The histone acetyltransferase GCN5 modulates stem cuticular wax accumulation by acetylating the key wax biosynthetic enzyme gene CER3 in Arabidopsis.
Keywords: Arabidopsis thaliana, CER3, GCN5, histone acetylation, stem, wax biosynthesis
Abstract
Cuticular wax is a major component of the surface cuticle of plants, which performs crucial functions in optimizing plant growth. Histone acetylation regulates gene expression in diverse biological processes, but its role in cuticular wax synthesis is not well understood. In this study, we observed that mutations of the Arabidopsis thaliana histone acetyltransferase GENERAL CONTROL NON-REPRESSED PROTEIN5 (GCN5) impaired the accumulation of stem cuticular wax. Three target genes of GCN5, ECERIFERUM3 (CER3), CER26, and CER1-LIKE1 (CER1-L1), were identified by RNA-seq and ChIP assays. H3K9/14 acetylation levels at the promoter regions of CER3, CER26, and CER1-L1 were consistently and significantly decreased in the gcn5-2 mutant as compared to the wild-type. Notably, overexpression of CER3 in the gcn5-2 mutant rescued the defect in stem cuticular wax biosynthesis. Collectively, these data demonstrate that GCN5 is involved in stem cuticular wax accumulation by modulating CER3 expression via H3K9/14 acetylation, which underlines the important role of histone acetylation in cuticular wax biosynthesis.
Introduction
Plant cuticular wax is a complex mixture of very-long-chain fatty acids (VLCFAs) and aldehydes, alcohols, alkanes, ketones, and esters, with predominant carbon chain-lengths ranging from C22 to C36 (Samuels et al., 2008; Li et al., 2016), and it forms one of the major lipid components of the cuticle that covers the outer surface of aerial plant tissues. The biosynthesis of cuticular wax is processed through two distinct pathways, termed the alcohol-forming and the alkane-forming pathways, which yield 17~18% and 80% of the total amount of wax, respectively (Bernard and Joubès, 2013). As a crucial adaptive characteristic, cuticular wax protects plants against biotic and abiotic stresses, such as pathogen attacks and water loss (Aharoni et al., 2004; Samuels et al., 2008). Therefore, elucidating the regulatory mechanisms controlling cuticular wax accumulation is of great interest for genetic engineering of agricultural crops.
To date, numerous candidate genes in the cuticular wax pathway have been identified, most of which encode enzymes, or work with enzymes, in the VLCFA biosynthesis and derivatization pathways (Koornneef et al., 1989; Bernard and Joubès, 2013). For example, CER2, and its homologues CER2-LIKE1/CER26 and CER2-LIKE2 have biological functions in two-carbon elongation of VLCFAs, from C28 to C30, C30 to C32, and C32 to C34, respectively (Haslam et al., 2012, 2015; Pascal et al., 2013; Lee and Suh, 2015). A member of the bifunctional wax ester synthase/diacylglycerol acyltransferase family, WSD1, is a key enzyme in wax ester synthesis of Arabidopsis stems (Li et al., 2008). As for the essential alkane-forming pathway, CER3/WAX2 plays an important role in synthesis of major wax components (Aarts et al., 1995; Chen et al., 2003; Rowland et al., 2007; Lee and Suh, 2015). The amount of wax found in the cer3 mutant was severely reduced compared with the wild-type in Arabidopsis stems, especially with regards to aldehydes, alkanes, secondary alcohols, and ketones (Rowland et al., 2007). Although the exact reactions catalysed by the CER3 enzyme remain unknown, it has been reported that CER3 may physically interact with CER1 and CYTOCHROME B5 ISOFORM (CYTB5) for the biosynthesis of very-long-chain alkanes (Bernard et al., 2012).
An increasing focus on the transcriptional regulation of the genes in cuticular wax biosynthesis has led to some transcription factors being reported in recent studies (Lee and Suh, 2015). In Arabidopsis, WAX INDUCER1/SHINE1 (WIN1/SHN1), an AP2-EREBP-type transcription factor, was the first to be identified and is a representative regulator of wax biosynthesis, regulating the CER1, CER2, and 3-KETOACYL-COA SYNTHASE1 (KCS1) genes (Aharoni et al., 2004; Broun et al., 2004). WIN1 overexpression lines exhibited enhanced drought tolerance compared with the wild-type (Aharoni et al., 2004). In addition, MYB transcription factors are important for wax biosynthesis under both biotic and abiotic stresses (Suh et al., 2005; Lee and Suh, 2015). For example, MYB96 controls wax biosynthesis by regulating the KCS1, KCS2, KCS6, BETA-KETOACYL REDUCTASE1 (KCR1), and CER3 genes under drought stress (Lee et al., 2016b), and KCS1, KCR1, CER2, and CER3 are the targets of MYB30 in response to pathogen attack (Raffaele et al., 2008).
With our increasing understanding of epigenetic mechanisms, recent reports have demonstrated that several epigenetic events are involved in wax biosynthesis (Lee and Suh, 2013, 2015). Recently, two RING E3 ligases, HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2, were demonstrated to be involved in wax biosynthesis by monoubiquitinating histone H2B proteins, which in turn activates the transcriptional levels of the wax biosynthetic genes LONG-CHAIN ACYL-COA SYNTHETASE2 (LACS2) and CER1 (Ménard et al., 2014). The Arabidopsis histone methyl transferases SET DOMAIN GROUP8 (SDG8) and SDG25 have been reported to contribute to wax accumulation through histone lysine methylation and/or H2B ubiquitination by targeting the key wax biosynthetic gene CER3 (Lee et al., 2016a). However, to the best of our knowledge, histone-acetylating events have not been reported in cuticular wax biosynthesis.
The reversible modulations of histone acetylation and de-acetylation are catalysed by histone acetyltransferases (HATs) and histone de-acetylases (HDACs), respectively (Pandey et al., 2002). As in yeast and mammals, the Arabidopsis HATs are grouped into four classes: GNAT (GCN5-related N-acetyltransferase), MYST (for ‘MOZ, Ybf2/Sas2 and Tip60’), p300/CBP (p300/CREB-binding protein), and TAF1 [for ‘TATA-binding protein (TBP)-associated factor’] (Servet et al., 2010). GCN5, a GNAT-type HAT that harbors a HAT domain and a bromodomain, has been well studied in extensive research (Benhamed et al., 2008; Servet et al., 2010). It has been observed that mutations of GCN5 result in various growth impairments, such as dwarfism, defects in terminal flower production, deformed seed development, and poor fertility (Bertrand et al., 2003; Vlachonasios et al., 2003). Moreover, GCN5 regulates a variety of biological processes in Arabidopsis, including cell differentiation, shoot and floral meristem formation, light and abiotic stress (e.g. heat and cold) responses, iron homeostasis, and fatty acid biosynthesis, by catalysing histone acetylation levels of target promoters at certain sites, including H3K14, H3K9, and H3K27 (Laux et al., 1996; Stockinger et al., 2001; Bertrand et al., 2003; Vlachonasios et al., 2003; Benhamed et al., 2006; Earley et al., 2007; Hu et al., 2015; Xing et al., 2015; Wang et al., 2016). In this current study, we found that the gcn5 mutants had significantly glossy (wax-deficient) stems compared with the wild-type. Analyses of chemical components coupled with scanning electron microscopy showed an obvious reduction in amounts of total wax and changes in their composition. Moreover, we found that GCN5 bound to the promoter of CER3 and this interaction was impaired in the gcn5-2 mutant. Taken together, we conclude that GCN5-mediated histone acetylation of CER3 regulates stem cuticular wax biosynthesis in Arabidopsis.
Materials and methods
Plant material and growth conditions
The Arabidopsis thaliana wild-types Col-0 and Ws, together with T-DNA insertion mutants involved in histone modifications were used in this study, as follows. (1) Histone acetylation: gcn5-1 and gcn5-2 (Ws background), hda2, hda2c, hda5, hda7, hda9, hda13, hda18, hda19, and srt2 (Col-0 background); and (2) histone methylation: ashh1, ashh2, ashh3, ashh4, ashr2, ashr3, atx1, atx2, atx4, atx5, atxr2, atxr3, and atxr4 (Col-0 background). The gcn5-1 and gcn5-2 mutants were both T-DNA insertion mutants in the bromodomain-coding region (Bertrand et al., 2003; Vlachonasios et al., 2003; Supplementary Fig. S3A at JXB online). Notably, the gcn5-2 mutation removes the entire bromodomain, which is required for binding to 11% of the GCN5 promoter targets (Servet et al., 2010). The other mutants were obtained in the homozygous state from ABRC (https://abrc.osu.edu/) or from individual donors. For germination, sterilized seeds were incubated at 4 °C for 3 d, and subsequently sown on Murashige and Skoog (MS) plates containing 1% sucrose and 0.6% agar. The seedlings were grown under 16/8 h light/dark conditions at 22 °C in a growth room.
RNA isolation and RNA-seq
Total RNA was extracted using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. RNA concentrations were measured using a NanoDrop 2000 spectrophotometer (ND-2000, ThermoFisher Scientific, Inc., MA, USA). RNA integrity was assessed using an Agilent 2100 Bioanalyser (Agilent Technologies, Inc., CA, USA). Paired-end sequencing libraries with an average insert size of 200 bp were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, USA) and sequenced using a HiSeq2500 platform (Illumina, San Diego, USA) according to the manufacturers’ standard protocols. Raw data obtained from Illumina sequencing were processed and filtered using the Illumina pipeline (http://www.illumina.com) to generate FastQ files. Approximately 12 G of high-quality 125-bp paired-end reads were generated from six libraries (Supplementary Table S1). The FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to evaluate the overall quality of the RNA-seq reads. Poor-quality bases were filtered out using Sickle (https://github.com/najoshi/sickle). High-quality RNA-seq reads from each library were mapped to The Arabidopsis Information Resource (TAIR10) version of the Arabidopsis genome using the splice-junction-aware short-read alignment suite TOPHAT v2.09 with default settings (Kim and Salzberg, 2011). The reads displaying unique alignment and not more than two nucleotide mismatches were kept for further analysis. The differentially expressed genes were identified by using the edgeR package (ver. 3.2.3) with an absolute value of log2-fold change ≥2 and a false-discovery rate <0.05 as cut-off (Robinson et al., 2010). The groups of differentially expressed genes identified by RNA-seq in this study are shown in Supplementary Table S2. Gene Ontology (GO) analysis was performed using agriGO v2.0 with a cut-off of P-value <0.05 (Tian et al., 2017), and the total enrichment categories are identified in Supplementary Table S3.
Quantitative real-time PCR
Real-time PCR was performed as previously described (Livak and Schmittgen, 2001) and ACTIN8 was used as the control gene, i.e. the expression levels of each gene were normalized to that of ACTIN8. The primer pairs used for real-time PCR are listed in Supplementary Table S4. The PCR analysis was performed using a CFX96 System (Bio-Rad) with SYBR Green. The following program was used for the real-time PCR: 95 °C for 3 min and 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s.
Plasmid construction and plant transformation
A DNA fragment containing a 2.0-kb fragment upstream of the CER3 coding sequence and full-length ORFs of GCN5 and CER3 were amplified by PCR-directed cloning based on the annotation from TAIR using the following primer pairs: CER3-P-F and CER3-P-R, GCN5-F and GCN5-R, CER3-F and CER3-R, respectively (Supplementary Table S4). The sequence-confirmed clones containing the ORFs of GCN5 and CER3 were then respectively cloned into the binary expression vector pCAMBIA1300 (driven by the CaMV35S promoter). The promoter region of CER3 was fused to the reporter gene encoding β-glucuronidase (GUS). The chimeric gene was then cloned into the binary expression vector pCAMBIA1300 to generate ProCER3::GUS. These vectors were transferred into the Agrobacterium tumefaciens strain GV3101. Transgenic plants were generated using the floral dip method and subsequently screened on solid plates containing 25 mg l–1 hygromycin (Clough and Bent, 1998). The hygromycin-resistant seedlings were then transferred to a mixture of soil and vermiculite (2:1). At least three independent T3 homozygous lines with a single T-DNA insertion were subjected to a detailed analysis. Because the gcn5-2 mutant exhibits low-fertility pollen that hinders the direct acquisition of transgenic plants in the gcn5-2 background (Bertrand et al., 2003; Xing et al., 2015), we initially generated transgenic plants in the Ws background, and three independent transgenic T3 lines were selected for crossing into the gcn5-2 mutant. The homologous transgenic lines in the gcn5-2 background were selected using the same method described above.
ChIP assay analysis
ChIP assays were performed using the Magna ChIP™ HiSens Kit (Catalogue No. 17-10460) combined with the ChIP method as previously described by Fiil et al. (2008). Six-week-old stems of Ws and gcn5-2 mutants were harvested and fixed in 1% formaldehyde for 15 min in a vacuum and subsequently neutralized using 0.125 M glycine for 5 min. After washing with sterilized water, the samples were dried with towels, and ground in liquid nitrogen. The resulting powders were resuspended in the Nuclei Extraction Buffer 1, which contained 0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mM PMSF (Sigma, P7626), and protease inhibitors (Roche, 11873580001), and mixed immediately. After incubation for 20 min at 4 °C with a rotator, the solutions were filtered through four layers of Miracloth into new tubes, and the filtrate was then centrifuged for 20 min at 3000 g at 4 °C. The nuclei pellets were resuspended in Nuclei Extraction Buffer 2, which contained 0.25 M sucrose, 10 mM Tris-HCl, pH8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors. The suspensions were transferred to microfuge tubes and centrifuged at 12 000 g for 10 min at 4 °C. The pellets were resuspended in Nuclei Extraction Buffer 2 and centrifuged at 12 000 g through a layer of Nuclei Extraction Buffer 3, which contained 1.7 M sucrose, 10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors, in microfuge tubes for 60 min at 4 °C. The nuclear pellets were lysed in Nuclei Lysis Buffer, which contained 50 mM Tris-HCl, pH8.0, 10 mM EDTA, 1% SDS, and protease inhibitors. The lysed nuclei were sonicated four times with a Bioruptor (UCD-200) in a water bath at 4 °C, with each sonication period consisting of 15 s on and 15 s off for 5 min and followed by centrifugation. The clear supernatants, which contained the sonicated chromatin, were transferred to new tubes. Immunoprecipitation (IP) was performed using 5 μl of chromatin, and the following IP steps were conducted using the Magna ChIP™ HiSens Kit. Aliquots of the dilution were used for the IP assays. The anti-GCN5 antibody was generated using two synthetic peptides (H2N-CARGADTDSDPDESED and H2N-SSRNTKLKTESSTVKLC); both peptide epitopes are located between amino acids 85 to 99 and between amino acids 136 to 150, which is the N-terminal region of the protein, provided by Prof. D-X Zhou, and the specificity of this GCN5 antibody was confirmed by protein gel blots (Benhamed et al., 2006, 2008). The anti-H3K14ac and anti-H3K9ac antibodies were purchased from Upstate Biotechnology. CHALCONE SYNTHASE (CHS) and AT4G03800 (gypsy-like retrotransposon family gene) were amplified as endogenous controls for the anti-GCN5 and anti-H3K9/H3K14 antibodies, respectively. The immunoprecipitated DNA was analysed by quantitative PCR in three biological replicates using the primer sets listed in Supplementary Table S4. Amplified DNA from the chromatin fractions prior to antibody incubation were used as the controls (inputs). The fold-enrichment was normalized to the chromatin inputs.
GUS histochemical and fluorometric assays
Three homologous transgenic T3 lines of ProCER3::GUS/Ws and the corresponding homologous ProCER3::GUS/gcn5-2 lines were used for GUS histochemical analysis. The seedlings were grown under strictly identical conditions. After pollination, Arabidopsis stems, siliques, flowers, and young leaves were vacuum-infiltrated with staining buffer (2 mM potassium ferricyanide, 10 mM phosphate buffer, 0.5% Triton X-100, and 1 mg ml−1 X-Gluc) and then incubated overnight. The tissue was then incubated in ethanol and acetic acid (1:1) for 4–8 h and cleared in 80% ethanol. The samples were observed with a stereomicroscope (Olympus SEX16).
For the quantification of GUS activity, we used the fluorometric assay based on the method of Jefferson et al. (1987). Total protein extracts from stems of three independent lines for each construct (ProCER3::GUS/Ws and ProCER3::GUS/gcn5-2) were determined using bovine serum albumin (BSA) as a standard according to the Bradford assay (Bradford, 1976). Fluorescence was measured using 4-methylumbelliferone (4-MU) as a substrate, with an excitation wavelength of 365 nm and an emission wavelength of 455 nm in a BioTek Synergy HT Multi-Mode Microplate Reader (BioTek, Vermont, USA). GUS activities of the extracts were calculated as nanomole 4-MU per minute per milligram protein.
Both GUS histochemical and fluorometric assays were conducted at least three times, and only the transgenic lines with stable GUS signals throughout different generations were selected for further analysis.
Scanning electron microscopy
Six-week-old stems were attached to double-sided carbon sealing tape. The specimens were examined under a Hitachi TM3000 Tabletop Scanning Electron Microscope at 15 kV, and the digital recordings were saved as TIFF files.
Cuticular wax analysis
Cuticular wax analysis was conducted as previously described by Chen et al. (2011). Because the environmental conditions could affect configuration and distribution of the surface wax structures, all the plants we used in this study were cultivated in strictly controlled temperature and humidity. Six-week-old stems were collected and pictures were taken (with an adjacent ruler) for later determination of area using ImageJ (https://imagej.nih.gov/ij/). Cuticular waxes were extracted by immersing the samples for 30 s in 1 ml of chloroform containing 10 μg of tetracosane (Fluka) as an internal standard. Three biological replicates per genotype were performed. Five individual stems were used for each replicate. The extracts were transferred to reactive vials, dried under nitrogen gas, and derivatized by adding 20 μl of N, N-bis-trimethylsilyltrifluoroacetamide (Macherey-Nagel) and 20 μl of pyridine, and incubated for 40 min at 70 °C. These derivatized samples were then analysed using a gas chromatography–flame ionization detector (GC-FID, Agilent, Technologies) and GC-MS (Agilent gas chromatograph coupled to an Agilent 5973N quadrupole mass-selective detector).
Consistent with previous studies (Haslam et al., 2015; Lee and Suh, 2015; Li et al., 2016), the content of the unidentified components (which showed no significant differences among genotypes) was excluded from the total wax load.
Statistical analysis
Statistical analyses of the phenotypic data and expression levels were performed using Student’s t-test in Excel. To assess the overall differences in the stem cuticular wax composition among genotypes, we compared the means using one-way ANOVA together with a Bonferroni adjustment test in R.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CER3, AT5G57800; GCN5, AT3G54610; CER26, AT4G13840; CER1-L1, AT1G02190; WSD1, AT5G37300; AT2, AT5G55370; FAR3, AT4G33790, ACTIN8, AT1G49240; CHS, AT5G13930; AT4G03800. The RNA-seq reads used for this study are deposited at the National Center for Biotechnology Information Short Read Archive under the accession number SRP093334.
Results
Mutation of Arabidopsis GCN5 impairs stem cuticular wax deposition
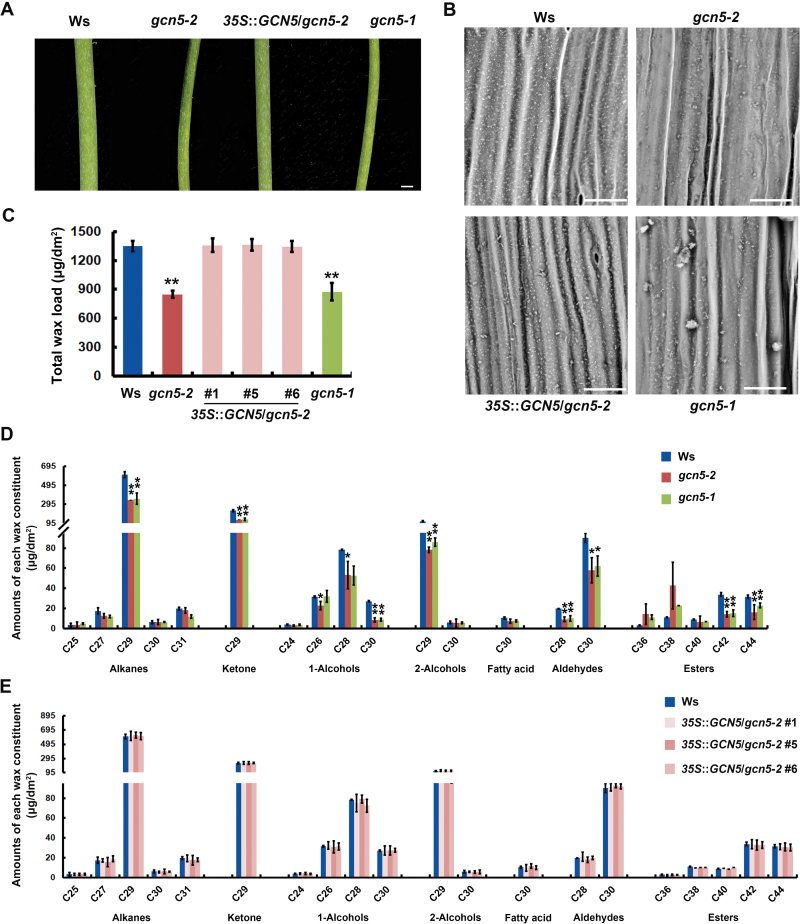
To investigate the potential roles of histone modification in cuticular wax accumulation, 23 T-DNA insertion mutants that harbor disruption in histone acetylation or methylation genes were selected for analysis, namely ashh1, ashh2, ashh3, ashh4, ashr2, ashr3, atx1, atx2, atx4, atx5, atxr2, atxr3, atxr4, gcn5-2, hda2, hda2c, hda5, hda7, hda9, hda13, hda18, hda19, and srt2. The stem cuticular wax of these mutants and the wild-type controls were observed and compared using SEM. As shown in Supplementary Figs S1 and S2, we found wide variations for cuticular wax crystal morphology and crystallization patterns in the mutants and wild-types. For example, atx4, atxr4, ashr2, and hda9 showed more cuticular wax crystals than the wild-type (Col-0), whereas less abundant wax was observed in atx5, ashh2, and hda18. Remarkably, fewer stem wax crystals were observed on the surface of the gcn5-2 mutant relative to the wild-type (Ws), which led to the appearance of a glossy stem (Fig. 1A, B). The compositions of the cuticular wax of the gcn5-2 mutant and wild-type were quantified by GC-FID and GC-MS analysis. The total cuticular wax content in the gcn5-2 mutant was approximately 63% of the control (Fig. 1C), which was attributable to notable decreases in the major wax constituents in the mutant stem, including alkanes (C29), ketone (C29), primary alcohols (C26, C28, and C30), secondary alcohols (C29), aldehydes (C28 and C30), and esters (C42 and C44; Fig. 1D and Table 1).
Fig. 1.
Mutations in GCN5 are responsible for defects in the stem cuticular wax in Arabidopsis. (A) Light reflectance and (B) SEM observations of 6-week-old stems from wild-type Ws, and gcn5-2, 35S::GCN5/gcn5-2 transgenic lines (#1, #5, #6), and gcn5-1 mutants. Scale bars: (A) 1 mm, (B) 30 μm. (C) Total cuticular wax content, calculated per unit area of 6-week-old stems from the four different genotypes. (D) Cuticular wax composition of 6-week-old stems for wild-type Ws, and gcn5-2 and gcn5-1 mutants, and (E) for Ws and 35S::GCN5/gcn5-2 transgenic lines (#1, #5, #6). The chemical classes and main chain-lengths of each constituent are indicated. Data are means (±SD) of three biological replicates. *P<0.05, **P<0.01; Student’s t-test.
Table 1.
Cuticular wax composition of stems in the wild-type Ws, gcn5-2, gcn5-1, and complementary transgenic lines
| Total | Alkanes | Ketones | 1-Alcohols | 2-Alcohols | Fatty acids | Aldehydes | Esters | |
|---|---|---|---|---|---|---|---|---|
| Ws | 1347.99 ± 54.38 | 650.81 ± 34.27 | 226.39 ± 12.94 | 140.44 ± 2.34 | 122.16 ± 6.91 | 10.39 ± 1.13 | 109.86 ± 4.70 | 87.93 ± 4.82 |
| gcn5-2 | 847.49 ± 35.42** | 375.94 ± 7.98** | 133.64 ± 2.16** | 87.01 ± 22.68* | 83.59 ± 4.50** | 7.03 ± 2.08 | 66.84 ± 14.77** | 93.45 ± 38.77 |
| #1 | 1357.28 ± 67.64 | 659.13 ± 34.75 | 230.33 ± 20.01 | 138.11 ± 17.45 | 122.64 ± 8.57 | 9.75 ± 3.34 | 111.81 ± 14.64 | 85.52 ± 7.39 |
| #5 | 1360.93 ± 61.58 | 666.21 ± 27.19 | 230.35 ± 19.85 | 141.23 ± 15.52 | 116.23 ± 12.49 | 11.59 ± 2.21 | 110.60 ± 8.14 | 84.72 ± 17.21 |
| #6 | 1344.06 ± 57.09 | 655.99 ± 27.72 | 228.46 ± 8.61 | 134.99 ± 11.51 | 117.54 ± 8.16 | 9.87 ± 1.85 | 111.51 ± 8.10 | 85.72 ± 14.12 |
| gcn5-1 | 875.47 ± 90.61** | 387.21 ± 41.12** | 136.54 ± 13.30** | 96.61 ± 17.24* | 96.91 ± 16.31* | 7.38 ± 1.03 | 72.00 ± 10.66** | 78.82 ± 7.08 |
Data are means (±SD; n=3) for the total wax load and each constituent class in μg dm–2. Five individual stems were used for each replicate. #1, #5, #6 represent three complementary transgenic lines (35S::GCN5/gcn5-2#1, #5, #6). *P<0.05, **P<0.01; Student’s t-test. 1-Alcohols, primary alcohols; 2-Alcohols, secondary alcohols.
To confirm that the observed defects in stem wax were indeed caused by the disruption of GCN5, another well-established T-DNA insertion mutant allele of GCN5 (gcn5-1) was used for further investigation. As expected, the gcn5-1 mutant also exhibited wax-deficient phenotypes (Fig. 1 and Table 1). Moreover, a genetic complementation experiment was performed by introducing a full-length GCN5 coding sequence driven by the CaMV35S promoter into the gcn5-2 mutant. Given the low-fertility pollen of the gcn5-2 mutant found in previous studies (Bertrand et al., 2003; Xing et al., 2015), we first generated three independent homologous 35S::GCN5/Ws transgenic plants harboring significantly high GCN5 expression levels, and crossed them with gcn5-2 mutants (Supplementary Fig. S3B). After three generations, three independent homozygous 35S::GCN5/gcn5-2 (#1, #5 and #6) transgenic lines were obtained (Supplementary Fig. S3B). No obvious phenotype differences were observed between 35S::GCN5/Ws transgenic plants and the wild-type (Supplementary Fig. S3C). However, 35S::GCN5/gcn5-2 transgenic lines exhibited similar phenotypes to the wild-type (Fig. 1, Table 1, and Supplementary Fig. S3C), indicating that constitutive expression of GCN5 could rescue the gcn5-2 wax deficiency. These results indicated that GCN5 is essential for the normal accumulation of cuticular wax on the stem surface of Arabidopsis.
RNA-seq analysis reveals significant alteration of lipid-related gene expression in the gcn5-2 mutant
To investigate whether the reduction of cuticular wax content in the gcn5-2 mutant was caused by decreased expression of wax-related genes, total RNA of 6-week-old stems of the gcn5-2 mutant and the wild-type Ws were isolated for high-throughput RNA sequencing and transcriptomic comparison. Three biological replicates per genotype were performed and the correlation coefficients of each genotype showed favorable reproducibility (Supplementary Fig. S4). The high reliability of the RNA-seq data was verified by qPCR of 10 randomly selected genes (Supplementary Fig. S5). For each sample, values for reads per kilobase of exon model per million mapped reads (RPKM) were calculated, and the genes with at least 2-fold change and a false discovery rate value P≤0.05 were selected. Compared with the control, we found that 54% (2616 genes) of the total of differentially expressed genes were down-regulated in gcn5-2 mutant stems. Because GCN5 usually positively regulates transcriptional processes (Servet et al., 2010) and mutation of GCN5 decreased the total wax load in Arabidopsis stems, these 2616 genes were expected to be direct or indirect GCN5 target genes and might be involved in cuticular wax accumulation.
We then used GO analysis of the 2616 candidate genes using agriGO v2.0 (Tian et al., 2017), and the categories showed considerably high enrichments in lipid metabolic process, cellular lipid metabolic process, and lipid biosynthetic process (P≤6.2 × 10–6). Moreover, a significant fraction of genes involved in metabolic and/or biosynthetic processes (including glycerolipid, neutral lipid, fatty acid, and unsaturated fatty acid), lipid catabolic process, and regulation of lipid metabolic and biosynthetic processes were enriched (Table 2), suggesting that GCN5 is involved in wax accumulation by modulating the transcription of lipid-related genes. For further screening, 145 non-redundant genes from three most-abundant lipid-related GO terms (GO:0006629, GO:0044255, and GO:0008610) were analysed together with their biological properties and functions in cuticular wax development as described in previous reports (Bernard and Joubès, 2013; Lee and Suh, 2015). Finally, we filtered five cuticular wax genes that are potential target genes of GCN5, namely WSD1, CER3, CER26, CER1-L1, and Long-chain-alcohol O-fatty-acyltransferase 2 (AT2; Table 3).
Table 2.
Lipid-related GO categories for the 2616 candidate genes down-regulated in the gcn5-2 mutant
| GO ID | Term | Query item | Query total | Bg item | Bg total | P-value |
|---|---|---|---|---|---|---|
| GO:0006629 | Lipid metabolic process | 145 | 2598 | 994 | 28362 | 2.7 × 10–7 |
| GO:0044255 | Cellular lipid metabolic process | 100 | 2598 | 649 | 28362 | 2.5 × 10–6 |
| GO:0008610 | Lipid biosynthetic process | 83 | 2598 | 522 | 28362 | 6.20 × 10–6 |
| GO:0046486 | Glycerolipid metabolic process | 24 | 2598 | 122 | 28362 | 0.0011 |
| GO:0045017 | Glycerolipid biosynthetic process | 15 | 2598 | 79 | 28362 | 0.011 |
| GO:0046460 | Neutral lipid biosynthetic process | 6 | 2598 | 18 | 28362 | 0.012 |
| GO:0006638 | Neutral lipid metabolic process | 7 | 2598 | 24 | 28362 | 0.013 |
| GO:0019216 | Regulation of lipid metabolic process | 11 | 2598 | 54 | 28362 | 0.019 |
| GO:0016042 | Lipid catabolic process | 31 | 2598 | 224 | 28362 | 0.024 |
| GO:0046890 | Regulation of lipid biosynthetic process | 9 | 2598 | 43 | 28362 | 0.028 |
| GO:0006636 | Unsaturated fatty acid biosynthetic process | 6 | 2598 | 24 | 28362 | 0.036 |
| GO:0006631 | Fatty acid metabolic process | 33 | 2598 | 252 | 28362 | 0.037 |
Query item: number of down-regulated genes in the gcn5-2 mutant annotated as the corresponding GO term. Query total: number of down-regulated genes in the gcn5-2 mutant. Bg item: number of genes in Arabidopsis whole genome annotated as the corresponding GO term. Bg total: number of genes in Arabidopsis whole genome.
Table 3.
Potential GCN5-regulated genes involved in cuticular wax synthesis
| Gene | Name | Annotation |
|---|---|---|
| AT5G37300 | WSD1 | Bifunctional wax synthase/acyl-CoA:diacylglycerol acyltransferase |
| AT5G57800 | CER3 | ECERIFERUM 3 |
| AT4G13840 | CER26 | ECERIFERUM 26 |
| AT1G02190 | CER1-L1 | Protein CER1-like 1 |
| AT5G55370 | AT2 | Long-chain-alcohol O-fatty-acyltransferase 2 |
ChIP assays identify target genes of GCN5 involved in cuticular wax biosynthesis
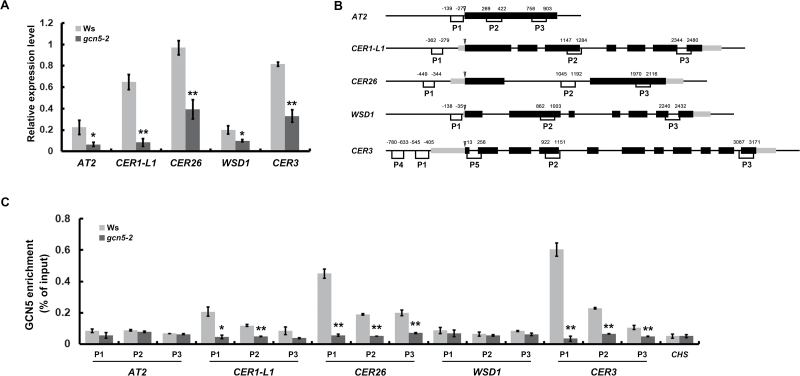
To confirm the GCN5-regulated target genes in stem cuticular wax biosynthesis, we first detected the transcript levels of the five candidate genes derived from RNA-seq data by qRT-PCR. As expected, their expression levels were significantly reduced in the gcn5-2 mutant (Fig. 2A). The candidate genes were then analysed by ChIP assays, using 6-week-old stems of the gcn5-2 and Ws plants and GCN5-specific antibodies (Benhamed et al., 2006, 2008). ChIP-qPCRs with three primer pairs spanning the promoter regions and gene body regions of each gene (WSD1, CER3, CER26, CER1-L1, and AT2) were conducted for binding tests (Fig. 2B). CHS was used as the negative control as its expression is not affected by GCN5 (Benhamed et al., 2006). As shown in Fig. 2C, significant decreases in enrichment in the gcn5-2 mutant were observed for most of the examined regions of the CER3, CER26, and CER1-L1 genes relative to the wild-type, especially in the transcription start-site region (P1) of CER3. By contrast, no significant changes in GCN5 enrichment were detected in any tested regions for the other two genes (WSD1 and AT2).
Fig. 2.
Identification of GCN5 target genes. (A) Expression levels of the GCN5 target genes as determined by qRT-PCR in 6-week-old stems of the wild-type Ws and the gcn5-2 mutant. (B) Diagram representing the genomic structure and primer sets (indicated by P1–P5) analysed for ChIP-qPCR in the AT2, CER1-L1, CER26, WSD1, and CER3 genes. The exact distance (in bp) of the primers from to the ATG start codon sites (indicated by triangles) are labeled. Black boxes represent exons and gray boxes represent untranslated regions (UTRs). (C) ChIP analysis with nuclei extracted from cross-linked, 6-week-old stems of Ws and the gcn5-2 mutant and antibody-specific for GCN5. The CHS gene was used as a negative control, which provided background level for the ChIP samples. Data are means (±SD) from at least three biological replicates. *P<0.05, **P<0.01; Student’s t-test.
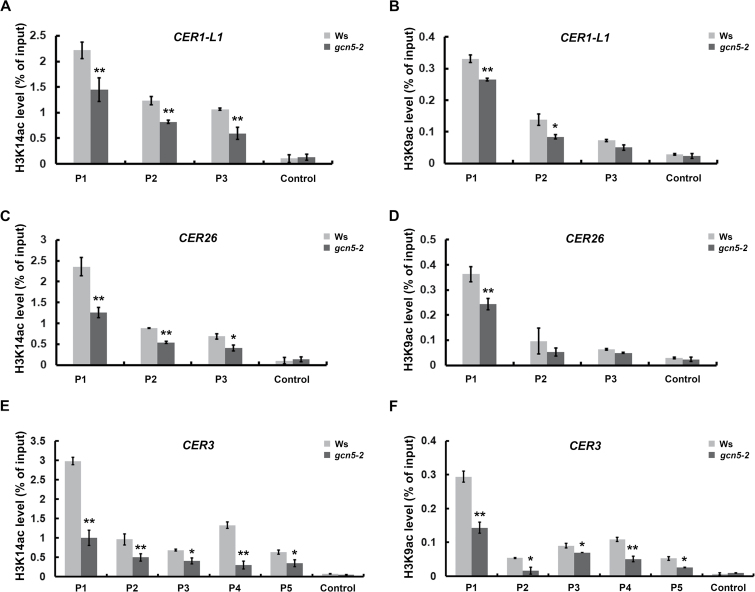
Previous studies have reported that GCN5 is specifically responsible for H3K14 acetylation (H3K14ac) and that it influences the H3K9ac and H3K27ac at the promoters of their targets, which are positively correlated with gene expression (Bertrand et al., 2003; Bhat et al., 2003; Earley et al., 2007). Thus, we analysed the acetylation levels of H3K14 and H3K9 at the CER3, CER26, and CER1-L1 loci using the primer sets indicated in Fig. 2B. Consistently, both the H3K14ac and H3K9ac levels of these candidate genes were significantly decreased in the gcn5-2 mutant compared to the wild-type, especially at the promoter regions (Fig. 3), which was in accordance with the expression profiles (Fig. 2A). Collectively, these data indicated that CER3, CER26, and CER1-L1 are the targets of GCN5, and their expression can be regulated by GCN5 by modulating their H3K14 and H3K9 acetylation.
Fig. 3.
H3K14 and H3K9 acetylation levels on GCN5 target genes. ChIP analysis of H3K14 and H3K9 acetylation on (A, B) CER1-L1, (C, D) CER26, and (E, F) CER3 genes. Nuclei extracted from cross-linked, 6-week-old stems of the wild-type Ws and gcn5-2 mutant and antibodies specific for H3K14ac and H3K9ac. AT4G03800 (gypsy-like retrotransposon family gene) was used as a negative control. Data are means (±SD) from at least three biological replicates. *P<0.05, **P<0.01; Student’s t-test.
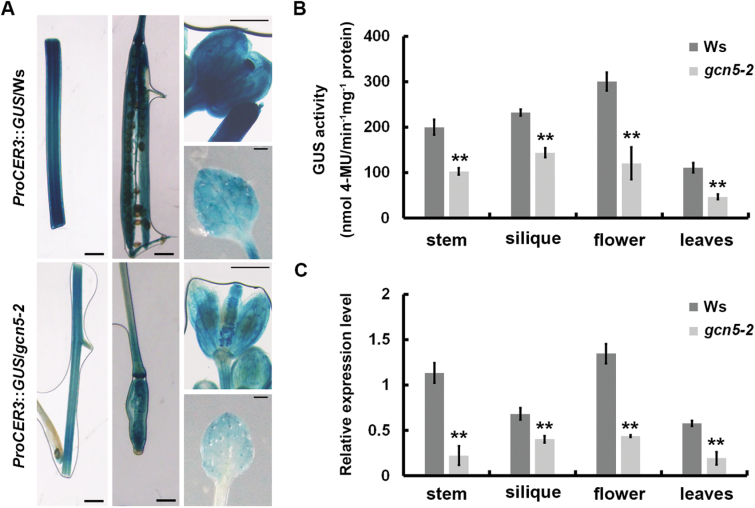
CER3 is a critical enzyme for cuticular wax synthesis (Aarts et al., 1995; Chen et al., 2003; Rowland et al., 2007). Interestingly, the cuticular wax composition of cer3 mutant stems was quite similar to that of the gcn5-2 mutant stem, especially for aldehydes, alkanes, (secondary alcohols, and ketone (Lai et al., 2007; Rowland et al., 2007). Therefore, we hypothesized that CER3 might be a critical target of GCN5 and decided to investigate the gene expression patterns of CER3 in ceriferous (i.e. wax-producing) tissues of gcn5-2 and Ws. Isogenic ProCER3::GUS/gcn5-2 and ProCER3::GUS/Ws lines were obtained in which each transgene was homozygous and correspondingly inserted into a single genomic locus. GUS staining revealed weaker signals in stems, siliques, flowers, and young leaves of the gcn5-2 mutant compared to that of the control (Fig. 4A). Quantification of GUS activity by fluorometric assay consistently revealed significantly lower activity in three independent homozygous ProCER3::GUS/gcn5-2 lines than in the corresponding ProCER3::GUS/Ws lines (Fig. 4B). qRT-PCR analysis also showed that the CER3 expression levels were decreased in different tissues of gcn5-2 mutant as compared to the wild-type (Fig. 4C). These results provided further evidence that the expression of CER3 is positively regulated by GCN5 in ceriferous tissues.
Fig. 4.
Expression patterns of CER3 in ceriferous tissues of the wild-type Ws and gcn5-2 mutant. (A) Spatial expression patterns of the CER3 gene in transgenic Ws and gcn5-2 plants harboring the CER3 promoters fused to the GUS gene. Promoter activity was visualized through histochemical GUS-staining in stems, siliques, flowers, and young leaves of 6-week-old plants. Scale bars: stems and siliques, 1.5 mm; flowers and leaves, 2 mm. (B) Quantification of the GUS activity using 4-methylumbelliferone (4-MU) as a substrate and (C) CER3 expression levels in stems, siliques, flowers, and young leaves of 6-week-old plants. Data are means (±SD) from three biological replicates. **P<0.01; Student’s t-test.
Overexpression of CER3 rescues the stem cuticular wax-deficient phenotype in the gcn5-2 mutant
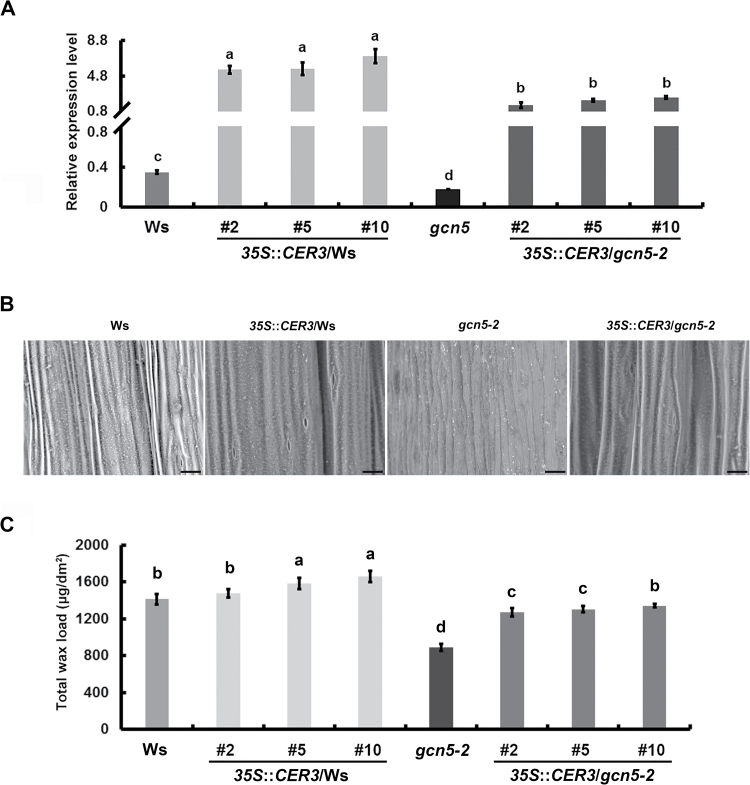
To assess the role of CER3 in GCN5-modulated biosynthesis of stem cuticular wax, we studied the accumulation and composition of the CER3 overexpression transgenic lines in both the Ws and gcn5-2 mutant backgrounds. Transcript levels of the CER3 in 35S::CER3/Ws and 35S::CER3/gcn5-2 transgenic lines increased significantly compared with the controls (Fig. 5A). SEM imaging showed that the deposition of wax crystals on the 35S::CER3/gcn5-2 stem was obviously increased compared with that of the gcn5-2 mutant and that it basically recovered to the level of Ws, but no significant differences were observed between 35S::CER3/Ws and Ws (Fig. 5B).
Fig. 5.
Overexpression of CER3 in the gcn5-2 mutant increases the cuticular wax component and restores it to the wild-type Ws level. (A) Relative expression levels of CER3 in plants of Ws, 35S::CER3/Ws (#2, #5, #10), gcn5-2, and 35S::CER3/gcn5-2 (#2, #5, #10). Total RNA was isolated from 6-week-old stems of Ws and the gcn5-2 mutant. ACTIN8 was used as an endogenous control. Data are means (±SD) from at least three biological replicates. (B) Scanning electron microscopy of the stems of Ws, 35S::CER3/Ws (#2, #5, #10), gcn5-2, and 35S::CER3/gcn5-2 (#2, #5, #10). Scale bars: 20 μm. (C) Total cuticular wax content was calculated over the unit area of 6-week-old stems in plants of Ws, 35S::CER3/Ws (#2, #5, #10), gcn5-2, and 35S::CER3/gcn5-2 (#2, #5, #10). The mean expression levels and total wax load were compared using one-way ANOVA together with a Bonferroni adjustment test in R. Different letters indicate significant differences among genotypes (P<0.05).
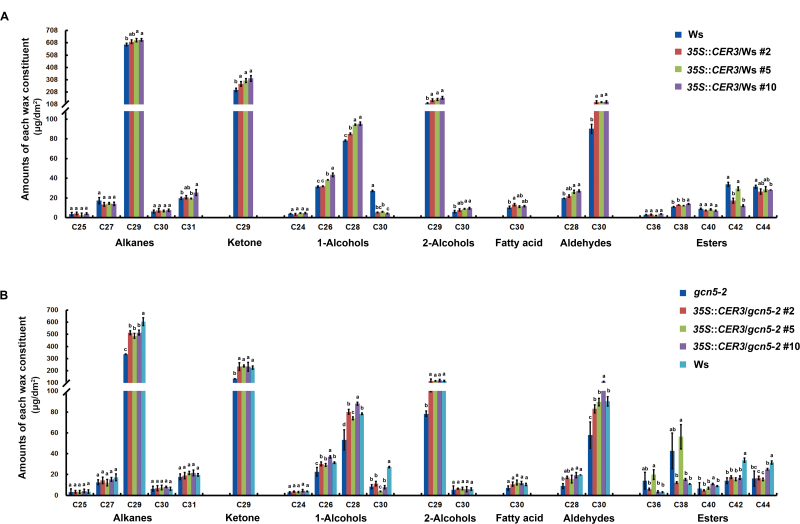
In addition, the total load and composition of stem cuticular wax of 35S::CER3/Ws, 35S::CER3/gcn5, Ws, and gcn5-2 plants were measured by GC-FID and GC-MS analysis. Although the wax crystal loading showed little increase under SEM observation (Fig. 5B), the total amount and individual components of stem cuticular wax in two of the three 35S::CER3/Ws transgenic lines (#5 and #10) were significantly increased as compared to Ws, especially for the amount of alkanes, primary alcohols, and ketone, which were consistent with the expression levels of the CER3 gene (Figs. 5C, 6A). Notably, the total wax load in 35S::CER3/gcn5-2 transgenic plants was obviously increased compared with that of the gcn5-2 mutant, and in the 35S::CER3/gcn5-2 #10 transgenic line it was even restored to the wild-type level (Fig. 5C). Analysis of the cuticular wax composition showed that the contents of C29 alkane, C29 ketone, C26 and C28 primary alcohols, C29 secondary alcohols, and C30 aldehyde significantly increased in the 35S::CER3/gcn5-2 transgenic lines compared with the control, thus providing genetic evidence that CER3 plays an important role in GCN5-regulated cuticular wax biosynthesis (Fig. 6B). It should be noted that overexpression of CER3 did not completely restore all wax component defects in the gcn5-2 mutant, for example C30 primary alcohol and some esters, and this may be attributed to other GCN5-regulated target genes in the wax biosynthetic pathway. FATTY ACID REDUCTASE3 (FAR3, also known as CER4), encodes an alcohol-forming fatty acyl-coenzyme A reductase, and is involved in the synthesis of primary alcohols (Rowland et al., 2006; Wang et al., 2015). Thus, we analysed the transcript levels of FAR3 in gcn5-2, Ws, and 35S::CER3 transgenic plants using qRT-PCR. However, the expression levels of FAR3 in these plants were not obviously changed (Supplementary Fig. S6).
Fig. 6.
Cuticular wax composition of the stems of wild-type Ws, 35S::CER3/Ws (#2, #5, #10), gcn5-2, and 35S::CER3/gcn5-2 (#2, #5, #10) from 6-week-old plants. The chemical classes and main chain-lengths of each constituent are indicated. Data are means (±SD) of three biological replicates. (A) Comparison between Ws and 35S::CER3/Ws (#2, #5, #10) and (B) among Ws, gcn5-2, and 35S::CER3/gcn5-2 (#2, #5, #10) transgenic plants. The mean values were compared using one-way ANOVA together with a Bonferroni adjustment test in R. Different letters indicate significant differences among genotypes (P<0.05).
Discussion
Lipids are an essential constituent of all plant cells, including fatty acids (FAs), waxes, sterols, and others (Li-Beisson et al., 2013). Post-translational modification of histone tails plays an important role in epigenetic regulation of gene expression, and this includes histone acetylation, methylation, phosphorylation, and ubiquitination (Pfluger and Wagner, 2007). As a member of the HAT enzymes, GCN5 is a versatile regulator of Arabidopsis development and stress responses (Bertrand et al., 2003; Benhamed et al., 2006; Kornet and Scheres, 2009; Hu et al., 2015; Xing et al., 2015). Recently, we found that GCN5 is involved in FA biosynthesis by affecting the acetylation levels of FAD3 (Wang et al., 2016). Here, we observed that GCN5 is essential for lipid metabolism in Arabidopsis stems. Firstly, mutation of GCN5 in Arabidopsis compromised the content of multiple lipid compounds (including very-long-chain alkanes, aldehydes, ketones, and alcohols), which resulted in a complete deficiency in stem cuticular wax accumulation. This wax deficiency could be fully rescued by complementation with 35S::GCN5. Secondly, GO analysis indicated that down-regulated genes in the gcn5-2 mutant were enriched in categories related to lipid synthesis, including the lipid biosynthetic process, neutral lipid biosynthetic process, glycerolipid biosynthetic process, and unsaturated fatty acid biosynthetic process. Thirdly, ChIP assays demonstrated that CER1-L1, CER26, and CER3, which encode proteins involved in VLCFA production and alkane-forming pathways of wax synthesis, are target genes of GCN5. Finally, enrichment of H3K9ac and H3K14ac at the promoters of CER1-L1, CER26, and CER3 was significantly decreased in the gcn5-2 mutant compared with the wild-type. Collectively, our previous data (Wang et al., 2016) and that from the present study have demonstrated that histone acetyltransferase GCN5 is involved in multiple lipid metabolic processes, from upstream de novo FA synthesis to subsequent wax production.
To determine the underlying mechanisms of GCN5-regulated biosynthesis of stem cuticular wax, three GCN5 target genes, CER3, CER26, and CER1-L1 were identified. CER26 is involved in the elongation of VLCFAs (from 30 C to 32 C) and has high specificity of tissue and substrate (Haslam et al., 2015; Pascal et al., 2013). Although little is known about CER1-L1 other than that it is a homolog of CER1 (Bernard et al., 2012), there is the possibility that, like CER1, it can physically interact with CER3 during the very-long-chain alkane biosynthesis process and contribute to the total wax load. Previous studies have reported that CER3, which catalyses redox-dependent alkane formation, is the key wax biosynthetic enzyme (Aarts et al., 1995; Chen et al., 2003; Rowland et al., 2007). The wax components of the Arabidopsis cer3 mutant are lacking in aldehydes, alkanes, secondary alcohols, and ketones in the stem compared with the wild-type (Rowland et al., 2007). Interestingly, our GC-FID and GC-MS analyses found significant reductions in total wax in the gcn5-2 mutant stem, especially for C30 aldehyde, C29 alkane, C29 ketone, and C29 secondary alcohols. Moreover, overexpression of CER3 in the gcn5-2 background significantly increased the levels of C29 ketone, C30 aldehydes, C29 alkanes, and C29 secondary alcohols, indicating that CER3 played a pivotal role in GCN5-regulated biosynthesis of stem cuticular wax. However, we cannot rule out the possibility that other unknown genes may also contribute, which may prove an interesting area for further investigation. For example, the ester component, which cannot be catalysed by CER3, also changed significantly in the gcn5-2 mutant stem. The gcn5-2 mutant deficiency of C30 primary alcohols was not rescued by overexpression of CER3. In addition, the expression of FAR3, an important enzyme in primary alcohol formation, was not regulated by GCN5.
Many reports have demonstrated that acetylation of histone tails induces the accessibility of transcription factors to the nucleosomal DNA, which subsequently influences the gene expression (e.g. Lee et al., 1993). As a key wax biosynthetic enzyme gene, CER3 is regulated by both transcription factors and epigenetic modulators (Lee and Suh, 2015). CER3 is positively regulated by two MYB transcription factors, MYB96 and MYB30, in response to drought and pathogen attack, respectively (Lee and Suh, 2013; Lee et al., 2016b). Based on our RNA-seq data (see Supplementary Table S2), the expression of MYB96 was up-regulated in the gcn5-2 mutant, suggesting that it does not contribute much to the changed expression of CER3 in the stem of this mutant. However, we cannot rule out the possibility that CER3 could be regulated by other unknown transcription factors that are deregulated in the gcn5-2 mutant. Recent studies showed that epigenetic modulators, namely the histone methyl transferases SDG8 (ASHH2) and SDG25 (ATXR7), were involved in wax accumulation through histone lysine methylation and/or indirectly through H2B ubiquitination by targeting CER3, and this was associated with diminished accumulation of lipids (Lee et al., 2016a). Here, our observations also showed that the ashh2 mutant exhibited mildly reduced wax crystal accumulation compared with Col-0. It might be of interest to further examine the relationships between GCN5, SDG8 (ASHH2), and SDG25 (ATXR7) in cuticular wax biosynthesis.
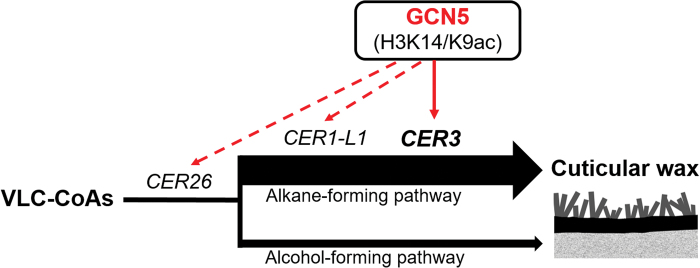
In conclusion, our results have demonstrated that, like histone ubiquitination and methylation, histone acetylation is also involved in the regulation of biosynthesis of stem cuticular wax, and we propose a working model to explain this process in Arabidopsis (Fig. 7). Briefly, the histone acetyltransferase GCN5 regulates the biosynthesis of stem cuticular wax by regulating the expression of CER3, CER1-L1, and CER26 via histone acetylation at the H3K9/14 sites. Thus, interruption of GCN5 dramatically reduces the total amount of cuticular wax and changes its composition, especially with regards to alkanes, aldehydes, and ketone, which are mainly synthesized in the alkane-forming process. Remarkably, overexpression of CER3 in the gcn5-2 mutant could rescue the cuticular wax deficiency, suggesting that it has an important role in GCN5-mediated cuticular wax biosynthesis. Our findings provide an insight into the epigenetic regulation of cuticular wax development through histone acetylation, which may contribute to wax-related stress responses in plants.
Fig. 7.
A model for the regulation of stem cuticular wax synthesis by GCN5-associated acetylation in Arabidopsis. As the wax precursors, the very-long-chain acyl-CoAs (VLC-CoA) can be processed through the alcohol-forming pathway and alkane-forming pathway, which yield 17~18% and 80% of the total cuticular wax mixture, respectively. Solid black arrows represent the cuticular wax biosynthesis pathway. The arrows from GCN5 indicate positive transcriptional regulation by GCN5 via H3K9/14ac modifications. GCN5 targets are marked at the positions where the enzymes they encode are required. CER3 is a key cuticular wax biosynthetic enzyme that catalyses the alkane-forming pathway in Arabidopsis stems (solid arrow), whereas CER26 and CER1-L1 might partially contribute to the total wax load but they were not verified functionally in this study (dashed arrows).
Supplementary data
Supplementary data are available at JXB online.
Table S1. Summary of the RNA-seq data and read mapping.
Table S2. Differently expressed genes in the RNA-seq data.
Table S3. GO analysis of the 2616 down-regulated genes in the stems of the gcn5-2 mutant compared with the wild-type Ws.
Table S4. Gene-specific primer pairs in this study.
Fig. S1. Stem cuticular wax phenotype of the histone-acetylation mutants.
Fig. S2. Stem cuticular wax phenotype of the histone-methylation mutants.
Fig. S3. GCN5 expression levels and phenotypes of the independent homologous 35S::GCN5 transgenic lines.
Fig. S4. Reproducibility of the RNA-seq biological replicates.
Fig. S5. Expression levels of 10 genes randomly selected to validate the accuracy of the RNA-seq data using qRT-PCR.
Fig. S6. FAR3 expression levels in wild-type Ws, gcn5-2, and 35S::CER3 transgenic lines.
Acknowledgements
The authors thank Prof. Jianxin Shi (Shanghai Jiao Tong University) for the GC-FID and GC-MS analyses in his laboratory. This work was supported by the Ministry of Agriculture of China for Transgenic Research (2016ZX08009002), the State Key laboratory of Agrobiotechnology Open Grant (2018SKLAB6-25) and the Fundamental Research Funds for the Central Universities (2412017QD016).
Glossary
Abbreviations:
- GCN5
general control non-repressed protein5
- CER3
eceriferum3
- VLCFA
very-long-chain fatty acid
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- GO
gene ontology
- GUS
β-glucuronidase
- GC-FID
gas chromatography–flame ionization detector.
References
- Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A. 1995. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. The Plant Cell 7, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. 2004. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. The Plant Cell 16, 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX. 2006. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. The Plant Cell 18, 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Martin-Magniette ML, Taconnat L, et al. . 2008. Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. The Plant Journal 56, 493–504. [DOI] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J. 2012. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. The Plant Cell 24, 3106–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J. 2013. Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Progress in Lipid Research 52, 110–129. [DOI] [PubMed] [Google Scholar]
- Bertrand C, Bergounioux C, Domenichini S, Delarue M, Zhou DX. 2003. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. The Journal of Biological Chemistry 278, 28246–28251. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Riehl M, Santandrea G, Velasco R, Slocombe S, Donn G, Steinbiss HH, Thompson RD, Becker HA. 2003. Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. The Plant Journal 33, 455–469. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. 2004. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D. 2011. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiology 157, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. 2003. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. The Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. 2007. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. The Plant Journal 52, 615–626. [DOI] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J. 2008. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protocols 2008, pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Haslam TM, Haslam R, Thoraval D, et al. . 2015. ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiology 167, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao L, Kunst L. 2012. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiology 160, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Song N, Zheng M, et al. . 2015. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. The Plant Journal 84, 1178–1191. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Bevan M, Kavanagh T. 1987. The use of the Escherichia coli beta-glucuronidase as a gene fusion marker for studies of gene expression in higher plants. Biochemical Society Transactions 15, 17–18. [DOI] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology 12, R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Thiel F. 1989. A genetic and phenotypic description of eceriferum mutants in Arabidopsis thaliana. The Journal of Heredity 80, 118–122. [Google Scholar]
- Kornet N, Scheres B. 2009. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. The Plant Cell 21, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Kunst L, Jetter R. 2007. Composition of alkyl esters in the cuticular wax on inflorescence stems of Arabidopsis thaliana cer mutants. The Plant Journal 50, 189–196. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72, 73–84. [DOI] [PubMed] [Google Scholar]
- Lee S, Fu F, Xu S, Lee SY, Yun DJ, Mengiste T. 2016a. Global regulation of plant immunity by histone lysine methyl transferases. The Plant Cell 28, 1640–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim HU, Suh MC. 2016b. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant & Cell Physiology 57, 2300–2311. [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC. 2013. Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Molecular Plant 6, 246–249. [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC. 2015. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Reports 34, 557–572. [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. 2008. Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiology 148, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang X, He S, et al. . 2016. CFLAP1 and CFLAP2 are two bHLH transcription factors participating in synergistic regulation of AtCFL1-mediated cuticle development in Arabidopsis. PLoS Genetics 12, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. . 2013. Acyl-lipid metabolism. The Arabidopsis Book 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ménard R, Verdier G, Ors M, Erhardt M, Beisson F, Shen WH. 2014. Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant & Cell Physiology 55, 455–466. [DOI] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger Da, Pickaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. 2002. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Research 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal S, Bernard A, Sorel M, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Domergue F, Joubès J. 2013. The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. The Plant Journal 73, 733–746. [DOI] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. 2007. Histone modifications and dynamic regulation of genome accessibility in plants. Current Opinion in Plant Biology 10, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D. 2008. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. The Plant Cell 20, 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L. 2007. The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Letters 581, 3538–3544. [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. 2006. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiology 142, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review of Plant Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Servet C, Conde e Silva N, Zhou DX. 2010. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Molecular Plant 3, 670–677. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Research 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. 2005. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology 139, 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. The Plant Cell 15, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Xing J, Liu X, et al. . 2016. Histone acetyltransferase general control non-repressed protein 5 (GCN5) affects the fatty acid composition of Arabidopsis thaliana seeds by acetylating fatty acid desaturase3 (FAD3). The Plant Journal 88, 794–808. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang M, Sun Y, Hegebarth D, Li T, Jetter R, Wang Z. 2015. Molecular characterization of TaFAR1 involved in primary alcohol biosynthesis of cuticular wax in hexaploid wheat. Plant & Cell Physiology 56, 1944–1961. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang T, Liu Z, et al. . 2015. GENERAL CONTROL NONREPRESSED PROTEIN5-mediated histone acetylation of FERRIC REDUCTASE DEFECTIVE3 contributes to iron homeostasis in Arabidopsis. Plant Physiology 168, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.