ABSTRACT
Arthropod-borne viruses, such as the members of the genus Alphavirus, are a significant concern to global public health. As obligate intracellular pathogens, RNA viruses must interact with the host cell machinery to establish and complete their life cycles. Despite considerable efforts to define the host-pathogen interactions essential for alphaviral replication, an unbiased and inclusive assessment of alphaviral RNA-protein interactions has not been undertaken. Moreover, the biological and molecular importance of these interactions, in the full context of their molecular function as RNA-binding proteins, has not been fully realized. The data presented here introduce a robust viral RNA-protein discovery method to elucidate the Sindbis virus (SINV) RNA-protein host interface. Cross-link-assisted mRNP purification (CLAMP) assessment revealed an extensive array of host-pathogen interactions centered on the viral RNAs (vRNAs). After prioritization of the host proteins associated with the vRNAs, we identified the site of protein-vRNA interaction by a UV cross-linking and immunoprecipitation sequencing (CLIP-seq) approach and assessed the consequences of the RNA-protein binding event of hnRNP K, hnRNP I, and hnRNP M in regard to viral infection. Here, we demonstrate that mutation of the prioritized hnRNP-vRNA interaction sites effectively disrupts hnRNP-vRNA interaction. Correlating with disrupted hnRNP-vRNA binding, SINV growth kinetics were reduced relative to wild-type parental viral infections in vertebrate and invertebrate tissue culture models of infection. The molecular mechanism leading to reduced viral growth kinetics was found to be dysregulated structural-gene expression. Collectively, this study further defines the scope and importance of the alphavirus host-pathogen vRNA-protein interactions.
IMPORTANCE Members of the genus Alphavirus are widely recognized for their potential to cause severe disease. Despite this recognition, there are no antiviral therapeutics, or safe and effective vaccines, currently available to treat alphaviral infection. Alphaviruses utilize the host cell machinery to efficiently establish and complete their life cycle. However, the extent and importance of host-pathogen RNA-protein interactions are woefully undercharacterized. The efforts detailed in this study fill this critical gap, and the significance of this research is 3-fold. First, the data presented here fundamentally expand the scope and understanding of alphavirus host-pathogen interactions. Second, this study identifies the sites of interaction for several prioritized interactions and defines the contribution of the RNA-protein interaction at the molecular level. Finally, these studies build a strategy by which the importance of the given host-pathogen interactions may be assessed in the future, using a mouse model of infection.
KEYWORDS: RNA-binding proteins, alphavirus, plus-strand RNA virus, posttranscriptional RNA-binding proteins, virus-host interactions
INTRODUCTION
The members of the genus Alphavirus are enveloped positive-sense RNA viruses recognized for their capacity to cause severe burdens on public health. Due to the ubiquity of competent vector mosquito species, the alphaviruses, as evidenced by the contemporary outbreak of chikungunya virus (CHIKV), have a high potential to cause epidemic emergences of arboviral disease (1–7). Broadly speaking, disease-causing alphaviruses can be classified into two groups based on the clinical manifestation of viral infection. While comparatively rare, encephalitic alphaviruses are capable of causing severe encephalitis, primarily in young children (8, 9). Survivors of alphaviral encephalitis are likely to be afflicted with neurological sequelae following primary convalescence (10). In contrast to the encephalitic alphaviruses, the arthritogenic alphaviruses, including CHIKV; Ross River virus; and the archetypical model arthritogenic alphavirus, Sindbis virus (SINV), produce demonstrably low mortality and morbidity despite being capable of causing severe disease (11–13). Clinical manifestations of infection by the arthritogenic alphaviruses range from mild febrile illness to incapacitating multijoint arthritis that may persist for prolonged periods (14–17). Collectively, the alphaviruses continue to be an emerging threat to global public health in developed and developing communities alike.
Despite causing varied symptomology and pathology in the human host, the members of the genus Alphavirus have molecular life cycles that are highly similar (18, 19). Viral entry through the endosomal pathway culminates, presumably, in the release of the nucleocapsid core into the cytoplasm of the newly infected host cell. Disassembly of the nucleocapsid core results in the release of the viral genomic RNA into the cytoplasmic milieu, where it may associate with the host translational machinery to initiate synthesis of the viral replicase complex. Recently, we have reported data that indicate that some viral capsid protein may remain associated with the viral genomic RNA, where it enhances early viral gene expression (20). Moreover, it should be noted that a large number of incoming genomic RNAs fail to engage with the host machinery and undergo rapid degradation following entry into the cell (20, 21). Regardless, the translation of the incoming viral RNA (vRNA) leads to the assembly of a functional replicase complex, resulting in the synthesis of nascent vRNAs (18, 19). Replication results in the formation of a minus-strand template vRNA, from which the progeny genomic vRNA and subgenomic vRNA are synthesized. Translation of the structural vRNA provides the structural components necessary for nucleocapsid assembly and virion release, effectively completing the viral life cycle.
As obligate intracellular pathogens, viruses are dependent upon the cellular machinery for the completion of their life cycles. As such, many viruses interact with host factors to ensure the efficient and successful completion of their life cycles. This is especially true for single-stranded RNA viruses, which lack the genomic capacity and complexity to provide viral surrogates of essential host factors. To date, there have been numerous studies seeking to identify and characterize the involvement of host factors in the alphaviral life cycle. Many of these efforts have focused on elucidating the role of host factors in viral RNA replication, and many others have sought to identify the protein-protein interactions of the alphaviral nonstructural proteins (22–31). While several studies have explored the RNA-protein interactions of other viruses, notably members of the genera Flavivirus and Picornavirus, a comprehensive examination of the vRNA-protein interface of the host-pathogen interaction of the alphaviruses has, to our knowledge, been overlooked (32–34). Despite this lack, there are several notable instances where host RNA-binding proteins have been implicated in the alphaviral life cycle (35–40). Nonetheless, many of these studies have not directly explored the role of the vRNA-protein interaction and utilized approaches that may have significant consequences for the host system. We hypothesize that vRNA-protein interactions are an essential interface of the host-pathogen relationship and that, by their nature as RNA-binding proteins, the binding of the host factors to the vRNA is an integral part of the regulation of vRNA function.
The primary goals of this study were 2-fold: (i) to develop and employ a method by which alphaviral vRNA-protein interactions could be identified in an unbiased manner during viral infection and (ii) to characterize the biological and molecular importance of the vRNA-protein interactions by site-directed mutagenesis of the RNA-protein interaction site. To this end, here, we describe the use of a robust vRNA-protein interaction discovery approach that utilizes cross-linking to preserve, and high-sensitivity mass spectrometry to detect, the genuine viral ribonucleoprotein (vRNP) complexes formed during viral infection. Second, we select a subset of host factors, namely, hnRNP K, hnRNP I, and hnRNP M, for UV cross-linking and immunoprecipitation (IP) sequencing (CLIP-seq) analysis and biological characterization following the disruption of the hnRNP-vRNA interaction by site-directed mutagenesis.
RESULTS
Identification of host-pathogen RNA-protein interactions using the CLAMP assay.
As mentioned in the introduction, a number of studies have identified RNA-binding proteins during studies designed to characterize alphaviral replication complexes (22–25, 27, 29, 30, 38–40). Nonetheless, for many of these factors, it is unknown whether they directly interact with the vRNAs. Moreover, to our knowledge, an extensive assessment of the alphavirus RNA-protein interactions has never been undertaken. To address this gap in knowledge, we sought to identify and characterize novel host-pathogen RNA-protein interactions. To elucidate such interactions in an unbiased manner, we adapted previous technologies to develop an approach that utilizes UV cross-linking in conjunction with streptavidin affinity purification to isolate the vRNP complexes formed during SINV infection. This method, which we have termed cross-link-assisted mRNP purification (CLAMP), is diagrammed in Fig. 1A, and its application is described below.
FIG 1.
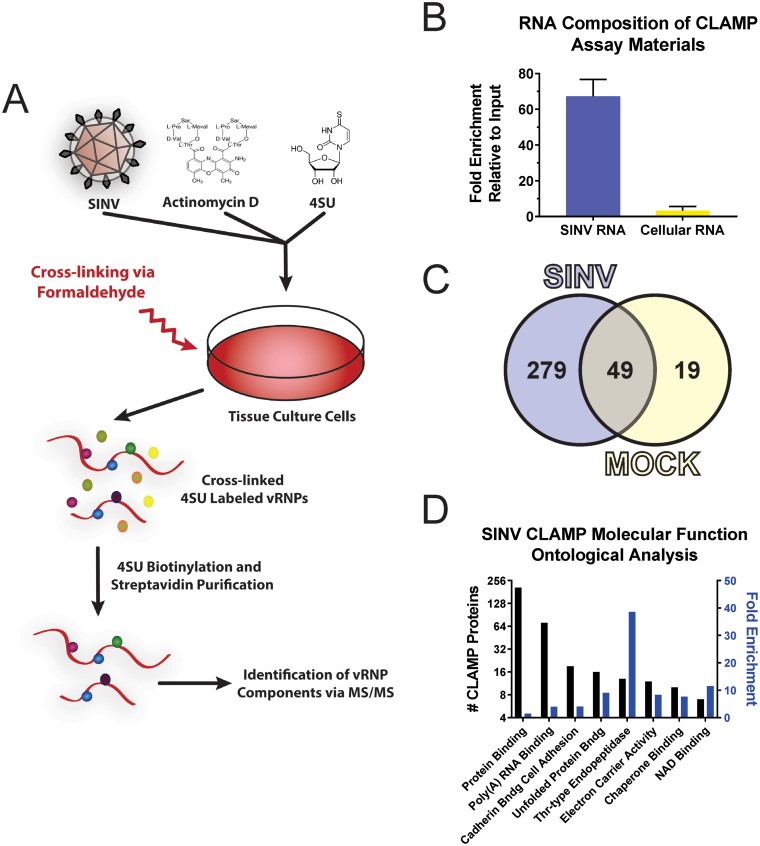
Identification of SINV vRNA-protein interactants by CLAMP assay. (A) Schematic of the CLAMP assay. Briefly, tissue culture cells are infected with SINV, and later, in the presence of actinomycin D, the vRNAs are cotranscriptionally labeled with the nucleoside analogue 4SU. At the desired time, the cells are cross-linked, and lysates are generated, from which vRNA is purified by biotinylation of the 4SU residues and streptavidin affinity purification. The components of the vRNP are identified by mass spectrometry. (B) Quantitative assessment of RNA species recovered following CLAMP assay. The data shown represent the fold enrichment of viral and cellular RNAs normalized to input RNA levels and are the means of three independent CLAMP purifications; the error bars represent standard deviations of the mean. (C) Venn diagram depicting the results of the comparative analysis of CLAMP-identified interactants for SINV-infected and mock-treated samples. (D) Ontological assignment of the SINV CLAMP-identified interactants in regard to molecular function. Shown on the left y axis is the number of individual CLAMP identified proteins, and on the right y axis is the fold enrichment relative to the DAVID Homo sapiens background list.
To identify alphavirus RNA-protein interactions, we infected 293HEK cells at a multiplicity of infection (MOI) of 10 PFU/cell to ensure total infection. After a 1-h adsorption period, the inoculum was removed and the unbound viral particles were removed by gentle washing prior to the addition of fresh whole medium. The tissue culture cells were then returned to the incubator and cultured under normal conditions. At 2 h postinfection (hpi), the culture medium was removed and replaced with fresh medium supplemented with actinomycin D at a concentration previously established to halt all cellular transcription (41). After 15 min, the medium was supplemented with the nucleoside analogue 4-thiouridine (4SU) at a final concentration of 100 μM in an excess volume, and the cells were further incubated for 4 h to establish a pool of cotranscriptionally labeled vRNAs (42). At the end of the incubation period, the supernatants were removed, and the cells were scraped into 1× phosphate-buffered saline (PBS) and harvested by gentle centrifugation. To stabilize the vRNA-protein interactions, the resulting cell pellets were dispersed/resuspended in a 1.0% formaldehyde solution and incubated under gentle agitation for a period of 7 min at room temperature (43). After cross-linking, the cells were re-collected by centrifugation for period of 3 min to obtain a cross-linking period of no more than 10 min in total. The supernatant was removed, and the cross-linked pellets were resuspended in 0.25 M glycine in PBS to quench residual formaldehyde. The cell pellets were harvested again and washed twice with PBS. The resulting pellets were stored at −80°C for later processing.
To generate whole-cell lysates for purification, the cross-linked cell pellets were resuspended in a minimal amount of radioimmunoprecipitation assay (RIPA) buffer, transferred to a single-use aerosol-tight vitrified tissue grinder system, and ground on ice. The lysates were then further diluted with RIPA buffer, collected into microcentrifuge tubes, and passed through a 30-gauge syringe to further homogenize the sample. The homogenized lysates were then clarified by centrifugation at 18,000 × g for 5 min, after which the supernatant was removed to a fresh microcentrifuge tube.
Each independent sample was then bound to streptavidin resin that had been prebound with HPDP-biotin, which consists of a biotin moiety and a reactive group capable of covalently linking to sulfhydryl residues, and incubated for a period of 1 h at 16°C under constant agitation. After the binding period, the vRNP complexes bound to the resin were harvested by centrifugation, and the supernatant was discarded. To attain a high degree of purity, the bound resin was washed several times with RIPA buffer supplemented with urea. After washing, the resin was buffer exchanged into PBS supplemented with 1.0% SDS prior to elution of the bound complexes.
The protein components of the purified vRNP complexes were eluted by heating the resin to 70°C for a period of 1 h. As formaldehyde-induced RNA-protein cross-links are sensitive to heat, we were able to selectively release the protein components of the vRNP complexes without inadvertently releasing contaminants that were either bound to the bead nonspecifically or inadvertently biotinylated by the HPDP-biotin. The released materials were collected into a fresh microcentrifuge tube and trichloroacetic acid (TCA) precipitated prior to in-liquid trypsin digestion. The resulting peptides were zip-tip exchanged and analyzed by mass spectrometry.
To verify the specificity of the above-described method, we utilized a quantitative-PCR (qPCR) approach to examine the enrichment of viral and cellular RNAs following CLAMP. Briefly, CLAMP materials were prepared as described above, with the exception that the RNA-protein complexes were released from the streptavidin bead by reversal of the HPDP-biotinylation reaction through the addition of excess dithiothreitol (DTT). The purified, unbound RNA-protein complexes were then un-cross-linked by heating as described above. The CLAMP-purified RNAs were extracted and used as the template for random-hexamer-primed cDNA synthesis by reverse transcription (RT). The relative enrichment of viral and cellular RNAs was then determined by comparative analysis of the CLAMP-purified RNAs relative to their respective inputs. As shown in Fig. 1B, SINV RNAs were enriched ∼70-fold while cellular RNAs were enriched ∼2.5-fold over input levels. Therefore, analysis of the bound RNAs by qRT-PCR indicated that the CLAMP process is highly selective for viral RNAs.
For all of the above-mentioned processes, parallel controls consisting of uninfected 4SU-treated lysates were developed and assessed by mass spectrometry. Control lysates consisting of un-cross-linked extracts, either infected or uninfected, failed to demonstrate the presence of appreciable amounts of protein by absorbance spectroscopy at 280 nm. Here, the CLAMP data sets SINV and mock refer to the 4SU-treated cross-linked lysates from infected and uninfected (mock-infected) 293HEK cells, respectively.
CLAMP identification of SINV-associated host factors.
SINV-associated host factors were identified by combinatorial analysis of paired SINV- and mock-infected samples. While the data shown in Fig. 1B indicate that the majority of CLAMP-identified proteins were likely to be genuinely associated with the viral RNAs, a degree of caution is important to prevent the erroneous assignment of potential interactants. Thus, in order to maintain analytical rigor, the proteins identified by mass spectrometry were triaged by a two-step process similar to that previously described (32). First, master lists of CLAMP-identified proteins were developed for SINV- and mock-infected samples by comparative analysis of the individual CLAMP assays (see Table S1 in the supplemental material). To be considered a genuine CLAMP identification, a particular protein must have been detected in both biological replicates and must have been represented by a minimum of three unique peptides in at least one of the data sets (see Tables S2 and S3 in the supplemental material). Second, to ensure specificity, the SINV and mock CLAMP-identified protein data sets were compared to identify proteins common to both treatments. As shown in Fig. 1C, CLAMP analysis of the SINV vRNP in 293HEK cells relative to mock-infected 293HEK cells revealed 279 host factors associated with the viral RNAs.
To facilitate the biological assessment of the SINV CLAMP data set, ontological analysis of molecular function was performed to group the identified host factors for further prioritization (44, 45). For these analyses, a false-discovery rate (FDR) statistical threshold of <0.05 was utilized. As presented in Fig. 1D, the proteins identified by the SINV CLAMP assay can be ontologically sorted into 8 subgroups based upon molecular function. The subgroup with the highest representation (207 individual proteins, or ∼75% of the SINV CLAMP data set) but the lowest fold enrichment (FE) (∼1.5-fold) was protein binding (Gene Ontology [GO] term, GO:0005515). This finding is, on the whole, unsurprising, as nearly half of the annotated Homo sapiens proteins are associated with this GO term. Regardless, these data imply that a significant number of SINV CLAMP-identified proteins may be the result of protein-protein interactions built upon an underlying scaffold of RNA-binding proteins. The subgroup with the highest FE of ∼39-fold was threonine-type endopeptidase activity (GO:0004298). Interestingly, over half of the human proteins associated with this GO term were observed in the SINV CLAMP data set. Further scrutiny revealed that the threonine-type endopeptidase activity-associated proteins identified by the SINV CLAMP assay constitute the majority of the catalytically active core particles of the 20S proteasome (with only proteasome subunit beta 2 missing from the SINV CLAMP data set). The precise importance of this interaction is currently unknown; however, the proteasome has been previously implicated in the alphaviral life cycle (46, 47). The GO term with the second greatest representation, and an FE of ∼4-fold, was poly(A) RNA binding (GO:0044822).
As the link between CLAMP-identified proteins and vRNA is, a priori, the most obvious for proteins ontologically identified as poly(A) RNA binding, we focused our efforts on further assessing the proteins classified in this subgroup. However, as GO terms contain inherent ambiguity [for instance, not all poly(A) RNA-binding proteins identified ontologically are genuine RNA-binding proteins], we elected to further analyze the poly(A) RNA-binding subset of proteins to determine the biological processes in which they were involved. For the ontological analysis of the biological processes of the poly(A) RNA-binding proteins, we utilized an FDR statistical threshold of <0.01. As shown in Fig. 2A, these analyses revealed that the poly(A) RNA-binding proteins were predominantly involved in four independent biological processes: protein folding (GO:0006457), gene expression (GO:0010467), mRNA splicing via the spliceosome (GO:0000398), and translational initiation (GO:0006413). While the majority of these biological processes were expected, the identification and apparent enrichment of host factors involved in the splicing of cellular mRNAs was, in general, unexpected, as alphaviral RNAs are not spliced.
FIG 2.
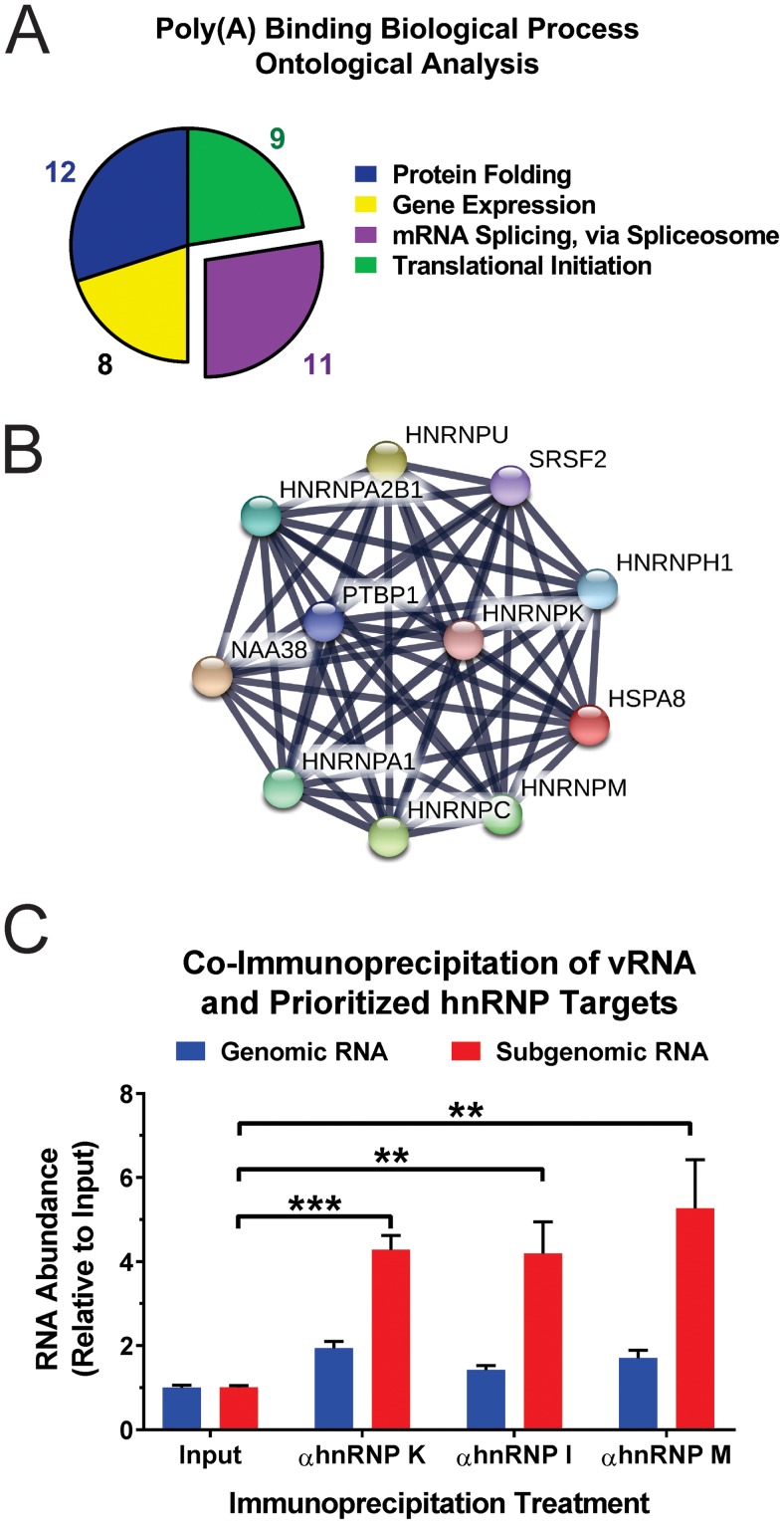
Ontological and interaction analysis of SINV CLAMP poly(A) RNA-binding proteins. (A) The SINV CLAMP proteins identified by DAVID as belonging to the poly(A) RNA-binding molecular function group were assessed to identify enriched biological processes. Shown is a pie graph of the 4 biological process ontological groups statistically enriched as determined by DAVID analyses. (B) Interaction map of the SINV CLAMP-identified interactants grouped into the mRNA splicing via spliceosome biological process ontological group. (C) Immunoprecipitation of cross-linked SINV vRNP complexes derived from 293HEK cells infected with wild-type SINV indicated that the prioritized hnRNP-vRNA interactants are direct interactions. Quantitative detection of the enrichment of the individual vRNA species relative to input levels was accomplished using qRT-PCR. The values shown are the means of at least three biological replicates, with the error bars representing standard deviations of the mean. Statistical significance was determined by Student's t test; **, P < 0.01; ***, P < 0.001.
Examination of the individual SINV CLAMP-identified vRNP components that fell into the mRNA splicing via the spliceosome ontological grouping indicated that a series of hnRNP proteins, namely, hnRNPs A1, A2/B1, C, H, I (PTBP1), K/J, M (NAGR1), and U, were associated with SINV vRNA during infection. STRING analysis of these proteins, as shown in Fig. 2B, revealed a high degree of interaction among these RNA-binding proteins (48, 49). For the purposes of this study, we elected to prioritize the SINV CLAMP interactants hnRNP K, hnRNP I, and hnRNP M for further analysis and characterization. This triaging of potential SINV hnRNP-vRNA interactants was performed primarily for two reasons. First, each of the factors has been previously identified as a potential interactant for an alphavirus, establishing a baseline of molecular impact through a clearly definable body of literature (22, 30, 35). Second, none of the hnRNPs has been evaluated at the level of the hnRNP-vRNA interaction, and as such, the biological roles of these prioritized factors have been, to date, incompletely assessed in light of their molecular function as RNA-binding proteins. Therefore, prioritizing the hRNP targets enabled us to assess the roles played by the hnRNP-vRNA interaction in terms of SINV biology.
To confirm that the hnRNP proteins were indeed acting as RNA-binding proteins in regard to the vRNAs, we assessed the RNA-protein interaction using quantitative immunoprecipitation. To this end, we compared the relative abundances of the SINV genomic and subgenomic RNAs of immunoprecipitated materials to those of their respective inputs. As shown in Fig. 2C, quantitative immunoprecipitation indicated that hnRNP K, hnRNP I, and hnRNP M interact predominantly with the subgenomic RNA during infection, as the quantities of the subgenomic RNA increased on average 4-fold over their relative input values following immunoprecipitation. Collectively, these data confirm the interaction of the hnRNP proteins with the viral RNAs and indicate preferential binding to the subgenomic RNA. Having identified and confirmed the interactions of cellular hnRNP proteins with the SINV vRNAs, we set out to characterize the role of the hnRNP-vRNA interactions in regard to viral infection.
Identification of hnRNP-vRNA interaction sites.
To identify the interaction sites for the prioritized hnRNP proteins with the viral RNAs, we utilized a CLIP-seq approach to develop cDNA libraries for each individual target (20, 42, 50, 51). Briefly, infected 293HEK cells were irradiated with short-wavelength radiation to form cross-linked RNA-protein complexes. The above-mentioned hnRNP proteins were immunoprecipitated using target-specific antibodies, and following RNA fragmentation, cDNA libraries were assembled and sequenced by next-generation sequencing technologies. Target-specific sites of interaction were identified by clustering of retained SINV sequences with a reference genome consisting of the parental SINV sequence (see Table S4 in the supplemental material).
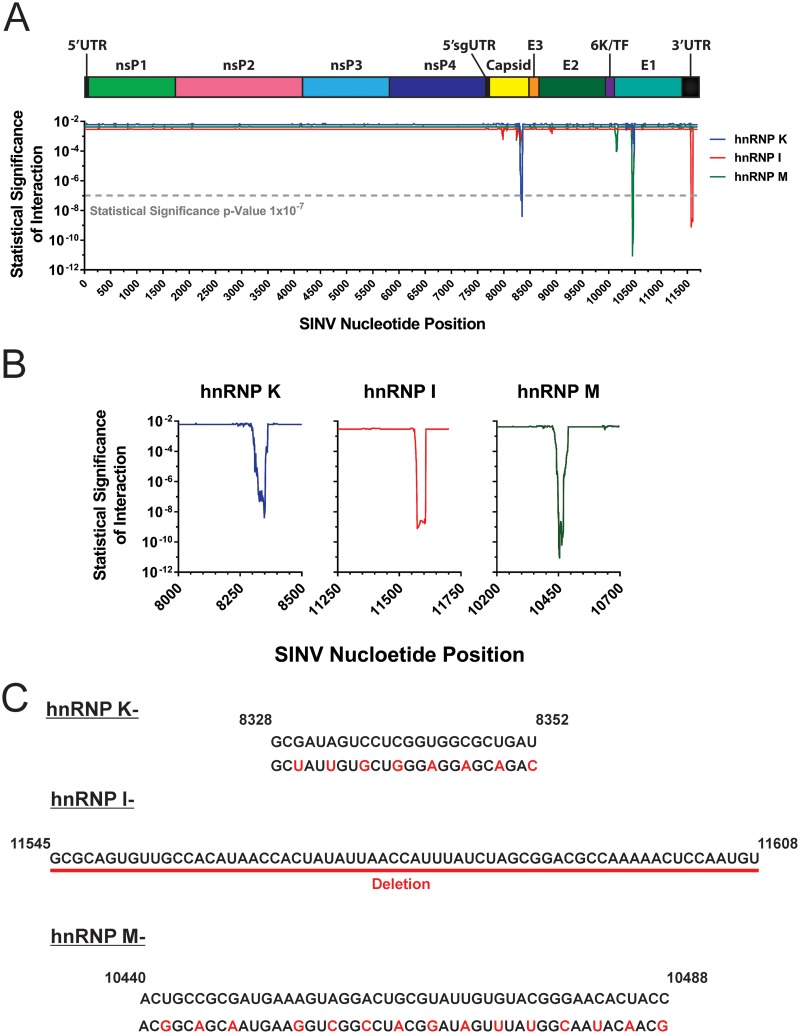
As shown in Fig. 3A and B, clustering of the cDNA libraries revealed that, in a target-specific manner, discrete sites within the viral genome were highly enriched. Statistical analysis indicated that each hnRNP protein had a single clearly identifiable interaction site. Interestingly, comparison of the CLIP-seq-identified interaction sites with previously established interaction motifs for each of the target hnRNP proteins indicated that there is little similarity at the nucleotide level between the consensus cellular and viral binding sites. The potential causes and biological ramifications of this phenomenon are discussed below.
FIG 3.
CLIP-seq identification of select hnRNP-vRNA interaction sites. (A) Map of the SINV genomic RNA displaying the nucleotide positions of the interaction sites of hnRNP K, hnRNP I, and hnRNP M. Above the graph is a scale depiction of the SINV genome with the relative positions of ORFs and UTRs with the 5′ subgenomic UTR indicated by 5′sgUTR. The y axis of the graph is representative of the statistical significance of the fold enrichment for a given library data set on a per-nucleotide basis; the x axis represents the nucleotide position relative to the SINV genomic RNA. The dashed line represents the statistical threshold of 1 × 10−7. (B) Magnified views of the individual hnRNP-vRNA interaction sites. (C) Depiction of the interaction site mutants developed by site-directed mutagenesis. All nucleotide numbers and sequences are derived from SINV Toto1101. Nucleotides colored red indicate the mutations introduced by site-directed mutagenesis.
The identification of the putative interaction sites for hnRNP K, hnRNP I, and hnRNP M on the viral RNAs enabled the mutational ablation of the interaction by site-directed mutagenesis. The interaction sites for hnRNP K and hnRNP M mapped to regions within the structural coding region, creating a restriction on the mutational approaches available for the characterization of hnRNP protein binding. To this end, silent mutations were incorporated across each individual putative interaction site, effectively altering the primary sequence of the nucleic acid while leaving the coding potential, and hence the viral protein, unaffected (Fig. 2C). The mutations were designed to maintain codon utilization to minimize or prevent nonspecific effects of the mutation process. In contrast, the interaction site for hnRNP I was found to be in the 3′ untranslated region (UTR), which is a noncoding region. Since there were no coding constraints in this region, the interaction site was simply deleted.
Mutation of the hnRNP interaction sites reduces hnRNP-vRNA binding.
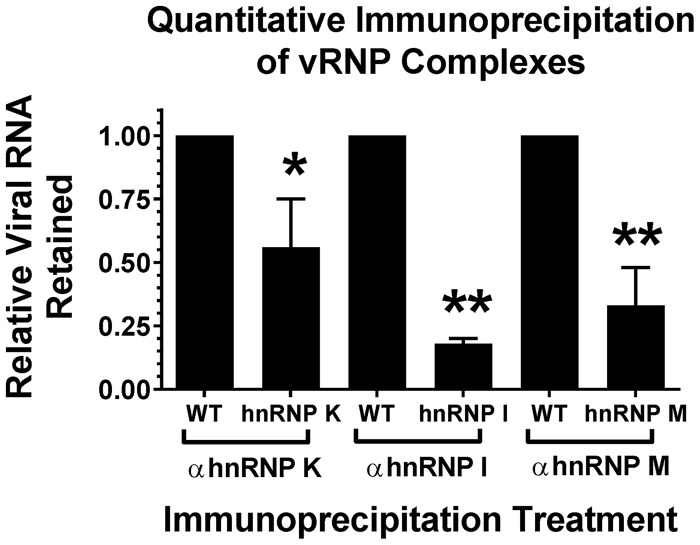
We next sought to characterize the extent to which hnRNP binding was affected by the mutation of the respective CLIP-seq-identified interaction sites. To this end, we infected 293HEK cells with either wild-type parental virus or the hnRNP interaction site mutants. At 16 hpi, the infected cells were cross-linked to form RNA-protein complexes, and the individual hnRNP proteins (and their covalently bound RNAs) were immunoprecipitated as described above. The quantity of coprecipitating vRNA was then detected by qRT-PCR. As shown in Fig. 4, mutation of the hnRNP interaction sites significantly reduced hnRNP-vRNA binding relative to that observed for the wild-type parental virus. Specifically, mutation of the hnRNP K, I, and M interaction sites reduced binding by ∼2-, 5-, and 3-fold, respectively. Importantly, this is a direct quantitative measure of the hnRNP-vRNA interaction, rather than a measurement of protein quantity (as is typically done during small interfering RNA [siRNA]-mediated knockdown).
FIG 4.
Mutation of the SINV hnRNP-vRNA interaction sites negatively affects hnRNP binding. Shown is quantitative immunoprecipitation of cross-linked SINV vRNP complexes derived from 293HEK cells infected with either wild-type (WT) parental virus or the individual hnRNP interaction site mutants, as indicated. On the x axis are the individual hnRNP immunoprecipitation conditions and virus pairs; on the y axis is the quantity of vRNA retained for each interaction site mutant relative to a wild-type parental control. The values shown are the means of at least three biological replicates, with the error bars representing the standard deviations of the mean. Statistical significance was determined by Student's t test; *, P < 0.05; **, P < 0.01.
Collectively, these data affirmed the use of interaction site mutation as a means by which the biological role of the hnRNP-vRNA interaction could be assessed during genuine viral infection of an unaltered host system. Moreover, they indicate that hnRNP-vRNA interaction is a genuine facet of alphavirus biology.
Mutation of hnRNP protein interaction sites negatively affects viral growth kinetics.
Following confirmation of decreased hnRNP-vRNA interaction, we evaluated the one-step growth kinetics of the hnRNP-vRNA interaction site mutant viruses in vertebrate and invertebrate tissue culture systems. Briefly, 293HEK or Aedes albopictus C6/36 cells were infected with either parental wild-type SINV or an hnRNP-vRNA interaction site mutant derivative at an MOI sufficient to ensure total infection of the cell monolayer. As shown in Fig. 5A, mutation of the hnRNP K, hnRNP I, and hnRNP M interaction sites, in general, resulted in decreased viral growth kinetics in vertebrate tissue culture models of infection. Mutation of the hnRNP K interaction site resulted in a statistically significant reduction of viral growth kinetics relative to wild-type parental SINV, with an average reduction in titer of ∼4-fold at 24 hpi. Similarly, mutation of the hnRNP I and hnRNP M interaction sites resulted in statistically significant reductions in viral growth kinetics in mammalian tissue culture cells, with ∼30- and ∼20-fold reductions in titer at 24 hpi, respectively.
FIG 5.
Disruption of the hnRNP-vRNA interactions negatively affects viral growth kinetics in mammalian and mosquito tissue culture systems. (A) One-step growth kinetics of the individual hnRNP-vRNA interaction site mutants and parental wild-type SINV as observed in 293HEK cells infected at an MOI of 10 PFU/cell. (B) Identical to panel A with the exception that the cell line used was the A. albopictus C6/36 cell line. All the data shown are the means of three independent biological samples, with the error bars representing standard deviations of the mean. Statistical significance was determined by area under the curve analysis; *, P < 0.05; **, P < 0.01.
The individual interaction site mutants, with the exception of the hnRNP K interaction site mutant, exhibited similarly decreased viral growth kinetic profiles in invertebrate tissue culture cells (Fig. 5B). Interestingly, the hnRNP K interaction site mutant displayed viral growth kinetics highly similar to those observed for the wild-type parental strain. Nonetheless, the hnRNP I and hnRNP M interaction site mutants exhibited decreased viral growth kinetics relative to the wild-type parental virus. The relative levels of reduction in viral titers at 24 hpi were ∼25-fold for hnRNP I and ∼8-fold for hnRNP M relative to the wild-type parental virus.
Collectively, these data indicate that the hnRNP-vRNA interaction is biologically important for viral infection in vertebrate and invertebrate tissue culture cells. Moreover, the fact that mutation of the hnRNP-vRNA interaction sites resulted in decreased viral growth kinetics in mammalian and mosquito host systems suggests that these regions of the vRNA are biologically important across the two host species. Precisely whether this is due to evolutionarily conserved RNA-protein interactions is, based on these data, unknown. The implications of this observation are a subject of later discussion below.
Mutation of the hnRNP-vRNA interaction sites does not affect vRNA abundance.
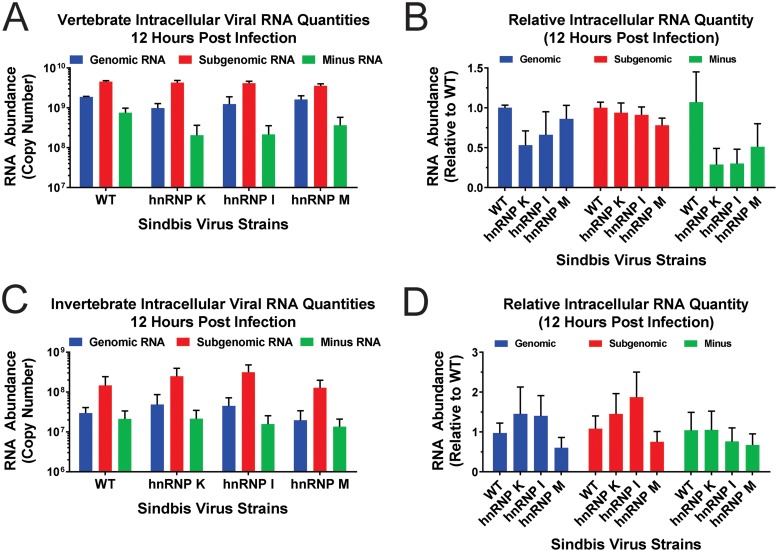
Decreased viral growth kinetics is indicative of defective completion of the viral life cycle. There are many stages of the viral life cycle that could be perturbed, resulting in the decreased viral growth kinetics observed with the hnRNP interaction site mutants. One such molecular process is viral RNA synthesis, including viral replication and transcription, which generate the minus-strand template intermediate and genomic RNA and the alphavirus subgenomic RNA, respectively (18, 19). To determine if mutation of the hnRNP-vRNA interaction sites decreased viral growth kinetics by altering viral RNA levels, we quantitatively examined the production of viral RNAs during vertebrate and invertebrate tissue culture infections. To this end, we assessed the levels of the individual viral RNAs of the hnRNP interaction site mutant viruses relative to those of the parental wild-type virus in 293HEK cells and C6/36 cells at 12 hpi.
As shown in Fig. 6, mutation of the hnRNP interaction sites did not significantly affect viral RNA accumulation during the infection of either vertebrate or invertebrate tissue culture cells. As shown in Fig. 6A and B, 293HEK cells infected with the hnRNP interaction site mutants accumulated the viral RNA species to more or less wild-type levels. Modest, statistically insignificant downward trends were observed between wild-type virus and the hnRNP interaction site mutants for the genomic and minus-strand viral RNAs. It is worthy of note that the downward trend observed with the minus-strand RNA was likely due to variation in wild-type minus-strand accumulation between the individual biological replicates, as the standard deviation between the samples was comparatively large. Nonetheless, increasing the number of samples to attain statistical significance is unwarranted, as the magnitude of the changes in viral RNA abundance are unlikely to be biologically impactful. Similar to what was observed in vertebrate cells, disruption of the prioritized hnRNP interaction site mutants did not alter viral RNA accumulation in mosquito tissue culture cells, as shown in Fig. 6C and D. Taken together, these data indicate that the interaction of the prioritized hnRNP proteins with the viral RNAs is not involved in the regulation of viral RNA synthesis or accumulation of the viral RNA species.
FIG 6.
Disruption of hnRNP-vRNA interactions does not affect the accumulation of SINV vRNAs. (A) 293HEK cells were infected with either wild-type parental virus or the individual hnRNP interaction site mutants at an MOI of 10 PFU/cell. At 12 h postinfection, the total cellular RNA was extracted and assessed for the absolute quantities of the genomic, subgenomic, and minus-strand vRNAs by qRT-PCR. (B) Comparative analyses of the individual viral RNA species relative to wild-type virus. (C and D) Identical to panels A and B, except that the A. albopictus C6/36 cell line was utilized. All the data shown are means of three independent biological samples, with the error bars representing standard deviations of the mean.
Disruption of hnRNP-vRNA interactions increases viral structural-gene expression.
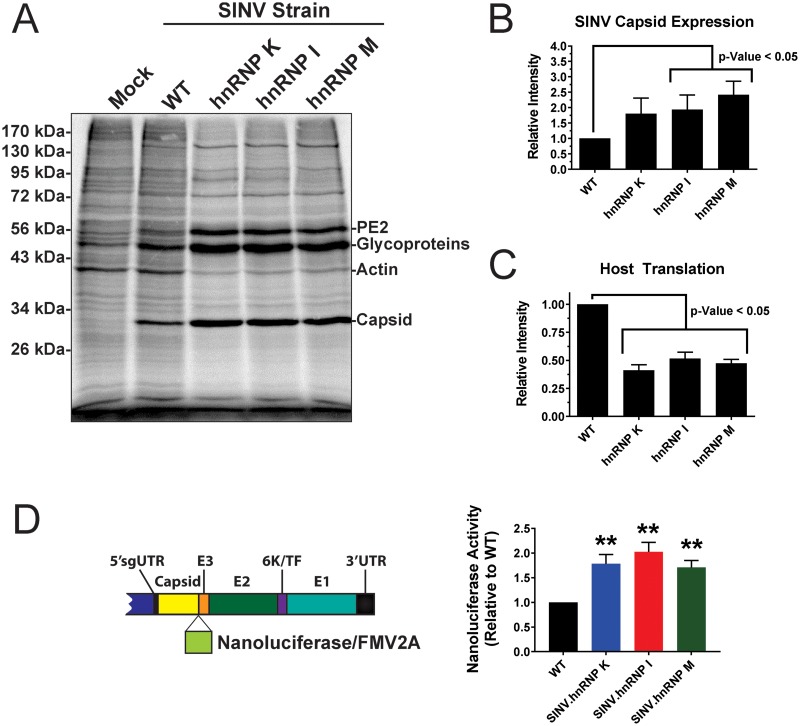
The cellular hnRNP proteins play a functional role in the regulation of the translational capacity of their bound cognate mRNAs (35, 52–54). As such, we hypothesized that the hnRNP-vRNA interactions might influence the rate of translation of their bound vRNAs. Since, as demonstrated by Fig. 2C, the target hnRNP proteins interact with the subgenomic RNA, we focused our efforts on primarily characterizing structural-gene expression. To this end, we assessed viral gene expression using a combinatorial set of approaches, including metabolic labeling, and a series of highly sensitive reporter viral strains that encode a nanoluciferase/FMV2A reporter between the SINV viral capsid and E3 proteins.
To observe viral and cellular protein synthesis during SINV infection, 293HEK cells were infected with either wild-type parental SINV or an hnRNP-vRNA interaction site mutant at an MOI of 10 PFU/cell. At 12 h postinfection, the cell culture medium was supplemented with [35S]methionine and [35S]cysteine to radiolabel the products of ongoing protein synthesis. After a 2-h labeling period, the cell monolayers were washed, and whole-cell lysates were generated by the addition of RIPA buffer. Equivalent amounts of lysate were resolved by SDS-PAGE. The radiolabeled protein species were detected by phosphorimaging, and the individual protein species of interest (host and viral) were assessed by densitometry. As shown in Fig. 7A, each of the individual hnRNP-vRNA interaction site mutants exhibited increased structural-gene expression relative to parental wild-type SINV, as evidenced by enhanced capsid and viral glycoprotein expression. Densitometric analysis of the SINV capsid protein expression indicated that the hnRNP interaction site mutants exhibited, on average, 2-fold greater translation than wild-type SINV (Fig. 7B). Interestingly, as evidenced by the intensity of the host actin band, the shutoff of host translation, a hallmark of alphaviral infection where host transcription and translation are directly inhibited by viral effectors, was enhanced in each of the hnRNP-vRNA interaction site mutants (Fig. 7C) (55, 56).
FIG 7.
Viral gene expression is altered following disruption of hnRNP-vRNA binding. (A) 293HEK cells were either mock treated or infected with wild-type parental virus or an individual hnRNP-vRNA interaction site mutant at an MOI of 10 PFU/cell, as indicated. At 10 h postinfection, the cells were pulsed for a period of 2 h with [35S]methionine and [35S]cysteine. After this period, the cells were harvested, and equal volumes of cell lysate were analyzed by SDS-PAGE and phosphorimaging. The data shown are representative of several independent biological replicates. (B) Densitometric quantification of the SINV capsid protein, with intensity relative to wild-type parental virus shown on the y axis. (C) Densitometric quantification of the host actin protein, with intensity relative to wild-type parental virus shown on the y axis. (D) Schematic diagram of the SINV nanoluciferase reporter used in this study. 293HEK cells were infected with either parental wild-type SINV or individual hnRNP-vRNA interaction site mutants/nanoluciferase reporters, and at 16 hpi, the level of nanoluciferase activity was quantified. All the quantitative data shown represent the means of three independent biological replicates, with the error bars representing standard deviations of the mean. Statistical significance was determined by Student's t test; **, P < 0.01.
Complementary assessment of SINV structural-gene expression using a nanoluciferase reporter construct confirmed what was observed by the metabolic labeling of infected cells. This approach utilizes a SINV strain that contains a nanoluciferase reporter between the viral capsid and E3 proteins, which is cleaved from the structural polyprotein by the autoproteolytic cleavage activities of capsid and a fused FMV2A protease at the C-terminal end of nanoluciferase (diagrammed in Fig. 7D, left). As shown in Fig. 7D, mutation of the hnRNP interaction sites differentially altered subgenomic viral genes late during infection. As observed by metabolic labeling, viral gene expression of the subgenomic RNA, as measured by a nanoluciferase reporter in frame with the structural open reading frame (ORF), was increased on average 1.5-fold relative to wild-type infection at 16 hpi.
Taken together, these data are indicative of a role for the hnRNP proteins in the regulation of viral structural-gene expression. Moreover, the disruption of the hnRNP-vRNA interactions differentially affects subgenomic gene expression. Additionally, disruption of the hnRNP-vRNA interaction sites increases the host translational shutoff observed during SINV infection in vertebrate cells.
DISCUSSION
The CLAMP assay: a new tool with versatile potential.
In this study, we developed and employed a robust vRNA-protein interaction discovery system. This method is highly adaptable and capable of expansion to any system that is insensitive to chemical inhibitors of transcription. Combining streptavidin affinity purification with highly sensitive peptide fingerprinting to identify vRNP components, the CLAMP method enables the discovery of viral RNA-protein interactions en masse for later validation and biological assessment.
The CLAMP discovery method operates largely on four principles: (i) that cellular RNA synthesis can be effectively inhibited by the addition of small molecules; (ii) that nucleoside analogues, such as 4SU, can be readily incorporated into RNA cotranscriptionally; (iii) that protein-RNA cross-linking (in the form of either short-wavelength UV irradiation or formaldehyde) can effectively secure RNPs in their biological context; and (iv) that the exceptional affinity of the streptavidin-biotin interaction enables purification of crude extracts under aggressive purification conditions. Together, these underlying principles form the crux of the CLAMP method, by which vRNP components can be identified in a highly selective and specific manner. Described below, and diagrammed in Fig. 1A, is the general purification scheme of the CLAMP process. Further details can be found in Materials and Methods.
Initially, actively growing tissue culture cell monolayers are infected with virus particles at an MOI appropriate for the goals of the study. At the desired time postinfection, the medium is discarded and replaced with whole medium supplemented with actinomycin D at a concentration capable of completely inhibiting cellular transcription. Shortly after the chemical inhibition of cellular transcription (∼15 min), the nucleoside analogue 4SU is added to the medium, and the tissue culture cells are incubated for a period to allow 4SU incorporation into the nascent viral transcripts. After the labeling period, the tissue culture supernatant is discarded and the monolayers are washed twice to remove contaminating medium. The cells are then cross-linked to preserve the bona fide vRNP complexes formed during infection.
The cross-linking method may vary depending on whether a “wide” or “narrow” elucidation of vRNA interactions is desired. For the wide discovery approach applied in this study, the vRNP complexes are cross-linked with formaldehyde, which results in reversible RNA-protein and protein-protein cross-linked complexes. In contrast, a narrow discovery approach would utilize short-wavelength UV irradiation to form RNA-protein cross-linked complexes. The use of short-wavelength UV irradiation serves two purposes. First, it forms RNA-protein complexes without incidentally introducing nucleotide bias. Second, it preserves the thiol group of the 4SU nucleoside analogue for chemical modification during the purification process. However, the use of a UV cross-linking approach comes at a cost, with a notable loss of efficiency. For the purposes of this study, a formaldehyde cross-linking approach was utilized to increase the number of interactants observed; importantly, preliminary studies using UV-mediated cross-linking yielded highly similar results (data not shown).
After the formation of cross-linked complexes, the tissue culture monolayers are rapidly harvested, and whole-cell lysates are generated by the addition of detergent and physical disruption by either grinding or sonication. The lysates are then further diluted and clarified by centrifugation. The vRNP complexes in the clarified supernatant are then captured onto streptavidin resin that has been preloaded with HPDP-biotin. Incubation of the 4SU-labeled vRNAs in the presence of HPDP-biotin results in the formation of a reversible covalent bond between the biotin moiety and the sulfur group of the uridine analogue. The resin is then washed several times under low- and high-stringency wash conditions to effectively isolate and purify the cross-linked vRNPs, which are bound by the streptavidin-biotin interaction. After sufficient washing, the vRNP complexes are released and prepared for protein identification. Release of the vRNP complexes can be accomplished by RNase treatment, which disassembles the RNA component of the cross-linked vRNP complex, thereby releasing the protein component into the solution. The released materials are transferred to a new tube and precipitated prior to protease digestion and mass spectrometric analysis/identification.
This approach is similar to the previously reported thiouracil cross-linking mass spectrometry (TUX-MS) approach; however, there are several key differences between the two systems (32). A primary difference is the utilization and functional purpose of the 4SU nucleoside analogue. For the TUX-MS process, the 4SU analogue is used as a photoactive cross-linking reagent. This approach, while widely used and highly efficient, introduces unintentional bias, as proteins that bind to regions of the target RNA with high uridine content will be selected/enriched. In contrast, the CLAMP approach uses the thiol-containing uridine analogue as a component of the purification process rather than a cross-linking aid, which effectively precludes the unintentional introduction of bias. Another key difference is the manner in which the RNP complexes are purified. The TUX-MS approach utilizes poly(A) selection to harvest the cross-linked RNP complexes prior to identification. This results in the purification of target and nontarget polyadenylated RNAs. While the purity of the RNP complexes is ensured by the specificity of 4SU incorporation and cross-linking, the amount of nontarget poly(A) RNAs is significant. Moreover, the technique is effectively limited to poly(A)-positive RNAs. Another drawback to the utilization of poly(A) selection is that it is highly sensitive to RNase degradation, as loss of the poly(A) tail abrogates selection. Once again, the CLAMP method relies upon selection of the target RNA by biotinylation of the cotranscriptionally incorporated 4SU. This difference effectively significantly reduces the amount of input material required to identify vRNP components (on average, ∼10- to 20-fold). In addition, the utilization of the highly resistant streptavidin-biotin interaction as a purification scheme allows rigorous washing conditions with varying detergent and ionic conditions, whereas the utilization of poly(A) selection is sensitive to the salt concentration. Collectively, these differences enable the CLAMP method to complement the TUX-MS approach and fundamentally expand the capacity of researchers to elucidate host-pathogen RNA-protein interactions.
An increasingly complete view of SINV vRNA-protein interactions.
As described above, there have been only a small number of studies that have characterized the interaction of host RNA-binding proteins with the alphaviral vRNAs during bona fide infection (22, 35–38). Indeed, the majority of similarly focused research has emphasized the elucidation of replicase complex components or host-pathogen protein-protein interactions. Seminal among these efforts are those reported by Cristea et al. (23, 24), Frolova et al. (25), Foy et al. (31), and Varjak et al. (22). In these studies, the composition of the viral replicase complex, using affinity purification of replication components, was assessed to identify host-pathogen interactions. Previous characterizations of alphavirus replication complex organelles indicated a potential role for host hnRNP interactions in viral infection, RNA synthesis, and host gene expression (22). However, the data presented here represent the first instance of the characterization of direct RNA-protein interactions for many of these host factors. We should note, however, that the interactions observed and reported here are not exhaustive; as proteins with either transient interactions or temporally regulated interactions may have been missed. Moreover, since a direct comparative analysis was used to discriminate and assign proteins identified as belonging to either the SINV or mock data set, it is likely that genuine interactants may have been triaged in favor of increased rigor.
Since the CLAMP approach as applied in this study relies on formaldehyde cross-linking, the observed host factors represent a complex representative mixture of proximal and distal host-pathogen interactions centered on the vRNA. The utilization of formaldehyde as the cross-linking agent undoubtedly results in the generation of both RNA-protein and protein-protein cross-linked complexes. However, purifying the cross-linked materials by the specific capture of vRNA results in a selective bias of host factors “closer” in interaction distance to the vRNA. While highly tempting, it is inappropriate, from the current data set, to assign representative distances to the individual host proteins detected by the CLAMP assay. Moreover, quantitative analysis of the data would likely be problematic, as this type of data set is often troubled by the varying absence or presence of specific peptide fragments due to low target concentrations.
Identifying vRNA-protein interactions by CLIP-seq.
The identification of the prioritized hnRNP interaction sites by CLIP-seq revealed that at least two hnRNP proteins, namely, hnRNP K and hnRNP M, interact with sequences assigned to a viral coding open reading frame. In contrast, hnRNP I was observed to interact with a region of the SINV 3′ UTR. Comparison of the candidate hnRNP proteins evaluated during this study and the consensus interaction motifs indicated limited conservation, if any, between cellular targets and the vRNAs. Regardless, as shown in Fig. 4, mutation of the hnRNP interaction sites resulted in a significant decrease in hnRNP-vRNA binding as observed by quantitative immunoprecipitation. It should also be noted that often the consensus binding motifs for the hnRNP proteins (and, truly, many RNA-binding proteins) are ambiguous and consist of homopolymers or dinucleotide repeats (52, 53, 57). For instance, the typical interaction motifs for hnRNP K are poly(C) rich, and examination of the KH domains using systematic evolution of ligands by exponential enrichment (SELEX) indicated a strong preference for U/C dinucleotide motifs (57, 58). The identified SINV hnRNP K interaction site reported in this study is not particularly C rich but does contains a limited U/C-rich pentamer. Similarly, the consensus binding sequence for hnRNP M is poly(G/U). Assessment of the identified SINV hnRNP M interaction site did not yield any readily identifiable poly(G/U) tracts and exhibited no significant specific enrichment of either G or U residues (59, 60). Likewise, the cellular hnRNP I protein binds to UCUU(C) motifs, which were not detected in the identified SINV hnRNP I interaction site (57, 61). In any case, despite the lack of readily apparent consensus sequences, we were able, from the data presented in Fig. 4, to conclude that mutating these regions negatively affected target hnRNP binding. It should be noted, however, that based on these data we cannot effectively rule out the possibility that the binding of other host factors may be negatively affected by these mutations. This is especially true for the hnRNP I mutant, where the mutational approach was gross deletion of the interaction site.
Furthermore, the lack of apparent conservation between the vRNA interaction site and host consensus sequences may be a consequence of alphaviral infection. Indeed, the localizations of many nuclear proteins, including the prioritized hnRNP proteins, are altered during alphaviral infection. This relocalization event, which is largely due to the shutoff of host translation, effectively changes the functional environment of the hnRNP proteins (22, 62). In addition, altered subcellular hnRNP localization can be a consequence of posttranslational modifications, including phosphorylation. At least one of the prioritized hnRNP proteins, specifically hnRNP K, has been previously reported to be posttranslationally modified during viral infection (35). The impact of this modification has yet to be determined; however, phosphorylation has been demonstrated to affect the RNA-binding properties of RNA-binding proteins.
In addition, the lack of consensus interaction motifs between cellular and SINV hnRNP interaction sites may be influenced by the dual-host system and cyclical transmission patterns common among arboviruses. The hnRNP proteins are generally well conserved across vertebrates and invertebrates, and in vitro binding assays strongly imply that significant conservation of structure results in the conservation of binding site preferences (57). For the hnRNP proteins, homologs with structural similarity of >70% often demonstrate nearly identical RNA-binding predilections; however, homologs with structural similarity of ∼50% start to exhibit differences in RNA-binding motifs, usually in flanking positions outside the core motifs. Sequence alignment comparisons of the human hnRNP proteins with their Aedes homologs revealed relatively low similarity (hnRNP K, P61978 versus AAEL014959-PA = 43.0%; hnRNP I, P26599 versus AAEL013723-RA = 57.4%; hnRNP M, P52272 versus AAEL003670-RA = 32.2%). Thus, it remains possible that the primary binding motifs for the individual prioritized hnRNP proteins have diverged between the two hosts, and significant evolutionary pressure on the pathogen could result in the formation of a single ensemble interaction site that is reminiscent of, but distinct from, the requirements for each host.
The observation that mutation of the hnRNP interaction sites results in similar phenotypes in vertebrate and invertebrate tissue culture models is highly suggestive that the hnRNP interaction sites are functionally important in both hosts. Nonetheless, based on the current data, it is impossible to determine precisely why this is so. While a possible mechanism is the existence of a common host factor interaction site between the host systems, it is also entirely possible that two (or more) unrelated host factors interact with the vRNA at proximal or overlapping sites in vertebrates and invertebrates. Additionally, while significant effort to avoid the introduction of deleterious mutations into the vRNA were made, it is impossible to rule out the possibility that the mutation scheme negatively affected an unforeseen aspect of the vRNA, including the formation of distal secondary structures. From the data presented in Fig. 6, we can assert that altered viral RNA function is not a significant consequence of interaction site mutation in either human or mosquito cells. Therefore, we can posit that mutations, in and of themselves, are likely not simply deleterious to viral RNA levels. Regardless, from a molecular and evolutionary biology standpoint, further assessment and refinement of this phenomenon is warranted.
Interestingly, the region identified as the SINV hnRNP M interaction site was previously identified as the interaction site for a SINV vRNA-binding protein. Recently, we have reported that the hnRNP M interaction site is also occupied by a novel cytoplasmic capsid-vRNA interaction site (20). Currently, it is unknown whether binding of these proteins is mutually exclusive; however, the data presented here, in conjunction with those previously reported, suggest that the SINV capsid and host hnRNP M protein interact with separate vRNAs, as capsid binds to the genomic RNA whereas hnRNP M engages primarily the subgenomic RNA. This finding is exciting, as it implies that the vRNA-protein interaction sites exist as molecular nexuses between the host and pathogen RNA-binding proteins. Precisely how a colinear sequence is capable of selectively associating with two RNA-binding proteins is unknown. However, there are contextual differences between the two RNA substrates, as in the genomic vRNA context the interaction sites are present in what essentially amounts to an extended untranslated region. In the subgenomic context, the interaction sites exist within a coding region and, as such, are regularly traversed by the translating ribosomes. Importantly, RNA-binding proteins have been previously demonstrated to be capable of binding to sequences within ORFs, whereby they regulate the translational efficiency of their cognate mRNAs (63–65).
A key advantage of the above-described approach is that by ablating the vRNA-protein interaction site the biological impact of the host-pathogen interaction may be assessed in an unperturbed host system. This enables the characterization of these interactions in wild-type murine models of viral infection, which has been elusive due to the essential nature of many RNA-binding proteins (66–70). It will be particularly interesting to assess the impact, i.e., what consequences disruption of the hnRNP-vRNA interactions have for viral pathology. Nonetheless, a caveat to this approach is the difficulty of making direct assignments of biological effects to the specific disruption of the target interaction. For instance, it is entirely possible that the disruption of the interaction site inadvertently affects an unknown aspect of viral infection or fundamentally alters the interaction of the pathogen with the host machinery in an unforeseen way. These effects may, and often do, cascade their impacts to other events of the viral life cycle, confounding the identification of the genuine cause and effect. As such, to further ascribe any biological differences directly to the disruption of a specific interaction, it is often prudent to pair mutational analyses with genetic approaches, including knockout, or knockdown, strategies. Unfortunately, given the relatively high concentration of the target hnRNP proteins and their ubiquitous involvement in the maturation of cellular RNAs, we were unable to utilize this confirmatory approach during this study. Therefore, based on the current data, it is difficult to disentangle precisely how the viral growth kinetics of the hnRNP-vRNA interaction site mutants are negatively affected. Ongoing research efforts in the laboratory are focused on further dissecting this phenomenon with the specific goal of better understanding the molecular nature of host-pathogen RNA-protein interactions.
Characterizing the biological consequences of select viral RNA and hnRNP protein interactions by binding site disruption.
As mentioned above, a number of studies have previously identified several hnRNP proteins as components of the viral life cycle (22, 35, 37, 38). To assess the importance of these host factors in regard to viral biology, many studies have relied on the use of RNA interference (RNAi)-mediated approaches to reduce the levels of the target protein. While generally successful at identifying a role(s) for each of the proteins, this approach overlooks several key factors. First, many RNA-binding proteins, including the hnRNP proteins, are essential contributors to RNA biology and the regulation of host gene expression. Indeed, knockdown of select hnRNP proteins has indicated essential roles for individual proteins in alternative splicing, transcriptional regulation, RNA stability, translatability, and RNA localization (52, 53, 71–73). As such, altering the host environment by RNAi would have unintended, and confounding, effects on viral infection. Further supporting this notion is the fact that efforts at developing individual hnRNP knockout animal models have often resulted in severe phenotypes with limited survival (66–70). Second, many hnRNP proteins are highly abundant in the host cell. Thus, a marked reduction in hnRNP protein levels may still result in an overabundant pool of proteins capable of maintaining a host-pathogen interaction. Hence, there is a distinct possibility that the magnitude of any observed effects has been effectively muted, or outright obscured, despite apparently successful knockdown. Finally, if the absence of the host factor from the normal system is a significant contributor to viral pathology, RNAi-based approaches may preclude the detection of such an effect. Therefore, the purpose of this study was to identify host-pathogen vRNA-protein interactions and to characterize the biological importance of these interactions by the disruption of the vRNA-protein interaction.
As demonstrated by the data presented, the disruption of the SINV hnRNP-vRNA interaction negatively affected multiple viral processes, and the relative magnitude of the effect correlated strongly with the extent to which the interaction was disrupted. Analysis of the one-step growth kinetics of the SINV hnRNP-vRNA interaction site mutants indicated that disruption of the hnRNP-vRNA interaction negatively affected viral infection in vertebrate and invertebrate tissue culture cells. These data are highly suggestive that the hnRNP-vRNA interaction may be conserved across the two host species. Nonetheless, given the current data, one cannot directly conclude that these effects are due to binding of a homologous, or orthologous, hnRNP protein across the two systems.
The decreased viral growth kinetics observed with hnRNP-vRNA interaction site mutants in vertebrate cells can largely be attributed to two distinct effects. First, the data presented in Fig. 7 indicate that disruption of the hnRNP-vRNA interaction is correlated with enhanced translation of the SINV subgenomic vRNA. These data, in conjunction with the observation that viral RNA accumulation is unaffected by the disruption of the hnRNP-vRNA interaction (Fig. 6), indicate that the inherent translatability of the subgenomic RNAs is repressed upon hnRNP binding. Second, the shutoff of host translation, as evidenced by the synthesis of host actin, is more robust when hnRNP-vRNA interactions are disrupted. It is important to note that the shutoff of host macromolecular synthesis during SINV infection is a two-part process involving the cessation of transcription and translation. Initially, through the activity of nsP2, host transcription is effectively abrogated (46, 55). Later during infection, cellular translation is inhibited by a mechanism that is less understood but that involves protein kinase R (PKR)-dependent and -independent mechanisms (74, 75). The observations described here are supportive of previous reports in which the translatability of the subgenomic RNA was positively correlated with the shutoff of host translation (56). In the prior study by Patel et al., it was shown that the translational activity of the subgenomic RNA was positively correlated with the reduction of host mRNA translation. A potential mechanism by which this could occur includes the possibility of vRNAs outcompeting the cellular mRNAs for the translationally competent host machinery following the mutation of the hnRNP-vRNA interaction sites. Nonetheless, based on these data, it is still unclear whether the increased translatability or the increased expression of a structural ORF product is primarily responsible for host shutoff. The precise consequences of these phenomena and how they may negatively affect viral infection are currently unclear.
One could envision a scenario in which enhanced protein expression and host shutoff might negatively affect particle quality. Since the processing and maturation of viral glycoproteins is highly complex, increasing the rate of expression may negatively impact their processing, leading to decreased infectivity. Likewise, early host shutoff may negatively affect viral infection by reducing or altering the cellular environment at key transitional stages of the intracellular environment during viral infection. Regardless, the metabolic-labeling and nanoluciferase assay data are indicative of a role for the hnRNP proteins in the regulation of viral gene expression by a tempering of structural-gene expression. In invertebrate cells, the precise mechanism through which the disruption of the hnRNP-vRNA interaction sites negatively affects the viral titer is even less clear. Simply put, the reduction in viral titer in invertebrate cells cannot be a function of host cell shutoff, as alphaviral infection does not affect mosquito macromolecular synthesis. Nonetheless, the possibility remains that enhancing the rate of translation may increase the likelihood of improper particle production.
A previous examination of hnRNP-alphavirus interactions by siRNA-mediated reduction of hnRNP proteins indicated that the effects of the hnRNP proteins on viral gene expression vary in an hnRNP-virus-dependent context (22). For instance, for Semliki Forest virus (SFV) replicons, the reduction of hnRNP proteins, with the exception of PCBP1 or hnRNP E, resulted in a net increase in viral gene expression. Nonetheless, parallel examinations using either CHIKV or SINV replicons exhibited differential behavior for individual hnRNP proteins; for instance, knockdown of hnRNP K in CHIKV and SINV modestly reduced viral gene expression. At first glance, these observations appear to be at odds with the data presented here; however, there are several key differences regarding experimental design that must be considered. Foremost, the Varjak et al. study (22) utilized replicon-based reporter systems to assess the role of the hnRNP proteins in regard to viral gene expression. Given the CLIP-seq data presented here, these replicon constructs, for SINV, lack the hnRNP-vRNA interaction site altogether. In addition, the considerations described above regarding the use of RNAi-based approaches to assess the roles of vital host factors in regard to viral infection likely influenced the results observed with previous studies.
Conclusions.
The data presented in this study reinforce the importance of the role that vRNA-protein interactions play in regard to the host-pathogen interface. Moreover, this study introduces a robust method by which the biologically genuine vRNA-protein interactions may be elucidated during bona fide viral infections of tissue culture cells. Further analysis of these data revealed that a number of host factors involved in the regulation of splicing interact with the nonspliced alphavirus genomic RNA during viral infection. Importantly, these data together also demonstrate that the disruption of select hnRNP-vRNA interactions is strongly correlated with impaired viral growth kinetics during infection in both vertebrate and invertebrate hosts. In addition, disruption of the hnRNP-vRNA interaction correlated with an increase in structural-gene expression and host macromolecular shutoff. Collectively, these data affirm the hypothesis that vRNA-protein interactions are essential to the regulation of viral RNA function and that the disruption of host-pathogen vRNA-protein interactions may be a viable strategy for the development of novel antiviral therapeutics or safe and effective vaccine candidates.
MATERIALS AND METHODS
Tissue culture cells.
BHK-21, 293HEK, and C6/36 cells were cultured in minimal essential medium (MEM) (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 1× antibiotic/antimycotic solution (Cellgro), 1× nonessential amino acids (NEAA) (Cellgro), and l-glutamine (Cellgro). All the cell lines were cultured in the presence of 5% CO2 in humidified tissue culture incubators. The temperature of culturing was 37°C for the BHK-21 and 293HEK cells and 28°C for the C6/36 cells.
Virus production.
Wild-type viruses, including SINV strain Toto1101, SINV p389 (a Toto1101 derivative with green fluorescent protein [GFP] in the coding frame of nsP3), and SINV TE12-nanoluciferase (a SINV TE12 derivative encoding a nanoluciferase reporter between the capsid and E3 proteins in lieu of the mCherry reporter) were prepared as previously described (20, 76). Briefly, 10 μg of in vitro-transcribed RNA was electroporated into BHK-21 cells by a single pulse from a Gene Pulser Xcell electroporation system (Bio-Rad). The electroporation conditions were as follows: one pulse of a square-wave discharge with a potential of 125 V held for a period of 12.50 ms. After the development of apparent cytopathic effect (typically 24 to 36 h postelectroporation), the tissue culture supernatants were harvested and clarified by centrifugation at 8,000 × g for 10 min at 4°C. The resulting P(0) stocks were aliquoted into small-volume samples and stored at −80°C for later use.
For all infections conducted during this study, the quantity of P(0) stock virus required to achieve a specific MOI was determined by titrations on the relevant tissue culture cell line, specifically including the titration of virus stocks on 293HEK and C6/36 cells. The endpoint analysis of viral growth kinetics assay samples was determined using BHK-21 cells, as the samples were not utilized as the infectious materials for further experiments.
CLAMP assay.
The CLAMP method utilized in the study was performed as follows. Per sample, a total of 1 × 108 cells were either mock treated or infected with SINV at an MOI of 10 PFU/cell. After a 1-h adsorption period, the inoculum was removed and the tissue culture monolayers were gently washed with 5 ml of 1× PBS prior to the addition of fresh growth medium (as described above). At 2 hpi, the medium was removed and replaced with preconditioned fresh medium supplemented with 10 μg/ml actinomycin D, and the cells were further incubated for a period of 15 min to effectively halt cellular transcription. Cotranscriptional labeling of the vRNAs was achieved by the addition of 15 ml of 4SU-containing medium for a final concentration of 100 μM in a volume of 20 ml. Actinomycin D levels were maintained throughout the labeling period at a concentration of 10 μg/ml. The cotranscriptional labeling period used in these studies was 4 h in length, at the conclusion of which the cells were harvested for cross-linking and lysate generation.
At the end of the labeling period, the labeling tissue culture medium was removed and the cells were washed once with 1× PBS to remove residual 4SU and growth medium prior to the removal of the cells by scraping. The cells were harvested, transferred to a 15-ml conical tube, and pelleted by centrifugation at 300 × g for 5 min. The cell pellets were then cross-linked by resuspension of the cell pellets in 10 ml of 1.0% formaldehyde diluted in 1× PBS. During the cross-linking period, the cells were gently rocked to prevent aggregation. After 7 min of incubation, the cells were repelleted by centrifugation at 1,000 × g for 3 min to yield a cross-linking period 10 min in duration. The supernatant was discarded, and the cell pellets were gently resuspended in 5 ml of 1× PBS supplemented with 0.25 M glycine to quench any residual formaldehyde. The cells were harvested by centrifugation and washed twice with 1× PBS. The cross-linked cell pellets were stored at −80°C until they were processed into lysates.
Whole-cell lysates were generated by the addition of buffer and mechanical disruption of the cross-linked cell pellets. Per sample, the cell pellets were resuspended in 0.5 ml of RIPA buffer (150 mM NaCl, 1.0% [vol/vol] NP-40, 0.5% [vol/vol] deoxycholic acid, 0.1% SDS, 50 mM Tris, pH 7.6), transferred to an aerosol-tight 15-ml vitrified tissue grinder system (Kimble Chase), and ground by hand for 1 min on ice. After mechanical grinding, the pestle was rinsed with 1× RIPA buffer, and the final volume of the lysate was brought to 1.25 ml. The lysate was collected in a fresh microcentrifuge tube and further homogenized by repeated extrusion through a 30-gauge syringe until the lysate was no longer viscous. The homogenized lysate was then clarified by centrifugation at 18,000 × g for 5 min, and the clarified supernatant was transferred to a fresh microcentrifuge tube containing preloaded capture resin.
The capture resin was prepared in a batch as follows. Per sample, 500 μl (packed volume) of washed Pierce Ultralink streptavidin agarose resin was saturated with an excess amount of HPDP-biotin at room temperature for a period of 30 min. The preloaded resin was washed twice with an excess volume of 1× PBS to remove unbound HPDP-biotin. The resin was resuspended in 1× RIPA buffer and aliquoted as required by the experimental setup.
Protein biotinylation was allowed to proceed for 1 h at 16°C under constant agitation. After the completion of the binding period, the resin was harvested by centrifugation and the supernatant was discarded. The resin was washed once with RIPA buffer and then three more times with RIPA buffer supplemented with 1 M urea (5 min per wash) to reduce nonspecific contaminants. After washing, the samples were exchanged into 1× PBS supplemented with 1.0% SDS by a two-wash process prior to reversing the formaldehyde cross-linking. Reversal of the formaldehyde-induced cross-links was achieved by heating the samples to 70°C for a period of 1 h prior to harvesting the supernatant following high-speed centrifugation of the resin. The recovered materials were TCA precipitated. This complex protein pellet was the end product of the CLAMP method.
Validation of CLAMP selectivity.
Validation of the selectivity of the CLAMP method for viral RNAs was performed using a qRT-PCR-based approach. The input materials were generated by the CLAMP approach described above with a single modification. Namely, the release of the CLAMP-purified materials was performed by the addition of excess reducing agent (DTT; 30 mM) to reverse the biotinylation of the 4SU residues. Following release of the cross-linked RNA-protein complexes, the formaldehyde-induced cross-links were reversed by heating at 70°C for 30 min. The RNAs were then extracted from the CLAMP products with TRIzol and used as the template for cDNA synthesis primed with random hexamers.
CLAMP RNA composition was assessed using qRT-PCR detection of total positive viral RNA, the highly abundant cellular actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs, and the cellular 18S rRNA. The specific primer sets used for the cellular mRNA targets were Actin.F (5′-CACCAACTGGGACGACAT-3′), Actin.R (5′-ACAGCCTGGATAGCAACG-3′), GAPDH.F (5′GGATTTGGTCGTATTGGG-3′), and GAPDH.R (5′-GGAAGATGGTGATGGGATT-3′). The sequences of the viral and rRNA primer sets were identical to those previously described (77). Quantitative analysis was performed by the standard ΔΔCT approach relative to input CLAMP materials using the level of actin as the normalization control; however, normalization with GAPDH gave essentially identical results. The data shown are the means of three independent CLAMP purifications, and the error bars represent standard deviations of the mean. The data for the 18S rRNAs were not specifically plotted, as they were de-enriched 20-fold relative to input levels.
Mass spectrometric analyses.
The CLAMP products (described above) were digested in liquid with mass spectrometry grade trypsin to yield a tryptic peptide library. Briefly, the pellets were resuspended in 100 μl of an aqueous solution containing 100 mM ammonium bicarbonate and 8 M urea. The pellets were vigorously vortexed until they were completely in solution prior to the addition of DTT to a final concentration of 10 mM. The samples were reduced by incubation at room temperature for 1 h. After reduction, the proteins were alkylated by the addition of freshly prepared iodoacetamide to a final concentration of 20 mM. Alkylation was performed in the dark (in a closed drawer) at room temperature for a period of 1 h. After alkylation, additional DTT was added to a final concentration of 40 mM to quench the iodoacetamide and prevent alkylation of the trypsin protease. The quenching reaction was allowed to progress for 10 min at room temperature, after which the samples were diluted with 100 mM ammonium bicarbonate solution to a final concentration of 1 M urea. Per sample, 1 μg of mass spectrometry grade trypsin was added, and the samples were digested overnight at 37°C to generate tryptic peptide library solutions for mass spectrometric analysis.
The tryptic libraries were then submitted to the Laboratory for Biological Mass Spectrometry at the Department of Chemistry of Indiana University—Bloomington. The resulting peptides were dried down and injected into an Eksigent high-performance liquid chromatography (HPLC) apparatus coupled to an LTQ Velos mass spectrometer (Thermo Fisher Scientific, Waltham MA) operating in “top eight” data-dependent tandem-MS (MS-MS) selection. The peptides were separated using a 75-μm by 15-cm column packed in-house with C18 resin (Michrom Bioresources, Auburn, CA) at a flow rate of 300 nl/min. A 2-h gradient was run from buffer A (2% acetonitrile, 0.1% formic acid) to 60% buffer B (100% acetonitrile, 0.1% formic acid). MS-MS peak lists were searched against the Swiss-Prot Homo sapiens database (20,274 entries; downloaded 16 December 2013) using Protein Prospector (v5.10.14). Carbamidomethylation of cysteine residues was set as a fixed modification. Acetylation of the protein amino terminus, oxidation of methionine, and pyroglutamine formation of peptide amino-terminal glutamine were set as variable modifications. A mass tolerance of 0.6 Da was used for both precursor and fragment ions. Peptide expectation values were set to <0.05. For proteins identified by a single peptide, this value was set to <0.01.
Analysis of CLAMP targets.
The peptide fragments detected by mass spectrometry were used to determine the composition of the CLAMP-purified materials and to identify the components of the vRNP through a series of comparative analyses. In order to be considered a genuine CLAMP-identified vRNA interactant, an individual protein must have been detected in both independent biological replicates and must have had a minimum of three unique peptides identified in at least one of the data sets (see Tables S2 and S3 in the supplemental material). The specificity and assignment of the CLAMP-identified factors were then ensured by the comparative analysis of SINV-infected and mock-treated CLAMP data sets. Proteins that were identified as present in the mock data sets were culled from the SINV interaction data set and removed from the SINV data set during further analysis (see Table S1 in the supplemental material).
The protein identities assigned as specific to SINV were further assessed ontologically. Initially, the SINV interactants were assessed based on their molecular functions by the bioinformatics tools available in the DAVID Bioinformatics Resources (version 6.8) (49). Briefly, the SINV interactant list was compared to the Homo sapiens background list to identify molecular function ontological categories that were enriched. Two criteria were used to identify enriched molecular function groups: (i) an individual molecular function group must contain a minimum of 5 individual CLAMP interactants and (ii) the enriched groups must have met the requirement of an FDR of <0.05. From the 8 molecular function ontological groups that were identified as statistically enriched, the poly(A) RNA-binding ontological group was selected for secondary analysis.
The SINV CLAMP interactants assigned to the poly(A) RNA-binding ontological group were assessed using DAVID to identify biological processes that were enriched relative to the Homo sapiens background list. Two criteria were used to identify enriched biological process groups: (i) the identified biological process must be enriched more than 10-fold over expected values and (ii) the enriched groups must have met the requirements of an FDR of <0.01.
STRING analysis of the protein-protein interactions of the SINV vRNA interactants associated with the mRNA splicing biological process group of the poly(A) RNA-binding ontological group was generated using the online version of the STRING database (48, 49). The parameters used to identify potential interactions included gene fusion, cooccurrence, experiments, databases, and text mining, and the confidence level was set to high.
The entire CLAMP data sets and ontological analyses are available as supplemental data files (see Tables S1 to S3 in the supplemental material).
CLIP-seq identification of hnRNP interaction sites.
The development and analysis of CLIP-seq library data sets was performed identically to that previously described for the analysis of SINV vRNA-capsid interactions (20). Per sample, a total of 2 × 107 293HEK cells were infected with SINV Toto1101 at an MOI of 5 PFU/cell. At 18 hpi, the tissue culture medium was removed and replaced with cold 1× PBS, and cell monolayers were irradiated with short-wave UV light at 5,700 × 100 μJ per cm2 in a Stratalinker. After cross-linking, the cells were harvested by scraping and centrifugation prior to being solubilized in RIPA buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and frozen at −80°C. Immediately preceding immunoprecipitation, the lysates were thawed on ice, vortexed until homogeneous, and clarified by centrifugation at 16,000 × g for 5 min in a 4°C refrigerated microcentrifuge.
Per sample, 500 μl of clarified lysate was transferred to a fresh microcentrifuge tube and predepleted with 50 μl (packed volume) of protein G Sepharose beads for 15 min at 4°C. The resin was removed from the clarified lysate by centrifugation at 5,000 × g for 5 min, and the depleted lysate was transferred to a fresh microcentrifuge tube. Per cDNA library, 40 μl of target-specific, or nonspecific, antibodies was added to individual aliquots of predepleted cross-linked extract. The target-specific antibodies used for the CLIP-seq process were selected based on their ubiquitous previous use and demonstrated specificity in similar experiments, including Western blotting (WB), immunofluorescence, IP, RNA immunoprecipitation, and CHIP-seq studies. The specific antibody reagents used were (i) anti-hnRNP K (D-6; SC-28380) (35, 78, 79), (ii) anti-hnRNP I (N-20; SC-16547) (80–82), and (iii) anti-hnRNP M (1D8; SC-20002) (83–85).
The extracts were incubated for a period of 2 h at 4°C under constant agitation. After the antibody binding period, the immunocomplexes were captured from the lysate by the addition of 100 μl (packed volume) of protein G Sepharose resin and an additional 2-h incubation period at 4°C under agitation. After binding, the RNAs present in the lysate slurry were fragmented by the addition of RNase T1 to each immunoprecipitation. Fragmentation was achieved by incubating the slurries for 15 min at room temperature prior to centrifugation at 5,000 × g for 2 min. The supernatant was discarded, and the immunocomplex-bound resin was washed three times with RIPA buffer and twice more with 1× PBS. The RNA components of the purified RNA-protein complexes were released by proteinase K digestion at 37°C in the presence of 1.0% SDS. The released RNA fragments were TRIzol extracted, and the purified RNA fragments were then used as the input material for cDNA library generation with the NEBNext small RNA sequencing kit, according to the manufacturer's directions. The resulting cDNA libraries were sequenced using the Illumina MiSeq platform to yield cDNA libraries by next-generation sequencing.
Prior to analysis, the cDNA reads were trimmed to remove indices and adaptors before alignment. Alignments were performed using the RNA-seq alignment package LAstZ with standard parameters, and the reference genome for alignment consisted of the sequence of the SINV Toto1101 genomic RNA. Only reads mapping to the positive-sense genomic RNA were assessed. Clustering of sequence coverage was used to identify enriched sequences at a nucleotide level of resolution for each individual cDNA library. To allow direct comparative analysis of specific and control cDNA libraries by a subtractive method, the depth of coverage for each individual cDNA library was converted to a percentage of the total aligned reads and multiplied by an arbitrary constant to normalize the intensity of each library. To identify uniquely enriched clusters for the hnRNP K, hnRNP I, and hnRNP M libraries, the control library intensities were subtracted from each individual hnRNP library to generate difference map data for further analysis. From the difference mapping data, the statistical significance was determined for each enriched nucleotide by calculating the z-score for each individual nucleotide position relative to the statistical mean and standard deviation of the cDNA library at large. A significance threshold of 1 × 10−7 was used to identify statistically enriched sequence clusters (which would limit potential type I error to 0.001 nucleotide per length of the SINV genome) and to assign putative hnRNP-vRNA interaction sites to each individual hnRNP protein. The analysis of the sequencing data can be found in Table S4 in the supplemental material.
SINV mutant generation.
After CLIP-seq analysis, the putative hnRNP-vRNA interaction sites were targeted for site-directed mutagenesis according to the instructions for the Q5 site-directed mutagenesis kit (NEB). The previously described parental wild-type strains were PCR amplified with the high-fidelity Q5 DNA polymerase using primer sets that incorporate the mutations depicted in Fig. 3. The PCR amplicons were assessed by agarose electrophoresis and diagnostic restriction digestion to confirm the product size prior to further selection. Individual mutants were identified and then further verified by sequencing prior to proceeding to subsequent analysis. Highly similar, if not identical, phenotypes were observed for a particular hnRNP mutant in all the virus backgrounds evaluated.
Quantitative immunoprecipitation.
To validate the interaction of the prioritized hnRNP proteins with the vRNAs and to identify which vRNAs were involved in the interaction, 293HEK cells were infected with wild-type SINV at an MOI of 5 PFU/cell. At 16 hpi, the cell monolayers were washed with PBS and cross-linked in their original tissue culture dishes by the addition of 1.0% formaldehyde diluted in 1× PBS for a period of 7 min. After this period, the formaldehyde solution was removed and the cells were scraped into 1× PBS supplemented with 0.25 M glycine prior to collection by centrifugation at 1,000 × g for 3 min. The cell pellets were resuspended in RIPA buffer and homogenized by sonication. The resulting lysates were clarified by centrifugation prior to immunoprecipitation.
Immunoprecipitation was performed as previously described (20). A total of 10 μl of either anti-hnRNP K (mouse monoclonal; Santa Cruz Biotechnology; SC-28380), anti-hnRNP I (goat polyclonal; Santa Cruz Biotechnology; SC-16547), anti-hnRNP M (mouse monoclonal; Santa Cruz Biotechnology; SC-20002), or nonspecific mouse IgG was added to 400 μl of lysate (generated from ∼1 × 106 cells, or roughly 1 well of a 6-well dish at ∼90% confluence). The antibody-lysate mixture was incubated at 4°C for 1 h prior to centrifugation to remove precipitated proteins from the lysate. The supernatant was transferred to a fresh microcentrifuge tube containing 50 μl (packed volume) of protein G agarose beads. The antibody-bound complexes were incubated under gentle mixing for a period of 1 h prior to pelleting of the resin at 1,000 × g for 5 min. After removal of the supernatant, the resin was washed three times with an excess volume of RIPA buffer supplemented with 1 M urea and buffer exchanged into RIPA buffer prior to reversal of the cross-linking. The cross-links were reversed by incubating at 70°C for 30 min, and the RNA component of the immunoprecipitated complexes was extracted by standard phenol chloroform extraction and ethanol precipitated in the presence of glycogen.
The recovered RNA was used as the template for cDNA synthesis using Protoscript II reverse transcriptase according to the manufacturer's instructions. The cDNA was then assessed by standard-curve qRT-PCR to determine the absolute quantity of the total positive-sense RNA and the genomic RNA. The quantity of the subgenomic RNA was determined by subtraction of the quantity of genomic RNA from the quantity of total positive-sense RNA, as previously described (20, 36, 86). To determine the relative enrichment of the individual RNA species, the quantities of the viral RNAs from the individual IPs were compared to their relative input levels following normalization to the abundance of the 18S rRNA present in each sample. Thus, the data presented in Fig. 2 represent the fold enrichment over input following immunoprecipitation.
In addition, quantitative immunoprecipitations were used to determine the effect of hnRNP-vRNA interaction site mutation on hnRNP binding. These immunoprecipitations were conducted identically to that described above, with the following modifications. Briefly, 293HEK cells were infected at an MOI of 5 PFU/cell with either wild-type virus or one of the hnRNP interaction site mutants. At 16 hpi, the cells were cross-linked, and following the generation of lysates, the immunoprecipitations were conducted as described above. After obtaining the immunoprecipitated RNA, cDNA was synthesized using random hexamers, and the total viral RNA was quantified using qRT-PCR.
To quantitatively assess the disruption of hnRNP binding, the relative recovery of viral RNA for each individual virus, wild type and mutant, was determined by comparing the immunoprecipitated levels with their paired input materials. The relative recoveries for each hnRNP interaction site mutant were then compared to paired target-specific wild-type immunoprecipitations. Hence, the data presented in Fig. 4 represent the normalized differences in viral RNA recovery between the individual hnRNP interaction site mutants and wild-type infections in a target-specific manner.
Growth kinetics.
Assessment of viral growth kinetics was performed as previously described (20). Briefly, 293HEK or C6/36 tissue culture cells were infected with either the wild-type parental virus or the individual hnRNP interaction site mutant viruses at an MOI of 10 PFU/cell. After a 1-h adsorption period, the inoculum was removed and the monolayers were washed twice with 1× PBS to remove unbound viral particles. Whole medium was added, and the infected tissue culture cells were incubated under their normal incubation conditions. At the indicated times postinfection, the tissue culture supernatants were harvested and then replaced with fresh growth medium. All the samples were frozen at −80°C until the completion of the experiment. The viral titer of each sample was determined using standard plaque assay protocols with BHK-21 cells and a 1% agarose overlay. Plaque assay mixtures were incubated until the plaques were readily visible prior to fixation with 3.7% formaldehyde in 1× PBS. Visualization of the plaques was by the common crystal violet staining method.
vRNA quantification.
To assess the quantities of vRNA produced during infection, 293HEK and C6/36 tissue culture cells were infected with either wild-type parental SINV or the individual hnRNP interaction site mutants at an MOI of 10 PFU/cell. At 12 hpi, the tissue culture supernatant was removed, and the tissue culture cells were washed twice with 1× PBS prior to the addition of TRIzol reagent. Total RNA was extracted according to the manufacturer's directions. Carrier glycogen was added to the precipitation reactions to assist in the visualization of RNA pellets. To synthesize cDNA, 0.5 μg of total RNA was utilized as the template for strand-specific cDNA synthesis, and the quantities of the individual vRNA species were detected by qRT-PCR comparison to known standard curves, as previously described (20, 86). Briefly, the absolute quantities of total positive-sense viral RNA are determined by the detection of sequences specific to the structural ORF, and similarly, the quantity of genomic RNA is determined by the detection of sequences specific to the nonstructural ORF. The abundance of the subgenomic RNA is then calculated by subtraction of the genome-specific signal from the total quantity of positive-sense RNA. Detection of the minus-strand RNA was performed using the same primer set and approach used to detect the genomic RNA. All qRT-PCR analyses were normalized to the level of 18S rRNA present in the sample.
Metabolic labeling.
293HEK cells were either mock treated or infected with wild-type parental SINV or an individual hnRNP-vRNA interaction site mutant at an MOI of 10 PFU/cell. The tissue culture cells were incubated for a period of 9 h under normal conditions. Twenty minutes prior to the radiolabeling period, the medium was removed and replaced with Dulbecco's modified Eagle's medium (DMEM) lacking methionine and cysteine (Corning) and supplemented with dialyzed FBS to deplete intracellular reserves of both amino acids. After the starvation period, the medium was replaced with methionine/cysteine-free DMEM supplemented with [35S]methionine and [35S]cysteine (Express 35S protein-labeling mix; PerkinElmer) at a final concentration of 50 mCi/ml. Radiolabeling of ongoing protein synthesis was performed for 2 h under normal incubation conditions. After the completion of the labeling period, the medium was removed, and the tissue culture monolayers were washed twice with 1× PBS. Whole-cell lysates were generated by the addition of RIPA buffer, and the samples were transferred to a microcentrifuge tube and stored at −80°C for later use.
After thawing, the samples were extensively vortexed and clarified by centrifugation at 18,000 × g for 5 min prior to being transferred to a fresh microcentrifuge tube. Protein-loading buffer was added to a final concentration of 1×, and the samples were prepared for resolution by electrophoresis by boiling. Equal volumes of lysate were loaded onto SDS-10% PAGE gels and resolved as appropriate. The gels were dried and exposed to phosphor screens for subsequent detection by a Typhoon Trio phosphorimager.
Luciferase assay.
293HEK cells were infected with either wild-type parental SINV-nanoluciferase/FMV2A or an individual hnRNP-vRNA interaction site mutant at an MOI of 10 PFU/cell on ice to block viral entry. After a 1-h adsorption period, the inoculum was removed and the medium was replaced with prewarmed whole medium (as described above). The infected cells were incubated under normal conditions, and at the indicated times postinfection, the medium was removed and the tissue culture cells were washed with 1× PBS. The cells were harvested into a crude lysate by the addition of 1× PBS supplemented with 0.1% Triton X-100 and scraping. The lysate was transferred to a microcentrifuge tube and frozen until the completion of the experimental time course. The samples were thawed and clarified by centrifugation at 16,000 × g for 5 min, and the luciferase samples were processed using the Nano-Glo nanoluciferase assay system (Promega) according to the manufacturer's instructions. Nanoluciferase readings were normalized to the protein content following Bradford analysis.
Statistical analyses.
Unless specifically noted, the quantitative data reported in this study are the means of a minimum of three independent biological replicates. The data presented in Fig. 5 were statistically assessed using an area under the curve approach to statistically assess differences in viral growth kinetics throughout the course of the growth assay. Where appropriate, the statistical analysis of comparative samples was performed using variable bootstrapping, as previously described (76). The error bars are indicative of the standard deviations of the mean. Where indicated, the P values associated with individual quantitative data sets are the result of Student's t test for the corresponding quantitative data.
Accession number(s).
All the RNA-sequencing data sets have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession number GSE103693.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Sokoloski laboratory, the Hardy laboratory, the Mukhopahdyay laboratory, and the Danthi laboratory for their input during the development of the project and the preparation/editing of the manuscript.
This work was funded by resources obtained from several sources, including R21 AI121450 (K.J.S. and R.W.H.), R01 AI090077 (R.W.H.), and F32 AI104217 (K.J.S.), all from the National Institute of Allergy and Infectious Diseases. Additional funding support was provided to K.J.S. from the University of Louisville as part of a generous startup package. The funders had no role in study design, implementation, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that no competing interests exist.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02171-17.
REFERENCES
- 1.Johansson MA, Powers AM, Pesik N, Cohen NJ, Staples JE. 2014. Nowcasting the spread of chikungunya virus in the Americas. PLoS One 9:e104915. doi: 10.1371/journal.pone.0104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kendrick K, Stanek D, Blackmore C, Centers for Disease Control and Prevention. 2014. Notes from the field: transmission of chikungunya virus in the continental United States–Florida, 2014. MMWR Morb Mortal Wkly Rep 63:1137. [PMC free article] [PubMed] [Google Scholar]
- 3.Ligon BL. 2006. Reemergence of an unusual disease: the chikungunya epidemic. Semin Pediatr Infect Dis 17:99–104. doi: 10.1053/j.spid.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renault P, Balleydier E, D'Ortenzio E, Baville M, Filleul L. 2012. Epidemiology of Chikungunya infection on Reunion Island, Mayotte, and neighboring countries. Med Mal Infect 42:93–101. doi: 10.1016/j.medmal.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Renault P, Solet JL, Sissoko D, Balleydier E, Larrieu S, Filleul L, Lassalle C, Thiria J, Rachou E, de Valk H, Ilef D, Ledrans M, Quatresous I, Quenel P, Pierre V. 2007. A major epidemic of chikungunya virus infection on Reunion Island, France, 2005–2006. Am J Trop Med Hyg 77:727–731. [PubMed] [Google Scholar]
- 6.Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis 49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 7.Weaver SC. 2014. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 8:e2921. doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calisher CH. 1994. Medically important arboviruses of the United States and Canada. Clin Microbiol Rev 7:89–116. doi: 10.1128/CMR.7.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Monte S, Castro F, Bonilla NJ, Gaskin de Urdaneta A, Hutchins GM. 1985. The systemic pathology of Venezuelan equine encephalitis virus infection in humans. Am J Trop Med Hyg 34:194–202. doi: 10.4269/ajtmh.1985.34.194. [DOI] [PubMed] [Google Scholar]
- 10.Ronca SE, Dineley KT, Paessler S. 2016. Neurological sequelae resulting from encephalitic alphavirus infection. Front Microbiol 7:959. doi: 10.3389/fmicb.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurkela S, Helve T, Vaheri A, Vapalahti O. 2008. Arthritis and arthralgia three years after Sindbis virus infection: clinical follow-up of a cohort of 49 patients. Scand J Infect Dis 40:167–173. doi: 10.1080/00365540701586996. [DOI] [PubMed] [Google Scholar]
- 12.Kurkela S, Manni T, Myllynen J, Vaheri A, Vapalahti O. 2005. Clinical and laboratory manifestations of Sindbis virus infection: prospective study, Finland, 2002–2003. J Infect Dis 191:1820–1829. doi: 10.1086/430007. [DOI] [PubMed] [Google Scholar]
- 13.Adouchief S, Smura T, Sane J, Vapalahti O, Kurkela S. 2016. Sindbis virus as a human pathogen—epidemiology, clinical picture and pathogenesis. Rev Med Virol 26:221–241. doi: 10.1002/rmv.1876. [DOI] [PubMed] [Google Scholar]
- 14.Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. 2009. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis 3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, Morrison TE. 2013. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J Virol 87:13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marimoutou C, Vivier E, Oliver M, Boutin JP, Simon F. 2012. Morbidity and impaired quality of life 30 months after chikungunya infection: comparative cohort of infected and uninfected French military policemen in Reunion Island. Medicine 91:212–219. doi: 10.1097/MD.0b013e318260b604. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 18.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokoloski KJ, Nease LM, May NA, Gebhart NN, Jones CE, Morrison TE, Hardy RW. 2017. Identification of interactions between Sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog 13:e1006473. doi: 10.1371/journal.ppat.1006473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokoloski KJ, Haist KC, Morrison TE, Mukhopadhyay S, Hardy RW. 2015. Noncapped alphavirus genomic RNAs and their role during infection. J Virol 89:6080–6092. doi: 10.1128/JVI.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varjak M, Saul S, Arike L, Lulla A, Peil L, Merits A. 2013. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J Virol 87:10295–10312. doi: 10.1128/JVI.01105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol 84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem 281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 25.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol 80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panas MD, Ahola T, McInerney GM. 2014. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 88:5888–5893. doi: 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J Virol 81:5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourai M, Lucas-Hourani M, Gad HH, Drosten C, Jacob Y, Tafforeau L, Cassonnet P, Jones LM, Judith D, Couderc T, Lecuit M, Andre P, Kummerer BM, Lotteau V, Despres P, Tangy F, Vidalain PO. 2012. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol 86:3121–3134. doi: 10.1128/JVI.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, Frolova EI. 2013. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol 87:1997–2010. doi: 10.1128/JVI.02853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenarcic EM, Landry DM, Greco TM, Cristea IM, Thompson SR. 2013. Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J Virol 87:8697–8712. doi: 10.1128/JVI.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viktorovskaya OV, Greco TM, Cristea IM, Thompson SR. 2016. Identification of RNA binding proteins associated with dengue virus RNA in infected cells reveals temporally distinct host factor requirements. PLoS Negl Trop Dis 10:e0004921. doi: 10.1371/journal.pntd.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnham AJ, Gong L, Hardy RW. 2007. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 367:212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin JY, Shih SR, Pan M, Li C, Lue CF, Stollar V, Li ML. 2009. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol 83:6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gui H, Lu CW, Adams S, Stollar V, Li ML. 2010. hnRNP A1 interacts with the genomic and subgenomic RNA promoters of Sindbis virus and is required for the synthesis of G and SG RNA. J Biomed Sci 17:59. doi: 10.1186/1423-0127-17-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardigon N, Strauss JH. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol 70:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardigon N, Lenches E, Strauss JH. 1993. Multiple binding sites for cellular proteins in the 3′ end of Sindbis alphavirus minus-sense RNA. J Virol 67:5003–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bensaude O. 2011. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 44.Huang Da W, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Akhrymuk I, Kulemzin SV, Frolova EI. 2012. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amaya M, Keck F, Lindquist M, Voss K, Scavone L, Kehn-Hall K, Roberts B, Bailey C, Schmaljohn C, Narayanan A. 2015. The ubiquitin proteasome system plays a role in Venezuelan equine encephalitis virus infection. PLoS One 10:e0124792. doi: 10.1371/journal.pone.0124792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. 2012. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spitzer J, Hafner M, Landthaler M, Ascano M, Farazi T, Wardle G, Nusbaum J, Khorshid M, Burger L, Zavolan M, Tuschl T. 2014. PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol 539:113–161. doi: 10.1016/B978-0-12-420120-0.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhury A, Chander P, Howe PH. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol 3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 55.Gorchakov R, Frolova E, Frolov I. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol 79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel RK, Burnham AJ, Gebhart NN, Sokoloski KJ, Hardy RW. 2013. Role for subgenomic mRNA in host translation inhibition during Sindbis virus infection of mammalian cells. Virology 441:171–181. doi: 10.1016/j.virol.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paziewska A, Wyrwicz LS, Bujnicki JM, Bomsztyk K, Ostrowski J. 2004. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett 577:134–140. doi: 10.1016/j.febslet.2004.08.086. [DOI] [PubMed] [Google Scholar]
- 59.Datar KV, Dreyfuss G, Swanson MS. 1993. The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res 21:439–446. doi: 10.1093/nar/21.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson MS, Dreyfuss G. 1988. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol 8:2237–2241. doi: 10.1128/MCB.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Blanco MA, Jamison SF, Sharp PA. 1989. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev 3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 62.Sanz MA, Garcia-Moreno M, Carrasco L. 2015. Inhibition of host protein synthesis by Sindbis virus: correlation with viral RNA replication and release of nuclear proteins to the cytoplasm. Cell Microbiol 17:520–541. doi: 10.1111/cmi.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barnes M, van Rensburg G, Li WM, Mehmood K, Mackedenski S, Chan CM, King DT, Miller AL, Lee CH. 2015. Molecular insights into the coding region determinant-binding protein-RNA interaction through site-directed mutagenesis in the heterogeneous nuclear ribonucleoprotein-K-homology domains. J Biol Chem 290:625–639. doi: 10.1074/jbc.M114.614735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26:5036–5044. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prokipcak RD, Herrick DJ, Ross J. 1994. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J Biol Chem 269:9261–9269. [PubMed] [Google Scholar]
- 66.Liu TY, Chen YC, Jong YJ, Tsai HJ, Lee CC, Chang YS, Chang JG, Chang YF. 2017. Muscle developmental defects in heterogeneous nuclear ribonucleoprotein A1 knockout mice. Open Biol 7:160303. doi: 10.1098/rsob.160303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye J, Beetz N, O'Keeffe S, Tapia JC, Macpherson L, Chen WV, Bassel-Duby R, Olson EN, Maniatis T. 2015. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci U S A 112:E3020–E3029. doi: 10.1073/pnas.1508461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin Z, Liang F, Yang J, Mei W. 2017. hnRNP I regulates neonatal immune adaptation and prevents colitis and colorectal cancer. PLoS Genet 13:e1006672. doi: 10.1371/journal.pgen.1006672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghanem LR, Kromer A, Silverman IM, Chatterji P, Traxler E, Penzo-Mendez A, Weiss MJ, Stanger BZ, Liebhaber SA. 2015. The poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol Cell Biol 36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaudreau MC, Grapton D, Helness A, Vadnais C, Fraszczak J, Shooshtarizadeh P, Wilhelm B, Robert F, Heyd F, Moroy T. 2016. Heterogeneous nuclear ribonucleoprotein L is required for the survival and functional integrity of murine hematopoietic stem cells. Sci Rep 6:27379. doi: 10.1038/srep27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krecic AM, Swanson MS. 1999. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol 11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 72.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623:123–147. [DOI] [PubMed] [Google Scholar]
- 73.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M Jr, Yeo GW. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sokoloski KJ, Hayes CA, Dunn MP, Balke JL, Hardy RW, Mukhopadhyay S. 2012. Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res 167:26–33. doi: 10.1016/j.virusres.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sokoloski KJ, Snyder AJ, Liu NH, Hayes CA, Mukhopadhyay S, Hardy RW. 2013. Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J Virol 87:12216–12226. doi: 10.1128/JVI.02437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang CK, Wagley Y, Law PY, Wei LN, Loh HH. 2017. Phosphorylation of poly(rC) binding protein 1 (PCBP1) contributes to stabilization of mu opioid receptor (MOR) mRNA by interaction with AU-rich element RNA-binding protein 1 (AUF1) and poly A binding protein (PABP). Gene 598:113–130. doi: 10.1016/j.gene.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee PT, Chao PK, Ou LC, Chuang JY, Lin YC, Chen SC, Chang HF, Law PY, Loh HH, Chao YS, Su TP, Yeh SH. 2014. Morphine drives internal ribosome entry site-mediated hnRNP K translation in neurons through opioid receptor-dependent signaling. Nucleic Acids Res 42:13012–13025. doi: 10.1093/nar/gku1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, Fave GD, Sette C. 2016. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene 35:2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bielli P, Bordi M, Di Biasio V, Sette C. 2014. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res 42:12070–12081. doi: 10.1093/nar/gku922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Motallebipour M, Rada-Iglesias A, Westin G, Wadelius C. 2010. Two polypyrimidine tracts in the nitric oxide synthase 2 gene: similar regulatory sequences with different properties. Mol Biol Rep 37:2021–2030. doi: 10.1007/s11033-009-9653-9. [DOI] [PubMed] [Google Scholar]
- 83.Marko M, Leichter M, Patrinou-Georgoula M, Guialis A. 2010. hnRNP M interacts with PSF and p54(nrb) and co-localizes within defined nuclear structures. Exp Cell Res 316:390–400. doi: 10.1016/j.yexcr.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 84.Marko M, Leichter M, Patrinou-Georgoula M, Guialis A. 2014. Selective interactions of hnRNP M isoforms with the TET proteins TAF15 and TLS/FUS. Mol Biol Rep 41:2687–2695. doi: 10.1007/s11033-014-3128-3. [DOI] [PubMed] [Google Scholar]
- 85.Jagdeo JM, Dufour A, Fung G, Luo H, Kleifeld O, Overall CM, Jan E. 2015. Heterogeneous nuclear ribonucleoprotein M facilitates enterovirus infection. J Virol 89:7064–7078. doi: 10.1128/JVI.02977-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. 2008. The 3′ untranslated region of Sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J Virol 82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.