Higuchi et al. identify recessive mutations in the mitochondrial gene, cytochrome c oxidase assembly factor 7 (COA7) in four unrelated patients with an axonal-type motor and sensory neuropathy with ataxia. Genetic, histopathological, radiological and functional data support a causative role for loss-of-function COA7 mutations in the observed phenotype.
Keywords: genetics, neuropathy, spinocerebellar ataxia, whole-exome sequencing, COA7
Abstract
Several genes related to mitochondrial functions have been identified as causative genes of neuropathy or ataxia. Cytochrome c oxidase assembly factor 7 (COA7) may have a role in assembling mitochondrial respiratory chain complexes that function in oxidative phosphorylation. Here we identified four unrelated patients with recessive mutations in COA7 among a Japanese case series of 1396 patients with Charcot-Marie-Tooth disease (CMT) or other inherited peripheral neuropathies, including complex forms of CMT. We also found that all four patients had characteristic neurological features of peripheral neuropathy and ataxia with cerebellar atrophy, and some patients showed leukoencephalopathy or spinal cord atrophy on MRI scans. Validated mutations were located at highly conserved residues among different species and segregated with the disease in each family. Nerve conduction studies showed axonal sensorimotor neuropathy. Sural nerve biopsies showed chronic axonal degeneration with a marked loss of large and medium myelinated fibres. An immunohistochemical assay with an anti-COA7 antibody in the sural nerve from the control patient showed the positive expression of COA7 in the cytoplasm of Schwann cells. We also observed mildly elevated serum creatine kinase levels in all patients and the presence of a few ragged-red fibres and some cytochrome c oxidase-negative fibres in a muscle biopsy obtained from one patient, which was suggestive of subclinical mitochondrial myopathy. Mitochondrial respiratory chain enzyme assay in skin fibroblasts from the three patients showed a definitive decrease in complex I or complex IV. Immunocytochemical analysis of subcellular localization in HeLa cells indicated that mutant COA7 proteins as well as wild-type COA7 were localized in mitochondria, which suggests that mutant COA7 does not affect the mitochondrial recruitment and may affect the stability or localization of COA7 interaction partners in the mitochondria. In addition, Drosophila COA7 (dCOA7) knockdown models showed rough eye phenotype, reduced lifespan, impaired locomotive ability and shortened synaptic branches of motor neurons. Our results suggest that loss-of-function COA7 mutation is responsible for the phenotype of the presented patients, and this new entity of disease would be referred to as spinocerebellar ataxia with axonal neuropathy type 3.
Introduction
Mitochondrial disorders are a clinically and genetically heterogeneous group of progressive multisystem disorders caused by impaired mitochondrial function. Affected patients often present with prominent neurologic features such as encephalopathy, seizures, dementia, deafness, ataxia, spasticity and neuropathy. To date, several genes related to mitochondrial functions, such as AFG3L2, AIFM1, COX6A1, COX10, COX20, COA3, DHTKD1, GDAP1, HK1, MFN2, MTATP6, PDK3, POLG, SLC25A46 and SURF1 have been identified as causative genes of neuropathy or ataxia. Particularly, MTATP6, SURF1, COX6A1 COX10, COX20 and COA3 were shown to be involved in assembly or in the activity of the mitochondrial respiratory chain (MRC) complex (Valnot et al., 2000; Pitceathly et al., 2012; Echaniz-Laguna et al., 2013; Szklarczyk et al., 2013; Tamiya et al., 2014; Ostergaard et al., 2015).
Our institution serves as a genetic testing centre for Charcot–Marie–Tooth disease (CMT) in Japan. We have performed comprehensive genetic testing for >1000 Japanese patients with clinically suspected CMT or other inherited peripheral neuropathy, including complex forms of CMT in which the neuropathy is combined with other features such as mental retardation, upper motor neuron signs, deafness, optic atrophy and cerebellar ataxia. As part of our work to identify novel causative genes for CMT, we recently identified MME as a causative gene for autosomal-recessive axonal CMT (MIM: 617017) using whole-exome sequencing (WES) analyses in patients with CMT with an overlap-based strategy (Higuchi et al., 2016). In the present study, we used the same strategy and identified recessive mutations in the cytochrome c oxidase assembly factor 7 (COA7) gene in four unrelated patients among many Japanese patients with CMT or other inherited peripheral neuropathies. The COA7 gene encodes cytochrome c oxidase assembly factor 7, which is also referred to as respiratory chain assembly factor 1 (RESA1), sel1 repeat-containing protein 1 (SELRC1), and C1orf163. Although the role of COA7 is still not completely understood, its potential role in assembly of the MRC complex I and complex IV was reported in vitro in cell culture studies (Kozjak-Pavlovic et al., 2014).
In the present study, we found that patients with COA7 mutations had characteristic neurological features of peripheral neuropathy with cerebellar ataxia. We also provide genetic, histopathological, and functional data to support the contention that the recessive mutations in COA7 are responsible for neurological impairment.
Materials and methods
Patient selection and mutation detection
A total of 1396 Japanese patients with CMT or other inherited peripheral neuropathies were enrolled in this study between 2005 and 2016. The genomic DNA of patients and family members was extracted as described previously (Higuchi et al., 2016).
First, in a Japanese case series of 372 patients with clinically diagnosed CMT, we excluded patients with pathogenic mutations in the known inherited peripheral neuropathy genes by an originally designed microarray resequencing DNA chip or WES as described previously (Zhao et al., 2012; Higuchi et al., 2016). Subsequently we selected 163 unrelated patients with presumed autosomal recessive inheritance or with sporadic inheritance. From WES data of these 163 patients, we identified biallelic novel mutations in the COA7 gene that were shared by two patients (Patients 1 and 2) from unrelated families, with the exome-based shared variants detection (ESVD) system using an overlap-based strategy, as previously described (Higuchi et al., 2016). Second, from an additional case series of 1024 Japanese patients with clinically diagnosed CMT or other inherited peripheral neuropathies, we identified another two patients (Patients 3 and 4) with a compound heterozygous mutation in the COA7 gene using WES or two methods, Illumina Miseq platform (Illumina Inc.) and a customized Ion AmpliSeq panel, as described previously (Higuchi et al., 2016). On Sanger sequencing, we reconfirmed the mutations revealed by next generation sequencing. Furthermore, segregation studies were performed in the available family members whenever possible. The study protocol was reviewed and approved by the institutional review board at Kagoshima University. All patients and family members provided written informed consent to participate in this study, including for the genetic analyses.
Multiplex ligation-dependent probe amplification
To detect partial deletions in any region of COA7, we performed customized multiplex ligation-dependent probe amplification (MLPA) using the MRC-Holland SALSA MLPA probe mix P300-A2 Human DNA Reference-2 (reference probe mix kit) in conjunction with synthetic custom probes for exons 1 to 3 of COA7. Probe sequences were obtained with the MLPA probe design tool (H-MAPD) (Zhi and Hatchwell, 2008) as recommended in the MRC Holland’s probe design guidelines. Oligonucleotides were purchased from Integrated DNA Technologies. Synthetic probe mixes were prepared in TE buffer. The MLPA reactions were carried out on the genomic DNA of Patient 2, his parents and three control samples. Fragment analysis was performed by capillary electrophoresis in an ABI PRISM 3130×l Genetic Analyzer (Applied Biosystems). The normalization of the data was performed using an originally prepared spreadsheet provided from FALCO Biosystems Ltd, which was prepared based on the General MLPA protocol. The threshold values (dosage quotient: DQ) for deletions and duplications were set to 0.65–1.3, respectively, which was also used for DNA analysis.
Histopathological examination
Sural nerve biopsy specimens obtained from Patients 1 and 2 were examined for morphometric changes under a light microscope and the findings compared with a disease control patient (female, 39 years old, polymyositis). Semi-thin sections from EPON™-embedded tissues were stained with toluidine blue. An immunohistochemical study was performed with anti-COA7 antibody (SIGMA, HPA029926) in tissue specimen of peripheral nerves of the control patient. In addition, frozen biopsies of the biceps brachii muscle specimens were obtained from Patient 1. Histochemical and immunohistochemical procedures were performed as described previously (Higuchi et al., 1999).
Respiratory chain enzyme assay
The enzymatic activities of MRC complexes I, II, III and IV were assayed in skin fibroblasts and muscle tissue as described previously (Kirby et al., 1999; Kishita et al., 2015). The activity of each complex was presented as a percentage of the mean value obtained from 35 healthy controls. For each patient, the percentage activities of complexes I, II, III and IV activity relative to that of citrate synthase as a mitochondrial enzyme marker were calculated (Rahman et al., 1996). Deficiency of each complex is confirmed when the citrate synthase ratio is <40% (fibroblasts) (Bernier et al., 2002).
Analysis with blue native polyacrylamide gel electrophoresis
Isolated mitochondrial fractions were stored in aliquots at −80°C until use. The amount and size of fully assembled complex I were studied by blue native polyacrylamide gel electrophoresis (BN-PAGE) and immunoblotting using mitochondria isolated from patients’ cells and solubilized with the detergent Triton™ X-100, as described previously (Kirby et al., 2003, 2004). Immunodecoration was performed using a monoclonal antibody specific for the 39 kD subunit of complex I, 70 kD subunit of complex II, core 1 subunit of complex III, and subunit 1 of complex IV (Molecular Probes). The percentage amount of each of fully assembled complex I, III or IV was estimated by densitometry (Bio-Rad GS-800 Calibrated Densitometer) of three independent BN-PAGE immunoblots using assembled complex II as a control. In each experiment, mitochondria from two controls were run in one gel simultaneously, and the mean relative value of these two controls was defined as 100%. In-gel enzyme staining of each complex was performed as described previously (Dubowitz et al., 1985; Dabbeni-Sala et al., 2001; Van Coster et al., 2001).
Complementary DNA cloning and expression constructs
MGC human COA7 complementary DNA (cDNA) (Clone ID: 4430419) was obtained from Thermo Fisher Scientific. The cDNA encoding COA7 with C-terminal FLAG epitope tag was cloned into pMRXIP (provided by Shoji Yamaoka, Tokyo Medical and Dental University, Tokyo, Japan) (Saitoh et al., 2002). Mutant COA7 constructs were made with the Q5 site-directed mutagenesis kit (New England Biolabs).
Cell culture and retroviral infections
HeLa cells were cultured in Dulbecco’s modified Eagle medium (Wako Pure chemical) supplemented with 10% foetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in an atmosphere containing 5% CO2. HeLa cells expressing Coa7-FLAG were generated using retroviral infection systems (Kitamura et al., 2003). Human Plat-E cells (provided by Toshio Kitamura, University of Tokyo, Tokyo, Japan) were co-transfected with pMRXIP-Coa7 and pCG-VSV-G using the transfection reagent Fugene HD (Promega) to generate recombinant retroviruses. HeLa cells were then infected with recombinant retroviruses and selected in a medium containing 3 μg/ml puromycin.
Immunocytochemistry
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were permeabilized with 0.1% Triton in PBS for 15 min and blocked with 5% normal donkey serum in PBS for 60 min. Cells were then incubated with indicated primary antibodies for 4 h. After washing, cells were incubated with secondary antibodies for 60 min. Mouse monoclonal anti-FLAG (M2, Sigma-Aldrich) and rabbit polyclonal anti-Tom20 (FL-145, Santa Cruz Biotech) were used as primary antibodies. Alexa Fluor® 488- or 594-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG) were used as secondary antibodies (Thermo Fisher Scientific). Images were acquired with an FV1000 confocal microscope as shown previously (Hara et al., 2014).
Fly stocks
We generated Drosophila models with dCOA7 (CG13865, the orthologue of human COA7) knockdown by RNA interference (RNAi). Fly stocks were maintained at 25°C on standard food as described previously (Takahashi et al., 1999). The flies carrying w; UAS-dCOA7 –IR143-253; + (CG13865) were obtained from the Vienna Drosophila RNAi Center (VDRC). The fly lines carrying w; UAS-dCOA7-IR11-17; + (CG13865), w; P[GAL4-elav.L]3 and w; P[UAS-GFPi]attP40; + (UAS-GFP-IR) were obtained from the Bloomington Drosophila Stock Center at the Indiana University. GMR-GAL4 used in this study was as described previously (Takahashi et al., 1999).
Comparison of amino acid sequences of human and Drosophila COA7
The amino acid sequence of Drosophila COA7 (dCOA7) was retrieved from Flybase (http://flybase.org). The identity and similarity of Drosophila COA7 and human COA7 were compared using BLAST (http://blast.genome.jp/) and FASTA (http://fasta.genome.jp/). FASTA was used for comparison of the whole sequences, and BLAST was used for comparison of each corresponding domain between dCOA7 and COA7.
Scanning electron microscopy
Adult flies were anaesthetized, mounted and examined under a scanning electron microscope (Keyence VE-7800) in high vacuum mode. In every experiment, the eye phenotypes of at least five adult male flies (3 to 5 days old) of each line were simultaneously examined by scanning electron microscopy. These experiments were repeated at least twice. No significant variation in eye phenotype was observed among the five individual flies.
Climbing assay
Climbing assays were performed as described previously (Feiguin et al., 2009; Kyotani et al., 2016). Flies carrying w; UAS-GFP-IR/+; elav-GAL4/+, w; UAS- dCOA7-IR143-253/+; elav-GAL4/+ and w; UAS-dCOA7-IR11-17/+; elav-GAL4/+ were placed at 28°C, and newly eclosed adult male flies were separated and placed in vials at a density of 20 flies per vial. Flies were transferred, without anaesthesia, to a conical tube. The tubes were then tapped to collect the flies to the bottom, after which they were given 30 s to climb the wall. The flies were collected at the bottom by tapping the tube and were again allowed to climb for 30 s. The same procedure, all of which was videotaped, was repeated five times in total. For all of the climbing experiments, the height to which each fly climbed was scored as follows [score (height climbed)]: 0 (<2 cm); 1 (2–3.9 cm); 2 (4–5.9 cm); 3 (6–7.9 cm); 4 (8–9.9 cm) and 5 (>10 cm). The climbing index of each fly strain was calculated as follows: the sum of the products of each score multiplied by the number of flies for which that score was recorded, was calculated, and this number was then divided by five times the total number of flies examined. These climbing assays were carried out at Days 3, 10, 17, 24, 31, 38 and 45 after eclosion.
Lifespan assay
Viability assays were conducted in a humidified, temperature controlled incubator at 25°C and 60% humidity on a 12 h light and 12 h dark cycle on standard fly food. Flies carrying w; UAS-GFP-IR/+; elav-GAL4/+, w; UAS-dCOA7-IR143-253/+; elav-GAL4/+ and w; UAS-dCOA7-IR11-17/+; elav-GAL4/+ were placed at 28°C, and newly eclosed adult male flies were separated and placed in vials at a low density (20 flies per vial). Every 3 days, they were transferred to new tubes containing fresh food and deaths were scored. Survival rate was determined by plotting a graph of the percentage of surviving flies versus days.
Visualization of the neuromuscular junction
For visualization of the neuromuscular junction (NMJ), third instar larvae carrying w; UAS-GFP-IR/+; elav-GAL4/+, w; UAS-dCOA7-IR143-253/+; elav-GAL4/+ and w; UAS-dCOA7-IR11-17/+; elav-GAL4/+ were dissected in HL3 saline (Feng et al., 2004) and fixed in 4% paraformaldehyde in PBS for 30 min at 25°C. After blocking with PBS containing 2% bovine serum albumin and 0.1% Triton™ X-100, FITC-conjugated goat anti-HRP (1:1000, MP biochemicals) was applied to the sample as the detection antibody. The stained samples were mounted and inspected under a confocal laser-scanning microscope (Olympus Flouview FV10i). MN4 (Ib) in muscle 4 in abdominal segment 2 was quantified. Nerve terminal branch lengths were measured using MetaMorph software (Molecular devices).
Data analysis
MetaMorph software (Molecular devices, Australia) was used for statistical analysis of NMJ length. P-values were calculated using Welch’s t-test and the error bars represent standard error from means (SEM). SPSS software (IBM) was used for statistical analysis of the results of climbing assay. All data are presented as means ± SEM. Viability assay results were assessed with GraphPad Prism version 6.02–Survival of Three groups–software.
Results
Identification of COA7 mutations
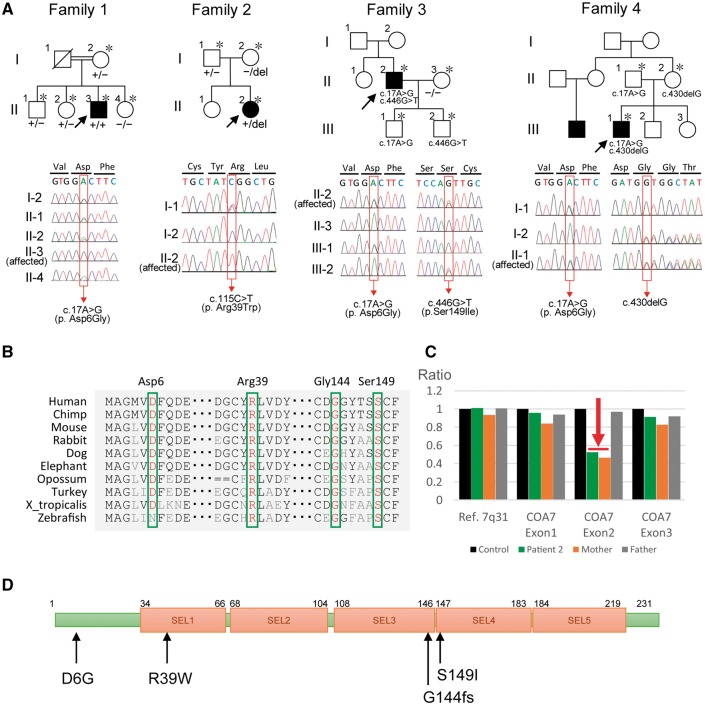
In a case series of 1396 Japanese patients with clinically diagnosed CMT or other inherited peripheral neuropathies, we extracted two homozygous and two compound heterozygous variants in the COA7 gene: [c.17A>G (p.Asp6Gly)], [c.115C>T (p.Arg39Trp)], [c.17A>G (p.Asp6Gly)/c.446G>T (p.Ser149Ile)] and [c.17A>G (p.Asp6Gly)/c.430delG, (p.Gly144fs)] in Patients 1 to 4, respectively (Fig. 1A and Table 1). These variants were located in a conserved region among different species (Fig. 1B), and were predicted to be deleterious in silico analysis including PolyPhen-2 (Adzhubei et al., 2010), SIFT (Kumar et al., 2009) and PROVEAN (Choi et al., 2012) (Supplementary Table 1). None of the variants were found in any of the public databases, with the exception of p.Asp6Gly, which was found as a rare variant in ExAC (allele count = 1/120 268) (Supplementary Table 1). Variants in Patients 1, 3 and 4 were found segregated with the disease in their families. On pedigree analysis of Patient 2, the c.115C>T heterozygous genotype was only detected in her father, but absent in her mother (Fig. 1A). We thus carried out an analysis for copy number variations using a custom-designed MLPA assay, which showed a deletion of COA7 exon 2 in a heterozygous state in Patient 2 and her mother (Fig. 1C). These findings suggest that Patient 2 has a compound heterozygous genotype that comprises a deletion of exon 2 (inherited from mother) and a missense variant (inherited from father).
Figure 1.
Family history, segregation analysis, conservation analysis and MLPA analysis. (A) Top: The pedigree structure and segregation analyses of Patients 1 to 4. Parents of Patient 1 are consanguineous; Patients 2–4 are sporadic. Bottom: Sequencing chromatograms shows homozygous or compound heterozygous mutation in the affected probands, while the unaffected members were heterozygous or wild-type carriers. A DNA sample was available for individuals marked with an asterisk. Squares represent males and circles represent females. Filled symbols represent those affected with a similar phenotype. Oblique lines represent deceased family members. Black arrows indicate the proband (Patient 1– Patient 4). Red box indicates the mutation site. +/+ = homozygous for mutation; +/− = heterozygous; −/− = homozygous for wild-type; del = deletion. (B) Comparison of COA7 from different species. Asp6, Arg39, Gly144 and Ser149 are highly conserved in many species. (C) MLPA analysis of Patient 2 and her parents. Histograms represent allelic dosage of COA7 and the reference gene. A single-copy deletion of the exon 2 in COA7 was detected in Patient 2 and her mother (red arrow). (D) Schematic representation of the COA7. Black arrows indicate the location of mutations. Arg39, Gly144 and Ser149 are located in the SEL1-like repeats.
Table 1.
Genetic, clinical and laboratory findings
| Patient | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Case from Martinez-Lyons et al. |
|---|---|---|---|---|---|
| Mutation | c.17A>G (p.D6G) | c.115C>T (p.R39W) exon 2 deletion | c.17A>G (p.D6G) c.446G>T (p.S149I) | c.17A>G (p.D6G) c.430delG (p.G144fs) | c.410A>G, (p.Y137C) c.287+1G>T |
| Genotype | Homozygous | CH | CH | CH | CH |
| Classification | Missense | Missense, deletion | Missense | Missense, deletion | Missense, splice |
| Sex/age | M/63 | F/21 | M/28 | M/27 | F/19 |
| Onset age | <5 | 4 | 15 | <5 | 1 |
| Inheritance pattern | AR | AR/sporadic | AR/sporadic | AR | AR/sporadic |
| Consanguinity | + | − | − | − | − |
| Initial symptom | Foot deformity | Gait disturbance | Difficulty walking | Poor manual dexterity | Psychomotor delay |
| MMTa | 4 | 0 | 1 | 4 | NA |
| Sensory disturbance | + | + | + | + | + |
| Decreased DTRs | + | + | + | + | + |
| Ataxia (limbs and truncal) | + | + | + | + | + |
| Dysarthria | + | + | + | − | + |
| Nystagmus | − | − | − | − | − |
| Romberg sign | + | + | + | + | − |
| Cognitive impairment | − | + | − | − | + |
| Cerebellar atrophy | + | + | + | + | − |
| Leukoencephalopathy | − | + | − | + | + |
| Spinal cord atrophy | − | NA | + | − | + |
| CK (IU/l)b | 189 | 262 | 577 | 272 | NA |
| Blood lactate (mg/dl)b | 22.1 | 12.1 | 14.0 | 8.2 | elevated |
| Blood pyruvate (mg/dl)b | 1.07 | 0.55 | 1.03 | 0.4 | NA |
| Blood L/P ratio | 20.7 | 22 | 13.6 | 20.5 | NA |
| CSF lactate (mg/dl)b | NA | 20.5 | 27.0 | NA | NA |
| CSF pyruvate (mg/dl)b | NA | 1.0 | 1.25 | NA | NA |
| CSF L/P ratio | NA | 20.5 | 21.6 | NA | NA |
| NCS | |||||
| Median | |||||
| DL | 3.4 | 3.1 | 3.8 | 3.6 | NA |
| MCV | 51.5 | 50.6 | 49.6 | 50.5 | NA |
| dCMAP | 12.5 | 4.0 | 4.7 | 14.0 | NA |
| SNAP | ND | ND | ND | ND | NA |
| Tibial | |||||
| dCMAP | ND | ND | ND | 1.5 | NA |
| Sural | |||||
| SNAP | ND | ND | ND | ND | NA |
aScores indicate manual muscle testing (MMT) grade in the distal lower limbs.
bNormal range: creatine kinase (CK), 12–170 IU/l; blood lactate, 3.0–17.0 mg/dl; blood pyruvate, 0.3–0.94 mg/dl; CSF lactate, 13.7–20.5 mg/dl; CSF pyruvate, 0.63–0.77 mg/dl.
AR = autosomal recessive; CH = compound heterozygous; dCMAP = distal compound muscle action potential (mV); DL = distal latency (ms); DTRs = deep tendon reflexes; L/P = lactate/pyruvate, MCV = motor conduction velocity (m/s); NA = not available (not scored or not examined); NCS = nerve conduction study; ND = not detected (evoked); SNAP = sensory nerve action potential (μV).
Clinical features of four unrelated patients with COA7 mutations
The clinical, genetic, laboratory and electrophysiological findings of patients with COA7 mutations are summarized in Table 1.
Patient 1, a 63-year-old male born to healthy consanguineous parents, had a pes cavus deformity and clumsy hands since his infancy. Drop foot was noticed during his school days and hammer toe was diagnosed by an orthopaedician when the patient was in his 30s. At the age of 55 years, he developed dysarthria along with gradually progressive unsteady gait, for which he was hospitalized at the age of 57 years. The neurological features included all-modality sensory loss, attenuated deep tendon reflexes in the four limbs, dysarthria and mixed sensory and cerebellar ataxia. He was rehospitalized owing to aggravation of symptoms at age 60; treatment with protirelin tartrate (TRH-T) resulted in slight transient improvement in ataxia. However, the symptoms progressively worsened and became refractory.
Patient 2 was a 21-year-old female born to healthy non-consanguineous parents. At 1 month after birth, she had a cardiac murmur, and was diagnosed with a ventricular septum deficiency. Because the foramen was gradually reducing, only periodical follow-up was necessary. She had gait abnormalities (steppage gait) at age 4. She was hospitalized because she had difficulty standing and an intention tremor at age 6. Neurological examination revealed truncal and limb ataxia along with predominant distal muscle atrophy/weakness of the four limbs. She also manifested mild intellectual disability: the Wechsler Preschool and Primary Scale of Intelligence (WPPSI) revealed a verbal score of 76, performance score of 71 and full-scale IQ of 68. She required a wheelchair at age 9. Currently, the muscle weakness is slowly progressive.
Patient 3 had difficulty walking at age 15 and subsequently developed slowly progressive gait disturbance. At age 26, he was admitted to the hospital because of progressive weakness and wasting of distal limb muscles and unsteady gait. Neurological examination revealed truncal ataxia, predominant distal muscle atrophy, weakness of the lower limbs, distal sensory loss and absent ankle reflexes.
Patient 4, a 27-year-old male born to healthy consanguineous parents, had poor manual dexterity and fine motor skills since infancy. He had difficulty in running at age 12 and was diagnosed with CMT at age 15. He was hospitalized at age 22 because of a slowly progressive unsteady gait. Neurological examination revealed a predominant distal muscle atrophy/weakness in all limbs, distal sensory loss, absent ankle reflexes and ataxic gait. He had no obvious cognitive impairment.
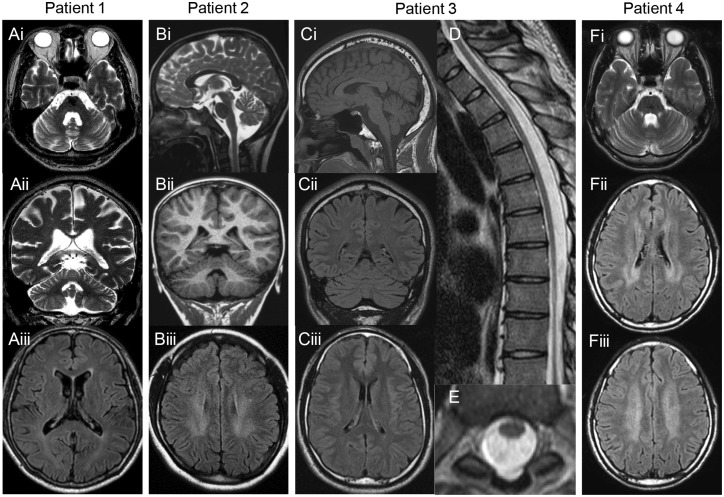
In all four patients, nerve conduction studies showed axonal sensorimotor neuropathy with mild decrease in motor conduction velocity (49.6 to 51.5 m/s) in the median nerve and absence or severe decrease of compound muscle action potential and sensory nerve action potential in lower limb nerves (Table 1). Brain MRI showed mild cerebellar atrophy (Fig. 2) in all four patients; Patient 2 and 4 had mild white matter lesions (Fig. 2F, M and N), while Patient 3 had spinal cord atrophy (Fig. 2J and K). Single-photon emission computed tomography (SPECT) revealed hypoperfusion in the cerebellum in Patients 1 and 3 (data not shown). Laboratory studies showed elevated lactate and pyruvate levels in blood or CSF with mild elevation of serum creatine kinase levels (Table 1).
Figure 2.
MRI findings. Brain or Spinal MRI of (A) Patient 1 at age 57, (B) Patient 2 at age 18, (C–E) Patient 3 at age 26 and (F). Axial and coronal T2-weighted images show mild cerebellar atrophy and enlarged fourth ventricle (Ai, ii and Fi). Sagittal T2-weighted and coronal T1-weighted images show mild cerebellar atrophy (Bi and ii). Sagittal T1-weighted and coronal fluid-attenuated inversion recovery (FLAIR) images show mild cerebellar atrophy (Ci and ii). FLAIR image of Patient 2 (Biii) and Patient 4 (Fii and iii) shows mild bilateral hyperintensity in the periventricular white matter, but not of Patients 1 (Aiii) and 3 (Ciii). Sagittal and axial T2-weighted images of the spine of Patient 3 show spinal cord atrophy with no intrinsic cord signal abnormality (D and E).
Histopathological findings and expression of COA7
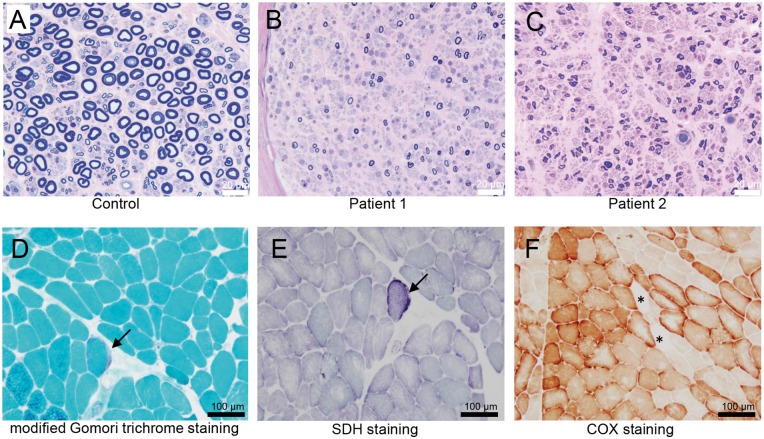
Sural nerve biopsy in Patients 1 and 2 showed marked loss of large and medium myelinated fibres as compared to that in sural nerve from the control patient, while a number of small myelinated fibres with thin myelin sheaths were observed (Fig. 3A–C). Inflammatory cells, onion-bulb formation and clusters of myelinated fibres were absent. These features are compatible with chronic axonal degeneration. Immunohistochemical staining with an anti-COA7 antibody showed positive expression of COA7 in the cytoplasm of Schwann cells of the sural nerve from the control patient (Supplementary Fig. 1), although it was little expressed in the myelin sheath and axon. Unfortunately, immunohistochemical staining of Patients 1 and 2 were not available because only EPON-embedded specimen blocks were stored. Muscle fibres in Patient 1 ranged from 4 to 100 µm in diameter. Some fibres were degenerating or regenerating, while necrotic fibres were not observed. A few ragged-red fibres (per low power field) are observed with the modified Gomori trichrome stain (Fig. 3D). Additionally, there were fibres with increased succinate dehydrogenase (SDH) activity (Fig. 3E) and reduced or absent cytochrome c oxidase (COX) activity (Fig. 3F), which was suggestive of mitochondrial abnormalities.
Figure 3.
Histopathological findings of the peripheral nerve and muscle. (A–C) Toluidine blue staining of a sural nerve. In comparison to a control patient (A), the density of large myelinated fibres is markedly decreased in Patients 1 (B) and 2 (C). (D–F) Histochemical analysis of the biceps brachii muscle from Patient 1. A typical ragged-red fibre (D, arrow) is seen in the section stained with modified Gomori trichrome stain; signs of increased subsarcolemmal enzyme activity (E, arrow) seen on succinate dehydrogenase staining; a few cytochrome c oxidase (COX)-negative fibres (F, asterisks) are seen on COX staining.
Characterization of respiratory chain enzymes in cultured fibroblasts and muscle tissue from patients
Respiratory chain enzyme assay of skin fibroblasts from Patient 1 showed a definite and significant decrease in complex I activity (37% of control), although the other assays, including the activity in muscle tissue, protein expression, and in-gel enzyme staining, revealed comparable results of complex I to control samples (Table 2 and Supplementary Fig. 2). Additionally, BN-PAGE immunoblotting and densitometry analysis in skin fibroblasts showed a slightly decreased amount of complex IV (60.7% remained) in Patient 1 (Supplementary Fig. 2A and Supplementary Table 2). With regard to Patients 2 and 3, MRC enzyme assay revealed a significant decrease in complex IV activity (24% and 9% of control, respectively) [<40% (Bernier et al., 2002)] (Table 2). BN-PAGE immunoblotting and densitometry analysis indicated dramatically reduced levels of fully assembled complex IV of Patients 2 and 3 (3.8% and 9.9% remained, respectively). Further, in-gel enzyme staining of Patients 2 and 3 showed reduced complex IV activity compared to control fibroblasts (Supplementary Fig. 2B and Supplementary Table 2).
Table 2.
Respiratory chain enzyme assay
| Tissue | Enzyme | % of control | Absolute values (mU/U CS) | Control range (mU/U CS) |
|---|---|---|---|---|
| Patient 1 | ||||
| Fibroblast | Co I | 37 | 159 | 257–792 |
| Co II | 146 | 795 | 299–1162 | |
| Co II + III | 66 | 347 | 346–1281 | |
| Co III | 106 | 100 | 22–281 | |
| Co IV | 193 | 37 | 14–64 | |
| Muscle tissue | Co I | 107 | 369 | 279–405 |
| Co II | 98 | 306 | 209–425 | |
| Co II + III | 130 | 388 | 131–488 | |
| Co III | 141 | 243 | 53–296 | |
| Co IV | 47 | 14 | 14–62 | |
| Patient 2 | ||||
| Fibroblast | Co I | 105 | 454 | 257–792 |
| Co II | 113 | 615 | 299–1162 | |
| Co II + III | 108 | 570 | 346–1281 | |
| Co III | 87 | 81 | 22–281 | |
| Co IV | 24 | 4.7 | 14–64 | |
| Patient 3 | ||||
| Fibroblast | Co I | 60 | 260 | 257–792 |
| Co II | 57 | 308 | 299–1162 | |
| Co II + III | 80 | 423 | 346–1281 | |
| Co III | 61 | 58 | 22–281 | |
| Co IV | 9 | 1.7 | 14–64 |
Activities (absolute values) of complex I, II, III, and IV are presented as milliunits per unit (mU/U) citrate synthase (CS). The per cent is calculated as percentage of absolute value in the mean activity value of normal controls. Bold indicates deficiency of the respective complex: <40% (fibroblasts) of citrate synthase ratio. Co I = complex I; Co II = complex II; Co III = complex III; Co IV = complex IV.
Wild-type and mutant COA7 localizes to the mitochondria
To clarify whether the reduction in MRC activities in patient fibroblasts is caused by abnormal subcellular localization of disease associated mutant COA7 or by loss-of-function, we performed immunocytochemical analysis using genes encoding wild-type or two mutant COA7 (Asp6Gly and Arg39Trp) with C-terminal FLAG tag. We generated HeLa cells with stable expression of these genes, and the subcellular localization of each protein was observed. Wild-type COA7 fusion proteins co-localized with Tom20, an outer mitochondrial membrane protein (Supplementary Fig. 3) as previously reported (Kozjak-Pavlovic et al., 2014). Notably, we also found both mutant COA7 proteins localized in mitochondria, which indicated that the disease-related mutation of COA7 did not affect mitochondrial recruitment (Supplementary Fig. 3).
Comparison of the amino acid sequence of human and Drosophila COA7
The amino acid sequence of Drosophila COA7 was retrieved from the Flybase and was compared with that of human COA7 using BLAST and FASTA (Supplementary Fig. 4). The identity and the similarity of the amino acid sequences of dCOA7 and human COA7 are 48% and 70%, respectively. With respect to conservation of specific COA7 domains, SEL1-like domain, which is believed to be involved in protein–protein interactions, is highly conserved between human COA7 and dCOA7 and showed 44% identity. The similarity of the human and Drosophila SEL1-like domain is as high as 78%. High conservation of amino acid sequences between dCOA7 and human COA7 suggests that dCOA7 is truly a Drosophila homologue of human COA7.
Knockdown of dCOA7 in eye imaginal discs induces morphologically aberrant rough eyes
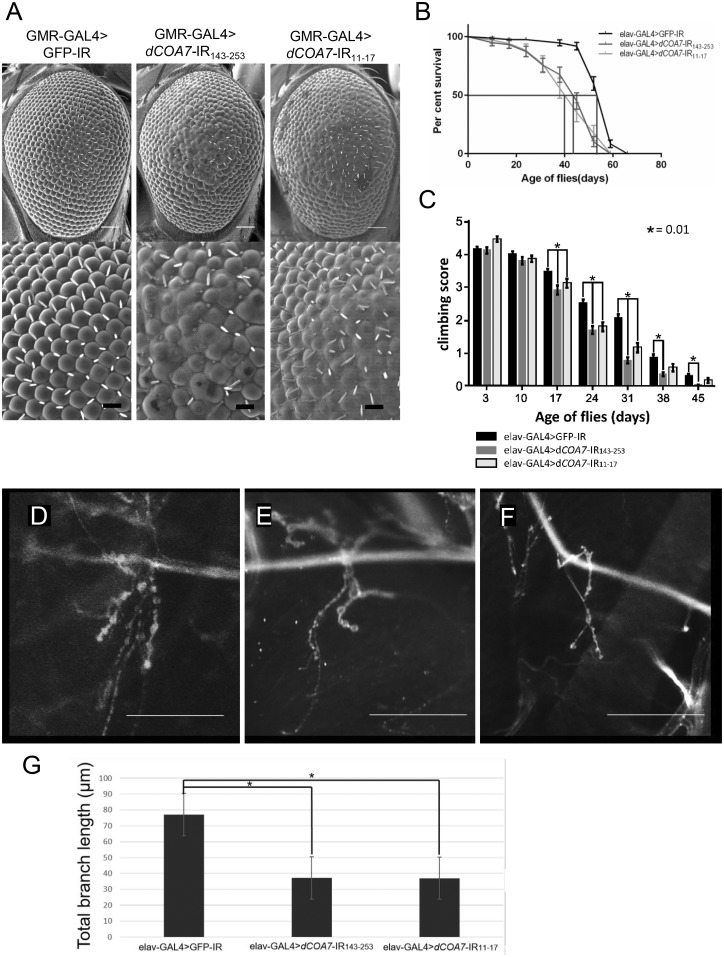
To investigate in vivo functions of dCOA7, we first generated model flies with eye-specific dCOA7-knockdown by the GMR-GAL4 driver lines. Knockdown of dCOA7 in eye imaginal disks by the GMR-GAL4 driver strain induced morphologically aberrant rough eyes. Two different dCOA7 knockdown flies carrying GMR-GAL4>dCOA7-IR143-253 and GMR-GAL4>dCOA7-IR11-17 showed rough eye phenotype, with fusion of ommatidia and lack of bristles, in comparison with control flies carrying GMR-GAL4>GFP-IR (Fig. 4A). Both of the two different RNAi strains targeted to different regions of the dCOA7 exhibited the rough eye phenotype, eliminating the possible off-target effects.
Figure 4.
Knockdown of dCOA7 affected eye phenotype, life span, locomotive ability and synaptic branches of motor neurons. (A) Knockdown of dCOA7 in eye imaginal discs induces rough eye phenotype. Scanning electron micrographs of adult compound eyes: (left) GMR-GAL4>GFP-IR (GMR-GAL4/y; UAS-GFP-IR/+); (middle) GMR-GAL4> dCOA7-IR143-253 (GMR-GAL4/y; UAS-dCOA7-IR143-253/+); (right) GMR-GAL4> dCOA7-IR11-17 (GMR-GAL4/y; UAS- dCOA7-IR11-17/+). Anterior is to the left and dorsal to the top. The white and black bars indicate 50 µm and 14.2 µm, respectively. (B) Life span assay. Percentage survivals of adult male flies carrying elav-GAL4> GFP-IR (w; UAS-GFP-IR/+; elav-GAL4/+, n = 102), elav-GAL4>dCOA7-IR143-253 (w; UAS-dCOA7-IR143-253/+; elav-GAL4/+, n = 113) and elav-GAL4>dCOA7-IR11-17 (w; UAS-dCOA7-IR11-17 /+; elav-GAL4/+, n = 105) are shown with SE bars. The median lifespan for each strain is indicated with red line. The reduction in life span of dCOA7-knockdown flies is statistically significant (P < 0.001). (C) Climbing assays. Five independent tests were performed for each genotype. The total number of flies counted was as follows: elav-GAL4>GFP-IR (n = 125); elav-GAL4>dCOA7-IR143-253 (n = 195); elav-GAL4>dCOA7-IR11-17 (n = 150). Flies carrying elav-GAL4>dCOA7-IR143-253 or elav-GAL4>dCOA7-IR11-17 showed significantly reduced ability to climb up as compared to that of control flies carrying elav-GAL4>GFP-IR. Horizontal bars indicate standard error of mean. *P < 0.01. (D–F) Confocal images of anti-HRP staining of muscle 4 synapse of third instar larvae. A representative image of anti-HRP staining of muscle 4 synapses in third instar larvae with control elav-GAL4>GFP-IR (D), elav-GAL4>dCOA7-IR143-253 (E) and elav-GAL4>dCOA7-IR11-17 (F) are shown. Scale bars = 50 µm. (G) Total branch length of the NMJ from muscle 4 for each of the indicated genotypes. Compared to the total length of synaptic branches of motor neurons in the control elav-GAL4>GFP-IR larvae, those in elav-GAL4>dCOA7-IR143-253 (*P < 0.05, n = 10) and elav-GAL4>dCOA7-IR11-17 larvae (*P < 0.05, n = 10) are significantly decreased.
Neuron-specific dCOA7 knockdown causes short life span and mobility defects in adults
We next investigated whether fly viability was affected by neuron-specific knockdown of dCOA7. Using an elav-GAL4 driver that expresses GAL4 in the nervous system of the fly, we determined the lifespan of flies carrying elav-GAL4>dCOA7-IR143-253, elav-GAL4>dCOA7-IR11-17 and the control elav-GAL4>GFP-IR (Fig. 4B). We examined adult flies until 120 days after eclosion. The median lifespan of the control flies (n = 102) was 51 days, whereas flies carrying elav-GAL4>dCOA7-IR143-253 (n = 113) or elav-GAL4>dCOA7-IR11-17 (n = 105) lived an average of 43 days and 40 days, respectively. The differences between control flies and two independent knockdown flies were 8 days and 11 days, respectively. The reduction in lifespan was statistically significant (P < 0.001). Lifespans of neuron-specific dCOA7-knockdown flies were thus significantly shorter than those of the control flies.
To evaluate the effects of neuron-specific dCOA7-knockdown on the locomotive function at different ages further, we then performed climbing assays of the dCOA7-knockdown adult flies (Fig. 4C). The flies carrying elav-GAL4>dCOA7-IR143-253 exhibited a significantly decreased climbing ability on Day 17 (by 16%), Day 24 (33%), Day 31 (63%), Day 38 (59%) and Day 45 (94%) compared with the control flies carrying elav-GAL4>GFP-IR. More prominent reductions in mobility were observed in aged flies. Similarly flies carrying elav-GAL4>dCOA7-IR11-17 showed a decreased climbing ability on Day 17 (by 10%), Day 24 (29%) and Day 31 (44%). These results indicate that dCOA7 is required for locomotion.
dCOA7 regulates the formation of motor neurons at presynaptic terminals at the neuromuscular junction
The mobility deficit observed in dCOA7-knockdown flies prompted us to analyse the morphology of motor neuron presynaptic terminals at the NMJ of these flies. Because most motor neurons of the adult fly originate from larval motor neurons, we compared the NMJ structure in muscle 4 of the larvae of elav-GAL4>dCOA7-IR143-253 and elav-GAL4>dCOA7-IR11-17 flies with that of control larvae carrying elav-GAL4>GFP-IR (Fig. 4D–F). Measurement of the total length of synaptic branches of motor neurons in these larvae indicated that the total branch length was significantly decreased in elav-GAL4>dCOA7-IR143-253 (38.2 ± 4.5 μm, P < 0.05) and elav-GAL4>dCOA7-IR11-17 (37.0 ± 6.0 μm, P < 0.05) flies compared to that of the control elav-GAL4>GFP-IR (77.0 ± 27.0 μm) flies (Fig. 4G). These results suggest that dCOA7 regulates the formation and/or maintenance of motor neurons at presynaptic terminals at the NMJ.
Discussion
In this study, we focused on gene variants shared among various unrelated patients with an overlap-based strategy and independently identified recessive mutations in COA7 in four unrelated patients, characterized by an axonal-type motor and sensory neuropathy with ataxia. We also provided genetic, histopathological, radiological and functional data to support the findings that COA7 gene is a novel causative gene for the phenotype of the presented patients.
Genetically, all four patients have different genotypes: homozygous (p.Asp6Gly) and compound heterozygous mutations (exon 2 deletion and p.Arg39Trp; p.Asp6Gly and p.Ser149Ile; p. Asp6Gly and p.Gly144fs). These mutations were segregated with the disease in each family and were predicted to be damaging or deleterious during in silico analysis; moreover, these were novel or extremely rare variants in public databases. Aspartic acid 6 (Asp6), arginine 39 (Arg39), glycine 144 (Gly144) and serine 149 (Ser149) in COA7 were highly conserved among species. Arg39 and Ser149 are hydrophilic amino acids located in the SEL1-like repeats (Fig. 1D), which are similar to tetratricopeptide repeats involved in protein–protein interactions (Blatch and Lassle, 1999). We predicted the 3D homology models of COA7 using SWISS-MODEL server (https://swissmodel.expasy.org/). Arg39 was exposed on the outer surface of the protein molecule (Supplementary Fig. 5), which suggested that Arg39Trp mutant may lead to abnormal assembly of the MRC complexes induced by an increase in hydrophobic interactions between the COA7 and its binding partners. These results indicated that the mutations could be loss-of-function mutations and affect the mitochondrial function.
Clinically, all four patients had similar electrophysiological, pathological and clinical findings characterized by an axonal-type motor and sensory neuropathy with ataxia. They showed predominant distal muscle atrophy/weakness of the lower limbs, distal sensory loss and decreased ankle reflexes. Moreover, neurological examination revealed gait ataxia and balance impairment with wide based gait, loss of vibration sense, positive Romberg’s sign and dysarthria (slurred speech) without nystagmus and oculomotor apraxia. These findings suggest that ataxia symptoms were attributable to mixed cerebellar and sensory ataxia with concomitant signs of the two basic types of ataxia. The clinical history of the patients suggests that peripheral neuropathy occurs early in the disease course, whereas cerebellar ataxia tends to develop late and lead to exacerbation of gait disturbance. The preceding neuropathy could be attributed to vulnerability to mitochondrial dysfunction of sensory and motor neurons in the peripheral nervous system, which have long axons and require large amounts of energy to maintain their own homeostasis. Cerebellar atrophy was observed on MRI in all four patients and cerebellar hypoperfusion was seen in two of the patients who underwent SPECT. Intriguingly, a recent study found that a 19-year-old female with compound heterozygous mutations in COA7 had early onset, progressive neurological impairments characterized by severe ataxia, peripheral neuropathy and mild cognitive impairment with leukoencephalopathy and spinal cord atrophy on MRI (Martinez Lyons et al., 2016). As shown in Table 1, the phenotypic characteristics of this case are similar to those in our patients, although some variations regarding the onset and severity of disease and the involvement of the CNS were observed. We confirmed expression of COA7 in the peripheral nervous system, particularly in the cytoplasm of Schwann cells (Supplementary Fig. 1). Additionally, the analysis of protein expression levels in the CNS from the Human Protein Atlas (available online: http://www.proteinatlas.org) showed that COA7 was highly expressed in the cerebellum. Therefore, the peripheral nervous system and the cerebellum are considered to be the main sites of lesions responsible for the neurological symptoms caused by COA7 dysfunctions (Fig. 5). This phenotype is clinically similar to that of SCAN1 (spinocerebellar ataxia with axonal neuropathy type 1, OMIM #607250; Takashima et al., 2002) and ataxia-ocular apraxia-2 (also called SCAN2, OMIM #606002; Moreira et al., 2004; Duquette et al., 2005), which are caused by TDP1 and SETX mutations, respectively. COA7 expression data obtained from the Human Protein Atlas showed that COA7 was expressed not only in the cerebellum but also in the cerebral cortex, basal ganglia and hippocampus. In addition, it is possible that patients with COA7 mutation have a subclinical myopathy as we observed the presence of a few ragged-red fibres and some COX-negative fibres on muscle biopsy as well as mildly elevated serum creatine kinase levels. Therefore, mutations in COA7 may also lead to heterogeneous neuromuscular involvement, manifesting phenotypic diversity in the future.
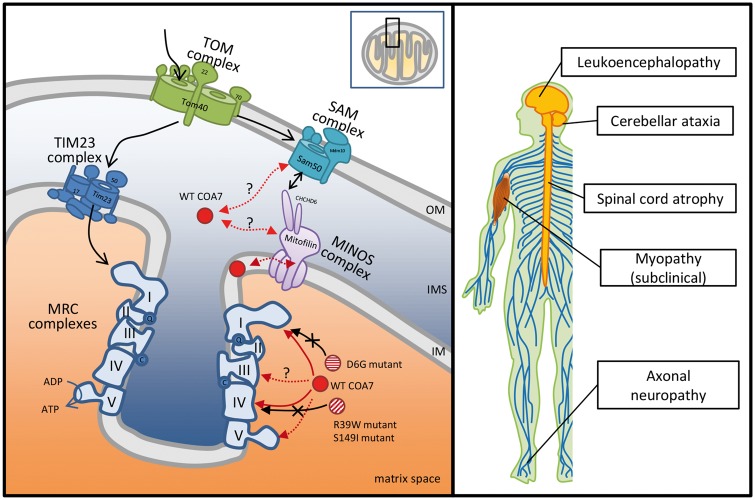
Figure 5.
Clinical spectrum of COA7 mutations and potential model for COA7 in mitochondria. Mitochondria consist of two membranes: the outer membrane (OM) and the inner membrane (IM). The intermembrane space (IMS) refers to the region between the outer and inner membrane. Wild-type (WT) COA7 has been reported to localize in mitochondrial intermembrane space in HeLa cells by swelling experiments combined with proteinase K treatment (Kozjak-Pavlovic et al., 2014). It is also suggested that COA7 may transiently associate with the inner membrane during its function because of the fact that a portion of the COA7 that remains in mitochondria after rupture of the outer membrane was protected by membrane lipids. However, Martinez-Lyons et al. reported that COA7 is localized in the mitochondrial inner compartment, mostly in the matrix, rather than in the intermembrane space or inner membrane based on their mitochondrial localization study with trypsin treatment in HEK293T cells (Martinez Lyons et al., 2016). With regard to its functions, Kozjak-Pavlovic et al. (2014) showed that (i) COA7 levels were affected by Sam50 and mitofilin, which is a subunit of the sorting and assembly machinery (SAM) complex in the outer membrane and a central component of the mitochondrial inner membrane organizing system (MINOS) complex, respectively; (ii) knockdown of COA7 lead to a reduction of some components of the mitochondrial respiratory chain (MRC) complexes; (iii) COA7 depletion affected the assembly of subunits of MRC complexes I (NDUFS1), III (Core I), IV (COX6a) and V (F1α). Especially, the assembly of the COX6a had the strongest effect; and (iv) COA7 depletion affected steady-state levels and activity of several MRC complexes, especially complex I and IV. Our results of the MRC enzyme assay and immunocytochemical study suggested that Asp6Gly (D6G) mutants might affect the MRC complex I, and Arg39Trp (R39W) and that Ser149Ile (S149I) mutants might affect the MRC complex IV.
Correspondingly, we observed varied MRC activity among three patients and the previous patient reported by Martinez Lyons et al. (2016). The respiratory chain enzyme assay of skin fibroblasts showed isolated deficiency of complex IV in Patients 2 and 3, in spite of the low activity of complex I in Patient 1. Fibroblasts of Patients 2 and 3 also showed decreased levels of fully assembled complex IV, suggesting an assembly defect as the primary cause of the decreased complex IV activity. On the other hand, skin fibroblasts in Patient 1 showed decreased activity of complex I in spite of normal levels of fully assembled complex I, which suggested that Asp6Gly mutation is likely to affect an active region of complex I rather than an assembly area. Additionally, also in Patient 1, a decreasing trend of complex IV activity in muscle tissue, COX-negative fibres, and slightly decreased amount of complex IV in fibroblasts were observed. These findings indicated that Asp6Gly mutation in COA7 may also lead to an assembly defect of complex IV. The molecular mechanism of decreased activity of complex I in Patient 1 is unclear, but a similar finding has been reported in a patient with mutations in the SURF1 gene, encoding the SURF1 protein, which is essential for COX assembly (Tay et al., 2005). Additionally, a previous study indicated that the depletion of COA7 could lead to an obvious reduction of activity not only in complex IV but also in complex I (Kozjak-Pavlovic et al., 2014). The variable effects on MRC activity and the clinical features may have been caused by a different interaction between COA7 and MRC complexes, possibly because of the difference in the site of mutation in the COA7 gene. Moreover, these findings suggest that COA7 may be involved in the assembly of MRC complex I in addition to that of complex IV. Further researches, such as immunoblot analysis to identify interaction partners of COA7 and complementation assay with wild-type COA7, are required to clarify the varied defects of MRC complexes caused by COA7 mutations.
On the other hand, we demonstrated localization of wild-type COA7 proteins in mitochondria in HeLa cells, which is consistent with previous reports (Kozjak-Pavlovic et al., 2014; Martinez Lyons et al., 2016). We also indicated that Asp6Gly and Arg39Trp COA7 mutants localize to the mitochondria, which suggests that the mutant COA7 does not affect the mitochondrial recruitment but rather reduces the activity of several MRC complexes by virtue of its effect on the stability or localization of COA7 interaction partners in the mitochondria. In a study by Kozjak-Pavlovic et al. (2014) COA7 depletion was shown to affect the assembly of some subunits of MRC complexes and especially that of COX6a into complex IV, which is a causative gene of autosomal recessive intermediate CMT D (OMIM #616039; Tamiya et al., 2014). Therefore, the degeneration of peripheral nerves in our patient may be explained by the reduction in the assembly of COX6a due to the loss-of-function of COA7. Further studies are needed to clarify the role of COA7 and the molecular pathomechanism that underlies the development of neurodegeneration. We showed a potential model for the interaction among proteins or complexes implicated with wild-type and mutant COA7 in mitochondria in Fig. 5.
In this study, we demonstrated that neuron-specific knockdown of dCOA7 in the Drosophila model decreased the life span, impaired locomotor function and shortened the synaptic branches of the motor neurons. These phenotypes were observed in two independent dCOA7 knockdown flies targeting different regions of dCOA7, excluding the possible off-target effects. However, because the observed defect in the NMJ may simply reflect a part of motor neuron degeneration, more detailed analyses on whole motor neurons would be necessary. A previous study indicated that mechanical motor axonal injury in Drosophila leads to degeneration at the NMJ (Lincoln et al., 2015). Additionally, it has been reported that NMJ morphological defects were also observed in the Drosophila model with neuron-specific knockdown of mitochondria related genes other than dCOA7, such as Marf [human Mitofusin 2 (MFN2) homologue responsible for CMT2A] (Sandoval et al., 2014) and DVAP-33A [human vesicle associated membrane protein (VAPB) homologue responsible for ALS8] (Chai et al., 2008). Therefore, we believe that the NMJ involvement in neuron-specific knockdown of dCOA7 in the Drosophila model reflects a secondary effect caused by a primary axonal change in the peripheral nervous system. These results suggest that loss-of-function of dCOA7 is sufficient to cause neurological impairment and dCOA7 plays a role in neurite outgrowth and/or its maintenance. The defects can be explained by a possible deficiency in MRC-mediated activities in the dCOA7-knockdown flies. The neuron-specific dCOA7-knockdown flies established in this study could be a useful model to search novel therapeutic target and candidate substances for therapy.
In conclusion, we identified recessive mutations in COA7, and showed that loss-of-function mutations in COA7 result impair MRC activities and could cause early onset, progressive axonal neuropathy and ataxia with or without leukoencephalopathy or spinal cord atrophy. We suggest that this new entity of SCAN be referred to as spinocerebellar ataxia with axonal neuropathy type 3 (SCAN3). Moreover, our studies on dCOA7 knockdown Drosophila models elucidated that disruption of the physiological functions of dCOA7 is critical to the development of neurodegeneration, which supports the contention that recessive mutations in COA7 are responsible for neurological impairment.
Supplementary Material
Acknowledgements
The authors thank the patients and their families for participating in this study and their physicians for submitting the clinical samples. Also the authors thank Mrs. Aya Ebina, Yukari Niwata of our department and Hiroko Harashima of Saitama Medical University for their excellent technical assistant, and Enago (www.enago.jp) for the English language review.
Funding
This study was supported, in part, by grants from the research on the Nervous and Mental Disorders and Research Committee for Charcot–Marie–Tooth Disease, Neuropathy, Ataxic Disease and Applying Health and Technology of Ministry of Health, Welfare and Labour, Japan. This research is supported by the research program for conquering intractable disease from Japan Agency for Medical Research and Development (AMED) and JSPS KAKENHI Grant Number 26461275, 22129001 and 22129002. This work was also supported in part by the Practical Research Project for Rare/Intractable Diseases from AMED to K.M. and Y.O. (http://www.amed.go.jp/en/) and the Project Promoting Clinical Trials for Development of New Drugs and Medical Devices, Japan Medical Association, from AMED to A.O.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- CMT
Charcot–Marie–Tooth disease
- MLPA
multiplex ligation-dependent probe amplification
- MRC
mitochondrial respiratory chain
- NMJ
neuromuscular junction
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology 2002; 59: 1406–11. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 1999; 21: 932–9. [DOI] [PubMed] [Google Scholar]
- Chai A, Withers J, Koh YH, Parry K, Bao H, Zhang B, et al. hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum Mol Genet 2008; 17: 266–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012; 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbeni-Sala F, Di Santo S, Franceschini D, Skaper SD, Giusti P. Melatonin protects against 6-OHDA-induced neurotoxicity in rats: a role for mitochondrial complex I activity. FASEB J 2001; 15: 164–70. [DOI] [PubMed] [Google Scholar]
- Dubowitz V, Sewry CA, Oldfors A, Lane R. Muscle biopsy: a practical approach. 2nd edn London: Bäilliere Tindall Co; 1985. [Google Scholar]
- Duquette A, Roddier K, McNabb-Baltar J, Gosselin I, St-Denis A, Dicaire MJ, et al. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann Neurol 2005; 57: 408–14. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Ghezzi D, Chassagne M, Mayencon M, Padet S, Melchionda L, et al. SURF1 deficiency causes demyelinating Charcot-Marie-Tooth disease. Neurology 2013; 81: 1523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett 2009; 583: 1586–92. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ueda A, Wu CF. A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J Neurogenet 2004; 18: 377–402. [DOI] [PubMed] [Google Scholar]
- Hara T, Hashimoto Y, Akuzawa T, Hirai R, Kobayashi H, Sato K. Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease. Sci Rep 2014; 4: 6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi I, Niiyama T, Uchida Y, Inose M, Nakagawa M, Arimura K, et al. Multiple episodes of thrombosis in a patient with Becker muscular dystrophy with marked expression of utrophin on the muscle cell membrane. Acta Neuropathol 1999; 98: 313–16. [DOI] [PubMed] [Google Scholar]
- Higuchi Y, Hashiguchi A, Yuan J, Yoshimura A, Mitsui J, Ishiura H, et al. Mutations in MME cause an autosomal-recessive Charcot-Marie-Tooth disease type 2. Ann Neurol 2016; 79: 659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Boneh A, Chow CW, Ohtake A, Ryan MT, Thyagarajan D, et al. Low mutant load of mitochondrial DNA G13513A mutation can cause Leigh's disease. Ann Neurol 2003; 54: 473–8. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 1999; 52: 1255–64. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Salemi R, Sugiana C, Ohtake A, Parry L, Bell KM, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest 2004; 114: 837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishita Y, Pajak A, Bolar NA, Marobbio CM, Maffezzini C, Miniero DV, et al. Intra-mitochondrial methylation deficiency due to mutations in SLC25A26. Am J Hum Genet 2015; 97: 761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 2003; 31: 1007–14. [PubMed] [Google Scholar]
- Kozjak-Pavlovic V, Prell F, Thiede B, Gotz M, Wosiek D, Ott C, et al. C1orf163/RESA1 is a novel mitochondrial intermembrane space protein connected to respiratory chain assembly. J Mol Biol 2014; 426: 908–20. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–81. [DOI] [PubMed] [Google Scholar]
- Kyotani A, Azuma Y, Yamamoto I, Yoshida H, Mizuta I, Mizuno T, et al. Knockdown of the Drosophila FIG4 induces deficient locomotive behavior, shortening of motor neuron, axonal targeting aberration, reduction of life span and defects in eye development. Exp Neurol 2016; 277: 86–95. [DOI] [PubMed] [Google Scholar]
- Lincoln BL II, Alabsi SH, Frendo N, Freund R, Keller LC. Drosophila neuronal injury follows a temporal sequence of cellular events leading to degeneration at the neuromuscular junction. J Exp Neurosci 2015; 9 (Suppl 2): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Lyons A, Ardissone A, Reyes A, Robinson AJ, Moroni I, Ghezzi D, et al. COA7 (C1orf163/RESA1) mutations associated with mitochondrial leukoencephalopathy and cytochrome c oxidase deficiency. J Med Genet 2016; 53: 846–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 2004; 36: 225–7. [DOI] [PubMed] [Google Scholar]
- Ostergaard E, Weraarpachai W, Ravn K, Born AP, Jonson L, Duno M, et al. Mutations in COA3 cause isolated complex IV deficiency associated with neuropathy, exercise intolerance, obesity, and short stature. J Med Genet 2015; 52: 203–7. [DOI] [PubMed] [Google Scholar]
- Pitceathly RD, Murphy SM, Cottenie E, Chalasani A, Sweeney MG, Woodward C, et al. Genetic dysfunction of MT-ATP6 causes axonal Charcot-Marie-Tooth disease. Neurology 2012; 79: 1145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Blok RB, Dahl HH, Danks DM, Kirby DM, Chow CW, et al. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann Neurol 1996; 39: 343–51. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Nakano H, Yamamoto N, Yamaoka S. Lymphotoxin-beta receptor mediates NEMO-independent NF-kappaB activation. FEBS Lett 2002; 532: 45–51. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Yao CK, Chen K, Jaiswal M, Donti T, Lin YQ, et al. Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. Elife 2014; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Wanschers BF, Nijtmans LG, Rodenburg RJ, Zschocke J, Dikow N, et al. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum Mol Genet 2013; 22: 656–67. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Hirose F, Matsukage A, Yamaguchi M. Identification of three conserved regions in the DREF transcription factors from Drosophila melanogaster and Drosophila virilis. Nucleic Acids Res 1999; 27: 510–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet 2002; 32: 267–72. [DOI] [PubMed] [Google Scholar]
- Tamiya G, Makino S, Hayashi M, Abe A, Numakura C, Ueki M, et al. A Mutation of COX6A1 causes a recessive axonal or mixed form of Charcot-Marie-tooth disease. Am J Hum Genet 2014; 95: 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SK, Sacconi S, Akman HO, Morales JF, Morales A, De Vivo DC, et al. Unusual clinical presentations in four cases of Leigh disease, cytochrome C oxidase deficiency, and SURF1 gene mutations. J Child Neurol 2005; 20: 670–4. [DOI] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, et al. A mutation in the human heme A: farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum Mol Genet 2000; 9: 1245–9. [DOI] [PubMed] [Google Scholar]
- Van Coster R, Smet J, George E, De Meirleir L, Seneca S, Van Hove J, et al. Blue native polyacrylamide gel electrophoresis: a powerful tool in diagnosis of oxidative phosphorylation defects. Pediatr Res 2001; 50: 658–65. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Hashiguchi A, Hu J, Sakiyama Y, Okamoto Y, Tokunaga S, et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. Neurology 2012; 78: 1644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi J, Hatchwell E. Human MLPA Probe Design (H-MAPD): a probe design tool for both electrophoresis-based and bead-coupled human multiplex ligation-dependent probe amplification assays. BMC Genomics 2008; 9: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.