ABSTRACT
Flaviviruses are arthropod-borne viruses that constitute a major global health problem, with millions of human infections annually. Their pathogenesis ranges from mild illness to severe manifestations such as hemorrhagic fever and fatal encephalitis. Type I interferons (IFNs) are induced in response to viral infection and stimulate the expression of interferon-stimulated genes (ISGs), including that encoding viperin (virus-inhibitory protein, endoplasmic reticulum associated, IFN inducible), which shows antiviral activity against a broad spectrum of viruses, including several flaviviruses. Here we describe a novel antiviral mechanism employed by viperin against two prominent flaviviruses, tick-borne encephalitis virus (TBEV) and Zika virus (ZIKV). Viperin was found to interact and colocalize with the structural proteins premembrane (prM) and envelope (E) of TBEV, as well as with nonstructural (NS) proteins NS2A, NS2B, and NS3. Interestingly, viperin expression reduced the NS3 protein level, and the stability of the other interacting viral proteins, but only in the presence of NS3. We also found that although viperin interacted with NS3 of mosquito-borne flaviviruses (ZIKV, Japanese encephalitis virus, and yellow fever virus), only ZIKV was sensitive to the antiviral effect of viperin. This sensitivity correlated with viperin's ability to induce proteasome-dependent degradation of NS3. ZIKV and TBEV replication was rescued completely when NS3 was overexpressed, suggesting that the viral NS3 is the specific target of viperin. In summary, we present here a novel antiviral mechanism of viperin that is selective for specific viruses in the genus Flavivirus, affording the possible availability of new drug targets that can be used for therapeutic intervention.
IMPORTANCE Flaviviruses are a group of enveloped RNA viruses that cause severe diseases in humans and animals worldwide, but no antiviral treatment is yet available. Viperin, a host protein produced in response to infection, effectively restricts the replication of several flaviviruses, but the exact molecular mechanisms have not been elucidated. Here we have identified a novel mechanism employed by viperin to inhibit the replication of two flaviviruses: tick-borne encephalitis virus (TBEV) and Zika virus (ZIKV). Viperin induced selective degradation via the proteasome of TBEV and ZIKV nonstructural 3 (NS3) protein, which is involved in several steps of the viral life cycle. Furthermore, viperin also reduced the stability of several other viral proteins in a NS3-dependent manner, suggesting a central role of NS3 in viperin's antiflavivirus activity. Taking the results together, our work shows important similarities and differences among the members of the genus Flavivirus and could lead to the possibility of therapeutic intervention.
KEYWORDS: ISG, viperin, NS3, flavivirus, proteasomal degradation, interferons
INTRODUCTION
The genus Flavivirus of the family Flaviviridae includes arthropod-borne viruses that cause millions of human infections annually. Despite the extensive research and concern for public health, there are no specific antiviral drugs for any flavivirus infection. Significant members of this genus include dengue virus (DENV) and yellow fever virus (YFV), which cause hemorrhagic fevers; tick-borne encephalitis virus (TBEV), Japanese encephalitis virus (JEV), and West Nile virus (WNV), which cause severe encephalitis (1); and the newly emerging Zika virus (ZIKV), which may cause microcephaly in neonates and Guillain-Barré syndrome in adults (2). Flaviviruses are small, spherical, enveloped viruses containing a positive-sense RNA genome of about 11 kb that has one open reading frame, which is translated into a polyprotein. The polyprotein is processed co- and posttranslationally by viral and cellular proteases to produce three structural proteins (capsid [C], premembrane [prM; the precursor form of M], and envelope [E]) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins form the virus particle, while the nonstructural proteins support viral RNA replication and virion assembly and counteract the antiviral immune response of the host (1, 3).
The innate immune system acts as the first line of defense against invading viruses, and interferons (IFNs) are key components of this defense. These cytokines are induced and secreted by infected cells when pathogens are recognized. After binding to cell surface receptors, they initiate a signaling cascade that leads to the upregulation of hundreds of IFN-stimulated genes (ISGs). Many ISGs act to limit viral replication, such as those encoding protein kinase R (PKR); the GTPase Mx1; the OAS/RNase L pathway; ISG15; the IFN-induced protein with tetratricopeptide repeats (IFIT) family; and virus-inhibitory protein, endoplasmic reticulum-associated, IFN-inducible (viperin) (4, 5). Viperin is an iron-sulfur protein (6, 7), the expression of which is highly upregulated upon viral infection. It has antiviral activity against a broad range of viruses, including influenza A virus (8, 9); human immunodeficiency virus (HIV) (10); chikungunya virus (11); Sindbis virus (12); respiratory syncytial virus (13); Junin virus (14); and also members of the family Flaviviridae such as hepatitis C virus (HCV), DENV, WNV, ZIKV, and TBEV (7, 15–19).
Viperin appears to be able to act at several stages of the viral life cycle and uses different mechanisms to inhibit viral replication. The antiviral activities can be attributed to viperin's interaction with several host factors and also with different viral proteins. Viperin inhibits the release of influenza A virus and human immunodeficiency virus (HIV) particles by disrupting lipid rafts (9, 10). Furthermore, it prevents replication of HCV RNA by interacting with HCV NS5A and host protein VAP-A, thus preventing the formation of replication complexes (18). In DENV-2-infected cells, viperin colocalizes with both NS3 protein and viral RNA, both of which are components of the flavivirus replication complex (15). TBEV is also highly sensitive to viperin. In this case, viperin selectively blocks positive-sense RNA amplification during the early stages of TBEV infection (7). However, it is not known whether viral proteins are targeted by viperin—and, if so, which of them are targeted. The antiviral mechanism is also unknown.
In this report, we show that viperin induces proteasome-dependent degradation of viral proteins as an antiviral mechanism. We identify several different viral proteins that viperin interacts with, the most important being NS3. This antiviral mechanism seems to be virus species specific, as viperin binds and degrades NS3 of ZIKV and TBEV but only binds to JEV and YFV NS3 without affecting viral replication.
RESULTS
Viperin interacts with and colocalizes with the prM, E, NS2A, NS2B, and NS3 proteins of TBEV.
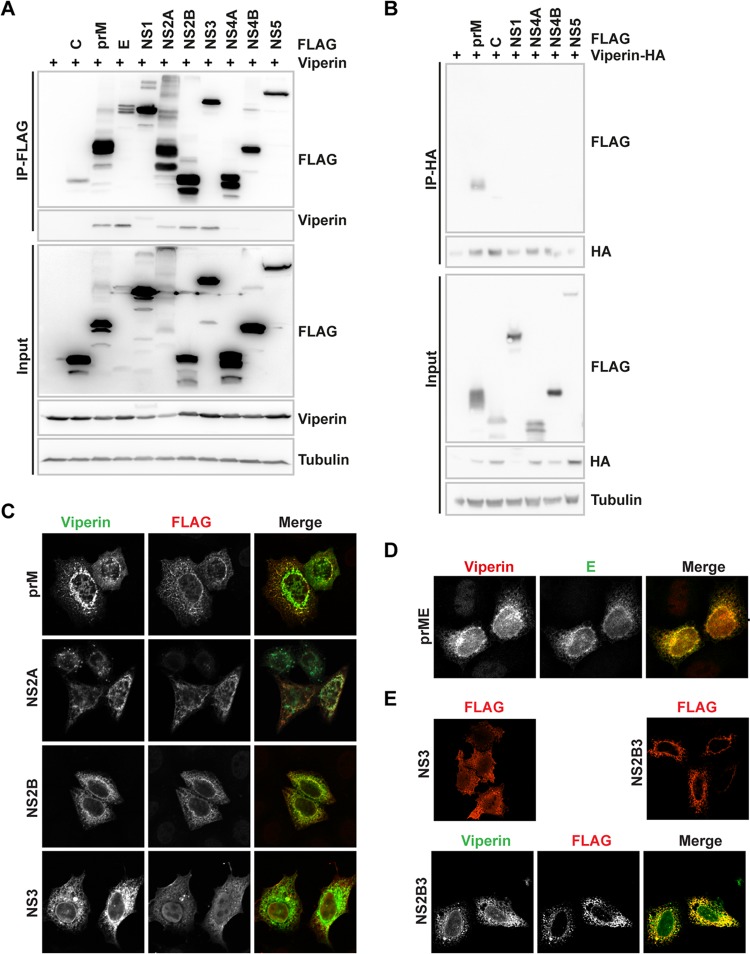
A key approach to understanding viperin's antiviral mechanism against flaviviruses is to identify the viral interactome. TBEV is highly sensitive to viperin and was therefore chosen as a model virus. HEK293T cells were transfected with plasmids encoding the TBEV proteins (C-terminally 3×FLAG tagged) along with viperin, which was followed by a coimmunoprecipitation (Co-IP) assay. Interestingly, five viral proteins (prM, E, NS2A, NS2B, and NS3) coprecipitated with viperin (Fig. 1A). Since weak bands were present also for NS4A and NS4B with long exposure, coimmunoprecipitations were performed in the reverse setup. Green fluorescent protein (GFP) and prM were used as negative and positive controls, respectively. In this setup, C, NS1, NS4A, NS4B, and NS5 could not coprecipitate with viperin (Fig. 1B). Next, the intracellular location of the viral proteins and viperin was investigated. HeLa cells were transfected with plasmids expressing the viral proteins and viperin, stained, and visualized using confocal microscopy. Viperin was found in close proximity to prM, E, NS2A, NS2B, and NS3 (Fig. 1C and D). The location of the viral proteins in the cell did not change in the presence of viperin (data not shown). In the case of NS3, a weak staining of nucleus was observed. Since NS3 is expressed as a polyprotein during virus infection, we compared the intracellular localization of NS3 expressed alone to that of NS3 expressed together with NS2B as a polyprotein. Clear cytoplasmic and nuclear staining was observed in NS3-expressing cells; however, when NS2B was coexpressed with NS3, no nuclear staining was observed for NS3 (Fig. 1E). Next, colocalization between viperin and NS2B3 was analyzed, and the signals for viperin and NS2B3 overlapped (Fig. 1E).
FIG 1.
Viperin interacts and colocalizes with TBEV prM, E, NS2A, NS2B, and NS3. (A) HEK293T cells were transiently transfected with plasmids encoding wild-type viperin and FLAG-tagged TBEV proteins and were subjected to anti-FLAG immunoprecipitation. Whole-cell lysates (input) and immunoprecipitated (IP) proteins were analyzed by immunoblotting using anti-FLAG and anti-viperin antibodies, including input levels of tubulin as a loading control. (B) HEK293T cells were transiently transfected with plasmids encoding HA-tagged viperin and FLAG-tagged TBEV prM, C, NS1, NS4A, NS4B, and NS5 and were subjected to anti-HA immunoprecipitation. Whole-cell lysates (input) and immunoprecipitated (IP) proteins were analyzed by immunoblotting using anti-FLAG and anti-HA antibodies, including input levels of tubulin as a loading control. (C and D) Immunofluorescence analysis of HeLa cells transiently transfected with plasmids expressing FLAG-tagged TBEV prM, NS2A, NS2B, and NS3 along with viperin (C) or prME with viperin (D). Proteins were detected using anti-FLAG, anti-viperin, and anti-E antibodies. (E) HeLa cells were transiently transfected with plasmids expressing FLAG-tagged TBEV NS3, NS2B3, or NS2B3 together with viperin. Representative images and blots are shown.
Viperin expression results in degradation of TBEV NS3.
To examine the effect of viperin on the expression levels of the TBEV proteins with which it interacted, HEK293T cells were cotransfected with plasmids encoding viperin and FLAG-tagged C, prM, E, NS2A, NS2B, and NS3. The cells were fixed 24 h posttransfection and stained, and the cellular fluorescence corresponding to protein expression was measured using a Trophos Plate Runner. Our results showed a significant reduction in NS3 expression in the presence of viperin, while the expression of the other viral proteins remained unaffected (Fig. 2A). To confirm these results, increasing amounts of viperin expression plasmid were transfected into cells, together with plasmid expressing NS3. Proteins were expressed for 24 h and analyzed by Western blotting followed by quantification of the bands. There was a dose-dependent reduction in NS3 protein levels (Fig. 2B). For the other viral proteins, no significant differences in the amount of protein were detected when viperin was present (Fig. 2C).
FIG 2.
Viperin facilitates degradation of TBEV NS3. (A) HEK293 T cells were transfected with FLAG-tagged TBEV C, prM, E, NS2A, NS2B, and NS3 in the presence of human viperin or empty vector. The cells were fixed 24 h posttransfection and stained using antibodies directed against FLAG tag and viperin and then stained with Alexa Fluor secondary antibodies, and fluorescent signal was quantified using a Trophos Plate Runner. Error bars represent means ± standard errors of the means (SEM) of data from three independent experiments performed in triplicate. (B) HEK293 T cells were transfected with plasmids expressing TBEV HA-NS3 and increasing amounts of viperin, and proteins were separated by SDS-PAGE and visualized by Western blotting. Band intensities were quantified, and the percentages of NS3-HA were calculated. Error bars represent means ± SEM of data from three independent experiments. (C) HEK293 T cells were transfected with plasmids expressing FLAG-tagged TBEV prM, E, NS2A, or NS2B with increasing amounts of viperin, protein amounts were analyzed with Western blotting, band intensities were quantified, and percentages were calculated. Total DNA amounts were always kept constant with addition of empty vector. Error bars represent means ± standard deviations (SD) of data from three independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (Student t test).
In summary, our data show that although viperin interacted and colocalized with the structural proteins prM and E, as well as with the nonstructural proteins NS2A, NS2B, and NS3, only the stability of NS3 was affected by viperin expression.
TBEV NS3 interacts with TBEV E, prM, NS2A, and NS2B and facilitates their degradation by viperin.
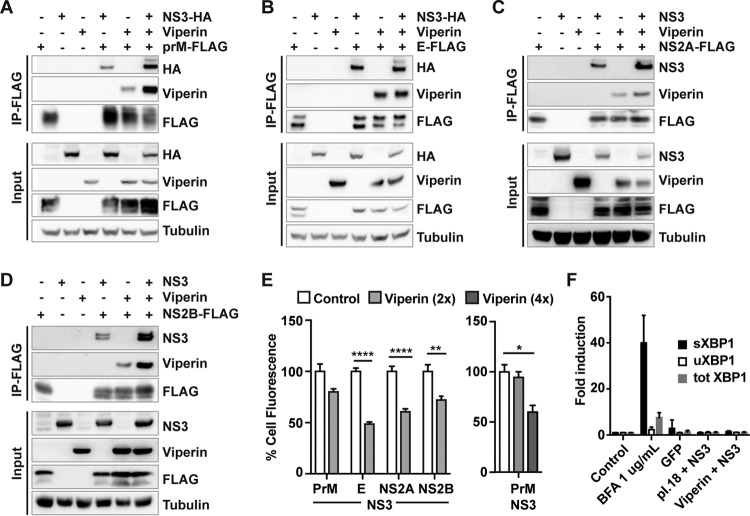
The viral NS3 protein is multifunctional. It is part of the flavivirus protease complex NS2B/NS3, which is involved in processing of the viral polyprotein. NS3 is the viral helicase that unwinds the double-stranded RNA before positive-strand RNA synthesis, and it also has nucleoside 5′-triphosphatase (NTPase) activity at its C terminus, important for viral RNA replication and virus particle formation (20, 21). In addition, flavivirus NS3 has been shown to have a role in the evasion of host innate immune responses (22–24). NS3 is essential for the life cycle of flaviviruses and interacts with most of the viral proteins. Thus, the interaction of viperin with NS3 could also affect other viral proteins. To investigate this, we performed a coimmunoprecipitation experiment. FLAG-tagged TBEV E, prM, NS2A, and NS2B were expressed in HEK293T cells along with hemagglutinin (HA)-tagged or nontagged NS3 and/or viperin. All the other viral proteins (prM, E, NS2A, and NS2B) interacted with NS3, and, interestingly, for prM, NS2A, and NS2B, the interaction with NS3 was enhanced in the presence of viperin (Fig. 3A to D). Since NS3 and viperin form a complex with the other viral proteins, we were interested to investigate the expression levels of the other viral proteins in the presence of NS3 and viperin. E, prM, NS2A, and NS2B were expressed together with NS3 and with or without viperin (with a ratio of viperin to TBEV plasmid of 2:1), and target protein levels were examined by fixing and staining the viral proteins and measuring the cell fluorescence. Interestingly, with expression of viperin, the expression levels of E, NS2A, and NS2B were significantly reduced in the presence of NS3 (Fig. 3E, left panel). No effect on prM was detected (Fig. 3E), but its expression levels were affected when the levels of viperin were increased further (Fig. 3E, right panel). Overexpression and accumulation of unfolded or misfolded proteins localized to the endoplasmic reticulum (ER) might induce ER stress responses that may cause unspecific degradation (25). As a marker for ER stress, XBP1 splicing can be measured. Cells were treated with ER stress inducer brefeldin A 1 (BFA1) or left untreated (control) or transfected with plasmids expressing GFP, empty plasmid (pI.18), or TBEV NS3 with or without viperin, and the levels of different XBP1 splice forms were measured with quantitative reverse transcription-PCR (RT-PCR) (26). The BFA1 positive control induced increased splicing of XBP1, whereas all other treatments did not (Fig. 3F).
FIG 3.
Viperin forms a complex with NS3 and TBEV prM, E, NS2A, and NS2B and reduces protein stability. (A to D) Co-IP analysis of HEK293 T cells expressing FLAG-tagged prM (A), E (B), NS2A (C), or NS2B (D) with either HA-tagged or untagged NS3 or viperin or both. Viral proteins were immunoprecipitated using a mouse monoclonal anti-FLAG antibody. The immunoblots show protein input and co-IP, including input levels of tubulin as a loading control. (E) (Left panel) HEK293 T cells were transfected with FLAG-tagged TBEV prM, E, NS2A, or NS2B along with HA-NS3 in the presence of viperin or empty vector (Control). The cells were fixed 24 h posttransfection and stained, and fluorescent signal was quantified using a Trophos Plate Runner. Error bars represent means ± SEM of data from three independent experiments performed in triplicate. (Right panel) HEK293 T cells were transfected with plasmids expressing FLAG-tagged prM along with HA-NS3 alone or with increasing amounts of viperin or empty vector (control). FLAG immunoblots were quantified, and the results are presented as percent FLAG-prM expression. (F) HEK293T cells were transfected with NS3 in the presence of viperin or empty expression vector or GFP or were treated with 1 μg/ml BFA; control cells were left untreated and untransfected. At 24 h after treatment, the amounts of spliced, unspliced, and total XBP1 were quantified using qPCR. Expression levels were normalized to the endogenous actin expression and are depicted as fold induction in comparison the results seen with untreated cells. Error bars represent means ± SEM of data from three independent experiments (n = 5). *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (Student t test).
Our results show that viperin did not interfere with the binding of NS3 to the other viral proteins but promoted NS3-dependent degradation of prM, E, NS2A, and NS2B.
Viperin interacts with and localizes close to flavivirus NS3.
The binding to and degradation of NS3 by viperin and the subsequent degradation of E, prM, NS2A, and NS2B in the presence of NS3 could be key steps in the antiviral mechanism of viperin, as the degraded viral proteins participate in several stages of viral infection. It was therefore important to show the interaction between NS3 and viperin during viral infection. HEK293T cells were infected with TBEV at a multiplicity of infection (MOI) of 2 and subsequently transfected with either viperin or empty plasmid vector. It was found that immunoprecipitation of viperin could coprecipitate NS3 (Fig. 4A), showing the relevance of the interaction. To determine whether this is a general mechanism against flaviviruses, we performed a coimmunoprecipitation screen to examine whether viperin interacts with the NS3 of TBEV (strain Torö, European subtype), ZIKV (strain MR766), YFV (strain Asibi), or JEV (strain Nakayama). HA-tagged NS3 of TBEV strain Torö, JEV, and YFV could efficiently pull down viperin, but the coimmunoprecipitation with ZIKA NS3 was quite inefficient (Fig. 4B). However, efficient pulldown of ZIKV NS3 was achieved using the reverse experimental setup (Fig. 4C). An immunofluorescence assay revealed that viperin was located close to the NS3 proteins of TBEV Torö, JEV, YFV, and ZIKV (Fig. 4D).
FIG 4.
Viperin binds to and is found in close proximity to flavivirus NS3. (A) HEK293T cells were infected with TBEV (MOI, 2) or mock infected for 1 h and were then transiently transfected with plasmid encoding FLAG-tagged wild-type viperin or empty vector (control). At 24 h postinfection, cells were lysed and immunoprecipitated using mouse monoclonal anti-FLAG antibody, as described before. The immunoprecipitated and whole-cell lysate (input) proteins were analyzed using immunoblots and antibody directed against the specific target proteins. (B and C) Protein-protein interaction analysis of flavivirus NS3 and viperin. Co-IP analysis of HEK293T cells expressing HA-tagged NS3 from TBEV Torö, JEV, YFV, and ZIKV with or without viperin was performed. Viral proteins were immunoprecipitated using anti-HA antibody (B), or viperin was immunoprecipitated with anti-viperin antibody (C). The immunoblots show protein input and co-IP, including input levels of tubulin as a loading control. Representative blots from three independent experiments are shown. (D) Colocalization analysis of viperin and flavivirus NS3. Immunofluorescence analysis of HeLa cells expressing HA-tagged NS3 from TBEV Torö, ZIKV MR766, YFV, and JEV along with viperin was performed.
Viperin specifically inhibits TBEV and ZIKV by degrading NS3 in a proteasome-dependent manner.
Next, we examined the stability of the different flavivirus NS3 proteins in the presence of increasing amounts of viperin, using Western blotting. Interestingly, although viperin interacted with all the flavivirus NS3 proteins tested, the effect on stability was not a general mechanism. Viperin expression strongly reduced the stability of the TBEV (Torö) and ZIKV (MR766) NS3 proteins, whereas only a 5-fold excess of viperin showed a moderate effect on YFV NS3 (Fig. 5A to D). To further analyze the effect of the antiviral mechanism on viral replication, FLP-IN T-Rex cells inducibly expressing viperin (27) were infected with different flaviviruses, progeny particle production was analyzed with a focus-forming assay, and the protein amount in cell lysate was analyzed by immunoblotting 24 h after infection (Fig. 5E and F). Interestingly, the antiviral effect of viperin correlated very well with its ability to reduce NS3 protein levels of TBEV and ZIKV. Viperin also showed a strong antiviral effect against both the laboratory-adapted ZIKV MR766 strain and low-passage-number ZIKV strain Brazil (Fig. 5E and F). Although NS3 of JEV was degraded in the presence of viperin (Fig. 5B), JEV has been shown to counteract the antiviral effects of viperin by targeting viperin for proteasomal degradation (28).
FIG 5.
Viperin specifically inhibits TBEV and ZIKV by degrading NS3 in a proteasome-dependent manner. (A to D) Stability of flavivirus NS3 in the presence of viperin. HEK293 T cells were transfected with expression plasmids expressing HA-tagged NS3 from TBEV Torö (A), JEV (B), YFV (C), and ZIKV (D) in the absence of or with increasing amounts of viperin, and total DNA levels were kept constant with empty vector. Immunoblots were quantitated, and the results are presented as percentages of NS3 compared to control. Error bars represent means ± SEM of data from three independent experiments. (E) Screening of the antiviral effect of viperin against different flaviviruses. FLP-IN T-Rex cells inducibly expressing FLAG-tagged viperin were treated for 24 h with 1 μg/ml Tet and infected with TBEV Torö, ZIKV MR766, ZIKV Brazil, YFV, or JEV at an MOI of 0.1. Viral replication was monitored 24 h postinfection (p.i.) using a focus-forming assay (Ffu). Means ± SEM are shown and represent data from three independent experiments performed in triplicate. (F) FLP-IN T-Rex cells inducibly expressing FLAG-tagged viperin were treated for 24 h with 1 μg/ml Tet and infected with TBEV Torö or ZIKV Brazil, and cell lysates were analyzed 24 h p.i. with immunoblotting using antibodies directed against viperin, NS3, and tubulin. (G and H) Cells transfected with plasmids expressing HA-NS3 of TBEV or ZIKV together with either viperin or GFP were treated with either proteasomal inhibitor MG132 or DMSO (as a control). Immunoblot analysis of cell lysates was performed with tubulin as the loading control (bottom panels). Results of quantification of band intensities of NS3 normalized to tubulin are shown as percentages of NS3. Error bars represent means ± SEM of data from four (G) or two (H) independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant (calculated with unpaired Student t test).
The proteasome is a major cellular pathway of protein degradation. To determine whether the proteasome was involved in viperin-mediated reduction of TBEV and ZIKV NS3 protein levels, we used proteasome inhibitor MG132. Transfected cells were treated with either MG132 or the solvent dimethyl sulfoxide (DMSO) as a control, and the amounts of NS3 and viperin were determined with Western blotting. Indeed, the amount of TBEV and ZIKV NS3 was restored when the proteasome was inhibited (Fig. 5G and H).
In conclusion, although the NS3 proteins of TBEV, JEV, YFV, and ZIKV interacted and colocalized with viperin, only TBEV and ZIKV were found to be sensitive to the antiviral action of viperin and this correlated with the efficient viperin-mediated degradation of their NS3 proteins via the proteasome.
Exogenous addition of NS3 to cells can restore viral growth in the presence of viperin.
The degradation of TBEV and ZIKV NS3 protein by viperin most likely contributes to the antiviral activity against these viruses. To test this hypothesis, HEK293T cells were transiently transfected with plasmids encoding viperin and increasing amounts of plasmid encoding TBEV NS3 and were subsequently infected with TBEV Hypr (MOI, 0.1). Alternatively, 293T FLP-IN T-Rex cells inducibly expressing viperin were induced by tetracycline and transfected with increasing amounts of plasmid encoding ZIKV NS3, followed by ZIKV infection (MOI, 0.1). The results showed that exogenous expression of NS3 could inhibit the antiviral effect of viperin against both TBEV and ZIKV, as determined by both viral RNA levels (data not shown) and the production of progeny virus particles (Fig. 6A and B). As NS3 of flaviviruses has been shown previously to be able to act only in cis and not in trans (29), the mechanism of action might be that exogenous NS3 acts as a decoy for viperin. To test this hypothesis, increasing amounts of NS3 of JEV or ZIKV were transfected together with viperin followed by TBEV infection. Both JEV NS3 expression and ZIKV NS3 expression could rescue the TBEV growth in the presence of viperin (Fig. 6C and D).
FIG 6.
Exogenous addition of NS3 completely restored viral growth in the presence of viperin. (A) HEK 293 cells were transiently transfected with plasmids expressing viperin and TBEV Hypr NS3, as indicated, and infected with Hypr (MOI, 0.1) for 24 h. Titers were analyzed using a focus-forming assay. (B) 293T FLP-IN T-Rex cells inducibly expressing viperin were induced with tetracycline as indicated, transfected with a plasmid expressing ZIKV NS3, and infected with ZIKV MR766 (MOI, 0.1). Supernatants were harvested 24 h p.i., and virus titers were determined using a focus-forming assay. (C and D) HEK 293 cells were transiently transfected with plasmids expressing viperin and JEV NS3 (C) or ZIKV MR766 NS3 (D), as indicated, and were infected with Hypr (MOI, 0.1) for 24 h. Titers were analyzed using a focus-forming assay. The total DNA amount was kept constant with empty vector. Means ± SEM of data from three independent experiments are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (calculated with unpaired Student t test).
Taken together, our results have revealed that viperin has a novel antiviral mechanism against TBEV and ZIKV. Viperin was able to inhibit the replication of both TBEV and ZIKV effectively, and there was a strong correlation between this antiviral effect and the ability of viperin to degrade their NS3 proteins; also, overexpression of NS3 efficiently reduced the antiviral effect. Furthermore, all the structural and nonstructural proteins of TBEV that interacted with viperin (prM, E, NS2A, and NS2B) were also degraded—but only in the presence of NS3.
DISCUSSION
Viperin is an IFN-stimulated gene product with a well-documented ability to inhibit a broad spectrum of viruses at different stages of their life cycle. However, the antiviral mechanism of viperin has been elucidated for only a few viruses. Viperin prevents HIV and influenza A virus from budding from the plasma membrane by affecting membrane fluidity and disrupting lipid rafts. Viperin achieves this effect by binding to and inhibiting farnesyl diphosphate synthase, an enzyme involved in cholesterol and isoprenoid metabolism (9, 10). Nevertheless, the ability of viperin to disrupt lipid rafts does not explain its activity against other viruses such as TBEV (7). In the case of respiratory syncytial virus, virus filament formation and transmission are inhibited by viperin (30). Viperin also inhibits several members of the family Flaviviridae, including HCV, DENV, WNV, ZIKV, and TBEV (7, 15–19). In HCV-infected cells, viperin binds to and interacts with the NS5A protein of HCV on the surface of lipid droplets and within the replication complexes. It also interacts with proviral host factor VAP-A in the replication complexes and impairs its association with NS5A, which is critical for HCV replication (18). For DENV-2, viperin reduces early RNA production by interfering with the replication complex and was found to interact with viral RNA and colocalize with NS3 (15). However, the exact mechanism of inhibition has not been clarified. Viperin also targets the early replication of TBEV, specifically, the positive-strand RNA replication (7). Since the production of flavivirus positive-sense RNA has an absolute requirement for the helicase activity of NS3, it is an attractive candidate for the viral target of viperin. Indeed, we were able to show that viperin can interact with the NS3 proteins of several different flaviviruses (TBEV, JEV, YFV, and ZIKV) and during TBEV infection. However, the antiviral activity of viperin against these viruses was strictly correlated to its ability to reduce the stability of NS3, indicating that although the binding motif is conserved in the NS3 proteins of different flaviviruses, the degradation is species specific. Although NS3 of JEV was degraded in the presence of viperin, viperin showed no antiviral activity against JEV; this can be explained by the fact that JEV counteracts viperin by downregulating its expression through proteasomal degradation (28). Remarkably, viperin's antiviral effect was abolished by exogenous addition of plasmid encoding NS3 of either TBEV or ZIKV. As the addition of NS3 in trans have been reported to not be able to complement the replication cycle of a flavivirus (29) and as adding NS3 from another flavivirus can restore virus replication in the presence of viperin, the mechanism is most likely due to a decoy mechanism.
We also found that the reduced stability of NS3 depended on the presence of a functional proteasome. Similar mechanisms have been reported for TRIM22 and ISG12a, two other antiviral ISGs. TRIM22 possesses an E3 ubiquitin ligase activity that ubiquitinates encephalomyocarditis virus 3C protease and mediates its degradation via the proteasome (31). In the case of ISG12a, it interacts with HCV NS5A and induces its proteasome-dependent degradation. In sharp contrast to the TRIM22 mechanism, ISG12a does not itself possess an E3 ligase activity but functions as an adaptor that promotes an E3 ligase, S-phase kinase-associated protein 2 (SKP2), to interact with and degrade NS5A (32). Viperin has not been shown to possess an E3 ubiquitin ligase activity like that of TRIM22, but it could possibly interact with one and function as an adaptor in a way similar to that seen with ISG12a. However, no possible E3 ubiquitin ligase has been identified yet. TRIM79α, on the other hand, has been shown to restrict TBEV replication by direct targeting of NS5, the viral RNA-dependent RNA polymerase, and a major IFN antagonist, for degradation via the lysosome (33).
NS3 is a multifunctional protein of approximately 70 kDa. In addition to its NTPase/helicase activity, it has serine protease activity, which, together with its cofactor protein NS2B, participates in processing of the polyprotein. NS3 also has an RNA 5′-triphosphatase at its C terminus that is important for 5′ capping of viral RNA (20, 21); it has been shown to play a role in virus assembly, and this function is not associated with the other enzymatic functions of NS3 (22, 23). Along with all the aforementioned functions, DENV NS3 recruits and stimulates fatty acid synthase, contributing to the remodeling of cytosolic membranes and the establishment of replication sites (34), while it also inhibits type I IFN production in infected cells by cleaving the adaptor protein STING (24, 35). To perform all these diverse functions, NS3 interacts with several viral proteins. It was interesting to see that viperin interacts not only with NS3 but also with the structural proteins prM and E (which constitute the viral shell), NS2A, and the protease cofactor NS2B. Although viperin induced degradation of NS3, these other viral proteins that interacted with viperin were degraded only in the presence of NS3. NS3 bound to all of these proteins irrespective of the presence of viperin. We and others have detected a strong antiviral effect on RNA replication early in infection (7, 15). This effect can be explained by the degradation of the viral proteins (NS2A, NS2B, and NS3) early, when the ratio of viperin to viral proteins is high. This might be important in cells expressing viperin at a baseline level, for example, in astrocytes in the brain (36). In addition to the effect on replication, with TBEV we have detected an effect on infectious particle release that is even stronger than that seen with RNA late in infection, in the presence of viperin (7). Recently, we showed that viperin interferes with assembly of TBEV by interacting with and inhibiting the function of the cellular protein Golgi brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF1) (37). GBF1 is important for cellular vesicular trafficking in cells, and viperin expression results in secretion of noninfectious capsid particles (37). It seems that particle formation might be targeted by additional mechanisms, as prM and E make up the viral coat and many of the viral proteins that interact with viperin are involved in both RNA replication and particle assembly. This suggests that targeting of the viral NS3 protein for proteasome degradation may be a universal antiviral strategy that might be exploited in the future for the purpose of controlling infection.
Overall, the present study has revealed a novel mechanism of viperin against both a tick-borne flavivirus (TBEV) and a mosquito-borne flavivirus (ZIKV). The molecular mechanism of restriction was the direct targeting of flavivirus NS3 protein for proteasomal degradation, and the fact that viperin and NS3 together induced degradation of other important structural and nonstructural proteins may indicate the potential antiviral activity of viperin.
MATERIALS AND METHODS
Cell culture, reagents, and viruses.
HEK293T and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 5% fetal calf serum (FCS; Gibco), 20 units/ml penicillin, and 20 μg/ml streptomycin (GE Healthcare) at 37°C in 5% CO2. Human 293T FLP-IN T-Rex cells inducibly expressing viperin were a kind gift from Ju-Tao Gou (27) and were cultured in DMEM with penicillin and streptomycin as described above but with 5% tetracycline-negative FCS (HyClone; Thermo Scientific). Cells were transfected using GeneJuice transfection reagent (Novagen) according to the manufacturer's instructions. Cell culture-grade MG132 and dimethyl sulfoxide (DMSO) were purchased from Sigma. TBEV strain Torö European subtype (an infectious clone sequenced and cloned from TBEV in a questing tick [38]), TBEV strain Hypr71, and ZIKV strain MR766 were kind gifts from G. Dobler (Bundeswehr Institute of Microbiology, Munich, Germany); low-passage-number Brazilian ZIKV strain ZIKV/H. sapiens/Brazil/PE243/2015 (GenBank accession number KX197192 [abbreviated ZIKV PE243; referred to only as ZIKV Brazil in this paper]) (39) was a kind gift of Alain Kohl; and JEV (strain Nakayama) was a kind gift from S. Vene (Folkhälsoinstitutet, Stockholm, Sweden). YFV (strain Asibi) was kindly provided by M. Niedrig (Robert Koch Institute, Berlin, Germany).
Antibodies.
Mouse monoclonal anti-FLAG M2 (F1804) and rabbit polyclonal anti-FLAG (F7425) were purchased from Sigma. Rabbit polyclonal anti-beta tubulin (6046), mouse monoclonal anti-HA tag (18181), rabbit polyclonal anti-HA tag (9110), mouse monoclonal anti-viperin (107359), and rabbit polyclonal anti-viperin (73864) were purchased from Abcam. Mouse monoclonal anti-TBEV E 1493.1 was a kind gift from M. Niedrig (Robert Koch Institute, Berlin, Germany [40]). Mouse monoclonal anti-flavivirus E for JEV (HB-112) and mouse monoclonal anti-YFV E (CRC1689) were from ATCC. Rabbit polyclonal anti-ZIKV NS3 was a kind gift from Andres Merits (University of Tartu, Tartu, Estonia) (41). Goat anti-mouse IgG (31430) (horseradish peroxidase [HRP] conjugated) was from Thermo Scientific; goat anti-rabbit IgG (31460) (HRP conjugated) was from Thermo Scientific; and rabbit anti-chicken IgG (AS101489) (HRP conjugated) was from Agrisera. Goat anti-mouse IgG Alexa Fluor 647 (A-21235), donkey anti-rabbit IgG Alexa Fluor 488 (A-21206), donkey anti-rabbit IgG Alexa Fluor 555 (A-31572), donkey anti-mouse IgG Alexa Fluor 488 (A-21202), and donkey anti-mouse IgG Alexa Fluor 555 (A-31570) were all purchased from Invitrogen. Chicken polyclonal anti-TBEV NS3 was raised against the peptide carboxyl-CEGRDIKEFVAYASGRR-amide; the antibodies were purified from eggs using standard immunization and purification protocols (Agrisera).
Plasmids.
Expression plasmids (backbone pI.18) encoding human viperin; GFP (7); C-terminally 3×FLAG-tagged TBEV Hypr C (without a C-terminal anchor); prM; E; NS1; NS2A; NS2B; NS3; NS2B3; NS4A; NS4B; NS5; and untagged TBEV NS3 were generated previously (42). N-terminally HA-tagged NS3 of TBEV (strains Hypr and Torö), JEV (Nakayama), YFV (Asibi), and ZIKV (MR766) were cloned into eukaryotic expression vector pI.18 (kindly provided by Jim Robertson, National Institute for Biological Standards and Control, Hertfordshire, United Kingdom) using standard PCR cloning methods. KOD Hot Start polymerase (Novagen), restriction endonucleases, and T4 DNA ligase (Thermo Fisher Scientific) were used according to the manufacturer's recommendations. All plasmids were sequenced to verify their correctness, and the oligonucleotide primer sequences are available upon request.
Coimmunoprecipitation (co-IP).
HEK293T cells were transfected with plasmids expressing the proteins of interest (24 h), washed with phosphate-buffered saline (PBS), and lysed in IP lysis/wash buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail [Roche]) for 20 min on ice. Cell lysates were centrifuged at 13,000 rpm for 10 min at 4°C, and the supernatants were precleared by addition of 50 μl of washed protein A agarose and fast-flow bead slurry (Millipore) and rotated for 2 h at 4°C. Beads were removed by centrifugation, and 50 to 100 μl of cell lysate was saved for input control. Antibody corresponding to the protein to be pulled down was added, and the reaction mixture was incubated overnight at 4°C. Antibody-protein complexes were precipitated with washed beads at 4°C for 90 min and purified by centrifugation and three washing steps with IP lysis/wash buffer. Proteins were eluted by incubation at 95°C in 1× Laemmli sample buffer prior to Western blotting.
Western blotting.
A 10-μg volume of cell lysate (input control) or co-IP samples was separated by SDS-PAGE using precast Bolt 4%-to-12% Bis-Tris gels (Invitrogen) and transferred onto an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore), followed by incubation in saturation buffer (PBS containing 5% nonfat dried milk and 0.05% Tween 20). The membrane was first incubated with primary antibody either overnight at 4°C or for 1 h at room temperature, washed three times with 0.05% PBS–Tween, and then incubated with HRP-conjugated secondary antibody. After washing steps, band detection was performed using a SuperSignal West Pico or Femto kit (Pierce) in a Fujifilm LAS-4000 luminescent image analyzer. Band quantification was performed using ImageJ software.
Viral infection and titrations.
Monolayers of cells were infected for 1 h at 37°C in 5% CO2. The inoculum was then replaced with DMEM supplemented with 2% fetal bovine serum (FBS), 20 U/ml penicillin, and 20 µg/ml streptomycin. Viral titers were determined 24 h postinfection (p.i.) by focus-forming assay, as previously described (42).
Immunofluorescence assay.
Cells were grown on coverslips to 20% to 40% confluence, transfected, and incubated for 24 h. They were fixed with 3% paraformaldehyde dissolved in PBS, quenched with 10 mM glycine, permeabilized with 0.5% Triton X-100 dissolved in PBS, and washed three times for 15 min with PBS containing 2% FCS. Primary antibodies were diluted in PBS containing 2% FCS. After incubation at room temperature for 1 h, the coverslips were washed with PBS containing 2% FCS, treated with secondary antibody, washed, and mounted. Images from single planes were taken using a Nikon A1R laser scanning confocal microscope with a 60× oil immersion lens (Plan Apochromat VC) under the control of NIS-Elements microscope imaging software (Nikon). Laser power and gain were chosen to be suitable to visualize the localization of both proteins for each sample. Linear histogram adjustment was done in ImageJ, and final image preparation was done in Adobe Illustrator.
Fluorescence-based quantification of TBEV protein degradation.
HEK293T cells were transfected with plasmids coding for TBEV C-FLAG, prM-FLAG, E-FLAG, NS2A-FLAG, NS2B-FLAG, or NS3-FLAG in the presence or absence of human viperin and NS3-HA. At 24 h after transfection, they were fixed and stained, and viral proteins were visualized as previously described using a Trophos Plate Runner HD instrument (Trophos SA, Marseille, France) (36).
XBP1 splicing assay.
HEK293T cells were transfected with TBEV NS3 in the presence of viperin or empty expression vector or with GFP or were treated with 1 μg/ml BFA; control cells were left untreated and untransfected. At 24 h posttreatment, RNA was isolated using a NucleoSpin RNA II kit (Macherey-Nagel). cDNA was synthesized from 400 ng total RNA with a Quantitect reverse transcription kit (Qiagen). Expression levels of spliced, unspliced, and total XBP1 were detected using previously described primers (26). Actin expression was determined using validated QuantiTect primer assays (Qiagen), a Kapa SYBR Fast quantitative PCR (qPCR) kit, and a StepOnePlus fast real-time PCR system.
Statistical analysis.
Data from focus-forming assays, protein quantifications from Western blots, and the fluorescence intensities measured by the Trophos Plate Runner HD instrument were analyzed with unpaired t tests using GraphPad Prism software.
ACKNOWLEDGMENTS
We thank Gerhard Dobler of the Bundeswehr Institute of Microbiology (Munich, Germany) for providing TBEV (strain Hypr) and ZIKV (strain MR766), S. Vene (Folkhälsoinstitutet, Stockholm, Sweden) for providing JEV (strain Nakayama), and M. Niedrig (Robert Koch Institute, Berlin, Germany) for providing YFV (strain Asibi) and the anti-TBEV E antibody. We are also grateful to the Biochemical Imaging Center, Umeå (BICU), part of the National Microscopy Infrastructure (NMI), for outstanding imaging support.
This work was supported by the Kempe Foundations, the Laboratory for Molecular Medicine Sweden (MIMS), the Umeå Center for Microbial Research (UCMR), Linneus Support, the Swedish Research Council (VR) (2011-2795, 2017-02438), the Swedish Foundation for Strategic Research (SSF) (ICA10-0059, FFL12-0089), and Västerbotten County Hospital (A.K.Ö.). The funding bodies had no role in study design, data collection or interpretation, or the decision to submit the work for publication.
C.P., R.L., C.K., K.V., J.P., K.E., and A.S.U. designed, performed, and evaluated the experiments. C.P. and R.L. wrote the manuscript; R.L. prepared the figures; and A.K.Ö. designed experiments and interpreted the data, wrote the manuscript, and provided funding. All authors analyzed the results.
We declare that we have no conflicts of interests.
REFERENCES
- 1.Gould EA, Solomon T. 2008. Pathogenic flaviviruses. Lancet 371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Lazear HM, Diamond MS. 2016. Zika virus: new clinical syndromes and its emergence in the Western Hemisphere. J Virol 90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. 2009. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoggins JW. 2014. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol 6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upadhyay AS, Stehling O, Panayiotou C, Rosser R, Lill R, Overby AK. 18 August 2017. Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin. J Biol Chem. doi: 10.1074/jbc.M117.780122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upadhyay AS, Vonderstein K, Pichlmair A, Stehling O, Bennett KL, Dobler G, Guo JT, Superti-Furga G, Lill R, Overby AK, Weber F. 2014. Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell Microbiol 16:834–848. doi: 10.1111/cmi.12241. [DOI] [PubMed] [Google Scholar]
- 8.Tan KS, Olfat F, Phoon MC, Hsu JP, Howe JL, Seet JE, Chin KC, Chow VT. 2012. In vivo and in vitro studies on the antiviral activities of viperin against influenza H1N1 virus infection. J Gen Virol 93:1269–1277. doi: 10.1099/vir.0.040824-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Hinson ER, Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, Wilkinson J, Bye CR, Wright TK, Rambukwelle D, Donaghy H, Beard MR, Cunningham AL. 2012. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120:778–788. doi: 10.1182/blood-2012-01-407395. [DOI] [PubMed] [Google Scholar]
- 11.Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NK, Koh EG, Chow A, Leo YS, Merits A, Chin KC, Ng LF. 2012. Viperin restricts chikungunya virus replication and pathology. J Clin Invest 122:4447–4460. doi: 10.1172/JCI63120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Burke CW, Ryman KD, Klimstra WB. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol 81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGillivary G, Jordan ZB, Peeples ME, Bakaletz LO. 2013. Replication of respiratory syncytial virus is inhibited by the host defense molecule viperin. J Innate Immun 5:60–71. doi: 10.1159/000342473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peña Cárcamo JR, Morell ML, Vázquez CA, Vatansever S, Upadhyay AS, Överby AK, Cordo SM, García CC. 2017. The interplay between viperin antiviral activity, lipid droplets and Junin mammarenavirus multiplication. Virology 514:216–229. doi: 10.1016/j.virol.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, Beard MR. 2013. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl Trop Dis 7:e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. 2011. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol 85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Wu X, Pan T, Song W, Wang Y, Zhang F, Yuan Z. 2012. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J Gen Virol 93:83–92. doi: 10.1099/vir.0.033860-0. [DOI] [PubMed] [Google Scholar]
- 19.Van der Hoek KH, Eyre NS, Shue B, Khantisitthiporn O, Glab-Ampi K, Carr JM, Gartner MJ, Jolly LA, Thomas PQ, Adikusuma F, Jankovic-Karasoulos T, Roberts CT, Helbig KJ, Beard MR. 2017. Viperin is an important host restriction factor in control of Zika virus infection. Sci Rep 7:4475. doi: 10.1038/s41598-017-04138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartholomeusz AI, Wright PJ. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch Virol 128:111–121. doi: 10.1007/BF01309792. [DOI] [PubMed] [Google Scholar]
- 21.Falgout B, Pethel M, Zhang YM, Lai CJ. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol 65:2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebhard LG, Iglesias NG, Byk LA, Filomatori CV, De Maio FA, Gamarnik AV. 2016. A proline-rich N-terminal region of the dengue virus NS3 is crucial for infectious particle production. J Virol 90:5451–5461. doi: 10.1128/JVI.00206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol 82:3342–3352. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JH, Walter P, Yen TS. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oslowski CM, Urano F. 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol 82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan YL, Chang TH, Liao CL, Lin YL. 2008. The cellular antiviral protein viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells. J Virol 82:10455–10464. doi: 10.1128/JVI.00438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol 76:10766–10775. doi: 10.1128/JVI.76.21.10766-10775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jumat MR, Huong TN, Ravi LI, Stanford R, Tan BH, Sugrue RJ. 2015. Viperin protein expression inhibits the late stage of respiratory syncytial virus morphogenesis. Antiviral Res 114:11–20. doi: 10.1016/j.antiviral.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Eldin P, Papon L, Oteiza A, Brocchi E, Lawson TG, Mechti N. 2009. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J Gen Virol 90:536–545. doi: 10.1099/vir.0.006288-0. [DOI] [PubMed] [Google Scholar]
- 32.Xue B, Yang D, Wang J, Xu Y, Wang X, Qin Y, Tian R, Chen S, Xie Q, Liu N, Zhu H. 2016. ISG12a restricts hepatitis C virus infection through the ubiquitination-dependent degradation pathway. J Virol 90:6832–6845. doi: 10.1128/JVI.00352-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. 2011. TRIM79alpha, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10:185–196. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindqvist R, Mundt F, Gilthorpe JD, Wolfel S, Gekara NO, Kroger A, Overby AK. 2016. Fast type I interferon response protects astrocytes from flavivirus infection and virus-induced cytopathic effects. J Neuroinflammation 13:277. doi: 10.1186/s12974-016-0748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonderstein K, Nilsson E, Hubel P, Nygard Skalman L, Upadhyay A, Pasto J, Pichlmair A, Lundmark R, Overby AK. 2017. Viperin targets flavivirus virulence by inducing assembly of noninfectious capsid particles. J Virol . doi: 10.1128/JVI.01751-17. [DOI] [PMC free article] [PubMed]
- 38.Asghar N, Lee YP, Nilsson E, Lindqvist R, Melik W, Kroger A, Overby AK, Johansson M. 2016. The role of the poly(A) tract in the replication and virulence of tick-borne encephalitis virus. Sci Rep 6:39265. doi: 10.1038/srep39265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donald CL, Brennan B, Cumberworth SL, Rezelj VV, Clark JJ, Cordeiro MT, Freitas de Oliveira Franca R, Pena LJ, Wilkie GS, Da Silva Filipe A, Davis C, Hughes J, Varjak M, Selinger M, Zuvanov L, Owsianka AM, Patel AH, McLauchlan J, Lindenbach BD, Fall G, Sall AA, Biek R, Rehwinkel J, Schnettler E, Kohl A. 2016. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl Trop Dis 10:e0005048. doi: 10.1371/journal.pntd.0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niedrig M, Klockmann U, Lang W, Roeder J, Burk S, Modrow S, Pauli G. 1994. Monoclonal antibodies directed against tick-borne encephalitis virus with neutralizing activity in vivo. Acta Virol 38:141–149. [PubMed] [Google Scholar]
- 41.Mutso M, Saul S, Rausalu K, Susova O, Zusinaite E, Mahalingam S, Merits A. 2017. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J Gen Virol 98:2712–2724. doi: 10.1099/jgv.0.000938. [DOI] [PubMed] [Google Scholar]
- 42.Overby AK, Popov VL, Niedrig M, Weber F. 2010. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J Virol 84:8470–8483. doi: 10.1128/JVI.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]