ABSTRACT
Human herpesvirus 8 (HHV-8) encodes four viral interferon regulatory factors (vIRF-1 to -4) that likely function to suppress innate immune and cellular stress responses through inhibitory interactions with various cellular proteins involved in these activities. It is notable that vIRF-1 and -4 have been reported to interact with the deubiquitinase ubiquitin-specific protease 7 (USP7), substrates of which include p53 and the p53-targeting and -destabilizing ubiquitin E3 ligase MDM2. Structural studies of vIRF-1 and vIRF-4 USP7 binding sequences in association with USP7 have been reported; both involve interactions with N-terminal-domain residues of USP7 via EGPS and ASTS motifs in vIRF-1 and vIRF-4, respectively, but vIRF-4 residues also contact the catalytic site. However, the biological activities of vIRF-1 and vIRF-4 via USP7 interactions are unknown. Here, we report that vIRF-3, which is latently, as well as lytically, expressed in HHV-8-infected primary effusion lymphoma (PEL) cells, also interacts with USP7—via duplicated EGPS motifs—and that this interaction is important for PEL cell growth and viability. The interaction also contributes to suppression of productive virus replication by vIRF-3, which we identify here. We further show that vIRF-1, which is expressed at low levels in PEL latency, promotes latent PEL cell viability and that this activity and vIRF-1-promoted productive replication (reported previously) involve EGPS motif-mediated USP7 targeting by vIRF-1. This study is the first to identify latent and lytic functions of vIRF-1 and vIRF-3, respectively, and to address the biological activities of these vIRFs through their interactions with USP7.
IMPORTANCE HHV-8 is associated with Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman's disease; both latent and lytic viral functions are believed to contribute. Viral interferon regulatory factors specified by HHV-8 are thought to be critically important for successful productive replication through suppression of innate immune and stress responses triggered by the lytic cycle. Latently expressed vIRF-3 contributes significantly to PEL cell survival. Here, we identify ubiquitin-specific protease 7 (USP7) deubiquitinase targeting by vIRF-3 (in addition to previously reported USP7 binding by vIRF-1 and vIRF-4); the importance of vIRF-1 and vIRF-3 interactions with USP7 for latent PEL cell growth and viability; and the positive and negative contributions, respectively, of USP7 targeting by vIRF-1 and vIRF-3 to HHV-8 productive replication. This is the first report of the biological importance of vIRF-1 in PEL cell latency, the modulation of productive replication by vIRF-3, and the contributions of vIRF-USP7 interactions to HHV-8 biology.
KEYWORDS: human herpesvirus 8, USP7, latency, lytic replication, viral interferon regulatory factors
INTRODUCTION
Human herpesvirus 8 (HHV-8) specifies a variety of latent and lytic activities that function to suppress innate immune, cell stress, and apoptotic signaling in response to primary, lytic, and latent infection. For example, with respect to apoptotic pathway inhibition, HHV-8 encodes lytically expressed homologues of Bcl-2, cIAP, chemokines, and chemokine receptors, among other proteins, that are able to promote cell survival, thereby likely contributing to virus production under lytic-cycle-induced stress (1–7). In latency, the viral replication protein latency-associated nuclear antigen (LANA) is likely to provide apoptosis-inhibitory activity through direct interactions with p53, in addition to proproliferative functions via inhibitory interactions with other key regulators, such as glycogen synthase kinase 3β and retinoblastoma protein (8–10). The viral interferon regulatory factors (vIRFs) specified by HHV-8 are believed to be major contributors to suppression of innate immune responses to virus infection and productive replication and to apoptotic suppression under these conditions. Of note is that deletion of the vIRFs in rhesus macaque rhadinovirus, which is closely related to HHV-8, leads to enhanced type I interferon responses during infection of cells in culture and to increased proinflammatory cytokine induction, accelerated antiviral T cell responses, and reduced viral loads in infected animals (11, 12). All four of the HHV-8 vIRFs are expressed during lytic replication and are therefore able to act at this phase of the virus life cycle, but vIRF-3 is also expressed as a bona fide latency gene in the context of HHV-8-infected primary effusion lymphoma (PEL) cells (13, 14). It is important to note that vIRF-1 is also expressed, at low levels, during latency in PEL cells, although it is induced during lytic reactivation (13, 15); therefore, vIRF-1 has the potential to contribute to latency in the setting of PEL (and possibly infection of untransformed B cells). We have reported such activity for predominantly lytically expressed viral interleukin 6 (vIL-6), which is expressed at low but functional and biologically significant levels in latently infected PEL cells (16).
With respect to mechanisms of vIRF function, there are several cellular interaction partners through which the vIRFs can operate (17). Most studies have been conducted on vIRF-1, which has been found to target, in inhibitory fashion, proapoptotic mediators including p53 (18, 19), the p53 activator kinase ATM (20), the BH3-only proteins Bim and Bid (21, 22), retinoic acid and interferon-inducible GRIM19 (23), the mitochondrial antiviral signaling adapter MAVS (24), and the transforming growth factor β (TGF-β)-activated transcription factors Smad3 and Smad4 (25), in addition to the transcription coactivators CBP and p300, which are required for cellular IRF induction of antiviral type I interferon genes (26–28). It is notable that vIRF-3 also has been reported to target and inhibit p53, and it is considered likely, although not established, that this property is important for vIRF-3 prosurvival activity in the context of PEL cell latency (14, 29). Also notable, and relevant to the present report, is that vIRF-1 and vIRF-4 target the deubiquitinase ubiquitin-specific protease 7 (USP7) (30, 31), substrates of which include p53 and the p53-targeting ubiquitin E3 ligase MDM2. We show here that vIRF-3 also interacts with USP7. Therefore, USP7 is targeted by three of the four HHV-8 vIRFs, in addition to other HHV-8 proteins (see below), indicating that it is an important cell regulatory protein that is modulated or usurped for the benefit of the virus.
It has generally been assumed that interactions of the vIRFs are inhibitory. However, although this makes sense teleologically, as the targets tend to induce cytostatic, apoptotic, or innate immune responses, this may not be the case for USP7. The deubiquitinase interacts with and stabilizes not only p53 and its E3 ligase MDM2, in addition to p53-activating Tip60 kinase, with overall proapoptotic effect under stress conditions, but also other key cellular regulators that have distinct functions. Such targets of USP7 include PTEN, which can function as a nuclear-localized tumor suppressor and which is excluded from the nucleus via USP7-mediated deubiquitination (32). USP7 can also inhibit the monoubiquitination and consequent nuclear translocation of the proapoptotic transcription factor FOXO4, and USP7 interacts with FOXO3, as well (33); perhaps related to this is the fact that vIRF-3 can promote phosphorylation-independent FOXO3 cytoplasmic sequestration (inactivation) via 14-3-3 protein interaction (34). These findings, among others (35), provide clear evidence that USP7 activities extend well beyond p53 regulation through interactions with p53 and MDM2 (36) and that modulation or redirection of USP7 activity by the vIRFs, rather than simple inhibition, may occur.
Herpesvirus targeting of USP7 is well established, although the biological outcomes are not well understood. The deubiquitinase was initially identified as a herpes simplex virus (HSV) ICP0-interacting protein (37). This interaction is important for stabilization of ICP0 (38), a key immediate-early protein of HSV required for lytic replication, but more recently it has also been reported to promote nuclear export of USP7 to effect deubiquitination and consequent inhibition of TRAF6 (tumor necrosis factor receptor-associated factor 6) and IKKγ (inhibitor of nuclear factor κB kinase subunit γ) and thereby inhibit antiviral Toll-like receptor signaling (39). The latent replication protein EBNA1 of Epstein-Barr virus also binds USP7, which can lead to destabilization of p53 via its competitive dissociation from USP7 (40). However, another consequence of this interaction is the corecruitment by EBNA1 of USP7 and the associated USP7 activator GMP synthase to latent viral episomes, resulting in H2A/B histone deubiquitination at the viral latent origin, OriP, potentially regulating OriP-directed replication (41). Counterintuitively, although USP7 appears to promote EBNA1-OriP binding, EBNA1-USP7 interaction leads to the inhibition of latent viral episome replication, and a similar phenomenon has been reported for HHV-8 LANA (42, 43). In addition to the vIRFs and LANA of HHV-8, USP7 is also targeted by the HHV-8 open reading frame 45 (ORF45)-encoded tegument protein, one consequence of which is stabilization of the cointeracting ORF33 tegument protein, which is important for virion assembly and release of infectious virus (44). Together, these findings highlight the central importance of viral targeting of USP7 and the variety of functional outcomes of viral protein-USP7 interactions. Of note for this report is the fact that although vIRF-1 and -4 are known to interact with USP7, the significance of these interactions with respect to function and HHV-8 biology have yet to be determined.
In this paper, we report that USP7 is targeted by vIRF-3, in addition to vIRF-1 and vIRF-4, and analyze the functions of vIRF-1 and vIRF-3 and their interactions with USP7 in HHV-8 latency and productive replication. Our data demonstrate that vIRF-1 promotes latent PEL cell viability, that vIRF-3 inhibits HHV-8 productive replication, and that vIRF-1 and vIRF-3 interactions with USP7 are important for these and previously reported proreplication and proviability activities of vIRF-1 and vIRF-3.
RESULTS
USP7-vIRF interactions.
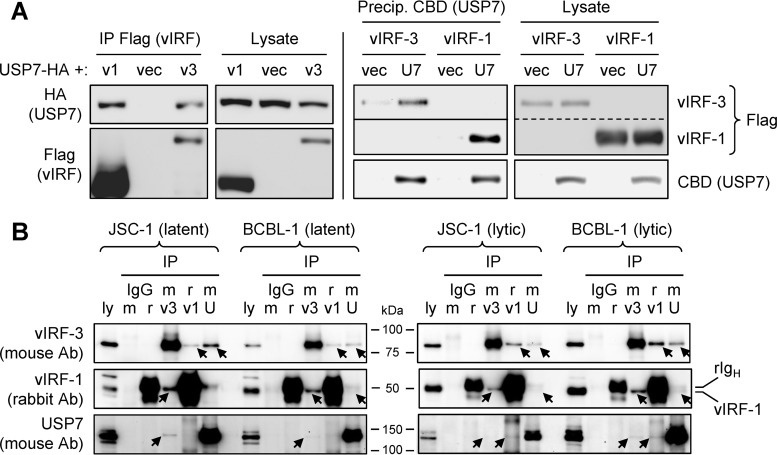
To survey for cellular proteins interacting with vIRF-1 and vIRF-3 in infected cells, we carried out tandem-affinity precipitations of the StrepII plus Flag (SF) affinity-tagged, lentivirus-transduced viral proteins from BCBL-1 PEL cell lysates and mass spectrometry analysis of coprecipitated cellular proteins. These initial experiments identified USP7 as a potential interaction partner of both vIRFs. It was subsequently reported by others that vIRF-1 binds USP7 via interaction of an EGPS motif in the viral protein with the TRAF domain of USP7 (30). Inspection of vIRF-3 revealed two EGPS motifs contained within residues 187 to 217. To confirm binding of USP7 by vIRF-3, we undertook coprecipitation experiments in appropriately transfected HEK293T cells, along with parallel experiments using vIRF-1. Flag epitope-tagged vIRF-1 and vIRF-3 were expressed together with hemagglutinin (HA) epitope-tagged USP7. Precipitation of vIRF-3 with Flag antibody beads and immunoblotting for USP7 revealed association of the two proteins, whereas no (nonspecific) binding of USP7 was detected when vIRF-3–SF was omitted (Fig. 1A, left). Evidence of vIRF-3 interaction with USP7 was also obtained from “reciprocal” precipitation of USP7–chitin-binding domain (CBD) and immunoblotting for vIRF-3 detection (Fig. 1A, right). In parallel vIRF-Flag and USP7-CBD precipitations, interaction of USP7 with vIRF-1 was detected, as expected. Therefore, USP7 can interact with both vIRF-1 and vIRF-3.
FIG 1.
Interactions of vIRF-1 and vIRF-3 with USP7. (A) HEK293T cells were transfected with plasmid vectors for expression of StrepII plus Flag (SF)-tagged vIRF-1 or vIRF-3 and HA-tagged USP7 (left) or CBD-fused USP7 (right). Lysates were prepared; either vIRF or USP7 proteins were precipitated with Flag antibody or chitin beads, respectively, and coprecipitated USP7-HA (left) or SF-tagged vIRF-1 and vIRF-3 (right) were detected by immunoblotting. The cell lysates were probed to ensure appropriate expression of the vector-encoded proteins. For the Flag blots on the right, the membrane for the precipitated samples was cut (indicated by the solid line) and exposed for different times for optimal detection of vIRF-1 and vIRF-3; the corresponding “Lysate” images were cropped (dashed line) from a single original image. U7, USP7; v1, vIRF-1; v3, vIRF-3; vec, empty vector. (B) Immunoprecipitation (IP) of vIRF-1, vIRF-3, or USP7 and reciprocal immunoblotting to detect coprecipitated binding partners were undertaken from latent or lytically induced BCBL-1 (TRExBCBL1-RTA) and JSC-1 PEL cell lysates harvested 24 h following treatment with doxycycline or TPA plus sodium butyrate (see Materials and Methods). Normal (nonimmune) rabbit (r) and mouse (m) IgGs were used in place of vIRF-1 rabbit antiserum and mouse antibodies (Ab) to vIRF-3 and USP7 to control for potential nonspecific interactions of target proteins. The arrows indicate coprecipitated vIRF-1, vIRF-3, and USP7 bands on the respective blots; the closely migrating positions of vIRF-1 and heavy-chain rabbit immunoglobulin (rIgH) are also indicated on the rightmost vIRF-1 blot. Precipitated proteins are indicated by v1 (vIRF-1), v3 (vIRF-3), and U (USP7) above the respective lanes. Samples of cell lysate (ly) (5% of that used for each precipitation) were loaded directly onto the gels to provide a positive control for immunoblotting and to identify the relevant bands.
Immunoprecipitation analyses were undertaken using lysates from latent and lytically induced BCBL-1 and JSC-1 PEL cells to assess the interactions of endogenously produced, virus-encoded vIRF-1 and vIRF-3 with endogenous USP7. Parallel immunoprecipitations using rabbit antiserum to vIRF-1 and mouse antibodies to vIRF-3 and USP7, followed by reciprocal immunoblotting for interaction partners, were carried out. Nonimmune rabbit and mouse immunoglobulins G were used to control for potential nonspecific association of proteins with antibodies and/or Fc-binding protein A-agarose beads. The data from these experiments (Fig. 1B) clearly identified vIRF-3 coprecipitation with immunoprecipitated USP7 from lytic and latent BCBL-1 and JSC-1 cell lysates (top blots); limited but specific coprecipitation of vIRF-1 with USP7 (middle blots); and, although faint, reciprocal coprecipitation of USP7 with both vIRF-1 and vIRF-3 (bottom blots). Interestingly, these data also revealed interaction of vIRF-1 and vIRF-3, as each could be coprecipitated with the other (top and middle blots). The immunoprecipitation data identified vIRF-1 and vIRF-3 interactions with USP7 in the context of HHV-8-infected cells, thereby providing evidence of biological relevance, in addition to confirming the interactions detected by mass spectrometry and transfection-based coprecipitation experiments (Fig. 1A) (30).
Characterization of vIRF-USP7 interactions.
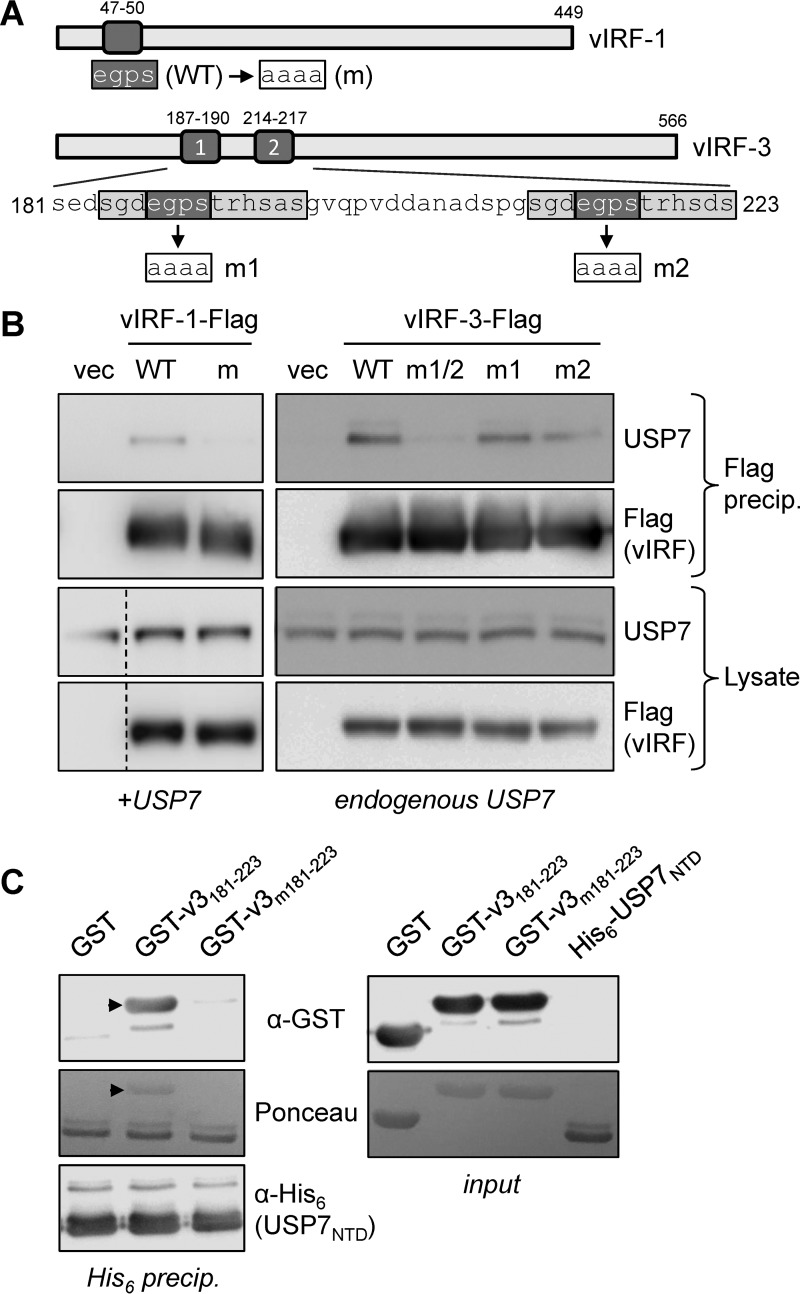
To determine the requirement for the EGPS motifs in vIRF-1 and vIRF-3 for interactions with USP7, we undertook further transfection-based coprecipitation experiments using vIRF-1 and vIRF-3 with mutations in the EGPS motifs (EGPS→AAAA), along with the wild-type vIRFs for comparison. For vIRF-3, which contains two EGPS motifs within a broader duplicated sequence (Fig. 2A), each motif was mutated individually, as well as together. Coprecipitation assays using versions of the wild-type and mutated vIRFs tagged with Flag epitope confirmed that the EGPS motif in vIRF-1 was required for USP7 association, as expected (30), and revealed the involvement of the equivalent motifs in vIRF-3, with abrogation of binding effected by dual mutation of these EGPS sequences (Fig. 2B). Therefore, for vIRF-1 and vIRF-3, intracellular interaction with USP7 is EGPS motif dependent.
FIG 2.
Requirement for vIRF-3 EGPS motifs in USP7 interaction. (A) Diagrammatic representation of the single and dual EGPS motifs (dark shading) in vIRF-1 and vIRF-3, respectively, along with the amino acid sequence of the corresponding region of vIRF-3 to show the broader sequence duplication (light shading) containing each of the EGPS motifs. WT, wild type. (B) Transfection-based coprecipitations similar to those outlined for Fig. 1 were carried out, in which Flag-tagged vIRF-1 or vIRF-3, or EGPS motif-mutated versions thereof, were precipitated with Flag antibody-conjugated beads. The precipitated material was immunoblotted with USP7-directed antibody for detection of vIRF-USP7 interactions and with Flag antibody to confirm vIRF-Flag immunoprecipitations. The lysates were analyzed by immunoblotting to ensure appropriate protein expression (the dashed lines in the left blots indicate deletion of a lane). vIRF-1 with the EGPS motif mutated (m) and singly (m1 and m2) and dually (m1/2) mutated vIRF-3 expressed in the respective transfected cultures are indicated above the lanes. +USP7, cotransfected USP7 expression plasmid. (C) Direct association between EGPS motif-containing vIRF-3 residues 181 to 223 and the N-terminal (TRAF) domain (NTD) (residues 52 to 204) of USP7 was verified by an in vitro binding assay using GST-fused vIRF-3 wild type (v3181–223) or EGPS-mutated (v3m181–223) residues 181 to 223 and His6-tagged USP7 NTD (His6-USP7NTD). (Left) His6-USP7NTD was precipitated with nickel beads, and coprecipitated GST-fused vIRF-3 peptides (arrowheads) were identified by anti-GST immunoblotting (top), in addition to Ponceau S staining (middle). The latter also detected precipitated His6-USP7NTD, the identity of which was confirmed by immunoblotting for His6 (bottom). (Right) Input material, visualized by immunoblotting for GST (vIRF-3 peptides) or Ponceau S staining.
To verify that the interaction of vIRF-3 with USP7 was direct, the USP7 binding region of vIRF-3 (residues 181 to 223) (vIRF-3181–223) and the N-terminal domain (NTD) (residues 52 to 204) of USP7 were bacterially expressed as glutathione S-transferase (GST)- and His6-fused recombinant proteins, respectively, and purified from bacterial lysates (see Materials and Methods). GST–vIRF-3181–223 could be coprecipitated with nickel bead-sedimented His6-NTD, whereas dually EGPS-mutated GST–vIRF-3m181–223 was essentially negative for coprecipitation, as was GST alone (Fig. 2C). Ponceau S staining of the input material showed that all the proteins were present at similar levels. Thus, vIRF-3, like vIRF-1 (30), interacts directly with NTD sequences of USP7, and this interaction is dependent on the EGPS motifs.
USP7 contributions to HHV-8 biology.
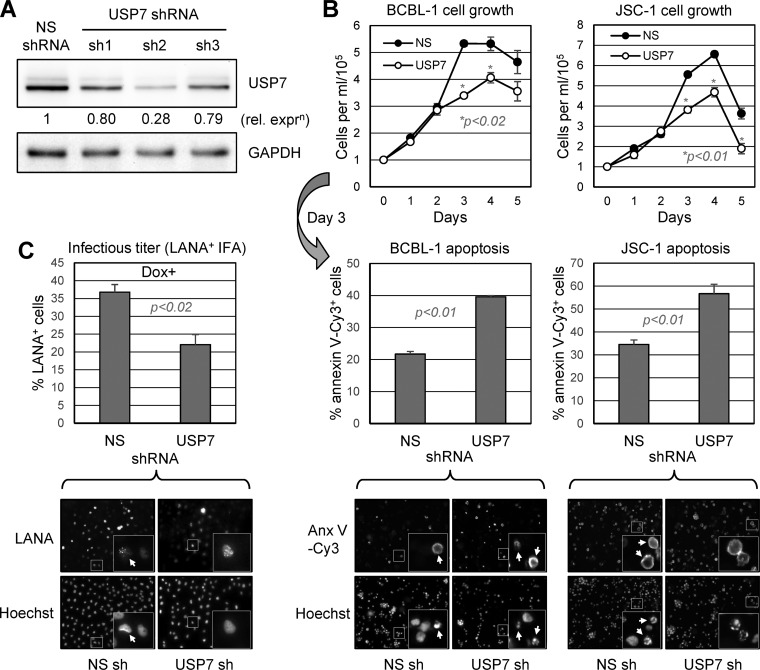
USP7 is targeted, not only by vIRF-1 and vIRF-3, but also by vIRF-4, ORF45 tegument protein, and LANA (43, 44). Although it has been demonstrated that the ORF45 protein interaction with USP7 is associated with stabilization and increased levels of cocomplexing ORF33 tegument protein (44), the functional significance of the other interactions and of USP7 more generally is unknown. It has been demonstrated that intracellular introduction of a peptide corresponding to the region of vIRF-4 interacting with the catalytic site of USP7 leads to loss of PEL cell viability, suggesting centrally important regulatory activities of USP7, possibly through modulation of p53 and downstream signaling (31). To assess independently the importance of USP7 for PEL cell growth and viability, we depleted USP7 using lentiviral vector-delivered short hairpin RNA (shRNA) and assayed for cell growth and rates of apoptosis. Prescreening of USP7 mRNA-directed shRNAs for their abilities to deplete USP7 in lentiviral vector-transduced BCBL-1 cells identified efficient USP7 depletion by one, shRNA-2 (sh2) (Fig. 3A), and it was used in subsequent experiments. The USP7 shRNA and nonsilencing (NS) shRNA control lentiviral vectors were used to infect BCBL-1 and JSC-1 PEL cells to achieve >90% transduction efficiency (as determined by lentiviral vector expressed green fluorescent protein [GFP] fluorescence), and 2 days posttransduction, cell culture densities were normalized and growth was monitored for 5 days by daily counting to determine viable (trypan blue-excluding)-cell densities. On day 3, a sample of cells was extracted from each culture for assessment of apoptosis by annexin V-Cy3 binding. USP7 depletion led to reduced BCBL-1 and JSC-1 cell growth (Fig. 3B, top), with increased rates of apoptosis detected in the cultures on the third (tested) day (Fig. 3B, bottom). These data provide evidence of a positive, proviability role of USP7 in latently infected PEL cells.
FIG 3.
Biological effects of USP7 depletion. (A) Three shRNAs directed to USP7 mRNA were tested for efficacy in PEL (TRExBCBL1-RTA) cells infected with the respective shRNA-expressing lentiviral vectors. USP7 and GAPDH immunoblotting of derived cell lysates made 4 days after lentiviral transduction revealed substantial USP7 depletion by shRNA no. 2 (sh2) relative to levels detected in NS shRNA-transduced control cultures. (B) USP7 versus NS shRNA-transduced TRExBCBL1-RTA (BCBL-1) and JSC-1 cultures were monitored for growth 48 h after shRNA transduction via lentiviral vector infection and following normalization of cell densities (day 0). Viable cells were identified by trypan blue exclusion, and cell densities were quantified by hemocytometric counting. On the third day, culture samples were extracted for analysis of apoptosis by annexin V-Cy3 staining and counterstaining with Hoechst 33342 dye for visualization of cell nuclei. The data were calculated from multiple random fields for each condition (examples are shown below each data set). For the growth and apoptosis assays, average values from three replicate cultures for each condition are shown, along with standard deviations from these values; NS versus USP7 depletion P values (unpaired, two-tailed t test) are shown. (C) Infectious-virus titers derived from doxycycline (Dox)-induced TRExBCBL1-RTA cultures transduced with either NS (control) or USP7-directed shRNA were determined by inoculations of naive iSLK cells with medium samples and immunofluorescence detection of LANA, along with Hoechst 33343 counterstaining to detect cell nuclei (example fields are shown). The data were derived from triplicate cultures and expressed as averages; standard deviations from the average values are indicated, along with P values (Student's t test). No infectious virus was detected in medium samples from uninduced cultures. The insets in the images of panels B and C are enlargements of the boxed areas; arrows indicate annexin V-Cy3-positive and LANA-positive cells in mixed populations.
USP7 depletion was also undertaken to determine the influence of the deubiquitinase on HHV-8 productive replication. Here, TRExBCBL1-RTA cells (45) were used, as they could be induced efficiently into a lytic cycle using doxycycline (see Materials and Methods), allowing ready detection and titration of derived infectious virus by inoculation and LANA staining of naive iSLK cells (46) (see Materials and Methods). TRExBCBL1-RTA cultures were infected with lentiviral vectors specifying USP7-specific or NS control shRNA 48 h prior to lytic induction, and culture media were harvested 4 days after lytic induction for titration of released virus. USP7 depletion led to ∼40% reduced infectious titers in the media of USP7-depleted cultures relative to the controls (Fig. 3C), demonstrating a positive role of USP7 in productive replication in this cell type.
vIRF-1 contributions to PEL latency.
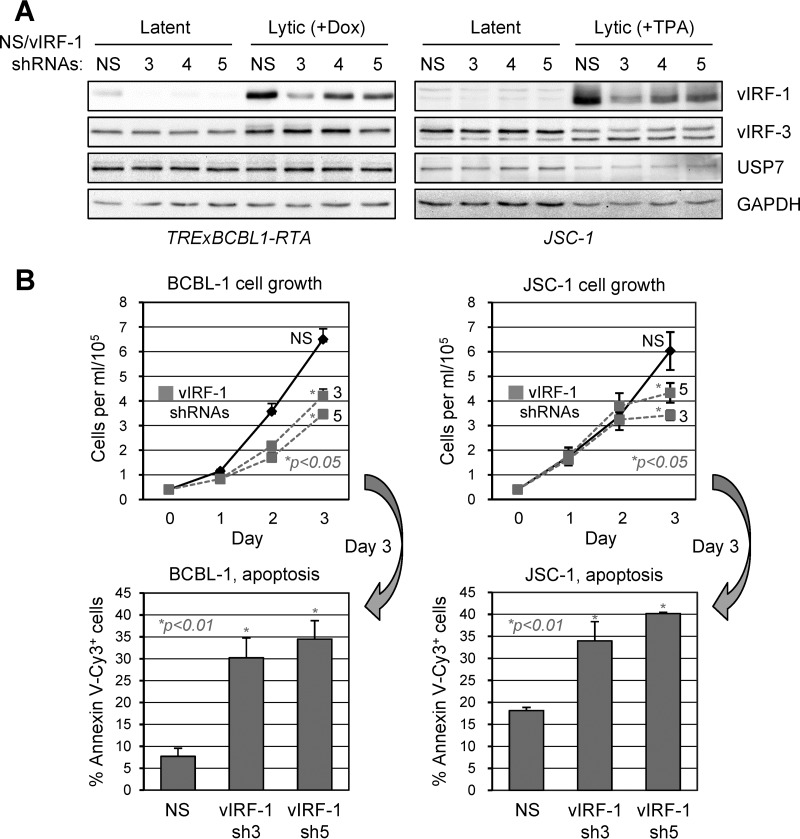
The role of vIRF-1 in productive replication has been demonstrated in endothelial and PEL cells (21, 24). However, in PEL cells, vIRF-1 is expressed not only in lytic replication, but also, at low levels, in latency (13, 15). To provide a means of testing the functional effects of vIRF-1, we generated and assessed the efficacy, in both latently infected and lytically reactivated BCBL-1 (TREx-RTA) and JSC-1 cells, of lentiviral vector-expressed vIRF-1 mRNA-directed shRNAs. All the shRNAs were able to deplete vIRF-1 in both cell types, including in lytically induced cells expressing higher levels of the viral protein (Fig. 4A). Depletion of vIRF-1 had no detectable effect on either vIRF-3 or USP7 levels, assayed along with vIRF-1, demonstrating lack of influence of vIRF-1 depletion on expression of these interaction partners. Lentiviral vectors specifying the two most effective vIRF-1-specific shRNAs (3 and 5) or NS control shRNA were used to infect BCBL-1 and JSC-1 cultures, and cell growth and apoptosis were determined via hemocytometric counting and annexin V-Cy3 staining, respectively. Each of the vIRF-1 shRNAs led to reduced PEL cell growth and to increased rates of apoptosis, tested on day 3 of the growth assay (initiated 2 days after shRNA transduction) (Fig. 4B). These data identified, for the first time, a latency function of vIRF-1, similar to that reported for vIRF-3 (47).
FIG 4.
Contribution of vIRF-1 to PEL latency. (A) Testing of vIRF-1 mRNA-directed shRNAs (designated 3, 4, and 5) for their vIRF-1 depletion efficacies in lentivirus-transduced PEL cells. Two days after lentiviral infection, cells were left untreated (latent) or treated with either doxycycline (Dox; 1 μg/ml) for TRExBCBL1-RTA cells or sodium butyrate (0.5 μM) and TPA (20 ng/ml) for JSC-1 cells to induce lytic replication; all the cells were harvested 48 h later. The cell lysates were immunoblotted to detect vIRF-1, vIRF-3, USP7, and GAPDH (loading control). (B) Lentiviral vectors expressing two of the vIRF-1-specific shRNAs (3 and 5) or NS control shRNA were used to infect TRExBCBL1-RTA and JSC-1 cultures, and after 48 h, cell densities were normalized and growth monitoring of triplicate cultures was initiated (day 0). Densities of viable, trypan blue-excluding cells were calculated daily for 3 days (1 to 3). On day 3, culture samples were analyzed by annexin V-Cy3 binding for detection of apoptotic cells and by Hoechst staining for detection of cell nuclei, and the percentage of Cy3+ cells within each population was calculated. The data were collected from multiple random fields (>100 total cells) for each sample. All growth and apoptosis data are shown as average values derived from triplicate cultures; the error bars show standard deviations from the mean values. P values (unpaired, two-tailed t test) are shown for NS versus vIRF-1 depletion.
vIRF-1 activities mediated through USP7 interaction.
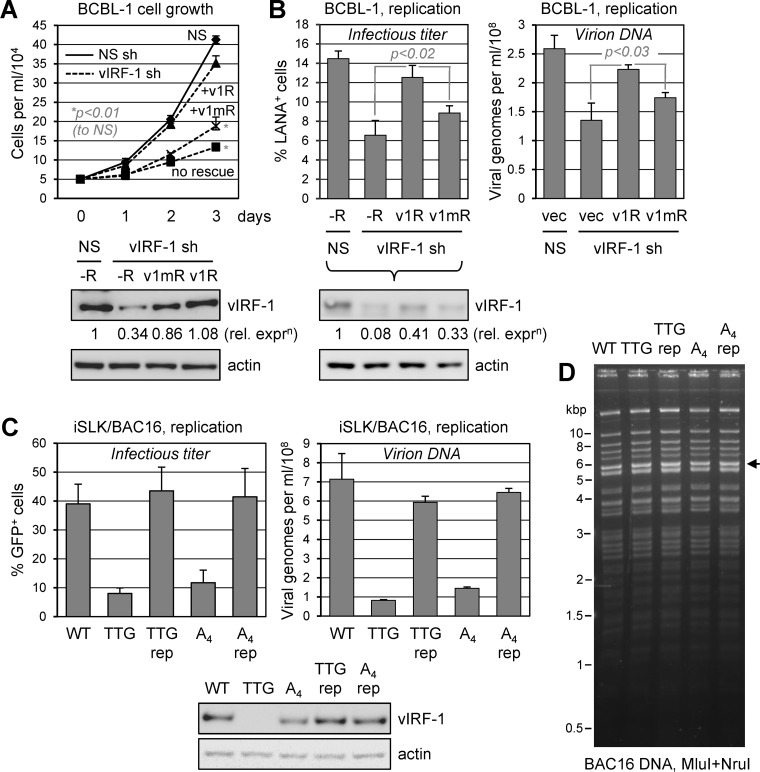
To determine the functional significance of USP7 targeting by vIRF-1, we first undertook the approach of vIRF-1 depletion and functional “rescue” with either wild-type vIRF-1 or vIRF-1 with the USP7 binding site mutated. The reintroduced vIRF-1 sequences were mutated (codon synonymously) to prevent recognition and cleavage by the selected shRNA used for endogenous vIRF-1 depletion. For growth/viability assays, we transduced BCBL-1 (TREx-RTA) cells with empty lentiviral vector (control) or vectors encoding either wild-type or USP7-refractory (EGPS→AAAA) vIRF-1 proteins via lentivirus infection and cointroduced vIRF-1 shRNA (carried on the same vectors) to selectively deplete virus-produced vIRF-1. Growth assays were performed as outlined for Fig. 4B. As before, vIRF-1 depletion alone (unrescued) led to substantially reduced rates of PEL cell growth. This could be prevented by exogenously introduced vIRF-1 (v1R), but not by EGPS-mutated vIRF-1 (v1mR), which only marginally relieved the effects of endogenous vIRF-1 depletion (Fig. 5A). These data reveal the biological significance, in latency, of USP7 targeting by vIRF-1.
FIG 5.
Roles of USP7 targeting by vIRF-1 in latent and lytic biology. (A) TRExBCBL1-RTA cells were transduced with either NS shRNA or vIRF-1 shRNA (no. 3), with the latter transduced either alone or together with shRNA-resistant expression cassettes for wild-type vIRF-1 (v1R) or USP7-refractory vIRF-1 (EGPS→AAAA mutated; v1mR). Lentivirus-infected cells were normalized (to 5 × 104 cells/ml) 3 days after transduction and monitored for growth daily over the next 3 days. The data are shown as average values from duplicate cultures for each condition; the error bars show deviations from the mean values. P values (two-tailed Student's t test) are indicated for “no rescue” and “v1mR” rescue relative to NS at day 3. Immunoblotting of terminal cell lysates (day 3) for vIRF-1 and β-actin (loading control) confirmed depletion of vIRF-1 (in the absence of rescue [−R]) and expression of complementing vIRF-1 proteins. (B) An equivalent depletion-rescue approach was used to assess the specific contribution of USP7–vIRF-1 interaction to HHV-8 productive replication in TRExBCBL1-RTA cells. Here, following shRNA transduction with or without coexpression of shRNA-resistant vIRF-1 (v1R) or USP7-refractory vIRF-1 (EGPS→AAAA; v1mR), culture media were harvested 96 h after doxycycline-induced lytic replication for quantitation of infectious virus (by LANA staining of inoculated iSLK cultures) (left) and encapsidated viral DNA (by qPCR) (right) (see Materials and Methods). Data from biological duplicates were compiled and analyzed as for panel A. Immunoblotting of PEL cell lysates confirmed appropriate depletion of endogenous vIRF-1 (vIRF-1 shRNA [sh] without rescue [−R]) and expression of complementing vIRF-1 proteins (relative to β-actin, used for normalization). (C) The contribution of USP7 targeting by vIRF-1 to productive replication was also assessed in iSLK cells infected with wild-type, vIRF-1-null (initiator ATG→TTG), USP7-refractory (EGPS→AAAA [A4]), or mutant-to-wild-type reverted (repaired; rep) BAC16 viruses. Lytic reactivation was induced by treatment of the cultures with doxycycline (1.9 nM) and sodium butyrate (1 mM). Media were harvested after 4 days for quantitation of infectious virus (percent GFP+, virus-infected, inoculated iSLK cells) and encapsidated viral DNA (qPCR-determined genome copy numbers) (see Materials and Methods). The data were derived from duplicate cultures; average values are shown, along with error bars indicating standard deviations from the mean values. Immunoblotting for vIRF-1 and β-actin (loading control) in terminal cell lysates confirmed appropriate knockout and expression of vIRF-1 in the mutated and repaired viruses. (D) Restriction analyses of wild-type and recombinant BAC16 genomes, showing identical restriction patterns and overall genome integrity. The arrow indicates the position of the restriction fragment (5,720 bp) containing the vIRF-1 gene; marker positions are indicated on the left.
The same depletion-rescue approach was used to assess the specific contribution of vIRF-1–USP7 interaction to vIRF-1 proreplication activity. TRExBCBL1-RTA cells were used to enable high-efficiency lytic induction. As expected, the exogenous introduction of native vIRF-1 (shRNA resistant) into vIRF-1 shRNA-cotransduced cultures led to recovery (∼86%) of replication titers suppressed (to <53%) by vIRF-1 depletion alone (Fig. 5B). However, USP7-refractory vIRF-1 could not effectively rescue this phenotype, although it was expressed at similar, albeit somewhat reduced, levels relative to wild-type vIRF-1 (Fig. 5B, immunoblots). These data indicate the importance of vIRF-1–USP7 interaction for the proreplication activity of vIRF-1 in this cell type.
To provide an independent means of assessing the contribution of vIRF-1 targeting of USP7 to HHV-8 productive replication, we used BAC16 (bacmid) HHV-8 (46), wild-type and vIRF-1 ORF-mutated, and infected iSLK cells for phenotypic analyses. EGPS→AAAA and initiator-methionine codon (ATG→TTG) mutations of the vIRF-1 ORF in the bacmid genome were individually introduced using established bacterium-based techniques of seamless recombinant engineering (48, 49) (see Materials and Methods). Each of the mutated viral genomes was repaired by reversion of the mutations to wild-type sequences to control for potential phenotypically significant genetic alterations that might have occurred during targeted mutagenesis. Viruses generated from BAC16 genome-transfected and reactivated (doxycycline- and sodium butyrate-induced) iSLK cells (46) were used to reinfect, using identical infectious doses, naive iSLK cells to produce “normalized” latently infected iSLK cultures to be used experimentally. These cultures were lytically induced, and virus yields produced over 4 days were assessed by infectious assay (expression of virus-encoded GFP in inoculated naive iSLK cells) and qPCR quantitation of encapsidated (DNase I-resistant) viral genomes released into the culture media (Fig. 5C). This experiment identified the importance of vIRF-1 and its interaction with USP7 for productive replication in iSLK cells, as in PEL (TRExBCBL1-RTA) cells. Thus, the proreplication activity of vIRF-1 and the biological importance of its interaction with USP7 appear to be general phenomena.
Influence of vIRF-3 on HHV-8 productive replication.
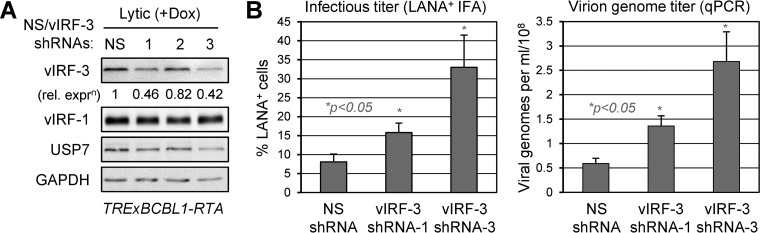
Although vIRF-3 has been reported to be required for PEL cell viability on the basis of depletion studies (47), its potential role in virus productive replication has not been investigated in this or other cell types. To do this, we utilized shRNA-expressing lentiviral vectors to deplete vIRF-3 in TRExBCBL1-RTA cells. The virus yields produced from cultures transduced with each of the two most effective vIRF-3-directed shRNAs (1 and 3) (Fig. 6A) were compared to titers of virus produced from cultures transduced with NS control shRNA. Depletion of vIRF-3 led to a 2- to 4-fold increase in the yield of infectious virus and nucleated virions released into the culture medium (Fig. 6B). Similar results, including the enhanced effects of shRNA-3, were obtained in two repeat experiments (data not shown). Our results indicate that vIRF-3 suppresses productive replication of HHV-8.
FIG 6.
Influence of vIRF-3 on productive replication. (A) Testing of lentiviral vector-transduced shRNA efficacies in lytically reactivated (doxycycline-treated) TRExBCBL1-RTA cells. Expression of vIRF-3 in lentivirus-infected cells was determined by immunoblotting of cell extracts derived from test (vIRF-3 shRNA) versus control (NS shRNA) transduced cultures following 48 h of lytic induction (48 h after lentiviral infection). GAPDH immunoblotting provided a protein-loading control, and vIRF-1 and USP7 levels were also monitored. (B) In a similar experiment, culture media were harvested 4 days after doxycycline treatment and analyzed for infectious virus and encapsidated virion DNA titers via inoculation of naive iSLK cultures and immunofluorescence detection of LANA (left) or viral genome-specific qPCR (right), respectively (see Materials and Methods). The data were derived from duplicate cultures, and average values are shown; the error bars indicate standard deviations from the mean values. Student's t test-derived P values (NS versus depletions) are indicated.
Latent and lytic functions of vIRF-3 via USP7 targeting.
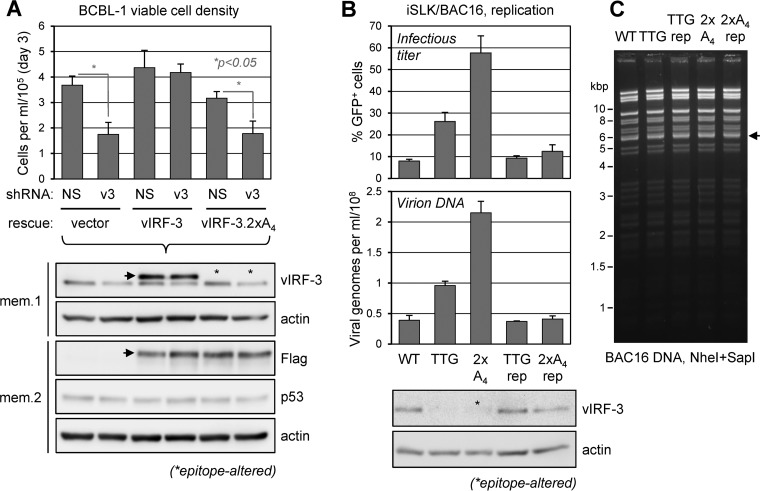
The significance of vIRF-3 interaction with USP7 in PEL cell latency was investigated using depletion-rescue, analogous to that used for vIRF-1 (see above). Consistent with previously published data (47), shRNA-mediated depletion of vIRF-3, in the absence of shRNA-resistant vIRF-3 complementation, led to a marked reduction of BCBL-1 viable-cell density (>50% at day 3 postnormalization), but this effect was abrogated in the presence of exogenously supplied vIRF-3 (Fig. 7A). In contrast, the USP7-refractory variant of vIRF-3 (2× EGPS→AAAA [2×A4]) was unable to rescue the vIRF-3 depletion phenotype. Lentivirus-transduced wild-type and USP7-refractory vIRF-3, both tagged with Flag (and StrepII), were expressed at equivalent levels and similarly to endogenous vIRF-3 (Fig. 7A, immunoblots). The data in Fig. 7A reveal that vIRF-3 interaction with USP7 contributes significantly to the proviability activity of vIRF-3 in latently infected PEL cells.
FIG 7.
USP7 targeting in latent and lytic activities of vIRF-3. (A) Depletion-rescue analysis similar to that for Fig. 5A was carried out. Here, shRNA-resistant wild-type and USP7-refractory (2×A4) vIRF-3–SF expression cassettes or empty vector was first transduced into separate TRExBCBL1-RTA cultures by lentiviral vector infection; after 48 h, the cultures were further transduced with lentiviral vectors expressing either NS (control) or vIRF-3 mRNA-directed (v3) shRNA. Following 48 h of rest, cell densities were normalized to 5 × 104 cells per ml and measured after 3 days by hemocytometric counting of trypan blue-excluding, viable cells. The data were derived from duplicate cultures; average values from the replicates are shown, along with standard deviations from these values. Student's t test-derived P values are shown for NS versus vIRF-3 depletion for the control (vector) and vIRF-3.2×A4-rescued samples; there were no significant differences between NS and vIRF-3 depletion for the wild-type vIRF-3-rescued samples. Samples of cell lysates were analyzed by vIRF-3 and Flag epitope immunoblotting to verify equivalent expression of complementing wild-type and EGPS-mutated vIRF-3–SF, their expression relative to virus-produced vIRF-3, and endogenous vIRF-3 depletion. Membranes 1 and 2 (mem.1 and mem.2) were probed for β-actin (loading control) and the latter also for p53. The arrows on the vIRF-3 and Flag blots indicate vIRF-3–SF; the asterisks on the former indicate that vIRF-3.2×A4 was not immunoreactive with the vIRF-3 antibody. (B) Analysis of vIRF-3 function in lytic replication was assessed in doxycycline/sodium butyrate-treated iSLK cultures infected with wild-type (WT), vIRF-3-null (TTG; initiator ATG mutant), USP7-refractory vIRF-3 mutant (2×A4), and corresponding revertant (repaired [rep]) HHV-8 BAC16 viruses. Infectious virus and encapsidated genome titers in culture media harvested 4 days postinduction were determined as for Fig. 5C. Average values from duplicate cultures and standard deviations from these values are shown. Knockout of vIRF-3 and appropriate repair of viral genomes with vIRF-3 ORF mutations were verified by immunoblotting. (C) Restriction analyses of wild-type versus recombinant BAC16 genomes showing identical restriction profiles. The arrow indicates the position of the vIRF-3 gene-containing restriction fragment (5,739 bp); migration positions of marker bands are indicated (left).
To test the effects of the USP7 binding by vIRF-3 on HHV-8 productive replication, we compared the replication of wild-type, vIRF-3-null (initiation codon ATG→TTG), and USP7-refractory (2× EGPS→AAAA) vIRF-3-substituted BAC16 HHV-8 genomes in the context of infected and reactivated iSLK cells. “Repaired” viruses were included to control for possible phenotypically significant genetic alterations arising spontaneously during the recombineering. The data from this experiment confirmed that vIRF-3 knockout in this system led to increased virus production, as observed previously in BCBL-1 cells (Fig. 6B), and demonstrated a marked further increase in virus yield by abrogation of USP7–vIRF-3 interaction (Fig. 7B). The two repaired control viruses were essentially phenotypically unaltered with regard to virus production, demonstrating specific effects of the ATG→TTG codon and EGPS→AAAA mutations. These data confirm the generality of vIRF-3-inhibtory effects on lytic replication, seen in both PEL (TRExBCBL1-RTA) and iSLK cells, and demonstrate the antagonistic effect specifically of vIRF-3–USP7 interaction on virus production. The more pronounced positive effect of vIRF-3–USP7 binding abrogation versus vIRF-3 deletion indicates the functional importance of USP7 targeting by vIRF-3 relative to other vIRF-3 interactions and activities, both positive and negative; thus, the positive effect of USP7 binding abrogation alone outweighs the net positive effect of removal of combined vIRF-3 activities.
Influence of USP7 on p53 and vIRF expression.
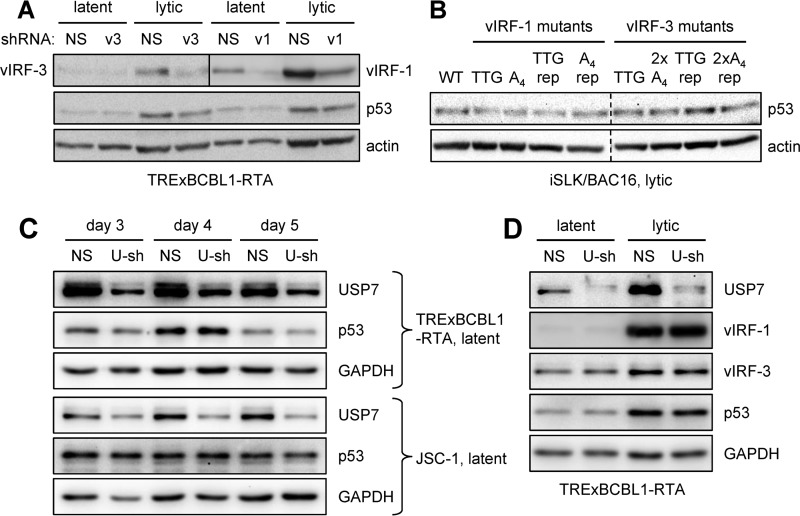
Given that USP7 is known to target both p53 and p53-destabilizing MDM2 (an E3 ubiquitin ligase of p53), resulting in increased or suppressed expression of p53, respectively (depending on conditions) (36); that USP7 is targeted by vIRF-1 and vIRF-3; and that vIRF-3 overexpression and suppression have been reported to modulate levels of active and total p53 in PEL cells (29), we wanted to investigate the effects on p53 levels of vIRF-1, vIRF-3, and USP7 depletion or knockout in PEL and infected iSLK cells. We first analyzed the effects of vIRF-1 and vIRF-3 depletion in latent and lytically induced TRExBCBL1-RTA cells on p53 expression, as determined by immunoblotting of extracts of cell cultures transduced with NS (control), vIRF-1, or vIRF-3 shRNA. This experiment provided no evidence of altered p53 expression (relative to an actin normalization control) as a function of either vIRF-1 or vIRF-3 suppression (Fig. 8A). Indeed, the data for vIRF-3 matched those obtained in our PEL cell viability experiment (Fig. 7A, immunoblots). The complementary approach of vIRF-1 or vIRF-3 deletion in the context of lytically reactivated BAC16-infected iSLK cells also revealed no p53 expression changes as a function of vIRF-1 or vIRF-3 (Fig. 8B). Data from this experiment, which incorporated EGPS motif-null forms of vIRF-1 and vIRF-3, also detected no changes in p53 expression due to abrogation of USP7 targeting by these vIRFs. Together, these data indicate that endogenously expressed vIRF-1 and -3 and their targeting of USP7 have no detectable effects on p53 expression, in contrast to biological phenotypes identified by the same experimental approaches.
FIG 8.
Expression of p53 and vIRFs in response to vIRF and USP7 depletion. (A) Depletion of either vIRF-1 or vIRF-3 in TRExBCBL1-RTA and JSC-1 cells was achieved through lentiviral vector-delivered shRNAs (v1 and v3, versions 3; see Fig. 4A and 6A). Forty-eight hours posttransduction, cell extracts were analyzed by immunoblotting for expression of p53, in addition to vIRF-1 and vIRF-3 (to confirm depletion) and β-actin (loading control). The upper membrane was cut (vertical line) for independent detection of vIRF-1 (right) and vIRF-3 (left). (B) Similar immunoblot analysis of p53 expression in cell extracts derived from lytically reactivated (doxycycline- and sodium butyrate-treated) BAC16-infected iSLK cells harvested 96 h postinduction. Wild-type, EGPS motif-mutated vIRF-1 (A4) and vIRF-3 (2×A4), and repaired (rep) viruses were used for parallel infections of iSLK cultures to allow comparisons. The dashed lines indicate flipping and repositioning of the vIRF-3 section of the blots to match the order of the vIRF-1 section. (C) TRExBCBL1-RTA and JSC-1 PEL cells were infected with lentiviral vectors expressing either NS (control) shRNA or USP7 mRNA-directed shRNA (U-sh). After 48 h, the cells were passaged and seeded to provide replicates, which were harvested at different times for analysis of p53 expression and verification of USP7 depletion by immunoblotting. GAPDH provided the protein-loading and membrane transfer control for the harvested samples. (D) Expression of vIRF-1, vIRF-3, and p53 as a function of USP7 depletion was analyzed in latently infected and lytically reactivated TRExBCBL1-RTA cells. Here, cells were transduced with either NS or USP7 shRNA, allowed to rest for 48 h, and then either left untreated or lytically induced with doxycycline and sodium butyrate. Latent and lytically reactivated PEL cell cultures were harvested after 2 days for analysis of vIRF-1, vIRF-3, and p53 expression and verification of USP7 depletion by immunoblotting; GAPDH provided the protein-loading and membrane transfer control.
To examine the influence of USP7 on p53 expression in PEL cells, TRExBCBL1-RTA and JSC-1 cells were transduced with USP7-specific shRNA, or NS shRNA control, by infections with the respective lentiviral vectors and allowed to rest for 2 days, and then USP7 depletion and p53 expression were monitored for a further 3 days. While USP7 depletion was effective in both cell lines, p53 expression was not detectably affected (Fig. 8C). These findings indicate that USP7-mediated regulation of p53 expression is unlikely to be the cause of the reduced cell growth and viability effected by USP7 depletion (Fig. 3B). We further investigated, in both latent and lytically reactivated TRExBCBL1-RTA cells, whether USP7 depletion affected expression of its interaction partners, vIRF-1 and vIRF-3. As both vIRFs support PEL cell viability (47) (Fig. 4), their possible destabilization by USP7 depletion (which conceivably could promote proteasomal degradation of vIRF-1 and/or -3) could account for the USP7 depletion phenotype we observed in latent PEL cells (Fig. 3B). Similarly, vIRF-1 destabilization could account for the lytic USP7 depletion phenotype (Fig. 3C). However, USP7 depletion caused little or no change in either vIRF-1 or vIRF-3 levels, or in p53 expression (as before) (Fig. 8D). Therefore, the expression of the USP7 binding partners vIRF-1 and vIRF-3, in addition to p53, was unaffected by USP7 depletion under the same conditions that led to latent and lytic phenotypes in these cells.
DISCUSSION
The data presented here show, for the first time, the interaction of HHV-8 vIRF-3 with USP7 and the biological significance to HHV-8 latency and productive replication of USP7 targeting by vIRF-3 and vIRF-1. An elegant structural analysis of vIRF-1 interaction with USP7 was reported previously, along with transfection-based assessment of the intracellular association of these proteins (30). We have shown here that vIRF-3 contains two copies of the central motif, EGPS, involved in USP7 targeting by vIRF-1 and certain other binding partners, including HHV-8 ORF45 protein and EBV EBNA1 (40, 44), and that each of the motifs in vIRF-3 can support USP7 interaction. The functional analyses here have focused on evaluation of the effects of mutation of the single EGPS motif in vIRF-1 or of both motifs in vIRF-3, sufficient to abrogate USP7 interactions by each vIRF, on latency and productive replication in the context of naturally infected PEL cells and HHV-8 BAC16-infected iSLK cells. Therefore, we have analyzed the biological consequences of vIRF-1/3–USP7 interactions, specifically, in addition to vIRF-1/3 depletion or knockout, affecting global activities of each of the vIRFs.
Previous investigations of vIRF–cellular-protein interactions have largely involved the analysis of binding and function in the context of transfected cells and overexpressed proteins. Examples of interactions identified and analyzed in this way for vIRF-1 and vIRF-3 are provided in the introduction. Of particular relevance here is the report by Chavoshi et al. (30) in which the effects of wild-type and USP7 binding-refractory vIRF-1 protein overexpression were assessed in U2OS osteosarcoma cells, revealing the suppression of endogenous p53 levels by wild-type vIRF-1, but not by a USP7-refractory variant (with residues 45 to 50 deleted); these data indicate that vIRF-1 interaction with USP7 is involved in p53 suppression. With regard to vIRF-3, it has been reported that overexpression of the protein in HCT-116 colon carcinoma cells leads to reduced p53 expression, and analyses in transfected HEK293 cells indicated that vIRF-3 expression correlates with increased polyubiquitination of p53 (29). The same study also found that vIRF-3 overexpression and vIRF-3 depletion in BC-3 PEL cells in the presence of doxorubicin-induced DNA damage led, respectively, to decreased and increased levels of total and active p53. It is notable, however, that our own data detected no effects on p53 levels of vIRF-1 or vIRF-3 depletion in latently or lytically infected BCBL-1 cells or of vIRF-1 or vIRF-3 deletion in lytically reactivated BAC16+ iSLK cells (Fig. 8). This was also evident in iSLK cells lytically infected with BAC16 mutants encoding USP7-refractory variants of vIRF-1 and vIRF-3 and in latent BCBL-1 and JSC-1 PEL cells depleted of USP7. These data suggest that regulation of p53 levels via vIRF-1 and vIRF-3 interaction with USP7, or other proteins (including p53 itself), may not be biologically significant, or at least universal. We have identified clear latent and lytic phenotypes associated with USP7 and its interactions with both vIRF-1 and vIRF-3, but our data indicate that they are unlikely to be mediated through regulation of p53 expression. Notably, neither can the detected proviability (latency) and replication phenotypes associated with vIRF-1 and vIRF-3 interactions with USP7 be attributed to USP7-regulated expression of vIRF-1 and vIRF-3. The vIRF protein levels were not detectably or substantially altered as a function of USP7 depletion in PEL cells or by mutation of the USP7 binding sites of vIRF-1 and vIRF-3, as assessed in transfected cells, in depletion-rescue experiments in PEL cells and, for vIRF-1, in BAC16 mutant-virus-infected iSLK cells (the untagged vIRF-3 EGPS variant was not immunoreactive and therefore was unable to be detected in the BAC16 system). However, it remains to be determined whether USP7 interactions with vIRF-1 and vIRF-3 affect the ubiquitination status of the proteins, which may alter their activities. Although proreplication activity of vIRF-1 has been reported previously, in the contexts of reactivated PEL cells and HHV-8-infected endothelial cells (21, 24), the present data identify vIRF-1 interaction with USP7, specifically, as important for efficient virus production in PEL and iSLK cells.
To our knowledge, our data are the first to show a function of vIRF-1 in latency and of vIRF-3 in productive replication. With respect to the former, it is known that vIRF-1 is expressed at low levels in latently infected PEL cells and induced upon lytic reactivation (13, 15). Immunofluorescence imaging has shown that in latency, vIRF-1 is predominantly nuclear and associated with promyelocytic leukemia (PML) bodies, but in lytic replication it exhibits more disseminated expression, including cytoplasmic localization (15). Therefore, the profiles of vIRF-1 targets and activities in latency and productive replication are likely to be distinct, although probably overlapping. It is notable that USP7 can localize to PML bodies and that USP7 is associated with PML body disruption, although independently of its deubiquitinase activity (50). However, whether our finding of vIRF-1 promotion of latent PEL cell growth and survival is related to PML body-localized or -related activities of USP7 remains to be determined. With respect to vIRF-3 activity in lytic replication, we have determined in the context of both PEL cells and BAC16-infected iSLK cells that vIRF-3 has inhibitory activity; depletion or knockout, respectively, led to notably increased virus yields from both cell types. This phenotype, at least in iSLK cells, involved USP7 targeting by vIRF-3, as a viral mutant encoding a USP7-refractory variant in place of wild-type vIRF-3 had increased virus yields. In fact, these yields substantially exceeded those derived from parallel cultures infected with vIRF-3 knockout virus. This can be explained if one considers that there are multiple interactions and activities of vIRF-3, both positive and negative with respect to productive replication, whose net effect is negative and to which USP7 contributes substantially. Thus, specific abrogation of this interaction and function of vIRF-3 could allow predominance of the positively acting vIRF-3 interactions over (remaining) replication-inhibitory interactions to effect a large increase in virus yield. Determining the individual contributions of the various vIRF-3 interactions and associated activities to lytic replication must await a more detailed understanding of and ability to specifically disrupt these interactions in the context of infection. Nonetheless, our data identify the negative influence of vIRF-3 and its targeting of USP7 on productive replication.
The importance of USP7 catalytic activity in latency was demonstrated previously by the elegant studies of Lee et al. (31). In that report, the USP7 catalytic-domain-interacting region of HHV-8 vIRF-4 was able to induce PEL cell death when introduced intracellularly as a cell-permeable peptide. Our data have confirmed, by the independent method of USP7 depletion, the importance of USP7 for PEL cell growth and viability and, further, demonstrate an overall positive role of USP7 (notwithstanding the negative effects of its targeting by vIRF-3) in HHV-8 productive replication. However, the underlying mechanisms involved in these prolatency and proreplication functions of USP7 have yet to be determined. There is evidence from the innovative studies of Gillen et al. (44) that ORF33 tegument protein stabilization through its interaction with USP7-bound ORF45 tegument protein may be one means by which USP7 contributes to productive replication. Noncatalytic functions of the protein, such as PML body disruption, could conceivably be involved. Our data show that p53 levels are not detectably altered by USP7 depletion, indicating that the corresponding phenotypic effects are independent of this pathway. Furthermore, vIRF-1 and vIRF-3 depletion in latent and lytic PEL cells or ablation in lytically reactivated BAC16+ iSLK cells also had no effect on p53 expression. Although the molecular bases of the biological activities of vIRF-1 and vIRF-3 via USP7 interaction are unknown, one possibility is that the viral proteins, like HHV-8 ORF45 protein (44), help to recruit USP7 substrates through vIRF-substrate and -USP7 complexing. Other possibilities include vIRF modulation of USP7 activity and/or substrate specificity or accessibility. For lytically expressed vIRF-4, which contacts and apparently inhibits the catalytic site of USP7, such modulation seems apparent, but it is unlikely that this would mirror the cell death-inducing activity exhibited by the isolated USP7-interacting region of vIRF-4 (31), which would be expected to be highly detrimental to lytic infection by HHV-8. Indeed, vIRF-4 also interacts with and stabilizes p53-destabilizing MDM2 E3 ligase, and this interaction, rather than USP7 binding, appears to dominate with respect to p53 regulation in PEL cells (31). It is possible that, in the context of full-length vIRF-4, the sequence binding to the catalytic domain of USP7 acts to fine tune its activity to provide conditions conducive to productive replication via prosurvival or other activities. Further analyses of vIRF-1, vIRF-3, and vIRF-4 are required to dissect the individual and combined roles of USP7 targeting by the virus-expressed proteins in the context of infected cells and the mechanisms underlying these activities.
In summary, here, we have presented analyses of vIRF-1 and vIRF-3 latent and lytic activities and the specific contributions of USP7 targeting by these vIRFs to latent cell viability and productive replication. The data have revealed proviability activity of vIRF-1 in latently infected PEL cells, the role of vIRF-1–USP7 interaction for this function, the negative influence of vIRF-3 on HHV-8 productive replication, and the significant contributions of USP7 targeting by vIRF-1 and vIRF-3 to the respective positive and negative effects of these vIRFs on lytic replication. This study extends our understanding of the biological activities of vIRF-1 and vIRF-3 and reveals the significance of their targeting of USP7, specifically, during infection.
MATERIALS AND METHODS
Cell culture and gene transduction.
TRExBCBL1-RTA (45) and JSC-1 (51) cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 10 μg/ml gentamicin. HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 10 μg/ml gentamicin. For transfections, HEK293T cells were passaged and plated at approximately 50% confluence. Transfections were conducted 16 to 18 h postseeding with mixtures of plasmid DNA and cationic polymer linear polyethylenimine (593215; Polysciences). Cells harvested 48 h posttransfection were lysed with immunoprecipitation (IP) buffer (50 mM Tris [pH 7.4], 5 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, protease inhibitor cocktails [P8340; Sigma]), and lysate supernatants were collected following centrifugation at 13,000 rpm for 15 min at 4°C. For production of lentiviruses, HEK293T cells were transfected with lentiviral vector together with psPAX2 and pMD2.G (Addgene; 12260 and 12259, respectively; deposited by D. Trono) encoding gag-pol and vesicular stomatitis virus G proteins. After 6 h of transfection, the culture media were replaced by 10% FBS-supplemented DMEM. Virus-containing media were harvested 48 h later and concentrated by ultracentrifugation at 80,000 × g to pellet the virus. The viral pellets were resuspended in RPMI 1640 medium supplemented with 10% FBS, and aliquots were stored at −80°C. Virus titers for gene knockdown were determined by assessing percentages of GFP+ cells at different inoculation doses prior to experimental use. Infectious titers of lentiviruses encoding Flag epitope-tagged versions of vIRF-1 and vIRF-3 were determined by Flag-based immunofluorescence assays.
Cell growth and apoptosis assays.
For depletion of vIRF-1, vIRF-3, and USP7 in TRExBCBL1-RTA and JSC-1 PEL cells, mRNA-directed or NS (control) shRNA-expressing lentiviral vectors were used for infection-mediated transduction. Lentiviral infections were carried out by incubation of PEL cultures with lentiviral inoculum in the presence of 5 μg/ml Polybrene for 6 h prior to replacement of the inoculation medium with fresh medium (1640 supplemented with 10% FBS). Transduction efficiencies of >90% were verified by UV microscopy for visualization of GFP, encoded by the lentiviral vector. The cells were allowed to rest for 2 to 3 days prior to cell density normalization and experimental use. For growth assays, PEL cells were seeded at a density of 0.5 × 105 to 1.0 × 105 cells per ml in wells of a 24-well plate, and cell densities were monitored daily by hemocytometric counting of trypan blue-excluding (viable) cells. For apoptosis assays, cells were pelleted by centrifugation and washed with annexin V binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, and 2.5 mM CaCl2), and annexin V-Cy3 reagent (BioVision; catalog number 1002-200) was added to the cells at a dilution of 1:500 in binding buffer for 5 min in the dark. The cells were then washed twice by suspension in binding buffer and pelleting by centrifugation prior to fixing of the cells and staining of nuclei by incubation in the dark for 15 min in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (PFA) and Hoechst 33342 stain (1 μg/ml). The cells were washed in PBS, resuspended in PBS containing 4% PFA, and transferred to a microscope slide; after desiccation, the cells were overlaid with 90% glycerol and a coverslip for visualization of Cy3 (annexin V; red) fluorescence and Hoechst-counterstained (blue) nuclei. Random fields (3 or 4; >100 total cells) were analyzed to calculate the percentage of annexin V+ cells among total (Hoechst stain-positive) cells.
Plasmid and lentivirus clones.
Plasmid pQHA-USP7 expressing HA epitope-tagged USP7 was obtained from G. Maertens and G. Peters through Addgene (plasmid 46753) (52). Lentiviral plasmid vector pCEBZ-USP7-CBD was generated by insertion of PCR-generated, NotI- and BamHI-flanked USP7 coding sequences into the corresponding cloning sites of pCEBZ-RFP-CBD (53), replacing the red fluorescent protein (RFP) ORF. Lentiviral vectors pCEBZ-vIRF-1-SF and pCEBZ-vIRF-3-SF expressing StrepII- plus Flag-tagged vIRF-1 and vIRF-3 were generated by PCR amplification of the respective vIRF genomic sequences from BAC36 using gene-specific forward and reverse primers and cloning of the terminally NotI- and BamHI-digested PCR products between the NotI and BamHI sites of lentiviral vector pCEBZ-SF (54). Sequences encoding shRNA-resistant vIRF-1 were cloned downstream of the Ubc promoter in a pCEBZ-based vector with the cytomegalovirus (CMV) promoter replaced by a Ubc promoter, pUbc-sEF1a-Puro. The resulting vector was used as a template for PCR amplification of Ubc promoter–vIRF-1 sequences for cloning as an NheI-SpeI fragment between the corresponding restriction sites of the lentiviral vector pYNC352 containing vIRF-1-specific shRNA (sh3) (see below). vIRF-1 and vIRF-3 sequences encoding proteins with mutated EGPS motifs were generated by overlapping PCR-based mutagenesis using primers containing the respective mutations. Sequences encoding the NTD of USP7 were cloned as a BamHI-EcoRI fragment into pET28b(+) (Novagen; catalog number 69865) to provide a vector for bacterial expression of USP7-NTD as an N-terminally His6-tagged fusion protein. Bacterial expression vectors for N-terminally GST-fused wild-type and EGPS→AAAA (2×A4) mutated residues 181 to 223 of vIRF-3 were generated by cloning the respective peptide-coding sequences as PCR-derived BamHI-EcoRI fragments between the BamHI and EcoRI cloning sites of the vector pGEX-4T-1 (GE Healthcare; catalog number 28-9545-49). Gene-specific shRNA sequences were designed with BLOCK-iT RNAi Designer software (Thermo Fisher Scientific) or siRNA (small interfering RNA) Wizard software (InvivoGen) and cloned into the BamHI and MluI sites of the pYNC352 lentiviral vector containing the H1 polII promoter for shRNA expression and a CMV promoter-driven GFP expression cassette. The target sequences incorporated into shRNAs for vIRF-3 depletion were as follows: GCGTTCAGTTACATGTGAT (sh1), GATCTACTGGAACATTTCTTG (sh2), and GCGATGAAGACCTCTTTGATC (sh3). The target sequences for vIRF-1 depletion were GGATAGTATGTCAAGTCAACA (sh3), GATTGGCAAAGGCATTTCTCA (sh4), and GGCATTCTGCTGACTAGCTCT (sh5). The target sequences for USP7 depletion were ACCCTTGGACAATATTCCT (sh1), AGTCGTTCAGTCGTCGTAT (sh2), and GGCAACCTTTCAGTTCACT (sh3). To generate shRNA-resistant vectors, codon-synonymous mutations were introduced into vIRF-1 and vIRF-3 ORFs corresponding to shRNA target sites (of vIRF-1 sh3 and vIRF-3 sh1) by overlapping PCR-mediated mutagenesis. For vIRF-1, the modified ORF-containing PCR product was inserted into the PacI and KpnI sites of pUBC-sEF1a-Puro and subsequently transferred, as a Ubc-vIRF-1 fragment, between the NheI and SpeI sites of pYNC532-vIRF-1sh3 (thereby generating a single vector for endogenous vIRF-1 depletion and coexpression of exogenous, shRNA-resistant wild-type or EGPS-mutated vIRF-1). For vIRF-3, the shRNA-resistant coding sequences were cloned between the NotI and BamHI sites of pCEBZ-SF for separate lentivirus-mediated transduction of wild-type or USP7-refractory shRNA-resistant expression cassettes.
HHV-8 mutagenesis.
Genetically altered HHV-8 genomes were generated by using the HHV-8 bacmid BAC16 (46) and Red recombinase-mediated recombination in the context of bacterial strain GS1783 (48). Two-step recombination involved the introduction of the desired mutation together with a kanamycin resistance (Kanr) gene cassette into BAC16 via Red-mediated recombination between the insertion fragment (PCR product) and the target locus (vIRF-1 or vIRF-3 gene) in BAC16 and then I-SceI restriction-induced intramolecular excision of the Kanr cassette by virtue of PCR primer-introduced flanking homologous sequences. GS1783 contains separately inducible expression of Red and I-SceI, effecting the first and second recombination steps, enabling seamless mutagenesis in the target genome through appropriate design of the PCR product used for mutagenesis (49). Recombination at each step was checked by PCR for insertion and excision, respectively, of the Kanr cassette, and locus-directed sequencing was performed to establish introduction of the appropriate mutations and the integrity of sequences flanking the locus of genetic recombination. Gross genome integrity was established through the use of restriction enzyme digestion and agarose gel analysis of digestion profiles, which were identical for engineered and wild-type BAC16 genomes. For introduction of initiator-ATG mutations (underlined) (ATG→TTG) in vIRF-1 and vIRF-3 ORF sequences, mutagenic primers spanning the ATG codons and containing common internal sequences, unique 5′ sequences homologous to HHV-8 sequences 5′ and 3′ of the common (overlapping) sequence, and 3′-terminal sequences corresponding to the 5′ end or complementary to the 3′ end of the Kanr gene in pEPkan-S (49) were generated according to the design of Tischer et al. (49). The forward and reverse primers used for ATG mutagenesis in vIRF-1 were as follows: vIRF-1(ATG→TTG) forward, 5′-CACTGGACATTGCGGCGCGAGCTAGTCTGGTTGCGGGACATTGGACCCAGGCCAAAGACCAGGATGACGACGATAAGTAGGG-3′; vIRF-1(ATG→TTG) reverse, 5′-CCAGGCGCCCCAAAAGGGTTCGGTCTTTGGCCTGGGTCCAATGTCCCGCAACCAGACTAGCCAACCAATTAACCAATTCTGATTAG-3′. Equivalent primers containing wild-type sequences were used for reversion of the mutation to generate the repaired control virus. Primers for ATG mutagenesis in the vIRF-3 ORF were vIRF-3(ATG→TTG) forward, 5′-GTGGGGTAATGCTCGGAGGCAGACCATTCTGACAGGTCAACTTGGCGGGACGCAGGCTTACCAGGATGACGACGATAAGTAGGG-3′, and vIRF-3(ATG→TTG) reverse, 5′-ACAATAAACTCAGAAATCCAGGTAAGCCTGCGTCCCGCCAAGTTGACCTGTCAGAATGGTCCAACCAATTAACCAATTCTGATTAG-3′ (boldface, introduced mutation; underlined, homologous to Kanr gene-flanking sequence). Corresponding primers containing a single base change were used for T→A reversion to the wild-type initiator ATG in the vIRF-3 ORF. Similar primers and strategies were used to introduce and repair alanine substitutions in the EGPS motif of vIRF-1 and in one or both of the EGPS motifs in vIRF-3, using motif codon-spanning mutagenic primers. Wild-type, mutated, and repaired BAC16 genomes were transfected into iSLK cells, and BAC16+ cells were selected by application of hygromycin (2.3 mM) to the cultures for 3 weeks. Virus was recovered from the cultures by treatment with doxycycline (1.9 nM) and sodium butyrate (1 mM), and virus-containing media were harvested after 5 days to provide virus stocks. Equivalent titers of virus were used to infect fresh iSLK cells to provide “normalized” cultures for phenotypic analyses.
Coprecipitation assays and recombinant proteins.
For CBD and Flag affinity precipitation experiments, pCEBZ-CBD-based and pCEBZ-SF-based vectors (5 μg of each) expressing CBD-fused USP7 and SF-tagged vIRF-1 or vIRF-3 (see above) or pCEBZ-USP7-CBD and pQHA-USP7 (52) (5 μg of each) were cotransfected into HEK293T cells in 10-cm dishes. Cells were harvested 2 days posttransfection for preparation of cell lysates via suspension in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 0.2% NP-40) supplemented with protease inhibitor cocktail (Sigma; catalog number P8340). The lysates were incubated with chitin beads (New England BioLabs; catalog number S6651) or Flag antibody beads (Sigma; catalog number M8823) at 4°C overnight, and then the beads and associated proteins were pelleted by microcentrifugation or magnetic capture (Flag) and washed in lysis buffer by repeated resuspension and pelleting prior to protein release by heating/denaturation (CBD-chitin) or addition of 100 ng/ml of 3×Flag peptide (Sigma; catalog number F4799) in lysis buffer (100 μl; ×2) to the beads and analysis of released proteins by SDS-PAGE and Western blotting. To generate recombinant proteins for in vitro coprecipitation assays, Escherichia coli BL21(DE3) cells (55) were transduced with appropriate expression plasmids, producing either GST- or His6-linked proteins. Log-phase cultures were induced for protein expression by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 24 h at 18°C. Bacteria were pelleted by centrifugation and then resuspended in either ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0, adjusted with NaOH), for His6-tagged protein, or PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 [pH 7.4]), for GST fusions. Cell lysis was mediated by addition of lysozyme (1 mg/ml) and sonication, and the lysates were cleared by centrifugation at 12,000 × g for 10 min at 4°C. The extracts were incubated with either Ni-nitrilotriacetic acid (NTA) (Qiagen; catalog number 30210) or glutathione Sepharose (GE Healthcare; catalog number 17-0756-01) beads at 4°C for 1 h prior to centrifugation-mediated sedimentation and washing with wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole; His6 precipitates) or PBS (GST precipitates) and release with His6 elution buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 250 mM imidazole) or glutathione buffer (50 mM Tris-Cl [pH 8.0], 10 mM reduced glutathione). The purified proteins were checked by SDS-PAGE and both Coomassie brilliant blue staining (for purity) and immunoblotting prior to their use in coprecipitation assays. For coprecipitation assays, each GST-tagged peptide or GST alone was incubated on ice for 2 h with His6-USP7NTD (residues 55 to 204) in His6 lysis buffer. The samples were then adjusted to a volume of 1 ml by adding ice-cold His6 lysis buffer containing 30 μl of Ni-NTA beads and incubated on ice for 2 h. The beads were then collected by centrifugation and washed 5 times with ice-cold wash buffer. Finally, the beads and associated proteins were boiled in SDS-PAGE loading buffer (10 min), and proteins were separated by SDS-PAGE and analyzed by Western blotting for detection of GST and His6 and by Ponceau S staining for global protein detection. For IPs of endogenous vIRF-1, vIRF-3, and USP7 proteins, TRExBCBL-1 and JSC-1 cells (latent or lytically reactivated by treatment for 24 h with doxycycline or 12-O-tetradecanoylphorbol-13-acetate [TPA] and sodium butyrate, respectively) were pelleted and washed in PBS prior to lysis in IP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.2% NP-40) containing protease inhibitors (Sigma; catalog number P8340) and sonication. After centrifugation at 14,000 × g for 15 min at 4°C, samples of the cleared lysates were incubated with the relevant primary antibodies (to vIRF-3 or USP7) or antisera (to vIRF-1) for 1 h at 4°C with rotation. Protein A/G agarose (Santa Cruz Biotechnology; catalog number sc-2003) was added, and incubation continued at 4°C overnight. The beads and associated antibodies, target proteins, and binding partners were pelleted by microcentrifugation, rinsed by resuspension and pelleting (three times) in washing buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.2% NP-40), and finally resuspended in denaturing sample buffer (125 mM Tris-HCl [pH 6.8], 1.3% SDS, 0.02% bromophenol blue, 6.7% glycerol, 83 mM dithiothreitol).
Antibodies.
Commercially obtained antibodies were as follows: LANA (Advanced Biotechnologies; catalog number 13-210-100); vIRF-3 (Novus Biologicals; catalog number CM-A807); USP7 (Bethyl Laboratories, catalog number A300-033A [rat], and GeneTex, catalog number GT481 [mouse]); Flag (Sigma; catalog number F1804); CBD (New England BioLabs; catalog number E8034S); β-actin (Sigma; catalog number A5316); GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam; catalog number ab9482); and HA, His6, GST, and p53 (Santa Cruz Biotechnology; catalog numbers sc-7392, sc-803, sc-138, and sc-126, respectively). Rabbit antiserum to vIRF-1 was provided by G. S. Hayward. Horseradish peroxidase (HRP)-conjugated secondary antibody and antibody-binding protein were as follows: enhanced chemiluminescence (ECL) anti-rabbit IgG (GE Healthcare; catalog number NA934V) and mouse IgG(κ) BP-HRP (Santa Cruz; catalog number sc-516102). Normal mouse and rabbit IgGs were obtained from Santa Cruz Biotechnology (catalog numbers sc-2025 and sc-2027, respectively).
HHV-8 lytic induction and replication assays.
Lytic replication was induced in PEL cells by treatment with either doxycycline (1 μg/ml) for TRExBCBL1-RTA cells or sodium butyrate (0.5 μM) and TPA (20 ng/ml) for JSC-1 cells. Media were changed after 24 h for TRExBCBL1-RTA cultures and after 6 h for JSC-1 cultures to remove doxycycline and sodium butyrate; TPA was retained in the replacement medium for JSC-1 cultures. Lytic replication in BAC16-infected iSLK cells was induced with doxycycline (1 μg/ml) and sodium butyrate (1 mM) applied continuously. For virus titrations, media from TRExBCBL1-RTA cultures were harvested 4 days after induction; 300- to 400-μl samples were used to inoculate iSLK cultures for determinations of relative infectious-virus titers by immunofluorescence assay (IFA) for LANA 1 day postinoculation. For LANA staining, cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS (20 min at room temperature), and permeabilized with 0.25% Triton X-100 in PBS (5 min; room temperature) before blocking with 2% bovine serum albumin and 5% normal goat serum in PBS for 2 h at room temperature. The treated cell monolayers were then rinsed three times with PBS prior to addition of LANA primary antibody (Advanced Biotechnologies; catalog number 13-210-100; 1:1,000 dilution) and incubation overnight at 4°C. Then, following 3 washes in PBS, Cy3-conjugated secondary antibody (Life Technologies; catalog number A10522; 1:400 dilution) was added for 1 h at room temperature. Following further washing and nuclear staining using Hoechst 33342 (see above), the cells were visualized by UV microscopy. Three random fields (>300 total cells) were analyzed to calculate the percentage of LANA+ cells among total (Hoechst stain-positive) cells. For BAC16-based replication experiments, infectious-virus titers were determined by calculation of the percentage of GFP+ cells in medium-inoculated naive iSLK cultures. To determine the released, encapsidated viral genome copy number, LANA ORF-directed quantitative PCR (qPCR) using primer sequences 5′-TACGGTTGGCGAAGTCACATC-3′ (forward) and 5′-CCTCGCAGCAGACTACACCTCCAC-3′ (reverse) was applied to DNA extracted from medium samples (300 μl) pretreated with DNase I (6 U/ml; 37°C; overnight) to digest any unencapsidated viral DNA. DNA was extracted by addition of silica gel (5 μg; Sigma; catalog number S5631) in 1 ml of 6 M guanidine thiocyanate at 55°C, incubation for 10 min, and pelleting of silica-bound DNA by centrifugation. The pellet was resuspended in 500 μl wash buffer (50% ethanol, 10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA), repelleted, and washed and pelleted again to isolate the clean, matrix-bound viral DNA. The DNA was released from the silica gel by addition of distilled water, and the eluate was analyzed by qPCR, essentially as outlined previously (22).
ACKNOWLEDGMENT
This study was supported by NIH grant R21-CA196348 to J.N.
REFERENCES
- 1.Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, Hardwick JM. 1997. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci U S A 94:690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med 3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 3.Montaner S, Sodhi A, Pece S, Mesri EA, Gutkind JS. 2001. The Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor promotes endothelial cell survival through the activation of Akt/protein kinase B. Cancer Res 61:2641–2648. [PubMed] [Google Scholar]
- 4.Choi YB, Nicholas J. 2008. Autocrine and paracrine promotion of cell survival and virus replication by human herpesvirus 8 chemokines. J Virol 82:6501–6513. doi: 10.1128/JVI.02396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang HW, Sharp TV, Koumi A, Koentges G, Boshoff C. 2002. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi's sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J 21:2602–2615. doi: 10.1093/emboj/21.11.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng P, Scott CW, Cho NH, Nakamura H, Chung YH, Monteiro MJ, Jung JU. 2004. Kaposi's sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol Cell Biol 24:3938–3948. doi: 10.1128/MCB.24.9.3938-3948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousins E, Nicholas J. 2014. Molecular biology of human herpesvirus 8: novel functions and virus-host interactions implicated in viral pathogenesis and replication. Recent Results Cancer Res 193:227–268. doi: 10.1007/978-3-642-38965-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friborg J Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894. [DOI] [PubMed] [Google Scholar]
- 9.Radkov SA, Kellam P, Boshoff C. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med 6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 10.Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat Med 9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 11.Robinson BA, O'Connor MA, Li H, Engelmann F, Poland B, Grant R, DeFilippis V, Estep RD, Axthelm MK, Messaoudi I, Wong SW. 2012. Viral interferon regulatory factors are critical for delay of the host immune response against rhesus macaque rhadinovirus infection. J Virol 86:2769–2779. doi: 10.1128/JVI.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson BA, Estep RD, Messaoudi I, Rogers KS, Wong SW. 2012. Viral interferon regulatory factors decrease the induction of type I and type II interferon during rhesus macaque rhadinovirus infection. J Virol 86:2197–2211. doi: 10.1128/JVI.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham C, Barnard S, Blackbourn DJ, Davison AJ. 2003. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J Gen Virol 84:1471–1483. doi: 10.1099/vir.0.19015-0. [DOI] [PubMed] [Google Scholar]
- 14.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol 75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozharskaya VP, Weakland LL, Zimring JC, Krug LT, Unger ER, Neisch A, Joshi H, Inoue N, Offermann MK. 2004. Short duration of elevated vIRF-1 expression during lytic replication of human herpesvirus 8 limits its ability to block antiviral responses induced by alpha interferon in BCBL-1 cells. J Virol 78:6621–6635. doi: 10.1128/JVI.78.12.6621-6635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D, Sandford G, Nicholas J. 2009. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J Virol 83:722–733. doi: 10.1128/JVI.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baresova P, Pitha PM, Lubyova B. 2013. Distinct roles of Kaposi's sarcoma-associated herpesvirus-encoded viral interferon regulatory factors in inflammatory response and cancer. J Virol 87:9398–9410. doi: 10.1128/JVI.03315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura H, Li M, Zarycki J, Jung JU. 2001. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 75:7572–7582. doi: 10.1128/JVI.75.16.7572-7582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo T, Park J, Lee D, Hwang SG, Choe J. 2001. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. J Virol 75:6193–6198. doi: 10.1128/JVI.75.13.6193-6198.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol 80:2257–2266. doi: 10.1128/JVI.80.5.2257-2266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YB, Nicholas J. 2010. Bim nuclear translocation and inactivation by viral interferon regulatory factor. PLoS Pathog 6:e1001031. doi: 10.1371/journal.ppat.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi YB, Sandford G, Nicholas J. 2012. Human herpesvirus 8 interferon regulatory factor-mediated BH3-only protein inhibition via Bid BH3-B mimicry. PLoS Pathog 8:e1002748. doi: 10.1371/journal.ppat.1002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo T, Lee D, Shim YS, Angell JE, Chidambaram NV, Kalvakolanu DV, Choe J. 2002. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus interacts with a cell death regulator, GRIM19, and inhibits interferon/retinoic acid-induced cell death. J Virol 76:8797–8807. doi: 10.1128/JVI.76.17.8797-8807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang KY, Choi YB. 2015. Modulation of mitochondrial antiviral signaling by human herpesvirus 8 interferon regulatory factor 1. J Virol 90:506–520. doi: 10.1128/JVI.01903-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo T, Park J, Choe J. 2005. Kaposi's sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res 65:1738–1747. doi: 10.1158/0008-5472.CAN-04-2374. [DOI] [PubMed] [Google Scholar]
- 26.Burysek L, Yeow WS, Lubyova B, Kellum M, Schafer SL, Huang YQ, Pitha PM. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol 73:7334–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung JU. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol 20:8254–8263. doi: 10.1128/MCB.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ Jr, Barber GN, Hiscott J. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800–811. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- 29.Baresova P, Musilova J, Pitha PM, Lubyova B. 2014. p53 tumor suppressor protein stability and transcriptional activity are targeted by Kaposi's sarcoma-associated herpesvirus-encoded viral interferon regulatory factor 3. Mol Cell Biol 34:386–399. doi: 10.1128/MCB.01011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chavoshi S, Egorova O, Lacdao IK, Farhadi S, Sheng Y, Saridakis V. 2016. Identification of Kaposi sarcoma herpesvirus (KSHV) vIRF1 protein as a novel interaction partner of human deubiquitinase USP7. J Biol Chem 291:6281–6291. doi: 10.1074/jbc.M115.710632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, Haas J, Toth Z, Jeon YH, Oh TK, Kim MH, Jung JU. 2011. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat Struct Mol Biol 18:1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. 2006. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 34.Munoz-Fontela C, Marcos-Villar L, Gallego P, Arroyo J, Da Costa M, Pomeranz KM, Lam EW, Rivas C. 2007. Latent protein LANA2 from Kaposi's sarcoma-associated herpesvirus interacts with 14-3-3 proteins and inhibits FOXO3a transcription factor. J Virol 81:1511–1516. doi: 10.1128/JVI.01816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson B, Suresh Kumar KG. 2011. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys 60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 36.Tavana O, Gu W. 2017. Modulation of the p53/MDM2 interplay by HAUSP inhibitors. J Mol Cell Biol 9:45–52. doi: 10.1093/jmcb/mjw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutell C, Canning M, Orr A, Everett RD. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J Virol 79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daubeuf S, Singh D, Tan Y, Liu H, Federoff HJ, Bowers WJ, Tolba K. 2009. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood 113:3264–3275. doi: 10.1182/blood-2008-07-168203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, Frappier L. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell 18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 41.Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. 2009. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog 5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem 278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 43.Jager W, Santag S, Weidner-Glunde M, Gellermann E, Kati S, Pietrek M, Viejo-Borbolla A, Schulz TF. 2012. The ubiquitin-specific protease USP7 modulates the replication of Kaposi's sarcoma-associated herpesvirus latent episomal DNA. J Virol 86:6745–6757. doi: 10.1128/JVI.06840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillen J, Li W, Liang Q, Avey D, Wu J, Wu F, Myoung J, Zhu F. 2015. A survey of the interactome of Kaposi's sarcoma-associated herpesvirus ORF45 revealed its binding to viral ORF33 and cellular USP7, resulting in stabilization of ORF33 that is required for production of progeny viruses. J Virol 89:4918–4931. doi: 10.1128/JVI.02925-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol 77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wies E, Mori Y, Hahn A, Kremmer E, Sturzl M, Fleckenstein B, Neipel F. 2008. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood 111:320–327. doi: 10.1182/blood-2007-05-092288. [DOI] [PubMed] [Google Scholar]
- 48.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 49.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 50.Sarkari F, Wang X, Nguyen T, Frappier L. 2011. The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS One 6:e16598. doi: 10.1371/journal.pone.0016598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cannon JS, Ciufo D, Hawkins AL, Griffin CA, Borowitz MJ, Hayward GS, Ambinder RF. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J Virol 74:10187–10193. doi: 10.1128/JVI.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. 2010. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J 29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen D, Nicholas J. 2015. Promotion of endoplasmic reticulum-associated degradation of procathepsin D by human herpesvirus 8-encoded viral interleukin-6. J Virol 89:7979–7990. doi: 10.1128/JVI.00375-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen D, Xiang Q, Nicholas J. 2017. Human herpesvirus 8 interleukin-6 interacts with calnexin cycle components and promotes protein folding. J Virol 91:e00965-17. doi: 10.1128/JVI.00965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]