Comparison of day-neutral temperate maize and photoperiod-dependent teosinte revealed divergent leaf gene expression networks that implicate different genes of the same gene families in response to flowering.
Keywords: Autonomous flowering, carbon sensing, circadian clock, floral induction, florigen, gene networks, maize migration, photoperiod
Abstract
Temperate maize was domesticated from its tropical ancestor, teosinte. Whereas temperate maize is an autonomous day-neutral plant, teosinte is an obligate short-day plant that requires uninterrupted long nights to induce flowering. Leaf-derived florigenic signals trigger reproductive growth in both teosinte and temperate maize. To study the genetic mechanisms underlying floral inductive pathways in maize and teosinte, mRNA and small RNA genome-wide expression analyses were conducted on leaf tissue from plants that were induced or not induced to flower. Transcriptome profiles reveal common differentially expressed genes during floral induction, but a comparison of candidate flowering time genes indicates that photoperiod and autonomous pathways act independently. Expression differences in teosinte are consistent with the current paradigm for photoperiod-induced flowering, where changes in circadian clock output trigger florigen production. Conversely, differentially expressed genes in temperate maize link carbon partitioning and flowering, but also show altered expression of circadian clock genes that are distinct from those altered upon photoperiodic induction in teosinte. Altered miRNA399 levels in both teosinte and maize suggest a novel common connection between flowering and phosphorus perception. These findings provide insights into the molecular mechanisms underlying a strengthened autonomous pathway that enabled maize growth throughout temperate regions.
Introduction
Temperate maize (Zea mays ssp. mays) is derived from its wild progenitor, teosinte (Zea mays ssp. parviglumis) (Doebley, 1990). Maize cultivation spread along a north–south axis from the Balsas river basin in Mexico, extending throughout the Americas. In addition to dramatic morphological modifications during domestication, growth in temperate climates required modern maize to respond to alternative stimuli to trigger flowering (Dorweiler et al., 1993; Studer et al., 2011; Hufford et al., 2012; Hung et al., 2012). The transition from vegetative to reproductive growth is a critical event in the plant life cycle that directly influences yield and fitness. Floral induction can be triggered by environmental signals, such as photoperiod, as well as internal autonomous signals. The integration of these inputs causes the floral transition at the shoot apical meristem (SAM). Unlike teosinte, which depends on short-day (SD) photoperiods, endogenous signals control flowering in temperate maize (Coles et al., 2010). Thus, flowering occurs at the same time regardless of day length, allowing maize to produce grain before a killing frost. The genetic mechanisms underlying autonomous signals in temperate maize remain poorly understood.
Study of model plants, notably Arabidopsis thaliana, has delineated floral induction mechanisms that are conserved across species (Koornneef et al., 1991). In Arabidopsis, long-day (LD) photoperiods promote early flowering via circadian clock-controlled production of florigen in leaves. A key component of the LD pathway is CONSTANS (CO), which encodes a transcription factor that cycles in a circadian rhythm within leaves (Putterill et al., 1995). Under LD photoperiods, CO protein accumulates and directly activates FLOWERING LOCUS T (FT) expression in the phloem (Suárez-López et al., 2001; Ayre and Turgeon, 2004; Tiwari et al., 2010). FT is a phosphatidylethanolamine-binding protein (PEBP) that acts as a mobile florigen (Araki et al., 1998; Corbesier et al., 2007). FT translocates through the phloem to the SAM where it interacts with FLOWERING LOCUS D (FD), a bZIP transcription factor (Abe et al., 2005; Wigge et al., 2005). The FT–FD complex directly activates expression of APETALA1 (AP1) and other targets to establish reproductive meristem identity (Mandel et al., 1992; Weigel et al., 1992). Loss of CO or FT function causes late flowering under LD conditions, but has little effect on flowering under non-inductive SD conditions (Koornneef et al., 1991).
FT homologues with florigenic function are also found in monocotyledonous SD plants (Kojima et al., 2002; Danilevskaya et al., 2010; Krieger et al., 2010). The maize PEBP gene family, known as Zea mays CENTRORADIALIS (ZCN), includes the experimentally validated florigen, ZCN8 (Lazakis et al., 2011; Meng et al., 2011). Maize possesses an orthologous bZIP transcription factor gene, DELAYED FLOWERING1 (DLF1), that likewise influences flowering time (Muszynski et al., 2006). In teosinte, ZCN8 is expressed at high levels upon SD induction, and some temperate inbreds retain a residual photoperiodic response (Lazakis et al., 2011; Meng et al., 2011). Loss of an obligate photoperiod requirement in temperate maize is partially attributed to a loss of LD-triggered circadian clock repression of flowering. Evidence suggests that loss of the negative floral regulator, Zea mays CO, CO-LIKE TIMING OF CAB1 PROTEIN DOMAIN (ZmCCT), contributed to the northward expansion of maize (Hung et al., 2012; Yang et al., 2013). Other clock-associated genes, including CONSTANS1 (CONZ1) and GIGANTEA1 (GI1), have minor effects on flowering in temperate maize (Miller et al., 2008; Bendix et al., 2013).
Small RNAs (sRNAs) are regulators of diverse components of plant growth and development, including flowering time. Conserved miRNAs regulate genes that influence autonomous control of flowering (Aukerman and Sakai, 2003; Wu and Poethig, 2006). miR156 and miR172 are intertwined antagonists of a conserved age-sensing network (Wu et al., 2009; Cho et al., 2012). During development, miR156 levels decrease and allow for elevated expression of miR172, which down-regulates APETELA2 (AP2) floral repressors (Aukerman and Sakai, 2003). A similar gene regulatory network has been described in maize. The dominant mutant Corngrass1 (Cg1) allele overexpresses a miR156 gene, and a miR172 homologue down-regulates AP2-like genes including GLOSSY15 and RELATED TO AP2.7 (RAP2.7) (Lauter et al., 2005; Chuck et al., 2007; Salvi et al., 2007). Interplay between miRNA-mediated gene regulation and other modes of floral induction remains unstudied in maize.
The predominant autonomous floral inductive pathway of temperate maize shares little similarity with characterized autonomous pathways in dicots (Colasanti and Coneva, 2009). The indeterminate1 gene (id1) encodes a transcriptional regulator that controls autonomous floral induction in maize (Colasanti et al., 1998). Loss of id1 function disrupts the autonomous pathway and severely delays the floral transition (Colasanti et al., 1998). The id1 gene is expressed exclusively in developing immature leaves (ILs), but loss of id1 function has no overt effect on leaf morphology. However, id1 mutant mature leaves (MLs) have increased levels of sucrose and starch, and modified energy metabolism (Coneva et al., 2012). Although the mechanism is unknown, ID1 is thought to be involved in sensing whole-organism carbon status and conveying readiness to flower (Coneva et al., 2007, 2012; Wong and Colasanti, 2007). Carbohydrate sensing is associated with autonomous control of flowering in other plants (Corbesier et al., 1998; Moore et al., 2003; Wahl et al., 2013).
The close genetic relationship between teosinte and temperate maize provides a unique opportunity to study how autonomous floral control evolved from an obligate photoperiodic ancestor. Photoperiod and autonomous pathways, despite responding to different stimuli, both rely on leaf-based signals. During maize domestication, an autonomous signal could have mimicked SDs, but could trigger the same downstream photoperiodic leaf inductive pathway. Alternatively, an independent and weak autonomous pathway could have been strengthened during domestication to allow for flowering in temperate climates. To investigate how the autonomous flowering pathway dominated the ancestral photoperiod pathway, leaf mRNA and sRNA profiles were compared pairwise between flowering and vegetative teosinte and between flowering and vegetative temperate maize. These comparisons show leaf expression overlap between photoperiodic and autonomously flowering Z. mays, revealing leaf events that were conserved during maize domestication. This overlap, however, does not include homologues of established floral regulators. Rather, distinct expression profiles of known floral regulators in maize and teosinte were observed. This suggests that the maize autonomous and photoperiodic gene networks act in parallel and only converge after signal perception at the SAM. These findings provide novel insights into mechanisms that favoured autonomous flowering that supplanted photoperiodic induction, fostering the widespread dissemination of maize.
Materials and methods
Plant materials and growth conditions
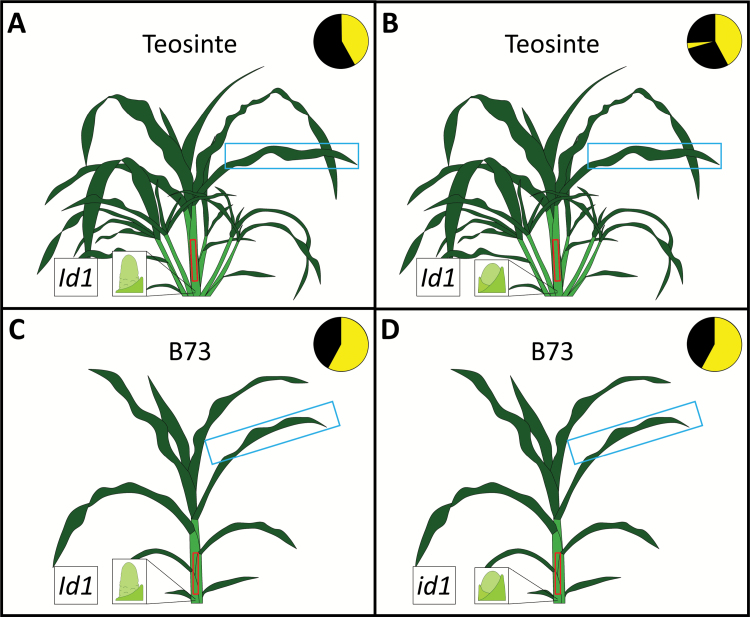
Maize and teosinte plants were grown as described previously (Mascheretti et al., 2015) (Fig. 1). Zea mays spp. parviglumis plants were grown under non-inductive LD conditions (14 h/10 h; light/dark) and then induced to flower by exposure to SD conditions (10 h light/14 h dark) or flowering was inhibited by a 1 h night-break (NB) treatment in the middle of the dark period. The id1-m1 allele was backcrossed 11 times into Z. mays spp. mays inbred B73, and segregating flowering Id1+ (WT) and non-flowering plants (id1-m1) at the V7 stage were genotyped and used for RNA isolation as previously described (Wong and Colasanti, 2007). Temperate maize plants were grown under LD photoperiods (14 h day/10 h night).
Fig. 1.
Tissue sampling scheme of reproductive and vegetative teosinte and B73. Teosinte was grown under inductive short-day (SD) conditions (A) and uninductive night-break (NB) conditions (B). B73 inbreds were grown under long-day conditions with either homozygous for (WT) Id1 (C) or homozygous for mutant id1 (D) to induce and repress the floral transition, respectively. RNA profiling was conducted on mature leaf (blue box) and the immature leaf whorl above the meristem (red box). Reproductive and vegetative expression changes were first compared (A versus B; C versus D) within subspecies, followed by a comparison of expression changes between teosinte and B73.
mRNA profiling and binding site prediction
Total RNA was extracted from three replicates of frozen tissues using the MirVana kit (Ambion) following the manufacturer’s instructions. Samples (10 µg) of total RNA were treated with 3 U of TURBO DNase (Ambion) for 1 h at room temperature in a final volume of 50 µl, followed by purification with RNA Clean and Concentration-5 column (Zymo Research). RNA quality and concentration were estimated by agarose gel electrophoresis and spectrophotometry. Libraries for next-generation Illumina system directional sequencing of total RNA were prepared with TruSeq Stranded Total RNA with a Ribo-Zero Plant kit (Illumina). Sequencing of 24 total RNA-seq (four sample types, each with three biological replicates) was performed at the Istituto di Genomica Applicata (Udine, Italy) on an Illumina Hiseq2500 platform. RNA-seq paired-end 2 × 100 bp sequences, producing ~20 million paired reads per sample, achieved a total of ~60 million 2 × 100 bp paired-end reads for each sample type (NCBI BioProject accession number PRJNA439244). FastQC was used to ensure reads were of appropriate quality (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Differential gene expression was analysed using the Tuxedo software suite (Bowtie2 v2.1.0; TopHat v2.0.11; cufflinks v2.2.1) and aligned against the B73 reference genome (V3.22) (Li et al., 2009; Schnable et al., 2009; Trapnell et al., 2012; Langmead and Salzberg, 2012). One B73 id1 immature leaf biological replicate produced low read alignment rates (27.85%) and was excluded from analysis. To quantify expression from the ZCN7 and ZCN8 loci, strand-specific reads were remapped using Bowtie2 independently for both the ZCN7 and ZCN8 coding sequence and for the unspliced sense and antisense transcripts. A reference gene (GRMZM2G161285) with stable FPKM (fragments per kilobase of transcript per million mapped reads) values with very low variance (6.94 ± 0.24) was selected and used to standardize reads across all libraries. Reads were called to either ZCN7 or ZCN8 based on MapQ values. Reads that mapped completely to exons were excluded in the ZCN7 and ZCN8 unspliced sense reads. All Gene Ontology (GO) enrichment was conducted using AgriGO (V5a) (Du et al., 2010). Venn diagrams were generated through Bioinformatics & Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/). Principal component analysis (PCA) was conducted through SPSS using transcripts within the 10th highest percentile of variance across all samples. Sequences upstream (≤2 kb) of genes that were differentially expressed in Id1+ versus id1-m1 leaves were employed for in silico motif discovery using the Find Individual Motif Occurrences (FIMO) tool and degenerate motif TTTGTCSYWWT (IUPAC nucleotide code) (Grant et al., 2011).
sRNA profiling
For sRNA-seq, total RNA was prepared and purified as described above and used as template for sRNA libraries prepared with the TruSeq small RNA kit (Illumina). Sequencing of the 24 sRNA-seq libraries (four sample types, each with three biological replicates) was performed on an Illumina Hiseq2500 platform. sRNA-seq single-end 1 × 50 bp read sequencing, producing ~10 million reads per sample, was done, thus obtaining a total of ~30 million 1 × 50 bp single-end reads for each sample type. Shortstack (V3.4) aligned sRNA reads to the B73 reference genome (V3.22) and created sRNA counts within sRNA clusters based upon physical position (default settings) (Axtell, 2013). These clusters were compared with annotated Z. mays miRNA loci (miRBase) to quantify miRNA expression (Kozomara and Griffiths-Jones, 2014). The R package ‘baySeq’ was used with the counts produced by Shortstack to determine differential sRNA cluster expression (Hardcastle and Kelly, 2010).
qPCR of mRNA and miRNA
Confirmation of expression trends was conducted on mRNA and miRNA using quantitative PCR (qPCR) and stem–loop qPCR, respectively. The qPCR and statistical analysis of data were performed as described previously (Mascheretti et al., 2015). Preparation of cDNA and stem–loop qPCR was performed as previously reported using the universal UPL28 probe (Roche) (Varkonyi-Gasic et al., 2007). Expression was normalized to miR166 for miRNA quantification and to both MEP and UBPC for mRNA quantification. Primers employed for qPCR and stem–loop qPCR are reported in Supplementary Tables S9 and S10 at JXB online.
Results
Comparing leaf transcriptomes of maize and teosinte at the floral transition
To examine similarities between photoperiod and autonomous floral inductive pathways, plants were grown in four groups (Fig. 1). Teosinte was grown under SDs to induce flowering and under SD photoperiods with a 1 h NB to maintain vegetative growth (Mascheretti et al., 2015). Plants introgressed into inbred background B73, and segregating normal Id1+ and mutant id1-m1 alleles were grown under LDs. Normal Id1+ plants (hereafter ‘WT’) and id1 homozygous mutants represent reproductive and vegetative treatments, respectively (Colasanti et al., 1998). Floral inductive signals produced in photosynthetic MLs are influenced by developmental patterning outside of the SAM in the ILs (Coneva et al., 2007; Mascheretti et al., 2015). Therefore, at the floral transition (V7) in WT B73 and SD teosinte, developing ILs and developed MLs were harvested as previously described and used for mRNA and sRNA expression profiling (Mascheretti et al., 2015). Pairwise comparisons in leaf expression were conducted between flowering and non-flowering groups. Teosinte SD was compared with teosinte NB, and B73 WT was compared with B73 id1. Finally, expression differences found in florally induced and uninduced teosinte (SD versus NB) were contrasted with those observed in florally induced and uninduced B73 (WT versus id1) to evaluate the overlap between the photoperiod and autonomous pathways in the leaf.
Teosinte leaf mRNA expression changes at the floral transition highlight alterations in circadian clock and florigen gene expression
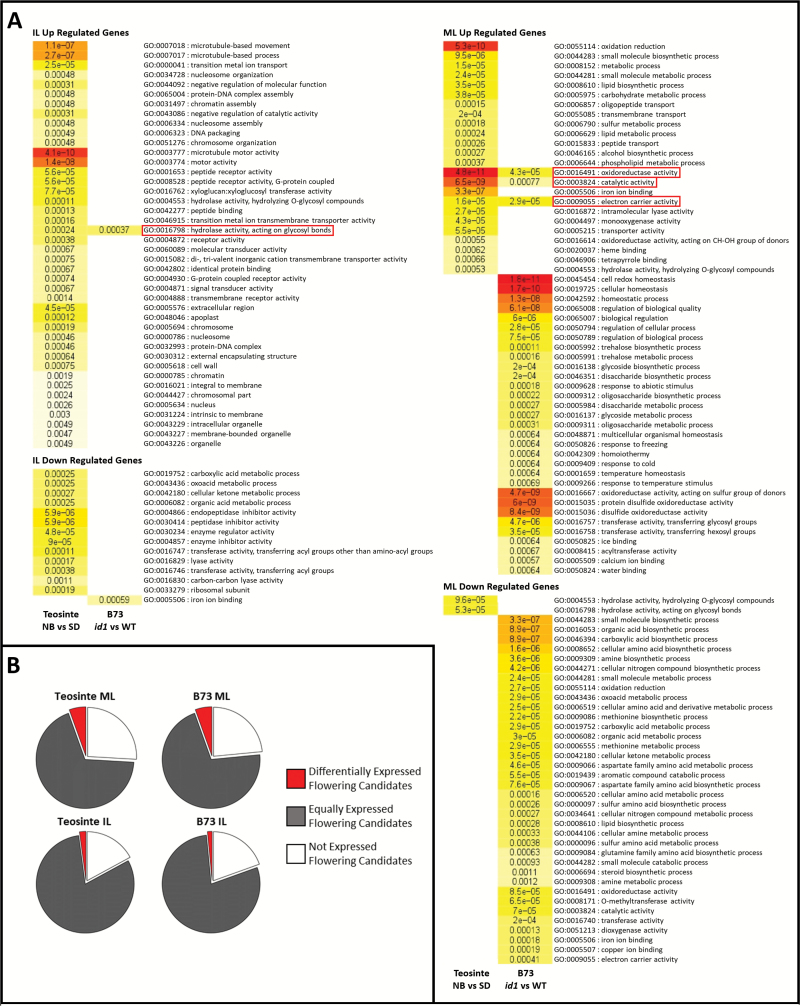
RNA sequencing from teosinte tissues produced an average of 10 million reads per sample. Through alignment to version v3.22 of the B73 genome, 25610 and 30057 expressed annotated transcripts were detected in MLs and ILs, respectively. From these, 1112 ML and 691 IL annotated genes showed differential expression between SD and NB treatments [false discovery rate (FDR)=0.05; Supplementary Table S1]. More than half of these transcripts, 756 MLs and 522 ILs, showed increased levels upon floral induction. GO term enrichment was conducted on these differentially expressed genes (Fig. 2). In MLs, this revealed up-regulation of genes involved in oxidation/reduction status, phospholipid metabolism and carbohydrate metabolism, and down-regulation of glycosyl hydrolase genes. In ILs, significant GO terms up-regulated included microtubule transport, xylosyl transferase activity, glycosyl hydrolase activity, and alterations to the cell wall and apoplast. In ILs, down-regulated GO terms included peptidase inhibition and organic acid metabolism.
Fig. 2.
The floral transition in teosinte and B73 results in transcriptional changes enriched in many Gene Ontology (GO) terms and contains candidate floral regulators. (A) Enriched GO terms up- and down-regulated in immature leaves (ILs; left) and mature leaves (MLs; right) in vegetative/reproductive teosinte (night-break; NB versus short-days; SD) and B73 (id1 versus WT). (B) Proportions of candidate floral regulators that were expressed (grey), differentially expressed (red), or not expressed (white) in vegetative/reproductive teosinte and B73. Shared GO terms are outlined in red, and P-values for each ontology term are displayed with a colour reflecting its level of significance.
Maize population genetics studies have revealed numerous small effect loci underpinning flowering time variation (Buckler et al., 2009; Coles et al., 2010; Li et al., 2016; Romero Navarro et al., 2017). A candidate gene list was created to compare putative floral regulators with observed differentially expressed genes (Supplementary Table S2) (Buckler et al., 2009; Coles et al., 2010; Dong et al., 2012; Li et al., 2016). Several genes implicated in the floral transition were detected in MLs and ILs, and a subset exhibited differential expression (Table 1; Fig. 2B). These included ZCN genes related to maize florigens. In MLs, up-regulation of ZCN7, ZCN8, ZCN12, and ZCN15, and down-regulation of ZCN18 and ZCN26 was observed in response to SDs. Genes associated with the circadian clock showed significant expression differences. In induced MLs, expression of CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and a CCT-like gene was increased, while an EARLY FLOWERING3-like (ELF3-like) homologue, and another CCT-like gene decreased in expression. A homologue of PHYTOCHROME INTERACTING FACTOR 3 (PIF3) showed decreased expression in SD ILs. Notable clock components that showed no altered expression in induced versus uninduced teosinte include ZmCCT and CONZ1.
Table 1.
Genes of interest differentially expressed in vegetative/reproductive (night-break versus short day) immature (IL) and mature (ML) teosinte leaves
| Gene identifier | Gene annotation | Log2 fold change (vegetative/ reproductive) |
|---|---|---|
| Teosinte ILs | ||
| GRMZM2G180406 | BHLH transcription factor | –0.608118 |
| GRMZM2G039996 | Chlorophyll A-B binding protein | –0.594586 |
| GRMZM2G082520 | Expansin precursor | 0.730048 |
| GRMZM2G095968 | Expansin-like | –1.57643 |
| GRMZM2G052616 | GATA27 | 0.74874 |
| GRMZM2G147716 | MADS67 | –3.32879 |
| GRMZM2G032339 | MADS4 (ZMM4) | –5.81219 |
| GRMZM2G115960 | PiF3 | 0.75819 |
| GRMZM2G125441 | Protodermal factor 1 homologue | –0.681943 |
| GRMZM2G700665 | Rap2.7 | 1.58665 |
| GRMZM2G052616 | GATA-27 | 0.74874 |
| GRMZM2G025783 | Kelch repeat protein | –1.64136 |
| GRMZM2G465091 | TCP family transcription factor | –1.71166 |
| GRMZM2G413006 | Xyloglucan GH16 | – 0.849902 |
| GRMZM2G392125 | Xyloglucan GH16 | – 2.291 |
| GRMZM2G026980 | Xyloglucan GH16 (XET1) | –1.54506 |
| GRMZM2G038898 | Xyloglucan GT14 | –1.47633 |
| GRMZM2G116079 | Zinc finger family protein | –0.828233 |
| Teosinte MLs | ||
| GRMZM2G026223 | Agamous-Like6 (SOC1 homologue) | 0.690749 |
| GRMZM2G369472 | AP2-EREBP172 | –1.69213 |
| GRMZM2G400714 | C2H2 zinc finger protein | –2.40909 |
| GRMZM2G134023 | C3HC4 zinc finger protein | –2.10963 |
| AC225718.2_FG006 | Calcium-binding EF-hand | –2.08808 |
| GRMZM2G340807 | Calcium-binding EF-hand | –1.85258 |
| GRMZM2G106945 | Calmodulin-related protein | –1.59218 |
| GRMZM2G014902 | CCA1 | –0.836844 |
| GRMZM2G155370 | CCT-Like | –1.09463 |
| GRMZM2G075562 | CCT-Like | 1.19754 |
| AC233870.1_FG003 | ELF3-Like | 0.605407 |
| GRMZM2G032339 | MADS4 (ZMM4) | –2.58678 |
| GRMZM2G347280 | TRPP1 (Trehalose6P phosphatase1) | –0.840243 |
| GRMZM2G425774 | Vernalization5 (VEL1-Like) | 0.588868 |
| GRMZM5G871347 | WRKY93 | –2.19404 |
| GRMZM2G141756 | ZCN7 | 0 expression NB |
| GRMZM2G179264 | ZCN8 | –5.2493 |
| GRMZM2G400167 | ZCN26 | 1.4505 |
Log2 fold change is reported, such that negative values represent an increase in reproductive plants. Transcripts in common with B73 vegetative/reproductive plants are shown, with bold and italics fold change values representing congruent and opposite expression changes, respectively.
Transcriptional regulators associated with photoperiodic flowering were altered in SD versus NB teosinte leaves. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) homologue AGAMOUS-LIKE6 (ZAG6 or MADS56) and a VERNALIZATION5 (VRN5) homologue were both down-regulated in MLs of induced teosinte. An established floral repressor, RAP2.7, showed decreased expression in ILs (Salvi et al., 2007; Castelletti et al., 2014). MADS4 (ZMM4), an AP1 homologue that promotes the floral transition in temperate maize SAMs, had higher expression in both SD leaf tissues (Danilevskaya et al., 2008). An uncharacterized gene, MADS67, had higher transcript accumulation in florally induced ILs. Differentially expressed genes were compared with putative domestication and improvement targets to find possible overlap between selection targets and genes altered by floral induction (Supplementary Table S3) (Hufford et al., 2012). Differentially expressed genes in teosinte ILs (P=0.60 domestication, P=0.81 improvement; χ2 goodness of fit) and MLs (P=0.86 domestication, P=0.13 improvement; χ2 goodness of fit) showed no significant enrichment in targets of historical selection. Altered expression of several genes was confirmed by independent qPCR analysis (Supplementary Fig. S1). Transcript profiling of teosinte grown under SD and NB conditions is congruent with photoperiod pathways in other plants, where perception of SDs changes the circadian clock output and increases florigen levels.
B73 leaf mRNA expression changes (WT versus id1) reveal alterations in circadian clock genes and carbohydrate utilization
Total RNA sequencing from B73 tissues produced an average of 10 million reads per sample and detected a similar number of expressed transcripts as detected in teosinte, namely 28115 and 29163 expressed annotated transcripts from MLs and ILs, respectively. In MLs and ILs, 1519 and 179 annotated genes, respectively, showed differential expression between the WT and id1 (FDR=0.05; Supplementary Table S4). Of these, 926 in MLs and 135 in ILs were up-regulated upon flowering. GO enrichment of ML genes down-regulated upon flowering included organic acid metabolism and nitrogen compound metabolism (Fig. 2). GO terms enriched within up-regulated ML genes include oxidation/reduction status, hexosyl-transferase activity, and trehalose metabolism. Of 20 trehalose metabolism genes expressed in leaves (Henry et al., 2014), seven were differentially expressed in MLs. TREHALOSE-6-PHOSPHATE SYNTHASE1 (TRPS1), which synthesizes trehalose 6-phosphate (T6P), and TREHALOSE-6-PHOSPHATE PHOSPHATASE1 (TRPP1), which degrades T6P, were increased and decreased, respectively, in WT MLs. Related to this finding, an SNF-RELATED KINASE1 (SnRK1) regulatory β subunit gene showed decreased expression in id1 IL tissues. Alterations in SnRK1 and T6P regulation are consistent with perturbed carbon partitioning in id1 tissues (Coneva et al., 2007, 2012; Wong and Colasanti, 2007). In IL samples, only two GO terms were enriched: glycosyl hydrolase activity and iron ion binding, which were up- and down-regulated, respectively. In agreement with a previous microarray study, a severe reduction in transcript abundance of DHURRINASE-like β-GLYCOSIDASES was observed in id1 ILs. Three GLYCOSYL-HYDROLASE FAMILY 16 (GH16) genes and a GLYCOSYL-TRANSFERASE FAMILY 14 (GT14) gene involved in hemicellulose remodelling showed altered expression in ILs. Transcriptome profiling of B73 flowering versus non-flowering leaves suggests changes in T6P metabolism, SNRK1 regulation, and hemicellulose metabolism. This is consistent with previously observed alterations in leaf carbon utilization and carbon signalling upon disruption of ID1-mediated autonomous induction (Coneva et al., 2012).
The list of candidate flowering time genes was compared with the differentially expressed genes (Fig. 2b; Table 2). Unlike teosinte, where six ZCN genes responded to floral induction, only ZCN26 was differentially expressed in the B73 ML transcriptome profile. In contrast to a florigenic signal, ZCN26 expression was down-regulated upon flowering. Expression analysis at the ZCN8 locus is complex due to the paralogue ZCN7 (94.3% amino acid identity) and previously detected sense and antisense unspliced transcripts (Meng et al., 2011; Mascheretti et al., 2013, 2015). Therefore, strand-specific reads were remapped and quantified for either ZCN7 or ZCN8 spliced transcripts and ZCN7 or ZCN8 unspliced transcripts (Supplementary Fig. S2). ZCN8 expression was reduced in B73 id1 MLs, as previously reported (Meng et al., 2011; Mascheretti et al., 2013, 2015). This confirmed that ZCN7 and ZCN8 expression from B73 Id1+ MLs was lower than in teosinte SD MLs, consistent with observed differences in the teosinte transcriptome profile.
Table 2.
Genes of interest differentially expressed in vegetative/reproductive (id1 versus WT) immature (IL) and mature (ML) B73 leaves
| Gene identifier | Gene annotation | Log2 fold change (vegetative/ reproductive) | ID1-binding site ≤2 kb upstream? |
|---|---|---|---|
| B73 ILs | |||
| GRMZM2G069146 | AP2-EREBP115 | –2.04479 | No |
| AC206951.3_FG017 | AP2-EREBP182 | –1.29962 | Yes |
| GRMZM2G177220 | ARR B6 | –0.6696 | No |
| GRMZM2G367834 | CCT-Like | –0.479088 | Yes |
| GRMZM2G162505 | Chitinase2 | 2.04718 | Yes |
| GRMZM2G076946 | Dhurrinase-like β-glucosidase | –9.89604 | Yes |
| GRMZM2G077015 | Dhurrinase-like β-glucosidase | 0 expression id1− | No |
| GRMZM2G014844 | Dhurrinase-like β-glucosidase | –0.744816 | No |
| GRMZM2G082520 | Expansin precursor | – 0.968866 | No |
| GRMZM2G052616 | GATA27 | –0.736017 | Yes |
| GRMZM2G011357 | ID1 | –3.06022 | No |
| GRMZM2G320287 | IDD7 | 1.26634 | Yes |
| GRMZM2G147716 | MADS67 | –5.10897 | Yes |
| GRMZM2G125441 | Protodermal factor 1 homologue | –0.600682 | Yes |
| GRMZM2G144782 | RING and CHY zinc finger protein | –1.70151 | No |
| GRMZM2G011078 | SCAMP | –2.75259 | No |
| GRMZM2G025459 | SNRK1 homolog | –0.912072 | No |
| GRMZM2G413006 | Xyloglucan GH16 | 0.839937 | Yes |
| GRMZM2G392125 | Xyloglucan GH16 | 0.903474 | No |
| GRMZM2G026980 | Xyloglucan GH16 (XET1) | –1.22364 | Yes |
| GRMZM2G038898 | Xyloglucan GT14 | –3.24967 | No |
| GRMZM2G116079 | Zinc finger family protein | –0.486814 | No |
| B73 MLs | |||
| GRMZM2G369472 | AP2-EREBP172 | –1.15875 | No |
| AC233935.1_FG005 | C2C2 zinc finger protein | 0.5426 | Yes |
| GRMZM2G400714 | C2H2 zinc finger protein | –0.900848 | No |
| GRMZM2G134023 | C3HC4 zinc finger protein | –0.635558 | No |
| AC225718.2_FG006 | Calcium-binding EF-hand | –0.631169 | No |
| GRMZM2G340807 | Calcium-binding EF-hand | –0.882565 | No |
| GRMZM2G106945 | Calmodulin-related protein | –1.27029 | No |
| GRMZM2G155370 | CCT-like | 0.519004 | Yes |
| GRMZM2G095598 | CCT-like | –0.481031 | No |
| GRMZM2G038783 | CCT-like | –0.580766 | No |
| GRMZM2G148772 | CCT-like | –0.706497 | No |
| GRMZM2G092363 | CCT-like | –0.926503 | Yes |
| GRMZM2G013398 | CCT-like | 0.756804 | No |
| GRMZM2G179024 | CCT-like | –0.466114 | No |
| GRMZM2G405368 | Conz1 | –0.872319 | Yes |
| GRMZM2G077015 | Dhurrinase-like β-glucosidase | –5.46369 | No |
| AC233870.1_FG003 | ELF3-like | –0.443462 | Yes |
| GRMZM2G171365 | MADS1 (SOC1 homologue) | 1.03822 | Yes |
| GRMZM2G347280 | TRPP1 (Trehalose6P phosphatase1) | 0.488956 | No |
| GRMZM2G068943 | TRPS1 (Trehalose6P synthase1) | –1.2737 | Yes |
| GRMZM5G871347 | WRKY93 | –0.93739 | No |
| GRMZM2G400167 | ZCN26 | 0.9708 | No |
Log2 fold change is reported, such that negative values represent an increase in reproductive plants. Transcripts in common with teosinte vegetative/reproductive plants are shown, with bold and italics fold change values representing congruent and opposite expression changes, respectively.
Significantly altered transcription factor genes included the floral regulator MADS1 (a SOC1 orthologue) that was down-regulated in MLs upon flowering. Additionally, MADS67 was expressed more highly in WT ILs. Unexpectedly, although plants were grown under identical photoperiods, significant expression changes of circadian clock-associated genes were observed (Table 2). These clock genes were different from those affected by floral induction in teosinte. Nine CCT-like genes showed altered expression in florally induced plants. Six of these, including the putative maize CONSTANS orthologue, CONZ1, had increased transcript levels in WT MLs, and two CCT-like genes and a homologue of the Arabidopsis clock gene LUX had decreased levels. Expression of one CCT-like gene increased in WT ILs. Other genes linked to the circadian clock with altered expression include ELF3-like, ELF4, and a PIF3 homologue. Like teosinte, ZmCCT genes showed no expression change. Expression of several genes was confirmed by qPCR (Supplementary Fig. S3). Genes differentially expressed upon floral induction were compared with putative domestication and improvement targets (Supplementary Table S3). Differentially expressed IL genes (P=0.58 domestication, P=0.88 improvement; χ2 goodness of fit) and ML genes (P=0.43 domestication, P=0.08 improvement; χ2 goodness of fit) showed no enrichment in putative selection targets. mRNA transcriptome changes associated with disruption of autonomous floral induction point to changes in circadian clock genes distinct from those observed in photoperiod induction.
To predict possible direct ID1 direct targets, sequences homologous to the consensus binding motif, TTTGTCSYWWT (IUPAC nucleotide code) were identified near differentially expressed genes (Supplementary Table S5; Kozaki et al., 2004). ID1 may directly bind to and change the epigenetic state of genes in the ILs to modulate later gene expression in MLs (Mascheretti et al., 2015). Therefore, a motif search was conducted ≤2 kb upstream of genes differentially expressed in both tissues (Supplementary Table S5). Several genes of interest contained putative binding sites near their promoters (Table 2). For IL genes, these included carbon metabolism genes such as a DHURRINASE-like gene, two GH16 genes, a CCT-like gene, and MADS67. For putative targets in ML genes, sites were found upstream of CONZ1, two other CCT-like genes, an ELF3-like gene, MADS1, and TRPS1. Thus carbohydrate utilization and clock output genes may be components of the autonomous pathway directly controlled by ID1.
Photoperiod-dependent teosinte and autonomously flowering maize share leaf transcriptional changes in response to flowering
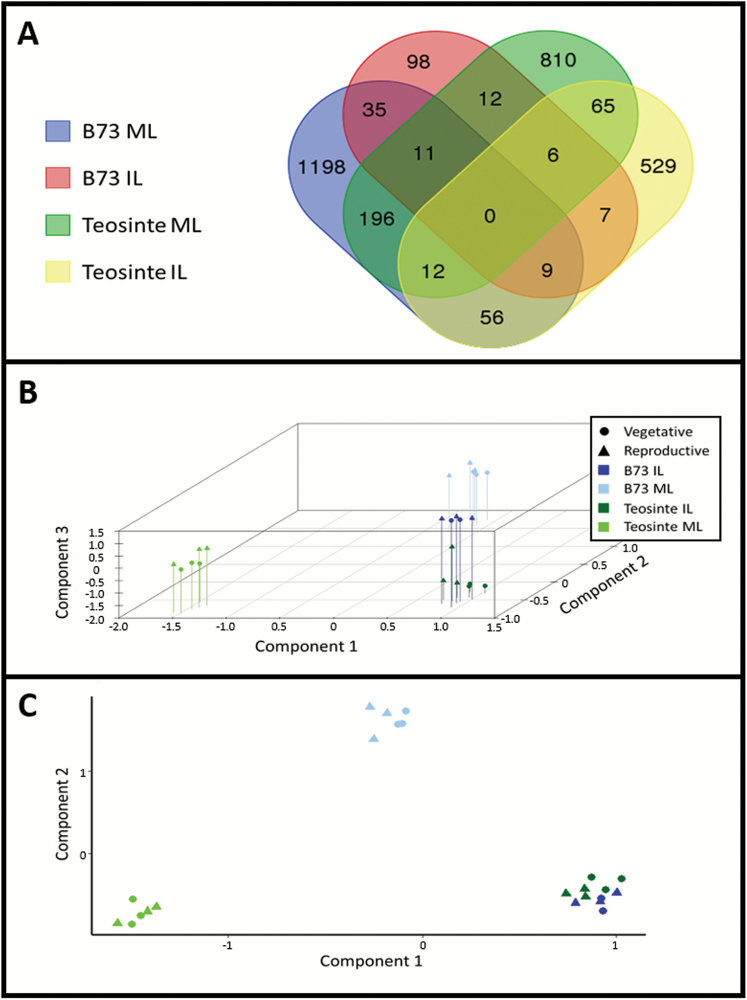
To examine universal changes accompanying the floral transition, transcripts altered in each group of florally induced versus uninduced tissue were compared (Fig. 3a). No genes were found to be differentially expressed in all treatment combinations. In MLs, 219 annotated transcripts were differentially expressed in teosinte and B73, which is more than expected due to chance (P<2.2E-16; χ2 goodness of fit). Of these, 141 transcripts show the same direction of change in reproductive and vegetative tissues. Some transcripts in common in MLs include ZCN26, AP2-EREBP172, WRKY93, Ca2+-sensitive genes, and two uncharacterized zinc finger genes. Of possible floral regulation genes, only ZCN26 was commonly expressed in induced MLs. In ILs, 22 transcripts were differentially expressed in both teosinte and B73, which is more than expected due to chance (P<2.2E-16; χ2 goodness of fit). Eleven transcripts showed common expression changes upon induction. These included MADS67 and a homologue of Arabidopsis PROTODERMAL FACTOR1 (PDF1). The four aforementioned GH16/GT14 hemicellulose remodelling genes were differentially expressed in both teosinte and maize ILs, comprising 18% of the overlap in this tissue. These genes are a part of the GO term ‘hydrolase activity, acting on glycosyl bonds’, which is up-regulated in both teosinte and B73 IL data sets. In MLs, the predominant common GO term is oxidative/reductive status, suggesting that large-scale leaf redox changes accompany the floral transition in domesticated maize and wild teosinte. These commonly regulated genes, however, include few of the canonical floral regulators differentially expressed in teosinte or B73 leaves. Even so, the comparison of gene expression in florally induced versus uninduced teosinte and B73 leaves unveiled a significant overlap in the transcriptional response to the floral transition.
Fig. 3.
Comparison of immature leaf (IL) and mature leaf (ML) transcriptomes from induced and uninduced teosinte and B73. (A) Venn diagram displaying the number of transcripts differentially expressed in both reproductive/vegetative B73 (WT versus id1) and teosinte (short-days versus night-break). Principal component analysis (PCA) of B73 (blue) and teosinte (green) mRNA mature leaf (ML; light shades) and immature leaf (IL; dark shades) transcriptomes in three (B) and two (C) dimensions, capturing 86.3% and 78.4% of the variance, respectively. Reproductive treatments are depicted as triangles, and vegetative treatments as circles.
PCA of all samples showed that, when three principal components are used, the groups separate into four clusters that correspond to each genotype–tissue combination (Fig. 3b). For two components, however, the teosinte IL and B73 IL transcriptomes cluster together (Fig. 3c), illustrating that differences between teosinte and B73 transcriptomes is greater in MLs than in ILs. No clustering is observed between reproductive or vegetative groups, showing that fewer transcriptional changes accompany the floral transition than those related to genotype and tissue. This is consistent with the relatively few transcripts (102–103) altered by induction in both genotypes.
Floral induction affects miR399 expression in both teosinte and B73 leaves
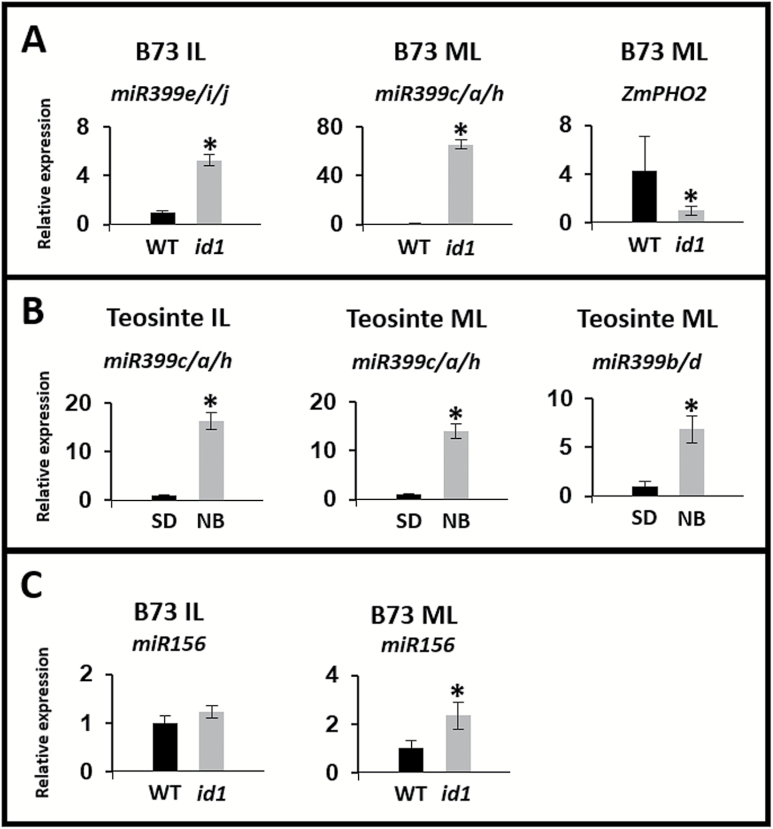
sRNA sequencing performed in tandem with mRNA sequencing revealed changes in sRNA species that may regulate genes controlling the floral transition in SD versus NB teosinte and WT versus id1 leaves. Ten million 50 bp single-end reads per sample were generated for each genotype and tissue. This analysis detected 119269 clusters producing unique sRNA from the B73 data sets and 469533 clusters of sRNA in teosinte (Supplementary Tables S7–S9). Annotated maize miRNA sequences were used to quantify miRNA expression levels (Supplementary Tables S7, S10). The only miRNAs with significant levels in id1 leaves were miR399c and miR399e in MLs and ILs, respectively. These miR399 isoforms were more abundant in non-flowering tissue than in the WT (Supplementary Table S7). In support of its established role in modulating gene expression, the previously confirmed in vivo target of miR399, GRMZM2G381709 (Lunardon et al., 2016), an Arabidopsis PHOSPHATE2 (PHO2) orthologue (hereafter ZmPHO2), had decreased mRNA abundance in id1 B73 IL and ML tissues (Supplementary Table S4). This was confirmed by qPCR (Fig. 4). Teosinte also had differentially expressed miR399 genes with increased accumulation in uninduced NB tissue. Differentially expressed miR399 isoforms included miR399c, miR399b, and miR399f in MLs, and miR399h in ILs. Stem–loop qPCR confirmed the differential expression of these miR399 isoforms in flowering and non-flowering tissues (Fig. 4). miR156, a known repressor of the reproductive transition, was among the differentially expressed annotated miRNAs in teosinte MLs and ILs (Supplementary Table S10; Wu and Poethig, 2006; Wu et al., 2009). Isoforms miR156e, miR156i, and miR156l had increased expression in teosinte NB ILs. Stem–loop qPCR also demonstrated a small, but significant increase of miR156 in id1 issues (Fig. 4c).
Fig. 4.
qPCR confirmation of miRNA and miRNA target gene expression changes upon floral induction. (A) Stem–loop qPCR quantification of miR399 isoforms and its target ZmPHO2 from immature (IL) and mature (ML) leaves of maize B73 WT and id1 plants. (B) Stem–loop qPCR quantification of miR399 isoforms in teosinte plants grown under inductive short days (SD) and inhibitory night breaks (NB). (C) Stem–loop qPCR quantification of miR156 expression in ILs and MLs from B73 WT and id1 plants. The primers used are not specific for a unique miRNA isoform, and a group of isoforms detected is reported. These graphs are representative of data obtained for one of the three biological replicates analysed. An asterisk indicates a statistically significant change (P≤0.05) when it was achieved across biological replicates.
Discussion
Teosinte and B73 leaf mRNA transcriptomes reveal responses to the floral transition by parallel pathways
During the domestication of temperate maize, autonomous floral inductive signals superseded the obligate photoperiod signals required by its tropical ancestor, teosinte. Here, leaf expression profiles of maize and teosinte at the floral transition were analysed to investigate central components of floral inductive pathways in Z. mays. These analyses revealed little overlap in the expression changes of many distinct floral regulators in teosinte and B73. This supports the hypothesis that teosinte possesses elements of an autonomous inductive pathway that act separately in the leaf, but are repressed under LDs. In this model, two events occurred during domestication: first, loss of dominant floral repressors, including ZmCCT, that allowed flowering without SD induction; and, secondly, potentiation of an extant autonomous inductive pathway (Yang et al., 2013). This is consistent with other models proposed in recent studies (Dong et al., 2012; Romero Navarro et al., 2017). In both teosinte and B73 temperate maize, for both tissues sampled, only one candidate floral regulator, ZCN26, shows the same expression profile. Other transcripts showing conserved expression changes are involved in the response to floral induction, and may contribute to physiological responses that prepare the plant for reproductive growth.
Expression of shared and distinct ZCN and MADS transcription factor genes is linked to photoperiodic and autonomous floral induction
In teosinte, the ZCN8 gene shows dramatic induction in leaves of SD-grown plants, analogous to expression changes in florigen genes of photoperiod-responsive rice and Arabidopsis (Lazakis et al., 2011; Meng et al., 2011). Similarly, leaf expression of ZCN7, ZCN12, and ZCN15 increases under inductive conditions in teosinte, demonstrating that these FT homologues also possess photoperiod sensitivity. The ZCN7 sequence and expression pattern are nearly identical to those of ZCN8, and ZCN15 is associated with floral timing in landrace populations (Romero Navarro et al., 2017). However, whether any of these uncharacterized ZCN genes are bona fide florigens requires further study.
In contrast, ZCN8 expression in leaves of B73 temperate maize grown under LD conditions showed more attenuated expression at the floral transition. Previous studies showed that ZCN8 expression levels are 3- to 4-fold higher in SD- than in LD-grown B73; however, this is relatively modest compared with the enormous up-regulation of ZCN8 in SD versus NB teosinte (Lazakis et al., 2011). Moreover, ZCN8 expression does not cycle in a diurnal pattern under LDs (Lazakis et al., 2011; Meng et al., 2011). ZCN8 shows a slight increase in expression in WT versus id1 MLs, downplaying its importance in the floral delay caused by id1. Transgenic overexpression or knock down of ZCN8 causes minor variations in flowering time (1–4 leaves), whereas loss of id1 causes an extreme flowering delay (Colasanti et al., 1998; Meng et al., 2011). If id1 acts by activating ZCN8 exclusively, loss of ZCN8 expression should produce a comparable extreme flowering delay. These findings suggest that temperate maize retains a residual photoperiod response; however, ZCN8 is not a major component of the autonomous pathway.
ZCN26 had an altered response at the floral transition in leaves of both teosinte and maize. ZCN26 expression is down-regulated in MLs in both cases, which is not indicative of florigen activity. Indeed, reduced ZCN26 expression in MLs is consistent with anti-florigen activity (Pin et al., 2010; Higuchi et al., 2013). However, overexpression of ZCN26 in transgenic maize was reported to have no effect on the floral transition (Meng et al., 2011). Therefore, it will be interesting to determine the role played by ZCN26 in leaves that accompanies flowering, but does not directly trigger the reproductive transition at the SAM.
MADS genes encode transcription factors that include members associated with flowering in diverse species (Mandel et al., 1992; Danilevskaya et al., 2008). MADS67 is up-regulated in ILs of both teosinte and maize at the floral transition. MADS67 has not been previously implicated in triggering reproductive growth, but there is evidence of selection during domestication and subsequent improvement of maize cultivars, indicating a possible role of agronomic importance (Hufford et al., 2012). Paralogous genes MADS1 and ZAG6 were significantly down-regulated at the floral transition in MLs of B73 and teosinte, respectively. MADS1 has an ID1 DNA-binding motif upstream of its transcription start site, perhaps indicating that MADS1 is a component of the autonomous pathway. A recent study identified MADS1 as a SOC1 orthologue that, when expressed in transgenic maize, altered flowering time in a bimodal fashion—low expression caused delayed flowering, but large increases resulted in earlier flowering by up to 2 weeks (Alter et al., 2016). Consistent with these studies, MADS1 showed a small increase in expression in id1 MLs. ZAG6 displayed expression changes in the same direction as MADS1, with a similar fold change in teosinte MLs under non-inductive NB conditions, suggesting that these paralogues have divergent functions, with both contributing to flowering but along different pathways. A maize whole-genome duplication event occurred between 5 and 12 million years ago (Blanc and Wolfe, 2004; Swigoňová et al., 2004). Duplication of a SOC1 precursor may have allowed for subfunctionalization of descendant genes into different floral inductive pathways, one of which was required for temperate growth habits.
miR399 is down-regulated at the floral transition in teosinte and maize
Expression of miR399 isoforms is higher in pre-transition leaves of both teosinte and B73. miR399 targets ZmPHO2, which is implicated in regulating root phosphorus acquisition (Lunardon et al., 2016). A genome-wide association study (GWAS) of flowering in maize landraces reported that ZmPHO2 is associated with the timing of male and female flowering, strengthening the argument for a role for miR399 and ZmPHO2 in maize reproduction (Romero Navarro et al., 2017). Phosphate deficiency is accompanied by increases in soluble sugars, and increased sugar concentrations subsequently induce phosphate starvation responses (Ciereszko et al., 2005; Karthikeyan et al., 2007; Dasgupta et al., 2014). Curiously, plants starved of phosphate and photosynthate do not accumulate miR399; thus, sugar levels supersede the miR399 expression response to phosphate (Liu et al., 2010). Therefore, miR399 levels may respond to altered abundance of leaf soluble sugars rather than phosphorus limitation. As previous studies have focused on root metabolism, the detection of miR399 expression differences in leaves upon floral induction may point to an undiscovered role for miR399 in leaves (Fujii et al., 2005; Pant et al., 2008).
Disruption of ID1-mediated autonomous floral induction supports a link between leaf carbon metabolism and flowering in temperate maize
Previous microarray and metabolomic studies suggest that id1 regulates a critical node that links leaf carbon status with long-distance signals that trigger the floral transition (Coneva et al., 2007, 2012; Wong and Colasanti, 2007). The present analysis supports these findings and provides new candidates downstream of id1. These include carbon metabolism and potential floral regulator genes with ID1 DNA-binding motifs in their putative promoters. Population genetic studies have uncovered loci showing signs of selection during domestication and improvement through classical breeding, but whether any of these loci were instrumental in transforming the reproductive timing mechanism is unknown. The GH16, MADS67, and TRPP1 genes are differentially expressed in both induced teosinte and maize, and show signs of historical selection (Hufford et al., 2012). Analysis of these genes may reveal additional molecular changes that facilitated the spread of maize to temperate regions.
T6P is a critical sensor of carbon availability that tunes growth and carbon utilization to sucrose availability (Figueroa and Lunn, 2016). In WT MLs, expression of the T6P biosynthesis gene, TRPS1, increased and expression of a gene involved in T6P catabolism, TRPP1, decreased. TRPS1 is a possible target of ID1 because it has a consensus ID1-binding motif in its putative promoter. GO term enrichment for up-regulated transcripts in WT MLs also supports changes in T6P metabolism. This agrees with a previous metabolomic study that suggested increased T6P levels in B73 leaves at the floral transition (Coneva et al., 2012). T6P levels are correlated with sucrose (Yadav et al., 2014), yet id1 MLs have higher sucrose levels and signatures of lower T6P, suggesting that id1 affects both carbon sensing and partitioning. Consistent with changes in carbon metabolism, a SnRK1 β subunit is down-regulated in id1 mutant ILs. SnRK1 activity is correlated positively with low cellular energy status and negatively with T6P levels (Bouly et al., 1999; Polge et al., 2008; Zhang et al., 2009). Developing ILs are major carbon sinks in both vegetative and reproductive shoots. Higher SnRK1 activity in induced ILs may lower energy utilization in maturing leaves, reducing leaf sink activity at a time when reproductive sinks are growing in strength. Carbohydrate sensing has a demonstrable effect on the floral transition in other plants (Corbesier et al., 1998; Moore et al., 2003; Wahl et al., 2013). Perception of sugar levels and subsequent id1 signalling would ensure that flowering occurs when energy levels are sufficient to support reproduction. In Arabidopsis, carbon signalling through T6P acts independently of photoperiod and miR156 to control flowering (Wahl et al., 2013). Interesting parallels are seen with id1 mutants, as loss of ID1 activity represses SD-induced florigen production and shows little change in miR156 expression (Lazakis et al., 2011; Meng et al., 2011; Mascheretti et al., 2015). This implies that autonomous carbohydrate signalling may be similarly retained across the monocot–dicot divide.
Developing leaves of Z. mays show altered expression of hemicellulose metabolism genes at the floral transition
Of the 22 genes showing altered expression at the floral transition in both B73 and teosinte ILs, four are predicted to remodel hemicellulose. Given heightened levels of carbohydrates in id1 mutant leaves, these remodelling genes may be involved in carbon metabolism. Hemicellulose is a major sink which, in some plants, is broken down for carbon remobilization (Hoch, 2007; Lee et al., 2007). Expanding maize leaves rapidly turn over select polymers during cell wall loosening (Gibeaut and Carpita, 1991). Aberrant polymer remodelling could limit carbon recycling during cell wall development, ‘locking’ carbon in the wall and preventing remobilization. Alternatively, changes in the maize apoplast may perturb IL function. Homologous cell wall remodelling genes in Arabidopsis are involved in phloem development (Bourquin et al., 2002). Maize ILs rely exclusively on apoplastic export of sugar from MLs via the phloem to support growth (Evert and Russin, 1993). Altering the cell walls of symplastically isolated phloem would have severe consequences for export of sugars and information molecules, including florigen (Colasanti and Sundaresan, 2000). A connection between walls and flowering has been described in maize and sorghum. Maize brown midrib (bm) mutants display both altered leaf lignin content and altered flowering time (Vermerris and Mcintyre, 1999; Vermerris et al., 2002). The mechanism underlying bm-mediated effects on flowering is unknown, but could involve events in the ILs. Mechanistic studies of monocot cell wall remodelling may unveil important physiological changes that accompany the floral transition.
Photoperiodic and autonomous floral induction alter expression of distinct genes linked to the circadian clock
Transcriptional changes in clock genes accompany the perception of inductive photoperiods and the transition to reproductive growth. Established photoperiodic models involve output from the circadian clock that triggers the expression of florigen (Tiwari et al., 2010; Dong et al., 2012; Hung et al., 2012; Romero Navarro et al., 2017). Photoperiod-sensitive maize, such as teosinte, are induced to flower by long nights. NB treatment disrupts the inductive dark period yet does not affect the day length. Expression of components of the maize circadian clock, including CCA1, ELF3-like, PIF3, and CCT-like genes, changes in response to SD induction. Several clock genes have been identified in previous studies of the genetic control of maize flowering time (Buckler et al., 2009; Coles et al., 2010; Hung et al., 2012; Yang et al., 2013; Li et al., 2016). Other clock components show no expression changes upon photoperiodic floral induction, suggesting that they are entrained to other stimuli. ZmCCT represses maize flowering under LDs and has been implicated in floral adaptation to temperate climates (Hung et al., 2012; Yang et al., 2013). This study finds that ZmCCT levels are unchanged under NB conditions in leaves, suggesting that ZmCCT expression is affected by day length, rather than night length, and that other repressors act upon flowering under NB treatment. The observed expression changes in ZCN genes upon exposure to SD supports a role for circadian clock output in triggering florigen expression.
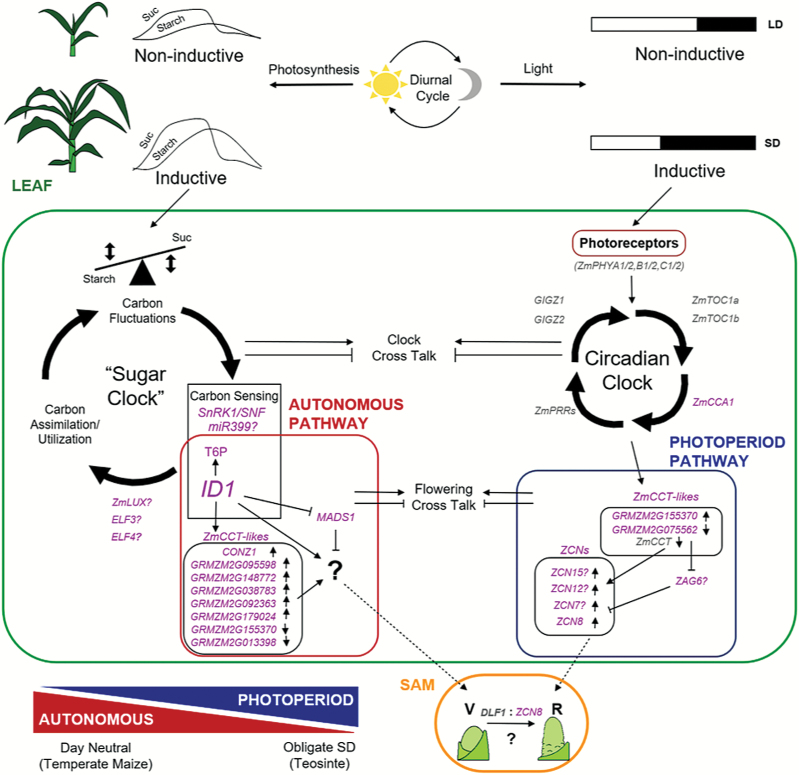
Interestingly, many circadian clock output genes show altered expression in the WT versus id1 comparison, including CONZ1, eight CCT-like genes, and an ELF3-like gene. Moreover, these clock-associated genes are different from those that are altered by photoperiod induction in teosinte. This was unexpected, as WT and id1 plants were grown under identical photoperiods. Part of the circadian rhythm is entrained to daily changes in sugars due to photosynthesis rather than the perception of light though photoreceptors (Haydon et al., 2013; Pattanayak et al., 2015). In maize, sucrose and hexoses cycle diurnally (Kalt-Torres et al., 1986). Consistent with altered carbon metabolism throughout the light/dark cycle, we propose that the cyclical sugar cycle, or ‘sugar clock’, is altered by loss of id1 function (Fig. 5). Changes in sucrose levels over the day accompany the late flowering id1 phenotype and, although a sugar cycle corresponding to the light/dark period persists, the magnitude of changes is significantly altered (Coneva et al., 2012). In this model, the magnitude of fluctuating photosynthate levels increases as plants grow, regardless of day length. Carbon sensors, including T6P, signal when a critical threshold is attained, relaying this information to ID1. ID1 acts as a key integrator that modulates the output from the ‘sugar clock’, such as CCT-like gene expression, to promote florigen production (Fig. 5). The nature of these autonomous florigens, whether ZCN-related genes, metabolites, or a multifactorial signal, remains to be determined. Notably, SD versus NB treatments do not alter leaf sucrose and starch levels in teosinte leaves (Coneva et al., 2012). Hence, gene expression changes observed in teosinte at the floral transition are in response to photoperiod and not altered sucrose levels. Distinct outputs from parallel internal oscillators may contribute to flowering by photoperiod or autonomous signalling.
Fig. 5.
Model: parallel clocks in maize leaf regulate autonomous and photoperiod pathways that converge at the shoot apical meristem (SAM) to cause flowering. Diurnal cycles of light and dark (top) are perceived by photoreceptors to entrain the circadian clock and induce flowering under short-day (SD) photoperiods (blue box, right). Similarly, in growing plants, daily fluctuations of fixed carbon levels (starch and Suc) attain a critical threshold and feed into a proposed ‘sugar clock’ to activate autonomous induction (red box, left). Output from each clock affects expression of distinct CCT-like genes that produce florigens, such as the ZCN genes. For autonomous flowering, ID1 modulates input from carbon sensors to signal expression of CCT-like regulators and other possible florigens (indicated by ‘?’). Crosstalk between clocks integrates environmental and endogenous signals to balance plant growth and determine the relative contribution of each flowering pathway (illustrated by the red and blue triangle graphic, lower left). Genes found to have altered expression in this study are shown in purple; key genes implicated in flowering from other studies are shown in grey. Dotted lines indicate movement of florigens from leaves. The green boxed area represents activity in leaf.
Conclusions
Transcriptome comparison of flowering pathways in obligate photoperiod-dependent teosinte and day-neutral autonomous temperate maize suggests that these two pathways operate through distinct leaf gene networks. Therefore, potentiation of the autonomous pathway combined with loss of LD floral repression accommodated the northward migration of maize. Intriguing shared responses were observed at the floral transition in teosinte and maize. Conserved miR399 down-regulation in leaves upon flowering alludes to novel roles for miR399 and ZmPHO2 beyond controlling phosphate uptake. Transcriptional regulators show conserved expression alterations in both temperate maize and teosinte. These include MADS67, a gene previously identified as a target of maize domestication and improvement. Furthermore, paralogues MADS1 and ZAG6 probably underwent subfunctionalization after prehistoric polyploidization, resulting in their involvement in the autonomous and photoperiodic pathways, respectively. Unique components of the circadian clock show mutually exclusive expression trends upon disruption of autonomous or photoperiodic pathways, suggesting that distinct components of the clock influence flowering via each pathway. This study illuminates genetic changes underpinning the evolution of temperate maize and provides new targets for single gene studies that will provide improved mechanistic understanding of the floral transition.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. qPCR assays in teosinte.
Fig. S2. Expression from paralogues ZCN7 and ZCN8.
Fig. S3. qPCR assays in maize B73.
Table S1. All annotated transcripts differentially expressed between teosinte treatment groups.
Table S2. Established candidate maize floral regulators.
Table S3. Genes common to historical selection events and differential expression upon floral induction.
Table S4. All annotated transcripts differentially expressed between B73 treatment groups.
Table S5. ID1-binding motifs ≤2 kb upstream of genes differentially expressed in id1 tissues.
Table S6. Differentially expressed small RNA clusters between B73 treatment groups.
Table S7. Differentially expressed small RNA clusters between teosinte treatment groups.
Table S8. Differentially expressed microRNAs between teosinte treatment groups.
Table S9. List of primers used in qPCR.
Table S10. List of primers used in stem–loop qPCR.
Acknowledgements
This research was supported by the Natural Sciences and Engineering Research Council of Canada (JC), the Italian Ministry of Education, University and Research (MIUR) and the National Research Council of Italy (CNR) for EPIGEN, the Epigenomics Flagship Project (VR). We thank Tannis Slimmon and Mike Mucci of the Guelph Phytotron for expert plant care. MAAM is a recipient of an NSERC Alexander Graham Bell Canada Graduate Doctoral scholarship.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Alter P, Bircheneder S, Zhou LZ, Schlüter U, Gahrtz M, Sonnewald U, Dresselhaus T. 2016. Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiology 172, 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Kobayashi Y, Kaya H, Iwabuchi M. 1998. The flowering-time gene FT and regulation of flowering in Arabidopsis. Journal of Plant Research 111, 277–281. [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre BG, Turgeon R. 2004. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiology 135, 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendix C, Mendoza JM, Stanley DN, Meeley R, Harmon FG. 2013. The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant, Cell and Environment 36, 1379–1390. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly JP, Gissot L, Lessard P, Kreis M, Thomas M. 1999. Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINalpha1, an SNF1-like protein kinase. The Plant Journal 18, 541–550. [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. 2002. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. The Plant Cell 14, 3073–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, et al. . 2009. The genetic architecture of maize flowering time. Science 325, 714–718. [DOI] [PubMed] [Google Scholar]
- Castelletti S, Tuberosa R, Pindo M, Salvi S. 2014. A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 4, 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Coruh C, Axtell MJ. 2012. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. The Plant Cell 24, 4837–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. 2007. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nature Genetics 39, 544–549. [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA. 2005. Interactive effects of phosphate deficiency, sucrose and light/dark conditions on gene expression of UDP-glucose pyrophosphorylase in Arabidopsis. Journal of Plant Physiology 162, 343–353. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Coneva V. 2009. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiology 149, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Sundaresan V. 2000. ‘Florigen’ enters the molecular age: long-distance signals that cause plants to flower. Trends in Biochemical Sciences 25, 236–240. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. 1998. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93, 593–603. [DOI] [PubMed] [Google Scholar]
- Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. 2010. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 184, 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Guevara D, Rothstein SJ, Colasanti J. 2012. Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition. Journal of Experimental Botany 63, 5079–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Zhu T, Colasanti J. 2007. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. Journal of Experimental Botany 58, 3679–3693. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G. 1998. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206, 131–137. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. . 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV. 2010. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiology 153, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG. 2008. Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiology 147, 2054–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta K, Khadilkar AS, Sulpice R, Pant B, Scheible WR, Fisahn J, Stitt M, Ayre BG. 2014. Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiology 165, 715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. 1990. Molecular systematics of Zea (Gramineae). Maydica 35, 143–150. [Google Scholar]
- Dong Z, Danilevskaya O, Abadie T, Messina C, Coles N, Cooper M. 2012. A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One 7, e43450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler J, Stec A, Kermicle J, Doebley J. 1993. Teosinte glume architecture 1: a genetic locus controlling a key step in maize evolution. Science 262, 233–235. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert RF, Russin WA. 1993. Structurally, phloem unloading in the maize leaf cannot be symplastic. American Journal of Botany 80, 1310–1317. [Google Scholar]
- Figueroa CM, Lunn JE. 2016. A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiology 172, 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. 2005. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology 15, 2038–2043. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. 1991. Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiology 97, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA. 2010. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. 2013. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Bledsoe SW, Siekman A, Kollman A, Waters BM, Feil R, Stitt M, Lagrimini LM. 2014. The trehalose pathway in maize: conservation and gene regulation in response to the diurnal cycle and extended darkness. Journal of Experimental Botany 65, 5959–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y, Narumi T, Oda A, Nakano Y, Sumitomo K, Fukai S, Hisamatsu T. 2013. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proceedings of the National Academy of Sciences, USA 110, 17137–17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch G. 2007. Cell wall hemicelluloses as mobile carbon stores in non-reproductive plant tissues. Functional Ecology 21, 823–834. [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, et al. . 2012. Comparative population genomics of maize domestication and improvement. Nature Genetics 44, 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-Y, Shannon LM, Tian F, et al. . 2012. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proceedings of the National Academy of Sciences, USA 109, E1913–E1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalt-Torres AW, Kerr PS, Usuda H, Huber SC, Carolina N. 1986. Diurnal changes in maize leaf photosynthesis: I. Carbon exchange rate, assimilate export rate, and enzyme activities. Plant Physiology 83, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. 2007. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J. 2004. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Research 32, 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D. 2010. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nature Genetics 42, 459–463. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. 2005. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proceedings of the National Academy of Sciences, USA 102, 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazakis CM, Coneva V, Colasanti J. 2011. ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. Journal of Experimental Botany 62, 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Matsumura Y, Soga K, Hoson T, Koizumi N. 2007. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant and Cell Physiology 48, 405–413. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Li C, Bradbury PJ, et al. . 2016. Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. The Plant Journal 86, 391–402. [DOI] [PubMed] [Google Scholar]
- Liu JQ, Allan DL, Vance CP. 2010. Systemic signaling and local sensing of phosphate in common bean: cross-talk between photosynthate and microRNA399. Molecular Plant 3, 428–437. [DOI] [PubMed] [Google Scholar]
- Lunardon A, Forestan C, Farinati S, Axtell MJ, Varotto S. 2016. Genome-wide characterization of maize small RNA loci and their regulation in the required to maintain repression6-1 (rmr6-1) mutant and long-term abiotic stresses. Plant Physiology 170, 1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mascheretti I, Battaglia R, Mainieri D, Altana A, Lauria M, Rossi V. 2013. The WD40-repeat proteins NFC101 and NFC102 regulate different aspects of maize development through chromatin modification. The Plant Cell 25, 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti I, Turner K, Brivio RS, Hand A, Colasanti J, Rossi V. 2015. Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiology 168, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. 2011. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. The Plant Cell 23, 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TA, Muslin EH, Dorweiler JE. 2008. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227, 1377–1388. [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. 2003. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. 2006. delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiology 142, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. 2008. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. The Plant Journal 53, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak GK, Lambert G, Bernat K, Rust MJ. 2015. Controlling the cyanobacterial clock by synthetically rewiring metabolism. Cell Reports 13, 2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O. 2010. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Polge C, Jossier M, Crozet P, Gissot L, Thomas M. 2008. Beta-subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities; physical interaction with nitrate reductase specifically occurs via AKINbeta1-subunit. Plant Physiology 148, 1570–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Romero Navarro JA, Willcox M, Burgueño J, et al. . 2017. A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nature Genetics 49, 476–480. [DOI] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, et al. . 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proceedings of the National Academy of Sciences, USA 104, 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. . 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nature Genetics 43, 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. 2001. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Swigoňová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. 2004. Close split of sorghum and maize genome progenitors. Genome Research 14, 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, et al. . 2010. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytologist 187, 57–66. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, McIntyre LM. 1999. Time to flowering in brown midrib mutants of maize: an alternative approach to the analysis of developmental traits. Heredity 83, 171–178. [DOI] [PubMed] [Google Scholar]
- Vermerris W, Thompson KJ, McIntyre LM, Axtell JD. 2002. Evidence for an evolutionarily conserved interaction between cell wall biosynthesis and flowering in maize and sorghum. BMC Evolutionary Biology 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wong AY, Colasanti J. 2007. Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. Journal of Experimental Botany 58, 403–414. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav UP, Ivakov A, Feil R, et al. . 2014. The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. Journal of Experimental Botany 65, 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li W, et al. . 2013. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proceedings of the National Academy of Sciences, USA 110, 16969–16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. 2009. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiology 149, 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.