Acquisition of secondary dormancy enables spatio-temporal control of germination. Evidence suggests that PHYD influences secondary dormancy induction by high temperature in Arabidopsis and correlates with removal of the germination repressor PIL5.
Keywords: Arabidopsis thaliana, germination, phenology, phytochrome, secondary dormancy, temperature, thermoinhibition
Abstract
Dormancy cycling controls the seasonal conditions under which seeds germinate, and these conditions strongly influence growth and survival of plants. Several endogenous and environmental signals affect the dormancy status of seeds. Factors such as time, light, and temperature influence the balance between abscisic acid (ABA) and gibberellic acid (GA), two phytohormones that play a key role in seed dormancy and germination. High temperatures have been shown to increase ABA level and prevent seed germination, a process known as thermoinhibition. High temperature can also cause the acquisition of secondary dormancy, preventing germination of seeds upon their return to favorable germination conditions. The mechanisms and conditions linking thermoinhibition and secondary dormancy remain unclear. Phytochromes are photoreceptors known to promote seed germination of many plant species including Arabidopsis thaliana. Here, we demonstrate a role for PHYD in modulating secondary dormancy acquisition in seeds exposed to high temperature. We found that a functional PHYD gene is required for the germination of seeds that experienced high temperature, and that ABA- and GA-related gene expression during and after pre-incubation at high temperatures was altered in a phyD mutant. We further show that the level of PHYD mRNA increased in seeds pre-incubated at high temperature and that this increase correlates with efficient removal of the germination repressor PIL5.
Introduction
Phenology, the timing of a plant’s life events such as germination and flowering, has a major impact on survival and fitness (Chuine and Beaubien, 2001; Walther et al., 2002; Parmesan, 2006; Bradshaw and Holzapfel, 2008; Donohue et al., 2010). Adaptive phenology ensures that each stage occurs during a season in which it can survive and progress to the next stage. For example, Arabidopsis thaliana (Arabidopsis) seeds can survive under hot, dry conditions, whereas the vegetative state cannot; rosettes are resistant to cold temperatures, whereas the reproductive stage is susceptible to freezing damage. To accommodate these vulnerabilities, seeds of winter annuals are dispersed in the spring but only germinate in the autumn, thus allowing seedlings to avoid summer drought. Similarly, flowering occurs after winter in the following spring, allowing reproductive buds to avoid freezing temperatures. These delays allow each life stage to avoid the seasonal conditions that could cause its mortality. Germinating at the appropriate time of year is the first crucial developmental transition that determines survival, and it is under strong natural selection in numerous plant species including Arabidopsis (reviewed in Donohue et al., 2010).
The timing of seed germination depends on the dynamics of dormancy induction and breakage, both of which are influenced by the environment. Dormancy is described as the absence of germination under conditions that would otherwise permit germination. It evolved to prevent germination under temporarily adequate but seasonably inappropriate conditions, such as a cool, wet day in an otherwise hot and dry summer (Baskin and Baskin, 1998; Fenner and Thompson, 2005; Finch-Savage and Leubner-Metzger, 2006). Primary dormancy is established during seed development and maturation (Hilhorst, 1995), and secondary dormancy refers to the reacquisition of dormancy in dispersed seeds that have lost their primary dormancy (reviewed in Bewley, 1997; Donohue, 2009). Dispersed seeds cycle between different depths of dormancy in response to several factors, including time (after-ripening) and environmental cues such as temperature, light, and nitrate concentration (Bewley et al., 2013; Footitt et al., 2013). Dormancy status influences the conditions under which germination can proceed, with less dormant seeds being able to germinate under a broader range of environmental conditions (Baskin and Baskin, 1998). In this manner, dormancy dynamics can precisely influence the timing and seasonal conditions of germination.
Two phytohormones—gibberellic acid (GA) and abscisic acid (ABA)—are the major regulators of dormancy. GA stimulates growth of the embryo by positively regulating processes such as cell division and elongation, mobilization of carbon and energy reserves, and weakening of seed barriers to facilitate radicle emergence (Ogawa et al., 2003; Finkelstein et al., 2008). In contrast, ABA inhibits GA action and is associated with the imposition and maintenance of primary and secondary dormancy (Yoshioka et al., 1998; Nambara and Marion-Poll, 2003; Ali-Rachedi et al., 2004; Gulden et al., 2004; Lin et al., 2007; Toh et al., 2008). Biosynthetic and catabolic enzymes dictate ABA and GA levels in seeds in response to developmental and environmental signals (Koornneef et al., 2002; Toh et al., 2012).
Much progress has been made in elucidating the genetic basis of primary dormancy established during seed maturation (reviewed in Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008), yet dormancy cycling, notably the induction of secondary dormancy, is less understood despite its critical role in the timing of germination under seasonally variable conditions. Both high and low temperatures are important regulators of dormancy in the seed bank (Baskin and Baskin, 1998; Probert, 2000; Fenner and Thompson, 2005). High temperature has a direct and indirect effect on seed germination: thermoinhibiton and secondary dormancy induction, respectively. Thermoinhibition refers to the inability of non-dormant seeds to germinate at high temperature. Thermoinhibited seeds can resume their germination program immediately following a return to favorable temperatures (Vidaver and Hsiao, 1975; Corbineau et al., 1988; Toh et al., 2008; Chiu et al., 2012). Secondary dormancy induction can be triggered by exposure to high temperature, but it is distinguished from thermoinhibition by the lack of germination following a return to favorable conditions. As such, exposure to thermoinhibitory temperature may or may not result in secondary dormancy induction. Conditions and molecular mechanisms linking thermoinhibition and secondary dormancy remain unclear. Oat (Avena sativa), barley (Hordeum vulgare), and Chenopodium bonus-henricus L. seeds that have been exposed to thermoinhibitory temperatures fail to germinate following the return of permissive temperatures (Khan and Karssen, 1980; Corbineau et al., 1988; Leymarie et al., 2008). In Arabidopsis, thermoinhibition occurs at temperatures >30 °C (Burghardt et al., 2016), and those temperatures have been shown to induce dormancy in partially dormant and completely non-dormant seeds (Auge et al., 2015). Thermoinhibition results from an increase in ABA level and is mediated by increased expression of ABA-related genes such as the transcription factor ABI5, the germination repressor SOMNUS (SOM) (Lim et al., 2013), and the ABA biosynthetic gene 9-cis-EPOXYCAROTENOID DIOXYGENASE9 (NCED) (Toh et al., 2008; Chiu et al., 2012; Lim et al., 2013).
In Arabidopsis and other species, non-dormant seeds require light and favorable temperature to initiate germination. Phytochromes are photoreceptors that sense red and far-red light, and are involved in numerous developmental processes, from germination to flowering (Schafer and Bowle, 2002; Casal et al., 2003). In non-dormant seeds, exposure to red light activates phytochromes, which degrade the basic helix–loop–helix (bHLH) repressor PIL5, thus promoting GA accumulation (increased biosynthesis and decreased catabolism) (Oh et al., 2006), and ABA reduction (Shinomura et al., 1994; Yamaguchi, 1998; Oh et al., 2004). In Arabidopsis, the phytochrome family is composed of five genes, PHYA–PHYE (Clack et al., 1994; Mathews and Sharrock, 1997), having both specific and overlapping activities. Each phytochrome has a different function in seed germination, with regard to responses to light quality (Ballaré et al., 1990; Casal and Sánchez, 2008) and temperature (Heschel et al., 2007, 2008; Donohue et al., 2008). Diversification of the function of individual members of a gene family can contribute to the precise environmental regulation of developmental processes. Elucidating the genetic pathways whereby specific members of gene families act on developmental responses to specific environmental cues contributes to an understanding of environmentally modulated development.
Here, we investigate the genetic pathways involved in secondary dormancy induction by high temperature and the contributions of individual members of the phytochrome gene family. Specifically, we aim to: (i) characterize the temperature dependence of secondary dormancy induction, and test whether disruption of different phytochromes alters secondary dormancy induction by high temperature; (ii) test the involvement of phytochromes in this response by measuring the expression of the different phytochromes at different temperatures of seed pre-incubation; (iii) determine the contributions of GA and ABA hormonal pathways to this response by measuring the expression of GA and ABA biosynthetic and catabolic genes at different temperatures of seed pre-incubation; and (iv) test whether the specific phytochrome identified in aims (i) and (ii), namely PHYD, influences the expression of dormancy-related genes, by measuring gene expression and the abundance of PIL5 protein in the wild-type and a phyD mutant.
Materials and methods
Plant growth conditions
Genotypes and sources of wild-type (wt) and phy mutant seeds were previously described (Donohue et al., 2008). All mutants were derived from the Landsberg erecta (Ler hereafter) background. phyA is phyA-1, phyB is phyB-1, phyD is phyD-1 (Aukerman et al., 1997), and phyE is phyE-1. The phyC mutant was not included in this study as it was not available to us, and PHYC was shown to be non-functional in the absence of other phytochromes (Hu et al., 2013).
Arabidopsis seeds of each genotype were planted in Metromix 360 (Scotts Sierra, Marysville, OH, USA) and cold-stratified for 5 d to induce synchronous germination. Twelve replicates of each genotype were randomly distributed within a growth chamber (EGC Model GC8-2 Plant Growth Chambers, Chagrin Falls, OH, USA) at 22 °C under long-day conditions (14 h light/10 h dark). When 70% of the seeds had matured, water was withheld for 2 weeks and seeds of all genotypes were harvested simultaneously. Seeds were collected, allowed to dry, and stored for 3 months (after-ripening) at ambient temperature before germination assays.
Germination assays
The first germination assay characterized temperature-dependent secondary dormancy induction in wt seeds. Wt Ler seeds that had after-ripened for 3 months were pre-incubated in the dark in the presence of water for 4 d at one of four temperatures: 4, 22, 32, or 34 °C. After pre-incubation in the dark, seeds were transferred to 22 °C in a 12 h photoperiod to assess germination.
The second germination assay tested wt and, phyA, phyB, phyD, and phyE mutants. Seeds were pre-incubated in the dark at 4, 22, or 32 °C before being transferred to a 12 h photoperiod at 22 °C to assess germination.
Germination assays were performed by placing 15 seeds in a Petri dish containing 0.5% agar. Each genotype×temperature assay used nine Petri dishes (three biological replicates×three technical replicates). Each Petri dish was sealed with parafilm, wrapped in aluminum paper, and incubated at the specified pre-incubation temperature in a germination chamber (Percival Scientific Inc., Perry, IA, USA) for 4 d. After the dark pre-incubation period, Petri dishes were unwrapped and transferred to a 22 °C chamber with a 12 h photoperiod. Seeds were examined once per day to assess radicle protrusion for up to 10 d, after which germination had plateaued. Seed viability was determined at the end of each experiment by assessing firmness to touch (Baskin and Baskin, 1998). The final germination percentage (number of germinants/number of viable seeds) of each Petri plate was used for all analyses.
Gene expression analysis
Seeds from three groups of 8–10 individual plants each were pooled into three biological replicates and after-ripened for 3 months. For each quantitative reverse transcription–PCR (qRT–PCR) time point analyzed, 25–50 mg of seeds of each replicate pool were deposited on a wet filter paper in 60 mm Petri dishes and incubated as described for the germination assays. Dry and pre-incubated seeds were collected at specific time points after the initiation of imbibition and flash-frozen in liquid nitrogen for RNA extraction. Seeds undergoing dark pre-incubation were collected in a dark room under green light. Frozen seeds were ground using a mortar and pestle in liquid nitrogen, and RNA was extracted following previously described protocols (Oñate-Sánchez and Vicente-Carbajosa, 2008), and further cleaned using a Qiagen RNeasy column following the manufacturer’s instructions (QIAGEN, CA, USA). qRT–PCR was performed using Applied Biosystems 1-step SYBR Green and the primers listed in Table 1. The transcript level was normalized to the TIP41-like gene (AT4G34270), whose expression was shown to remain stable throughout seed germination (Czechowski et al., 2005; Footitt et al., 2011).
Table 1.
Sequences of primers used for qRT–PCR analysis
| Name | TAIR | Orientation | Sequence (5'→3') |
|---|---|---|---|
| ABI5 | AT2G36270 | F R |
GGGAAGGAAAAGAGTAGTGGAT CCACTGTATATGCTTGTTTTCTT |
| TIP41-like | AT4G34270 | F R |
GTGAAAACTGTTGGAGAGAAGCAA TCAACTGGATACCCTTTCGCA |
| GA2ox2 | AT1G30040 | F R |
AATAACACGGCGGGTCTTCAAATCT TCCTCGATCTCCTTGTATCGGCTAA |
| GA3ox1 | AT1G15550 | F R |
CGACCACCCGGACGCGACTA TGCGTTGGACAGGTAGCCCG |
| GA3ox2 | AT1G80340 | F R |
ATTGGCTCTCCCCTCCACGATTT CCCGGCCCATTGTATGTCCTTTTC |
| NCED6 | AT3G24220 | F R |
TGATCTTCCTTACCAAGTGAAGA CTTAGGATGCGCTATCACTGAA |
| NCED9 | AT1G78390 | F R |
TAATGGTGTTCGTTCACGAC CTAACACAAAGCTTGCTTCG |
| PHYA | AT1G09570 | F R |
ACGCATTAGAAGGAACTGAGGAG GTAAGATCATGGGCTACAAAACACA |
| PHYB | AT2G18790 | F R |
GCAGTCCAACGGAGGCA CTTTAGCACAAATGAACCGTCTTC |
| PHYD | AT4G16250 | F R |
AGAGGAGGCAGGAGAGTGAA AATGAACCGTCATCTATGCTTTTG |
| PHYE | AT4G18130 | F R |
AAGCATTGAGGAAGGCAAG CATTGAGAGGCAGAGTTTTGA |
| PIL5 | AT2G20180 | F R |
TGAATCCCGTAGCGAGGAAACAA TTCCACATCCCATTGACATCATCTG |
| RD29A | AT5G52310 | F R |
AGTGATCGATGCACCAGGCGT CGGAAGACACGACAGGAAACACC |
| SOMNUS | AT1G03790 | F R |
AGCAATCAGCGTCTCCATCTCCAG TCAAGTCAAGAGATCATTGACCCATCC |
ABA-dependent transcript level
A 25–50 mg aliquot of Ler wt seeds (3 months after-ripened) was placed on a filter paper imbibed with 0, 10, or 100 µM ABA (mixed isomers, Sigma-Aldrich, St. Louis, MO, USA). ABA was dissolved in ethanol to a concentration of 100 mM, and subsequent dilutions were performed in water. The control treatment contained 0.0001% ethanol, equivalent to the amount of ethanol present in the 10 µM ABA solution. Plates were sealed with parafilm and incubated at 22 °C under a 12 h photoperiod. Seeds were collected 24 h later and RNA was extracted as described above. Transcript levels of PHYD and RD29a were quantified by qRT–PCR as described above.
Statistical analysis
To test for significant effects of pre-incubation temperature on germination rates and proportions, germination proportions at each time point were analyzed using logistic regression (PROC LOGISTIC in SAS 9.4; SAS Institute) using Fisher’s scoring optimization (ML) algorithm with Type-III likelihood ratio tests. The Firth’s penalized likelihood was used to accommodate issues of quasi-separation caused by extreme germination proportions (0 or 100%) in some treatments. The total number of germinants (successes)/the total number of viable seeds (trials) was the dependent variable for all analyses of germination proportions. To test whether phytochrome disruption altered germination responses to pre-incubation temperature, we tested for genotype×temperature interactions in logistic regression models of germination proportions, with genotype, temperature, and their interaction as factors; the wt was the reference genotype. To test whether pre-incubation temperature altered gene expression in wt seeds, normalized gene expression levels were analyzed with ANOVA, with time, temperature, and their interactions as fixed factors. To test whether ABA concentration altered gene expression of PHYD and RD29a, normalized gene expression was compared across ABA concentrations. To test whether wt and phyD genotypes differed in gene expression, normalized gene expression was compared between genotypes over time using ANOVA, with genotype, time, and their interaction included as fixed factors.
Protein extraction and western blot
Ler and phyD seeds were pre-incubated in the dark at 32 °C for 4 d as described in the germination assay, then transferred to permissive germination conditions (22 °C, 12 h photoperiod). Samples were collected at 0, 1, 2, 3, and 4 d following transfer to germination conditions. Seeds were ground using a mortar and pestle in liquid nitrogen. Proteins were extracted as described previously (Galvão et al., 2012; Qiu et al., 2015). Protein extracts were loaded on an SDS–polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad, CA, USA). Anti PIL5 (PIF1) polyclonal antibody (Qiu et al., 2015) was used to detect PIL5 protein levels. PIL5 protein band intensities were quantified using ImageJ software.
Results
Phytochromes B and D are necessary to prevent high temperature-induced secondary dormancy
To examine the effect of pre-incubation temperatures on secondary dormancy induction, seeds that had after-ripened for 3 months were pre-incubated in the dark at different temperatures, then transferred to permissive germination conditions (22 °C with a 12 h photoperiod) and monitored for germination by measuring radicle emergence. Ler wt seeds that had after-ripened for 3 months were used throughout these experiments since they have little residual primary dormancy compared with fresh seeds (see Supplementary Fig. S1 at JXB online).
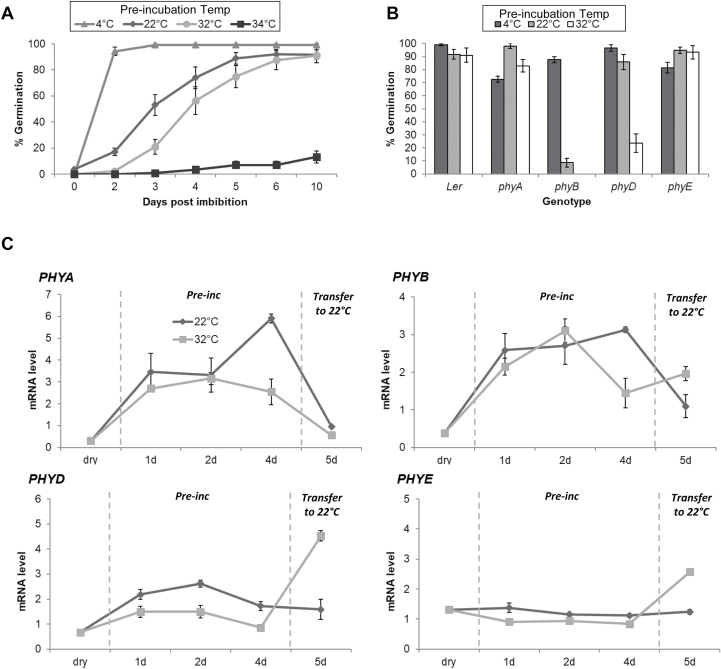
Ler seeds eventually germinated to almost 100% following pre-incubation at 4, 22, and 32 °C, but only to 10% following pre-incubation at 34 °C (Fig. 1A). This indicates that pre-incubation at 34 °C induced secondary dormancy in Ler wt seeds. A closer examination of the germination time course reveals that seeds pre-incubated at 4 °C germinated the fastest (within 2 d of light treatment), whereas seeds pre-incubated at 32 °C showed a 1 d delay in germination compared with seeds pre-incubated at 22 °C (Fig. 1A).
Fig. 1.
Role of phytochromes in germination at different temperatures. (A) Effect of temperature on germination. Ler wt seeds that had after-ripened for 3 months were pre-incubated on agar in the dark for 4 d at the indicated temperature (4, 22, 32, or 34 °C). After 4 d, seeds were transferred to 22 °C with a 12 h photoperiod, and radicle protrusion was monitored daily. Error bars represent the SE of nine replicates of 15 seeds each. The speed of germination depended on temperature (germination×temperature interaction: Wald χ2=36.46, P=0.006, df=18). (B) Effect of phytochrome mutants on germination. Ler wt seeds and phytochrome mutant seeds (after-ripened for 3 months) were pre-incubated on agar in the dark for 4 d at the indicated temperature (4, 22, or 32 °C). After 4 d, seeds were transferred to 22 °C and a 12 h photoperiod, and radicle protrusion was monitored after 10 d. Error bars represent the SE of nine replicates of 15 seeds each. Differences among genotypes depended on temperature (genotype×temperature interaction: Wald χ2=76.41, P<0.001, df=8; see Supplementary Table S1 for comparisons between Ler wt and mutant genotypes at each temperature). (C) Effect of temperature on PHY gene expression. Relative expression level of PHY genes in Ler wt seeds at different time points: dry seeds, 96 h pre-incubation (Pre-inc), and 24 h after transfer to permissive germination conditions (22 °C, 12 h photoperiod). mRNA levels were normalized to the AT4G34270 (TIP41-like) transcript. Error bars indicate the SE of three biological replicates. See Supplementary Table S2 for analysis of differences in transcript levels between temperature treatments.
We next compared the germination ability of seeds from different phytochrome mutants after dark pre-incubation at 4, 22, and 32 °C and subsequent transfer to 22 °C in the light. As expected from previous studies (Heschel et al., 2007; Donohue et al., 2008), phyB mutant seeds failed to germinate in all treatments except 4 °C (Fig. 1B; Supplementary Table S1). The phyA and phyE mutants behaved like wt Ler seeds under the conditions examined. Interestingly, the phyD mutant showed a <25% germination proportion after a 32 °C pre-incubation compared with >90% germination for wt seeds under the same conditions. Therefore, PHYB is necessary for germination across many temperatures, whereas PHYD is required for germination specifically after a 32 °C pre-incubation treatment.
Pre-incubation in the dark at 32 °C triggers PHYD expression
To test whether the contribution of PHYD to germination after pre-incubation at 32 °C in the dark could be mediated by an increase in PHYD gene expression, we compared PHYD mRNA transcript levels during and after pre-incubation at 22 °C and 32 °C. We also measured the transcript level of other phytochromes for comparison. Temperature had a small influence on PHYD transcript levels during dark pre-incubation, but a 4- to 5-fold up-regulation was observed following transfer to permissive germination conditions in seeds that had been pre-incubated at 32 °C (Fig. 1C; Supplementary Table S2). This increase in transcript level was not seen in seeds pre-incubated at 22 °C. Therefore, PHYD is specifically up-regulated after a hot, dark pre-incubation period upon transfer to permissive germination conditions (in the light). A second round of qRT–PCR focusing on later germination time points was performed to further characterize PHYD expression following hot pre-incubation. This assay confirmed that PHYD transcription increased following pre-incubation at 32 °C in the dark, peaking 24 h after transfer to permissive germination conditions (Supplementary Fig. S2). In contrast, PHYA and PHYB transcript levels increased during pre-incubation in the dark at both 22 °C and 32 °C, with seeds pre-incubated at 22 °C showing a >2-fold increase in transcript level in the dark compared with seeds pre-incubated at 32 °C (Fig. 1C; Supplementary Table S2). Interestingly, the PHYE transcript level also increased following hot dark pre-incubation, even though PHYE disruption did not alter germination after hot pre-incubation.
Combined, these results indicate that the transcription of phytochromes changes during dark pre-incubation at 32 °C. PHYA and PHYB are up-regulated in the dark at both temperatures, whereas PHYD expression is strongly up-regulated after pre-incubation at 32 °C and transfer to light and permissive temperature. Therefore, the involvement of PHYD in germination after hot pre-incubation appears to be, at least in part, influenced by its temperature-dependent expression.
Dark pre-incubation at 32 °C increases mRNA transcript levels of ABA-related genes and decreases those of GA-related genes
To identify the major hormonal pathways involved in germination responses to pre-incubation at high temperature, we used qRT–PCR to examine the transcript level of representative germination- and dormancy-associated genes within the ABA and GA pathways in wt seeds. For subsequent analysis, we compared transcript levels in seeds that were pre-incubated in the dark at 22 °C versus 32 °C and then transferred to permissive germination conditions for up to 4 d. As described above, PHYD expression increased upon transfer to light and 22 °C if seeds were pre-incubated in the dark at 32 °C, but it remained low in seeds pre-incubated in the dark at 22 °C (Figs 1C, 2A; Supplementary Table S3). ANOVAs revealed significant effects of temperature, time, and time×temperature interaction on transcript levels of most genes (Supplementary Table S3).
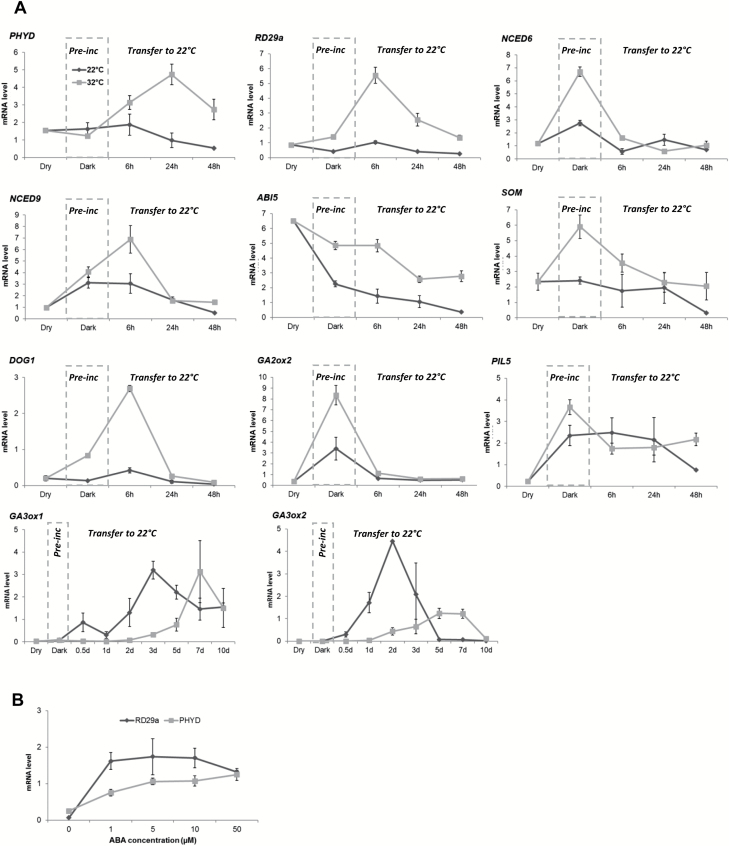
Fig. 2.
mRNA levels during and after pre-incubation in the dark at high temperature. (A) Effect of pre-incubation at 32 °C in the dark on mRNA levels in Ler wt seeds. The relative mRNA level was measured in dry seeds, 96 h after dark pre-incubation (Pre-inc) at 22 °C or 32 °C, and at 6, 24, and 48 h following transfer to permissive germination conditions at 22 °C and a 12 h photoperiod. mRNA levels of the focal genes were normalized to transcript levels of the reference gene TIP41-like (AT4G34270). For GA3ox1 and GA3ox2, time points (0.5d–10d) correspond to days after transfer to permissive germination conditions. See Supplementary Table S3 for analysis of differences in mRNA levels between temperature treatments. (B) The PHYD mRNA level is regulated by ABA. PHYD and RD29a mRNA levels in Ler wt seeds incubated at 22 °C for 24 h (12 h photoperiod) in the presence of 0, 1, 5, 10, and 50 µM ABA. mRNA levels were normalized to the reference gene, TIP41-like (AT4G34270) transcript. Error bars indicate the SE of three biological replicates. Test for effect of ABA concentration: PHYD F-value=11.95, P<0.001, df=4; RD29a F-value=7.47, P=0.005, df=4.
Because thermoinhibitory temperatures have been shown to induce a strong increase in ABA levels (Toh et al., 2008), we tested whether dark pre-incubation at 32 °C triggered an increase in ABA levels that persisted after transfer to permissive conditions, by measuring the transcript levels of the ABA reporter gene RD29a (Nakashima et al., 2006). As shown in Fig. 2A, we observed a >5-fold increase in RD29a transcript level following a 32 °C dark pre-incubation, consistent with an increase in ABA levels. Up-regulation of the ABA biosynthetic genes NCED6 and NCED9 could contribute to the increase in ABA (Fig. 2A). NCED6 showed increased expression during the dark pre-incubation phase and returned to the basal level after transfer to permissive germination conditions. This pattern contrasted with that of NCED9, whose expression increased only after transfer to permissive germination conditions following a period of pre-incubation at 32 °C. Nonetheless, both ABA biosynthesis genes were up-regulated by pre-incubation at 32 °C, consistent with the higher transcript level of RD29a at that pre-incubation temperature.
ABI5 encodes a transcription factor that mediates the effects of ABA. Its expression was high in dry seeds and gradually declined during pre-incubation and germination at both temperatures tested, but the decline was more pronounced in seeds pre-incubated at 22 °C compared with 32 °C, suggesting that ABA responsiveness would be higher in seeds pre-incubated at 32 °C. SOMNUS (SOM) is a key repressor of seed germination in the dark and has recently been shown to be up-regulated in thermoinhibited seeds by ABI3, ABI5, and the DELLA transcription complex (Lim et al., 2013). SOM gene expression was up-regulated in seeds pre-incubated at 32 °C in the dark. SOM then returned to basal levels following transfer to permissive germination conditions in seeds pre-incubated at both 22 °C and 32 °C.
Expression of DOG1 correlates with both primary (Teng et al., 2008; Chiang et al., 2011; Mortensen et al., 2011; Mortensen and Grasser, 2014) and secondary dormancy (Footitt et al., 2011, 2013). Recently, DOG1 was shown to be involved in an ABA-independent thermoinhibition (Huo et al., 2016). DOG1 expression was briefly up-regulated >2-fold following pre-incubation at 32 °C, peaking at 6 h after transfer to permissive germination conditions before returning to basal levels by 24 h.
The GA-catabolizing gene GA2ox2 showed increased expression during dark pre-incubation at 32 °C compared with 22 °C, suggesting that pre-incubation at high temperature leads to decreased GA content via GA degradation. GA3ox1 and GA3ox2 enzymes promote the synthesis of bioactive GA and thereby promote seed germination (Yamaguchi, 1998; Mitchum et al., 2006). Since seeds pre-incubated at 32 °C showed delayed germination (Fig. 1A), we extended our time course to 10 d after transfer to permissive germination conditions in order to capture full GA3ox1/2 expression patterns (Fig. 2A). Transcript levels of GA3ox1 increased around the seventh day after transfer to permissive germination conditions in seeds pre-incubated at 32 °C compared with the third day for seeds pre-incubated at 22 °C. A similar delay in GA3ox2 transcript accumulation was also observed, with a higher expression between the fifth and seventh day in seeds pre-incubated at 32 °C compared with the second day in seed pre-incubated at 22 °C. The shift in GA3ox1/2 expression levels reflects the delayed germination phenotype observed in seeds pre-incubated at 32 °C (Fig. 1A).
The major germination repressor PIL5 was up-regulated upon pre-incubation at both 22 °C and 32 °C, and its expression remained similar between both seed populations for the first 24 h following transfer to permissive germination conditions. At 48 h, PIL5 expression dropped in seeds pre-incubated at 22 °C but remained high in seeds pre-incubated at 32 °C, although this difference was not significant (Supplementary Table S3).
Taken together, these results suggest that pre-incubation at 32 °C enhanced ABA biosynthesis and GA degradation in the dark and led to delayed GA3ox1 and GA3ox2 expression upon transfer to permissive germination temperature. Thus, wt seeds pre-incubated at 32 °C seem to have acquired a weak secondary dormancy that was broken by transfer to light and permissive germination temperature.
ABA concentration correlates with PHYD expression level
Comparison of gene expression patterns of seeds pre-incubated at 32 °C, described above, suggests that an increase in ABA levels could induce PHYD expression that could then oppose secondary dormancy induction in wt seeds. To test this possibility, we measured PHYD transcript levels in the presence of different concentrations of ABA under permissive germination conditions (22 °C, 12 h photoperiod). As shown in Fig. 2B, the RD29a transcript level increased in the presence of ABA, indicating that exogenous ABA could penetrate the imbibing seeds and influence transcription. Interestingly, the PHYD transcript level correlated with ABA concentration, indicating that ABA can up-regulate PHYD expression independently of temperature.
PHYD modulates PIL5 protein level and germination-associated gene expression
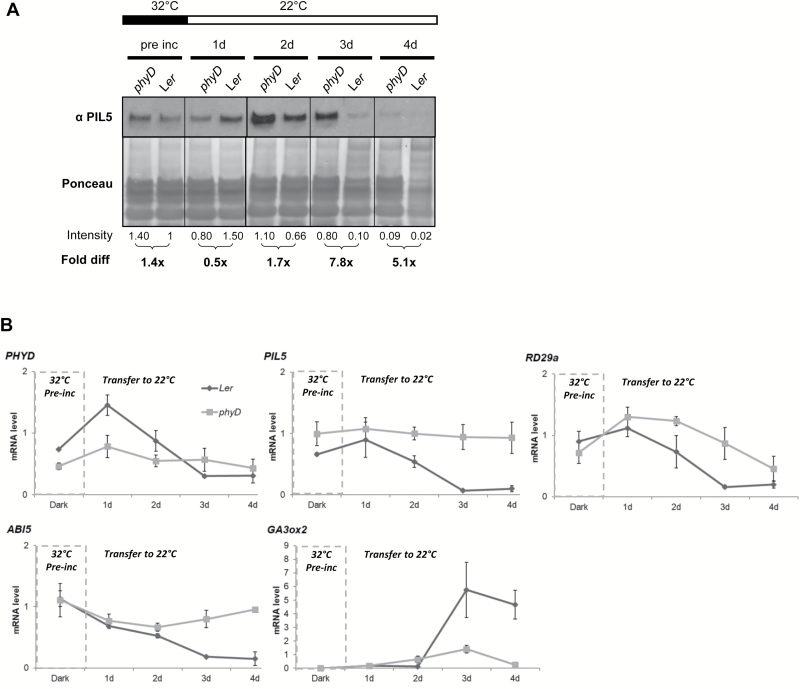
PHYA and PHYB promote germination by interacting with and inducing the degradation of the germination repressor PIL5 (Oh et al., 2004). To test if increased PHYD expression increased PIL5 degradation, we compared PIL5 protein levels in wt and phyD seeds pre-incubated in the dark at 32 °C. Since PHYD expression peaked 24 h after transfer to permissive germination conditions (Fig. 2A), we measured PIL5 protein levels during dark pre-incubation and after 1, 2, 3, and 4 d following transfer to permissive germination conditions. The PIL5 protein level was more slowly degraded in phyD seeds compared with wt seeds (Fig. 3A). PIL5 protein was still detected up to 3 d after transfer to permissive germination conditions in the phyD mutant, whereas it was barely detectable in wt seeds at a corresponding time point. This decrease in PIL5 protein in wt seeds after 3 d in permissive conditions is consistent with a pronounced increase in germination proportion of wt seeds at that time (Fig. 1A). Thus, functional PHYD appears to promote the degradation of PIL5 after pre-incubation at 32 °C.
Fig. 3.
Effect of phyD mutation on PIL5 protein stability and mRNA levels after 32 °C pre-incubation. (A) PHYD influences PIL5 levels. PIL5 levels in Ler and phyD mutant seeds during pre-incubation in the dark at 32 °C and 1, 2, 3, and 4 d after transfer to permissive germination conditions (22 °C, 12 h photoperiod). Intensity: relative intensity of PIL5 protein level compared with Ler pre-incubated sample (measured by ImageJ software). Fold diff: fold difference in PIL5 protein levels between Ler and phyD samples at each time point. (B) Effect of phyD mutation on mRNA levels. mRNA levels in Ler wt and phyD mutant seeds during pre-incubation in the dark at 32 °C and 1, 2, 3, and 4 d after transfer to permissive germination conditions (22 °C, 12 h photoperiod). mRNA levels were normalized to the TIP41-like (AT4G34270) transcript. Error bars indicate the SE of three biological replicates. See Supplementary Table S4 for results of analysis of differences in gene expression between Ler wt and phyD mutant genotypes.
To test whether PHYD influences the expression of germination-associated genes when seeds experienced pre-incubation at 32 °C, we examined the expression level of PHYD, PIL5, and a subset of downstream targets of PIL5 in Ler wt and phyD mutant seeds using qRT–PCR. PHYD expression was lower in the phyD mutant, as expected (Fig. 3B; Supplementary Table S4) (Aukerman et al., 1997). Interest ingly, the PIL5 transcript level remained higher in the phyD mutant compared with wt seeds after transfer to permissive germination conditions, suggesting that PHYD may directly or indirectly repress PIL5 expression. The expression of ABI5, a direct target of PIL5 (Oh et al., 2009), decreased over time in Ler wt seeds, but remained high in phyD mutant seeds, consistent with higher levels of PIL5 gene expression and higher levels of PIL5 protein in phyD seeds. ABA levels or sensitivity to ABA, as detected by the reporter gene RD29a, decreased more rapidly in wt than in phyD mutant seeds, also consistent with higher levels of PIL5 protein in phyD seeds. GA3ox2 expression increased at 3 d in wt seeds but remained very low in phyD seeds, consistent with the absence of germination in those seeds. GA3ox1 remained low in both wt and phyD seeds at the time points examined (Supplementary Fig. S3).
Taken together, these results suggest that increased PHYD expression influences ABA and GA responses by influencing PIL5 protein levels in seeds pre-incubated at high temperature, following their return to favorable germination conditions.
Discussion
Thermoinhibition is a process that occurs at high temperature to prevent germination of seeds under unfavorable temperatures. Secondary dormancy, in contrast, correspond to a lack of germination under favorable conditions. We show that exposure to high temperature triggered the establishment of secondary dormancy, impeding seed germination even after a return to permissive germination conditions. The depth of secondary dormancy in wt seeds was proportional to the temperature experienced. Pre-incubation at 32 °C appeared to induce a shallow dormancy that was overcome by permissive germination conditions, but pre-incubation at 34 °C induced a deeper dormancy that was not removed by transfer to permissive conditions. We showed that the ability to overcome shallow 32 °C-induced dormancy required a functional PHYD gene, as seeds with non-functional PHYD remained in a dormant state even after transfer to permissive conditions. PHYD reduced the amount of the germination repressor PIL5 protein following transfer to permissive conditions. When PHYD was non-functional (in phyD mutant seeds), PIL5 protein persisted in seeds pre-incubated at 32 °C following transfer to permissive germination conditions, and such seeds maintained higher ABA biosynthesis and decreased GA synthesis, leading to lower germination. Therefore, PHYD influences the upper temperature seeds can experience without entering deep secondary dormancy.
Effect of high temperature pre-incubation on gene expression
Previous studies have focused on thermoinhibition, in which seed germination at high temperature is prevented (Toh et al., 2008; Lim et al., 2013). To expand on these studies, we examined seeds transiently exposed to high temperatures, but subsequently returned to permissive germination conditions. This allowed the effect of high temperature on secondary dormancy induction to be studied. It is unclear how thermoinhibition and secondary dormancy processes are related and whether they are controlled by the same genes.
One may expect genes involved in thermoinhibition to be influenced by high temperature, but that their expression would return to previous levels once seeds are returned to favorable conditions. In contrast, genes involved in secondary dormancy induction and maintenance should remain or become differentially expressed even after seeds are returned to permissive germination conditions. We found that several genes previously associated with dormancy and thermoinhibition had altered gene expression in wt Ler seeds after exposure to high temperatures. Because wt seeds did eventually germinate after pre-incubation at 32 °C, it is unclear whether the observed delay in germination of wt seeds represents weak secondary dormancy that was broken by permissive conditions, or whether it represents thermoinhibition that persisted even after transfer to permissive conditions. The observation that wt seeds were induced into persistent secondary dormancy by pre-incubation at only a slightly higher temperature (34 °C) suggests that shallow dormancy is a likely possibility, and the continuum between persistent effects of thermoinhibition and secondary dormancy itself suggests that these processes are closely related.
After seeds were pre-incubated at high temperature, we detected an up-regulation of the ABA biosynthetic gene NCED6, the ABA signaling gene ABI5, the GA-degrading gene GA2ox2, and the dormancy-related genes SOM and DOG1. Interestingly, GA2ox2, NCED6, and SOM expression returned to basal levels as soon as seeds were placed under permissive germination conditions, suggesting a role in thermoinhibition but not in maintaining secondary dormancy. In contrast, ABI5 expression remained high during both the dark pre-incubation and germination phases in seeds pre-incubated at 32 °C, consistent with ABA’s role in controlling both thermoinhibition and secondary dormancy. Barley seeds induced into secondary dormancy by high temperatures were also shown to accumulate high levels of ABA both during high temperature (30 °C) exposure and following transfer to permissive germination conditions (20 °C) (Leymarie et al., 2008).
NCED6 and NCED9 are members of a multigene family regulating the key step in ABA biosynthesis. Although both genes have been shown to be expressed in Arabidopsis seeds, only the expression of NCED9 was shown to increase during thermoinhibition by Toh et al. (2008). Our results show that NCED6 expression is also up-regulated in response to high temperature in the dark. Up-regulation of NCED6 in dark-imbibed Cvi seeds had previously been correlated with increased dormancy (Cadman et al., 2006; Seo et al., 2006; Footitt et al., 2011). This pattern suggests that NCED6 is involved in dormancy cycling in the seed bank; that is, in buried seeds that have restricted light, whereas NCED9 is involved in the regulation of seed ABA levels in the light.
PIL5 is an important negative regulator of germination. We found similar levels of PIL5 transcript during dark pre-incubation and the first day after transfer to light, regardless of pre-incubation temperature, but a slightly (but not significantly) higher transcript at 48 h after transfer to permissive conditions. This observation, coupled with the observed accumulation of PIL5 protein in the phyD mutant (Fig. 3, and discussion below), suggests that PIL5 protein levels could be critical in bridging thermoinhibition and secondary dormancy acquisition.
Expression of the GA biosynthetic genes GA3ox1 and GA3ox2 is positively modulated by light and precedes radicle emergence (Yamaguchi, 1998). We found a delay in the expression of GA3ox1 and GA3ox2 in seeds pre-incubated at 32 °C compared with seeds pre-incubated at 22 °C following their transfer to light. This delay correlates with the delay in germination observed in those seeds (Fig. 1A) and suggests that 32 °C induces a shallow dormancy that is overcome upon transfer to permissive conditions, whereas pre-incubation at higher temperatures (34 °C) leads to induction of a deeper dormancy that cannot be broken by light and permissive temperature.
In sum, dormancy induction by high temperature involves both the up-regulation of ABA-mediated dormancy pathways and up-regulation of the major gene involved in GA catabolism, resulting in a delay in germination-promoting GA biosynthesis.
Role of PHYD
We found that a functional PHYD allele influenced the ceiling temperature that seeds can experience before acquiring secondary dormancy. Indeed, phyD mutant seeds failed to germinate after exposure to 32 °C, whereas wt seeds failed to germinate only after exposure to 34 °C, indicating that PHYD either prevents the establishment of secondary dormancy at 32 °C or can overcome the shallow secondary dormancy induced by this treatment. PHYD transcript remained low during dark pre-incubation regardless of temperature, but increased almost 3-fold in seeds pre-incubated at 32 °C after transfer to permissive germination conditions. This suggests that PHYD contributes to the removal of the shallow secondary dormancy established during pre-incubation at 32 °C. Previous studies have also reported a correlation between PHYD expression and dormancy levels in Cvi seeds (Cadman et al., 2006; Finch-Savage et al., 2007; Footitt et al., 2013).
The mechanism by which PHYD influences germination after pre-incubation at high temperature could be mediated by its influence on the germination repressor PIL5. PIL5 protein was degraded faster in Ler wt than in phyD mutant seeds pre-incubated at 32 °C following transfer to permissive germination conditions. Since wt seeds germinated to higher proportions (Fig. 1A), they are expected to contain lower levels of PIL5 protein because of the germination process (Lee et al., 2014), partially explaining the lower levels of PIL5 protein observed in wt seeds. However, quantification of the PIL5 protein level indicates that (i) a difference in PIL5 intensity is observed as early as day 2 when very few wt seeds had germinated; and (ii) the difference in PIL5 protein intensity at day 3 between the wt and phyD is six times higher than expected if the only difference were the percentage of non-germinated seeds. Indeed, only ~20% of wt seeds had germinated by day 3 (see Fig. 1A), which translates into a 1.25-fold increase in ungerminated seed compared with the wt. The difference in PIL5 protein intensity between phyD and wt samples at day 3, however, reveals a 7.8-fold increase in the former (i.e. more than six times that expected from germination alone). This suggests that although a portion of the decrease in PIL5 protein level can be attributed to seed germination per se, most of the difference is attributable to the genotypic difference between the seeds.
PHYA and PHYB proteins directly interact with and promote the degradation of PIL5 (Oh et al., 2004; Shen et al., 2008). PHYD is the closest homolog of PHYB, and its ability to promote PIL5 degradation alongside PHYA and PHYB has already been reported (Shen et al., 2008). We suggest that pre-incubation at high temperature leads to increased expression of PHYD, which is then able to induce the degradation of PIL5 protein, most probably by promoting its phosphorylation and subsequent ubiquitination. It is also possible that the higher PIL5 protein levels observed in phyD mutant seeds is caused by altered PIL5 gene expression, since PIL5 was expressed to slightly higher levels in phyD mutant seeds (Fig. 3). Interestingly, PIL5 protein eventually decreased to very low levels in phyD mutant seeds, even though the seeds never germinated, indicating that lack of germination is not solely due to the persistence of PIL5 protein. This is consistent with the low expression of PIL5 observed in deeply dormant seeds during winter time in the field (Footitt et al., 2011, 2013) and suggests that PIL5 provides a temporary block on germination but is not necessary for maintenance of the secondary dormant state.
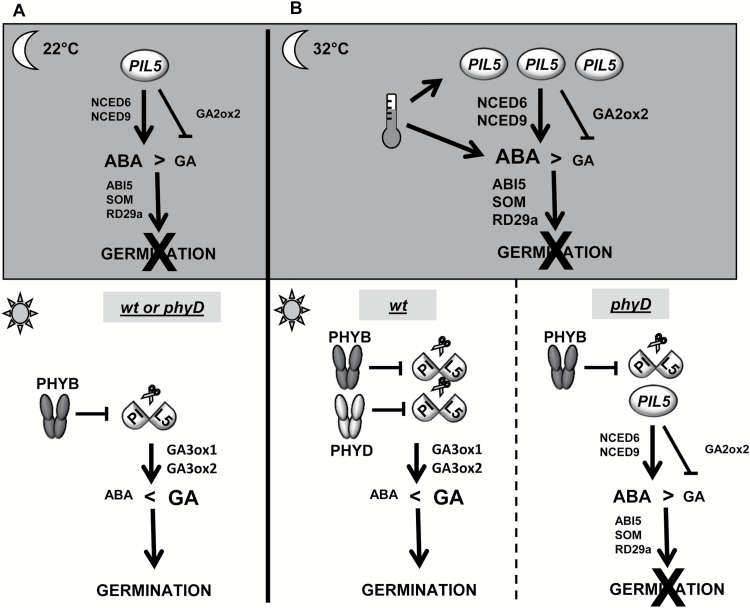
We propose the following model to explain the role of PHYD in secondary dormancy induction by high temperature (Fig. 4). Under a permissive temperature (22 °C), seed pre-incubation leads to the accumulation of the germination repressor PIL5, a mechanism believed to prevent premature germination in the dark. Upon transfer to light, PIL5 is degraded by PHYB, thereby reducing ABA biosynthesis and ABA response, and impeding GA catabolism, ultimately allowing germination. Seed pre-incubation at 32 °C leads to higher and/or more stable PIL5 proteins that cannot be entirely degraded by PHYB under permissive conditions. Increased PHYD expression following 32 °C pre-incubation provides additional phytochrome activity to fully overcome PIL5-imposed germination repression by increasing PIL5 protein degradation and potentially through direct or indirect repression of PIL5 gene expression, therefore allowing germination to proceed. PHYD could function as a homodimer or could heterodimerize with PHYB (Sharrock and Clack, 2004) to target PIL5, since PHYB is also necessary for germination following pre-incubation at 32 °C (Fig. 1B). In the absence of functional PHYD protein, the higher PIL5 protein level can delay germination until the establishment and/or consolidation of secondary dormancy. Whether PIL5 itself is necessary for secondary dormancy acquisition or maintenance of seeds pre-incubated at 32 °C or only allows a PIL5-independent pathway to complete secondary dormancy establishment remains to be tested. In addition, further study of interactions between PHYD and other genes involved in dormancy induction and maintenance would provide valuable information on the pathways whereby PHYD may contribute to the regulation of secondary dormancy. Additional studies of gene expression in phyD mutants and double mutant analysis would be especially informative.
Fig. 4.
Germination model. (A) Dark pre-incubation at 22 °C leads to accumulation of PIL5 protein, which leads to higher ABA levels (via NCED6 and NCED9 expression) and responsiveness (via ABI5 and SOM) and lower GA (via GA2ox2), which prevents germination. Upon transfer to light, PHYB degrades PIL5, allowing the accumulation of GA (via GA3ox1 and GA3ox2), which promotes germination. (B) Pre-incubation in the dark at high temperature (32 °C) leads to increased accumulation of PIL5 and/or PIL5 with higher stability, which strongly impedes germination via ABA accumulation and GA catabolism. Upon transfer to favorable germination conditions, PHYB and PHYD degrade PIL5, allowing GA accumulation and germination. PHYD might also repress the PIL5 gene expression level. In the absence of PHYD (phyD mutant), PIL5 remains active and blocks germination.
PHYD transcriptional regulation
We found that ABI5 expression was higher in seeds pre-incubated at 32 °C than in those pre-incubated at 22 °C at time points preceding PHYD expression. The ABI3/ABI5/DELLA protein complex was implicated in gene regulation in response to high temperature, including the expression of the transcription factor gene SOM (Lim et al., 2013). Although PHYD was not reported by Lim et al. (2013) as a putative target, it would be interesting to test whether the ABI3/ABI5/DELLA transcription complex is involved in its expression.
We found that ABA is directly implicated in the regulation of PHYD expression. We showed a positive correlation between ABA concentration and PHYD expression in seeds pre-incubated under favorable germination conditions, thus demonstrating that ABA can increase PHYD expression even in the absence of elevated temperature. This result is consistent with that of Okamoto et al. (2010), who reported an increase in PHYD and PHYE expression in seeds of genotypes that overproduced ABA.
PHYD expression increased only after transfer to permissive germination conditions in the light, suggesting that light is also needed for its up-regulation. A previous study suggests that light has little influence on phytochrome gene expression, including PHYD (Clack et al., 1994). This study was performed on seedlings, and it remains possible that light regulation of phytochrome expression during seed germination was overlooked. In our experiments, the transfer to permissive germination conditions entailed altering both light (dark to light) and temperature (32 °C to 22 °C) simultaneously. Specifically, decoupling light from permissive germination temperature would permit a test of whether PHYD expression in seeds pre-incubated at high temperature requires light.
Regulation of environment-dependent germination by duplicated genes
This study shows that phytochromes are involved in the ability to germinate after pre-incubation at high temperatures. PHYD is the most similar phytochrome gene to PHYB (Clack et al., 1994; Mathews and Sharrock, 1997), yet it exhibits its own distinctive pattern of gene expression and temperature-dependent phenotypic effects. Whereas both PHYB and PHYD are required for germination after pre-incubation at 32 °C, only PHYB contributes to germination at other temperatures; PHYD disruption does not alter germination at lower temperatures (Heschel et al., 2007, 2008; Donohue et al., 2008). PHYD could be a major determinant of dormancy cycling dynamics in temperate climates in which hot summers are frequent, as it influences secondary dormancy induction by high temperatures. In this study, we showed that different phytochromes have distinctive patterns of gene expression in response to environmental cues, and that their contribution to the phenotype of germination depends on environmental conditions. We did not investigate the role of PHYC in high temperature germination as it was shown to be non-functional in the absence of other phytochromes (Clack et al., 2009; Hu et al., 2013). PHYC has since been shown to modulate PHYA activity during far-red-mediated germination (Arana et al., 2014), and it would be interesting to test whether PHYC also influences germination regulation following high temperature exposure. Additional studies are necessary to compare the pathways whereby specific phytochromes may independently influence germination. This study, however, demonstrates that part of the diversification of germination regulation by specific phytochromes occurs at the level of gene expression.
In conclusion, we showed that PHYD is required to relieve the shallow dormancy imposed by exposure to 32 °C. Induction of secondary dormancy following exposure to high temperature shares a partial genetic basis with thermoinhibition, in that increased ABA biosynthesis, increased expression of an ABA response gene, and increased GA catabolism appear to be involved in both responses. Seeds’ ability to germinate after a return to permissive temperatures is associated with the degradation of PIL5 protein. PHYD has a distinctive pattern of gene expression in response to pre-incubation at high temperature and appears specifically to impede the maintenance of dormancy after hot pre-incubation. This highlights a functional diversification of phytochrome expression and function that can contribute to fine-tuned responses of germination to complex environmental scenarios.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Tests for differences between Ler wild-type and mutant phytochrome genotypes at different temperatures.
Table S2. Tests for effect of temperature on transcript level over time.
Table S3. Tests for effect of temperature on transcript level during and after high temperature pre-incubation in the dark.
Table S4. Tests for differences between Ler wild-type and the phyD mutant in transcript levels over time.
Fig. S1. Primary dormancy in fresh and 3 months after-ripened (AR) seeds of Ler wild-type and phyD mutant genotypes.
Fig. S2. PHYD transcript level in Ler wild-type seeds pre-incubated at two temperatures.
Fig. S3. GA3ox1 transcript level following high temperature pre-incubation.
Acknowledgements
We thank Dr Meina Li, Dr Meng Cheng, and Katherine Kovach for technical support, and the staff of the Duke Phytotron Facility for care of plants. This work was funded by grants NSF-DEB-1020963 and NSF-IOS-11-46383 to KD.
References
- Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M. 2004. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219, 479–488. [DOI] [PubMed] [Google Scholar]
- Arana MV, Sánchez-Lamas M, Strasser B, Ibarra SE, Cerdán PD, Botto JF, Sánchez RA. 2014. Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant, Cell and Environment 37, 2014–2023. [DOI] [PubMed] [Google Scholar]
- Auge GA, Blair LK, Burghardt LT, Coughlan J, Edwards B, Leverett LD, Donohue K. 2015. Secondary dormancy dynamics depends on primary dormancy status in Arabidopsis thaliana. Seed Science Research 25, 230–246. [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. 1997. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. The Plant Cell 9, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Sánchez RA. 1990. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 247, 329–332. [DOI] [PubMed] [Google Scholar]
- Baskin CC, Baskin JM. 1998. Seeds: ecology, biogeography, and evolution of dormancy and germination. London: Academic Press. [Google Scholar]
- Bewley JD. 1997. Seed germination and dormancy. The Plant Cell 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H. 2013. Dormancy and the control of germination. In: Seeds: physiology of development, germination and dormancy, 3rd edn New York: Springer, 247–295. [Google Scholar]
- Bradshaw WE, Holzapfel CM. 2008. Genetic response to rapid climate change: it’s seasonal timing that matters. Molecular Ecology 17, 157–166. [DOI] [PubMed] [Google Scholar]
- Burghardt LT, Metcalf CJ, Donohue K. 2016. A cline in seed dormancy helps conserve the environment experienced during reproduction across the range of Arabidopsis thaliana. American Journal of Botany 103, 47–59. [DOI] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. 2006. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal 46, 805–822. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Luccioni LG, Oliverio KA, Boccalandro HE. 2003. Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Dedicated to Professor Silvia Braslavsky, to mark her great contribution to photochemistry and photobiology particularly in the field of photothermal methods. Photochemical and Photobiological Sciences 2, 625. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Sánchez RA. 2008. Phytochromes and seed germination. Seed Science Research 8, 317–329. [Google Scholar]
- Chiang GC, Bartsch M, Barua D, et al. . 2011. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Molecular Ecology 20, 3336–3349. [DOI] [PubMed] [Google Scholar]
- Chiu RRS, Nahal H, Provart NNJ, et al. . 2012. The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biology 12, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I, Beaubien EG. 2001. Phenology is a major determinant of tree species range. Ecology Letters 4, 500–510. [Google Scholar]
- Clack T, Mathews S, Sharrock RA. 1994. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Molecular Biology 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. 2009. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix–loop–helix transcription factor. The Plant Cell 21, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbineau F, Rudnicki RM, Come D. 1988. Induction of secondary dormancy in sunflower seeds by high temperature. Possible involvement of ethylene biosynthesis. Physiologia Plantarum 73, 368–373. [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K. 2009. Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society B: Biological Sciences 364, 1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Heschel MS, Butler CM, Barua D, Sharrock RA, Whitelam GC, Chiang GC. 2008. Diversification of phytochrome contributions to germination as a function of seed-maturation environment. New Phytologist 177, 367–379. [DOI] [PubMed] [Google Scholar]
- Donohue K, Rubio de Casas R, Burghardt L, Kovach K, Willis CG. 2010. Germination, postgermination adaptation, and species ecological ranges. Annual Review of Ecology, Evolution, and Systematics 41, 293–319. [Google Scholar]
- Fenner M, Thompson K. 2005. Seed dormancy. In: The ecology of seeds. Cambridge: Cambridge University Press, 97–109. [Google Scholar]
- Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. 2007. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. The Plant Journal 51, 60–78. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–23. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. 2011. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proceedings of the National Academy of Sciences, USA 108, 20236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Huang Z, Clay HA, Mead A, Finch-Savage WE. 2013. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. The Plant Journal 74, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M. 2012. Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes and Development 26, 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden RH, Thomas AG, Shirtliffe SJ. 2004. Secondary dormancy, temperature, and burial depth regulate seedbank dynamics in canola. Weed Science 52, 382–388. [Google Scholar]
- Heschel MS, Butler CM, Barua D, Chiang GCK, Wheeler A, Sharrock RA, Whitelam GC, Donohue K. 2008. New roles of phytochromes during seed germination. International Journal of Plant Sciences 169, 531–540. [Google Scholar]
- Heschel MS, Selby J, Butler C, Whitelam GC, Sharrock RA, Donohue K. 2007. A new role for phytochromes in temperature-dependent germination. New Phytologist 174, 735–741. [DOI] [PubMed] [Google Scholar]
- Hilhorst HWM. 1995. A critical update on seed dormancy. I. Primary dormancy. Seed Science Research 5, 61–73. [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC. 2013. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proceedings of the National Academy of Sciences, USA 110, 1542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H, Wei S, Bradford KJ. 2016. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proceedings of the National Academy of Sciences, USA 113, E2199–E2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Karssen CM. 1980. Induction of secondary dormancy in Chenopodium bonus-henricus L. seeds by osmotic and high temperature treatments and its prevention by light and growth regulators. Plant Physiology 66, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. 2002. Seed dormancy and germination. Current Opinion in Plant Biology 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Lee N, Kang H, Lee D, Choi G. 2014. A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. The Plant Journal 78, 282–293. [DOI] [PubMed] [Google Scholar]
- Leymarie J, Robayo-Romero ME, Gendreau E, Benech-Arnold RL, Corbineau F. 2008. Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds. Plant and Cell Physiology 49, 1830–1838. [DOI] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, et al. . 2013. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. The Plant Cell 25, 4863–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. 2007. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiology 143, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA. 1997. Phytochrome gene diversity. Plant, Cell and Environment 20, 666–671. [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP. 2006. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. The Plant Journal 45, 804–818. [DOI] [PubMed] [Google Scholar]
- Mortensen SA, Grasser KD. 2014. The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Letters 588, 47–51. [DOI] [PubMed] [Google Scholar]
- Mortensen SA, Sønderkær M, Lynggaard C, Grasser M, Nielsen KL, Grasser KD. 2011. Reduced expression of the DOG1 gene in Arabidopsis mutant seeds lacking the transcript elongation factor TFIIS. FEBS Letters 585, 1929–1933. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Molecular Biology 60, 51–68. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2003. ABA action and interactions in seeds. Trends in Plant Science 8, 213–217. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. 2003. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. 2009. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. The Plant Cell 21, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. 2004. PIL5, a phytochrome-interacting basic helix–loop–helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. The Plant Cell 16, 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G. 2006. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. The Plant Journal 47, 124–139. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, et al. . 2010. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. The Plant Journal 62, 39–51. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C. 2006. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology Evolution and Systematics 37, 637–669. [Google Scholar]
- Probert RJ. 2000. The role of temperature in the regulation of seed dormancy and germination. In: Fenner M, ed. Seeds: the ecology of regeneration in plant communities. Wallingford, UK: CABI Publishing, 261–292. [Google Scholar]
- Qiu Y, Li M, Pasoreck EK, et al. . 2015. HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. The Plant Cell 27, 1409–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Bowle C. 2002. Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Reports 3, 1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, et al. . 2006. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. The Plant Journal 48, 354–366. [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. 2004. Heterodimerization of type II phytochromes in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 11500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. The Plant Cell 20, 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. 1994. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiology 104, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Rognoni S, Bentsink L, Smeekens S. 2008. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. The Plant Journal 55, 372–381. [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, et al. . 2008. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiology 146, 1368–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y. 2012. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant and Cell Physiology 53, 107–117. [DOI] [PubMed] [Google Scholar]
- Vidaver W, Hsiao AI. 1975. Secondary dormancy in light-sensitive lettuce seeds incubated anaerobically or at elevated temperature. Canadian Journal of Botany 53, 2557–2560. [Google Scholar]
- Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJ, Fromentin JM, Hoegh-Guldberg O, Bairlein F. 2002. Ecological responses to recent climate change. Nature 416, 389–395. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. 1998. Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. The Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Endo T, Satoh S. 1998. Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant and Cell Physiology 39, 307–312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.