ABSTRACT
Antibodies are routinely used to study the activity of transcription factors, using various in vitro and in vivo approaches such as electrophoretic mobility shift assay, enzyme-linked immunosorbent assay, genome-wide method analysis coupled with next generation sequencing, or mass spectrometry. More recently, a new application for antibodies has emerged as crystallisation scaffolds for difficult to crystallise proteins, such as transcription factors. Only in a few rare cases, antibodies have been used to modulate the activity of transcription factors, and there is a real gap in our knowledge on how to efficiently design antibodies to interfere with transcription. The molecular function of transcription factors is underpinned by complex networks of protein-protein interaction and in theory, setting aside intra-cellular delivery challenges, developing antibody-based approaches to modulate transcription factor activity appears a viable option. Here, we demonstrate that antibodies or an antibody single-chain variable region fragments are powerful molecular tools to unravel complex protein-DNA and protein-protein binding mechanisms. In this study, we focus on the molecular mode of action of the transcription factor SOX18, a key modulator of endothelial cell fate during development, as well as an attractive target in certain pathophysiological conditions such as solid cancer metastasis. The engineered antibody we designed inhibits SOX18 transcriptional activity, by interfering specifically with an 8-amino-acid motif in the C-terminal region directly adjacent to α-Helix 3 of SOX18 HMG domain, thereby disrupting protein-protein interaction. This new approach establishes a framework to guide the study of transcription factors interactomes using antibodies as molecular handles.
KEYWORDS: SOX18 transcription factor, antibody, scFv, protein-protein interaction, transcriptional activation.
Introduction
Forward and reverse genetics approaches to identify new pathways critical for developmental and pathological processes have reached a high level of routine and standardization due to advances in next-generation sequencing and gene editing methods. As a research field, developmental biology has contributed the most to identification of key molecular switch transcription factors (TFs), in the major SOX, FOX and HOX classes.1-4 In contrast, study of the modes of action of TFs is still in its infancy, involving complex, empirical, multipronged cross disciplinary approaches, in need of refinement before they can gain a similar momentum. Twenty years ago, TFs were defined in a simple canonical manner, as proteins that bind to genomic DNA transactivating target genes expression. This definition was enough to allow Venter and collaborators to estimate the total number of transcription factors to 1500, later refined to a little less than 2000 in the sequenced human genome.5 In the restrictive context of this canonical definition (DNA binding and target gene transactivation), however, this number of TFs seems small to account for life's complexity. While the definition is still used to differentiate TFs from other nuclear proteins, new insights have been gathered on their features and functionalities, i.e., TFs do not work alone, but instead they are embedded into complex protein-protein interaction networks involving other transcription factors and nuclear proteins (chromatin remodellers, cofactors).6 Transcription factors are packed with modular protein-protein interfaces and display intrinsically disordered structural characteristics allowing flexibility and interchangeability in elicited protein-protein interactions.7-10
The study of this complex system is extremely challenging and any new investigational tool added to the current toolbox is a worthy improvement.11,12 Many developmental TFs have their expression resumed in adults under pathological conditions,9,13-15 while not required to maintain cell phenotypes under normal conditions. One such class of developmental factors, the SOX (SRY-related HMG-box) TFs, have emerged as pivotal aetiological components in cancer-related conditions.16-21 Structural biology studies22-24 and attempts at modulating their activity24-28 have provided useful insights in their mode of action, paving the way for the conceptual development of new therapeutics applications. Study of TFs using antibodies is one attractive option because of the intrinsic selectivity and affinity of antibodies, but also for their ability to help elucidate the structure, trafficking and molecular functions of TFs in one single concerted approach.29,30 Antibodies are already fundamental tools heavily used in the study of TFs' DNA binding activity, via techniques such as electrophoretic mobility shift assay (EMSA), ELISA-based DNA-binding assays and ChIP-seq.31 In a few rare instances, as early as 1994, monoclonal antibodies (mAbs) were even used to interfere with transcription factors activity (DNA binding), to elucidate their mode of action.32 More recently, some studies have focused on “the use of antibody fragments as crystallization chaperones to aid the structural determination of otherwise ‘uncrystallizable’ or ‘undruggable’ target proteins”, such as TFs.30 Others have focussed on developing antibodies against TFs for the academic community, using an automated phage display approach.29 In this study, we used a phage display library screening to develop an antibody that was used to explore the molecular mode of action and the functional protein-protein interaction network of the TF SOX18, which has been at the center of recent pharmacomodulation attempts with small molecules.27,28
Results
To select antibodies specific to the SOX18 protein, we screened a human phage display library of antibody scFv using a 109-amino-acid (aa) peptide of mouse SOX18 HMG domain (Fig. 1A), centered on α-Helix 3 and its direct surroundings. Several binders were isolated, and, after sequencing, two unique binders were identified, F5 and B11 (Suppl. to Fig. 1, panels A and B). These two binders were expressed as scFvs and also reformatted into complete human IgG1 antibodies (Suppl. to Fig. 1, panel C). The binder F5, selected for its higher affinity than B11 (Suppl. to Fig. 1, panel D), showed no cross reactivity with the human SOX2 protein, since a counter-screen against this related SOX transcription factor was performed in parallel (Fig. 1C). ScFv and full-length antibodies selectively recognized mouse 109aa SOX18 HMG fragment and full-length human and mouse SOX18 (Fig. 1B-C, and Suppl. to Fig. 1, panel D). These results were cross-validated using a surface plasmon resonance (SPR)-based approach using histidine-tagged mouse full-length SOX18 immobilized on a nitrilotriacetic acid (NTA) sensor chip (Fig. 1D). Taken together these data confirm that the F5 binding epitope is present and accessible in both SOX18 HMG fragment and full-length SOX18 and is not displayed by SOX2 protein.
Figure 1.
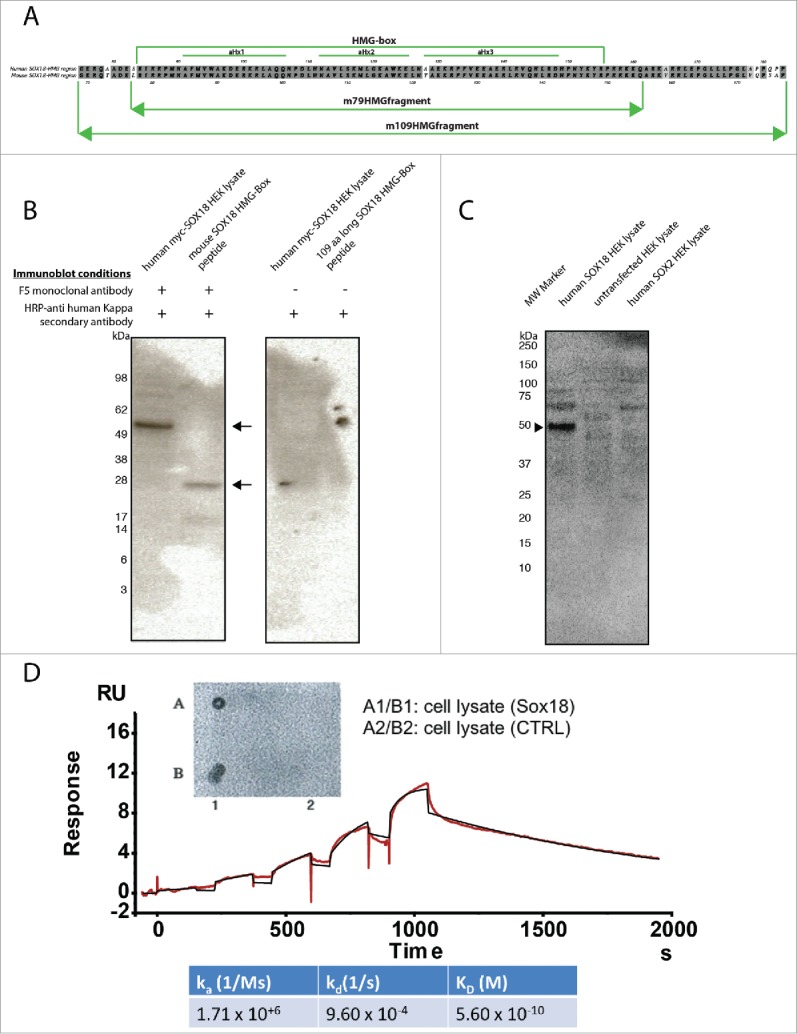
F5 anti-sox18 MAb selectively recognizes full-length human and mouse SOX18 and mouse 109-aa SOX18 HMG-Box peptide but not SOX2. A. Alignment of 109-aa mouse Sox18 fragment (109mSOX18-HMG) corresponding to residues 69 to 177 of full-length protein with its human counterpart corresponding to residues 75 to 183 of full-length protein. A 93% ClustalW similarity score was measured between the two fragments. A 99% ClustalW similarity score was measured between the HMG-box domains of each fragments. Green lines indicate the position of HMG-box domain, and its alpha-helices 1, 2 and 3. B. Lysate of HEK cells overexpressing human SOX18 and a peptide representative of the mouse SOX18 HMG-Box were run on PAGE, transferred to PVDF membrane and probed with F5 mAb and HRP-anti-human kappa secondary antibody. The F5 mAb binds to the hSOX18 protein and the mouse SOX18 HMG-Box peptides (monomer and dimer) as indicated by the arrows. Negative controls with HRP anti-human kappa secondary antibody alone have also been performed. C. Western blot analysis of F5 mAb reactivity to human SOX18 and SOX2 transiently expressed in HEK cell lysates. Samples were resolved on 4–12% NuPAGE Bis-TRIS Gel (ThermoFisher) and subsequently transferred onto Trans-Blot® PVDF membranes and probed with the F5 mAb followed by detection with the anti-kappa light chain HRP. The F5 mAb binds to the hSOX18 protein as indicated by the arrowhead, but does not react with human SOX2. D. Insert – Dot-blot ELISA of full-length F5 mAb on lysate of naïve Sf9 insect cells or expressing full-length mouse Sox18. Main panel/ Estimation of antibody binding affinity to full-length mouse Sox18 by Surface Plasmon Resonance. Binding affinity of the F5 mAb was estimated by single cycle kinetics on a Biacore T200 (GE, US). 100 nM murine Sox18 engineered with a 6xHis tag was immobilised onto one channel of a nitrilotriacetic acid (NTA) sensor chip activated with 0.5 mM NiSO4. The sensorgram of binding for five antibody concentrations increasing from 0.1 nM to 11 nM was corrected against two blank runs before curve fitting using a 1:1 surface binding model. Association constant (ka) and dissociation constant (kd) were calculated with a standard error of approximately 1% and then used to determine the affinity constant (KD). Goodness of fit as measured with Chi2 (mean square of the residual profile) and uniqueness value (U-value, uniqueness of the calculated rate constants and Rmax) indicate reliable data (Chi2 = 0.105, U-value = 2).
SOX18 is a small (44 kDa) yet complex protein, with only one structurally well-defined functional DNA binding domain, the HMG-box. When expressed as a separate fragment, the HMG-box is easy to crystallize and conserves its native ternary structure and functions. This enabled detailed biochemical analyses and in-silico crystal molecular dynamics simulations on SOX18 HMG-box.24,28 Conversely, other SOX18 functional domains, such as C-terminal transactivation domain(s) were arbitrarily suggested based on phylogenetic studies, and the presence of consensus sequences or local enrichments in specific amino acids.33 The boundaries of these functional domains are therefore hypothetical, at best ill-defined.
Therefore, to gain a better understanding of the possible mode of action of the putative blocking antibody, we first assessed the roles and level of conservation of the three α-helical structural components of SOX18 DNA-binding domain (HMG-box, Figure 1A), using in silico analysis based on variant databases and conservation across evolution. As shown in the box plot of Figure 2A, comparing frequency of cancerous somatic mutation (COSMIC variants) to exome variants (EVS database), somatic mutations of human SOX HMG domains are strongly linked to cancer, and human HMG domains display very little “tolerance” for exome variations. Both evidences point towards the HMG domain's functional importance (Fig. 2A, and Suppl. to Fig. 2A). Based on 10 nanoseconds molecular dynamic simulations performed on HMG models of 20 human SOX, in the presence or absence of DNA, amino acids displaying significant differences in carbon alpha root-mean-square deviation (RMSD) of atomic position (Å) were observed in α-Helix 1 and 2 and accounted for two-thirds of all COSMIC variants identified in human SOX HMG domains (Fig. 2B, Suppl. to Fig. 2A, and Suppl. to Fig. 2B). While α-Helix 1 and 2 seem mostly involved in DNA binding and HMG structural configuration, α-Helix 3′s functional importance could not clearly be linked to either (Fig. 2B-C, and Suppl. to Fig. 2C). Further, an open reading frame (ORF)-based natural selection analysis revealed that α-Helix 3 and its direct surroundings were by far the most conserved region in vertebrate SOX18 HMG domains (Fig. 2D, and Suppl. to Fig. 2D, panels A and B), yet did not interact with DNA (Fig. 2B). This last observation suggests that α-Helix 3 is likely involved in protein-protein interactions. In summary, comparison of human SOX variants and computational analysis of phylogenetic, functional and structural features of SOX18 protein suggest that protein-protein interaction might be driven by a region conserved in the α-Helix 3 or in its vicinity. This in silico approach enabled us to predict that the α-Helix 3 region of the SOX18 protein is likely to contain the F5 binding epitope.
Figure 2.
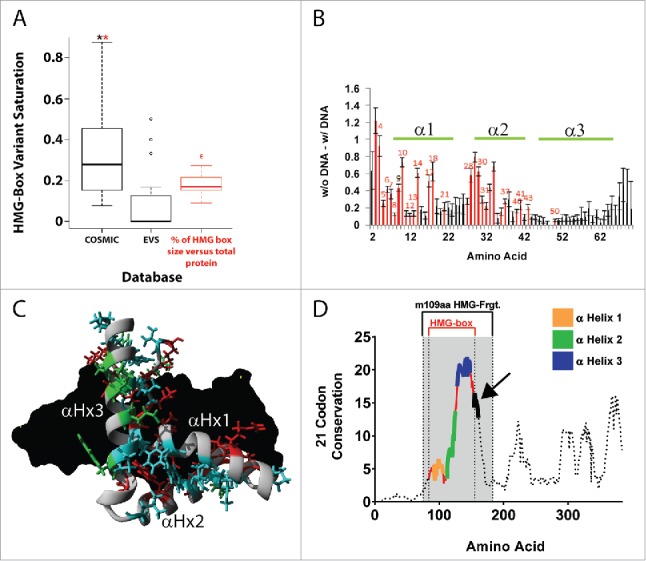
human SOX HMG domains have a very low incidence of exome variants and somatic mutations are strongly linked to cancer. Highly-conserved vertebrate SOX18 α-Helix 3 does not interact with DNA. A. Box plot for amino acid variant saturation in the HMG-box domain of 20 human SOX proteins in COSMIC database (Catalogue Of Somatic Mutations In Cancer, http://cancer.sanger.ac.uk/cosmic, UK) and EVS database (Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/, Seattle, WA), relative to the amino acid percentage of the HMG-box for SOX proteins in red. A significant difference of P < 0.05 using repeated measures ANOVA followed by post-hoc Wilcoxon signed rank test or Bonferroni corrected pairwise t-test was determined for the COSMIC dataset to EVS (black *) and for the COSMIC dataset to the percentage of HMG box in total protein (red *). A table of the sequence alignment of the human SOX proteins across groups A-H, and the variant details for COSMIC and EVS can be found in Supplemental to Figure 2A. B. 10 nanoseconds (ns) molecular dynamic simulations on 20 human SOX HMG protein models in presence or absence of DNA. Average difference in movement of each amino acid in is displayed with SEM error bars across all 20 simulations in presence or absence of DNA. Amino acids in red show significant (t-test, P < 0.01) difference between simulations with DNA and without DNA. Amino acids with red number labels are those with identified COSMIC variants that are significantly different. Green lines indicate HMG-box alpha-helices 1, 2 and 3 positions. Details of Molecular dynamic simulations can be found in Supplemental to Figure 2B. C. Location of the amino acids with proposed functions: cyan = contributes to the structural fold of the HMG box, red = contacts DNA based on known structures, green = known or potential site for protein interaction. DNA is shown in black. The fold of the HMG box is homologous in all SOX proteins as shown by structural alignment of all known SOX protein structures (Supplemental to Figure 2C). Amino acids involved in DNA contacts contribute to sequence specific binding of SOX transcription factor proteins to DNA element ACAAT. D. SOX18 Sequence Conservation using ORFs in 102 vertebrate species: Maximum Likelihood analysis of natural selection codon-by-codon. Computational details can be found in Supplemental to Figure 2D (panels A and B).
To establish experimentally the mechanism by which the antibody likely interferes with SOX18 protein activity, we first used a fluorescence polarisation (FP)-based assay able to measure SOX18 DNA binding activity to a specific oligonucleotide harbouring a SOX binding site previously reported in the Prox1 gene.34 FP-based binding competition assay demonstrated that neither antibody nor scFv competed with the binding of HMG domain to its Prox1 DNA consensus element, instead forming a “SOX18:DNA:mAb” ternary complex (Fig. 3A). The difference in plateau height (Fig. 3A) is due to the fact that the mP index value increases with the molecular weight of the final assembly.35 In one condition, it reached ∼ 215 kDa for SOX18:DNA:mAb (40 kDa SOX18 + 25 kDa DNA + 150 kDa IgG), but only 90 kDa for SOX18:DNA:scFv (40 kDa SOX18 + 25 kDa DNA + 25 kDa scFv). Here, we took advantage of reformatting the scFv into IgG to show that a specific increase of the mP index value correlates with a change in molecular weight. Further, these concentration-dependent data fitted very well to a Hill slope monovalent (1:1) model (R2 > 0.9), returning dissociation constants in the low- to mid-nanoMolar range (Suppl. to Fig. 3, panel B). This finding suggests that while SOX18-HMG domain binds to the DNA via α-Helix 1 and 2, it may remain capable of eliciting protein-protein interaction in a DNA-bound state via its α-Helix 3 region, as predicted by our in-silico analysis.
Figure 3.
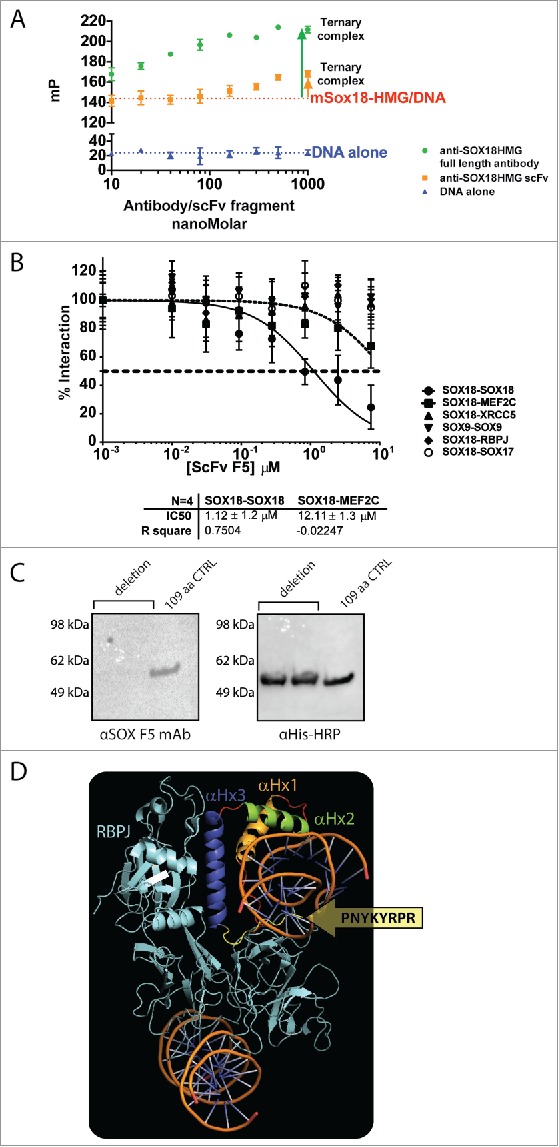
F5 scFv and mAb binding does not compete with DNA and selectively disrupts SOX18 homodimerization. A. Fluorescence polarisation-based measurement of F5 mAb concentration-dependent binding with 109-aa Sox18 fragment used for phage library affinity screening, denotes the formation of a ternary “DNA-Sox-mAb” complex. Despite binding to the HMG domain or in its vicinity, the F5 mAb does not compete with DNA binding to Sox18 HMG-box. Experimental data used for the fitting were obtained from independent triplicates. B. Representative ALPHA-Screen concentration-response curve for SOX18 Protein-Protein Interaction disruption by F5 scFv. Data shown are mean ± SEM. ALPHAScreen was performed as previously described.38,39 The assay for disruption of protein-protein interaction (IC50) was conducted by expressing the protein pairs in Leishmania tarentolae cell-free extract and incubating with a dilution range of tested scFv (0.01 to 7.5 µM) for 1h. Percentage of interaction was calculated as: from 3 independent experiments. C. F5 MAb does not bind to the 101-aa YRPRRKKQ deletion mutant of 6HIS-MBP-SOX18-109 (Left panel, lanes 1 and 2), compared to unchanged 109-aa control in lane 3. The right panel corresponds to expression controls with HRP-coupled anti-His tag Mab. D. Docking of the SOX18/DNA structure 24 into the structure of the Notch transcription complex. To investigate possible protein-protein interaction sites of SOX18, we used in silico protein-protein docking, in combination with MD simulations, to build a complex model of SOX18/DNA with its protein partner RBPJ. For RBPJ we used the X-ray crystal structure of a section of the human Notch transcription complex, elucidated in 2012.23 This section contains the ankyrin (ANK) repeat domain, the RBPJ-J-associated molecule (RAM) domain of the Notch intracellular domain, bound to coactivator MAML1, and the transcription factor RBPJ bound to its consensus DNA. Docking the SOX18/DNA structure into the structure of this Notch transcription complex with subsequent MD simulation for optimization resulted in a RBPJ/SOX18 interaction mediated by the HMG domain. The interaction between SOX18 and RBPJ (cyan) is provided by the C-terminal part of α-Helix 3 depicted in purple (residues Gln138, Arg141, Asp142, and His143) and was refined to exclude amino acid residues from the C-terminal tail of the HMG domain part of F5 Mab epitope.
To further investigate the effect of the F5 scFv antibody on SOX18 protein partner recruitment, we next took advantage of an ALPHAScreen-based technology. This approach is founded on the analysis of pairwise interaction in a cell-free protein expression system36-39 using a human proteome library. Protein-protein interaction (PPI) measurement of full-length SOX18 with itself and various protein binding partners showed that the scFv antibody was able to selectively disrupt SOX18 homodimerization, but none of SOX18 heterodimerizations with known protein partners such as MEF2C, XRCC5, RBPJ or SOX1727,28 (Fig. 3B). The homodimerization of SOX9, a well-characterized dimer40 in the SOX family, was not affected either, suggesting different interaction modalities for SOX9 and SOX18 homodimers (Fig. 3B). The scFv antibody failed, however, to recognize a 31-aa peptide corresponding to just the α-Helix 3 of the HMG box (Suppl. to Fig. 3, panel C). This could be due to the weak helicity of α-Helix 3 peptide when removed from the remainder of the HMG structure, but even in that case, some “weak” recognition would still be expected (Suppl. to Fig. 3, panel C). Altogether, these results suggest that the antibody and scFv, which were generated from a 109-aa peptide of mouse SOX18 HMG domain, most likely interact with the C-terminal region directly adjacent to SOX18 HMG domain α-Helix 3 (Suppl. to Fig. 3, panel A).24
To pinpoint the exact binding location of the scFv, we next performed an epitope mapping approach using the mouse SOX18 109-aa fragment used for the initial biopanning of the human scFv phage library. We used various truncated versions of the same 6HIS-MBP-tagged 109-aa construct, concentrating on α-Helix 3 and directly adjacent C-terminal region (Suppl. to Fig. 3, panel D). This approach confirmed that the antibody and scFv antibody fragment interact with an 8-aa motif, directly adjacent to SOX18 HMG domain (Fig. 3C, Suppl. to Fig. 3, panels E-G).
The fact that F5 antibody disrupts SOX18 homodimer but not SOX18 heterodimer formation suggests that this 8-aa motif is likely to be involved in protein self-recruitment. To gain a better understanding of the difference between homo- and hetero-dimer formation, a SOX18:RBPJ complex was investigated by in silico docking using ClusPro (Fig. 3D). This analysis takes advantage of the SOX18/DNA:Notch-RBPJ transcription complex28 generated from SOX18/DNA crystal,24 and Notch-RBPJ transcription complex crystal.23 In this model, the epitope sequence binding to the scFv is located outside the SOX18:RBPJ interface, exposed to the solvent. This spatial configuration might enable the SOX18/RBPJ complex to recruit the antibody without interfering with its own heterodimer interface, mainly involving α-Helix 3. This in silico analysis further defines a hot spot interface that could be targeted to disrupt a specific PPI, essential to mediate the activity of this transcriptional complex (Fig. 3B-D).
We previously demonstrated using small molecule inhibitors, that modulation of SOX18 protein-protein interactions disrupts its transcriptional activity.27,28 Here, we tested whether the scFv antibody co-expressed with full-length SOX18 and a luciferase reporter construct was able to inhibit SOX18-mediated transcriptional activation, in a cell-based assay. In this assay, we used a luciferase gene placed under the control of SOX18-dependent Vcam-1 gene promoter fragment (Fig. 4).34 Results reveal that SOX18 activity is repressed in the presence of increasing levels of transfected plasmid expressing the F5 scFv blocking antibody, further validating the use of the scFv as a potential blocker of SOX18-mediated transcription. One of the main limitation of the transient transfection assay is that the apparent EC50 (potency) cannot be estimated accurately, since the level of F5 scFv antibody intracellular expression is unknown, except for the amount of expression vector transfected. In summary, we report the first in class SOX18 blocking antibody, which might prove useful as therapeutics when efficient trans-membrane delivery methods become available for mAbs or antibody scFv.41
Figure 4.
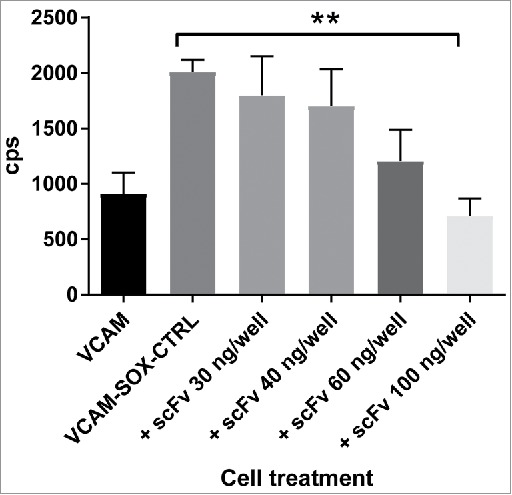
scFv F5 mAb expressed in situ in fibroblastic cells inhibits Sox18-mediated luciferase expression. Luciferase reporter assay performed in fibroblast cells (COS-7) transiently transfected with SOX18 and a vector containing 1889 bp of the proximal Vcam1 promoter construct fused to the firefly luciferase reporter gene. Cells were transfected for 7 hours with aforementioned vectors along with an empty pcDNA 3.1 vector or the same vector containing the ORF of C-terminal myc-tagged scFv F5 mAb. Following an 18 hour-recovery and expression period, Sox18-mediated luciferase activity was measured as depicted in bar graph. Data are corresponding to three independent experiments with 6 internal replicates, error bars are SD of the mean.
Discussion
Here, we report the discovery and characterization of a new biologic that proves to be useful for deciphering the molecular mode of action of a transcriptional regulator, the SOX18 transcription factor.27,28 The novel human antibody recognizes a highly conserved 8-aa motif directly positioned on the C-terminal extremity of the HMG-Box of the SOX18 protein. This antibody displays selective disruption of SOX18 self-assembly and inhibits SOX18-mediated transcriptional activation in cells. As expected, the reformatting into a complete human IgG1 antibody (Suppl. to Fig. 1, panel C) substantially improved affinity as measured by the dissociation constant (Fig. 1D, Suppl. to Fig. 3, panel B). However, the overall effect of reformatting on SOX18-blocking efficacy remains to be evaluated in vitro and in vivo. If the F5 mAb affinity level can be preserved during affinity maturation, its efficacy in blocking SOX18 homodimerization could be further improved by decreasing its concentration-independent off rate, currently estimated at 10−3 s−1 (Fig. 1D). Despite dissociation constants in the low to mid nanoMolar range, denoting strong affinity from full-length antibody or scFv for their epitope, neither were able to competitively displace the HMG-box from its DNA binding site (Fig. 3A, and Suppl. to Fig. 3, panel B). These data combined with our results obtained on protein partner recruitment using ALPHAScreen assay (Fig. 3B) strongly suggest that the mode of action of this antibody is via disruption of SOX18′s protein-protein interaction. The possibility that the epitope was located on α-Helix 1 or 2 was rapidly dismissed, as these two helixes are largely involved in protein-DNA interaction (Suppl. to Fig. 3, panel A, red crosses indicate regions involved in protein-DNA binding), which would not fit with observed non-competitive binding.24 One potential position for the F5 epitope was α-Helix 3, involved only in a limited manner in protein-DNA interaction (Suppl. to Fig. 3, panel A), and already described in literature as engaged in protein-protein interactions.42,43 However, a α-Helix 3 peptide was not recognized by the scFv F5 antibody (Suppl. to Fig. 3, panel C). This pointed towards an epitope located in the N-terminal region outside the SOX18 HMG-box or in the C-terminal region adjacent to the HMG-box, near α-Helix 3. On the 109-aa peptide, the N-terminal region adjacent to the HMG domain consists of only 9 amino acids, compared to the 28-aa C-terminal region. In addition, SOX9 homodimer is not disrupted by the scFv F5 antibody (Fig. 3B), and a SOX9 dimerization domain has been identified on the N-terminal side of SOX9 HMG.40 Taken together, this evidence prompted us to prioritize epitope mapping on the C-terminal region of SOX18-HMG.
Across all SOX proteins, the HMG-box shares at least 46% of sequence homology.33 Just outside of this domain, however, SOX homology diverges due to decreased selection pressure.33 In this context, the antibody did indeed recognize both human and mouse SOX18 (Fig. 1B-D), which share 93% homology in this region (Calculated ClustalW similarity score, Fig. 1A), while it did not recognize divergent human SOX2. Interestingly, codon sequence conservation analysis showed that the 8-aa epitope coincides with a highly conserved region (Fig. 2D, black arrow), suggesting an important role in the binding of some SOX18 protein partner.27
The protein SOX18 is a key molecular switch driving angio- and lymphangio-genesis during development, and a molecular target in various pathophysiological conditions. We hereby demonstrated that it can be pharmacologically modulated using an antibody that inhibits its transcriptional activity by disrupting its ability to recruit other proteins (in this case homodimerization) required to activate gene trans-activation. TF protein-protein interactions do not occur exclusively between transcription factors and cofactors in the nucleus. In 2010, Malki et al. identified nucleocytoplasmic shuttling canonical sequences in SOX protein sequences, especially in and around the HMG domain.44 This study identified an Importin-β binding motif, which precisely overlaps the 8 aa epitope reported in this study. It is therefore possible that blockade of SOX18 activity occurs either via interference with protein nuclear trafficking or co-factor recruitment. Further evidence of the sub-cellular localization of the scFv antibody will help to pinpoint the exact mode of action. The control of SOX proteins access to their target genes is potentially a powerful approach to modulate specific genetic programs. Further, the disruption of specific transcriptional complex is an alternative methodology that would allow subtle targeting of gene subsets. Both avenues open new attractive molecular strategies to study TF mechanisms.
Material and methods
In silico analysis of SOX18 natural selection and human SOX variants
Codon selection analysis of SOX18
For each codon, estimates of the number of inferred synonymous (s) and nonsynonymous (n) substitutions are presented along with the number of sites that are estimated to be synonymous (S) and non-synonymous (N). These estimates are produced using the joint Maximum Likelihood reconstructions of ancestral states under a Muse-Gaut model45 of codon substitution and Tamura-Nei model46 of nucleotide substitution. Maximum Likelihood computations of dN and dS were conducted using HyPhy software package.47 The analysis involved 102 nucleotide sequences with a total of 384 positions in the final dataset using MEGA5.48
Analysis of genetic variants in 20 human SOX proteins
Amino acid variant saturation was compared in the HMG-box domain of 20 human SOX proteins between COSMIC database49 and EVS database (Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/, Seattle, WA), relative to the amino acid percentage of the HMG-box for SOX proteins. A significant difference of P < 0.05 using repeated measures ANOVA followed by post-hoc Wilcoxon signed rank test or Bonferroni corrected pairwise t-test was determined for the COSMIC dataset to EVS and for the COSMIC dataset to the percentage of HMG box in total protein. A table of the sequence alignment of the human SOX proteins across groups A-H, and the variant details for COSMIC and EVS can be found in material supplemental to Fig. 2A.
Molecular dynamic simulation across 20 human SOX HMG models
Molecular dynamic simulations were performed for 10 nanoseconds (ns) on all 20 human SOX HMG protein models with or without DNA. Difference in movement (root-mean squared fluctuation, RMSF) was analysed for each amino acid in presence or absence of DNA across all 20 models. Amino acids displaying significant difference between simulations with DNA and without DNA were identified (t-test, P < 0.01).
SOX18-RBPJ in silico docking
The protein-protein docking between Notch1 transcription complex and SOX18 was performed with ClusPro online server version (cluspro.bu.edu), using pdb: 3V79 and pdb: 4Y60 for the structures of Notch1 transcription complex and SOX18-HMG, respectively. DNA molecules were removed before docking, as ClusPro is unable to process them, and restored after docking. Docking solutions clashing with DNA molecules were rejected.28 The model was further refined by rejecting docking solutions clashing with ScFv epitope location considering its inability to inhibit SOX18-RBPJ interaction. The resulting top docking pose was used as starting conformation in a 50 ns MD simulation to optimize the docking pose, and validate the stability of the new multi protein complex.
SOX18 fragments preparation
SOX18 peptide fragment preparation for phage library panning
The 109-aa mouse SOX18 HMG fragment used for phage library panning was BP cloned from cDNA templates (IMAGE cDNA clone IDs, Sox18: 3967084) into a pDONR™221 pENTRY vector, sequenced and recombined into a pETG20A or a pHisMBP expression plasmid using Gateway®LR Technology.50 Constructs were transformed into Escherichia coli BL21(DE3) cells (Luria-Bertani, 100 μg/ml Ampicillin).
His-MBP-Sox18-109 and His-MBP-Sox18-79 fusion peptide fragments expression and preparation for Western blot analysis
The 109 aa residues corresponding to the full HMG Box region of mouse Sox18 (Sox18-109) and a shorter 79 residue version of the HMG Box of mouse Sox18 (Sox18-79) were cloned into the pDest-HisMBP gateway vector for expression with an N-terminal His-MBP fusion partner. The His-MBP-Sox18-109 and His-MBP-Sox18-79 gene constructs were transformed into BL21 DE3 Star chemically competent cells (Invitrogen, ThermoFisher Scientific) using standard heat shock protocol and plated on LB-Agar with 100 µg/ml Ampicillin. Colonies were inoculated into 3 ml LB medium with 100 µg/ml ampicillin and grown for 2–4 hrs until OD600 of 0.4–0.6 was obtained. Fusion protein expression was induced by the addition of IPTG to the cultures to a final concentration of 1 mM and cultures were induced at 20°C for 20hrs. IPTG-induced cultures for His-MBP-Sox18-79 and His-MBP-Sox18-109 were mixed with PAGE loading dye with reducing agent, heated at 95°C for 5 mins and samples run on 4–12% NuPAGE Bis-Tris PAGE (Invitrogen, ThermoFisher Scientific). Proteins were transferred to PVDF membranes, the membranes washed with 0.05% Tween-20 in 1x phosphate-buffered saline (PBST) and blocked in 2% (w/v) skim milk in PBST for 30 mins at room temperature (RT). The membranes were probed for 60 mins at RT with the anti-Sox18 MAb F5 (50 µg/ml in block solution) to map its Sox18 binding site. The membranes were then washed 3xPBST/5 mins and probed for 60 mins at RT with horseradish peroxidase (HRP)-labelled anti-human kappa antibody (The Binding Site, Ltd, Birmingham, UK, Cat# AP015) diluted 1/1000 in blocking solution. The proteins were probed on separate PVDF membranes with HRP anti-His (Miltenyi Biotech AG, Germany, Cat# 130-092-785) diluted 1/2000 in block solution to confirm fusion protein expression. The membranes were washed 3xPBST/5 mins and the proteins detected by chemiluminescence with ECL substrate (Invitrogen, ThermoFisher Scientific). The binding of F5 MAb to human SOX18 and human SOX2 expressed in HEK lysates (Origene) was also determined by immunoblot with HEK lysate alone as a control.
SOX18-109 truncations and deletion for F5 mAb epitope mapping
Site Directed Mutagenesis PCR (SDM-PCR) was used to generate variants of Sox18-109 to determine the epitope recognized by the anti-sox18 MAb F5. For initial epitope mapping studies, SDM-PCR with specific oligonucleotides was used to introduce stop codons at specific sites in the gene encoding Sox18-109 to create C-terminal truncation variants. This included Sox18 truncated at residues 20, 41, 56, 63 and 72. A variant with a deletion of residues 80–87 “YRPRRKKQ” was also generated for the epitope mapping of F5 mAb. PCR products representing the variants of His-MBP-Sox18-109 were gel-purified, treated with Dpn1 (New England Biolabs, Inc.) for 3 hrs at 37°C and was transformed into BL21 DE3 Star chemically competent cells (Invitrogen, ThermoFisher Scientific). The fusion protein expression and immunoblots were performed similarly to His-MBP-Sox18-109 and His-MBP-Sox18-79 as already outlined.
Phage library panning
A human naive scFv antibody library in phagemid vector pHEN1, with a reported diversity of 6.7 × 109, was kindly provided by Prof. James Marks (University of California, San Francisco, USA).51 Biopanning was conducted against the mSOX18 109-aa fragment for three rounds based on the standard method.52 In brief, immunotubes (Nunc Maxisorp, ThermoFisher Scientific) were coated overnight with 1 mL of truncated mSox18 in PBS at 10 µg/mL, followed by a series of washes with PBS. The tubes were subsequently blocked with 2% skim milk PBS for 1 hour at room temperature. In parallel, 1013 phage particles from the naïve scFv antibody library were blocked in 2% skim milk PBS for one hour and subsequently transferred to the immunotubes and incubated for a further 1 hour. Unbound phage was removed by washing the tubes three times with 0.1% PBST and three washes with PBS alone. Bound phage was eluted with 200 mM glycine pH 2.5 and neutralized with the addition of 1 M Tris-HCl pH 7.4. The eluate was then used to infect log phase XL1-blue cells grown in 2YT medium supplemented with 3 ug/mL tetracycline for 30 minutes. The cells were centrifuged, resuspended in 2x YT media and spread onto 2YT agar plates supplemented with 100 µg/mL ampicillin and 2% glucose (2YT-AmpGlu). Plates were incubated at 30°C overnight. Cells were detached from the plate with 2YT AmpGlu media and 20% glycerol and stored at −80°C.
Phage particles were rescued from the E. coli glycerol stocks and grown to log phase at 37°C in 2YT-AmpGlu followed by an addition of 1011 M13KO7 helper phage particles and incubated for 30 minutes at 30°C. The cells were centrifuged, resuspended in 2YT supplemented with 100 µg/mL ampicillin and 30 µg/mL kanamycin (2YT-AmpKana) and incubated overnight at 30°C. Phage particles were recovered from the culture supernatant by two rounds of precipitation with 20% PEG, 2.5M NaCl, and stored at −80°C in PBS-20% glycerol. This stock was then used for the second round of panning, using 1012 particles, and similarly a third round of panning using 1011 particles. Enrichment of the library was assessed by ELISA in 96-well plates (Nunc Maxisorp, ThermoFisher Scientific) coated with 5 µg/mL truncated Sox18 and blocked with 2% MPBS. Serial 10x dilutions of purified phage pools from each round of panning were incubated in the wells prior to washing and detection of phage by HRP-labelled anti-M13 mAb (GE Healthcare, Cat # 27-9421-01).
Analyses of individual clones were done by randomly selecting individual phage infected E. coli colonies and rescuing the phage as previously described, with the omission of the precipitation step. The supernatants containing the secreted phage particles were then assessed for reactivity to the mSO18 109 aa fragment through standard ELISA as described above. The scFv DNA sequence was then determined from the positive clones by Sanger sequencing at the Australian Genome Research Facility, using pHEN1-specific primers flanking the scFv sequence.53
Production of scFvs
DNA from two clones (B11 and F5) selected from the Sheets library panning was recovered by (miniprep) and used as templates in PCR with primers to incorporate a C-terminal cysteine residue, a 6xHis tag and HindIII and XbaI sites. PCR products were subsequently cloned into pcDNA3.1(+) (Life Technologies, Inc., CA, USA). ScFvs were produced in CHO-S by incubating the plasmid DNA with PEI Max in OptiPro media.54 The DNA-PEI complex was subsequently added to the CHO-S cells at 3.0 × 106 cell/mL and incubated for 4 hours at 37°C. Efficient Feed A and Feed B were added at a final concentration of 7.5%, and anti-clumping agent at 0.4% followed by a further incubation at 32°C for 10 days. Cells were removed by centrifugation and His-tagged scFv protein was purified from the culture supernatant by His Trap FF affinity chromatography, eluting the product with 350 mmol L−1 imidazole in phosphate buffer, pH 7.4. This was followed by a buffer exchange into PBS pH 7.4, using a GE Hi Prep 26/10 desalting column. The final product was filtered through 0.22 µm membrane and stored at −20°C.
Reformatting of IgG molecules
The two selected clones (B11 and F5) were reformatted to complete human IgG1 as previously described.55 Briefly the heavy and light chain variable fragments were individually amplified from the phagemid DNA, using primers specific to the variable regions and containing 15 bp overhangs at the 5′ ends to allow ligation-independent cloning into heavy and light chain ReformAb mammalian expression vectors (Acyte Biotech, Brisbane Australia) using the “In Fusion” system™ (Clontech, Inc, CA, USA). The heavy and light chain plasmids for each antibody were co-transfected, at 1:1 ratio, into CHO-S cells using PEI-Max as described above. Cells were removed by centrifugation and IgG was purified from the culture supernatant by protein A HiTrap affinity chromatography, eluting with 0.1 M glycine pH 3.0 followed by buffer exchange into PBS pH 7.4, using a GE Hi Prep 26/10 desalting column and the final product was filtered through 0.22 µm membrane and stored at −20°C.
Homogeneous assays
Surface plasmon resonance
A single cycle kinetic and affinity study of full-length mAb F5 dissociation from a murine His-StrepII-TEV-SOX18 (N-term) immobilized onto a nitriloacetic acid (NTA) sensor chip was performed using a Biacore T200 instrument (Biacore Life Sciences, GE Healthcare, Uppsala, Sweden). NTA sensor chip was activated with 0.5 mM NiSO4 and immobilization was performed with a 100 nM Sox18 solution. The sensorgram of association and dissociation of antibody F5 consisted in a series of five analyte concentrations ranging from 0.1 to 11 nM. Data were corrected against two blank runs before curve fitting using a 1:1 surface binding model. Association constant (ka) and dissociation constant(kd) were calculated using BIAevaluation software (Biacore Life Sciences, GE Healthcare, Uppsala, Sweden), with a standard error of 1% to determine the affinity constant (KD).
Fluorescence polarization-based DNA binding assay
Fluorescence polarization (FP) was used as a homogenous in vitro assay to assess protein / DNA interaction using light polarization as a readout.56 We used an E coli recombinantly expressed 109-aa peptide, corresponding to the HMG domain of mouse SOX18 and a 40bp-long double-stranded oligonucleotides harboring SOX responsive elements and labeled with 5′ fluorescein amidite (FAM) label (GeneWorks).57 The FP assay was run in 384-well black plates (BD Biosciences, USA) in a 25–30 μl final volume using 18FP assay buffer (30 mM HEPES pH 7.5, 100 mM KCl, 40 mM NaCl, 10 mM NH4OAc, 10 mM guanidinium, 2 mM MgCl2, 0.5 mM EDTA, 0.01% NP-40). All data are displayed in milliP [mP] fluorescence polarization index (mP index).
Briefly, 100 nM mSOX18-HMG was preincubated in 384-well plates for 10–15 minutes with full-length antibody or corresponding scFv at concentrations ranging from 10 to 1000 nM. Labeled DNA probe was then added at 5 nM, and the mix briskly agitated for 5 minutes at room temperature protected from ambient light. 384-well plates were sealed (TopSeal®-A, PerkinElmer, USA) and following a further 30-minute incubation at room temperature, fluorescence polarization was read on a Tecan M1000 Pro (λexc = 480 nm, λem = 535 nm). All experiments were performed in internal as well as independent triplicate. Data analysis and plotting were performed with Prism 6 for Mac OS X (version 6.0d, GraphPad Software, Inc., CA, USA) using a basic one site specific binding model;
ALPHAScreen based protein-protein interaction assay
Plasmid preparation and cell-free expression
Transcription factor proteins were genetically encoded with enhanced green fluorescent protein (GFP), mCherry and cMyc (myc) tags, and cloned into cell-free expression Gateway destination vectors: N-terminal GFP tagged (pCellFree_G03), N-terminal Cherry-cMyc (pCellFree_G07) and C-terminal Cherry-cMyc tagged (pCellFree_G08).58 Human RBPJ (BC020780) and MEF2C (BC026341) ORFs were sourced from the Human ORFeome collection, version 1.1 and 5.1, and the Human Orfeome collaboration OCAA collection (Open Biosystems), as previously described38 and cloned at the ARVEC facility, UQ Diamantina Institute. The entry clones pDONOR223 or pENTR201 vectors were exchanged with the ccdB gene in the expression plasmid by LR recombination (Life Technologies, Australia). The full-length human SOX18 gene was synthesized and the transfer to vectors was realized using Gateway PCR cloning. Translation competent Leishmania tarentolae extract (LTE) was prepared as previously described.36,37 Protein pairs were co-expressed by adding 30 nM of GFP template plasmid and 60 nM of Cherry template plasmid to LTE and incubating for 3 hours at 27 °C.
ALPHAScreen
ALPHAScreen was performed as previously described,38,39 using the cMyc detection kit and Proxiplate-384 Plus plates (PerkinElmer). The LTE lysate co-expressing the proteins of interest was diluted in buffer A (25 mM HEPES, 50 mM NaCl). For the assay, 12.5 μL (0.4 μg) of anti-cMyc coated Acceptor Beads in buffer B (25 mM HEPES, 50 mM NaCl, 0.001% NP40, 0.001% casein) were aliquoted into each well. This was followed by the addition of 2 μL of diluted sample, at different concentration, and 2 μL of biotin-labelled GFP-Nanotrap in buffer A. The plates were incubated for 45 min at room temperature, before adding 2 μL (0.4 μg) of streptavidin-coated Donor Beads diluted in buffer A, and incubation in the dark for 45 min at room temperature. The ALPHAScreen signal was measured on an Envision Plate Reader (PerkinElmer), using manufacturer's recommended settings (λexc = 680/30 nm for 0.18 s, λem = 570/100 nm after 37 ms). The resulting bell-shaped curve is an indication of a positive interaction, while a flat line reflects a lack of interaction between the proteins. The measurement of each protein pair was repeated in triplicate.
I is the highest signal level (top of the hook effect curve) and Ineg is the lowest (background) signal level. The signals were normalized to the Iref signal obtained for the strongest interaction. The assay for disruption of protein-protein interaction (IC50) was conducted by expressing the protein pairs in LTE and incubating with a dilution range of tested scFv antibody (0.01 to 7.5 µM) in buffer B for 1h. Percentage of interaction was calculated as: Data from 3 independent experiments were fitted in GraphPad Prism version 6.0 using 3-parameter non-linear regression.
Cell-based assay
Monkey kidney fibroblast-like cells (COS-7) were cultured at 37 °C, 5% CO2 in DMEM (Life technologies, 11995) with fetal bovine serum (FBS), sodium pyruvate, L-glutamine, penicillin, streptomycin, non-essential amino acids and HEPES. Cells were grown in 96-well plates to 80% confluency, and transfected with mouse plasmids pGL2 Vcam-1 promoter construct (VC1889), pSG5 Sox18, and either an empty pcDNA 3.1 vector or containing the ORF of scFv F5. Transfection was performed using X-tremeGENE 9 DNA transfection reagent (Roche, 06365787001).34,57 After 7 h transfection, cells could recover in 0.5% FBS medium for another 18 h, before lysis and luciferase assay (Perkin Elmer, 6016711).
Supplementary Material
Funding Statement
This work was supported by grants from the NHMRC (APP1107643 to MF) and the ARC (ARC IC_160100027 to MF, SM, SK, FF, MJ and CH). MF is supported by a NHMRC Career Developmental Fellowship (APP1111169). National Institutes of Health Office of the Director award K01ES025435 to Jeremy Prokop.
Abbreviations
- aa
amino-acid
- FOX
Forkhead box
- HOX
Homeo-box
- RBPJ
Recombining binding protein suppressor of hairless
- scFv
single-chain variable fragment
- SOX
Sry-related HMG-box
- TFs
Transcription Factors
References
- 1.Francois M, Caprini A, Hosking B, Orsenigo F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456:643–7. doi: 10.1038/nature07391. PMID:18931657 [DOI] [PubMed] [Google Scholar]
- 2.Koopman P. Sry, Sox9 and mammalian sex determination. EXS. 2001;91:25–56. PMID:11301599 [DOI] [PubMed] [Google Scholar]
- 3.Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–78. doi: 10.1016/S0092-8674(00)81511-1. PMID:10499794 [DOI] [PubMed] [Google Scholar]
- 4.Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–7. doi.org/ 10.1242/dev.133900. PMID:27246709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venter JC. et al.. The sequence of the human genome. Science. 2001;291:1304–51. doi.org/ 10.1126/science.1058040. PMID:11181995 [DOI] [PubMed] [Google Scholar]
- 6.Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–8. doi.org/ 10.1038/nature15518. PMID:26550823 [DOI] [PubMed] [Google Scholar]
- 7.Xia K, Fu Z, Hou L, Han JD. Impacts of protein-protein interaction domains on organism and network complexity. Genome Res. 2008;18:1500–8. doi.org/ 10.1101/gr.068130.107. PMID:18687879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. PMID:25531225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi.org/ 10.1038/nrg2538. PMID:19274049 [DOI] [PubMed] [Google Scholar]
- 10.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–4. doi.org/ 10.1038/nature12294. PMID:23783631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi.org/ 10.1016/j.cell.2010.01.044 PMID:20211142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium F, Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi.org/ 10.1038/nature13182. PMID:24670764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyadjiev SA, Jabs EW. Online Mendelian Inheritance in Man (OMIM) as a knowledgebase for human developmental disorders. Clin Genet. 2000;57:253–66. doi: 10.1034/j.1399-0004.2000.570403.x. PMID:10845565 [DOI] [PubMed] [Google Scholar]
- 14.Darnell JE., Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi.org/ 10.1038/nrc906 PMID:12360277 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Bigas N, Blencowe BJ, Ouzounis CA. Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics. 2006;22:269–77. doi.org/ 10.1093/bioinformatics/bti781 PMID:16287936 [DOI] [PubMed] [Google Scholar]
- 16.Jethon A, Pula B, Olbromski M, Werynska B, Muszczynska-Bernhard B, Witkiewicz W, Dziegiel P, Podhorska-Okolow M. Prognostic significance of SOX18 expression in non-small cell lung cancer. Int J Oncol. 2015;46:123–32. doi.org/ 10.3892/ijo.2014.2698. PMID:25310193 [DOI] [PubMed] [Google Scholar]
- 17.Pula B. Olbromski M, Wojnar A, Gomulkiewicz A, Witkiewicz W, Ugorski M, Dziegiel P, Podhorska-Okolow M. Impact of SOX18 expression in cancer cells and vessels on the outcome of invasive ductal breast carcinoma. Cell Oncol (Dordr). 2013;36:469–83. doi.org/ 10.1007/s13402-013-0151-7. PMID:24065215 [DOI] [PubMed] [Google Scholar]
- 18.Eom BW, Jo MJ, Kook MC, Ryu KW, Choi IJ, Nam BH, Kim YW, Lee JH. The lymphangiogenic factor SOX 18: a key indicator to stage gastric tumor progression. Int J Cancer. 2012;131:41–48. doi.org/ 10.1002/ijc.26325. PMID:21796627 [DOI] [PubMed] [Google Scholar]
- 19.Young N, Hahn CN, Poh A, Dong C, Wilhelm D, Olsson J, Muscat GE, Parsons P, Gamble JR, Koopman P. Effect of disrupted SOX18 transcription factor function on tumor growth, vascularization, and endothelial development. J Natl Cancer Inst. 2006;98:1060–7. doi.org/ 10.1093/jnci/djj299. PMID:16882943 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Huang S, Dong W, Li L, Feng Y, Pan L, Han Z, Wang X, Ren G, Su D, Huang B, Lu J. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009;277:29–37. doi.org/ 10.1016/j.canlet.2008.11.014. PMID:19108950 [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Lee S, Lee S, Kim K, Yang Y, Kim JH, Adams RH, Wells JM, Morrison SJ, Koh GY. Sox17 promotes tumor angiogenesis and destabilizes tumor vessels in mice. J Clin Invest. 2013;123:418–31. doi.org/ 10.1172/JCI64547. PMID:23241958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett KL, Hass M, McArthur DG, Ilagan MX, Aster JC, Kopan R, Blacklow SC. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat Struct Mol Biol. 2010;17:1312–7. doi.org/ 10.1038/nsmb.1938. PMID:20972443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SH, Wales TE, Nam Y, O'Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–9. doi.org/ 10.1016/j.str.2011.12.011. PMID:22325781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaus M, Prokoph N, Girbig M, Wang X, Huang YH, Srivastava Y, Hou L, Narasimhan K, Kolatkar PR, Francois M. Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction. Nucleic Acids Res. 2016;44:3922–35. doi.org/ 10.1093/nar/gkw130. PMID:26939885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimhan K, Micoine K, Lacôte E, Thorimbert S, Cheung E, Hasenknopf B, Jauch R. Exploring the utility of organo-polyoxometalate hybrids to inhibit SOX transcription factors. Cell Regen (Lond). 2014;3:10. doi: 10.1186/2045-9769-3-10. PMID:25678957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narasimhan K, Pillay S, Bin Ahmad NR, Bikadi Z, Hazai E, Yan L, Kolatkar PR, Pervushin K, Jauch R. Identification of a polyoxometalate inhibitor of the DNA binding activity of Sox2. ACS Chem Biol. 2011;6:573–81. doi: 10.1021/cb100432x. PMID:21344919 [DOI] [PubMed] [Google Scholar]
- 27.Overman J, Fontaine F, Moustaqil M, Mittal D, Sierecki E, Sacilotto N, Zuegg J, Robertson AA, Holmes K, Salim AA. Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice. Elife. 2017;16;24:346–359. doi.org/ 10.7554/eLife.21221 PMID:28137359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontaine F, Overman J, Moustaqil M, Mamidyala S, Salim A, Narasimhan K, Prokoph N, Robertson AAB, Lua L, Alexandrov K. Small-Molecule Inhibitors of the SOX18 Transcription Factor. Cell Chem Biol. 2017;24(3):346–359. doi.org/ 10.1016/j.chembiol.2017.01.003 PMID:28163017 [DOI] [PubMed] [Google Scholar]
- 29.Hornsby M, Paduch M, Miersch S, Sääf A, Matsuguchi T, Lee B, Wypisniak K, Doak A, King D, Usatyuk S, et al. A High Through-put Platform for Recombinant Antibodies to Folded Proteins. Mol Cell Proteomics. 2015;14:2833–47. doi.org/ 10.1074/mcp.O115.052209. PMID:26290498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffin L, Lawson A. Antibody fragments as tools in crystallography. Clin Exp Immunol. 2011;165:285–91. doi.org/ 10.1111/j.1365-2249.2011.04427.x. PMID:21649648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi.org/ 10.1126/science.1141319. PMID:17540862 [DOI] [PubMed] [Google Scholar]
- 32.Orten DJ, Strawhecker JM, Sanderson SD, Huang D, Prystowsky MB, Hinrichs SH. Differential effects of monoclonal antibodies on activating transcription factor-1 and cAMP response element binding protein interactions with DNA. J Biol Chem. 1994;269:32254–63. PMID:7528210 [PubMed] [Google Scholar]
- 33.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–55. doi.org/ 10.1006/dbio.2000.9883. PMID:11071752 [DOI] [PubMed] [Google Scholar]
- 34.Hosking BM, Wang SC, Downes M, Koopman P, Muscat GE. The VCAM-1 gene that encodes the vascular cell adhesion molecule is a target of the Sry-related high mobility group box gene, Sox18. J Biol Chem. 2004;279:5314–22. doi.org/ 10.1074/jbc.M308512200. PMID:14634005 [DOI] [PubMed] [Google Scholar]
- 35.Pope AJ, Haupts UM, Moore KJ. Homogeneous fluorescence readouts for miniaturized high-throughput screening: theory and practice. Drug Discov Today. 1999;4:350–62. doi: 10.1016/S1359-6446(99)01340-9. PMID:10431145 [DOI] [PubMed] [Google Scholar]
- 36.Kovtun O, Mureev S, Jung W, Kubala MH, Johnston W, Alexandrov K. Leishmania cell-free protein expression system. Methods. 2011;55:58–64. doi.org/ 10.1016/j.ymeth.2011.06.006. PMID:21704167 [DOI] [PubMed] [Google Scholar]
- 37.Mureev S, Kovtun O, Nguyen UT, Alexandrov K. Species-independent translational leaders facilitate cell-free expression. Nature biotechnology. 2009;27:747–52. doi.org/ 10.1038/nbt.1556. PMID:19648909 [DOI] [PubMed] [Google Scholar]
- 38.Sierecki E, Giles N, Polinkovsky M, Moustaqil M, Alexandrov K, Gambin Y. A cell-free approach to accelerate the study of protein-protein interactions in vitro. Interface Focus. 2013;3:20130018. doi.org/ 10.1098/rsfs.2013.0018. PMID:24511386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sierecki E, Stevers LM, Giles N, Polinkovsky ME, Moustaqil M, Mureev S, Johnston WA, Dahmer-Heath M, Skalamera D, Gonda TJ. Rapid mapping of interactions between Human SNX-BAR proteins measured in vitro by AlphaScreen and single-molecule spectroscopy. Mol Cell Proteomics. 2014;13:2233–45. doi.org/ 10.1074/mcp.M113.037275. PMID:24866125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang YH, Jankowski A, Cheah KS, Prabhakar S, Jauch R. SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci Rep. 2015;5:10398. doi.org/ 10.1038/srep10398. PMID:26013289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buecheler JW. et al.. Development of a protein nanoparticle platform for targeting EGFR expressing cancer cells. Journal of Chemical Technology & Biotechnology. 2015;90:1230–6. doi.org/ 10.1002/jctb.4545 [DOI] [Google Scholar]
- 42.Merino F, Ng CKL, Veerapandian V, Schöler HR, Jauch R, Cojocaru V. Structural basis for the SOX-dependent genomic redistribution of OCT4 in stem cell differentiation. Structure. 2014;22:1274–86. doi.org/ 10.1016/j.str.2014.06.014. PMID:25126959 [DOI] [PubMed] [Google Scholar]
- 43.Prokop JW, Leeper TC, Duan ZH, Milsted A. Amino acid function and docking site prediction through combining disease variants, structure alignments, sequence alignments, and molecular dynamics: a study of the HMG domain. BMC bioinformatics. 2012;13(Suppl 2):S3. PMID:22536866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malki S, Boizet-Bonhoure B, Poulat F. Shuttling of SOX proteins. The international journal of biochemistry & cell biology. 2010;42:411–6. doi: 10.1016/j.biocel.2009.09.020 doi.org/ 10.1016/j.biocel.2009.09.020 [DOI] [PubMed] [Google Scholar]
- 45.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–24. PMID:7968485 [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. PMID:8336541 [DOI] [PubMed] [Google Scholar]
- 47.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079 doi.org/ 10.1093/bioinformatics/bti079. PMID:15509596 [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092 doi.org/ 10.1093/molbev/msm092. PMID:17488738 [DOI] [PubMed] [Google Scholar]
- 49.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi.org/ 10.1093/nar/gku1075. PMID:25355519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012;40:4933–41.doi.org/ 10.1093/nar/gks153. PMID:22344693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, Hemingsen G, Wong C, Gerhart JC, Marks JD. Efficient construction of a large nonimmune phage antibody library: The production of high-affinity human single-chain antibodies to protein antigens. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6157–62. doi.org/ 10.1073/pnas.95.11.6157. PMID:9600934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nature biotechnology. 2005;23:1105–16. doi: 10.1038/nbt1126. PMID:16151404 [DOI] [PubMed] [Google Scholar]
- 53.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization: Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–97. doi: 10.1016/0022-2836(91)90498-U. PMID:1748994 [DOI] [PubMed] [Google Scholar]
- 54.Codamo J, Hou JJC, Hughes BS, Gray PP, Munro TP. Efficient mAb production in CHO cells incorporating PEI-mediated transfection, mild hypothermia and the co-expression of XBP-1. Journal of Chemical Technology & Biotechnology. 2011;86:923–34. doi: 10.1002/jctb.2572 doi.org/ 10.1002/jctb.2572 [DOI] [Google Scholar]
- 55.Jones ML, Seldon T, Smede M, Linville A, Chin DY, Barnard R, Mahler SM, Munster D, Hart D, Gray PP. A method for rapid, ligation-independent reformatting of recombinant monoclonal antibodies. Journal of Immunological Methods. 2010;354:85–90. doi.org/ 10.1016/j.jim.2010.02.001. PMID:20153332 [DOI] [PubMed] [Google Scholar]
- 56.Kim J. et al.. Development of a fluorescence polarization assay for the molecular chaperone Hsp90. J. Biomol. Screen. 2004;9:375–81. doi.org/ 10.1177/1087057104265995. PMID:15296636 [DOI] [PubMed] [Google Scholar]
- 57.Duong T, Koltowska K, Pichol-Thievend C, Le Guen L, Fontaine F, Smith KA, Truong V, Skoczylas R, Stacker SA, Achen MG. VEGFD regulates blood vascular development by modulating SOX18 activity. Blood. 2014;123:1102–12. doi.org/ 10.1182/blood-2013-04-495432. PMID:24269955 [DOI] [PubMed] [Google Scholar]
- 58.Gagoski D, Mureev S, Giles N, Johnston W, Dahmer-Heath M, Škalamera D, Gonda TJ, Alexandrov K. Gateway-compatible vectors for high-throughput protein expression in pro- and eukaryotic cell-free systems. J Biotechnol. 2015;195:1–7. doi.org/ 10.1016/j.jbiotec.2014.12.006. PMID:25529348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


