Abstract
Motivation
Accurately mapping and annotating genomic locations on 3D protein structures is a key step in structure-based analysis of genomic variants detected by recent large-scale sequencing efforts. There are several mapping resources currently available, but none of them provides a web API (Application Programming Interface) that supports programmatic access.
Results
We present G2S, a real-time web API that provides automated mapping of genomic variants on 3D protein structures. G2S can align genomic locations of variants, protein locations, or protein sequences to protein structures and retrieve the mapped residues from structures. G2S API uses REST-inspired design and it can be used by various clients such as web browsers, command terminals, programming languages and other bioinformatics tools for bringing 3D structures into genomic variant analysis.
Availability and implementation
The webserver and source codes are freely available at https://g2s.genomenexus.org.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
With extensive recent large-scale genome sequencing projects such as The Cancer Genome Atlas (TCGA) (Weinstein et al., 2013), and 1000 Genomes Project (Auton et al., 2015), a great number of germline and somatic genomic variants are being detected. Protein structure changes due to genome variation in protein-coding regions are interesting for genetic marker discovery and interpretation of disease mechanisms. Mapping genomic variants onto the specific 3D protein structures is the first critical step to analyze the variants in the context of protein structures.
Several resources have been developed to address the need of mapping protein positions to protein structures. SIFTS (Velankar et al., 2013) maps UniProt (Bateman et al., 2017) entries to Protein Data Bank (PDB) (Berman et al., 2000) entries and provides XML and flat files for download. PDB utilizes SIFTS and provided a user web interface for accessing the mapping (http://www.rcsb.org/pdb/chromosome.do) (Berman et al., 2000; Prlic et al., 2016). G23D (Solomon et al., 2016) also provides a user web interface for mapping genomic variants onto 3D protein structures. However, none of them provides a web API for programmatic access of the sequence alignments and residue mapping.
Here, we present G2S, a web API that supports programmatic mapping and annotation of genomic variants on 3D protein structures. The following functionalities were implemented: (i) retrieving protein structure chains aligned to a primary protein sequence (a UniProt/Ensembl entry or a user-defined sequence); (ii) retrieving mapping between genomic positions and structural positions; and (iii) retrieving mapping between amino acid positions and structural positions. G2S provides a RESTful API. The pre-computed alignments are automatically updated weekly to keep up to date with the PDB structure archive. G2S API and source codes are publicly available at https://g2s.genomenexus.org/.
2 G2S pipeline
The G2S backend pipeline collects and aligns UniProt and Ensembl protein sequences along with carefully parsed PDB sequences using BioJava (Holland et al., 2008). Raw protein sequences were retrieved from the atom records of PDB files directly to avoid flaws and inconsistences from SEQRES and DBREF records in PDB files. The alignments of the protein sequences against the PDB sequences by BLASTP (Altschul, 1997) were stored in a relational database. The pipeline updates the pre-computed alignments weekly as new PDB structures added to RCSB PDB. The workflow and architecture of G2S API are shown in Supplementary Figure S1.
3 G2S API
The G2S API accepts UniProt names, UniProt Isoform names, Human Ensembl names, genomic positions (GRCh37 and 38) and user-defined protein sequences. G2S API returns high confidence pre-calculated mapping of the protein/residue aligned to the protein structures. For user-provided protein sequences, G2S API calculates alignments against structure sequences on the fly. Several alignment quality metrics as E-value and bit-score can be used as parameters in the API request to refine alignment results. G2S API is a RESTful service and all API endpoints provide fast real-time responses in JSON format. The details of API endpoints, use cases and additional design details are provided on the web site (https://g2s.genomenexus.org) and in Supplementary Material.
4 Use cases: cBioPortal and 3DHotspots
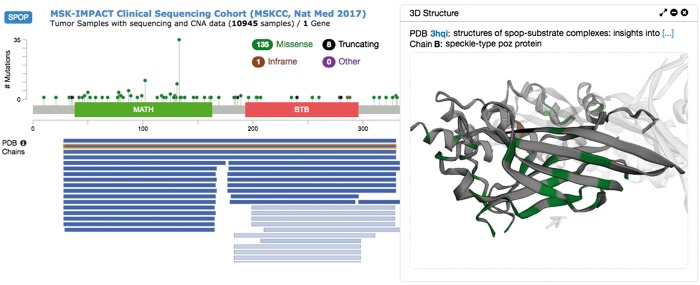
The cBioPortal for Cancer Genomics (http://cbioportal.org), a widely used resource for studying cancer genomics (Cerami et al., 2012; Gao et al., 2013), utilizes the G2S API to retrieve updated sequence-structure alignments and residue mapping to visualize cancer mutations in protein structures (see Fig. 1 for an example). The G2S API is also being used for detecting 3D mutational hotspots in cancer https://github.com/knowledgesystems/mutationhotspots (Gao et al., 2017).
Fig. 1.
Protein structure visualization of SPOP mutations in the MSK-IMPACT study in the cBioPortal. The mutations were plotted along the primary sequence (up left); alignments from primary sequence to PDB chains were plotted underneath (bottom left); the protein structure of the selected alignment was displayed with mutations highlighted in the structure (right)
5 Discussion
The G2S API provides an auto-updated real-time resource for retrieving residue-level sequence-structure alignments. It fills the critical gap that no existing resources provide programmatic access to up-to-date protein structure alignments and residue mapping. The API was designed with high performance and real-time access in mind so that third-party tools such as cBioPortal can achieve smooth user experience when mapping their variants against up-to-date protein structures, and supporting visualization and analysis of variants in the context of protein structures.
Funding
This work has been supported by Google Summer of Code 2016 [JW], National Institutes of Health Grant (R33-GM078601 and R01-GM100701) [DX], the Marie-Josée and Henry R. Kravis Center for Molecular Oncology [NS, JG], a National Cancer Institute Cancer Center Core Grant (P30-CA008748), the Fund for Innovation in Cancer Informatics from the Brown Performance Group (www.BrownPerformance.com/ici) [NS, JG] and the Robertson Foundation [NS].
Conflict of Interest: none declared.
Supplementary Material
References
- Altschul S.F. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res., 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M. et al. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. (2017) 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med., 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland R.C. et al. (2008) BioJava: an open-source framework for bioinformatics. Bioinformatics (Oxford, England), 24, 2096–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A. et al. (2016) Integrating genomic information with protein sequence and 3D atomic level structure at the RCSB protein data bank. Bioinformatics (Oxford, England), 32, 3833–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon O. et al. (2016) G23D: online tool for mapping and visualization of genomic variants on 3D protein structures. BMC Genomics, 17, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar S. et al. (2013) SIFTS: Structure Integration with Function, Taxonomy and Sequences resource. Nucleic Acids Res., 41, D483–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.N. et al. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.