug target further, we tested its effect in modulating Huntington’s disease-associated phenotypes in vivo in a knock-in mouse model, which expresses mHtt (indicating the mouse mutant HTT protein) from its endogenous locus. We then discovered a novel Gpr52-specific small molecule antagonist E7, and tested the possibility of lowering soluble mHtt levels and treating Huntington’s disease via targeting Gpr52 by E7. Our data provide the proof-of-concept evidence of treating Huntington’s disease by reducing soluble mHtt via Gpr52 blockade with compound drugs.
Materials and methods
Experimental design
The overall objective of this study was to test the possibility of targeting Gpr52 for Huntington’s disease treatment and drug discovery by in vivo experiments.
To this end, we used Huntington’s disease in vivo knock-in mouse models and Huntington’s disease in vivo Drosophila models. In addition, we used the HEK293 stable cell line expressing hGpr52 was used for compound screening of hGpr52 antagonists.
For validation of Gpr52 per se, Huntington’s disease or wild-type mice with different genotypes of Gpr52 were tested for Huntington’s disease-relevant phenotypes and HTT levels, the animals were allocated by their genotypes and no randomization was necessary (Wang et al., 2014). For validation of Gpr52 antagonist E7, cellular, Drosophila and mouse Huntington’s disease models were used. For cellular experiments, cells were resuspended and randomly distributed during plating for each cell type. For Drosophila experiments, the flies were randomly sorted in the testing tubes for each type of fly. For mouse experiments, a random number between 0 and 1 was generated for each mouse by Microsoft Excel to determine the E7 versus dimethyl sulphoxide (DMSO) intracerebroventricular injection (E7: ≥ 0.5, DMSO: <0.5). The mouse behavioural experiments were all performed blind, and the mouse genotypes or drugs delivered were not revealed before data analysis. For statistical analysis, sufficient samples/replicates were collected (power > 0.8) and the sample sizes are comparable or higher than similar studies (Park et al., 2013). All representative images were repeated multiple times, and the replication numbers are indicated in the quantification and/or figure legends.
For detailed experimental procedures and information of materials in cellular and animal models, refer to the Supplementary material.
Results
Knockout of Gpr52 rescues behavioural phenotypes in a Huntington’s disease knock-in mouse model
We have previously demonstrated that lowering Gpr52 rescues Huntington’s disease-associated phenotypes in cellular and Drosophila models (Yao et al., 2015). To investigate the therapeutic potential of Gpr52 as a Huntington’s disease drug target further, we tested whether heterozygous or homozygous knockout of Gpr52 rescues Huntington’s disease-associated behavioural phenotypes in vivo in mice. We crossed the Gpr52 knockout mice to a well-established Huntington’s disease knock-in mouse model expressing endogenous mHtt proteins with 140Q (HdhQ140/Q140; the wild-type HTT protein has 7Q) (Menalled et al., 2003). This model expresses mHtt from its original genomic locus, and thus has high fidelity to human Huntington’s disease patients in the aspects of disease genetics and mHtt protein levels. In addition, mHtt expression in this model does not lead to weight changes (Menalled et al., 2003), minimizing potential artefacts due to weight differences.
Consistent with other groups (Menalled et al., 2003; Hickey et al., 2012), we observed several motor function-related deficits in the homozygous Huntington’s disease (HdhQ140/Q140) mice, including less activity (measured by the frequency of rearing) in a pen holder with mashed surface, locomotion deficits in the open-field, abnormal gait behaviours and rotarod deficits (Figs 1 and 2). Meanwhile, using the heterozygous Huntington’s disease mice (HdhQ7/Q140) at similar ages, the Huntington’s disease-associated behavioural phenotypes were much less reliable and failed to give us a sufficient window to test potential rescue effects (Yu et al., 2017), and thus we tested Gpr52’s effects mainly in homozygous Huntington’s disease mice.
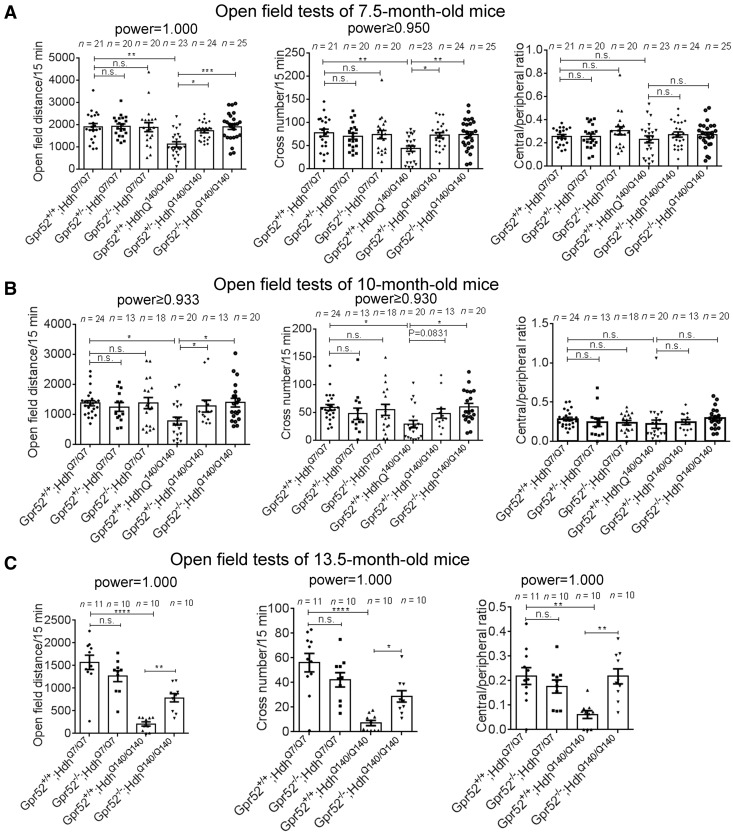
Figure 1.
Knockout of Gpr52 rescued Huntington’s disease-associated open-field phenotypes in a knock-in Huntington’s disease mouse model. (A) Travel distances, cross numbers and central/peripheral ratios in the open-field tests of 7.5-month-old mice of the indicated genotypes. Each dot represents an individual mouse, and each corresponding bar represents mean ± standard error of the mean (SEM). The statistical analysis was performed by one-way ANOVA and post hoc Bonferroni’s tests for the indicated comparisons. n.s. = not significant = P > 0.1, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. P-values between 0.05 and 0.1 are presented by exact values. Huntington’s disease mice showed significant lowering in the travel distance and cross number, which were rescued by Gpr52+/− or Gpr52−/−. (B and C) As in A, but using 10-month-old or 13.5-month-old mice, respectively.
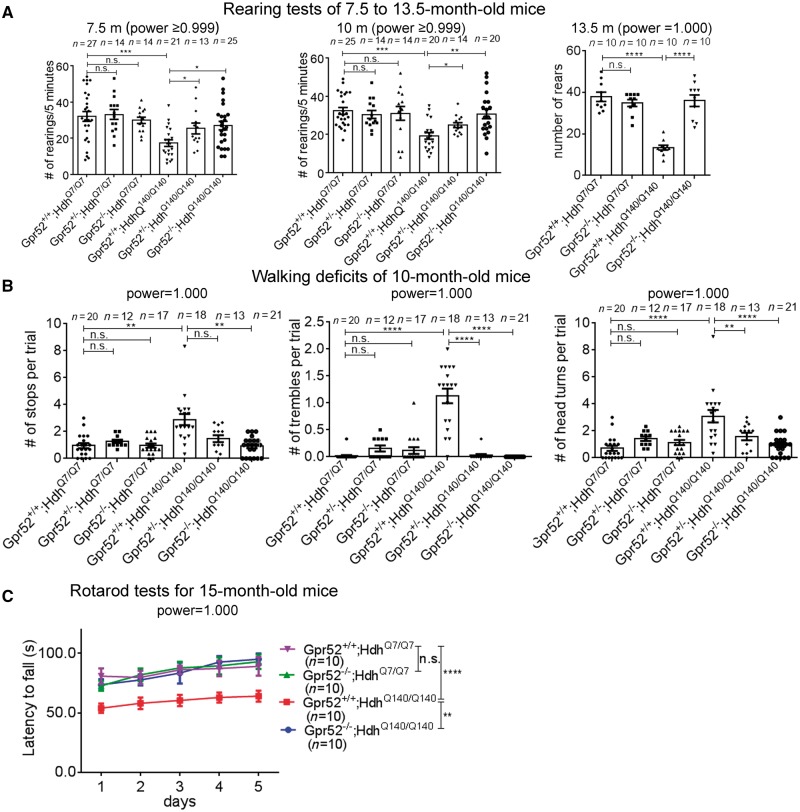
Figure 2.
Knockout of Gpr52 rescued Huntington’s disease-associated rearing, gait and rotarod phenotypes in a knock-in Huntington’s disease mouse model. (A) Rearing number per 5 min in the mouse of indicated genotypes (x-axis) at the indicated ages (above each graph). Each dot represents an individual mouse, and each corresponding bar represents mean ± SEM. (B) In the CatWalk gait analysis tests, the number of stops, trembles and head turns of each trial were quantified based on the video captured. The averaged numbers per trial were calculated based on three repeated trials for each mouse, and were then summarized based on the genotypes indicated in the x-axis. Each dot represents an individual mouse, and each corresponding bar represents mean ± SEM. (C) Rotarod test results for 15-month-old mice of indicated genotypes. Each mouse was trained for 3 days and then tested for five consecutive days. The mean ± SEM of the latency to fall (y-axis) of each genotype (colour) was plotted. The statistical analyses were performed by one-way ANOVA tests for A and B and two-way ANOVA tests for C. n.s. = P > 0.1, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001.
We performed power analysis based on the effect size and variation estimated by our preliminary and previously published studies, and determined that at least five mice were needed for each group to reach a statistical power > 0.8. We also calculated the post-experiment powers for all the experiments showing significance to ensure sufficient statistical power (indicated above each figure panel). The number of mice used for each group is similar or higher than similar studies from other groups (Park et al., 2013; Wang et al., 2014; Ochaba et al., 2016).
By crossing the Gpr52 knockouts to the Huntington’s disease mice for several generations, we obtained both Gpr52 heterozygous (Gpr52+/−) and homozygous (Gpr52−/−) knockout versus housemate controls (Gpr52+/+) in the Huntington’s disease (HdhQ140/Q140) as well as wild-type (HdhQ7/Q7) background. In the open field tests, Gpr52 homozygous knockout significantly (P < 0.05) rescued deficits in the travel distance and the cross-number measurements at the age of 7.5 and 10 months, and the Gpr52 heterozygous knockout also had a similar effect (Fig. 1A and B, the right three bars of each panel). At the age of 13.5 months, Huntington’s disease mice also developed a significant lowering of the ratio between the travel distance in the central versus the peripheral region (Fig. 1C, right), suggesting an increased anxiety level of Huntington’s disease mice compared to the wild-type mice. This phenotype was not observed at younger ages (Fig. 1A and B, right), suggesting that Huntington’s disease mice develop psychological phenotypes in addition to motor deficits at later ages, consistent with Huntington’s disease human patients. At the age of 13.5 months, Gpr52 knockout significantly rescued the Huntington’s disease-associated phenotypes in the travel distance, cross number and central/peripheral ratio in the open-field tests (Fig. 1C, the right two bars of each panel). Thus, lowering Gpr52 may rescue the Huntington’s disease-associated phenotype in the open-field tests, and the effects may persist at older ages. Noticeably, Gpr52 knockout had no effect in the wild-type mice (Fig. 1A–C), confirming that the rescue effects in Huntington’s disease mice were disease-relevant. Similarly, Gpr52 heterozygous or homozygous knockout rescued the rearing phenotype in the Huntington’s disease mice at all the ages tested (Fig. 2A).
Another disease-relevant phenotype of Huntington’s disease patients that influences the life quality of many Huntington’s disease patients is abnormal walking behaviour (Daneault et al., 2015), and thus we tested potential walking phenotype of these Huntington’s disease mice by the CatWalk gait analysis system, which captured videos of walking behaviours and gaits of mice passing through a lane. While Huntington’s disease mice exhibited some gait and movement abnormalities around 7 months, the phenotype was much more obvious and robust at 10 months. The Huntington’s disease mice showed significantly more frequent stops, trembles, head turns and irregular steps compared to the wild-type mice. We thus quantified the frequencies of these abnormal walking behaviours of 10-month-old mice as readout for Huntington’s disease walking deficits (Fig. 2B). Gpr52+/− or Gpr52−/− significantly rescued these walking deficits in Huntington’s disease mice without influencing the wild-type mice (Fig. 2B). We also quantified the variation of the speed of each foot and time spent passing the test lane, and Huntington’s disease mice exhibited larger variation of speed and longer passing time, reflecting gait irregularities and deficits. Gpr52−/− significantly rescued these phenotypes in Huntington’s disease mice, although Gpr52+/− had little effect (Supplementary Fig. 1), likely because Gpr52−/− may lead to stronger rescue effects on walking and gait abnormalities than Gpr52+/−. Similar trends could be observed in the open-field and rearing tests (Figs 1A, B and 2A).
The rotarod performance is another widely accepted readout for motor functions influenced in Huntington’s disease. At the age of 15 months, Huntington’s disease mice exhibit evident motor function deficits in the rotarod tests quantified by the latency to fall (Fig. 2C). The Gpr52 knockout Huntington’s disease mice (Gpr52−/−; HdhQ140/Q140) showed significant improvement of the rotarod performance (Fig. 2C), confirming the rescue of rotarod phenotype that exhibited at late ages. Similar to other behavioural tests, Gpr52 knockout had no effect on the wild-type mice (Fig. 2C), suggesting that the rescue effects were Huntington’s disease-specific.
Knockout of Gpr52 rescues Huntington’s disease-associated biomarkers in vivo
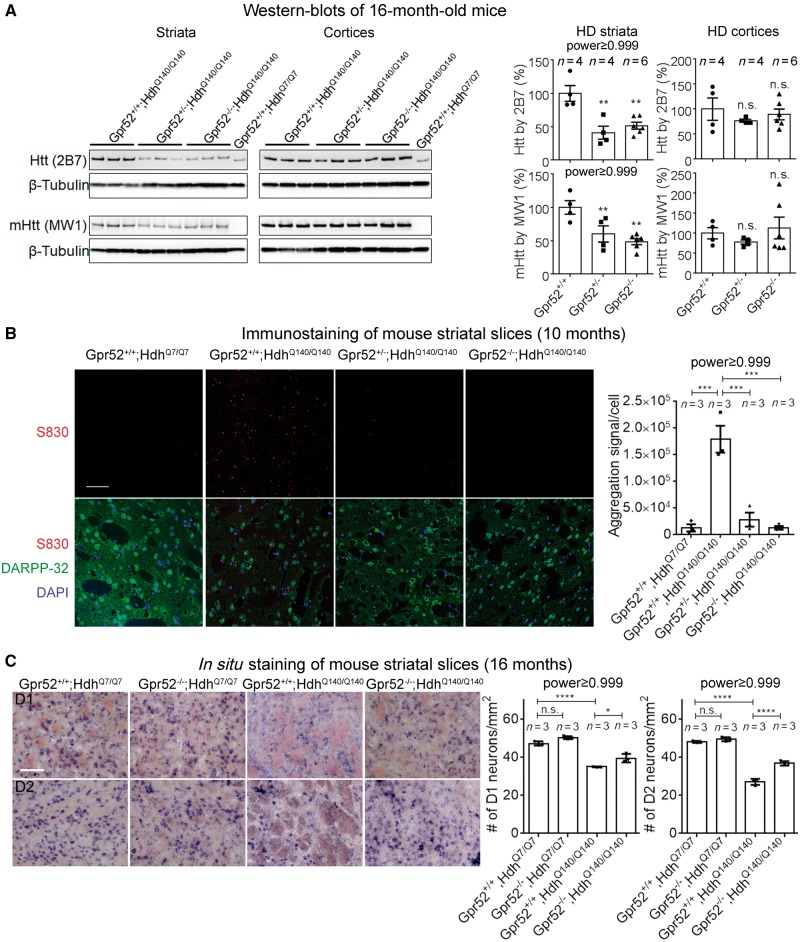
We then investigated whether Gpr52 knockout rescued Huntington’s disease-associated molecular phenotypes, which reflect Huntington’s disease cytoxicity in vivo. Huntington’s disease neurodegeneration mainly influences the medium spiny neurons in the striatal region, and the level of medium spiny neuron marker DARPP-32 reflects Huntington’s disease neurotoxicity (Hodas et al., 2012). At 16 months of age, Huntington’s disease mice striata exhibited a decreased level of DARPP-32, whereas the heterozygous or homozygous knockout of Gpr52 significantly increased the DARPP-32 level (Supplementary Fig. 2A), suggesting a rescue effect at the molecular level. Meanwhile, Gpr52−/− only slightly increased the DARPP-32 level in the wild-type mouse striata, and the increase was not significant after normalizing to the loading control (β-tubulin), suggesting that the effect is Huntington’s disease-specific.
Besides neurons, glial cells were also altered in Huntington’s disease and significant astrogliosis and microgliosis are detected in the caudate nucleus and internal capsule of Huntington’s disease patients, but not in the normal brain (Dieterich et al., 2006). This phenomenon is likely associated with the elevated neuroinflammation responses in Huntington’s disease. While its functional significance in Huntington’s disease remains unclear, the relevant glia activation markers have been widely used as indicators of Huntington’s disease pathology (Kirkin et al., 2009; Yang et al., 2017). We thus investigated GFAP and Iba1, which are markers for astrocytes and microglia, respectively (Kirkin et al., 2009). Consistent with previous studies, Huntington’s disease mice exhibited elevated numbers of GFAP+ and Iba1+ cells in the striata at 16 months of age (Supplementary Fig. 2B), likely due to increased neuroinflammation in the context of mHtt expression. Knocking-out Gpr52 significantly reduced the GFAP+ cells and the Iba1+ cells in the Huntington’s disease but not wild-type striata (Supplementary Fig. 2B), suggesting a rescue effect.
Knockout of Gpr52 reduces soluble and aggregated mHtt level in the striata in vivo
We previously demonstrated that lowering GPR52 reduces soluble mHtt protein levels in cellular Huntington’s disease models and in the striata of HdhQ7/Q140 knock-in mice at early ages (∼2 months) (Yao et al., 2015).
To test if the lowering mHtt effect persists in the homozygous Huntington’s disease mice (HdhQ140/Q140) at old age, we tested the soluble mHtt levels of mouse brain tissues at 16 months of age. Consistently, heterozygous or homozygous knockout of Gpr52 significantly reduced soluble mHtt levels in the striata but not the cortices (Fig. 3A). In addition, the total fluorescent signal of mHtt macro-aggregates in Huntington’s disease mice at 10 months of age were significantly reduced by heterozygous knockout of Gpr52, and almost absent in Gpr52 homozygous knockout mouse striatal slices (Fig. 3B), probably as a consequence of the reduction of soluble mHtt protein and prevention of aggregate formation. This is also consistent with previous Htt-targeting antisense oligonucleotide studies, which showed ablation of mHtt aggregates after antisense oligonucleotide treatment (Kordasiewicz et al., 2012). In summary, lowering Gpr52 reduced both soluble and aggregated Htt levels in vivo.
Figure 3.
Knockout of Gpr52 reduced soluble and aggregated mHtt levels in striata of a knock-in Huntington’s disease mouse model. (A) Representative western blot and quantification of mHtt protein levels in striata and cortices from 16-month-old mice of indicated genotypes. Bars represent mean ± SEM. Statistical analysis was performed by one-way ANOVA and post hoc Dunnett tests. n.s. = P > 0.1, **P < 0.01. (B) Representative immunostaining and quantification of mHtt aggregates in striata slices from 10-month-old mice of indicated genotypes. Scale bar = 50 μm. Bars represent mean ± SEM; n indicates the number of different mice of each genotype. The image capture and analyses were performed blindly before annotating the genotypes. Aggregation signal per cell was analysed by particle analysis in ImageJ and calculated by: the number of aggregates × aggregate size × mean aggregate fluorescent intensity / cell number counted by DAPI. The statistical analysis was performed by one-way ANOVA and post hoc Dunnett tests for the indicated comparisons: ****P < 0.0001. (C) Representative in situ images and quantification of D1 or D2 positive neurons in striata slices from 16-month-old mice of indicated genotypes. Scale bar = 10 μm. Bars represent mean ± SEM; n indicates the number of different mice of each genotype. The statistical analysis was performed by one-way ANOVA and post hoc Bonferroni’s tests for the indicated comparisons: *P < 0.05, ****P < 0.0001.
We then assayed the Huntington’s disease neurodegeneration by counting the density of D1 and D2 dopamine receptor-expressing neurons identified by in situ staining. Significant decrease of D1 and D2 neuron density was observed in disease striatal slices, and knocking-out Gpr52 significantly rescued this phenotype (Fig. 3C), consistent with the lowering mHtt.
Striatal expression of human Gpr52 restored mHtt levels and Huntington’s disease-associated phenotypes
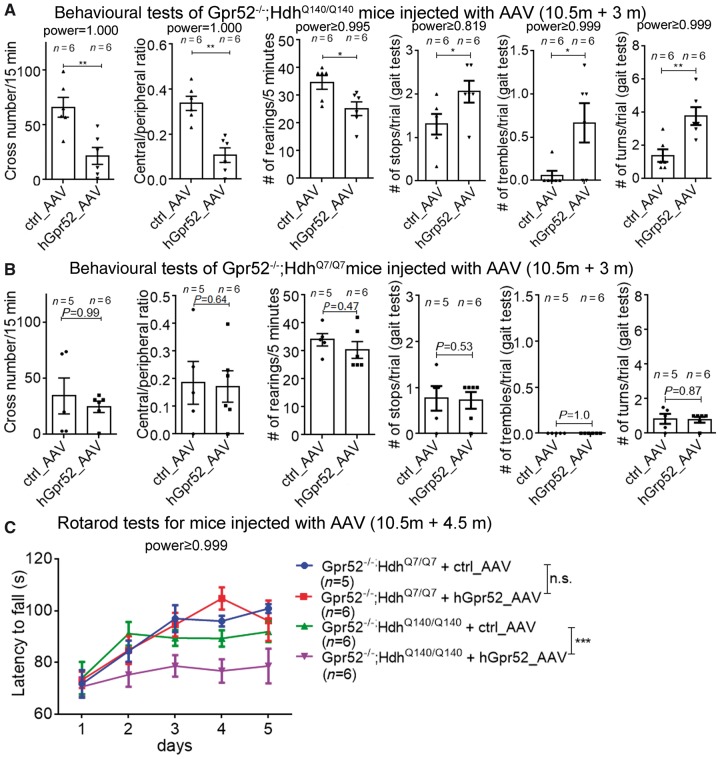
To confirm that the effect of Gpr52 knockout was mediated by the loss of Gpr52, we tested whether the expression of Gpr52 cDNA in the striatum is able to restore mHtt levels and Huntington’s disease-associated phenotypes. We injected adeno-associated viruses (AAV) expressing human GPR52 (hGpr52) cDNA driven by a neuronal promoter (the human synapsin 1 promoter) into both left and right striata of the Gpr52 knockout Huntington’s disease mice (Gpr52−/−, HdhQ140/Q140), and managed to express hGpr52 in the striata (Supplementary Fig. 3A and B). Compared to the control AAV (expressing EGFP) injected mice, the hGpr52 expressing AAV significantly increased the soluble mHtt level 4.5 months after injection (Supplementary Fig. 3A). Mutant Htt aggregate signals were not significantly changed (Supplementary Fig. 3C). The number of GFAP+ cells and the number of Iba1+ cells in the Huntington’s disease striata were increased (Supplementary Fig. 4A) whereas the DARPP-32 level remained the same (Supplementary Fig. 4B), suggesting that the Huntington’s disease-associated gliosis phenotype was partially restored by increasing mHtt levels.
We then tested the hGpr52 AAV-injected Gpr52−/−, HdhQ140/Q140 mice in the open-field, rearing, gait and rotarod tests, and examined different motor function-relevant parameters. Three to 4.5 months after injection, the hGpr52 cDNA AAV-injected mice exhibited Huntington’s disease-associated behavioural deficits in almost all the parameters tested (Fig. 4 and Supplementary Fig. 5), compared to the control AAV-injected mice. Only the travel distance in the open-field test and the passing time in the gait analysis were not significantly influenced by the hGpr52 cDNA AAV-injection in the Gpr52−/−, HdhQ140/Q140 mice (Supplementary Fig. 5A and B). In summary, our data suggest that most of the Huntington’s disease-associated behavioural phenotypes were restored by expressing hGpr52 via AAV injection, likely due to restoration of the mHtt levels in the striata. The restoration of the Huntington’s disease-associated phenotypes was not due to toxicity of hGpr52, because none of the behavioural parameters were significantly influenced in the Gpr52−/−, HdhQ7/Q7 mice (Fig. 4B, Supplementary Fig. 5A and C), suggesting that the effect was Huntington’s disease-specific.
Figure 4.
hGpr52 cDNA AAV injection in the striatum restored Huntington’s disease-associated phenotypes in a knock-in Huntington’s disease mouse model. (A) hGpr52 cDNA AAV injected Gpr52 knockout Huntington’s disease mice (Gpr52−/−; HdhQ140/Q140) were tested in the indicated behavioural phenotypes and the parameters were analysed. Each dot represents an individual mouse, and each corresponding bar represents mean ± SEM. The statistical analysis was performed by two-tailed unpaired t-tests: *P < 0.05, **P < 0.01. For most behavioural test parameters, hGpr52 cDNA AAV injection significantly restored the Huntington’s disease-associated behavioural deficits. (B) As in A, but using Gpr52 knockout wild-type mice (Gpr52−/−; HdhQ7/Q7). (C) hGpr52 cDNA or control AAV injected Gpr52 knockout Huntington’s disease (Gpr52−/−; HdhQ140/Q140) and wild-type (Gpr52−/−; HdhQ7/Q7) mice were tested in the rotarod tests. The statistical analysis was performed by two-way ANOVA tests: n.s. = P > 0.1, ***P < 0.001.
The data above confirm that the rescue of Huntington’s disease-associated phenotypes by Gpr52 knockout was mediated through the deletion of Gpr52, because the expression of hGpr52 in the striatum is able to restore mHtt expression and Huntington’s disease deficits. In addition, the human and mouse Gpr52 have a conservative role in regulating Htt.
A novel Gpr52 antagonist E7 decreases soluble mHtt levels
GPR52 regulates the mHtt level via its GPCR function (Yao et al., 2015), and thus small molecule antagonists blocking GPR52 activity may lower mHtt levels and treat Huntington’s disease. Unfortunately, GPR52 antagonists are completely unknown, although GPR52 compound agonists have been reported previously (Setoh et al., 2014). Thus, we carried out a compound screen to identify novel GPR52 antagonists.
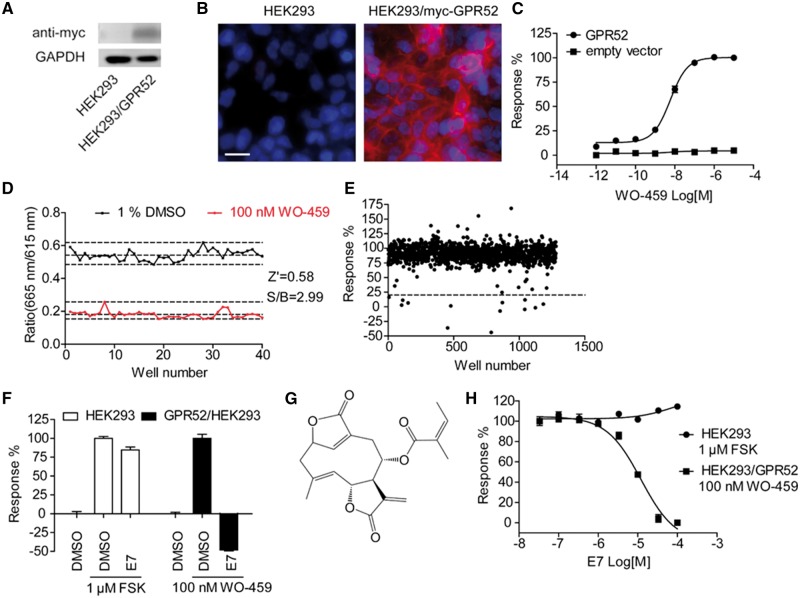
We generated a HEK293 cell line stably expressing myc-tagged human GPR52. Western blots and immunofluorescent staining with anti-myc antibody indicated the proper expression of GPR52 (Fig. 5A and B). GPR52 is a Gαs-coupled GPCR, and a reported GPR52 agonist WO-459 {N-(2-hydroxyethyl)-3-methyl-1-[2-[3-(trifluoromethyl)benzyl]benzothiophen-7-yl]-1H-pyrazole-4-carboxamide} (Setoh et al., 2014) induced dose-dependent elevation of intracellular cAMP in HEK293/GPR52 cells with EC50 value of 6.0 ± 0.7 nM, but not in the parental HEK293 cells (Fig. 5C).
Figure 5.
Identification of E7 as a novel GPR52 antagonist. (A) Western blot analysis of HEK293 cells transfected with empty vector or stably expressing myc-hGPR52. (B) Immunofluorescent staining of HEK293 cells transfected with empty vector or stably expressing myc-hGPR52 with anti-myc antibody (red) and nuclear staining by Hoechst 33342 (blue). Scale bar = 10 μm. (C) Intracellular cAMP response (measured by HTRF) in HEK293 cells stably expressing myc-hGPR52 or transfected with empty vector after WO-459 (a reported GPR52 agonist) stimulation. Plots represent mean ± SEM, n = 4, independently plated and treated wells. (D) Z′ factor determination. Replicates of positive (WO-459) and negative (DMSO) signals were studied. Dashed lines indicate means ± 3 SD of 40 data points. Z′ value for the assay was 0.58 and the S/B ratio was 2.99. (E) Representative results of the primary screening of 40 000 compounds. HEK293/GPR52 cells were treated with test compounds at 30 μM concentration and then stimulated with 100 nM WO-459. Intracellular cAMP levels were measured, and the change by WO-459 in each well was normalized to the average change in the DMSO pretreated group (response %). The dashed line (20%) indicates the threshold used for the primary screen. (F) Effect of E7 (100 μM) on WO-459 (100 nM)-stimulated cAMP in HEK293/GPR52 cells, or forskolin (FSK, 1 μM)-stimulated cAMP in HEK293 cells. Bars represent mean ± SEM, n = 3, independently plated and treated wells. (G) Chemical structure of E7. (H) Dose-response of purchased E7 on WO-459 (100 nM)-stimulated cAMP in HEK293/GPR52 cells, or forskolin (1 μM)-stimulated cAMP in HEK293 cells. Plots represent mean ± SEM, n = 3, independently plated and treated wells.
The initial screen was carried out with the cAMP assay using HEK293/GPR52 cells. The cells were treated with test compounds (30 μM) or DMSO (negative control) for 30 min, and then stimulated by WO-459 (100 nM). Thirty minutes later, cells were lysed for intracellular cAMP measurements. We assessed the robustness of the screening assay by Z′ factor, the normalized 3 × SD window between the negative and positive controls (Zhang et al., 1999). The Z′ value for the assay was 0.58, and the signal-to-background ratio was 2.99, indicating that the assay was adequately optimized (Fig. 5D). Approximately 40 000 compounds from the Chinese National Compound Library were screened and the representative data points were shown in Fig. 5E. Among those compounds that showed inhibitory effect in the primary screen, a compound designated AD31E7 (E7) showed significant inhibitory effect on WO-459-induced cAMP elevation in HEK293/GPR52 cells, but did not affect forskolin-induced cAMP accumulation in HEK293 cells, indicating a GPR52-specific effect (Fig. 5F).
The chemical structure of E7 is presented in Fig. 5G. E7 is a natural product with the chemical name (3S,4R,8R,9E)-10-methyl-5-methylidene-6,14-dioxo-7,13-dioxatricyclo[10.2.1.0⁴,⁸]pentadeca-1(15),9-dien-3-yl (2Z)-2-methylbut-2-enoate. Since compound collections are inherently unstable, new E7 was purchased from Analyticon Discovery (#NP-012321) and used for the further characterizations. The newly purchased E7 displayed dose-dependent inhibition of WO-459 (100 nM)-stimulated cAMP in HEK293/GPR52 cells with an IC50 value of 12.0 ± 0.7 μM, and did not affect forskolin-induced cAMP in HEK293 cells (Fig. 5H), suggesting that E7 modulates cAMP levels via inhibiting Gpr52. The specificity of E7 was further tested against several Gαs-coupled GPCRs, including β2AR, GCGR and GLP-1R (Supplementary Fig. 6). E7 did not block cAMP elevation induced by the activation of those GPCRs, confirming that E7 does not act on or downstream of heterotrimeric Gαs. To validate the experimental system, we tested the reported antagonists of those receptors, and they displayed dose-dependent inhibition of their target GPCRs (Supplementary Fig. 6).
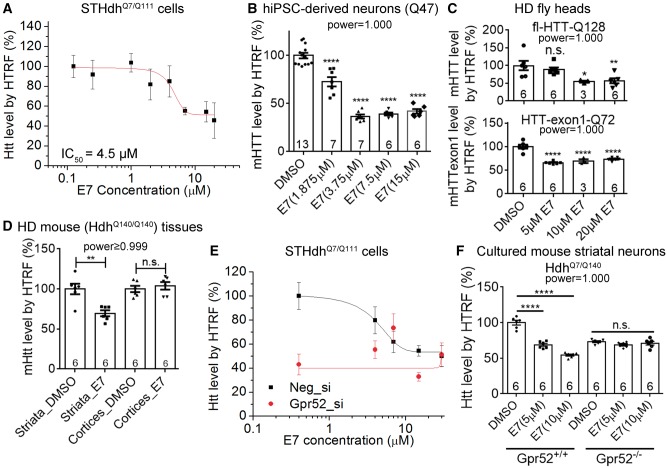
We then tested E7’s effect on HTT levels in several Huntington’s disease models. To achieve accurate quantification of HTT levels of cells treated with E7 at a number of different concentrations, we measured HTT levels by the well-established homogeneous time resolved fluorescence (HTRF) assay (Weiss et al., 2009), and then confirm the results by western blots. The HTRF assay uses a terbium-conjugated antibody (donor) and a D2-conjugated antibody (acceptor) targeting the same protein, and the time resolved-fluorescence resonance energy Transfer (TR-FRET) (Mathis, 1993) occurs when the two antibodies come to close proximity by binding with the same protein molecule. As a result, the HTRF signals are proportional to the target protein concentration (Paganetti et al., 2009). The technology has been successfully applied to the measurement of human or mouse HTT levels in many studies (Weiss et al., 2009, 2012). In the mouse striatal cell line (STHdhQ7/Q111) (Trettel et al., 2000), E7 reduced Htt levels and the IC50 value was close to those of Gpr52 inhibition (IC50 = 4.5 μM, Fig. 6A). We utilized the 2B7/2166 antibody pair, which detects both mHtt and wild-type HTT. The antibody pairs that detect mHtt specifically, such as the 2B7/MW1 and the 2B7/3B5H10 antibody pairs, do not work properly in these cells (Liang et al., 2014). In Huntington’s disease patient iPSC-derived striatal neurons generated as previously described (Yao et al., 2015), we used the 2B7/3B5H10 antibody pair to measure mHTT levels. E7 treatment significantly reduced the mHTT level in these neurons, with a maximum reduction achieved at 3.75 μM (Fig. 6B). The observed reduction is not due to cell loss, because we controlled the total protein concentration for the HTT measurement. In addition, the CellTiter-Glo® measurement indicates slight increase of the cell number by the compound treatment (Supplementary Fig. 7A and B).
Figure 6.
A novel Gpr52 antagonist E7 reduced mHtt levels. (A) HTT levels in STHdhQ7/Q111 treated for 2 days with different doses of E7 were measured by 2B7/2166 HTRF. Six independently plated and treated wells were tested for each dose of each compound. The plots (mean ± SEM) were fitted with the Boltzmann curve. (B) mHTT levels in Huntington’s disease patient iPSC-derived neurons (Q47) treated with the indicated concentrations of E7 were measured by 2B7/3B5H10 HTRF. The number in each bar represents the number of independently plated and treated wells. (C) mHTT levels in fly head lysates from elav-GAL4 driven transgenic Drosophila expressing full-length human mHTT with 128Q (full-length HTT-Q128) or mHTT-exon1 fragments with 72Q (HTT-exon1-Q72) fed for 6 days with E7 or the DMSO control at the indicated concentrations in the food. mHTT levels were measured by 2B7/3B5H10 HTRF. n indicates the number of independent vials containing 15 virgin female flies in each vial. (D) Mutant Htt levels in the E7 versus vehicle control (DMSO) intracerebroventricularly-injected mouse brain tissues. Mutant Htt levels were measured by 2B7/2166 HTRF. Both the striata and the cortices were tested after 9 days of injection. Six mice in each group were measured. (E) Similar to the E7 treatment in A, but the cells were transfected with the non-targeting (Neg_si) or the Gpr52 siRNA (Gpr52_si) 2 days before E7 treatment. Gpr52 knock-down abolished E7’s effect. (F) The cultured primary striatal neurons from neonatal mice with the indicated genotypes were treated with the indicated concentrations of E7, and their Htt levels were measured by the 2B7/2166 HTRF 2 days after treatment. Six independently plated and treated wells are tested. All bar plots represent mean ± SEM. Statistical analyses were performed by two-tailed unpaired t-tests (D) or one-way ANOVA and Dunnett’s post hoc tests (B, C and F) for indicated comparisons: n.s. = P > 0.1, *P < 0.05; **P < 0.01; ****P < 0.0001.
We then investigated E7’s effects in vivo. The behavioural phenotype in Huntington’s disease Drosophila model was rescued by loss-of-function of the Drosophila homologue of Gpr52 (CG18314, dGpr52) (Yao et al., 2015). Consistent with this, the transgenic mHTT level was reduced by dGpr52 loss-of-function (Supplementary Fig. 7C), suggesting a conserved regulatory pathway that justifies the potential of using Drosophila models for E7 validation in vivo. We thus fed the Huntington’s disease Drosophila with E7-containing foods versus the DMSO-containing controls. For each genotype and treatment group, the flies were distributed randomly in a number of different glass tubes containing 15 flies per tube. Treatment of 10 or 20 μM E7 for 6 days significantly reduced mHTT levels in both the full-length and the exon 1 Huntington’s disease models (full-length HTT-Q128 and HTT-exon1-Q72, Fig. 6C). Treatment of 5 μM E7 for 6 days significantly reduced mHTT levels in the exon1 Huntington’s disease model, but not in the full-length model (Fig. 6C). In summary, E7 can reduce mHtt levels in both models when sufficient dose is given.
Encouraged by the fly data, we tested E7’s effect in Huntington’s disease mice. Since it is unclear whether E7 can penetrate the mouse blood–brain barrier, we delivered E7 directly into the mouse brain by intracerebroventricular injection. We inserted and fixed a capped plastic tunnel through the skull of each mouse so that the injection could be made repeatedly (see ‘Materials and methods’ section and Supplementary Fig. 7F). E7 injection for 9 days with one dose (2 μl at 2.5 mM) per day significantly reduced mHtt levels in the striatum but not the cortices based on the HTRF measurements (Fig. 6D). The reduction in Huntington’s disease cells and tissues was also validated by western blots (Supplementary Fig. 7D–F). E7 reduced both mHtt and wild-type HTT level (Supplementary Fig. 7D and E), consistent with the effect of Gpr52 knockout.
To confirm that the E7’s effect was mediated specifically via Gpr52, we knocked-down Gpr52 in STHdhQ7/Q111 cells, and found that the E7’s effect on Htt largely disappeared (Fig. 6E). Consistently, in cultured striatal neurons from Gpr52 knockout Huntington’s disease mice (Gpr52−/−, HdhQ7/Q140), the E7’s effect on Htt disappeared as well (Fig. 6F). The data above confirm that the E7’s effect in reducing Htt was mediated by Gpr52.
Our previous data suggested that Gpr52 modulates HTT protein levels but not mRNA levels (Yao et al., 2015). However, this conclusion was not adequately tested in vivo. More importantly, we did not examine the level of an aberrant incomplete transcription of mutant HTT that results in expression of mHTT-exon 1 protein particularly in the striatum (Sathasivam et al., 2013). We thus tested both the full-length and the incomplete exon 1–intron 1 transcripts in the striata from E7 versus control-treated mice by real time-quantitative PCR, and observed no significant change of these transcripts by E7 (Supplementary Fig. 8), suggesting that E7 regulates the mHTT level at the post-transcriptional level.
The Gpr52 antagonist E7 rescues Huntington’s disease relevant phenotypes in human and mouse models
We then investigated whether E7 may have beneficial effects on Huntington’s disease-associated phenotypes by lowering mHTT. If successful, E7 may provide the proof-of-concept evidence for treating Huntington’s disease via targeted small molecule compounds that lowers mHTT, or even a candidate lead compound that could be developed into a Huntington’s disease drug.
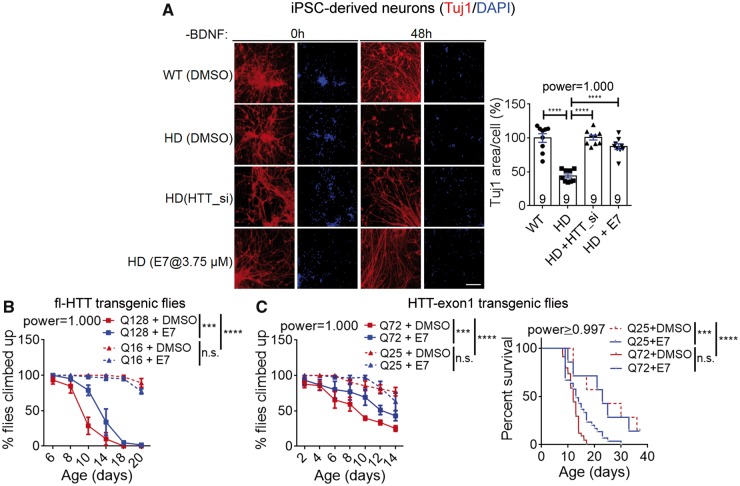
Huntington’s disease patient iPSC-derived neurons exhibit elevated apoptosis, shrinkage and loss of processes under stress conditions such as withdrawn of the brain-derived neurotrophic factor (BDNF) (HD-iPSC-Consortium, 2012; Lu et al., 2013). These phenotypes are mHTT-dependent and could be used as readout for assaying Huntington’s disease-associated cytotoxicity (Lu and Palacino, 2013; Yao et al., 2015). In the Huntington’s disease patient iPSC-derived striatal neurons (Q47), the apoptosis phenotype could be measured by assaying the caspase-3 activity with the fluorescent dye (Supplementary Fig. 9A), and the neuronal shrinkage and processes loss phenotype could be measured by immunostaining for Tuj1, a neuronal-specific-tubulin that captures neuronal morphology (Fig. 7A). Treatment of E7 at the optimal dose significantly rescued both phenotypes (Fig. 7A and Supplementary Fig. 9A), confirming E7’s effect on Huntington’s disease-associated phenotypes at the cellular level.
Figure 7.
A novel Gpr52 antagonist E7 rescued Huntington’s disease-associated phenotypes in cells and flies. (A) Images: representative immunostaining results of neuronal specific tubulin marker Tuj1and DAPI showing neuronal morphology of patient iPSC-derived striatal neurons (Huntington’s disease: Q47; wild-type: Q19). Loss of processes and shrinkage of neurons could be observed in Huntington’s disease neuronal after BDNF removal. Scale bar = 100 μm. Bar plots: quantification of the Tuj1 signal covered area (Tuj1 area) normalized to the nuclei counts. The lower Tuj1 area per cell reflects neuronal processes shrinkage and loss. Data were normalized to the wild-type control. The statistical analysis was performed by one-way ANOVA with post hoc Dunnett’s tests: ****P < 0.0001. (B) Virgin female flies with indicated genotypes and treatment were placed in different vials and tested in the climbing behavioural assay. Q128: full-length HTT-Q128 driven by elav-GAL4, n = 4 for both DMSO and E7 (10 μM) treated groups; Q16: full-length HTT-Q16 driven by elav-GAL4, n = 3 for both DMSO and E7 (10 μM) treated groups. n indicates the number of different batches of flies, which were place in different vials at 15 flies (virgin female) per vial. Data were plotted as mean ± SEM, and the statistical analysis was performed by two-way ANOVA tests. ***P < 0.001, ****P < 0.0001. (C) Left: As in B, but using flies expressing HTT-exon1 fragments with indicated Q lengths. The statistical analysis was performed by two-way ANOVA tests. ***P < 0.001, ****P < 0.0001. Right: survival curves of the flies expressing HTT-exon1 fragments treated with the indicated compounds. The statistical analysis was performed by the log-rank test. ***P < 0.001, ****P < 0.0001.
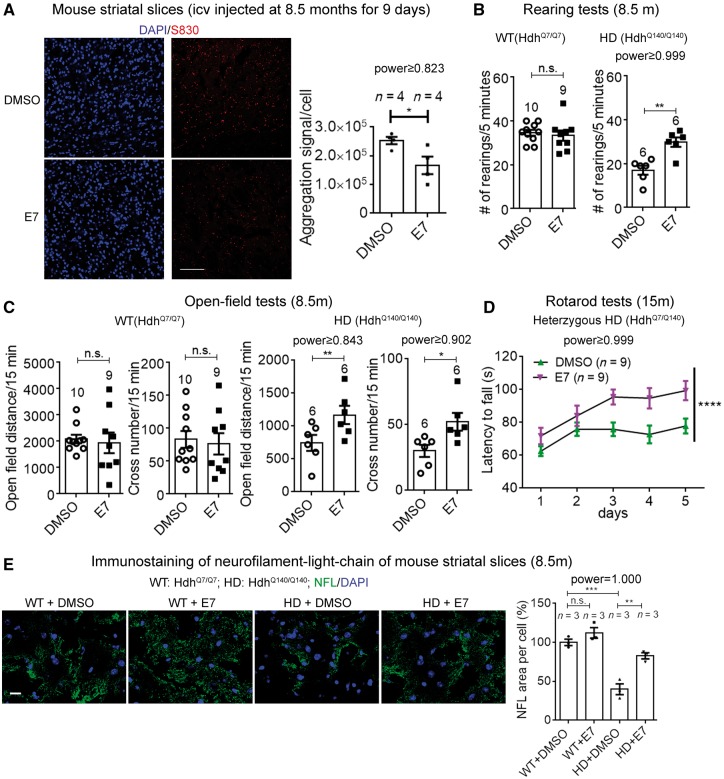
To confirm E7’s effect in vivo, we first examined the climbing behavioural phenotype in the Huntington’s disease transgenic flies expressing full-length mHTT. Feeding with foods containing 10 μM E7 significantly rescued the climbing deficits in Huntington’s disease flies and had no effect in the flies expressing wild-type HTT (Fig. 7B), confirming E7’s rescue effect at the behavioural level. Flies expressing the mHTT-exon 1 fragment exhibited similar climbing deficits and a shorter lifespan, and these phenotypes were also rescued by feeding with E7-containing food (Fig. 7C). We thus further examined the cellular and motor function phenotypes of E7-treated mice. In 8.5-month-old Huntington’s disease mice (HdhQ140/Q140), E7 injection for 9 days significantly reduced mHtt aggregates in the striatum (Fig. 8A). The number of GFAP+ and the number of Iba1+ cells were both reduced as well (Supplementary Fig. 9B), suggesting the Huntington’s disease-associated gliosis phenotype was partially rescued by E7 injection. In the rearing and open-field tests, intracerebroventricular injection of E7 for 9 days with one dose per day to 8.5-month-old mice significantly improved the motor function performance in the Huntington’s disease mice, and had no influence on the wild-type mice (Fig. 8B and C), confirming E7’s rescue effect in vivo. In all the experiments described above, we investigated the rescue of mouse behavioural phenotypes in homozygous Huntington’s disease mice (HdhQ140/Q140), because their phenotypes were more robust at earlier ages (before 12 months old). To confirm the effect in heterozygous Huntington’s disease mice (HdhQ7/Q140), we further tested E7’s effect in HdhQ7/Q140 mice at the age of 15 months, because these mice had the genotype closest to the real Huntington’s disease patients. Intracerebroventricularly-injected E7 significantly rescued the rotarod deficits of these mice (Fig. 8D), confirming the therapeutic potential of targeting Gpr52.
Figure 8.
A novel Gpr52 antagonist E7 rescued Huntington’s disease-associated phenotypes in mice. (A) Representative immunofluorescent images (by the antibody S830) (scale bar = 100 μm), and quantifications of mHtt aggregates in Huntington’s disease mouse striatal slices from mice intracerebroventricular-injected with E7 or vehicle control. The quantification was performed in the same way as in Fig. 3B. The statistical analysis was performed by two-tailed unpaired t-tests: *P < 0.05. (B) Rearing test results of the E7 versus vehicle control intracerebroventricularly-injected wild-type (left) or Huntington’s disease mice (right). The mice (8.5 months of age) were intracerebroventricularly injected for 9 days before the behavioural tests and brain tissue extraction (Fig. 6D). Each dot represents an individual mouse. Bar plots represent mean ± SEM. Statistical analysis was performed by two-tailed unpaired t-tests: n.s. = P > 0.1, **P < 0.01. E7 had no effect in the wild-type mice and significantly rescued Huntington’s disease mice. (C) As in B, but for open-field tests. The statistical analysis was performed by two-tailed unpaired t-tests: n.s. = P > 0.1, *P < 0.05, **P < 0.01. (D) Rotarod test results of the E7 versus vehicle control intracerebroventricularly-injected 15-month-old heterozygous Huntington’s disease mice (HdhQ7/Q140 mice injected for 7 days, trained for 3 days and then tested for 5 days; the one injection per day was continued during training and testing days). The statistical analysis was performed by two-way ANOVA tests. ****P < 0.0001. (E) Representative immunofluorescent images of NFL (scale bar = 10 μm) and quantifications of neurofilament light chain (NFL) in wild-type and Huntington’s disease mouse striatal slices from mice intracerebroventricularly-injected with E7 or vehicle control. The NFL signals were normalized by the number of cells (counted by DAPI). The statistical analysis was performed by one-way ANOVA and Bonferroni’s post hoc tests for indicated comparisons: n.s. = P > 0.1, **P < 0.01, ****P < 0.001.
Finally, we assayed Huntington’s disease neuronal damage by immunostaining of the neurofilament light chain (NFL), which exhibited significant loss in disease striatal slices, and this was partially rescued by E7 injection (Fig. 8E).
Discussion
Gpr52 as a Huntington’s disease drug target
Our current study provides two major pieces of evidence for in vivo establishing Gpr52 as a Huntington’s disease drug target: lowering mHtt and rescuing Huntington’s disease-associated phenotypes. We confirmed the effect by both the genetic deletion and the compound antagonist. In addition, we showed that expressed of hGpr52 restored the mHtt levels and Huntington’s disease-associated phenotypes, confirming that the knockout effect is on target.
From 40 000 compounds, we identified one relatively specific Gpr52 antagonist (Fig. 5 and Supplementary Fig. 6), E7, which functioned as expected in lowering mHtt and rescuing Huntington’s disease-associated phenotypes (Figs 6–8). In addition, E7 likely modulates mHtt levels via inhibiting Gpr52: knocking-down or knocking-out Gpr52 abolished the effect of E7 in lowering Htt (Fig. 6E and F). Given the EC50 of WO-459 (Fig. 5C) and the IC50 of E7 in inhibiting Gpr52 with treatment of agonist (100 nM WO-459, Fig. 5H), the Ki of E7 for Gpr52 was roughly estimated to be ∼0.9 μM using the Cheng-Prussof equation (Lazareno and Birdsall, 1993). The range is largely consistent with the effective concentrations of E7 in lowering mHtt levels (Fig. 6). E7 was active mainly in the Huntington’s disease genetic background (Figs 6–8), and this might reflect that Gpr52 might have enhanced activation in Huntington’s disease, possibly due to an increase in the concentration of its agonist.
Our previous studies have demonstrated that Gpr52 modulates Htt levels via a cAMP-dependent but PKA-independent pathway (Yao et al., 2015), and this is consistent with previous chemical–genetics studies (Williams et al., 2008). In addition, we demonstrated that the effect was mediated by enhanced proteasomal degradation of mHtt via inactivation of the small GTPase Rab39B (Yao et al., 2015). While the impact of cAMP pathway in Huntington’s disease is complicated and we cannot exclude other mechanisms mediating Gpr52’s effect, the fact that targeting Gpr52 by genetic deletion or an antagonist significantly lowers mHtt and rescues Huntington’s disease phenotypes is convincing to justify Gpr52 as a potential Huntington’s disease target.
Regarding safety, the physiological function of endogenous Gpr52 remains unclear. In our study, we observed no behavioural phenotypes in Gpr52 knockouts in the wild-type background. We observed no change in the animal size, weight and major organ morphology by Gpr52 knockout in both the wild-type and Huntington’s disease background (not shown). The previously reported Gpr52 homozygous knockout generated in another background (129SvEv) exhibited no obvious phenotypes and no change in brain morphology (Komatsu et al., 2014). There was no change in the travel distance in the open-field tests (Komatsu et al., 2014), consistent with our observation (Fig. 1). They exhibited higher frequency of startle but not prepulse inhibition behaviours when treated with the NMDA receptor antagonist MK-801 (Komatsu et al., 2014), and enhanced the locomotor-stimulating effect of the ADORA2A antagonist istradefylline but not methamphetamine MK-801 (Hancock et al., 2012). The only behavioural phenotypes of the 129SvEv Gpr52 knockout model without compound treatment were the increased time staying in the central region in the open-field test (Komatsu et al., 2014) and the increased novelty-induced locomotor activity (Hancock et al., 2012). These two phenotypes might be somewhat inconsistent with our observation (Figs 1–2), possibly due to different genetic background and test conditions (age, equipment, etc.). In addition, the method to generate the Gpr52 knockout was different, and the Gpr52 knockout mice in the previous Gpr52 knockout study have kept the LacZ and neomycin-resistant gene cassette, which may induce artefacts. Nonetheless, inhibiting Gpr52 by E7 or other antagonist may interfere with the dopamine circuits via influencing intracellular cAMP and induce phenotypes under certain conditions such as schizophrenia. Special cautions should be given for patients with these conditions when treated with Gpr52 antagonists. Meanwhile, E7 treatment in the Huntington’s disease mice did not influence prepulse inhibition or new-objective recognition phenotypes (Supplementary Fig. 10), suggesting that E7 treatment may not lead to psychotic or cognitive defects.
Lowering mHtt as a potential strategy for Huntington’s disease treatment
Lowering mHtt protein levels is considered as the most promising strategy for Huntington’s disease treatment. In fact, a clinical trial aiming at lowering Htt levels by antisense oligonucleotides has already been launched. While such strategies using RNA-like molecules targeting mRNA are certainly promising and of great importance in the field, their delivery is still highly challenging and their cost are unacceptable for most Huntington’s disease patients. Thus, small molecule compound drugs are highly desired. Gpr52 and its antagonist E7 provide new avenues for discovery of such drugs that may modify Huntington’s disease progression via lowering mHtt protein level.
Mutant Htt lowering per se is likely the major mechanism that mediates the effect of Gpr52 on Huntington’s disease-associated phenotypes. Compelling evidence has shown that reducing mHtt by many different approaches can rescue the Huntington’s disease-associated phenotypes (Yu et al., 2014), and Gpr52 significantly reduces mHtt levels in all striatal Huntington’s disease models tested. Importantly, Gpr52 or E7 does not influence the phenotypes in the wild-type mice (Figs 1, 2, 4 and 8). In addition, knocking-down Gpr52’s downstream modulator Rabgap1l largely abolished its rescue effect in patient iPSC-derived striatal neurons (Yao et al., 2015), confirming that the rescue was mediated through lowering mHtt, at least at the cellular level. Interestingly, expression of hGpr52 cDNA using a neuron-specific promoter restored the increase of Iba1+ or GFAP+ glial cells in the Huntington’s disease mice (Supplementary Fig. 4A). Injection of the Gpr52 antagonist E7 for 9 days significantly reduced Iba1+ or GFAP+ glial cells in the Huntington’s disease mice as well (Supplementary Fig. 9). The data suggest that a short-term lowering of the neuronal mHtt level is sufficient to reduce neuroinflammation related gliosis, potentially through non-cell autonomous mechanisms.
Contribution of striatal mHtt to Huntington’s disease
One common characteristic of Huntington’s disease and most other neurodegenerative disorders is that the neurodegeneration occurs quite selectively in certain brain regions. This regional selectivity is especially intriguing in Huntington’s disease, because the disease-causing protein mHtt is widely expressed, while the neurodegeneration mainly influences in the striatum, especially at early stages (Raymond et al., 2011). One potential possibility is that striatal cells express specific genes that enhance mHtt toxicity or stabilizes mHtt (Blum et al., 2003; Thomas, 2006; Subramaniam et al., 2009). In fact, the mHtt turnover rate is slower in the striatal neurons compare to cortical neurons (Tsvetkov et al., 2013), likely because of striatal-enriched mHtt stabilizers.
Gpr52 expression is striatal-enriched in mice (Komatsu et al., 2014). The microarray data from Allen Brain Atlas (www.brain-map.org) and our immunohistochemistry results from post-mortem brain slices confirmed the striatal-enriched expression of Gpr52 in the human brain as well (Supplementary Fig. 11). Our study used a knock-in Huntington’s disease model expressing endogenous mHtt, and targeting Gpr52 mainly modulates striatal mHtt (Fig. 3). The lowering of striatal mHtt by targeting Gpr52 rescued Huntington’s disease-associated behavioural phenotypes (Figs 1 and 2), whereas hGpr52 cDNA expression restored striatal mHtt levels and restored Huntington’s disease-associated phenotypes (Fig. 4 and Supplementary Fig. 3), confirming contributions of striatal mHtt in Huntington’s disease pathogenesis at least in the knock-in model. Our data were also consistent with recent genome-editing study, which showed that lowering striatal mHtt by CRISPR/Cas9 significantly rescued behavioural phenotypes of the knock-in Huntington’s disease mice (Yang et al., 2017).
The Gpr52 antagonist E7 and its delivery by intracerebroventricular injection
Importantly, the HTT-lowering effects and the rescue of Huntington’s disease-associated phenotypes by targeting Gpr52 have been validated cross-species in fly, mouse and human models (Figs 6–8). The cross-species validation may increase the success rate of targeting Gpr52 in human patients, which are impossible to test in preclinical studies.
While RNA-targeting molecules such as antisense oligonucleotides are also promising drug candidates to lower mHTT, their delivery is highly challenging and their costs are expensive. This is hard to be fundamentally improved because of their intrinsic nature of being RNA-like large molecules. In contrast, small molecule compound antagonists for Gpr52 have much higher potential to be structurally modified to a compound for easier delivery, although E7 per se may not be ready such purpose yet, because structural–activity relationship studies and pharmacokinetics/pharmacodynamics studies are needed. Meanwhile, E7 provides a tool for the proof-of-concept study to demonstrate the potential of targeting Gpr52.
In our current study, we delivered E7 by intracerebroventricular injection, which we believe is an ideal approach for proof-of-concept studies of novel compounds. Delivery by other approaches such as intraperitoneal injection and intravenous injection are closer to clinical use, but the results could be hard to interpret. Negative outcome could be due to failure of passing the blood–brain barrier and/or pharmacokinetics/pharmacodynamics issues. In addition, a much larger amount of compound is needed for these approaches compared to intracerebroventricular injection, because the former approaches deliver the compound to the whole body.
Many further steps are needed to develop Gpr52 antagonists for clinical use. E7 is currently active only in the micromolar range, and the structure–activity relationship studies of E7 and its structural analogues are desirable to discover better compounds. Noticeably, E7 is a racemate containing two enantiomers. Revealing the Gpr52-antagonizing and mHTT-lowering activity of both enantiomers is highly desired for drug discovery purposes, although this is technically challenging at this point, because neither of these enantiomers with sufficient purity and quality are available. Additional screening with larger libraries may be desired for better compounds.
Once a high-efficacy compound has been identified by optimizing the structure of E7 or additional screening, the compound will be tested for its penetration of the blood–brain barrier, its pharmacokinetics/pharmacodynamics properties and its safety profiles for further drug discovery purposes.
Supplementary Material
Acknowledgments
The authors wish to thank Dr Chenqi Lu (biostatistician) for the statistical analysis, Dr Lixiang Ma and Saiyin Hexige for sharing the patient fibroblast lines and help with immunohistochemistry, Drs Min Jiang, Linyun Liu and Qian Huang for their technical support of mouse behavioural experiments, Drs Chengyong Shen, Chen Zhang, Jisong Guan and Zhenggang Yang for providing some of the tested commercial antibodies, Dr Yanai Mei for help of the novel object recognition experiments, and Dr Lin Mei for help of the prepulse inbition experiments.
Funding
This work is supported by the National Key Research and Development Program of China (2016YFC0905100), National Natural Science Foundation of China (91649105, 81425024), and the Fudan-SIMM Joint Research Fund (FU-SIMM 20174013).
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- DMSO
dimethyl sulfoxide
- GPCR
G protein-coupled receptor
- hGpr52
human Gpr52
- HTRF
homogeneous time resolved fluorescence
- iPSC
induced pluripotent stem cell
- mHTT
mutant HTT
References
- Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem 2012; 287: 1406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum D, Hourez R, Galas MC, Popoli P, Schiffmann SN. Adenosine receptors and Huntington's disease: implications for pathogenesis and therapeutics. Lancet Neurol 2003; 2: 366–74. [DOI] [PubMed] [Google Scholar]
- Daneault JF, Carignan B, Sadikot AF, Duval C. Inter-limb coupling during diadochokinesis in Parkinson's and Huntington's disease. Neurosci Res 2015; 97: 60–8. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci USA 2006; 103: 9482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA 2007; 104: 17204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock MK, Hermanson SB, Dolman NJ. A quantitative TR-FRET plate reader immunoassay for measuring autophagy. Autophagy 2012; 8: 1227–44. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA 2005; 102: 5820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HD-iPSC-Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 2012; 11: 264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey MA, Zhu C, Medvedeva V, Lerner RP, Patassini S, Franich NR, et al. Improvement of neuropathology and transcriptional deficits in CAG 140 knock-in mice supports a beneficial effect of dietary curcumin in Huntington's disease. Mol Neurodegener 2012; 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodas JJ, Nehring A, Hoche N, Sweredoski MJ, Pielot R, Hess S, et al. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT). Proteomics 2012; 12: 2464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34: 259–69. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Maruyama M, Yao S, Shinohara T, Sakuma K, Imaichi S, et al. Anatomical transcriptome of G protein-coupled receptors leads to the identification of a novel therapeutic candidate GPR52 for psychiatric disorders. PLoS One 2014; 9: e90134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron 2012; 74: 1031–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareno S, Birdsall NJ. Estimation of competitive antagonist affinity from functional inhibition curves using the Gaddum, Schild and Cheng-Prusoff equations. Brit J Pharmacol 1993; 109: 1110–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Yao Y, Lu M, Hou J, Yu S, Lu B. TR-FRET assays for endogenous Huntingtin protein level in mouse cells. J Huntingtons Dis 2014; 3: 253–9. [DOI] [PubMed] [Google Scholar]
- Lu B, Al-Ramahi I, Valencia A, Wang Q, Berenshteyn F, Yang H, et al. Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat Neurosci 2013; 16: 562–70. [DOI] [PubMed] [Google Scholar]
- Lu B, Palacino J. A novel human embryonic stem cell-derived Huntington's disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J 2013; 27: 1820–9. [DOI] [PubMed] [Google Scholar]
- Mathis G. Rare earth cryptates and homogeneous fluoroimmunoassays with human sera. Clin Chem 1993; 39: 1953–9. [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol 2003; 465: 11–26. [DOI] [PubMed] [Google Scholar]
- Ochaba J, Monteys AM, O'Rourke JG, Reidling JC, Steffan JS, Davidson BL, et al. PIAS1 regulates mutant huntingtin accumulation and Huntington's disease-associated phenotypes in vivo. Neuron 2016; 90: 507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov 2006; 5: 993–6. [DOI] [PubMed] [Google Scholar]
- Paganetti P, Weiss A, Trapp M, Hammerl I, Bleckmann D, Bodner RA, et al. Development of a method for the high-throughput quantification of cellular proteins. Chembiochem 2009; 10: 1678–88. [DOI] [PubMed] [Google Scholar]
- Park J, Al-Ramahi I, Tan Q, Mollema N, Diaz-Garcia JR, Gallego-Flores T, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature 2013; 498: 325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond LA, Andre VM, Cepeda C, Gladding CM, Milnerwood AJ, Levine MS. Pathophysiology of Huntington's disease: time-dependent alterations in synaptic and receptor function. Neuroscience 2011; 198: 252–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther 2005; 12: 618–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA 2013; 110: 2366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoh M, Ishii N, Kono M, Miyanohana Y, Shiraishi E, Harasawa T, et al. Discovery of the first potent and orally available agonist of the orphan G-protein-coupled receptor 52. J Med Chem 2014; 57: 5226–37. [DOI] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci 2003; 4: 49–60. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science 2009; 324: 1327–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993; 726: 971–83. [DOI] [PubMed] [Google Scholar]
- Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res 2006; 84: 1151–64. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 2000; 9: 2799–809. [DOI] [PubMed] [Google Scholar]
- Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol 2013; 9: 586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Gray M, Lu XH, Cantle JP, Holley SM, Greiner E, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington's disease. Nat Med 2014; 20: 536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Abramowski D, Bibel M, Bodner R, Chopra V, DiFiglia M, et al. Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington's disease. Anal Biochem 2009; 395: 8–15. [DOI] [PubMed] [Google Scholar]
- Weiss A, Trager U, Wild EJ, Grueninger S, Farmer R, Landles C, et al. Mutant huntingtin fragmentation in immune cells tracks Huntington's disease progression. J Clin Invest 2012; 122: 3731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol 2008; 4: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 2000; 101: 57–66. [DOI] [PubMed] [Google Scholar]
- Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J Clin Invest 2017; 127: 2719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Cui X, Al-Ramahi I, Sun X, Li B, Hou J, et al. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. Elife 2015; 4: 1–19. doi: 10.7554/eLife.05449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Fu Y, Liang Y, Song H, Yao Y, Wu P, et al. Suppression of MAPK11 or HIPK3 reduces mutant Huntingtin levels in Huntington's disease models. Cell Res 2017; 27: 1441–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Liang Y, Palacino J, Difiglia M, Lu B. Drugging unconventional targets: insights from Huntington's disease. Trends Pharmacol Sci 2014; 35: 53–62. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang S, Binari R, Zhou R, Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics 2010; 184: 1165–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.