ABSTRACT
Previous studies indicated that hepatitis B virus (HBV) stimulates autophagy to favor its production. To understand how HBV co-opts autophagy as a proviral machinery, we studied the roles of key autophagy proteins in HBV-replicating liver cell cultures. RNA interference-mediated silencing of Atg5, Atg12, and Atg16L1, which promote autophagophore expansion and LC3 membrane conjugation, interfered with viral core/nucleocapsid (NC) formation/stability and strongly diminished virus yields. Concomitantly, the core/NC membrane association and their sorting to envelope-positive compartments were perturbed. A close inspection of the HBV/autophagy cross talk revealed that the virus depended on Atg12 covalently conjugated to Atg5. In support of this finding, HBV required the E2-like enzymes Atg10 and Atg3, which catalyze or facilitate Atg5-12 conjugation, respectively. Atg10 and Atg3 knockdowns decreased HBV production, while Atg3 overexpression increased virus yields. Mapping analyses demonstrated that the HBV core protein encountered the Atg5-12/16L1 complex via interaction with the intrinsically disordered region of the Atg12 moiety that is dispensable for autophagy function. The role of Atg12 in HBV replication was confirmed by its incorporation into virions. Although the Atg5-12/16L1 complex and Atg3 are essential for LC3 lipidation and, thus, for autophagosome maturation and closure, HBV propagation did not require LC3. Silencing of LC3B, the most abundant LC3 isoform, did not inhibit but rather augmented virus production. Similar augmenting effects were obtained upon overexpression of a dominant negative mutant of Atg4B that blocked the lipid conjugation of the LC3 isoforms and their GABARAP paralogues. Together, our data indicate that HBV subverts early, nondegradative autophagy components as assembly scaffolds, thereby concurrently avoiding autophagosomal destruction.
IMPORTANCE Infections with the hepatitis B virus (HBV), an enveloped pararetrovirus, cause about 1 million deaths per year, as current therapies rarely achieve a cure. Understanding the HBV life cycle and concomitant host cell interactions is instrumental to develop new antiviral concepts. Here, we proceeded to dissect the roles of the autophagy machinery in virus propagation. By using RNA interference and overexpression studies in HBV-replicating cell lines, we identified the autophagic Atg5-12/16L1 elongation complex along with Atg10 and Atg3 to be an essential scaffold for HBV nucleocapsid assembly/stability. Deficits in Atg5-12/16L1 and Atg10/Atg3, which normally drive autophagophore membrane expansion, strongly impaired progeny virus yields. HBV gained access to Atg5-12/16L1 via interaction of its core protein with the Atg12 moiety of the complex. In contrast, subsequent autophagosome maturation and closure events were unnecessary for HBV replication, as evidenced by inhibition of Atg8/LC3 conjugation. Interfering with the HBV/Atg12 cross talk may be a tool for virus control.
KEYWORDS: Atg12-Atg5/Atg16L1 complex, Atg8/LC3 conjugation, autophagy, hepatitis B virus, virus assembly
INTRODUCTION
Autophagy is a physiological catabolic pathway that delivers autophagic substrates to lysosomes via double-membrane vesicular structures called autophagosomes. It is orchestrated by more than 30 specific autophagy (Atg) proteins that mediate the formation and elongation of a double-membrane sack (the isolation membrane or phagophore), cargo engulfment, autophagophore closure, and autophagosome fusion with the lysosome (1, 2). Although autophagy is an essential defense mechanism of the cell to confront viral invasion, many viruses, including the hepatitis B virus (HBV), evolved strategies to evade autophagy and even exploit the autophagy machinery for their own benefits (3–6).
HBV chronically infects about 250 million people worldwide and can ultimately lead to liver failure and hepatocellular carcinoma (HCC). HBV infection remains an important public health problem, as current therapeutics for chronic carriers cannot eradicate the virus completely. HBV is an enveloped DNA virus that replicates by protein-primed reverse transcription and exclusively infects human hepatocytes by using a bile acid transporter, the sodium taurocholate cotransporting polypeptide (NTCP), as a receptor (7). With about 3 kb, the HBV genome is one of the smallest viral genomes known and encompasses four overlapping open reading frames encoding the polymerase/reverse transcriptase (Pol), the capsid-forming core protein, three related envelope proteins, and the regulatory X protein (8–10). Due to the tiny size of its genome, HBV likely depends on a close interplay with host factors for the generation of new viral particles. The assembly of progeny virions begins with the formation of icosahedral nucleocapsids (NC) within the cytoplasm that package the pregenomic RNA (pgRNA) together with the covalently linked viral Pol. Inside the capsids, composed of 180 or 240 copies of the single core protein, the progeny DNA genome is synthesized through reverse transcription of the pgRNA (8–10). Mature NCs can then be enclosed by the viral envelope, composed of cellular lipids and three viral glycoproteins, the small (S), middle (M), and large (L) envelope proteins, that originate at the endoplasmic reticulum (ER) membrane (10, 11). HBV budding occurs at intracellular membranes, presumably at multivesicular bodies (MVBs), and requires functions of the endosomal sorting complex required for transport (ESCRT) machinery (12–17). Aside, autophagy pathway functions have been implicated in HBV morphogenesis (18–20).
Macroautophagy, referred to here as autophagy, requires the activity of two ubiquitin-like (UBL) conjugation systems that are recruited to the forming phagophore. In the first system, LC3 (microtubule-associated protein 1 light chain 3) and GABARAP (γ-aminobutyric acid receptor-associated protein) family members, the mammalian orthologues of Saccharomyces cerevisiae yeast Atg8, are conjugated to the membrane lipid phosphatidylethanolamine (PE) (1, 2, 21). LC3 is first cleaved by a specific cysteine protease, Atg4, to expose its C-terminal cysteine. Analogous to ubiquitination, LC3 is conjugated to PE by Atg7, an E1-like protein, and Atg3, an E2-like enzyme, which transform cytosolic LC3-I to the membrane-bound LC3-II form. In the second UBL system, the ubiquitin-like protein Atg12 is covalently conjugated to Atg5, which requires the activity of Atg7 (E1-like) and Atg10 (E2-like). The Atg5-12 conjugate in turn associates noncovalently with Atg16L1, which together scaffold the maturing phagophore (1, 2, 19). In addition, the Atg5-12/16L1 complex promotes the recruitment and activation of the Atg7/Atg3 proteins, thereby acting as an E3-like protein during LC3 lipidation (1, 22). After completion of autophagosome formation, the Atg5-12/16L1 complex dissociates from the phagophore, indicating that it participates primarily in the membrane expansion step.
The autophagic machinery had also been implicated in HBV replication, as the virus enhances autophagy and as targeted knockdowns (KD) of Beclin1, Atg5, and Atg7 impaired virion release (18–20). However, the underlying mechanisms are a matter of controversy. One set of experiments indicates that the viral regulatory X protein is essential for autophagy induction (19, 20), while the other identified the small envelope protein to be an inducer (18). The step(s) of HBV replication affected by autophagy is also less clear. One study indicated that reverse transcription within the NCs was found to be impaired upon autophagy inhibition (20), whereas another study hinted to defects in the NC envelopment reaction (18). Consistently, however, most studies reported that HBV stimulates autophagy without ending up in lysosomes. This is possibly mediated by the HBV X protein, which has been shown to inhibit autophagic degradation (23).
HBV morphogenesis is accompanied by the production and release of subviral empty envelope particles and subviral nonenveloped capsids/NCs (10, 11, 16, 24). There is increasing evidence that HBV exploits distinct cellular pathways and host factors to release its particle types (11, 13, 15, 24). Because the nonlytic release pathway of naked capsids is uncommon (24), we previously employed a small interfering RNA (siRNA)-based screen to identify host proteins guiding naked capsid export. Thereby, we discovered the Rab33B GTPase in conjunction with its autophagic effector, the Atg5-12/16L1 complex, to be dependency factors (25). Originally, we interpreted the necessity of Rab33B and its effector as a prosurvival mechanism specific for HBV naked capsids (25). By expanding these analyses, we recently showed that Rab33B is likewise crucial for the production of HBV particles by guiding core transport to NC assembly sites and/or NC transport to budding sites (26). Inspired by these observations, we here investigated the roles of autophagy pathway steps in HBV morphogenesis, with a special emphasis on the Atg5-12/16L1 elongation complex.
RESULTS
Silencing of Atg5, Atg12, and Atg16L1 impairs HBV formation and release.
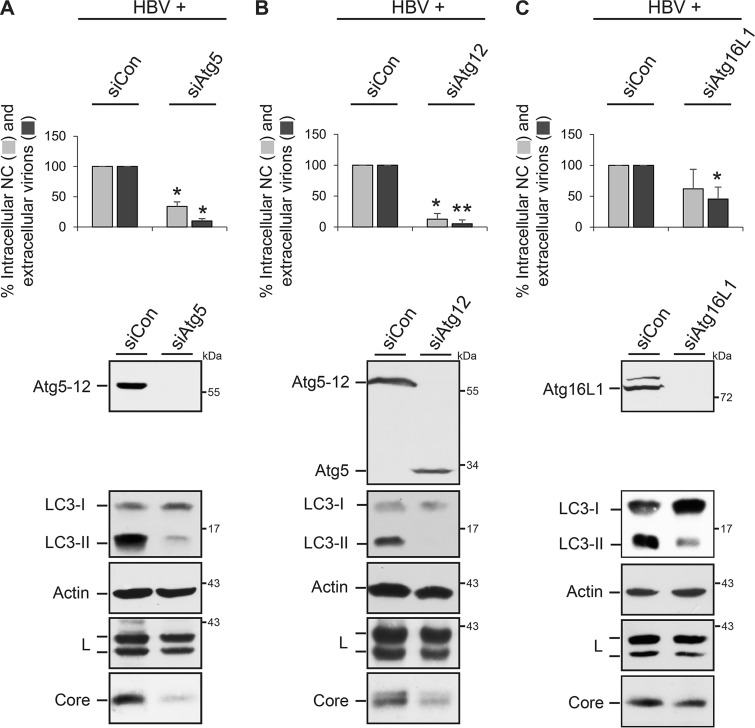
To investigate the potential role of the autophagy machinery in the life cycle of HBV, we used a transient replication system by transfecting human hepatoma HuH-7 cells with a replication-competent HBV replicon plasmid. This cell line is not susceptible to HBV infection because it expresses very low levels of the NTCP receptor (7) and is therefore a useful model to study the production and release of the virus rather than infection. To perturb Atg5, Atg12, or Atg16L1 in HuH-7 cells, the autophagy factors were depleted with single siRNAs targeting Atg5 (siAtg5) or Atg12 (siAtg12) or an Atg16L1-specific pool containing four different duplexes. After 48 h, cells were retransfected with the HBV replicon, and cell lysates and supernatants were harvested after an additional 72 h. Lysates, prepared with the detergent Triton X-100, were subjected to specific Western blotting (WB), which showed an almost complete lack of the Atg5-12 conjugate in siAtg5- and siAtg12-treated cells and of Atg16L1 in siRNA targeting Atg16L1 (siAtg16L1)-treated cells (Fig. 1). Notably, in siAtg12-treated cells, unconjugated Atg5 that was absent in control siRNA (siCon)-transfected cells appeared (Fig. 1B). Similarly, trace amounts of unconjugated Atg12 were detectable in cells depleted of Atg5 (data not shown). Because the Atg5-12/16L1 complex is essential for LC3 lipidation (1, 21) and, hence, for the conversion of the LC3-I form to the LC3-II form, the lysates were also examined by anti-LC3-specific WB. The depletion of either Atg protein largely reduced the levels of the lipidated LC3-II isoform, indicative of a high siRNA transfection rate and of the functional ablation of the Atg5-12/16L1 complex (Fig. 1). The production and release of HBV particles were analyzed by immunoprecipitation (IP) of intracellular NC and extracellular virions with capsid- or envelope-specific antibodies, respectively, followed by particle disruption and real-time PCR measurement of the number of HBV genomes. As shown in Fig. 1, the knockdown (KD) of Atg5, Atg12, or Atg16L1 significantly decreased the amounts of both intracellular NC and extracellular virions compared to those in siCon-treated cells. To gain mechanistic insights, lysates were probed for the expression and stability profiles of the envelope and core proteins by WB. The intracellular steady-state levels of the L envelope protein, synthesized in nonglycosylated p39 and single-glycosylated gp42 forms, were largely unaffected, while the level of core was dramatically reduced in Atg5- and Atg12-KD cells and slightly diminished in Atg16L1-depleted cells (Fig. 1). These results indicate that HBV progeny production requires the action of Atg5, Atg12, and Atg16L1. Since unconjugated Atg5 survived in siAtg12-treated cells and vice versa without enabling HBV propagation, virus maturation seemingly depends on Atg12-conjugated Atg5, likely in complex with Atg16L1.
FIG 1.
HBV formation and release require Atg5, Atg12, and Atg16L1. HuH-7 cells were treated with control siRNA (siCon) or siRNAs targeting Atg5 (A), Atg12 (B), or Atg16L1 (C) for 48 h and retransfected with pHBV (HBV). After an additional 72 h, lysates and supernatants were harvested. HBV release was monitored by envelope-specific immunoprecipitation of supernatants and real-time PCR measurement of the viral genomes (virions). Nonenveloped intracellular nucleocapsids (NCs) were immunoprecipitated with anticapsid antibodies and analyzed by PCR. PCR results were demonstrated in percent amounts relative to those for siCon-transfected cells. Error bars indicate the standard deviations of the means from four experiments measured in duplicate. *, P < 0.05 compared to the control; **, P < 0.01 compared to the control. To monitor depletion, cell lysates, prepared with 0.2% Triton X-100, were examined by Atg5-specific (A, B) and Atg16L1-specific (C) WB. The same samples were analyzed for the ratio of LC3-I and LC3-II by anti-LC3 WB and for uniform sample loading by anti-β-actin WB. RNAi effects on the synthesis/stability of L and core were analyzed by specific immunoblotting of lysates. Numbers to the right refer to molecular mass markers (in kilodaltons).
Impacts of Atg5, Atg12, and Atg16L1 silencing on HBV transcription, core solubility, and cell vitality.
To gain clues into how deficits in Atg5, Atg12, and Atg16L1 impaired the synthesis/stability of core and, hence, HBV formation, we first analyzed the HBV transcription profile in corresponding KD cells. Because the foreign human metallothionein IIa (hMTIIa) promoter preceding the viral core/Pol promoter within the HBV replicon plasmid may hamper transcriptional analyses, a modified HBV replicon (pHBVΔHP) with a deletion of the hMTIIa promoter was used (14). To investigate the degree of HBV transcription in cells depleted of Atg5, Atg12, or Atg16L1, total mRNA was extracted and measured by quantitative reverse transcription-PCR (qRT-PCR) using either a primer pair targeting the HBV pgRNA and the core/Pol- and envelope-specific transcripts or a primer pair specific for the pgRNA and the core/Pol transcript. As shown in Fig. 2A, no gross changes in the levels of the HBV transcripts were evident. In addition, we performed Northern blot analysis of total cellular RNA isolated from transfected cells using an HBV-specific RNA probe. Consistent with the qRT-PCR results, this analysis revealed no significant effects of the applied siRNAs on the HBV transcript levels (Fig. 2B).
FIG 2.
Effects of Atg5, Atg12, and Atg16L1 silencing on HBV transcription, core protein solubility, and cell viability. (A) For HBV gene expression profiling, HuH-7 cells were treated with the indicated siRNAs and were retransfected with the pHBVΔHP replicon. For qRT-PCR, total mRNAs were isolated, reverse transcribed, and analyzed by PCR using HBV-specific primer sets PP#1 and PP#2. PP#1 targets the pgRNA and the core/Pol- and envelope-specific transcripts, while PP#2 is specific for the pgRNA and core/Pol transcript. Error bars indicate the standard deviations of the means from two experiments measured in duplicate. (B) Northern blot analysis of HBV RNA transcripts. Total RNA was isolated from cells transfected as described above, separated on a 1.5% agarose gel, and transferred to a nylon membrane, which was reacted with a DIG-labeled HBV-specific RNA probe. The positions of HBV 3.5-kb, 2.4-kb, and 2.1-kb RNAs are indicated. The results for rRNAs (18S), used as loading controls, are also presented. (C) HuH-7 cells treated with the indicated siRNA duplexes were retransfected with pHBV. At 120 h after transfection, cells were lysed under denaturing conditions using SDS, and the lysates were probed by core- and β-actin-specific WB. In parallel, lysates were prepared with Triton X-100, and total protein levels were determined using a Bradford assay and are demonstrated in percent amounts relative to those for control cells (n = 2). To probe for cell lysis, the supernatants of transfected cells were assayed for LDH activity (n = 2).
Next, we inspected the solubility profile of cell-associated core. In order to measure the formation of progeny NC by IP and PCR analysis, cells were disrupted with mild detergent (0.2% Triton X-100) (Fig. 1). Because less soluble proteins could get lost under these conditions, lysis of cells was performed in more stringent buffer using the denaturing detergent SDS. However, as described above, the amounts of core were strongly reduced in Atg5- or Atg12-KD cells, while the depletion of Atg16L1 induced a modest decline in core levels compared to those in control cells (Fig. 2C). To account for this, we assumed that the loss of the autophagic factors might misdirect proper NC formation and/or cause NC destabilization concomitantly with total core protein reduction. Notably, in WB analyses core migrated as a doublet (Fig. 2C), with the upper-migrating form possibly representing a modified, e.g., phosphorylated or methylated, core species (27) and/or the precore protein. The precore protein is identical to core, save for its N-terminal ER signal sequence, composed of 29 residues, that directs precore to the ER, where 19 amino acids are cleaved off from the N terminus (10). The precore protein is able to retrotranslocate from the ER to the cytoplasm (28), where it can oligomerize with core into heterocapsids (29, 30). Hence, the silencing of the proviral Atg5, Atg12, or Atg16L1 proteins might simultaneously downregulate precore. Although the unaffected HBV L protein content and transcript levels argued against possible harmful effects of the applied RNA interference (RNAi) strategy, we nonetheless performed cytotoxicity studies. The analyses of the total protein concentrations of cell lysates and the lactate dehydrogenase (LDH) activity of cell supernatants revealed no differences in protein expression or evidence of cell damage upon transient depletion of Atg5, Atg12, or Atg16L1 (Fig. 2C). From these findings together, we infer that that the viral core is the main target for Atg5/Atg12/Atg16L1 intervention.
Atg12 silencing alters the intracellular distribution of HBV.
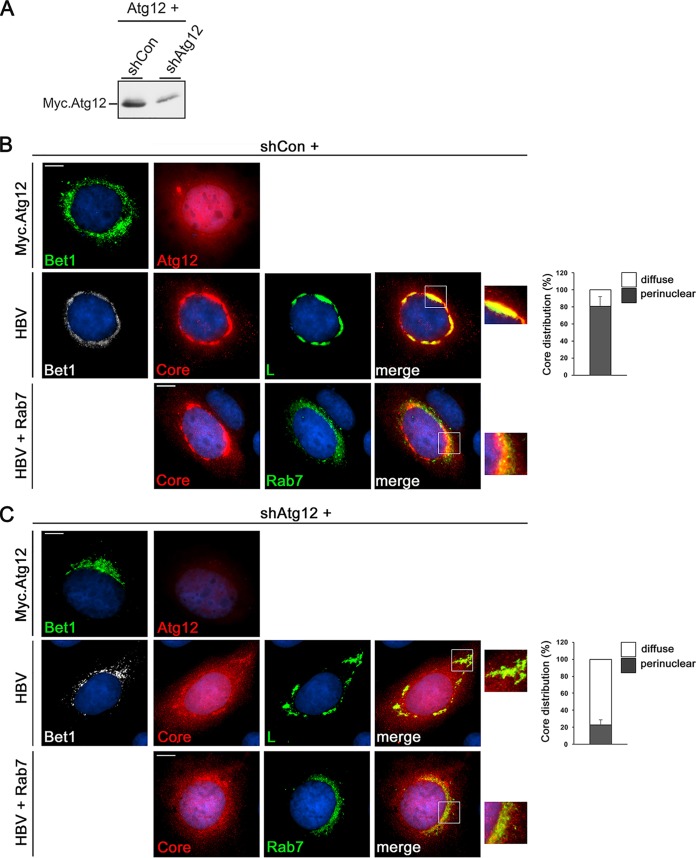
To study the fate of HBV in Atg12-inactivated single cells, a short hairpin RNA (shRNA)-mediated Atg12 knockdown was applied using a plasmid carrying an Atg12-specific shRNA sequence along with a hemagglutinin (HA)-tagged Bet1 marker that allows the identification of KD cells. The silencing efficacy of the applied Atg12 shRNA sequence had been proven in two reports (31, 32). For validation, the Atg12-specific shRNA (shAtg12) or control shRNA (shCon) plasmids were cotransfected with a cloned, Myc-tagged Atg12 gene, and cells were costained with Myc- and HA-specific antibodies. In parallel, cell lysates were analyzed by Myc-specific WB. As shown in Fig. 3, the shAtg12 construct almost completely reduced the Atg12-specific Myc signal in HA-positive cells. The WB analysis also showed a reduction of the Myc.Atg12 content, albeit less complete, likely because not every cell received both plasmids (Fig. 3A). Next, the shRNA constructs were cotransfected with the HBV replicon and immunostained with HA, core, and L antibodies. In shCon-transfected cells, core yielded its typical diffuse staining dispersed throughout the nucleus and cytoplasm along with a prominent perinuclear accumulation in a ring-like structure in about 80% of inspected HA.Bet1-positive cells. The perinuclear core structure nearly completely overlapped the L staining pattern (Fig. 3B), implicative for virus assembly sites. For further assessment, cells were cotransfected using a plasmid encoding a green fluorescent protein (GFP)-tagged Rab7 that had been reported to colocalize with HBV during virus assembly and egress (33). Consistent with this, we found that the ring-like core structure partly overlapped the Rab7-positive vesicular structures (Fig. 3B). Upon Atg12 silencing, core contents were found to be reduced, wherefore images were captured with automatic, i.e., optimal adjusted, exposure times (Fig. 3C). Even so, significantly less core accumulated in the perinuclear, ring-like structure, concomitant with a significant reduction in the degree of core/L and core/Rab7 colocalization (Fig. 3C). From these data, we infer that Atg12 silencing may hamper proper core trafficking to NC assembly/envelopment sites, thereby rendering core/NC sensitive to destabilization and destruction.
FIG 3.
Atg12 silencing impairs HBV core/L colocalization. HuH-7 cells were cotransfected with shCon- or shAtg12-expressing plasmids carrying an HA-tagged Bet1 reporter plus the constructs indicated on the left of the panels. At 4 days posttransfection, cells were fixed with PFA and permeabilized. To monitor depletion, cells were transfected with the shRNA constructs plus Myc-tagged Atg12 and stained with HA-specific (Bet1; green) and Myc-specific (Atg12; red) antibodies. Images were captured with fixed exposure times (B, C, top). In parallel, cell lysates were analyzed by Myc-specific WB (A). Upon cotransfection with the HBV replicon, cells were reacted with antibodies against HA (Bet1; white), core (red), and L (green). Merged images of core and L staining are displayed in the right panels (yellow), which show enlargements of the areas outlined in the merge panels. Images were captured with automatic exposure times (B, C, middle). For quantification, 50 HA.Bet1-positive cells per coverslip (n = 3) were inspected for the perinuclear accumulation of core in the ring-like structure overlapping L (graphs and B, C, middle). To probe for the degree of HBV colocalization with Rab7, shRNA constructs, the HBV replicon, and GFP-tagged Rab7 were cotransfected into cells. Core staining is in red, and Rab7 autofluorescence is in green. Images were captured with automatic exposure times (B, C, bottom). Experiments were repeated three times, and representative pictures are shown. Bars, 10 μm.
Silencing of Atg5, Atg12, and Atg16L1 impairs the membrane association of HBV core.
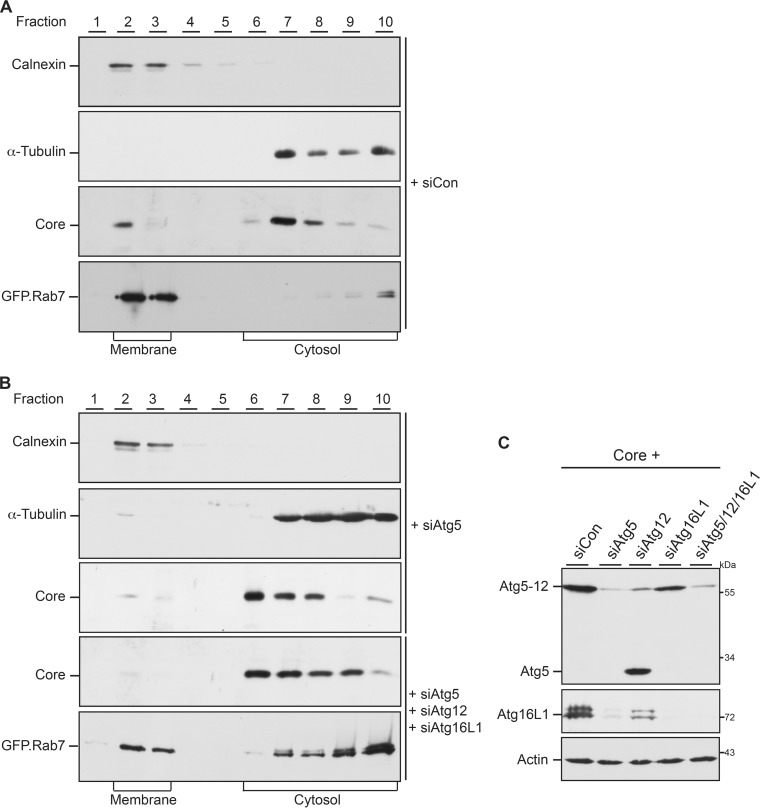
The faulty trafficking of core in Atg12-KD cells prompted us to study the core-membrane association. Previously, we identified a membrane-targeting site within the basic C terminus of core that may confer its targeting to budding sites (26). For membrane flotation analyses, HuH-7 cells were treated with control siRNA duplexes, the Atg5-specific siRNA, or a mixture of Atg5-, Atg12-, and Atg16L1-specific siRNAs prior to transfection with an expression plasmid carrying only core (pCore). This strategy was chosen because the downregulation of core/capsids induced by a deficit of Atg5/12/16L1 is less prominent in cells expressing only core (25) than in virus-replicating cells (this study). Flotation fractions were first analyzed by calnexin- and α-tubulin-specific WB, in which calnexin and α-tubulin served as references for membrane-associated and soluble proteins, respectively (Fig. 4). The distribution of these markers was coincident in control and Atg5-depleted cells, indicating that Atg5 silencing did not affect the separation of membranes (Fig. 4). Core-specific WB showed that core partitioned between the soluble and membrane fractions in control cells. Upon expression of the GFP-tagged Rab7 construct, Rab7 could also be detected almost exclusively in the membrane fractions (Fig. 4A). Conversely, in Atg5-depleted cells the portion of membrane-associated core was diminished concomitantly with its accumulation in the cytosolic fractions (Fig. 4B). An even enhanced reduction in core-membrane association was observed upon a simultaneous KD of Atg5/12/16L1 (Fig. 4B). Under these conditions, the portion of membrane-associated GFP.Rab7 also declined (Fig. 4B). Since Rab7 is recruited toward maturating autophagosomes (33), the inhibition of autophagy in Atg5/12/16L1-KD cells likely diminished its membrane association. To control the efficiency of the mixture of Atg5-, Atg12-, and Atg16L1-specific siRNAs, Atg5- and Atg16L1-specific WB analyses were performed. As shown in Fig. 4C, this mixture clearly depleted the Atg5-12 conjugate, unconjugated Atg5, and Atg16L1. Consistent with our previous work (25), we observed cross-depletion effects, as the Atg16L1 signal was decreased in siAtg5- and siAtg12-treated cells, while the siAtg16L1-specific siRNAs diminished the Atg5-12 conjugate (Fig. 4C). Hence, the Atg5-12/16L1 complex likely loses its compactness upon depletion of one subunit. Together, these data indicate that the autophagic elongation complex is involved in guiding core to membranes.
FIG 4.
HBV core-membrane association involves Atg5, Atg12, and Atg16L1. HuH-7 cells were treated with control siRNA (A), the Atg5-specific siRNA, or a mixture of Atg5-, Atg12-, and Atg16L1-specific RNA duplexes (B) and were retransfected with pCore. In the bottom panels of panels A and B, cells were transfected with GFP-tagged Rab7. Cells were lysed by sonication, and membrane flotation was performed. Ten fractions were collected from the top of the gradient and analyzed by core-specific WB. The blots were reprobed with antibodies specific for α-tubulin and calnexin to determine the cytosolic and membrane fractions, respectively. The blots shown in the bottom panels of panels A and B were reacted with GFP-specific antibodies. (C) For assessment of depletion efficiencies, Atg5-, Atg12-, and Atg16L1-specific siRNAs were individually applied or transfected in combination prior to retransfection of cells with pCore. Cell lysates were immunoblotted with antibodies against Atg5 (top), Atg16L1 (middle), and β-actin (bottom). Numbers to the right refer to molecular mass markers (in kilodaltons).
HBV core associates with the intrinsically disordered region of Atg12.
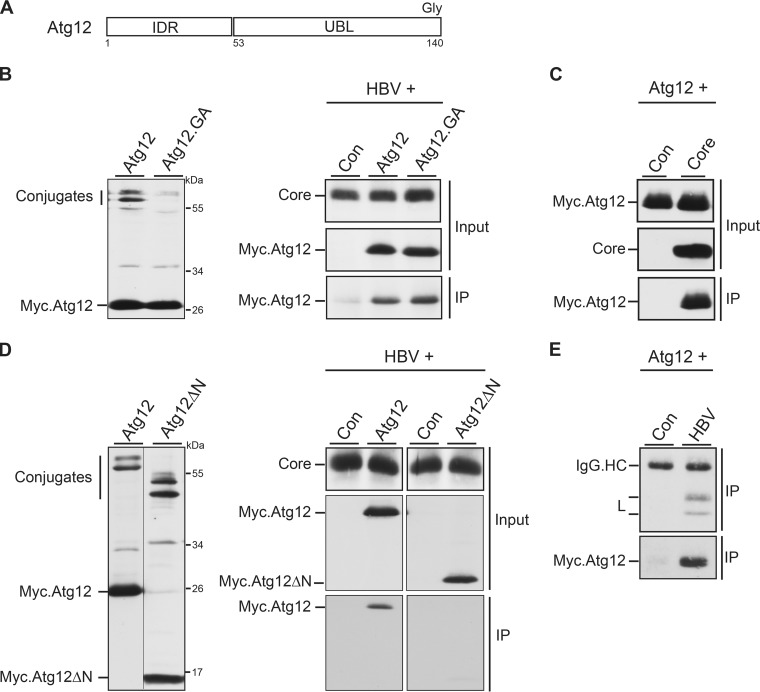
Previously, we reported that core, synthesized in the absence of an ongoing HBV replication, interacted with Atg5 (25). However, so far it has been unclear whether Atg5 or the conjugated Atg12 moiety is the target for core. We therefore tested for an Atg12-core interaction both in the presence and in the absence of ongoing HBV replication. In addition, we analyzed an Atg12.GA mutant that is defective in conjugation due to a mutation of the C-terminal glycine (Fig. 5A) (21). Because anti-Atg12 antibodies proved to be less suitable for IP analyses, ectopic expression of the wild-type (wt) or mutant Atg12 proteins fused with N-terminal Myc tags was used. Both proteins were easily detectable by Myc-specific WB of lysates of transfected cells (Fig. 5B). Atg12 appeared in unconjugated and two conjugated species, representing Atg5-12 and Atg12 linked to Atg3 (34), whereas Atg12.GA lacked conjugated forms (Fig. 5B). The upper-migrating Atg3-12 conjugate disappeared in Atg3-depleted cells (data not shown), indicating that the mouse Atg3 is a functional substrate for conjugation in human cells. These constructs were next cotransfected with pHBV, and lysates were subjected to a core-specific IP followed by Myc-specific WB. Thereby, core specifically brought down Atg12 as well as its conjugation-deficient mutant (Fig. 5B). Of note, in this experiment a faint signal appeared in the negative control. This was likely due to unspecific staining of immunoglobulin G light chains, used in IP, that have a molecular mass (25 kDa) similar to that of Myc.Atg12. Antibody cross-reactions with immunoglobulin heavy chains (55 kDa; see also Fig. 5E) hampered detection of whether core might also associate with the 56-kDa Atg5-12 conjugate. Immunocapture analyses of cells expressing only core, instead of HBV, yielded identical results (Fig. 5C). Since the conjugation ability of Atg12 was dispensable for core binding, we searched for other interaction domains within Atg12, which is composed of a ubiquitin-like (UBL) fold domain and a consensus intrinsically disordered region (IDR) (Fig. 5A). IDRs are natively unstructured regions that lack stable secondary and tertiary structures and are predicted to facilitate protein-protein interactions (35). The analysis of an IDR-lacking Atg12 mutant (Atg12ΔN) revealed that it retained its conjugation capacity but failed to efficiently associate with core (Fig. 5D). Finally, we wondered whether Atg12 might even be incorporated into the mature virus particle. To this aim, supernatants of HBV/Myc.Atg12-cotransfected cells were immunoprecipitated with envelope-specific antibodies under detergent-free conditions. L- and Myc-specific WB showed virion precipitation and, importantly, the presence of Atg12 in the secreted virion (Fig. 5E). To control the specificity of the precipitation, supernatants of cells expressing Myc.Atg12 in the absence of pHBV were likewise reacted with the rabbit antienvelope antibodies. Thereby, no specific signal was detectable (Fig. 5E). We thus conclude that HBV core/NCs associate with Atg12 and presumably with the Atg5-12/16L1 complex in a productive manner.
FIG 5.
Atg12 associates with HBV core via its IDR domain and is incorporated into HBV particles. (A) Schematic depiction of Atg12 with its N-terminal IDR and C-terminal UBL domains. The terminal glycine (Gly) residue, required for Atg5 conjugation, is indicated. (B) Expression profile of Myc-tagged Atg12 and its conjugation-defective Atg12.GA mutant in transiently transfected HuH-7 cells. Cell lysates were probed by Myc-specific WB. Unconjugated and conjugated Atg12 forms are indicated (left). For co-IP, the HBV replicon was cotransfected with the control, Atg12, or Atg12.GA expression construct at a 3:1 DNA ratio (right). Synthesis of core, ATG12, and ATG12.GA is shown by WB of lysates with anticore and anti-Myc antibodies (Input). For IP, lysates were incubated with anticore antibodies followed by Myc-specific WB. (C) IP analysis of cells cotransfected with Myc-tagged Atg12 and a control or core-expressing plasmid was done as described in the legend to panel B. (D) Expression analysis of Myc-tagged Atg12 and the Atg12ΔN mutant, lacking the IDR domain, in transfected HuH-7 cells. Unconjugated and conjugated Atg12 forms are depicted. The co-IP analysis was done as described in the legend to panel B. (E) Cellular supernatants of HuH-7 cells, cotransfected with the Myc-tagged Atg12 construct plus a control or pHBV, were subjected to an L-specific IP to precipitate extracellular virions. Immunoprecipitates were then immunoblotted with anti-L (top) or anti-Myc (bottom) antibodies. IgG.HC, unspecifically stained immunoglobulin G heavy chains.
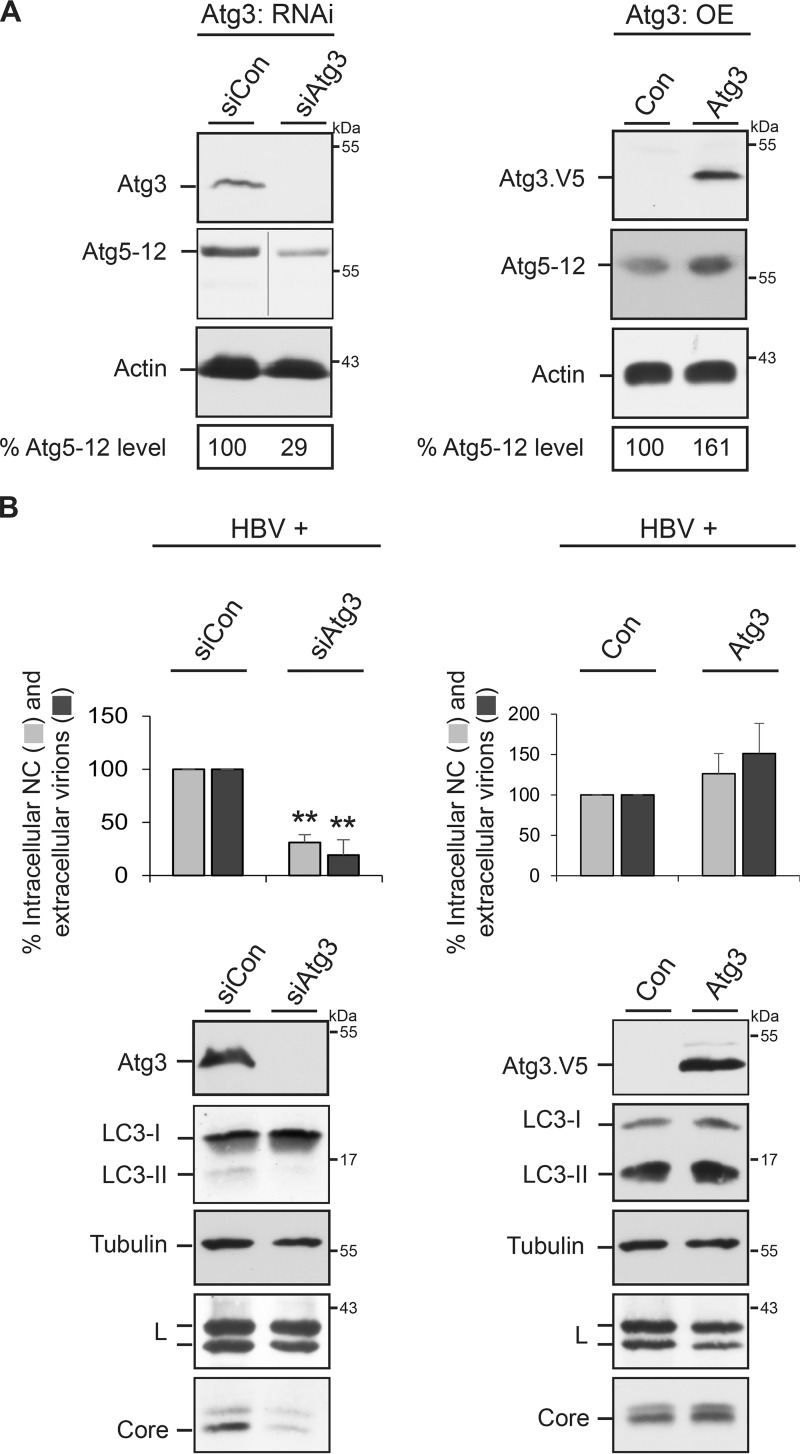
Atg3 positively regulates HBV production and Atg5-12 conjugation.
Because HBV maturation apparently depends on the action of Atg5-12 conjugate (Fig. 1), we next focused on Atg3. Atg3 is essential for the formation of preautophagosomal membranes and is the E2-like enzyme that conjugates LC3/GABARAP to PE (1, 2, 21). Moreover, Atg3 also affects the degree of Atg5-12 conjugation, as evidenced in Atg3-deficient mice (36). For assessment in our experimental setting, we inspected the extent of Atg5-12 conjugation in Atg3-KD and -overexpressing HuH-7 cells. For depletion, an siRNA pool with four duplexes was used, while ectopic overexpression was achieved by transfection with a V5-tagged mouse Atg3 gene. WB analyses of cell lysates with Atg3/V5- and Atg12-specific antibodies revealed that the lack of Atg3 reduced Atg5-12 conjugation, while its overexpression increased the level of Atg5-12 (Fig. 6A). This prompted us to analyze the fate of HBV in Atg3-KD and -overexpressing cells. Upon Atg3 silencing, the amounts of intracellular NC and extracellular virions were strongly reduced (Fig. 6B), reminiscent of the phenotypes observed in Atg5/12/16L1-KD cells. As would be expected from the E2-like role of Atg3, its inactivation blocked LC3 lipidation. Under these conditions, the synthesis/stability of the L protein was not considerably affected, while the intracellular core level strongly declined (Fig. 6B). When Atg3 was overexpressed, reverse effects were observed, in that excess Atg3 enhanced the yields of intracellular NC and extracellular virions (Fig. 6B). Concomitantly, core levels were not reduced but, rather, were slightly increased. We deduce from these data that Atg3 is essential for HBV propagation and promotes virus replication, likely by regulating the degree of Atg5-12 conjugation.
FIG 6.
Impact of Atg3 on Atg5-12 conjugation and HBV production. (A) The Atg3 protein level affects the degree of Atg5-12 conjugation. HuH-7 cells were treated with control siRNA (siCon) or siRNA duplexes targeting Atg3 (Atg3: RNAi, left) or were transfected with a control plasmid (Con) or V5-tagged Atg3 (Atg3: OE [overexpression], right). Lysates of Atg3-KD cells were reacted with Atg3- or Atg12-specific antibodies (left), whereas extracts of Atg3-overexpressing cells were probed with V5- and Atg12-specific antibodies (right). The uniformity of sample loading is shown by anti-β-actin WB. The Atg5-12 levels were quantitated and demonstrated in percent amounts relative to those for control cells (n = 2). (B) The Atg3 protein content affects the degree of HBV formation and release. HuH-7 cells were treated with control siRNA or Atg3-specific siRNAs prior to transfection with the HBV replicon (left). For Atg3 overexpression, pHBV was transfected with control DNA or V5-tagged Atg3 at a 1:1 DNA ratio (right). Lysates and supernatants were harvested and assayed for the production of intracellular NCs and extracellular virions by immunocapture and real-time PCR quantification of progeny genomes, as described in the legend to Fig. 1. Error bars indicate the standard deviations of the means from three experiments measured in duplicate. **, P < 0.01 compared to the control. Atg3 KD and overexpression were analyzed by Atg3- and V5-specific WB, respectively. The same lysate samples were probed by LC3-, β-actin-, L-, and core-specific antibodies.
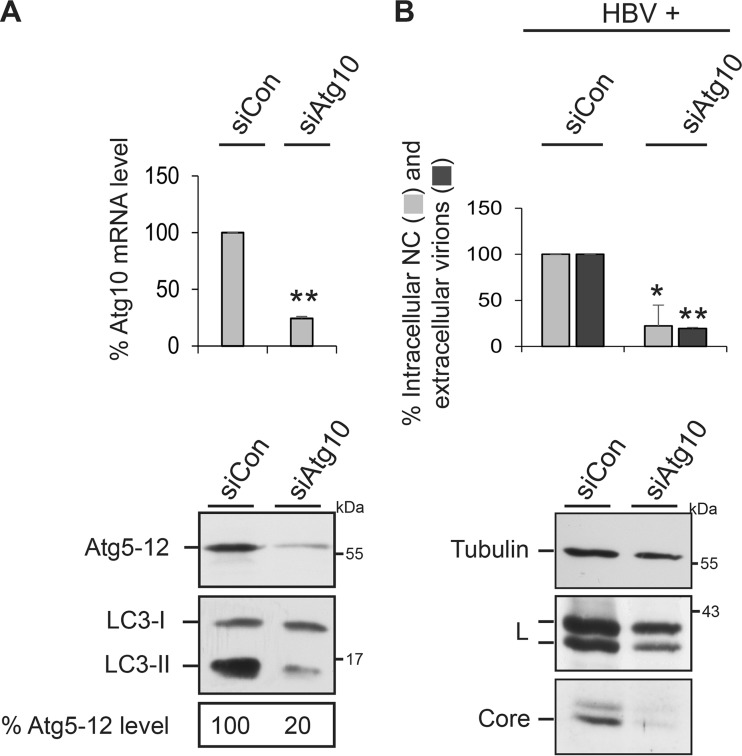
Silencing of Atg10 impairs HBV production and Atg5-12 conjugation.
To corroborate the pivotal role of the Atg5-12 conjugate in HBV propagation, we finally studied the impacts of an inactivation of Atg10, the E2-like enzyme that catalyzes the conjugation reaction. Upon treatment of cells with an Atg10-specific siRNA pool, Atg10 mRNA levels dropped down to about 25%, as measured by qRT-PCR of reverse-transcribed total mRNA (Fig. 7A). Concomitantly, the levels of the Atg5-12 conjugate and of the lipidated LC3-II form substantially declined (Fig. 7A), implicating an efficient Atg10 knockdown. Consistent with the results obtained in Atg3-, Atg5-, and Atg12-KD cells, Atg10 silencing impeded HBV nucleocapsid formation and virus release, which were accompanied by a strong reduction in intracellular core levels (Fig. 7B).
FIG 7.
Impact of Atg10 on Atg5-12 conjugation and HBV production. (A) Atg10 silencing decreases the Atg5-12 conjugate level. HuH-7 cells were treated with control siRNA (siCon) or Atg10-specific siRNAs, followed by isolation of total cellular mRNA. qRT-PCR runs were performed in duplicate, and error bars indicate the standard deviations of the means from three experiments. In parallel, cell lysates were assayed by Atg12- and LC3-specific WB. The Atg5-12 levels were quantitated and demonstrated in percent amounts relative to those for control cells (n = 2). (B) Atg10 silencing blocks HBV propagation. HuH-7 cells were treated with control siRNA or Atg10-specific siRNAs, followed by transfection with the HBV replicon. Lysates and supernatants of cells were analyzed for HBV production and release by PCR analysis (n = 3), exactly as described in the legend to Fig. 1. *, P < 0.05 compared to the control; **, P < 0.01 compared to the control. The same lysate samples were probed by anti-α-tubulin-, L-, and core-specific WB.
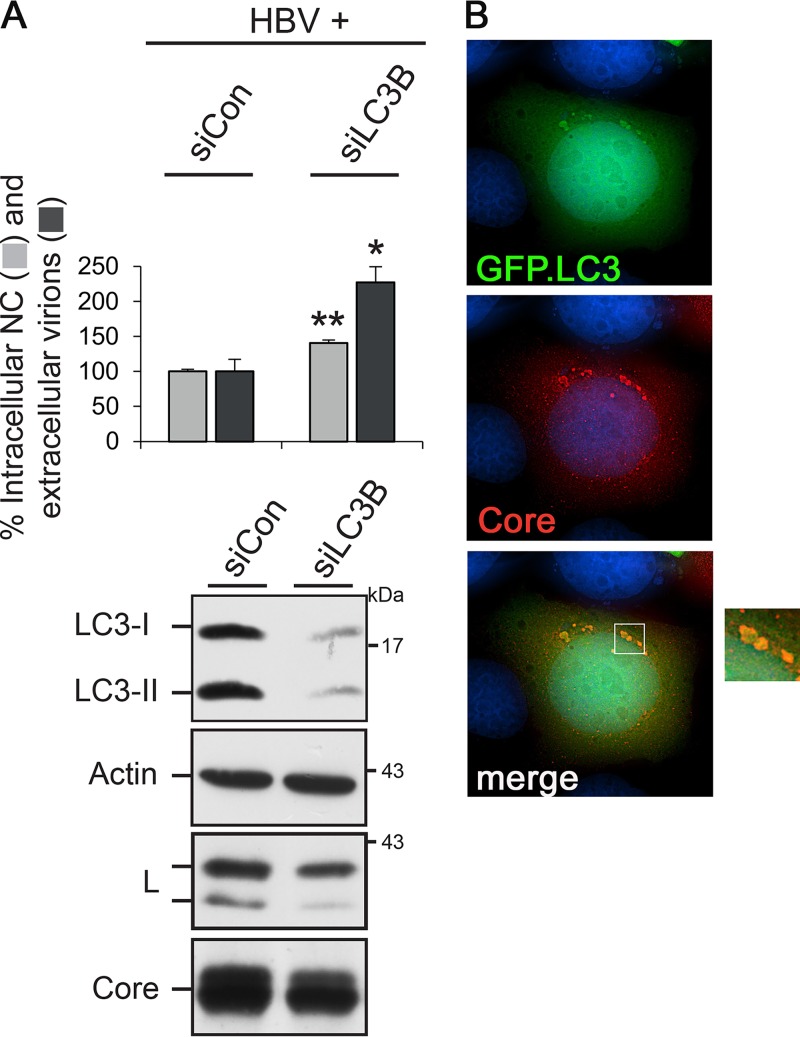
Silencing of LC3B augments HBV formation and release.
Since Atg3 and the Atg5-12/16L1 complex initiate autophagosome biogenesis and are together required for LC3 lipidation, we next probed for a role of LC3 in HBV formation. Mammalian cells contain three LC3 isoforms, referred to as A, B, and C, with LC3B being the most abundant molecule in many cell types (21, 37). Again, an RNAi approach was applied using an siRNA targeting LC3B (siLC3B) that effectively depleted both LC3 forms (Fig. 8A). This treatment did not impair intra- or extracellular NC/virion levels. Rather, LC3B silencing repeatedly enhanced virus yield and egress, thereby leading to a slight decline in intracellular envelope and core protein contents (Fig. 8A). The dispensability of LC3B together with the requirements of Atg3 and Atg5/12/16L1 implicate that HBV formation needs only part of the autophagy machinery. Nevertheless, we tested for spatial contacts between HBV and autophagic structures using a GFP-tagged LC3B construct as a genuine autophagic reporter. Thereby, we observed that core clearly colocalized with LC3B-positive, dot-like structures and appeared to decorate their surfaces (Fig. 8B). Pulldown experiments yielded no hints for an interaction between core and LC3B (data not shown), suggesting that core/LC3B colocalization may, rather, be mediated through Atg5-12 tethered to the phagophore.
FIG 8.
HBV formation and release do not require LC3B. (A) For RNAi, HuH-7 cells were treated with control- or LC3B-specific duplexes and retransfected with pHBV. Cell lysates and supernatants were assayed for the production and release of progeny viral particles via PCR analysis, exactly as described in the legend to Fig. 1. Error bars indicate the standard deviations of the means from four experiments measured in duplicate. *, P < 0.05 compared to the control; **, P < 0.01 compared to the control. To monitor depletion, lysates were assayed by LC3-specific WB. The same lysate samples were probed by anti-β-actin-, L-, and core-specific WB. (B) Representative images of cells coexpressing HBV (core, red) and GFP-tagged LC3B (green). The overlay of the deconvoluted fluorescent pattern is shown in the right panel, which is an enlargement of the boxed region in the bottom left panel, and indicates colocalization of core with surface structures of LC3B-positive dot-like vesicles.
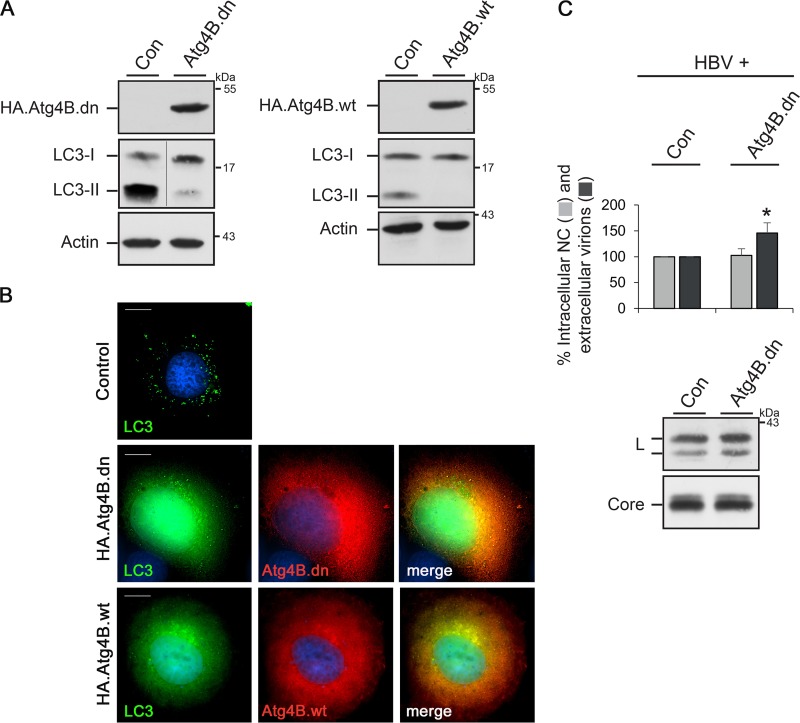
Inhibition of LC3/GABARAP lipidation does not impair HBV formation and release.
Besides the LC3 family of proteins, mammalian cells contain at least three further LC3 paralogues belonging to the GABARAP subfamily that all have overlapping functions (21, 37). For simultaneous inactivation of all LC3/GABARAP members, we took use of overexpression of a dominant negative (dn) Atg4B mutant (38). Atg4B is a cysteine protease that processes pro-LC3/GABARAP paralogues for subsequent lipidation and has a broad specificity for all LC3/GABARAP paralogues. Its enzymatic activity can be blocked by a cysteine-to-alanine substitution at position 74, resulting in a block of LC3/GABARAP lipidation and autophagy inhibition (1, 2, 38). Notably, the overexpression of the wild-type (wt) Atg4B protein has been reported to likewise inhibit autophagy, indicating that the negative effect is independent of the enzymatic activity (38). For assessment, HuH-7 cells were transiently transfected with HA-tagged versions of the Atg4B.dn and Atg4B.wt constructs and investigated by WB and immunofluorescence (IF) analyses. As shown in Fig. 9A, the level of lipidated LC3-II strongly decreased. Moreover, upon IF microscopy, the typical dot-like LC3 puncta, observed in starved control cells, were not detectable in Atg4B.dn- and Atg4B.wt-overexpressing cells (Fig. 9B). Rather, GFP-tagged LC3B dispersed throughout the cytoplasm, likely due to its sequestration in stable complexes with Atg4B (38). Aside, GFP-LC3B was also found within the nucleus, likely as a consequence of the blocked punctum formation in the cytoplasm, which is consistent with a previous report (38). The functionality of the mutant led us to analyze HBV formation in Atg4B.dn-overexpressing cells. Consistent with the results obtained for the sole depletion of the LC3B isoform, the Atg4B.dn-induced abrogation of LC3/GABARAP lipidation did not impact HBV formation and release (Fig. 9C). Instead, the amounts of extracellular virions increased (Fig. 9C), implicating that the prevention of LC3/GABARAP processing and, thus, of autophagosome closure appears to benefit the virus.
FIG 9.
HBV formation and release do not require lipidation of LC3/GABARAP paralogues. To impair autophagy concomitantly with the lipidation of LC3/GABARAP paralogues, overexpression of HA-tagged versions of an Atg4B.dn mutant or Atg4B.wt was used. (A) HuH-7 cells were transiently transfected with control (Con), HA.Atg4B.dn, or HA.Atg4B.wt plasmids, and cell extracts were investigated by HA-, LC3-, and β-actin-specific WB. (B) Cells were cotransfected with pEGFP.LC3B plus control, HA.Atg4B.dn, or Atg4B.wt vectors. After starvation for 2 h, cells were permeabilized with methanol and stained with anti-HA antibodies, followed by staining with secondary antibodies. The autofluorescence of LC3B is shown in green, and the staining pattern of HA.Atg4B.dn or Atg4B.wt is in red. The overlay of the deconvoluted fluorescent pattern is shown in the right panel, with the yellow color indicating colocalization. Bars, 10 μm. (C) HuH-7 cells were cotransfected with pHBV plus the control (Con) or HA.Atg4B.dn construct at a 1:1 DNA ratio. At 3 days posttransfection, lysates and supernatants were harvested and subjected to immunocapture and PCR analysis (n = 5). *, P < 0.05 compared to the control. To inspect HBV protein synthesis/stability, lysates were probed by anti-core- and anti-L-specific WB.
DISCUSSION
The current study adds further proof that HBV subverts autophagic pathway functions to enhance its own replication. By dissecting the different steps of autophagy, we found that early, nondegradative stages of autophagy promoted HBV yields, while late stages were not mandatory.
One important outcome of this work was the finding that HBV strictly depends on Atg12 conjugated to Atg5. Analogously to ubiquitination, the UBL Atg12 protein is conjugated to Atg5 by Atg7 and Atg10, acting as E1-like and E2-like enzymes, respectively. Atg5 further interacts with Atg16L1 to generate the Atg12-5/16L1 complex, which localizes to the developing phagophore membrane, promotes its expansion, and facilitates LC3 lipidation (21, 22, 37). Our observation that unconjugated Atg5 survived in Atg12-depleted cells but failed to assist in virus replication underscored the HBV dependency on the Atg5-12 conjugate. In support of this finding, the silencing of the E1-like Atg7 enzyme, essential for Atg5-12 conjugation, had previously been shown to inhibit HBV replication (19). Moreover, herein we identified Atg10, the E2-like enzyme of the conjugation reaction, to be crucial for HBV propagation. Together, these findings substantiate that HBV exploited the Atg5-12 conjugate. Aside, we discovered that Atg3 is a positive player in HBV formation, as its silencing blocked virus replication, while its overexpression had augmenting effects. In the canonical pathway, Atg3 is responsible for PE conjugation of LC3 and, thus, for phagophore extension and closure. Studies in knockout mice revealed that Atg3 also affected the degree of Atg5-12 conjugation (36), which we proved herein in the HuH-7 cell culture model. Given the dispensability of LC3/GABARAP lipidation but the necessity of Atg3 in HBV production, we inferred that its effect on Atg5-12 conjugation rather than its E2-like enzymatic activity regulated virus replication. The important role of the Atg5-12 conjugate in the HBV life cycle is supported by a report demonstrating an upregulation of Atg5 and Atg12 protein levels in stably HBV-replicating cell lines and in tumor liver tissues of HCC patients with HBV infection (39). In the same settings, no upregulation of the Atg4B protease was observed (39), which matched our result that Atg4B was not essential for HBV propagation.
We identified the HBV core protein to be the prime target, being acutely Atg5-12 dependent. The silencing of the Atg5-12/16L1 complex and of Atg3 all reduced intracellular core contents at a posttranscriptional level, while L envelope amounts only marginally declined. Core content reduction impeded the assembly and/or stability of intracellular NCs and, consequently, virus yields. In principle, our findings are consistent with those of previous works in which the effects of Atg5 and Atg7 silencing on HBV replication were studied in cell culture systems and transgenic mice (18–20). The outcomes of these studies are still a matter of debate, as one set of experiments indicated that NC maturation was found to be impaired (19, 20), whereas another report pointed to the NC envelopment reaction (18). Our results favor the NC assembly reaction being the prime target of autophagic intervention, which ultimately may lead to compromised NC envelopment, as indicated by the poor colocalization of core and L in Atg12-KD cells.
The formation of the HBV capsid was viewed as a spontaneous self-assembly process, as it could be recapitulated in cell-free systems (40). More recent reports, however, indicate that viral capsid assembly is an active process sustained by host factors that may act as assembly machines (41). In the case of HBV capsids, the host serine-arginine protein kinase had been shown to act like a molecular chaperone to prevent the core protein from assembling at the wrong time and place (42). When HBV core particle formation was inhibited by chemical compounds, like heteroaryldihydropyrimidines (HAPs), unassembled core proteins were shunted to increased degradation (43). Accordingly, it is tempting to speculate that the Atg5-12 conjugate is another important player chaperoning proper HBV capsid/NC assembly, with the consequence that its deficit may favor core/NC destabilization and degradation, a phenotype reminiscent of the action of HAPs.
The proviral function of the Atg5-12 conjugate in HBV biogenesis is supported by our observation that core physically interacted with the autophagic initiation machinery, which is in agreement with our previous report (25). The continuative analyses of this interaction now reveal that core associates with the Atg12 moiety of the elongation complex, as evidenced by the binding of core to a conjugation-defective Atg12.GA mutant. Atg12 interacted with core both in the absence and in the presence of ongoing virus replication, indicating that core per se rather than the associated pgRNA/Pol complex was recognized. This is different from the findings for hepatitis C virus (HCV), which likewise requires autophagy pathway functions for replication but engages the pathway via a transient interaction of the viral NS5B polymerase with Atg5 (6, 44, 45). In the case of HBV, the Atg12/core interaction appeared to be stable, as deduced from Atg12 incorporation into the mature extracellular virion. In this respect, HBV shares features with other autophagy-dependent viruses, like Epstein-Barr virus, that have been shown to package lipidated LC3-II into viral particles (32). Our mapping analysis of the HBV core/Atg12 interaction identified the so-called IDR of Atg12 to be a binding module. IDRs are protein domains lacking stable secondary and tertiary structures and are predicted to promote protein-protein interactions (35). Intriguingly, the Atg12-specific IDR was reported to be dispensable for its conjugation to Atg5 (Fig. 5D) and autophagy (35, 46), implicating that core gained access to Atg12 without constricting autophagic functions. A recent study identified an interaction between Atg12 and cellular Alix, an ESCRT-associated adaptor (34). Since HBV budding is known to depend on ESCRT functions (12–15), it is conceivable that autophagy and ESCRT machineries could have a close interplay in HBV propagation.
Our results pose the question of whether HBV may use the Atg5-12/16L1 conjugate in a noncanonical manner (i.e., as a distinct function mediated by individual Atg proteins) or a canonical (i.e., autophagy-like) manner, with the latter involving autophagic membranes. Previously, we showed that the HBV core is able to associate with intracellular membranes (26), albeit the functional relevance is yet unknown. In the case of many other viruses, like retroviruses, membrane targeting of viral components helps to concentrate and drive the assembly reaction (47). For HBV, the spatial and temporal requirements for NC formation, i.e., the packaging of the pgRNA/Pol complex into the forming capsid, are still elusive but have been suggested to involve membrane platforms (48). The depletion of the Atg5-12/16L1 complex, which has an intrinsic membrane-tethering activity (49), led to a reduction in core-membrane association. This decline may be a consequence of an impaired phagosome expansion and/or the lack of Atg12, piggybacking core to membranes. Previously, it was shown that HBV is able to activate the endocytic Rab7 GTPase, thereby altering its own secretion by inducing changes in the morphology of autophagic and MVB compartments. Concomitantly, HBV core was found to colocalize with the autophagosomal marker LC3 (33). Consistent with this, we detected a clear colocalization of core with the surfaces of LC3-positive, dot-like structures, reminiscent of autophagosomal membranes/vesicles. We thus hypothesize that HBV may co-opt the Atg5-12/16L1 conjugate in a phagophore-tethered state. As supportive evidence, we recently showed that the Rab33B GTPase, an effector of the Atg5-12/16L1 conjugate, is a positive regulator of HBV assembly, guiding NC assembly/stability (26, 50). Like all Rab proteins, Rab33B exerts its function in a membrane-tethered form and had been implicated in the regulation of autophagy via fusion of Rab33B-decorated, Golgi apparatus-derived vesicles with the Atg5-12/16L1 complex in order to supply lipids for the expanding phagophore (50).
Several RNA viruses, including coxsackievirus, dengue virus, influenza A virus, and HCV, have evolved strategies to use autophagic vesicles as scaffolds for replication and assembly (3–6). In the case of picornaviruses, like poliovirus, remodeled autophagic intracellular membranes serve as a structural platform for replication, assembly, and nonlytic virus egress (5, 51). For HCV, numerous studies indicated that autophagy is important to improve its infection and replication (6, 31, 44, 45, 52). Interestingly, a recent work demonstrated that the Atg5-12/16L1 complex positively regulates HCV replication in an LC3-independent manner (45). HCV is known to induce intracellular membrane rearrangements, the membrane web (MW) structures, in which viral replication and assembly proceed. MW formation was found to rely on the Atg5-12/16L1 conjugate, implicating that it plays a role in the construction of the HCV replication scaffold (45). Despite being unrelated viruses, HCV and HBV seemingly co-opt and omit identical autophagic proteins/pathway functions to improve their propagation. Moreover, both viruses have been shown to increase the expression of Rab33B for benefits (26, 53), suggesting that the conjoint Atg5-12/16L1-Rab33B complex may play a general role in autophagy-assisted virus assembly reactions.
As a novel finding, we observed that HBV did not require LC3B or LC3/GABARAP lipidation and, thus, did not depend on phagophore maturation and closure. The dispensability of the late degradative stages of autophagy may explain why HBV induces autophagy without ending up in the destructive autophagolysosomes. To account for the proviral effects of missing LC3B and inhibited LC3/GABARAP lipidation, one simple interpretation could be that more functional Atg5-12 may be available for HBV purposes, because the natural LC3/GABARAP substrates for the E3 activity of Atg5-12/16L1 are lacking or sequestered by inactive Atg4B.dn. Such a competing mechanism has been described for Kaposi's sarcoma-associated herpesvirus (KSHV), albeit with different consequences. KSHV encodes an antiapoptotic Fas-associated death domain-like interleukin-1β-converting enzyme-inhibitory protein (vFLIP) that interacts with Atg3, the E2-like enzyme of the LC3 conjugation system. vFLIP competes with LC3 for binding to Atg3, thereby suppressing LC3 conjugation and autophagy (54).
In summary, we identified the Atg5-12 conjugate, likely in conjunction with Atg16L1, to be the prime autophagic component scaffolding HBV assembly in an LC3/GABARAP-independent manner. Future studies should elucidate whether perturbations of the HBV core interaction with the functionally irrelevant IDR of Atg12 may be tools to control virus multiplication.
MATERIALS AND METHODS
Plasmids.
For HBV replication, the vector pCEP4ΔCMVΔSV40/1.1×HBV (pHBV) was used. It carries a 1.1×-unit-length HBV genome (genotype D; GenBank accession number V01460) in which the viral core/Pol promoter is preceded by the hMTIIA promoter (14). For HBV transcription analyses, a modified version of the replicon was engineered (pHBVΔHP) in which the hMTIIa promoter region was removed (14). For solitary expression of the HBV core protein, plasmid pNI2.C, which carries the core gene under the control of the hMTIIa promoter, was employed (24). For in vitro transcription, an HBV DNA fragment encompassing nucleotides (nt) 129 to 1986 (numbering is in reference to the HBV genotype D DNA genome) was cloned between the SP6 and T7 promoter regions of plasmid pGEM2 (Promega). Plasmid pCMV-myc-Atg12 contains the long isoform of human Atg12 (APG12L) with an N-terminal Myc tag and was provided by Toren Finkel (National Institutes of Health, Bethesda, MD, USA) via Addgene (plasmid number 24923) (55). To generate the small isoform of Atg12 (APG12, GenBank accession number NM_004707.3) that presents the canonical sequence, a DNA fragment encoding the N-terminal 69 amino acids (aa) of APG12L was removed and is termed Atg12 herein. To attenuate Atg12 expression, a truncation was introduced within the cytomegalovirus (CMV) promoter region of pCMV-myc-Atg12, as described previously (26). The conjugation-defective Atg12 mutant (Atg12.GA), in which the C-terminal glycine at position 140 was replaced by alanine, was created by mutagenesis using a Q5 site-directed mutagenesis kit (New England BioLabs). To delete the IDR domain of Atg12, the N-terminal 42-aa-encoding sequence was removed by cloning (Atg12ΔN). A SureSilencing shRNA plasmid (pGeneCliphMGFP) carrying a control shRNA sequence under the control of the U1 promoter and a humanized monster green fluorescence protein (hMGFP) reporter gene was purchased from Qiagen. For the construction of an Atg12-specific shRNA construct, the control shRNA sequences were replaced by sense sequence 5′-CCAAGGACTCATTGACTTCAT-3′ and antisense sequence 5′-ATGAAGTCAATGAGTCCTTGG-3′. Because the expression of hMGFP turned out to be harmful for HBV replication, the hMGFP gene was replaced by an HA-tagged Bet1 (blocked early in transport; GenBank accession number NM_005868.5) reporter gene (shAtg12.Bet1). Plasmid pBABEpuroWT.Atg3, carrying the mouse Atg3 gene (GenBank accession number NM_026402) with a C-terminal V5 tag, was obtained from Jayanta Debnath (University of San Francisco, San Francisco, CA, USA) via Addgene (plasmid number 26926) (56). Notably, the mouse and human Atg3 gene products are strongly homologous and differ in only 7 out of 314 residues. Plasmid pmStrawberry-Atg4B.C74A expresses the mouse Atg4B gene (GenBank accession number NM_174874) with an N-terminal mStrawberry tag and was provided by Tamotsu Yoshimori (Osaka University, Osaka, Japan) via Addgene (plasmid number 21076). The Atg4B.dn mutant carries a cysteine-to-alanine substitution at position 74 (38). To exchange the mStrawberry tag for an HA tag, Atg4B.C74A was subcloned into the vector pCMV.HA (Clontech). To generate a wt Atg4B expression construct (Atg4B.wt), the cysteine-to-alanine substitution at position 74 was reversed by mutagenesis. A mammalian expression vector expressing the Canis lupus Rab7 protein (GenBank accession number NM_001003316.1) fused to the C terminus of the enhanced GFP (EGFP) gene was a gift from Gilles Spoden (University Medical Center Mainz, Mainz, Germany). A vector carrying an EGFP-tagged human LC3B gene (GenBank accession number NM_022818) was obtained from Karla Kirkegaard (Stanford Medicine, Stanford, CA, USA) via Addgene (plasmid number 11546) (51). All constructs were verified by sequencing, and cloning details are available on request.
Antibodies.
Polyclonal antisera against recombinant native (K45) or denatured (K46) core particles were raised in rabbits, as described previously (24). For IF studies, a commercially available polyclonal rabbit antiserum against the core antigen (catalog number B0586; Dako) was used. For immunodetection of the L protein, either the MA18/7 mouse antibody (a gift from Dieter Glebe, University of Gießen, Gießen, Germany) or a rabbit antibody raised against a recombinant peptide encoding aa 1 to 42 of L fused to glutathione S-transferase (catalog number K1350; Eurogentec) were employed. Other commercially available antibodies are listed by the suppliers. An antibody against the V5 tag was purchased from Bio-Rad (antibody MCA1360GA). Antibodies obtained from Cell Signaling were anti-Atg3 (antibody 3415), anti-Atg5 (antibody D5F5U), anti-Atg12 (antibody 2010), and anti-Atg16L1 (antibody D6D5). Antibodies obtained from Covance were mouse anti-HA (antibody 16B12) and anti-Myc (antibody 9E10). The anticalnexin antibody (antibody SPA-860) was purchased from Enzo Life Sciences. A rat anti-HA antibody (antibody 3F10) was from Roche Diagnostics. Antibodies obtained from Sigma-Aldrich were anti-β-actin (antibody AC15), anti-α-tubulin (antibody B-5-1-2), and anti-LC3A/B (antibody L8918). Peroxidase-labeled, secondary antibodies were obtained from Dianova, and fluorophore-labeled antibodies were purchased from Thermo Fisher Scientific.
siRNAs, cell culture, and transfection.
The human hepatocellular carcinoma cell line HuH-7 was obtained from the Japanese Collection of Research Bioresources. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 5 μg/ml ciprofloxacin. Where indicated, cells were subjected to starvation by incubation in serum-free Hanks' balanced salt solution for 2 h. Transfections of HuH-7 cells with plasmid DNAs were performed with the Lipofectamine Plus reagent (Thermo Fisher Scientific). Unless otherwise indicated, 2 × 105 cells per well of a six-well plate were transfected with a final concentration of 4 μg plasmid DNA. For transfection of HuH-7 cells with siRNAs, the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) was used. Briefly, 1.3 × 105 cells per well of a six-well plate were transfected with 20 pmol siRNA according to the protocol of the supplier. siRNAs were obtained from Dharmacon or Sigma-Aldrich, and oligonucleotide sequences are provided in Table 1. As a control, a nonsense siRNA with no known homology to mammalian genes was used (Qiagen). After 48 h, cells were retransfected with plasmid DNA using Lipofectamine Plus and harvested after an additional 72 h.
TABLE 1.
siRNAs used in this study
| Gene description | siRNA target sequence |
|---|---|
| Atg3 | GCAAACAGAUGGAAUAUUC |
| Atg3 | CAAGACACUUCACAAUGUA |
| Atg3 | GCGGAUGGGUAGAUACAUA |
| Atg3 | UGAAGAGAGUGGAUUGUUG |
| Atg5 | CAUCUGAGCUACCCGGAUA |
| Atg10 | GAAUCUACCUCUGAGUUAU |
| Atg10 | GGACCAUGGGACACUAUUA |
| Atg10 | CCAACGUUAUUGUGCAGAA |
| Atg10 | CAGCGUCCGAAGUGAUUAA |
| Atg12 | GCAGCUUCCUACUUCAAUU |
| Atg16L1 | GAGCAUAGUUCGAGAGAUG |
| Atg16L1 | UGUGGAUGAUUAUCGAUUA |
| Atg16L1 | GAUCUACGCAGCAAAGUCU |
| Atg16L1 | GUACCAAACAGGCACGAGA |
| LC3B | AGGAGACGUUCGGGAUGAA |
Cell protein analyses, viral particle analysis, and multiplex real-time PCR.
The production of HBV particles was determined by a TaqMan chemistry-based, multiplex real-time PCR, as described previously (14). Cell supernatants were harvested, and cells were lysed with 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and 0.2% Triton X-100, followed by centrifugation for 5 min at 15,000 × g and 4°C. Where indicated, cells were lysed with 1× Laemmli buffer (50 mM Tris-HCl [pH 6.8], 10% β-mercaptoethanol, 10% glycerol, 2% SDS, 0.1% bromphenol blue), scraped from the plates, and boiled for 10 min prior to centrifugation for 5 min at 13,000 × g and 4°C. To probe for protein expression, aliquots of the lysates were analyzed by WB. Intracellular NC and extracellular virions were isolated by immunomagnetic separation using PureProteome protein G magnetic beads (Millipore) coated with capsid- and envelope-specific antibodies, respectively. The immunoprecipitated particles were next subjected to a treatment with 100 U/ml Benzonase (Merck) for 2 h at 37°C in the presence of 0.2% Triton X-100 to remove the residual plasmid DNA used for transfection. The viral DNA was isolated with a High Pure viral nucleic acid kit (Roche Diagnostics) according to the manufacturer's instructions. PCR analyses were performed with a 7500 real-time PCR system and sequence detection software (version 4.0; Applied Biosystems). Because the transfected HBV replicon plasmid DNA (pHBV) and progeny virus DNA are genetically identical, two primer/probe sets targeting either the HBV genome or the hygromycin resistance gene of the pCEP plasmid backbone were used, as described previously (14). In parallel, cells were inspected for the presence of damage and toxicity. LDH activity in the culture media was determined using a colorimetric quantification assay (Cytotoxicity Detection KitPlus; Roche Diagnostics). To measure total protein concentrations in cell extracts, a Bradford protein assay (Bio-Rad) with bovine serum albumin (BSA) standards (New England BioLabs) was used.
qRT-PCR analysis.
Total mRNAs were isolated from cells using the TRIzol reagent (Life Technologies) and a Direct-zol RNA miniprep kit (Zymo Research), according to the protocols of the suppliers. The mRNA was treated with 5 U RNase-free, recombinant DNase I (Roche Diagnostics), and cDNA synthesis was performed by using a qScript cDNA synthesis kit (Quanta Biosciences). qRT-PCRs were performed as described previously (26). For data analysis, the comparative cycle threshold (CT) method was used, and data are reported as the fold change normalized to an endogenous reference gene (β-actin). For HBV transcription analyses, the sense (5′-TGTCCTCCAACTTGTCCTGGTT-3′) and antisense (5′-AGGCATAGCAGCAGGATGAAGA-3′) primer pair, which targets the 3.5-kb pgRNA, the 3.4-kb core/Pol-, and the 2.4- and 2.1-kb envelope-specific transcripts (primer pair #1 [PP#1]), was used. The sense (5′-ATTCGCACTCCTCCAGCTT-3′) and antisense (5′-GGGACCTGCCTCGTCGTCTA-3′) primer pair (PP#2) is specific for the pgRNA and core/Pol transcript. To measure the degree of Atg10 depletion by qRT-PCR, the primer pair 5′-GGTGATAGTTGGGAATGGAGACC-3′ and 5′-GTCTGTCCATGGGTAGATGCTC-3′ was used.
Northern blot analysis.
Total cellular RNA was extracted as described above. Five micrograms of RNA was resolved in a 1.5% agarose gel containing morpholinepropanesulfonic acid and formaldehyde and transferred to a positively charged nylon membrane (Roche Diagnostics) with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). A digitonin (DIG)-labeled plus-strand-specific RNA probe corresponding to nt 1374 to 1986 of the HBV genome was generated by T7-driven in vitro transcription from the HBV DNA template cloned into pGEM2. The probe preparation and subsequent DIG detection were conducted with a DIG Northern starter kit (Sigma-Aldrich) according to the manufacturer's instructions.
Coimmunoprecipitation assays.
To probe for intracellular protein interactions, cells were lysed with a 2% solution of the nondenaturing detergent CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}-HBS (50 mM HEPES-KCl, pH 7.5, 200 mM NaCl), 20 mM N-ethylmaleimide, supplemented with 1× protease inhibitor mixture (Serva) for 20 min on ice. After centrifugation, the lysates were immediately subjected to immunoprecipitation using tosyl-activated, superparamagnetic polystyrene beads (Dynabeads Sheep anti-rabbit IgG; Dynal) that had been precoated with the core antibody K45 as described previously (24). After incubation for 3 h at 4°C with agitation, the immune complexes were washed three times with 0.5% CHAPS-HBS and once with phosphate-buffered saline (PBS) prior to SDS-PAGE and immunoblotting. For immunocapture of extracellular viral particles, the magnetic beads were coated with the anti-L antibody K1350, and precipitation was performed in the absence of CHAPS.
Fluorescence microscopy.
For IF, cells grown on coverslips were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature and permeabilized with 0.2% Triton X-100 for 10 min. Alternatively, cells were fixed and permeabilized with ice-cold methanol containing 2 mM EGTA. Cells were blocked in PBS containing 1% BSA, incubated with the primary antibodies indicated above for 1 h at 37°C, rinsed with PBS, and then incubated with Alexa Fluor-tagged secondary antibodies for 1 h at 37°C. DNA was stained with Hoechst 33342 (Sigma-Aldrich). Images were acquired separately for each channel using a Zeiss Axiovert 200 M microscope equipped with a Plan-Apochromat 100× objective lens (numerical aperture, 1.4) and a Zeiss Axiocam digital camera. Where indicated, z-stack images were optically deconvoluted using the software supplied by Zeiss. AxioVision software (version 4.7.1) was used for the merging of pictures. Tiff images were assembled into figures using Adobe Photoshop CS6 software.
Membrane flotation analysis.
The density flotation assay has been described previously (26). Briefly, cells were broken by Dounce homogenization in homogenization buffer, and extracts were centrifuged at 1,000 × g for 10 min at 4°C to sediment nuclei and debris. The postnuclear supernatant was adjusted to 40% OptiPrep (Sigma-Aldrich), placed on the bottom of a SW60 centrifuge tube, and overlaid with 30% and 5% OptiPrep solutions. The step gradient was centrifuged for 3 h at 100,000 × g and 4°C in an SW60 rotor (Beckman), and fractions were collected from the top.
Statistics.
Statistically significant differences between groups and statistical graphs were assessed with a two-tailed, unpaired t test using Microsoft Office Excel 2016 software. Differences between groups were considered significant when the P value was <0.05 or <0.01.
ACKNOWLEDGMENTS
We thank Jayanta Debnath, Toren Finkel, Dieter Glebe, Karla Kirkegaard, Gilles Spoden, and Tamotsu Yoshimori for generously providing antibodies, cell lines, and plasmid DNA constructs.
This work was supported by grants to R.P. from the Deutsche Forschungsgemeinschaft (PR 305/3-1/3-2) and by grants to C.B. from the Stipendienstiftung Rheinland-Pfalz.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Noda NN, Inagaki F. 2015. Mechanisms of autophagy. Annu Rev Biophys 44:101–122. doi: 10.1146/annurev-biophys-060414-034248. [DOI] [PubMed] [Google Scholar]
- 2.Weidberg H, Shvets E, Elazar Z. 2011. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 3.Dong X, Levine B. 2013. Autophagy and viruses: adversaries or allies? J Innate Immun 5:480–493. doi: 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- 5.Jackson WT. 2014. Dangerous membranes: viruses that subvert autophagosomes. EBioMedicine 1:97–98. doi: 10.1016/j.ebiom.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ploen D, Hildt E. 2015. Hepatitis C virus comes for dinner: how the hepatitis C virus interferes with autophagy. World J Gastroenterol 21:8492–8507. doi: 10.3748/wjg.v21.i28.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W. 2015. The hepatitis B virus receptor. Annu Rev Cell Dev Biol 31:125–147. doi: 10.1146/annurev-cellbio-100814-125241. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Seeger C. 2015. Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med 5:a021386. doi: 10.1101/cshperspect.a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassal M. 2008. Hepatitis B viruses: reverse transcription a different way. Virus Res 134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Blondot ML, Bruss V, Kann M. 2016. Intracellular transport and egress of hepatitis B virus. J Hepatol 64:S49–S59. doi: 10.1016/j.jhep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Prange R. 2012. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol 201:449–461. doi: 10.1007/s00430-012-0267-9. [DOI] [PubMed] [Google Scholar]
- 12.Lambert C, Doring T, Prange R. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol 81:9050–9060. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A 104:10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stieler JT, Prange R. 2014. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS One 9:e91279. doi: 10.1371/journal.pone.0091279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou SF, Tsai ML, Huang JY, Chang YS, Shih C. 2015. The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PLoS Pathog 11:e1005123. doi: 10.1371/journal.ppat.1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patient R, Hourioux C, Roingeard P. 2009. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol 11:1561–1570. doi: 10.1111/j.1462-5822.2009.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo JT, Guo H. 2015. The interferon-inducible protein tetherin inhibits hepatitis B virus virion secretion. J Virol 89:9200–9212. doi: 10.1128/JVI.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. 2011. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol 85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A 107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. 2011. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol 85:13453–13456. doi: 10.1128/JVI.06064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng J, Klionsky DJ. 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, Liu W. 2014. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 10:416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardens A, Doring T, Stieler J, Prange R. 2011. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol 13:602–619. doi: 10.1111/j.1462-5822.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doring T, Prange R. 2015. Rab33B and its autophagic Atg5/12/16L1 effector assist in hepatitis B virus naked capsid formation and release. Cell Microbiol 17:747–764. doi: 10.1111/cmi.12398. [DOI] [PubMed] [Google Scholar]
- 26.Bartusch C, Doring T, Prange R. 2017. Rab33B controls hepatitis B virus assembly by regulating core membrane association and nucleocapsid processing. Viruses 9:E157. doi: 10.3390/v9060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubyova B, Hodek J, Zabransky A, Prouzova H, Hubalek M, Hirsch I, Weber J. 2017. PRMT5: a novel regulator of hepatitis B virus replication and an arginine methylase of HBV core. PLoS One 12:e0186982. doi: 10.1371/journal.pone.0186982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia PD, Ou JH, Rutter WJ, Walter P. 1988. Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: after signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J Cell Biol 106:1093–1104. doi: 10.1083/jcb.106.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaglioni PP, Melegari M, Wands JR. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol 71:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duriez M, Thouard A, Bressanelli S, Rossignol JM, Sitterlin D. 2014. Conserved aromatic residues of the hepatitis B virus precore propeptide are involved in a switch between distinct dimeric conformations and essential in the formation of heterocapsids. Virology 462-463:273–282. doi: 10.1016/j.virol.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, Munz C. 2014. Macroautophagy proteins assist Epstein Barr virus production and get incorporated into the virus particles. EBioMedicine 1:116–125. doi: 10.1016/j.ebiom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue J, Krueger EW, Chen J, Cao H, Ninomiya M, McNiven MA. 2015. HBV secretion is regulated through the activation of endocytic and autophagic compartments mediated by Rab7 stimulation. J Cell Sci 128:1696–1706. doi: 10.1242/jcs.158097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrow L, Malhotra R, Debnath J. 2015. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. 2014. Intrinsically disordered regions in autophagy proteins. Proteins 82:565–578. doi: 10.1002/prot.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slobodkin MR, Elazar Z. 2013. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem 55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 38.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunanopparat A, Kimkong I, Palaga T, Tangkijvanich P, Sirichindakul B, Hirankarn N. 2016. Increased ATG5-ATG12 in hepatitis B virus-associated hepatocellular carcinoma and their role in apoptosis. World J Gastroenterol 22:8361–8374. doi: 10.3748/wjg.v22.i37.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marreiros R, Muller-Schiffmann A, Bader V, Selvarajah S, Dey D, Lingappa VR, Korth C. 2015. Viral capsid assembly as a model for protein aggregation diseases: active processes catalyzed by cellular assembly machines comprising novel drug targets. Virus Res 207:155–164. doi: 10.1016/j.virusres.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Wang JC, Zlotnick A. 2011. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog 7:e1002388. doi: 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 44.Guevin C, Manna D, Belanger C, Konan KV, Mak P, Labonte P. 2010. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahmy AM, Labonte P. 2017. The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci Rep 7:40351. doi: 10.1038/srep40351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popelka H, Klionsky DJ. 2015. Post-translationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J 282:3474–3488. doi: 10.1111/febs.13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorizate M, Krausslich HG. 2011. Role of lipids in virus replication. Cold Spring Harb Perspect Biol 3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lingappa JR, Newman MA, Klein KC, Dooher JE. 2005. Comparing capsid assembly of primate lentiviruses and hepatitis B virus using cell-free systems. Virology 333:114–123. doi: 10.1016/j.virol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 49.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. 2012. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. 2008. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Ou JH. 2015. Hepatitis C virus and autophagy. Biol Chem 396:1215–1222. doi: 10.1515/hsz-2015-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. 2010. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol 84:5404–5414. doi: 10.1128/JVI.02529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, Jung JU. 2009. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee IH, Finkel T. 2009. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. 2010. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]