ABSTRACT
Defective viral genomes (DVGs) of hepatitis C virus (HCV) exist, but their biological significances have not been thoroughly investigated. Here, we analyzed HCV DVGs circulating in patient sera that possess deletions in the structural protein-encoding region. About 30% of 41 HCV clinical isolates possess DVGs that originated from the full-length genome in the same patients. No correlation between DVGs, viremia, and alanine aminotransferase (ALT) levels was found. Sequencing analysis of DVGs revealed the existence of deletion hot spots, with upstream sites in E1 and downstream sites in E2 and NS2. Interestingly, the coding sequences for the core protein and the C-terminal protease domain of NS2 were always intact in DVGs despite the fact that both proteins are dispensable for HCV genome replication. Mechanistic studies showed that transmembrane segment 3 (TMS3) of NS2, located immediately upstream of its protease domain, was required for the cleavage of NS2-NS3 and the replication of DVGs. Moreover, we identified a highly conserved secondary structure (SL750) within the core domain 2-coding region that is critical for HCV genome packaging. In summary, our analysis of serum-derived HCV DVGs revealed novel viral cis elements that play important roles in virus replication and assembly.
IMPORTANCE HCV DVGs have been identified in vivo and in vitro, but their biogenesis and physiological significances remain elusive. In addition, a conventional packaging signal has not yet been identified on the HCV RNA genome, and mechanisms underlying the specificity in the encapsidation of the HCV genome into infectious particles remain to be uncovered. Here, we identified new viral cis elements critical for the HCV life cycle by determining genetic constraints that define the boundary of serum-derived HCV DVGs. We found that transmembrane segment 3 of NS2, located immediately upstream of its protease domain, was required for the cleavage of NS2-NS3 and the replication of DVGs. We identified a highly conserved secondary structure (SL750) within the core-coding region that is critical for HCV genome packaging. In summary, our analysis of serum-derived HCV DVGs revealed previously unexpected novel cis elements critical for HCV replication and morphogenesis.
KEYWORDS: hepatitis C virus, defective genome, cis elements, core, NS2, virus packaging
INTRODUCTION
Hepatitis C virus (HCV), an enveloped virus of the Flaviviridae family, is one of the major causes of chronic liver disease, liver cirrhosis, and hepatocellular carcinoma (HCC) (1). Direct-acting antivirals (DAAs) targeting key viral enzymes/proteins can cure over 90% of hepatitis C patients (2, 3). However, potential drug resistance, high therapy expenses, and a lack of access to DAAs in countries with high HCV prevalence rates impose great challenges to the global eradication of the pathogen, which requires the development of an effective prophylactic vaccine (1). HCV's positive-sense single-stranded RNA genome is approximately 9.6 kb long and encodes a single open reading frame (ORF), flanked by 5′ and 3′ untranslated regions (UTRs). Cap-independent HCV protein translation is mediated by the internal ribosomal entry site (IRES), a critical viral cis element located in the 5′ UTR and the N-terminal core-coding sequences (4–6). The synthesized viral polyprotein precursor is processed by host and viral proteases into structural proteins (core, E1, and E2) and nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (1). The processing of core-NS2 is carried out by the endoplasmic reticulum (ER)-associated host signal peptidase, while the processing of NS3-NS5B is catalyzed by the viral NS3 serine protease with the help of its cofactor NS4A (1). The cleavage of the NS2/NS3 junction is carried out by the NS2-3 autoprotease. The catalytic activity of this autoprotease mainly resides in the C-terminal half of the NS2 cysteine protease domain and the N-terminal 177 amino acids (aa) of NS3 (7).
HCV possesses very high genetic diversity due to its error-prone RNA polymerase and highly active genome replication, resulting in the diversification of virus genomes into 7 major genotypes (GTs) and many more subgenotypes (1, 8). HCV also exists in an infected individual as quasispecies that are derived from common ancestors but possess distinct viral genomes. Viruses with a deletion mutation in their genome have been identified as defective interfering (DI) particles for many virus species. The synthesis of defective viral genomes (DVGs) during HCV replication further increases HCV genetic diversity. However, the biological and physiological significances of HCV DVGs have not been thoroughly investigated. HCV DVG variants characterized by large genomic deletions can be detected in sera or liver biopsy specimens of chronic hepatitis C patients (9–15). HCV DVGs typically contain a deletion within the core-NS2 region but retain the genome portions that are essential for autonomous HCV replication (the 5′ UTR, NS3-NS5B, and the 3′ UTR) (13, 15). Using a cell culture model for HCV infection (HCVcc), it was demonstrated previously that DVG RNAs can replicate in cells and can be packaged into infectious viral particles when coexpressed together with the full-length viral genome (FLG) (14, 16).
We recently discovered a large HCV cohort in Northern China with thousands of chronic hepatitis C patients, likely due to needle sharing during 1970s and 1980s, (17). This cohort of chronic hepatitis C patients provides an invaluable opportunity to study the genetic evolution of DVGs. Here, we analyzed HCV DVGs circulating in patient sera that possess deletions in the structural protein-encoding region. No correlation between DVGs, viremia, and alanine aminotransferase (ALT) levels was found. Remarkably, genetic analysis of these DVGs revealed novel viral cis elements in the core-coding region and the NS2 protein that play important roles in virus assembly and replication, respectively.
RESULTS
Identification of DVGs in clinical HCV isolates.
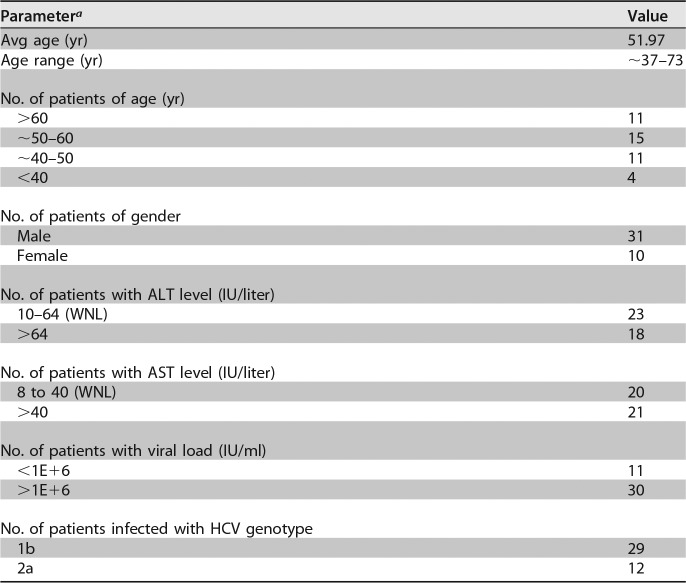
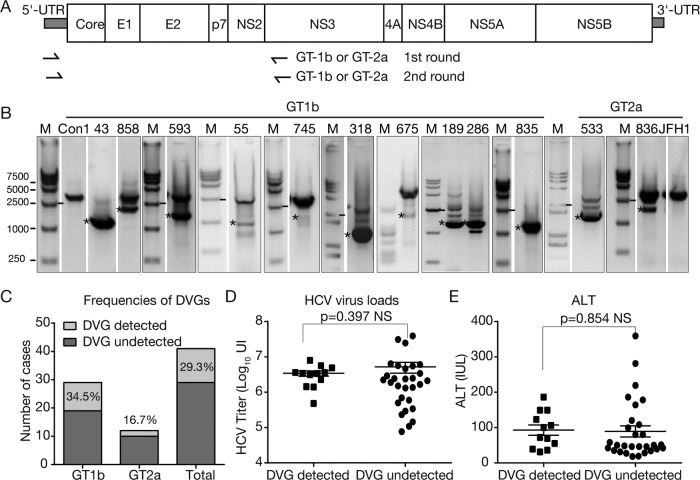
A total of 41 chronic hepatitis C patient sera were screened for the occurrence of an internal deletion within HCV genomic sequences between core and NS2, which are not required for HCV genome replication. All the sera were collected during the treatment-naive stage, and the clinical and virological characteristics of these patients are summarized in Table 1. Twenty-nine of the 41 patients were infected with HCVs of genotype 1b (GT-1b) (70.7%), and 12 were infected with GT-2a (29.3%). Nested PCRs with GT-1b- or GT-2a-specific primer sets spanning the 5′ UTR and the N terminus of NS3 were conducted to amplify the HCV sequences from core to NS2 (Fig. 1A). The FLGs of the con1 strain (GT-1b) and the JFH1 strain (GT-2a) were included as positive controls for GT-1b and GT-2a, respectively. Most sera had one PCR product amplified from the FLG (data not shown); however, PCR of 10 GT-1b (patients 43, 55, 189, 286, 318, 593, 675, 745, 835, and 858) and 2 GT-2a (patients 533 and 836) clinical specimens resulted in products with a size smaller than the one from the FLG (marked with asterisks in Fig. 1B). Sequencing analysis indicated that these smaller PCR products were indeed derived from HCV genomes harboring internal deletions. Of note, other smaller PCR products shown in Fig. 1B were due to nonspecific amplification or failed to be sequenced. In summary, about 29.3% of these 41 HCV clinical isolates possessed DVGs, and the frequencies of DVGs in GT-1b and GT-2a isolates were 34.5% and 16.7%, respectively (Fig. 1C). Notably, no correlation of DVGs with viral loads (Fig. 1D) or with ALT levels (Fig. 1E) in patient sera was found.
TABLE 1.
Cohort of chronic hepatitis C patients used in this study
a AST, aspartate aminotransferase; WNL, within normal limits.
FIG 1.
Identification of HCV DVGs from clinical isolates. (A) Schematic of the HCV genome and positions of primers used to detect DVGs in this study. (B) Agarose gel electrophoresis of PCR products to amplify DVGs. HCV prototype strains con1 (GT-1b) and JFH1 (GT-2a) were included as controls. The PCR products potentially derived from DVGs are labeled with asterisks. The gel graph was modified so that only the lanes with shorter PCR products are shown. The original gel graph is available upon request. M, molecular weight marker. (C) Frequencies of DVGs in 41 clinical isolates. (D) Correlation of DVGs with viral loads. (E) Correlation of DVGs with ALT levels. NS, nonsignificant.
DVGs originate from a full-length genome that exists in the same patient.
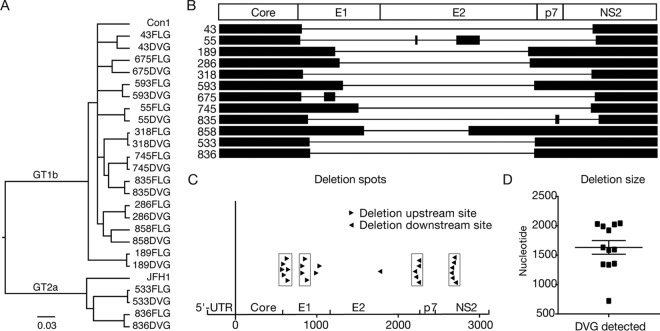
Next, we examined whether DVGs and FLGs can be detected in the same patient sera. As shown in Fig. 1B, the PCR products from the both FLGs and DVGs were detected in all samples except for the sample from patient 835, which had weak FLG amplification by nested PCR, but its FLG PCR product became visible after the third round of PCR amplification. Next, to determine whether the DVGs and FLGs in the same clinical specimen were related phylogenetically, the PCR products from both the DVGs and FLGs were sequenced. As shown in Fig. 2A and Table 2, the PCR products of DVGs and FLGs derived from the same patients shared high sequence identity and were clearly related to each other by phylogenetic analysis, suggesting that these DVGs likely originated from FLGs that exist in the same patient.
FIG 2.
Identification of hot deletion spots within DVGs. (A) Phylogenetic analysis of DVGs and FLGs. HCV prototype strains con1 (GT-1b) and JFH1 (GT-2a) were included for references. (B) Illustration of the deleted region within DVGs. Lines and black boxes represent deleted and remaining sequences, respectively. (C) Location of deletion sites on DVGs. The upstream and downstream deletion sites are labeled with black triangles pointing to the right and left, respectively. The hot deletion spots are grouped with boxes. The x axis indicates nucleotide positions starting from the beginning of core. (D) Lengths of the deleted sequences in DVGs. The average length of the deletions was 1,632 nt.
TABLE 2.
DVGs originating from a full-length genome that exists in the same patient
| Patient | FLG GT | DVG GT | Identity (%) |
|---|---|---|---|
| 43 | 1b | 1b | 96.135 |
| 55 | 1b | 1b | 98.992 |
| 189 | 1b | 1b | 98.864 |
| 286 | 1b | 1b | 98.660 |
| 318 | 1b | 1b | 99.517 |
| 533 | 2a | 2a | 99.465 |
| 593 | 1b | 1b | 97.751 |
| 675 | 1b | 1b | 94.789 |
| 745 | 1b | 1b | 99.378 |
| 835 | 1b | 1b | 98.460 |
| 836 | 2a | 2a | 98.337 |
| 858 | 1b | 1b | 94.225 |
Identification of frequently occurring deletion sites within DVGs.
Next, we analyzed the pattern of internal deletions that occurred in these 12 DVGs. As shown in Fig. 2B, 9 DVGs (from patients 43, 189, 286, 318, 593, 745, 858, 533, and 836) had a single deletion between core and NS2, while 2 DVGs (patients 675 and 835) had two deletions and 1 DVG (from patient 55) had three deletions within this region. The DVG in patient 675 retained a 73-nucleotide (nt) sequence in E1 between its two truncations, while the DVG in patient 835 retained a 19-nt sequence in p7 between its two truncations. The DVG in patient 55 had three deletions separated by two interval sequences of 8 and 152 nt, both in E2. Interestingly, the deletions occurred frequently in two upstream and two downstream sites (Fig. 2C). Six and four upstream deletion sites were found to be located within two small regions in E1 (aa 189 to 212 and aa 280 to 291), respectively, while five and six downstream deletion sites were found in the C terminus of E2 (aa 726 to 744) and the N terminus of NS2 (aa 876 to 893), respectively. In total, 87.5% of DVG deletions occurred in these four hot spots. The average size of deletions was 1,632 nt, and most of the DVGs had a deletion ranging between 1,350 and 2,050 nt long, except for the DVG in patient 858, which had a deletion of 723 nt (Fig. 2D and Table 3).
TABLE 3.
DVGs are in-frame deletions and contain the intact core and the C-terminal protease domain of NS2
| DVG from patient | Upstream deletion position | Downstream deletion position | Deletion length (nt) | Protein status |
||||
|---|---|---|---|---|---|---|---|---|
| Core | E1 | E2 | p7 | NS2 | ||||
| 43 | 582 | 2636 | 2,043 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 55a | 569 | 2661 | 1,923 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 189 | 814 | 2181 | 1,356 | Intact | Deletion | Deletion | Intact | Intact |
| 286 | 848 | 2194 | 1,335 | Intact | Deletion | Deletion | Intact | Intact |
| 318 | 589 | 2631 | 2,031 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 533 | 636 | 2231 | 1,596 | Intact | Deletion | Deletion | Intact | Intact |
| 593 | 872 | 2227 | 1,344 | Intact | Deletion | Deletion | Intact | Intact |
| 675a | 814 | 2652 | 1,989 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 745 | 982 | 2625 | 1,632 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 835a | 625 | 2676 | 2,022 | Intact | Deletion | Deletion | Deletion | Protease domain |
| 836 | 641 | 2227 | 1,587 | Intact | Deletion | Deletion | Intact | Intact |
| 858 | 1023 | 1757 | 723 | Intact | Deletion | Deletion | Intact | Intact |
DVG with multiple intern deletions. The remaining internal sequences within DVGs were at E2 positions 1333 to 1340 and 1677 to 1834 for patient 55, E1 positions 742 to 814 for patient 675, and p7 positions 2376 to 2394 for patient 835.
DVGs contain an in-frame deletion with an intact core and the protease domain of NS2.
Next, we analyzed the impact of the deletions in the DVGs on viral protein translation. Interestingly, all 12 DVGs had an in-frame deletion (Table 3) so that the translation of downstream NS3-NS5B proteins was not disrupted, likely because the downstream nonstructural proteins essential for viral genome replication were also required in cis for the replication of DVGs. All 12 DVGs had deletions in E1- and E2-coding sequences, thus destroying the expression of viral envelope proteins. In contrast, the core-coding sequences were always intact in these DVGs. Half of the 12 DVGs had a deletion of the p7-coding sequences. In addition, half of the DVGs had intact NS2, and the other half had a deletion in the N terminus of NS2 but still retained its C-terminal protease domain (Fig. 2B and Table 3).
Virus production of DVGs can be rescued by the expression of core-NS2 in trans.
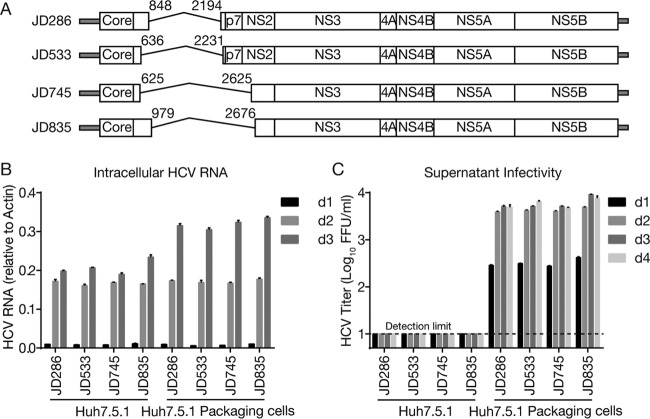
Next, we examined whether these identified DVGs can be packaged into virus particles if the missing structural proteins were provided in trans. Because of the difficulty in constructing infectious cell culture-derived HCV (HCVcc) from clinical isolates, we utilized a previously developed JFH1-based HCVcc system (18–20) to generate four DVGs (denoted JD286, JD533, JD745, and JD835) according to the deletion patterns of DVGs found in patients 286, 533, 745, and 835, respectively (Fig. 3A). Of note, JD286 and JD533 had deletions from E1 to E2, and JD745 and JD835 had deletions from E1 to the N terminus of NS2. To facilitate the detection of virus production, JFH1 containing a deletion of 5 amino acids (EEDDT) at the C terminus of NS5A (positions 2435 to 2439), which we previously showed enhances JFH1 virus production and accelerates virus expansion kinetics (21), was used. The four JFH1 DVG RNAs were prepared by in vitro transcription and then electroporated into naive Huh7.5.1 cells and Huh7.5.1 packaging cells that stably express core-NS2. The intracellular HCV RNA levels at days 1, 2, and 3 posttransfection were determined by reverse transcription-quantitative PCR (RT-qPCR), and the infectivity titers in culture supernatants were determined at days 1, 2, 3, and 4 posttransfection. As shown in Fig. 3B, all JFH1 DVGs replicated well in both cell types. Importantly, infectious viruses were detected in the supernatants of the packaging cells but not naive cells by as early as day 1 and reached peak titers of >103 focus-forming units (FFU)/ml at day 2 posttransfection (Fig. 3C). These results demonstrated that these representative DVG deletion patterns identified in clinical isolates can replicate in cells and can be packaged into infectious virions by ectopically expressed structural proteins.
FIG 3.
Packaging of DVGs by ectopically expressed structural proteins. (A) Schematic of four JFH1-based DVGs, JD286, JD533, JD745, and JD835, that possess representative deletion patterns of the clinical DVGs (patients 286, 533, 745, and 835). The residual p7 portion in DVG from patient 835 was deleted in JD835. The numbers indicate the HCV nucleotide sequence starting from the beginning of the 5′ UTR of the viral genome. (B and C) Intracellular HCV RNA levels (B) and extracellular infectivity titers (C) in naive Huh7.5.1 cells and Huh7.5.1 packaging cells transfected with RNA of JD286, JD533, JD745, and JD835. The HCV RNA levels were determined by RT-qPCR and expressed as values relative to the cellular actin mRNA levels. The dotted line is the detection limit of the titration assay. The error bars represent standard deviations from three independent experiments.
The transmembrane domain upstream of the NS2 cysteine protease domain is essential for DVG replication.
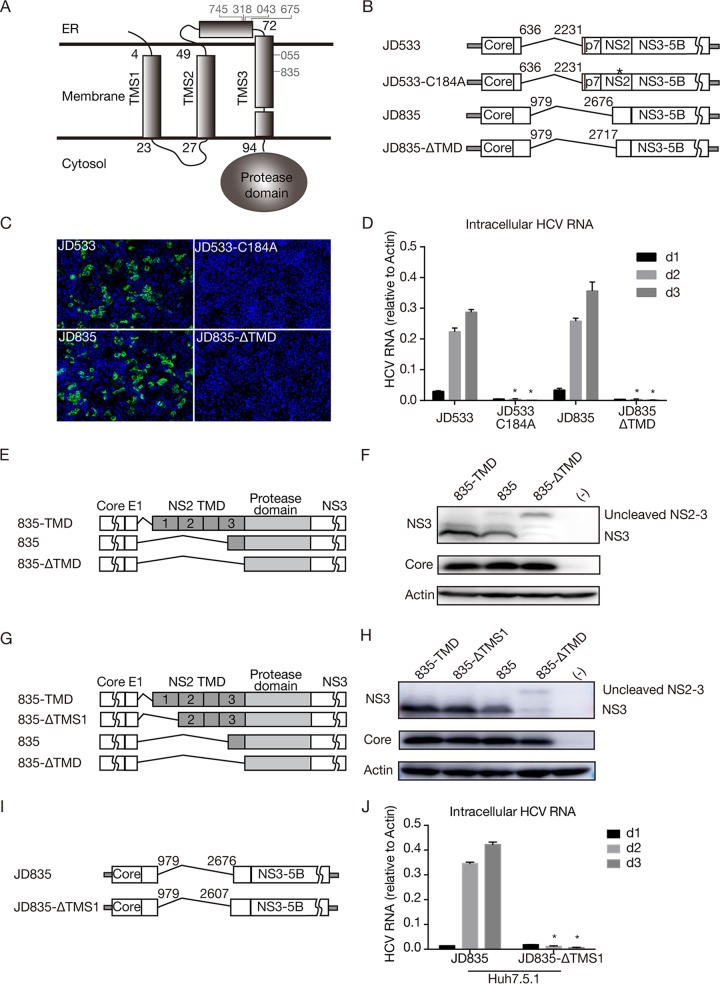
Previous studies have shown that NS2 is not required for HCV genome replication, and a subgenomic replicon without NS2 can replicate well in cells (4). In addition, although NS2 is indispensable for HCV virion assembly, it could still be deleted from the viral genome, and its essential role in virus assembly can be complemented by expressing NS2 in trans (4, 22). NS2 consists of a highly hydrophobic N-terminal membrane binding domain (MBD) and a C-terminal cysteine protease domain essential for cleaving off the N terminus of NS3. As shown in Fig. 2B and Table 3, half of the DVGs had intact NS2, and the other half had a deletion in the N-terminal MBD but still retained its C-terminal protease domain, suggesting that the NS2 protease activity needs to be retained in DVGs to ensure the proper cleavage of NS2-NS3.
The N-terminal MBD of NS2 consists of three transmembrane segments (TMSs) (TMS1 to TMS3), each of which enables the anchorage of downstream NS2 to the membrane (Fig. 4A) (23). Analysis of 6 DVGs with a partial NS2 deletion showed that 4 DVGs (from patients 745, 318, 43, and 675) had the deletion site between TMS2 and TMS3, while 2 DVGs (from patients 55 and 835) had the deletion site in TMS3 (Fig. 4A). Of note, the 835 DVGs had the largest NS2 deletion but still retained 14 aa of TMS3. To test whether the entire TMS3 of NS2 can be deleted without impairing DVG replication, all three TMSs (positions 1 to 94) were deleted in JD835 (JD835-ΔTMD) (Fig. 4B). In addition, C184A, a point mutation at the NS2 protease active site known to disrupt the NS2 autoprotease activity (24), was introduced into JD533 to serve as a control (Fig. 4B). The JD835, JD835-ΔTMD, JD533, and JD533-C184A RNAs were prepared by in vitro transcription and transfected into naive Huh7.5.1 cells. NS5A immunostaining of transfected cells was performed on day 2 posttransfection (Fig. 4C), and the intracellular HCV RNA levels on days 1, 2, and 3 posttransfection were determined by RT-qPCR (Fig. 4D). Neither NS5A-positive cells nor replicating HCV RNAs were detected for JD835-ΔTMD and JD533-C184A, suggesting that intact TMS3 may be required for the function of the downstream NS2-NS3 autoprotease. Consistently, no infectivity titer was detected for Huh7.5.1 packaging cells transfected with JD835-ΔTMD and JD533-C184A (data not shown).
FIG 4.
The transmembrane domain upstream of the NS2 cysteine protease domain is essential for DVG replication. (A) Schematic of three transmembrane segments of NS2. The numbers start with the first amino acid of NS2. Six DVGs that have a deletion site in NS2 are marked. (B) Schematics of the JFH1-based DVG JD533 and its NS2 protease-defective mutant (JD533-C184A) and of the JFH1-based DVG JD835 and its mutant with a deletion of the entire TMD (JD835-ΔTMD). (C) NS5A (green) immunostaining and nuclei (blue) in Huh7.5.1 cells transfected with in vitro-transcribed DVG RNA at day 2 posttransfection. (D) HCV RNA levels in Huh7.5.1 cells transfected with in vitro-transcribed DVG RNAs. The HCV RNA levels were determined by RT-qPCR and are expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. *, P < 0.05. (E) Schematic of plasmids expressing core-NS3 of JD835 (835), JD835 with wild-type NS2 (835-TMD), or JD835 with TMD-deleted NS2 (835-ΔTMD). (F) Western blot analysis of NS2-NS3 cleavage in HEK293 cells. (G) Schematic of plasmids expressing core-NS3 of JD835 (835), JD835 with wild-type NS2 (835-TMD), JD835 with TMS1-deleted NS2 (835-ΔTMS1), or JD835 with TMD-deleted NS2 (835-ΔTMD). (H) Western blot analysis of NS2-NS3 cleavage in HEK293 cells. (I) Schematic of JFH1-based DVG JD835 and its mutant with TMS1-deleted NS2 (JD835-ΔTMS1). (J) HCV RNA levels in Huh7.5.1 cells transfected with in vitro-transcribed DVG RNAs. The HCV RNA levels were determined by RT-qPCR and are expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. *, P < 0.05.
Next, we determined the impact of the TMS3 deletion on the cleavage of NS2/NS3 in JD835. The plasmid expressing core-NS3 of JD835, JD835 with wild-type NS2 (835-TMD), or JD835 with TMD-deleted NS2 (835-ΔTMD) (Fig. 4E) was transfected into HEK293 cells, and cleaved NS3 and uncleaved NS2-NS3 were analyzed by Western blotting. As shown in Fig. 4F, the cleavage of NS2-NS3 was significantly reduced by the deletion of TMS3, confirming the important role of TMS3 in downstream NS2/3 cleavage.
Interestingly, no DVG deletion site was found in TMS1 and TMS2 of NS2 (Fig. 4A), possibly reflecting the requirement for the proper membrane topology of NS2 for its biological functions. To test this possibility, TMS1 was deleted to change the membrane orientation of NS2 such that the mutant NS2 protease containing only TMS2 and TMS3 will translocate from the cytosolic side to the ER lumen side. First, a plasmid expressing core-NS3 of JD835 with TMS1-deleted NS2 (835-ΔTMS1) (Fig. 4G) was transfected into HEK293 cells for analysis of NS2-NS3 cleavage by Western blotting. As shown in Fig. 4H, the deletion of TMS1 did not significantly affect NS2-NS3 cleavage, suggesting that NS2-NS3 cleavage is independent of the membrane orientation of NS2. Next, we examined the impact of the TMS1 deletion on HCV replication (JD835-ΔTMS1) (Fig. 4I). As shown in Fig. 4J, the TMS1 deletion severely abolished HCV genome replication, likely because the downstream nonstructural viral proteins translocated to the ER lumen side are unable to exert their biological functions. Altogether, these results suggested that the transmembrane domains upstream of the NS2 protease are critical for its proteolytic activity and membrane orientation and, thus, for HCV genome replication.
The core-coding sequence contains a critical cis element for DVG packaging.
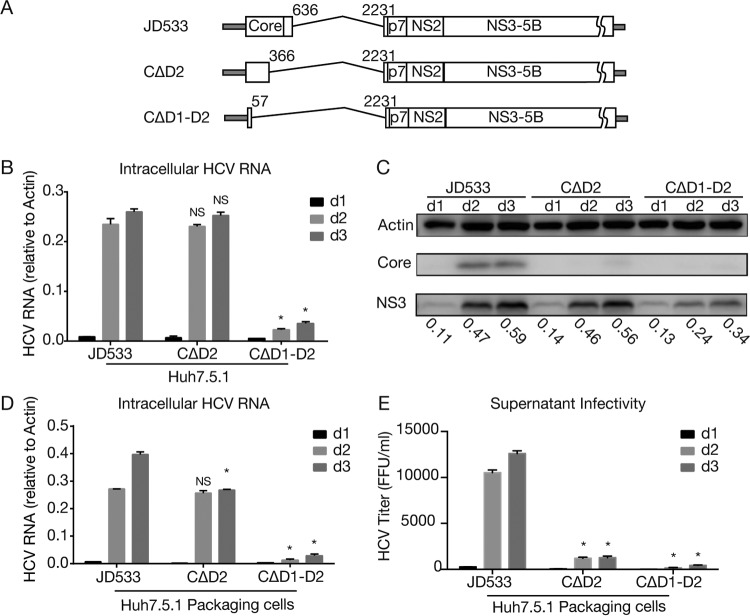
Previous studies have shown that the HCV core protein is dispensable for viral genome replication, and the major portions of the core-coding sequence can be deleted without impairing viral genome replication, except for its N-terminal 57-nt sequence, which is a part of the internal ribosomal entry site (IRES) required for cap-independent viral protein translation (4–6). In addition, the function of the core protein in encapsidating the viral genome can be complemented in trans, as an HCV genome with a defect in core could be packaged into infectious virions by expressing the missing proteins in trans (25, 26). Interestingly, as shown in Fig. 2B and Table 3, the core-coding sequences were always intact in these DVGs, raising the possibility that either the core protein or core-coding sequences play important roles in the replication or packaging of HCV DVGs. The mature HCV core protein consists of 2 functional domains: N-terminal basic hydrophilic domain 1 (D1) (aa 1 to 118), involved in RNA binding and homo-oligomerization, and C-terminal hydrophobic domain 2 (D2) (aa 119 to 173), involved in targeting core proteins to lipid droplets (27). To evaluate the impact of the deletion of these core domains on DVG replication, we constructed two JD533 mutants, one with a DVG deletion starting from core domain 2 (denoted CΔD2) and another one with a DVG deletion starting from domain 1 but retaining the N-terminal IRES sequence for protein translation (denoted CΔD1-D2). (Fig. 5A). The in vitro-synthesized RNAs of JD533, CΔD2, and CΔD1-D2 were transfected into naive Huh7.5.1 cells, and HCV RNA and NS3 protein levels were determined on days 1, 2, and 3 posttransfection by RT-qPCR and Western blotting, respectively. As shown in Fig. 5B and C, comparable amounts of HCV RNA and NS3 proteins were detected in cells transfected with JD533 and CΔD2 RNAs, suggesting that domain 2 of core was dispensable for DVG replication, consistent with the core protein not being required for HCV replication. Notably, the domain 2-deleted core protein cannot be detected by the anti-core antibody used because the epitopes that the antibody recognizes were located in domain 2 (Fig. 5C). In contrast, the deletion of both domains 1 and 2 severely impaired HCV genome replication (Fig. 5B and C), consistent with previous reports that several stem-loop structural elements (SL388, SL427, and SL588) located in core domain 1 are important for HCV genome replication/translation (28–31).
FIG 5.
The core domain 2-coding region is critical for DVG packaging. (A) Schematic of JD533, CΔD2, and CΔD1-D2. Domain 2 of core was deleted in CΔD2, and both domains 1 and 2 were deleted in CΔD1-D2. The numbers indicate the HCV nucleotide sequence starting from core. (B and C) HCV RNA (B) and viral protein (C) levels in Huh7.5.1 cells transfected with JD533, CΔD2, and CΔD1-D2. The HCV RNA levels were determined by RT-qPCR and are expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. The NS3 protein levels were quantified by using ImageJ and normalized against the internal actin control and are presented below the blot. (D and E) Intracellular HCV RNA levels (D) and extracellular infectivity titers (E) in Huh7.5.1 packaging cells transfected with JD533, CΔD2, and CΔD1-D2. The HCV RNA levels were determined by RT-qPCR and expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. NS, nonsignificant (P > 0.05); *, P < 0.05.
Next, we examined whether core domain 2 was involved in the packaging of DVGs. The in vitro-synthesized RNAs of JD533, CΔD2, and CΔD1-D2 were transfected into Huh7.5.1 packaging cells stably expressing core-NS2. The intracellular HCV RNA levels and extracellular HCV infectivity titers were determined by RT-qPCR and a titration assay. Remarkably, while there was no significant differences in HCV RNA levels on day 2 between JD533 and CΔD2 (Fig. 5D), the infectivity titers of CΔD2 were significantly reduced compared to those of JD533 on days 2 and 3 (Fig. 5E), suggesting that domain 2 of core contributed to virus production. Consistently, RT-qPCR analysis indicated that CΔD2 had less HCV RNA in the culture supernatants (data not shown). The lower intracellular HCV RNA levels in CΔD2 on day 3 than those of JD533 likely reflected the reduced reinfection rate of CΔD2 due to lower-level virion production.
The core domain 2-coding region contains a structural element critical for DVG packaging.
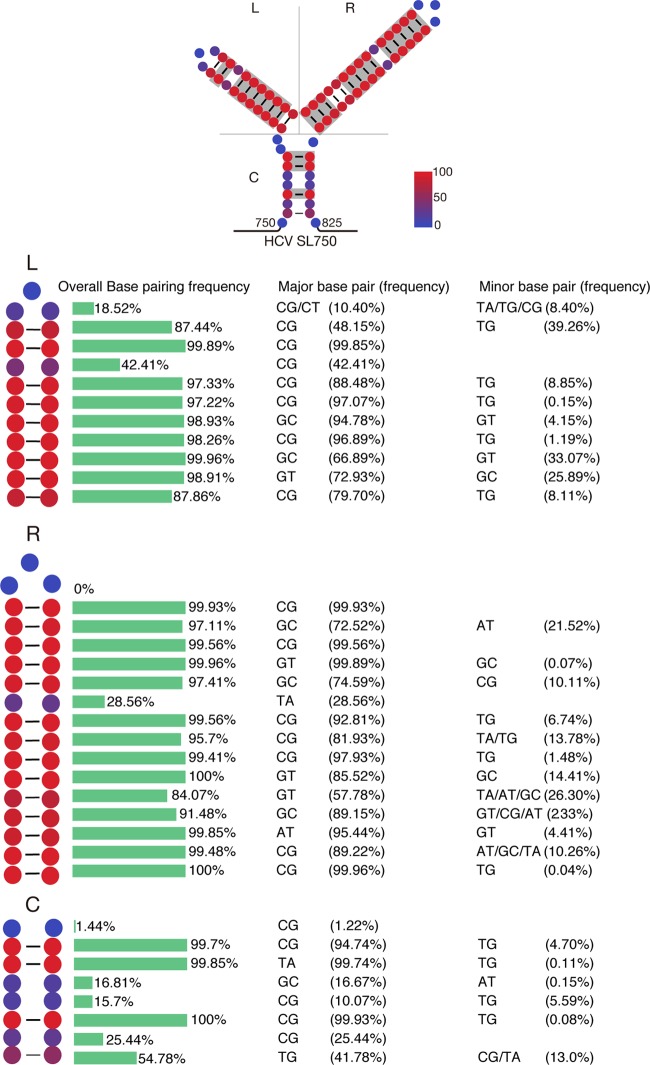
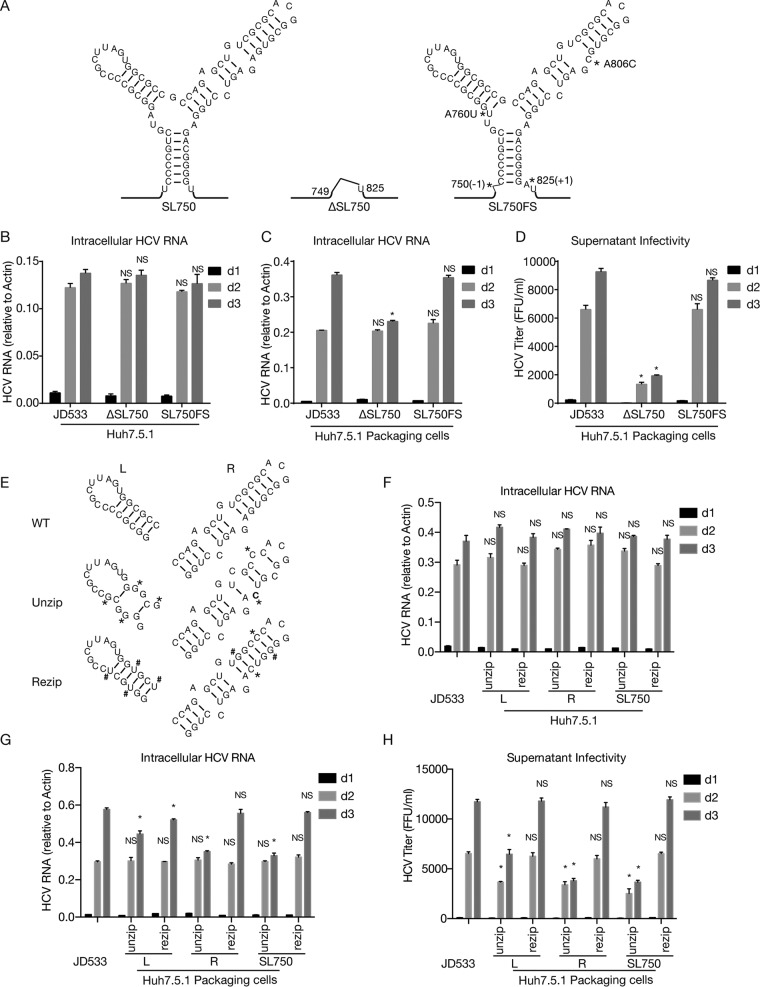
We speculated that the core domain 2-coding region may contain a cis element required for DVG packaging. RNA secondary structure analysis predicted a highly conserved RNA structure (denoted SL750) located between nt 750 and nt 825 of the core domain 2-coding region (Fig. 6). Analysis of 3,326 sequences of HCV strains across 7 genotypes showed that this Y-shaped SL750 motif consisted of two arms of stem-loops, which were highly conserved among all genotypes (Fig. 6). To examine the potential role of SL750 in DVG packaging, we made two mutants in SL750. The first one was ΔSL750, in which the entire SL750 motif was deleted from JD533. Another mutant (SL750FS) had a 1-nucleotide (U) deletion at the first nucleotide of SL750 (nt 750) and a 1-nucleotide (A) insertion at the last nucleotide of SL750 (nt 825), producing a frameshift in the translation of the SL750-coding sequence but still maintaining the Y-shaped RNA structure. To avoid two stop codons introduced by this frameshift, two more substitution point mutations were also engineered into SL750FS (A760U and A806C), which were predicted to have little effect on the RNA structure of SL750 (Fig. 7A). The JD533, ΔSL750, or SL750FS RNAs were prepared by in vitro transcription and transfected into naive Huh7.5.1 cells, and the HCV RNA levels on days 1, 2, and 3 posttransfection were determined by RT-qPCR. As shown in Fig. 7B, all three groups had comparable levels of HCV RNA, suggesting that SL750 was not required for HCV genome replication. Next, we transfected JD533, ΔSL750, or SL750FS RNA into Huh7.5.1 packaging cells and analyzed intracellular HCV RNA levels and extracellular infectivity titers. Consistent with the data shown in Fig. 7B, comparable levels of intracellular HCV RNA on days 1 and 2 posttransfection were detected among the three groups (Fig. 7C). However, the infectivity titers of ΔSL750 were significantly reduced, whereas SL750FS had infectivity titers similar to those of JD533 (Fig. 7D), strongly suggesting that the SL750 RNA structure was critical for DVG packaging.
FIG 6.
SL750 is a highly conserved structural element. A total of 3,326 HCV sequences from the HCV database of the U.S. Los Alamos National Laboratory (www.lanl.gov) were analyzed. (Top) Each nucleotide of SL750 was labeled with color according to its conservation in base pairing, and base-pairing nucleotides in more than 90% of HCV strains are highlighted in gray. (Bottom) Frequencies of base pairing at each position of the left arm (L), the right arm (R), and the central arm (C) of SL750 as well as detailed information on major and minor base pairings.
FIG 7.
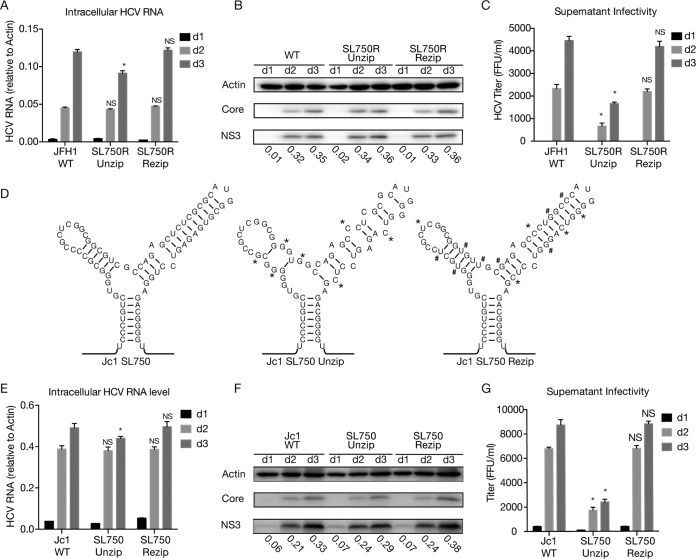
SL750 within domain 2 of the core-coding region contains a cis element critical for HCV genome packaging. (A) Secondary structure of SL750 in JFH1 (left), ΔSL750 (middle), and SL750FS (right). SL750FS had a 1-nucleotide deletion upstream of SL750 (nt 750) and a 1-nucleotide insertion downstream of SL750 (nt 825), which led to a frameshift of the SL750-coding sequence but maintained the Y-shaped RNA structure. To avoid stop codons introduced by the frameshift, A760U and A806C were also engineered into SL750FS. (B and C) HCV RNA levels in naive Huh7.5.1 cells (B) or Huh7.5.1 packaging cells (C) transfected with JD533, ΔSL750, and SL750FS. HCV RNA levels were determined by RT-qPCR and are expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. (D) Infectivity titers in the supernatants of Huh7.5.1 packaging cells transfected with JD533, ΔSL750, and SL750FS. The error bars represent standard deviations from three independent experiments. NS, nonsignificant (P > 0.05); *, P < 0.05. (E) Schematic of unzipping and rezipping mutations on the left and right arms of SL750. Unzipping and rezipping mutations are marked with * and #, respectively. WT, wild type. (F and G) HCV RNA levels in Huh7.5.1 cells (F) or Huh7.5.1 packaging cells (G) transfected with JD533 and unzipping and rezipping mutants. The HCV RNA levels were determined by RT-qPCR and are expressed as values relative to the cellular actin mRNA levels. The error bars represent standard deviations from three independent experiments. (H) Infectivity titers in the supernatants of Huh7.5.1 packaging cells transfected with JD533 and unzipping and rezipping mutants. The error bars represent standard deviations from three independent experiments. NS, nonsignificant (P > 0.05); *, P < 0.05.
Next, we sought to determine which cis element within SL570 was involved in DVG packaging. We introduced 4 mutations in the left arm of SL750 (SL750L, previously known as SL761 [30, 31]) to disrupt 4 base pairings on this stem while maintaining the amino acid sequence (unzip) (Fig. 7E). As a control, we introduced different nucleotide changes in these 4 positions of SL750L to maintain base pairing (rezip) (Fig. 7E). Similarly, 2 synonymous mutations were introduced in the right arm of SL750 (SL750R, previously known as SL783 [30, 31] or SL443 [29]) to disrupt 2 base pairings on the stem (unzip), and two additional complementary mutations were made to restore base pairing (rezip) (Fig. 7E). Finally, we made constructs that harbor these unzipping or rezipping mutations on both arms of SL750. The in vitro-synthesized RNAs were transfected into naive Huh7.5.1 cells to assess how the mutations that unzipped or rezipped the left, right, or both arms of SL750 affected HCV RNA replication. As shown in Fig. 7F, these mutations had little effect on DVG replication. Next, we transfected these DVG RNAs into Huh7.5.1 packaging cells and assessed the impact of these mutations on DVG packaging. While comparable levels of intracellular HCV RNA on day 2 posttransfection were detected among the three groups (Fig. 7G), the mutations unzipping the left or right arm of SL750 significantly reduced extracellular infectivity titers, while this defect could be rescued by rezipping mutations to restore the base pairing of SL750L and SL750R (Fig. 7H). In addition, SL750R seemed to play a more critical role than SL750L in virion production. Taken together, these results demonstrated that both the left and right arms of SL750 are cis elements critical for DVG packaging.
SL750 is also critical for HCV FLG packaging.
Finally, we assessed whether SL750 was also involved in HCV FLG packaging. The same unzipping and rezipping mutations in the right arm of SL750 (SL750R) were introduced into the full-length JFH1 genome with an adaptive mutation of a 5-amino-acid deletion (EEDDT) in NS5A (21). The in vitro-synthesized RNAs were transfected into naive Huh7.5.1 cells, and the intracellular HCV RNA levels, viral protein levels, and extracellular infectivity titers were determined. Consistent with the observations of DVGs, the unzipping mutations had little effect on FLG replication (Fig. 8A and B) but significantly reduced extracellular infectivity titers (Fig. 8C), which could be rescued by the mutations that rezip SL750R.
FIG 8.
SL750 is critical for HCV FLG packaging. Huh7.5.1 cells were transfected with in vitro-transcribed wild-type JFH1, mutant JFH-1 containing unzipping and rezipping mutations in SL750R, wild-type Jc1, or mutant Jc1 containing unzipping and rezipping mutations in SL750. (A to C and E to G) Intracellular HCV RNA levels (A and E), viral protein levels (B and F), and extracellular infectivity titers (C and G) on days 1, 2, and 3 posttransfection were determined by RT-qPCR, Western blotting, and a titration assay, respectively. NS3 protein levels were quantified by using ImageJ and normalized against the internal actin control and are presented below the blot. The error bars represent standard deviations from three independent experiments. NS, nonsignificant (P > 0.05); *, P < 0.05. (D) Schematic of wild-type and unzipping and rezipping mutations on the left and right arms of SL750 in the Jc1 strain. Unzipping and rezipping mutations are marked by * and #, respectively.
Next, we examined the role of SL750 in HCV FLG packaging using the Jc1 strain that consists of the structural proteins of the J6 strain and nonstructural proteins of JFH1 (32). The unzipping and rezipping mutations in the left and right arms of SL750 were introduced into the full-length Jc1 genome (Fig. 8D). Consistently, the unzipping mutations had little effect on FLG replication (Fig. 8E and F) but significantly reduced the extracellular infectivity titers (Fig. 8G), which could be rescued by mutations that rezip SL750. These results demonstrated that SL750 was also a critical cis element for HCV FLG packing.
DISCUSSION
In this study, we analyzed HCV DVGs in serum samples obtained from a cohort of patients chronically infected with GT-1b or GT-2a HCVs. The screening designed to detect the deletion mutations in the region from the 5′ UTR to NS3 revealed the presence of viruses carrying large in-frame deletions, located in the E1-NS2 region, in about 30% of patient sera. Our finding was consistent with many previous studies that reported that 11 to 33% of clinical isolates contain DVGs (10, 11, 13–15). Due to the PCR detection limit, the actual frequencies of DVGs in patient sera should be even higher. The potential clinical impacts of HCV DVGs have not been vigorously explored. A previous study showed that the presence of DVGs was correlated with older patient age, higher viral loads, increased liver inflammation, and possibly virological relapse of interferon-treated patients (15). In our cohort, we observed that the presence of HCV DVGs was not associated with viral loads and liver inflammation levels. In addition, no correlation between the deletion sites or frequency of DVGs and clinical manifestation was observed in our study (data not shown). It is important to point out that our failure to identify a possible clinical relevance of HCV DVGs could be due to the small sample size and other unexplored clinical manifestations in our study.
Sequencing analysis of DVGs revealed the existence of deletion hot spots. The occurrence of deletion hot spots within DVGs could be explained by two possibilities. First, these hot spots may reside in certain specific RNA structures that are intrinsically prone to internal deletion during the replication of the HCV RNA genome. Alternatively, the deletion of the HCV RNA genome at these hot spots may result in DVGs that bear selection advantages, for example, favoring more-efficient virus replication and packaging. More research will be needed to explore these hypotheses.
Our results showed that DVGs originate from a full-length genome that exists in the same patient. Because DVGs found in blood circulation are likely being encapsidated inside virions, DVGs and their corresponding FLGs should be present in the same hepatocytes so that the packaging of DVGs into virions by the structural proteins of FLGs could occur. The high sequence identity between DVGs and FLGs present in the same patients not only reveals the nature of the origins of DVGs but also may reflect the stringent requirements for compatibility between nonstructural proteins and structural proteins that work concertedly during HCV virion morphogenesis.
Our results demonstrated that DVGs always retain the C-terminal cysteine protease domain of NS2. This is consistent with the finding that the NS2 autoprotease activity is required for cleavage at the NS2-NS3 junction such that functional NS3 would be produced for DVG replication. Interestingly, we found that TMS3, an α-helix transmembrane segment immediately upstream of the cysteine protease domain, is required for DVG replication. It has been reported that green fluorescent protein (GFP) fused with TMS3 (aa 60 to 99) is anchored to the intracellular membrane. We speculated that TMS3 is required for the membrane association of the uncleaved NS2 precursor so that the autoproteolytic cleavage of NS2-NS3 can occur efficiently. More studies will be needed to test this hypothesis. In addition, we demonstrated that the transmembrane domains upstream of the NS2 protease dictate the membrane orientation of NS2, which is also essential for HCV genome replication.
Our results showed that serum-derived HCV DVGs possess the intact core-coding sequences. A previous study reported that DVGs found in the liver tissues of HCC patients contain deletions in the core-coding region, resulting in truncated core proteins (11). In contrast to DVGs in liver tissues, DVGs derived from patient sera always maintain intact core (14), which is consistent with our results. Previous studies identified several stem-loop structural elements in the core domain 1-coding region that are crucial for the HCV life cycle (31). For example, SLIV (positions 331 to 354) is a part of the IRES structure required for cap-independent viral protein translation (33). SL388 (29), SL427 (28, 29), and SL588 (30) are involved in HCV protein translation or viral genome replication. A long-range kissing-loop interaction between SL427 and SL588 has been identified to play a critical role in viral genome replication (30). In agreement with data from those studies, we showed that the deletion of these cis elements in domain 1 (CΔD1-D2) severely diminished DVG replication (Fig. 5B and C). It remains intriguing why these cis elements in domain 1 are crucial for HCV replication while they are dispensable for an HCV subgenomic replicon lacking almost the entire core-coding region. Importantly, we found that SL750, a cis element in the core domain 2-coding region, needs to be retained in the HCV genome so that HCV virions can be efficiently produced. Since SL750 is the structural element closest to the C terminus of core, this cis element would best explain the retention of full-length core in DVGs.
In this study, we found that the core domain 2-coding sequence may contain a critical cis element for HCV genome packaging. The specificity of virus packaging can be conferred by the recognition of sequences or structures on viral genomes, termed packaging signals. However, a typical packaging signal has not yet been identified for HCV, and mechanisms underlying the specificity in the encapsidation of the HCV genome into infectious particles remain to be uncovered. The 3′ UTR was previously reported to be a cis-acting element for HCV genome encapsidation (34). In another report, multiple RNA motifs scattered on the viral genome were thought to act as cooperative packaging signals to drive specific RNA encapsidation during HCV assembly (35). Our results showed that a highly conserved RNA structure, SL750, located within the core domain 2-coding region plays a critical role in packaging HCV genomic RNA. The deletion of SL750 had no apparent effect on HCV RNA levels, suggesting that SL750 is not involved in HCV genome replication. Importantly, mutations disrupting the base pairing of SL750 significantly decreased the virion production of DVGs and FLGs, which could be rescued by complementary mutations that restore the unzipped base pairs (Fig. 7 and 8). The base pairings of the SL750 stem are highly conserved across all 7 genotypes (Fig. 6), consistent with the critical roles of this evolutionarily conserved structure in HCV packaging. It is important to point out that the HCV RNA genome lacking SL750 can still be packaged into infectious virions with a low efficiency (Fig. 5E). Previous reports also indicated that HCV subgenomic replicon RNA, which lacks SL750, could be encapsidated into infectious virus-like particles in cell culture (4, 36). These data suggested that SL750, although important, may not be essential for HCV virion assembly, thus ruling out the possibility that SL750 serves as a classical packaging signal for HCV assembly. It is also important to point out that although the impact of SL750 on the packaging of JFH1/Jc1 is limited, SL750 may impose a greater impact on the packaging of HCV strains that are less efficient in virion production, particularly clinical strains. Interestingly, a conflicting role of SL750 in the HCV life cycle was reported in several previous studies. While two studies showed that the unzipping mutations of the right arm of SL750 (SL750R, also designated SL783 or SL443 in the two previous studies) had little effect on virus replication and production (29, 30), another study showed that disruption of both the left and right arms of SL750 significantly reduced virion production (37). The exact role of SL750 and the underlying mechanisms need more investigations.
In conclusion, our analysis of HCV DVGs circulating in patient sera identified novel cis elements critical for the HCV life cycle.
MATERIALS AND METHODS
Clinical samples and measurements.
Sera of 29 HCV genotype 1b-infected and 12 genotype 2a-infected patients were used in this study. The patients were among a cohort established previously in Fuyu city, Jilin province, China (38). All samples were stored at −80°C until use. Serum ALT levels were analyzed by using a Synchron LX20 autoanalyzer (Beckman-Coulter, Brea, CA, USA). The serum HCV RNA levels were quantified by using Roche Cobas AmpliPrep/Cobas TaqMan HCV assays (Roche Diagnostics, Grenzach, Germany).
Detection of HCV DVGs.
Total RNA was isolated from 140 μl of plasma by using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) and dissolved in 60 μl of RNase-free water. Reverse transcription was performed on 8 μl of RNA by using random hexamers and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The nested PCR primers were designed based on the conserved regions in the 5′ UTR or NS3 of HCV GT-1b or GT-2a. The sequences of these primers were as follows: RCTAGCCGAGTAGYGTTG and CAARTARAGGTCCGARCT (GT-1b) or GAGTGACTGTRGACAGGA (GT-2a) for the first round and TTGTGGTACTGCCTGATA and CGCAGGTGCAYGGTGT (GT-1b) or CATGCTCACCACTATGG (GT-2a) for the second round. All PCR products were analyzed by electrophoresis in 1% agarose gels stained with ethidium bromide. The desired PCR products were gel purified and sequenced. The deletions were determined by comparing the sequencing results to those for reference strains con1 (GT-1b; GenBank accession number AJ238799) and JFH1 (GT-2a; GenBank accession number AB047639).
Cell culture, plasmid construction, and HCV RNA transfection.
The hepatic cell line Huh7.5.1 (obtained from Francis Chisari at The Scripps Research Institute, USA) and derivative packaging cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 10 mM HEPES buffer, 100 U/ml penicillin, and 100 mg/ml streptomycin.
JFH1 DVG plasmids, which contain different sizes of in-frame deletions and additional point mutations, were constructed by PCR amplification to generate mutations and insertions of the PCR products into pUC-JFH1 (18). All constructed plasmids were verified by DNA sequencing. In vitro-transcribed HCV RNAs were transfected into Huh7.5.1 cells by electroporation as described previously (18, 39).
Indirect immunofluorescence.
Intracellular immunostaining was performed as described previously (18). HCV NS5A was stained by using a rabbit polyclonal anti-NS5A antibody (40). Bound primary antibodies were detected by using Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes, Eugene, OR, USA). Nuclei were stained with Hoechst dye.
Western blot analysis.
The assay was performed as described previously (21). Membranes were probed with a primary antibody against HCV NS3 (8G-2; Abcam, Cambridge, United Kingdom), anti-core antibody (C7-50; Abcam), or actin antibody (Sigma, St. Louis, MO, USA) and then probed with alkaline phosphatase-conjugated goat anti-mouse secondary antibody (Promega, Madison, WI, USA). Proteins were visualized by a BCIP (5-bromo-4-chloro-3-indolylphosphate)–nitro blue tetrazolium kit (Promega).
Quantification of HCV infectivity titers and genomic RNA.
HCV infectivity titers were determined by a titration assay on Huh7.5.1 cells using endpoint dilution and immunostaining as described previously (18). HCV RNA levels were determined by RT-qPCR as described previously (18).
RNA secondary structure prediction.
The HCV core-coding region was analyzed by using CG v2.0 software (41) to predict the formation of putative stem-loops.
Ethics statement.
Serum sample collection and experiments were approved by the ethics committee of the First Hospital of Jilin University. Samples were collected from a cohort established previously in Fuyu city, Jilin province, China, and all samples were anonymized (38).
Accession number(s).
The sequences of all DVGs and FLGs have been deposited in the NCBI GenBank database (accession numbers KY930675 to KY930698).
ACKNOWLEDGMENTS
We thank Jie Lu, Tanyu Gan, Yannan Zhao, Yisha Liang, and Tao Yu for technical assistance.
This study was supported by grants from the National Natural Science Foundation of China (31670172) to J.Z., the Chinese National 973 Program (2015CB554300) to J.Z. and J.N., the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB03) to J.Z., and the National Natural Science Foundation of China (31400160) to Y.X. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no conflict of interest.
REFERENCES
- 1.Li D, Huang Z, Zhong J. 2015. Hepatitis C virus vaccine development: old challenges and new opportunities. Nat Sci Rev 2:285–295. doi: 10.1093/nsr/nwv040. [DOI] [Google Scholar]
- 2.Klibanov OM, Gale SE, Santevecchi B. 2015. Ombitasvir/paritaprevir/ritonavir and dasabuvir tablets for hepatitis C virus genotype 1 infection. Ann Pharmacother 49:566–581. doi: 10.1177/1060028015570729. [DOI] [PubMed] [Google Scholar]
- 3.Hayes CN, Chayama K. 2015. Emerging treatments for chronic hepatitis C. J Formos Med Assoc 114:204–215. doi: 10.1016/j.jfma.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 5.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J Virol 66:1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 7.Reed KE, Grakoui A, Rice CM. 1995. Hepatitis C virus-encoded NS2-3 protease: cleavage-site mutagenesis and requirements for bimolecular cleavage. J Virol 69:4127–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2013. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment Web resource. Hepatology 59:318–327. doi: 10.1016/j.jhep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh C-T, Lu S-C, Chu C-M, Liaw Y-F. 1997. Molecular cloning of a defective hepatitis C virus genome from the ascitic fluid of a patient with hepatocellular carcinoma. J Gen Virol 78:2761–2770. doi: 10.1099/0022-1317-78-11-2761. [DOI] [PubMed] [Google Scholar]
- 10.Yagi S, Mori K, Tanaka E, Matsumoto A, Sunaga F, Kiyosawa K, Yamaguchi K. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J Med Virol 77:399–413. doi: 10.1002/jmv.20469. [DOI] [PubMed] [Google Scholar]
- 11.Iwai A, Marusawa H, Takada Y, Egawa H, Ikeda K, Nabeshima M, Uemoto S, Chiba T. 2006. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J Viral Hepat 13:523–531. doi: 10.1111/j.1365-2893.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu YK, Hijikata M, Oshima M, Shimizu K, Yoshikura H. 2006. Detection of a 5′ end subgenome of hepatitis C virus terminating at nucleotide 384 in patients' plasma and liver tissues. J Viral Hepat 13:746–755. doi: 10.1111/j.1365-2893.2006.00745.x. [DOI] [PubMed] [Google Scholar]
- 13.Noppornpanth S, Smits SL, Lien TX, Poovorawan Y, Osterhaus ADME, Haagmans BL. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J Virol 81:12496–12503. doi: 10.1128/JVI.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugiyama K, Suzuki K, Nakazawa T, Funami K, Hishiki T, Ogawa K, Saito S, Shimotohno KW, Suzuki T, Shimizu Y, Tobita R, Hijikata M, Takaku H, Shimotohno K. 2009. Genetic analysis of hepatitis C virus with defective genome and its infectivity in vitro. J Virol 83:6922–6928. doi: 10.1128/JVI.02674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheroni C, Donnici L, Aghemo A, Balistreri F, Bianco A, Zanoni V, Pagani M, Soffredini R, D'Ambrosio R, Rumi MG, Colombo M, Abrignani S, Neddermann P, De Francesco R. 2015. Hepatitis C virus deletion mutants are found in individuals chronically infected with genotype 1 hepatitis C virus in association with age, high viral load and liver inflammatory activity. PLoS One 10:e0138546. doi: 10.1371/journal.pone.0138546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier C, Duverlie G, Castelain S. 2013. Are trans-complementation systems suitable for hepatitis C virus life cycle studies? J Viral Hepat 20:225–233. doi: 10.1111/jvh.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Yu G, Sun H, Lv J, Wang M, Kong F, Zhang M, Chi X, Wang X, Wu R, Gao X, Zhong J, Sun B, Jiang J, Pan Y, Niu J. 2015. Use of parenteral caffeinum natrio-benzoicum: an underestimated risk factor for HCV transmission in China. BMC Public Health 15:928. doi: 10.1186/s12889-015-2299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Tong Y, Chi X, Yang W, Zhong J. 2017. Functional analysis of hepatitis C virus (HCV) envelope protein E1 using a trans-complementation system reveals a dual role of a putative fusion peptide of E1 in both HCV entry and morphogenesis. J Virol 91:e02468-. doi: 10.1128/JVI.02468-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii K, Murakami K, Hmwe SS, Zhang B, Li J, Shirakura M, Morikawa K, Suzuki R, Miyamura T, Wakita T, Suzuki T. 2008. Trans-encapsidation of hepatitis C virus subgenomic replicon RNA with viral structure proteins. Biochem Biophys Res Commun 371:446–450. doi: 10.1016/j.bbrc.2008.04.110. [DOI] [PubMed] [Google Scholar]
- 23.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Tang H, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog 6:e1001233. doi: 10.1371/journal.ppat.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831–835. doi: 10.1038/nature04975. [DOI] [PubMed] [Google Scholar]
- 25.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 26.Pacini L, Graziani R, Bartholomew L, De Francesco R, Paonessa G. 2009. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J Virol 83:9079–9093. doi: 10.1128/JVI.00308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 28.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A 104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol 82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. 2016. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol Cell 62:111–120. doi: 10.1016/j.molcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams RL, Pirakitikulr N, Pyle AM. 2017. Functional RNA structures throughout the hepatitis C virus genome. Curr Opin Virol 24:79–86. doi: 10.1016/j.coviro.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset F-L, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A 103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. 2011. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi G, Ando T, Suzuki R, Matsuda M, Nakashima K, Ito M, Omatsu T, Oba M, Ochiai H, Kato T, Mizutani T, Sawasaki T, Wakita T, Suzuki T. 2016. Involvement of the 3′ untranslated region in encapsidation of the hepatitis C virus. PLoS Pathog 12:e1005441. doi: 10.1371/journal.ppat.1005441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart H, Bingham RJ, White SJ, Dykeman EC, Zothner C, Tuplin AK, Stockley PG, Twarock R, Harris M. 2016. Identification of novel RNA secondary structures within the hepatitis C virus genome reveals a cooperative involvement in genome packaging. Sci Rep 6:22952. doi: 10.1038/srep22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 37.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. 2015. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci U S A 112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, Zhang P, Chi X, Jiang Y, Gao Y, Zhong J, Sun B, Xu D, Jiang J, Niu J. 2012. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One 7:e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao W, Xu C, Ding Q, Li R, Xiang Y, Chung J, Zhong J. 2009. A single point mutation in E2 enhances hepatitis C virus infectivity and alters lipoprotein association of viral particles. Virology 395:67–76. doi: 10.1016/j.virol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Weng L, Li R, Li L, Toyoda T, Zhong J. 2012. The N-terminal helix α(0) of hepatitis C virus NS3 protein dictates the subcellular localization and stability of NS3/NS4A complex. Virology 422:214–223. doi: 10.1016/j.virol.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Andronescu M, Bereg V, Hoos HH, Condon A. 2008. RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinformatics 9:340. doi: 10.1186/1471-2105-9-340. [DOI] [PMC free article] [PubMed] [Google Scholar]