Abstract
Motivation
With the increasing amount of genomic and epigenomic data in the public domain, a pressing challenge is to integrate these data to investigate the role of epigenetic mechanisms in regulating gene expression and maintenance of cell-identity. To this end, we have implemented a computational pipeline to systematically study epigenetic variability and uncover regulatory DNA sequences.
Results
Haystack is a bioinformatics pipeline to identify hotspots of epigenetic variability across different cell-types, cell-type specific cis-regulatory elements, and associated transcription factors. Haystack is generally applicable to any epigenetic mark and provides an important tool to investigate the mechanisms underlying epigenetic switches during development. This software is accompanied by a set of precomputed tracks, which may be used as a valuable resource for functional annotation of the human genome.
Availability and implementation
The Haystack pipeline is implemented as an open-source, multiplatform, Python package called haystack_bio freely available at https://github.com/pinellolab/haystack_bio.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Epigenetic patterns are highly cell-type specific, and influence gene expression programs (Jenuwein and Allis, 2001). Recently, a large amount of epigenomic data across many cell types has been generated and deposited in the public domain, in part thanks to large consortia such as Roadmap Epigenomics Project (Bernstein et al., 2010), and ENCODE (Dunham et al., 2012). These data sources offer unprecedented opportunities for systematic integration and comparison. In an earlier work (Pinello et al., 2014), we developed and validated a computational strategy to systematically evaluate cross-cell-type epigenetic variability and to identify the underlying regulatory factors of such variability. Here we provide an implementation of this strategy that automatically integrates multiple data types in an easy-to-use command line software. Our goal is to facilitate biologists’ efforts at analyzing epigenetic data without the burden of coding, and to enable researchers to integrate their own sequencing data with information from the public domain.
2 Description
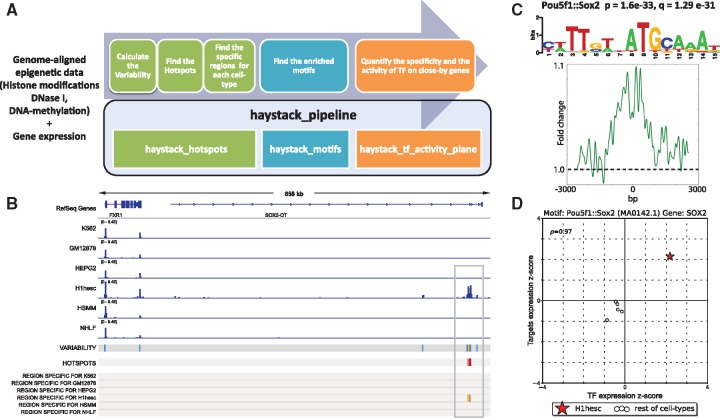
Haystack takes as input the genome-wide distributions of an epigenetic mark across multiple cell types or subjects—measured by ChIP-seq, DNase-seq, ATAC-seq or similar assays—as well as gene expression profiles quantified by microarray or RNA-seq. Users can start with publicly available preprocessed data or integrate their own data in the pipeline by providing BAM or bigWig files that can be generated by existing tools such as the ENCODE Uniform Processing Pipelines. Haystack’s entire computational pipeline can be executed with a single command (i.e. haystack_pipeline). The pipeline is composed of three modules: haystack_hotspots, haystack_motifs, and haystack_tf_activity_plane. Each module is designed to carry out a distinct but related task (Fig. 1A), as described below.
Fig. 1.
(A) Haystack overview: modules and corresponding functions. (B) Hotspot analysis on H3k27ac: signal tracks, variability track and hotspots of variability are computed from the ChIP-seq aligned data; the regions specific for a given cell type are also extracted. (C) Motif analysis on the regions specific for the H1hesc cell line: Pou5f1:: Sox2 is significant; P- and q-value, motif logo and average profile are calculated. (D) TF activity for Sox2 in H1esc (star) compared to the other cell types (circles), x-axis specificity of Sox2 expression (z-score), y-axis effect (z-score) on the gene nearby the regions containing the Sox2 motif
2.1 Module 1. Discovery of hotspots and cell-type specific regions
haystack_hotspots identifies the hotspots of epigenetic variability, i.e. those regions that are highly variable for a given epigenetic mark among different cell types. The algorithm for identifying the hotspots was described previously in Pinello et al. (2014). Briefly, the input for the pipeline is a set of genome-aligned sequencing tracks for a given epigenetic mark in different cell types, in BAM or bigWig format. The haystack_hotspots module first quantifies the sequence reads to non-overlapping bins of predetermined size (500 bp by default), and normalizes data using a variance stabilization method followed by quantile normalization. It then quantifies the variability of the processed data signal in each bin using the variance-to-mean ratio. The most variable regions, accordingly to this measure, are selected as hotspots (originally termed as Highly Plastic Regions in Pinello et al. [2014]). The subsets of hotspot regions that have specific activity in a particular cell type are next identified, based on a z-score metric. Finally, an IGV (http://www.broadinstitute.org/igv/) XML session file is created to enable easy visualization of the results (Fig. 1B, Supplementary Fig. S1).
2.2 Module 2. Analysis of transcription factor motif
haystack_motifs identifies transcription factors (TFs) whose binding sequence motifs are enriched in a cell-type specific subset of hotspots. This module takes the output of haystack_hotspots as its input. Alternatively, the input may be a generic set of genomics regions; e.g. promoters for a set of genes of interest or cell-type specific enhancers. A motif database can also be specified (JASPAR [Mathelier et al., 2016] by default) to look for motif enrichment (the basic counting of each motif is based on the FIMO software; Grant et al., 2011), with use of random or C + G content matched genomic sequences as background. We find that the latter option is more appropriate for histone modifications. The output of this module consists of an HTML page (Supplementary Fig. S2) that reports each enriched motif, a series of informative parameters including the target/background ratio, the P-value (calculated with the Fisher’s exact test) and q-value, the motif logo, the central enrichment score, the average profile in the target regions containing the motif, and the closest genes for each region (Fig. 1C, Supplementary Fig. S2).
2.3 Module 3. Integration of gene expression data
Because different TFs may share similar sequence binding patterns, the exact regulator cannot be determined by motif enrichment analysis alone. Spurious association may also occur due to, e.g. over-abundance of motif sequences. haystack_tf_activity_plane provides an additional filter to select for the most relevant TFs by further integrating gene expression data; it is based on the assumption that the expression level of a functional TF is correlated with the expression level of the target genes of hotspot regions. Such a relationship is visualized with the use of an activity plane representation (Fig. 1D). A detailed description of the tf activity plane plot and how it is generated is provided in Supplementary Material Section 3. Briefly, for each cell type, an activity plane plot (Supplementary Fig. S3) is generated for each enriched motif identified in that cell type by the haystack_motifs. In this representation, the cell-type of interest is marked with a red star. The furthest is the star from the origin the more cell-type specific is either the expression of the TF (x-axis) or its effect on nearby genes (y-axis). This allows us to capture how informative (as measured by the gene expression level of the TF) a particular TF is for a given cell type compared with other cell types. However, not all possible plots are generated by default; Only those passing the following filters are reported: (i) the activity of the TF recapitulates changes in gene expression such that the value of the correlation of the TF with nearby genes exceeds a given threshold (default rho = 0.3) and (ii) the average gene expression is greater in the considered cell type such that the standardized gene expression values are positive (i.e. default z-score > 0). Earlier we showed that these filters are important for identifying factors that truly play a key role in mediating poised enhancer activities (Pinello et al., 2014).
3 Related methods
Several epigenomics software packages already exist that share Haystack’s goals of identifying functional regulatory sequences or regulators involved in gene regulation. The main contribution of Haystack can be summarized by the following three general aspects of the pipeline: (i) Haystack takes as input not just epigenomic data, but also genomic and transcriptomics data. The majority of available epigenomic tools are designed to work with one or two types of data. DeepChrome instead (Singh et al., 2016) is an example of an integrative deep learning method that takes in histone modification signal as input and gene expression as output to be predicted. However, DNA sequence is not incorporated, and the histone signal is constrained to a small window around the transcription start site. (ii) Haystack takes as input epigenomic data for a single epigenetic mark across multiple cell types and generates cell-type specific hotspot annotation tracks. In contrast, chromatin state annotation methods such as ChromHMM (Ernst and Kellis, 2012), Segway (Hoffman et al., 2012), diHMM (Marco et al., 2017) and Spectacle (Song and Chen, 2015) take as input epigenomic data for a single cell type across multiple epigenetic marks and annotate genomic regions into discrete chromatin states (e.g. enhancers, promoters) based on the patterns of marks in a single cell type. These generated annotated regions are not necessarily variable across cell types. (iii) By computing cell-type specific enriched motifs using a central enrichment filter and incorporating gene expression data, Haystack generates a list of cell-type specific TFs. In contrast, Homer (Heinz et al., 2010) can find enriched or de novo motifs from a set of sequences but cannot perform central enrichment filtering and DREME (Bailey, 2011) can be used only for de novo motif discovery but cannot calculate enrichment of known motifs. Neither method incorporates gene expression data. A detailed comparison of related methods is presented in Supplementary Table S1.
4 Results
4.1 Analysis of H3K27ac data
To demonstrate Haystack’s utility, we analyzed 6 ChIP-seq datasets from the ENCODE project (Dunham et al., 2012) for the histone modification H3K27ac (Fig. 1B). H3K27ac often marks active enhancers that promote the expression of nearby genes. We also integrated six RNA-seq assays, to quantify gene expression for the same cell types. Figure 1 shows the output of the pipeline: Haystack not only recovers regions that are highly dynamic (variability and hotspots tracks in Fig. 1), but also regions that are specifically active in each cell type. Additionally, Haystack detects several TFs that are likely to play an important regulatory role in those regions (Supplementary Fig. S3). For example, for regions that are specifically active in the embryonic stem cell line (H1hesc), we found that the Pou5f1:: Sox2 composed motif was highly enriched, and the expression of Sox2—a fundamental TF for embryonic stem cell identity—was highly specific and positively correlated with activity of the target genes.
4.2 Analysis of roadmap epigenomics project
We applied the Haystack pipeline to data from the Roadmap Epigenomics Project using the maximal number of non-redundant cell-types for which gene expression and epigenetic data was available (Supplementary Material Section 4). We provide precomputed analysis for H3k27ac (41 cell types), H3K27me3 (41 cell types), H3K4me3 (41 cell types) and DNase I hypersensitivity (25 cell types). These precomputed tracks provide a valuable resource for researchers interested in identifying functional elements in the human genome, exploring how epigenetic variability is controlled in different cell types, and uncovering regulatory sequences.
4.3 Reproducible results through Cloud and Docker support
To facilitate the use of Haystack without the need to access an intensive computational facility, we provide detailed instructions in the Supplementary Material on how to deploy and test Haystack on the Amazon Web Services cloud or similar services. We also provide a Docker image to make our tool more user-friendly and reproducible (see Supplementary Material Section 5).
5 Usage
The entire pipeline can be executed simply by running a single command. By default, the users need only to create a single description file that contains information about the data file paths (e.g. samples_names.txt) and the reference genome used in the analysis (e.g. hg19):
haystack_pipeline samples_names.txt hg19
If the specified genome information and annotations are not available locally, they will be automatically downloaded from the internet. The haystack_pipeline command is equivalent to running haystack_hotspots followed by haystack_motifs and haystack_tf_activity_plane. A detailed description of the settings is provided in Supplementary Material Section 9. To illustrate the Haystack workflow, we also provide a walk-through example (see Supplementary Material Section 3) that reproduces the results described in Section 4.
Supplementary Material
Acknowledgements
We would like to thank Dr Stuart Orkin, Dr Jian Xu, Dr Nadin Rohland, Dr Kimberly Glass, Dr Eugenio Marco Rubio, Dr Jialiang Huang, Dr Assieh Saadatpour, Dr Jennifer Wu and Dr Kendell Clement for their helpful discussions and/or for testing the software.
Funding
This work was supported by National Institutes of Health award [R00HG008399 to L.P. and R01HG009663 to G.-C.Y.].
Conflict of Interest: none declared.
References
- Bailey T.L. (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics, 27, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E. et al. (2010) The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol., 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I. et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M. (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E. et al. (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics, 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.M. et al. (2012) Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods, 9, 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Mathelier A. et al. (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res., 44, D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E. et al. (2017) Multi-scale chromatin state annotation using a hierarchical hidden Markov model. Nat. Commun., 8, 15011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L. et al. (2014) Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc. Natl. Acad. Sci. USA, 111, E344–E353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. et al. (2016) DeepChrome: deep-learning for predicting gene expression from histone modifications. Bioinformatics, 32, i639–i648. [DOI] [PubMed] [Google Scholar]
- Song J., Chen K.C. (2015) Spectacle: fast chromatin state annotation using spectral learning. Genome Biol., 16, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.