ABSTRACT
Determinants and mechanisms of cell attachment and entry steer adeno-associated virus (AAV) in its utility as a gene therapy vector. Thus far, a systematic assessment of how diverse AAV serotypes engage their proteinaceous receptor AAVR (KIAA0319L) to establish transduction has been lacking, despite potential implications for cell and tissue tropism. Here, a large set of human and simian AAVs as well as in silico-reconstructed ancestral AAV capsids were interrogated for AAVR usage. We identified a distinct AAV capsid lineage comprised of AAV4 and AAVrh32.33 that can bind and transduce cells in the absence of AAVR, independent of the multiplicity of infection. Virus overlay assays and rescue experiments in nonpermissive cells demonstrate that these AAVs are unable to bind to or use the AAVR protein for entry. Further evidence for a distinct entry pathway was observed in vivo, as AAVR knockout mice were equally as permissive to transduction by AAVrh32.33 as wild-type mice upon systemic injection. We interestingly observe that some AAV capsids undergo a low level of transduction in the absence of AAVR, both in vitro and in vivo, suggesting that some capsids may have a multimodal entry pathway. In aggregate, our results demonstrate that AAVR usage is conserved among all primate AAVs except for those of the AAV4 lineage, and a non-AAVR pathway may be available to other serotypes. This work furthers our understanding of the entry of AAV, a vector system of broad utility in gene therapy.
IMPORTANCE Adeno-associated virus (AAV) is a nonpathogenic virus that is used as a vehicle for gene delivery. Here, we have identified several situations in which transduction is retained in both cell lines and a mouse model in the absence of a previously defined entry receptor, AAVR. Defining the molecular determinants of the infectious pathway of this highly relevant viral vector system can help refine future applications and therapies with this vector.
KEYWORDS: AAV, virus, virus receptor, adeno-associated virus, attachment, gene therapy, vector, viral entry, viral gene transfer
INTRODUCTION
Adeno-associated virus (AAV) is a small, nonenveloped, single-stranded DNA virus that is not associated with any known human pathogenesis. As such, AAV is an attractive vehicle for therapeutic gene transfer. AAV gene therapy has been used safely and effectively for multiple indications, such as hemophilia (1, 2), in clinical trials. An AAV drug developed for the treatment of lipoprotein lipase deficiency (3–5) was the first licensed AAV-based therapy (Glybera) in Europe; however, it was taken off the market. Other AAV gene therapies, such as those for spinal muscular atrophy (6–8), are more promising, with Luxterna, an AAV gene therapy for the treatment of a type of inherited retinal degeneration, being recently approved by the U.S. Food and Drug Administration (9–12). Although great strides have been made in the translation of AAV gene therapy into the clinic, there are still large voids in our understanding of the host factors determining AAV entry and tropism.
AAV is known to attach to specific glycans at the plasma membrane in a serotype-dependent manner (13–18). Several reports have identified putative protein receptors; however, the roles of these receptors were more incremental, with little demonstration of their relevance in vivo (19–26). After attachment, the vector undergoes endocytosis and microtubule-dependent trafficking (27) to the nucleus. It remains unclear whether endocytosis is clathrin dependent (28, 29) or clathrin independent (30). After internalization, the AAV particle exits the endosome by using the phospholipase domain located in the VP1 portion of the capsid (31). AAV nuclear import is thought to be mediated by importin-β, but small interfering RNA (siRNA) knockdown causes only a moderate decrease in the transduction efficiency (32). Of note, most of our mechanistic understanding comes from studies of only a few serotypes, most notably AAV2, so it is unclear which serotype-specific entry pathway differences dictate the unique biodistribution patterns observed in vivo.
Recently a multiserotype AAV receptor (AAVR) was identified by using an unbiased genome-scale genetic screen (33). AAVR directly interacts with AAV particles via Ig-like polycystic kidney disease (PKD) domains present in the ectodomain of AAVR (33, 34). It is required for the efficient transduction of multiple AAV serotypes (AAV1, -2, -3B, -5, -6, -8, and -9) in all cell lines that were tested. Furthermore, AAVR is essential for in vivo transduction, as demonstrated with AAV9 using an AAVR knockout (KO) mouse model. Because AAVR is a rapidly endocytosed receptor that traffics to the Golgi network, AAVR could play a role in cell attachment, the trafficking of AAV particles to the Golgi network, or the escape of AAV out of the Golgi network. However, the exact role of AAVR in these processes is still unknown, as is its relationship with the glycan receptors that are important for cell surface attachment.
In the present study, we examine from an evolutionary perspective how different AAV capsid lineages require AAVR and further define the role of AAVR in facilitating cell attachment. We found that the majority of the serotypes are strongly dependent on AAVR for transduction and that AAVR likely acts at a postattachment step. Our analysis includes in silico-reconstructed ancestral AAV capsids, which differ significantly from the extant strains, suggesting that AAVR dependence arose early. Strikingly, we identified a single AAV4-related evolutionary lineage that has evolved independently from AAVR and several capsids that are able to enter cells through an alternate pathway in the absence of AAVR. Representative members of this AAV4 lineage efficiently transduce AAVR knockout cell lines derived from different cell types as well as mice that lack AAVR. Our work highlights the conserved nature of AAVR among most primate AAVs yet identifies several scenarios in which transduction is achieved in the absence of AAVR.
RESULTS
Identification of AAVR-independent serotypes.
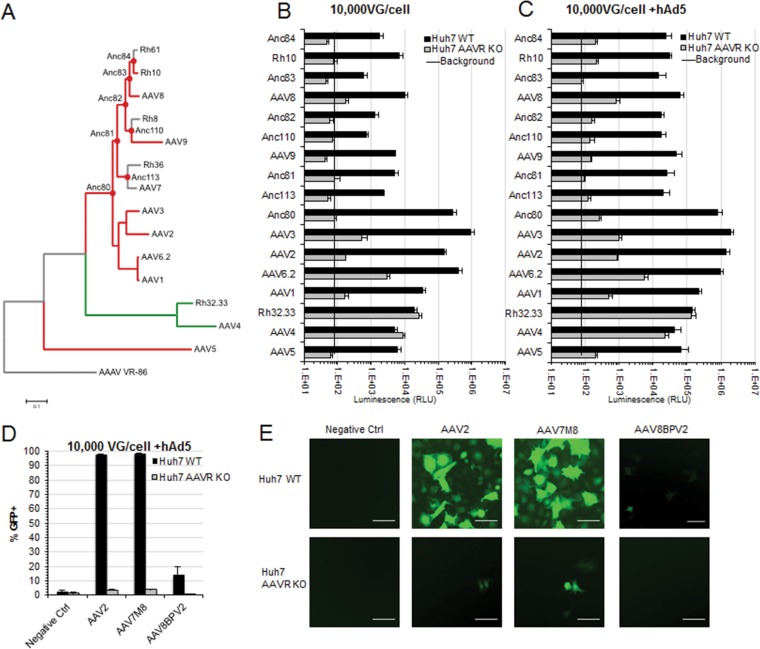
Since the initial discovery of AAV, there have been over a hundred different naturally occurring isolates and countless other capsid variants identified through either rational design, error-prone PCR, or capsid shuffling, each with their own unique cell and tissue tropism profiles (35, 36). We examined AAVR usage in Huh7 cells with (wild type [WT]) or without (KO) AAVR expression in a transduction assay using replication-defective AAV expressing a luciferase transgene and assessed a total of 9 natural isolates, 2 directed-evolution variants, and 7 putative ancestral variants, most of which had not been previously tested (Fig. 1A). Most of the AAVs tested demonstrated a strong requirement for AAVR, illustrated by a >10-fold decrease in the transduction of KO versus WT cells (Fig. 1B). Interestingly, however, a chimera of two rhesus isolates, AAVrh32.33 (37, 38), and an African green monkey AAV4 isolate (39, 40) were uniquely able to transduce AAVR KO cells as efficiently as WT cells. These data illustrate that the majority of, yet not all, AAV serotypes primarily use AAVR for entry.
FIG 1.
Transduction of a panel of extant, ancestral, and peptide insertion AAV capsids demonstrates an AAVR-independent capsid lineage. Shown are data for in vitro transduction of extant and putative ancestral AAVs. (A) Evolutionary lineage of AAVR-dependent (red) and AAVR-independent (green) extant and putative ancestral AAV capsids and avian AAV (AAAV) as an outgroup. (B and C) Transduction of 104 viral genomes per cell of the CMV.Luciferase.SVPA (AAVrh10, AAV8, AAVAnc82, AAV9, AAVAnc81, AAVAnc80, AAV3, AAV6.2, AAV1, AAVrh32.33, AAV4, and AAV5) or CMV.eGFP.T2A.Luciferase.SVPA (AAVAnc84, AAVAnc83, AAVAnc110, AAVAnc113, and AAV2) transgene with (C) or without (B) hAd5 coinfection. RLU, relative light units. (D) Flow cytometry analysis of Huh7 WT or AAVR KO cells transduced with 105 viral genomes per cell of the CMV.eGFP.WPRE transgene with hAd5 coinfection. (E) Representative fluorescence images of CMV.eGFP.WPRE-transduced cells quantified in panel D. Bar, 100 μm. Data presented are means ± standard errors of the means of results from at least 4 independent experiments.
AAV4 and AAVrh32.33 represent a distinct evolutionary lineage that is independent of AAVR.
Since AAV4 and AAVrh32.33 are highly divergent from many other currently circulating serotypes, we decided to interrogate a panel of capsids predicted to be evolutionary intermediates (41) in an attempt to identify where AAVR independence may have arisen (Fig. 1A). Although many of the putative ancestral capsids are highly divergent from the extant serotypes, they also demonstrated a high level of AAVR usage in the same assay, with some serotypes, such as AAVAnc80, having a >1,000-fold decrease in transgene expression (Fig. 1B). These data demonstrate that AAV4 and AAVrh32.33 represent a distinct evolutionary lineage of AAV capsid that can undergo AAVR-independent entry (Fig. 1A).
Peptide engraftment does not alter AAV receptor usage.
Regions of the capsid in hypervariable region IV, near the heparan sulfate binding motif of AAV2, have been shown to tolerate a small peptide insertion, which often leads to a unique cell or tissue type specificity. This begs the question of whether peptide insertion allows the usage of a secondary entry receptor. We examined two peptide insertion mutant capsids, AAV7M8, which has demonstrated increased outer retinal transduction via intravitreal injection (42), and AAV8BPV2, which transduces retinal bipolar cells (43). Flow cytometry of Huh7 WT and AAVR KO cells transduced with AAV expressing a CMV.eGFP.WPRE transgene was performed, and both peptide insertion mutants required AAVR for efficient entry, demonstrating that AAVR is required to complete the viral entry process in both parental and peptide insertion capsids (Fig. 1D). Although transduction was highly decreased in AAVR KO cells, we were still able to observe the transduction of some cells by both AAV7M8 and its AAV2 parental capsid (Fig. 1E). Similar levels of transduction in these two serotypes (about 3%) (Fig. 1D) suggest that peptide insertion may influence another aspect of cellular targeting or entry rather than engagement of its protein receptor.
AAVR dependence is transgene and helper virus independent.
Since AAVR has been shown to directly bind intact capsid, it is not surprising that AAV2 capsids expressing different transgenes, either luciferase (Fig. 1B) or enhanced green fluorescent protein (eGFP) (Fig. 1D), were both defective for transduction in AAVR KO cells. This further demonstrates that AAVR-mediated entry is an inherent property of the capsid. Since the presence of a helper virus increases AAV transduction, we wanted to know whether coinfection with a helper virus would alter AAVR usage. Huh7 WT or Huh7 AAVR KO cells were preincubated with 200 PFU/cell of WT human adenovirus type 5 (hAd5) for 2 h prior to AAV transduction with the CMV.Luciferase or CMV.eGFP.T2A.Luciferase transgene, and transduction efficiency was assessed by a luciferase assay. While overall transduction increased under all conditions (Fig. 1C), the AAVR usage trends remained the same, with all serotypes aside from AAV4 and AAVrh32.33 maintaining a high level of dependence on AAVR. However, highly reduced yet detectable transduction was seen for AAV3 and AAV6.2 in the absence of adenovirus coinfection and for AAV1, -2, -3, -6.2, -8, and -7m8 and some other AAVs at lower levels (Fig. 1B, C, and E), indicating the possibility of a low level of AAVR-independent entry for these serotypes. These data suggest that AAVR usage is inherent in the capsid and that it is independent of a transgene or helper virus.
AAVR-independent entry is a pathway that exists in multiple cell lines.
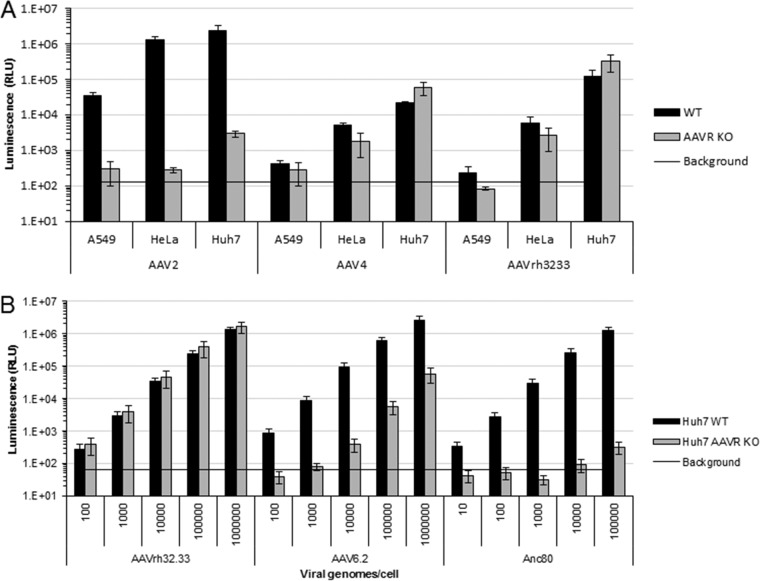
Previous studies demonstrated that AAVR is required for the entry of many AAV serotypes, an observation that was confirmed in multiple cell lines (33). Using a luciferase assay to assess AAV entry and subsequent transgene expression, we confirmed that AAV2 entry is abolished in A549 and HeLa AAVR KO cells (Fig. 2A). A highly reduced yet low level of luciferase expression in Huh7 AAVR KO cells was retained, demonstrating the strong dependency of AAV2 on AAVR expression; however, a possible alternate entry pathway is available in some cells for some serotypes. In all tested cell lines, there were comparable levels of transduction by AAV4 and AAVrh32.33 in WT and KO cells (Fig. 2A). The entry of AAV4 and AAVrh32.33 into AAVR KO A549, HeLa, and Huh7 cells derived from different tissues (human lung carcinoma, human cervical cancer, and human hepatocellular carcinoma, respectively) suggests that these diverse cell types express an alternate protein receptor that these vectors are able to use for virus entry.
FIG 2.
AAVR-independent entry is serotype specific and exists in a variety of cell types. (A) In vitro transduction of 10,000 VG/cell AAV2, 10,000 VG/cell AAV4, and 100,000 VG/cell AAVrh32.33 with the CMV.Luciferase.SVPA transgene in A549, HeLa, and Huh7 WT or AAVR KO cells. Data presented are means ± standard errors of the means of results from 3 independent experiments. (B) Dose response of cells transduced with AAVrh32.33, AAV6.2, or AAVAnc80 CMV.Luciferase.SVPA over a 5-log range of viral genomes per cell in Huh7 WT or Huh7 AAVR KO cells.
AAVR independence is an alternate AAV entry pathway.
Previous studies of AAV2 entry produced conflicting reports on receptor usage and endosomal pathway requirements, differences in which are thought to be due to different multiplicities of infection (MOIs) used for these studies (28–30). Due to this and the low level of transduction observed for some serotypes in AAVR KO cell lines (Fig. 1A), we wanted to test whether this observation was due to capsid-specific differences in receptor usage or whether it was due to the nonspecific uptake of highly potent vectors. To test this, we assessed the transduction of several serotypes over a 5-log range in viral genomes (VG) per cell. Interestingly, we observed three distinct scenarios. First, our previously identified AAVR-independent serotype, AAVrh32.33, was able to undergo similar levels of transduction in AAVR KO and WT cells at all virus doses tested (Fig. 2B). Second, we observed an intermediate phenotype in which some vectors, such as AAV6.2, could undergo significant levels of transduction in AAVR KO cells at high virus doses. Third, many vectors, such as AAVAnc80, were unable to transduce AAVR KO cells at any virus dose tested, despite very high levels of transduction in WT cells. These different phenotypes suggest that there is an alternate entry receptor used by AAVrh32.33 and that some serotypes are able to use this alternate pathway, although for these serotypes, it is much less efficient than that in AAVR-expressing cells. These data are in line with previously presented data suggesting that some AAV serotypes such as AAV2.5 may be able to undergo entry in the absence of AAVR on polarized airway epithelial cells (44).
AAVR expression is not required for cellular attachment.
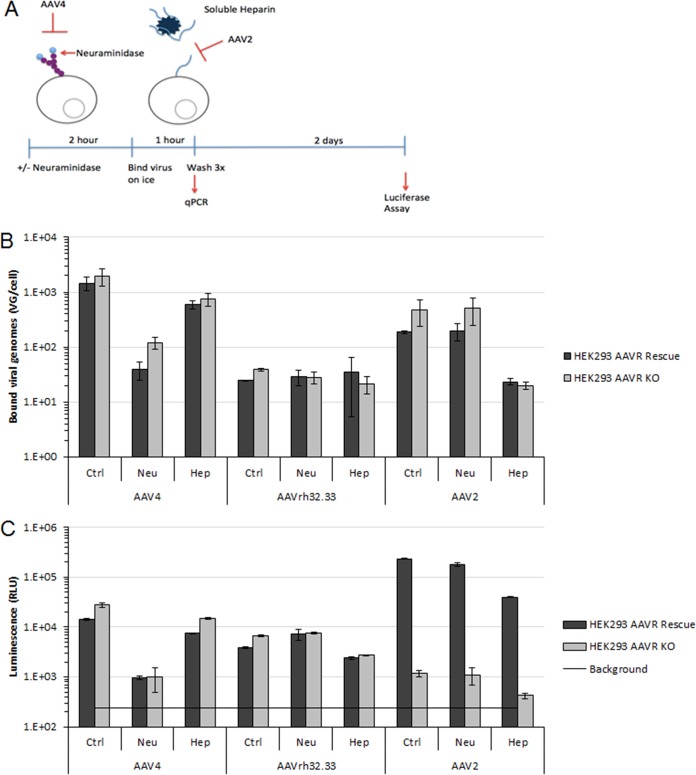
To distinguish between a role of AAVR in AAV attachment and a role in postattachment steps such as endocytosis, trafficking, or endosomal escape (33), we employed a cellular binding assay in AAVR-overexpressing cells and AAVR knockout cells. To further assess the role of AAVR in the absence of potentially dominant glycan interactions, we pretreated the cells with neuraminidase to cleave the terminal sialic acid from cellular glycans, or virions were pretreated with soluble heparin, which competes for heparan sulfate proteoglycan binding (Fig. 3A). As expected for the AAVR-independent serotypes AAV4 and AAVrh32.33, no differences were observed between AAVR-overexpressing and knockout cells in binding (Fig. 3B) or transduction (Fig. 3C). In agreement with data from previous studies and validating data from our binding assay, AAV4 binding was reduced by neuraminidase treatment (Fig. 3B) (17). Interactions with heparan sulfate or sialic acid do not play a major role in AAVrh32.33 binding (Fig. 3B) (45) or are redundant. It is of note that both AAV4 and AAVrh32.33, which interact differently with glycans, are independent of AAVR. Importantly, we did not observe differences in the binding of AAV2, which uses AAVR, in AAVR-overexpressing versus knockout cells, even when transduction was performed in the presence of soluble heparin, which decreased AAV2 binding and transduction by 10-fold. The transduction of AAV2, assessed in parallel, showed a >100-fold decrease in the transduction of KO cells (Fig. 3C), demonstrating that AAVR expression is not a requirement for AAV2 attachment. Although we observed similar levels of AAV2 binding in both WT and KO cells, AAV2 may still first interact with AAVR at the cell surface; however, this interaction would be indistinguishable in this assay.
FIG 3.
AAVR is not required for attachment or entry of AAVR-independent serotypes. (A) Diagram of the in vitro cell binding assay. (B and C) Paired binding (B) and transduction (C) assays for 104 VG/cell AAV4, AAVrh32.33, and AAV2 in HEK293 AAVR overexpression (rescue) or HEK293 AAVR KO cells. Data presented are from one representative experiment of three independent trials and standard deviations of technical replicates.
AAV4 is unable to bind purified AAVR domains or AAVR expressed in cell lysates.
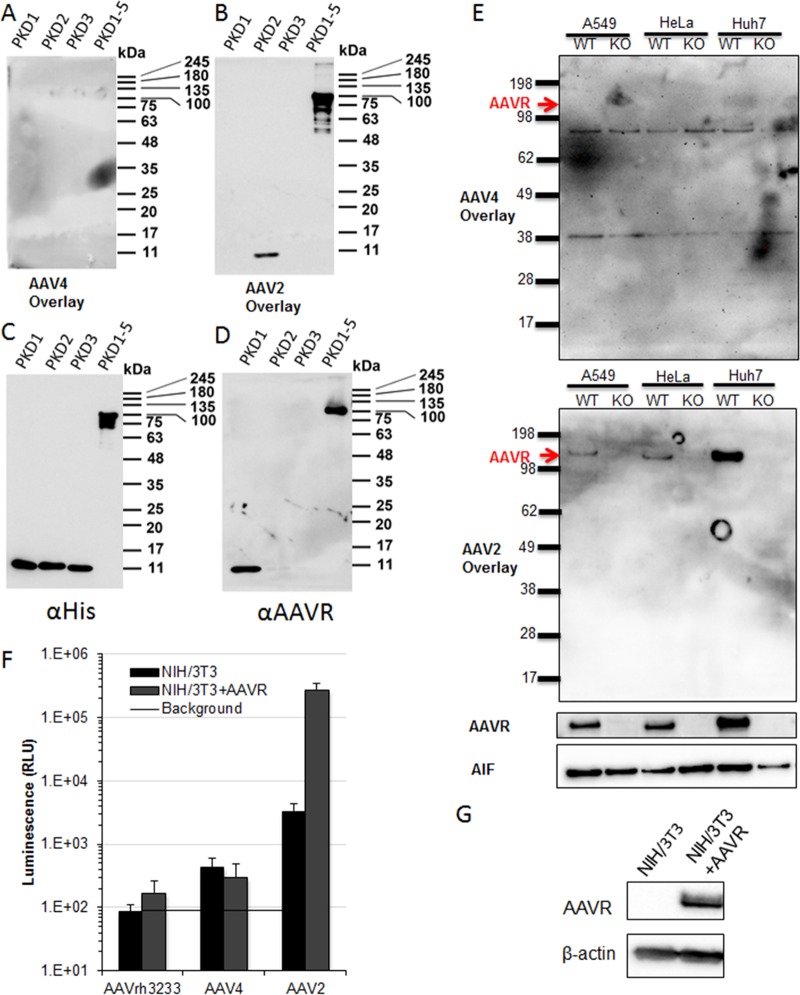
Although there is no observable role for AAVR in the cellular attachment of AAV4 and AAVrh32.33, we wanted to determine whether these vectors are able to bind to AAVR even if they are unable to functionally engage it for cellular entry. We used a previously reported virus overlay assay (34, 46) to interrogate the binding of AAV4 to purified AAVR or AAVR expressed in cell lysates. The AAV4 virus overlay on purified AAVR domains illustrates that under the same conditions as those that demonstrated AAV2 binding to AAVR, no detectable binding of AAV4 to AAVR PKD1, PKD2, PKD3, or PKD1-5 is observed (Fig. 4A). Conversely, AAV2 is able to bind purified PKD2 as well as PKD1-5 (Fig. 4B), as previously reported (34). Anti-His (Fig. 4C) and anti-AAVR (Fig. 4D) Western blots demonstrate that these constructs are efficiently detected on polyvinylidene difluoride (PVDF) membranes, although it appears the AAVR antibody recognizes only constructs containing PKD1. In an attempt to identify a membrane protein that AAV4 may be binding to and using as a receptor, we performed a virus overlay assay on membrane preparations from three different WT and AAVR KO paired cell lines. AAV4 appears to bind to two proteins smaller than AAVR, of about 85 and 40 kDa, in all cell lysates tested (Fig. 4E). Although we observed binding of AAV4 to something in these membrane preparations, this may or may not be a protein receptor, as these denaturing gels may alter a conformational epitope to which AAV4 and AAVrh32.33 bind. These observations are confirmatory for AAV2 to demonstrate direct binding to AAVR (Fig. 4E) (33, 34) and are consistent with our findings from AAV4 and AAVrh32.33 binding (Fig. 3B) and transduction (Fig. 3C) experiments, in which AAVR binding could not be demonstrated.
FIG 4.
AAV4 and AAVrh32.33 are unable to bind AAVR or use AAVR for entry. (A to D) SDS-PAGE (10%) and virus overlay or Western blotting of purified AAVR subunits. (E) SDS-PAGE (4 to 20%) and virus overlay or Western blotting of WT or AAVR KO A549, HeLa, or Huh7 cell lysates. (F) NIH 3T3 cells stably expressing AAVR-flag or AAVRΔC-tail transduced with 105 viral genomes per cell of AAVrh32.33, AAV4, or AAV2 CMV.Luciferase.SVPA. (G) Western blot demonstrating AAVR expression in stably transduced NIH 3T3 cells. Data presented are means ± standard errors of the means of results from 3 independent experiments.
AAV4 and AAVrh32.33 are unable to use AAVR as an entry receptor.
In addition to determining whether AAV4 and AAVrh32.33 can bind AAVR, we wanted to test whether AAV4 and AAVrh32.33 are able to use AAVR as a receptor by reintroducing AAVR into a nonpermissive cell line. To do this, we stably reintroduced full-length AAVR via a lentiviral vector into nonpermissive NIH 3T3 cells. AAVR was able to rescue AAV2 transduction, as expected, but not AAV4 or AAVrh32.33 transduction (Fig. 4F), despite high levels of AAVR expression, as determined by Western blotting (Fig. 4G). An inability to rescue transduction upon overexpression demonstrates that AAVR expression is not sufficient for the entry of AAV4 or AAVrh32.33 in these cell lines.
AAVR is not required for AAVrh32.33 transduction in vivo.
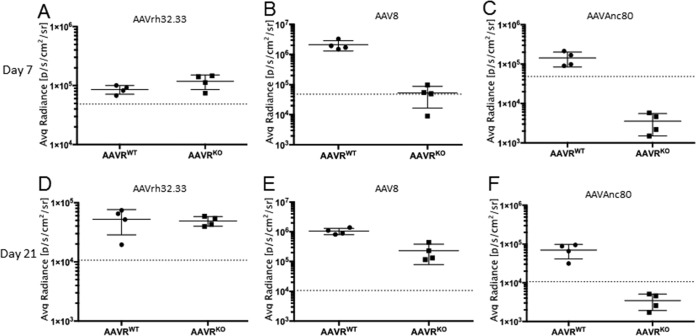
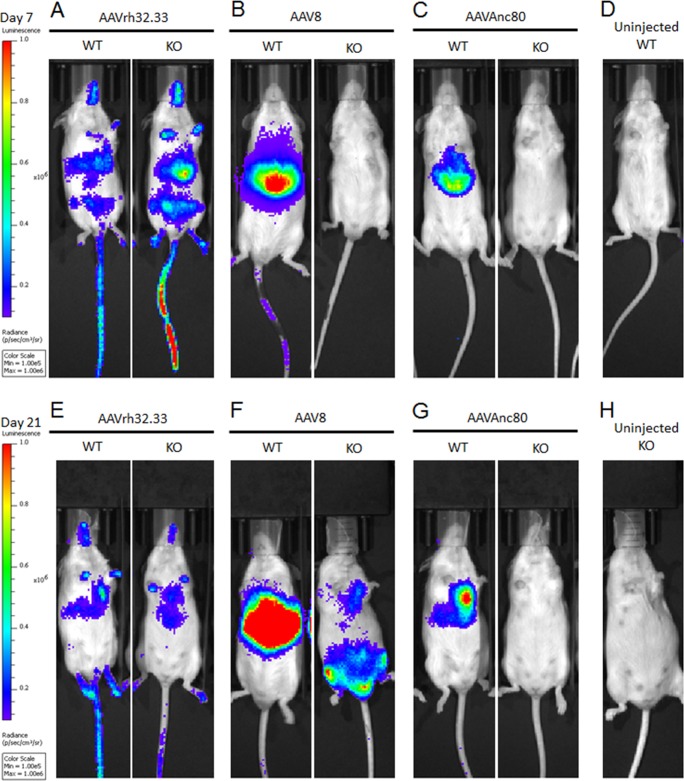
Aavr KO and WT control mice were injected intravenously via retro-orbital injection with 1011 genome copies (GC) per mouse of AAV8, AAVAnc80, or AAVrh32.33 expressing CMV.eGFP.T2A.Luciferase. As expected, at day 7 postinjection, AAVrh32.33 showed similar levels of luciferase expression in WT and KO mice (Fig. 5A). High levels of transduction of AAV8 (Fig. 5B) and AAVAnc80 (Fig. 5C) were observed in WT mice, but background levels were observed in Aavr KO mice. Similar levels of AAVrh32.33 transduction were observed in WT and KO mice at 21 days postinjection as well (Fig. 5D). Interestingly, at 21 days postinjection, we began to see luciferase expression in Aavr KO mice transduced with AAV8 (Fig. 5E), suggesting that, when forced, AAV8 can undergo AAVR-independent entry in vivo although with a delayed onset compared to that when AAVR is present. Anc80, which also uses AAVR (Fig. 4C), did not demonstrate such delayed AAVR-independent transduction at the time points tested (Fig. 5F). AAVrh32.33 showed a distinctly wider biodistribution pattern based on in vivo bioluminescence imaging (Fig. 6A) than that of AAV8 or AAVAnc80, which, as previously observed, is dominated by liver (Fig. 6B and C). Interestingly, AAV8 transduction in KO mice at 21 days postinjection had an altered biodistribution compared to that in WT mice (Fig. 6E). These data demonstrate that there is an alternate AAV entry pathway in vivo, which AAVR-independent serotypes such as AAVrh32.33 and, to some extent, AAVR-dependent serotypes such as AAV8 can use for entry in vivo.
FIG 5.
AAV capsids can undergo AAVR-independent entry in vivo. Shown is quantification of bioluminescence from mice injected retro-orbitally with 1011 VG/mouse AAVrh32.33 (A and D), AAV8 (B and E), or AAVAnc80 (C and F). Whole-body luminescence was imaged and quantified on day 7 (A to C) and day 21 (D to F) postinjection.
FIG 6.
Bioluminescence demonstrates altered biodistribution upon AAVR independent entry. Bioluminescence images demonstrate the biodistribution of AAVrh32.33 (A and E)-, AAV8 (B and F)-, and AAVAnc80 (C and G)-injected or uninjected control (D and H) WT and AAVR KO mice at 7 days (A to C) or 21 days (D to F) postinjection.
DISCUSSION
The recent development and implementation of genome-scale genetic knockout screens have allowed the identification of a multiserotype AAV protein receptor (47), which previous methods were unable to define (46). Our data suggest that there are at least two predominant entry mechanisms within the primate Dependoviridae, namely, a canonical entry pathway by which most AAV serotypes enter cells and which requires AAVR as well as an alternate, AAVR-independent pathway used by AAV4 and AAVrh32.33. Recently, Pillay et al. (34) demonstrated that the most evolutionarily distinct serotype, AAV5, primarily uses a different domain of AAVR (PKD1) than the one used by the other tested, naturally isolated serotypes (PKD2). From the AAV capsid lineage, it appears that two distinct branches of AAV capsids have emerged. One branch is entirely AAVR independent, uses a currently unidentified receptor, and is composed of AAV4 and AAVrh32.33. The other distinct branch is composed of all serotypes whose putative evolutionary ancestor is AAVAnc80 and who have evolved to predominantly use PKD2 of AAVR for binding and efficient transduction. It is curious that these two monkey isolates have evolved to be AAVR independent, given the high level of conservation between human AAVR and AAVRs of all nonhuman primates. Human and rhesus AAVRs share 98% sequence homology, suggesting that the evolution of AAVR-independent serotypes may have arisen to allow a unique tissue specificity for these AAVs, rather than having coevolved with their host. Interestingly, although AAVAnc80 and AAV5 are also highly divergent from other naturally circulating serotypes, we found that they both are dependent on AAVR. Investigating the sequence space occupied by the most common ancestor of the AAV4 and AAVrh32.33 branch and AAV5 or AAVAnc80 may provide insight into what dictates receptor usage by different AAV serotypes.
Since we were able to observe a low level of transduction by some serotypes both in KO cells in vitro (AAV6.2, AAV3, and AAV2) and in KO mice in vivo (AAV8), it is possible that these serotypes are also able to use an alternate entry pathway. It remains to be determined if these serotypes could be using the same alternate receptor as the one used by AAV4 and AAVrh32.33. Additionally, the peptide insertion mutants AAV7M8 and AAV8BPV2 are highly dependent on AAVR, demonstrating that peptide insertion does not change the requirement for AAVR. Since peptide insertions do not alter AAVR usage, we hypothesize that peptide insertion may alter another aspect of the entry pathway, such as (glycan) attachment.
Similar levels of binding and transduction by AAV4 and AAVrh32.33 in overexpression and AAVR KO cells demonstrate that AAVR does not play a role in AAV4 or AAVrh32.33 attachment. Due to the high sequence similarity of AAV4 and AAVrh32.33, it may be expected that they would use the same glycan attachment factor. However, we demonstrated that neither sialic acid nor heparan sulfate appears to play a dominant role in the attachment of AAVrh32.33. Alternatively, AAVrh32.33 may have redundant glycan usage. There is currently no predominant glycan known that AAVrh32.33 uses for attachment. A striking observation was the similar levels of AAV2 binding in AAVR overexpression and AAVR KO cells. This is in contrast to previously reported data that suggested a role for AAVR at the plasma membrane (33). In those experiments, cells incubated on ice with either soluble AAVR (ectodomain without a transmembrane domain) or an anti-AAVR antibody showed decreased AAV2 transduction in a dose-dependent manner (33). However, those experiments did not directly assess AAV particle binding at the cell surface, and it cannot be excluded that the antibodies blocked a step after AAVR was endocytosed or that it interfered with the formation of AAVR homodimers or a multimeric receptor complex (47). Although our data suggest that the predominant role for AAVR is at a postattachment step, they do not preclude the possibility that AAV2 comes into contact with AAVR at the plasma membrane. It is also possible that the relative affinities of the AAV2/heparan sulfate proteoglycan (HSPG) interaction (dissociation constant [Kd] of 0.1 to 3.7 nm) (48–50) versus that of the AAV2/AAVR interaction (Kd of 150 nm) (33) allow high levels of AAV2 attachment to occur whether or not AAVR is present at the plasma membrane, such that the relative contribution of AAVR to AAV attachment is undetectable in this assay.
A virus overlay on purified AAVR domains confirms the previously reported interaction of AAV2 with AAVR (Fig. 4B), yet we were unable to detect the binding of AAV4 to these purified proteins (Fig. 4A). AAV4 appears to bind the same two membrane proteins in a virus overlay on three different cell types (Fig. 4E), although the identity of these proteins and the significance for AAV4 entry remain to be determined.
Although the stable overexpression of AAVR in nonpermissive NIH 3T3 cells is able to rescue AAV2 transduction, it is unable to rescue AAV4 or AAVrh32.33 transduction, suggesting that AAV4 and AAVrh32.33 use a different protein entry receptor. Since NIH 3T3 cells are a mouse-derived cell line, it is possible that other cellular cofactors are required for AAVR usage by AAV4 and AAVrh32.33 and that these cofactors from other species are unable to interact with human AAVR. However, since AAVR is neither necessary nor sufficient for the transduction of AAV-independent serotypes, we hypothesize that there is an entirely separate protein receptor used by these serotypes. What this receptor is and whether these two serotypes use the same receptor for entry are currently unknown.
Our study demonstrates that AAV4 and AAVrh32.33 may be attractive for use where AAVR is rate limiting. For example, although AAVR expression is fairly ubiquitous, a large amount of the vector injected intravenously is trapped in the liver. This is in stark contrast to the wide biodistribution observed from the intravenous injection of AAVrh32.33. Such evenly distributed transgene expression suggests that the entry receptor required for AAVrh32.33 entry is expressed in a wide range of tissues. Interestingly, 21 days after injection, we observed low levels of luciferase expression in AAV8-injected Aavr KO mice although with an altered distribution. AAV8-injected Aavr KO mice seem to have liver-detargeted transduction. The observed distribution pattern is similar to what was reported previously for the intravenous injection of an AAV2 R484E;R585E mutant (51), suggesting that residues in this region, which has long been thought to play a role in receptor binding, may influence not only heparan sulfate binding but also the usage of a secondary receptor. It is undoubtedly of interest to identify the AAVR binding domain(s) on the AAV capsid in order to develop tissue-specific novel AAV vectors.
The data presented here demonstrate that AAVR is the major determinant for the efficient entry of most, but not all, known serotypes and furthermore demonstrate the immense complexity underlying the AAV entry pathway. By elucidating AAV entry mechanisms, we aim to further inform targeting and delivery aspects of AAV-based gene therapies.
MATERIALS AND METHODS
Phylogeny generation.
To generate the phylogeny, 15 representative isolates of AAV were chosen, including one avian AAV for use as an outgroup. Amino acid sequences of the VP1 proteins from each of these strains were aligned by using ClustalOmega as implemented on the EMBL-EBI Web server (52, 53). A maximum likelihood phylogeny was constructed by using PhyML 3.0, using the LG+I+G+F substitution model and nearest-neighbor interchange (NNI) tree improvements (54). The phylogeny was rendered by allowing AAAV isolate VR-865 to act as an outgroup and rooting on its branch.
Cell lines.
All cell lines were maintained in Dulbecco's modified Eagle's minimal medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS) (GE Healthcare) and 100 IU/ml penicillin-streptomycin (Corning) in a humidified incubator with 5% CO2 at 37°C. Parental cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, VA), and AAVR KO cell lines were generated as previously described (33).
AAV production and purification.
All vectors were produced, purified, and titrated by the MEEI/SERI Gene Transfer Vector Core (http://vector.meei.harvard.edu/). Large-scale vector preparations were generated by polyethylenimine (catalog number 24765-2; Polysciences) triple transfection of pHelp; pAAVector2[Cap]; and pCMV.Luciferase.SVPA, pCMV.eGFP.T2A.Luciferase, or pCMV.eGFP.WPRE.bGH transgenes at a 2:1:1 ratio. A total of 520 μg total DNA was transfected in 10-layer hyperflasks by using a PEI Max/DNA ratio of 1.375:1 (wt/wt). Three days after transfection, vectors were concentrated by tangential-flow filtration and purified by iodixanol gradient ultracentrifugation as previously described (55).
AAV genome titration.
DNase I-resistant viral genomes were quantified by using TaqMan quantitative PCR (qPCR) (catalog number 4304449; ThermoFisher), using a primer/probe set detecting the cytomegalovirus (CMV) promoter. Vector purity was assessed by SDS-PAGE.
AAV transduction.
All luciferase transduction assays excluding those done for the binding assay in Fig. 3 were done by seeding 10,000 cells per well in poly-l-lysine (catalog number P4707; Sigma-Aldrich)-coated black-bottom 96-well plates overnight. All flow cytometric transduction assays were done by seeding 1 × 105 cells per well of a 12-well plate overnight. When indicated, cells were preincubated with 200 PFU/cell of WT hAd5 (University of Pennsylvania Vector Core) in DMEM supplemented with 10% FBS (D10) for 2 h, and hAd5-containing medium was then removed prior to transduction. Cells were transduced with AAV at 1 × 104 VG/cell in serum-free DMEM for 1 h at 37°C, D10 was then added, and transduction levels were analyzed by either a luciferase assay or flow cytometry at 48 h postransduction.
Luciferase assay.
At 2 days postransduction, cell culture medium was removed, and cells were lysed in 20 μl per well of 1× reporter lysis buffer (Promega) and then frozen at −80°C. After thawing, firefly luciferase (ffLuc) expression was measured in relative light units per second on a Synergy H1 Hybrid Multi-Mode microplate reader using 100 μl luciferin buffer (200 mM Tris [pH 8], 10 mM MgCl2, 300 μM ATP, 1× firefly luciferase signal enhancer [catalog number 16180; Thermo], and 150 μg/ml d-luciferin).
Flow cytometry.
Cells were harvested for flow cytometry analysis using phosphate-buffered saline (PBS) without Ca2+ and Mg2+ supplemented with 5 mM EDTA at 37°C for 10 min and then fixed in 4% paraformaldehyde for 20 min at room temperature. All flow cytometric analyses were done at the Massachusetts General Hospital Flow Cytometry Core (Simches Research Building), using an Amnis ImageStream mkII imaging flow cytometer. Live cells were distinguished by propidium iodide staining. Green fluorescent protein (GFP)-positive cells were counted, and the mean fluorescence intensity was quantified by using FlowJo v8.8.6 software.
Fluorescence imaging.
Live cells were imaged at 2 days postransduction. GFP expression in transduced cells was imaged by using an Evos FL cell imaging system (ThermoFisher), using a 10× objective and 50% GFP intensity. Images were analyzed by using ImageJ.
Virus binding assay.
Black-bottom 96-well plates were coated with poly-l-lysine (catalog number P4707; Sigma-Aldrich) by incubation at 37°C for 30 min. HEK293 overexpression or HEK293 AAVR KO cells were plated onto poly-l-lysine-coated plates at 100,000 cells per well overnight. Cells were incubated with 50 mU/ml neuraminidase from Vibrio cholerae type III (catalog number N7885; Sigma-Aldrich) in serum-free DMEM for 2 h, or untreated wells were incubated with serum-free DMEM alone at 37°C with 5% CO2. Vectors were preincubated in either serum-free DMEM or serum-free DMEM supplemented with 10 μg/ml soluble heparin (catalog number H3149; Sigma) for 1 h at 37°C and then prechilled on ice for 10 min prior to addition to cells. After neuraminidase incubation, cells were placed on ice for 10 min, and 109 VG per well of the prechilled vector (with or without heparin preincubation) were then added in a total volume of 50 μl per well. Vectors were allowed to bind cells on ice on an orbital shaker platform for 1 h. Following binding, cells were washed three times with ice-cold PBS with Mg2+ and Ca2+, and either 50 μl PBS was then added and cells were frozen immediately (“binding”) (Fig. 3) or 200 μl prewarmed D10 was then added and cells were cultured for 2 days prior to the luciferase assay (“transduction”) (Fig. 3). Binding assay plates underwent 3 freeze-thaw cycles, prior to resuspension and viral genome quantification by qPCR as described above using a CMV primer/probe set.
Western blotting and antibodies.
Whole-cell Western blot samples were prepared by lysing 1 × 105 cells per sample in 0.2% Triton X and 10 U/ml DNase I (catalog number M0303; New England BioLabs) in 1× DNase I buffer for 30 min on ice. After 30 min, samples were adjusted to contain 62.5 mM Tris (pH 7), 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 100 mM dithiothreitol (DTT), and 0.001% bromophenol blue. Samples were boiled at 100°C for 2 min prior to running on a NuPage 4 to 12% Bis-Tris gel (ThermoFisher) and transfer to PVDF membranes. Membranes were blocked at room temperature for 45 min by using 5% blotting-grade nonfat dry milk in PBS-Tween (PBST) (0.02%). Primary antibodies were incubated overnight in a 0.05% block solution. Antibodies used are as follows: mouse polyclonal anti-KIAA0319L (catalog number H00079932-B01; Novus Biologicals) at a 1:2,000 dilution, mouse monoclonal antiactin (catalog number MA1-744; ThermoFisher) at 1:10,000, and a sheep anti-mouse IgG–horseradish peroxidase (HRP) conjugate (catalog number GENXA931; GE Healthcare) at 1:2,000.
Virus overlay assay.
Individual His-tagged AAVR domains were expressed and purified as previously described (34), and recombinant proteins were loaded onto 10% or 6% SDS-PAGE gels at 0.5 μg/lane. Cellular membrane proteins were isolated by using detergent-based cellular fractionation (catalog number 9038; Cell Signaling). The virus overlay assay was performed as previously described (34). A20 intact capsid antibody (catalog number 03-61055; American Research Products, Inc.) was used for AAV2 overlay, and ADK4 intact capsid antibody (catalog number 03-651147; American Research Products, Inc.) was used for AAV4 overlay.
Lentivirus production.
Lentiviral constructs encoding AAVR-flag and AAVRΔC-tail were previously described (33). Lentivirus was produced from HEK293T cells (ATCC, Manassas, VA) by transient transfection using PolyJet in vitro DNA transfection reagent (catalog number SL100688; SignaJen) according to the manufacturer's protocol for lentivirus production. HEK293T cells were seeded overnight at 4 × 106 cells per 10-cm dish. One hour prior to transfection, medium was changed to fresh prewarmed D10, followed by transfection of psPAX2, pLenti-CMV-AAVR-flag-Puro or pLenti-CMV-AAVRΔC-tail-Puro, and pCMV-VSV-G at a 10:10:1 ratio. Medium was changed to fresh D10 6 h after transfection, and the supernatant virus was harvested 48 h later, clarified by centrifugation at 2,000 rpm for 5 min in a Sorvall tabletop centrifuge, and filtered through a 0.45-μm filter.
Generation of stable cell lines.
NIH 3T3 cells (ATCC, Manassas, VA) were seeded at 1 × 106 cells per well of a 6-well plate the night prior to transduction. Cells were transduced by centrifugation during infection (spinfection) for 30 min at 25°C at 2,500 rpm in a tabletop centrifuge, using 1 ml per well of the supernatant lentivirus in the presence of 10 μg/ml Polybrene (catalog number TR1003G; ThermoFisher Scientific). Medium was changed to fresh D10 following spinfection, and 1 day later, stably transduced cells were selected by using 5 μg/ml puromycin (catalog number P9620; Sigma-Aldrich) for 2 days.
Ethics statement.
All animal studies were carried out under conditions approved by the Stanford University Institutional Animal Care and Use Committee (IACUC), and mice were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Experimental protocols were approved under Stanford IACUC Asia Pacific Laboratory Accreditation Cooperation (APLAC) protocol number 28856. This protocol adheres to the guidelines set by the Public Health Service Policy on Humane Care and Use of Laboratory Animals (56) and the Guide for the Care and Use of Laboratory Animals, 8th ed. (57).
Animal studies.
Aavr KO mice carrying a 1-bp deletion (CCCGCTTCC—GGGTTTTGCCAGGA, where — marks the deletion) resulting in a frameshift mutation in Aavr (33) or WT cage mates were intravenously injected with 1 × 1011 VG/mouse via the retro-orbital route. Bioluminescence using the whole mouse as a region of interest was quantified at 7 and 21 days postinjection. Mice were anesthetized with 2% isoflurane and oxygen. A total of 3.3 μg of the d-luciferin substrate per mouse was injected intraperitoneally, and 10 min after injection, bioluminescent images were taken by using an IVIS 100 cryogenically cooled charge-coupled-device camera (Xenogen, Alameda, CA). Bioluminescence was recorded at 1, 10, 60, and 100 s, with the visual output representing the average radiance in photons emitted per second per square centimeter as a false color image where the maximum is red and the minimum is dark blue. The region of interest for radiance quantification (photons per second per square centimeter per radian) was designated the whole mouse.
ACKNOWLEDGMENTS
We thank members of the Vandenberghe and Carette laboratories for discussions and support and Eric Zinn for phylogenetic reconstruction analysis. We thank MEEI/SERI Gene Transfer Vector Core members Ru Xiao, Trisha Barungi, and Eva Andres-Mateos for viral vector production as well as David Dombkowski (MGH) for flow cytometry analysis.
A.M.D., S.P., A.S.P., J.E.C., and L.H.V. were responsible for experimental design. A.M.D. performed transduction experiments, generated stable cell lines, conducted virus binding assays, and confirmed virus overlay assays in cell lysates. S.P. generated AAVR KO cell lines. F.C. and J.Q. purified and conducted virus overlays on AAVR-purified proteins. A.M.D. conducted virus overlay assays on membrane preparations. A.S.P. and C.M.N. conducted the in vivo mouse study. A.M.D. wrote the manuscript, with intellectual input from S.P., A.S.P., C.M.N., F.C., J.Q., J.E.C., and L.H.V.
Funding for these studies was provided by National Institutes of Health (https://www.nih.gov/) grant DP2AI104557, National Institute of Allergy and Infectious Diseases (https://www.niaid.nih.gov/) grant U19AI109662 (J.E.C.), the Weston Havens Foundation (A.S.P. and S.P.), Giving/Grousbeck, and Lonza Houston (L.H.V.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We have the following competing interests. S.P., A.S.P., and J.E.C. are inventors on a patent describing the utilization of AAVR to modulate AAV transduction. L.H.V. is an inventor on a number of patents related to AAV gene transfer that are licensed to various biotechnology and pharmaceutical entities, including AAV9 and ancestral AAVs (AncAAVs). L.H.V. receives research support from Lonza Houston and Selecta Biosciences, both licensees of AncAAV technology. L.H.V. is a consultant to several companies pursuing AAV gene therapy, including Lonza Houston. L.H.V. is a cofounder of GenSight Biologics, an AAV gene therapy company.
REFERENCES
- 1.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. 2011. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, Della Peruta M, Lheriteau E, Patel N, Raj D, Riddell A, Pie J, Rangarajan S, Bevan D, Recht M, Shen YM, Halka KG, Basner-Tschakarjan E, Mingozzi F, High KA, Allay J, Kay MA, Ng CY, Zhou J, Cancio M, Morton CL, Gray JT, Srivastava D, Nienhuis AW, Davidoff AM. 2014. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stroes ES, Nierman MC, Meulenberg JJ, Franssen R, Twisk J, Henny CP, Maas MM, Zwinderman AH, Ross C, Aronica E, High KA, Levi MM, Hayden MR, Kastelein JJ, Kuivenhoven JA. 2008. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler Thromb Vasc Biol 28:2303–2304. doi: 10.1161/ATVBAHA.108.175620. [DOI] [PubMed] [Google Scholar]
- 4.Gaudet D, de Wal J, Tremblay K, Dery S, van Deventer S, Freidig A, Brisson D, Methot J. 2010. Review of the clinical development of alipogene tiparvovec gene therapy for lipoprotein lipase deficiency. Atheroscler Suppl 11:55–60. doi: 10.1016/j.atherosclerosissup.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Carpentier AC, Frisch F, Labbe SM, Gagnon R, de Wal J, Greentree S, Petry H, Twisk J, Brisson D, Gaudet D. 2012. Effect of alipogene tiparvovec (AAV1-LPL(S447X)) on postprandial chylomicron metabolism in lipoprotein lipase-deficient patients. J Clin Endocrinol Metab 97:1635–1644. doi: 10.1210/jc.2011-3002. [DOI] [PubMed] [Google Scholar]
- 6.Benkhelifa-Ziyyat S, Besse A, Roda M, Duque S, Astord S, Carcenac R, Marais T, Barkats M. 2013. Intramuscular scAAV9-SMN injection mediates widespread gene delivery to the spinal cord and decreases disease severity in SMA mice. Mol Ther 21:282–290. doi: 10.1038/mt.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nizzardo M, Simone C, Rizzo F, Salani S, Dametti S, Rinchetti P, Del Bo R, Foust K, Kaspar BK, Bresolin N, Comi GP, Corti S. 2015. Gene therapy rescues disease phenotype in a spinal muscular atrophy with respiratory distress type 1 (SMARD1) mouse model. Sci Adv 1:e1500078. doi: 10.1126/sciadv.1500078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armbruster N, Lattanzi A, Jeavons M, Van Wittenberghe L, Gjata B, Marais T, Martin S, Vignaud A, Voit T, Mavilio F, Barkats M, Buj-Bello A. 2016. Efficacy and biodistribution analysis of intracerebroventricular administration of an optimized scAAV9-SMN1 vector in a mouse model of spinal muscular atrophy. Mol Ther Methods Clin Dev 3:16060. doi: 10.1038/mtm.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying GS, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JI, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. 2009. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, Windsor EA, Conlon TJ, Sumaroka A, Pang JJ, Roman AJ, Byrne BJ, Jacobson SG. 2009. Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum Gene Ther 20:999–1004. doi: 10.1089/hum.2009.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL, Rossi S, Marshall K, Banfi S, Surace EM, Sun J, Redmond TM, Zhu X, Shindler KS, Ying GS, Ziviello C, Acerra C, Wright JF, McDonnell JW, High KA, Bennett J, Auricchio A. 2010. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther 18:643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett J, Ashtari M, Wellman J, Marshall KA, Cyckowski LL, Chung DC, McCague S, Pierce EA, Chen Y, Bennicelli JL, Zhu X, Ying GS, Sun J, Wright JF, Auricchio A, Simonelli F, Shindler KS, Mingozzi F, High KA, Maguire AM. 2012. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med 4: 120ra15. doi: 10.1126/scitranslmed.3002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. 2010. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol 84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. 2006. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol 80:9093–9103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 72:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol 76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol 75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. 2011. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem 286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med 5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 20.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. 2005. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol 79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol 80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurzeder C, Koppold B, Sauer G, Pabst S, Kreienberg R, Deissler H. 2007. CD9 promotes adeno-associated virus type 2 infection of mammary carcinoma cells with low cell surface expression of heparan sulphate proteoglycans. Int J Mol Med 19:325–333. [PubMed] [Google Scholar]
- 23.Blackburn SD, Steadman RA, Johnson FB. 2006. Attachment of adeno-associated virus type 3H to fibroblast growth factor receptor 1. Arch Virol 151:617–623. doi: 10.1007/s00705-005-0650-6. [DOI] [PubMed] [Google Scholar]
- 24.Ling C, Lu Y, Kalsi JK, Jayandharan GR, Li B, Ma W, Cheng B, Gee SW, McGoogan KE, Govindasamy L, Zhong L, Agbandje-McKenna M, Srivastava A. 2010. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum Gene Ther 21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med 9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 26.Weller ML, Amornphimoltham P, Schmidt M, Wilson PA, Gutkind JS, Chiorini JA. 2010. Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat Med 16:662–664. doi: 10.1038/nm.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao PJ, Samulski RJ. 2012. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol 86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhrig S, Coutelle O, Wiehe T, Perabo L, Hallek M, Buning H. 2012. Successful target cell transduction of capsid-engineered rAAV vectors requires clathrin-dependent endocytosis. Gene Ther 19:210–218. doi: 10.1038/gt.2011.78. [DOI] [PubMed] [Google Scholar]
- 29.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. 1999. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol 73:10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonnenmacher M, Weber T. 2011. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe 10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 32.Nicolson SC, Samulski RJ. 2014. Recombinant adeno-associated virus utilizes host cell nuclear import machinery to enter the nucleus. J Virol 88:4132–4144. doi: 10.1128/JVI.02660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, Deng X, Wosen JE, Davulcu O, Yan Z, Engelhardt JF, Brown KE, Chapman MS, Qiu J, Carette JE. 5 July 2017. AAV serotypes have distinctive interactions with domains of the cellular receptor AAVR. J Virol doi: 10.1128/JVI.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, Calcedo R, Sanmiguel J, Abbas Z, Wilson JM. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A 100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Calcedo R, Vandenberghe LH, Bell P, Somanathan S, Wilson JM. 2009. A new genetic vaccine platform based on an adeno-associated virus isolated from a rhesus macaque. J Virol 83:12738–12750. doi: 10.1128/JVI.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandenberghe LH, Breous E, Nam HJ, Gao G, Xiao R, Sandhu A, Johnston J, Debyser Z, Agbandje-McKenna M, Wilson JM. 2009. Naturally occurring singleton residues in AAV capsid impact vector performance and illustrate structural constraints. Gene Ther 16:1416–1428. doi: 10.1038/gt.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blacklow NR, Hoggan MD, Rowe WP. 1968. Serologic evidence for human infection with adenovirus-associated viruses. J Natl Cancer Inst 40:319–327. [PubMed] [Google Scholar]
- 40.Chiorini JA, Yang L, Liu Y, Safer B, Kotin RM. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J Virol 71:6823–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinn E, Pacouret S, Khaychuk V, Turunen HT, Carvalho LS, Andres-Mateos E, Shah S, Shelke R, Maurer AC, Plovie E, Xiao R, Vandenberghe LH. 2015. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep 12:1056–1068. doi: 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, Flannery JG, Schaffer DV. 2013. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med 5: 189ra76. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 43.Cronin T, Vandenberghe LH, Hantz P, Juttner J, Reimann A, Kacso AE, Huckfeldt RM, Busskamp V, Kohler H, Lagali PS, Roska B, Bennett J. 2014. Efficient transduction and optogenetic stimulation of retinal bipolar cells by a synthetic adeno-associated virus capsid and promoter. EMBO Mol Med 6:1175–1190. doi: 10.15252/emmm.201404077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamilton B, Zabner J. 2017. An alternative route of infection for a novel AAV, abstr 507. Abstr 20th Annu Meet Am Soc Gene Cell Ther, Washington, DC, 10 to 13 May 2017. [Google Scholar]
- 45.Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K, Agbandje-McKenna M, Wilson JM. 2011. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest 121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizukami H, Young NS, Brown KE. 1996. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glycoprotein. Virology 217:124–130. doi: 10.1006/viro.1996.0099. [DOI] [PubMed] [Google Scholar]
- 47.Summerford C, Samulski RJ. 2016. AAVR: a multi-serotype receptor for AAV. Mol Ther 24:663–666. doi: 10.1038/mt.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu J, Handa A, Kirby M, Brown KE. 2000. The interaction of heparin sulfate and adeno-associated virus 2. Virology 269:137–147. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- 49.Negishi A, Chen J, McCarty DM, Samulski RJ, Liu J, Superfine R. 2004. Analysis of the interaction between adeno-associated virus and heparan sulfate using atomic force microscopy. Glycobiology 14:969–977. doi: 10.1093/glycob/cwh118. [DOI] [PubMed] [Google Scholar]
- 50.Zhang F, Aguilera J, Beaudet JM, Xie Q, Lerch TF, Davulcu O, Colon W, Chapman MS, Linhardt RJ. 2013. Characterization of interactions between heparin/glycosaminoglycan and adeno-associated virus. Biochemistry 52:6275–6285. doi: 10.1021/bi4008676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller OJ, Leuchs B, Pleger ST, Grimm D, Franz WM, Katus HA, Kleinschmidt JA. 2006. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res 70:70–78. doi: 10.1016/j.cardiores.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 55.Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z, Wilson JM. 2010. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther 21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.National Institutes of Health. 2015. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 57.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]