ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication and constant primary infection of fresh cells are crucial for viral tumorigenicity. The virus-encoded bZIP family protein K8 plays an important role in viral DNA replication in both viral reactivation and de novo infection. The mechanism underlying the functional role of K8 in the viral life cycle is elusive. Here, we report that K8 is an RNA binding protein that also associates with many other proteins, including other RNA binding proteins. Many protein-protein interactions involving K8 are mediated by RNA. Using a UV cross-linking and immunoprecipitation (CLIP) procedure combined with high-throughput sequencing, RNAs that are associated with K8 in BCBL-1 cells were identified, including both viral (PAN, T1.4, T0.7, etc.) and cellular (MALAT-1, MRP, 7SK, etc.) RNAs. An RNA binding motif in K8 was defined, and mutation of the motif abolished the ability of K8 to bind to many noncoding RNAs, as well as viral DNA replication during de novo infection, suggesting that the K8 functions in viral replication are carried out through RNA association. The functions of K8 and associated T1.4 RNA were investigated in detail, and the results showed that T1.4 mediates the binding of K8 to ori-Lyt DNA. The T1.4-K8 complex physically bound to KSHV ori-Lyt DNA and recruited other proteins and cofactors to assemble a replication complex. Depletion of T1.4 abolished DNA replication in primary infection. These findings provide mechanistic insights into the role of K8 in coordination with T1.4 RNA in regulating KSHV DNA replication during de novo infection.
IMPORTANCE Genomewide analyses of the mammalian transcriptome revealed that a large proportion of sequence previously annotated as noncoding regions is actually transcribed and gives rise to stable RNAs. The emergence of a large number of noncoding RNAs suggests that functional RNA-protein complexes, e.g., ribosomes or spliceosomes, are not ancient relics of the last ribo-organism but would be well adapted to a regulatory role in biology. K8 has been puzzling because of its unique characteristics, such as multiple regulatory roles in gene expression and DNA replication without DNA binding capability. This study reveals the mechanism underlying its regulatory role by demonstrating that K8 is an RNA binding protein that binds to DNA and initiates DNA replication in coordination with a noncoding RNA. It is suggested that many K8 functions, if not all, are carried out through its associated RNAs.
KEYWORDS: 7SK RNA, K8, Kaposi's sarcoma-associated herpesvirus (KSHV), PAN RNA, T0.7 RNA, T1.4 RNA, viral lytic DNA replication, human herpesvirus 8 (HHV-8), noncoding RNA, ori-Lyt
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), also referred to as human herpesvirus 8 (HHV-8), is the causative agent of several AIDS-associated malignancies, including Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1–3). KSHV has two alternative life cycles, namely, latent and lytic replication cycles (4, 5). In order to evade host immune surveillance, KSHV establishes latent infection in most infected cells. However, in KS lesions, latent viral episomes can be quickly lost as cells divide (6). To sustain the virus-infected cell population, a small percentage of infected cells undergo spontaneous lytic replication and produce infectious progeny virions (7–9). Thus, unlike other oncogenic viruses, where the latent life cycle is primarily responsible for the oncogenic activities of the virus, KSHV lytic replication and constant de novo infection of fresh cells are essential for viral pathogenesis and tumorigenesis (6).
During de novo infection, a set of viral genes are expressed and participate in immune evasion, epigenetic modification, and transcriptional reprogramming before the latent infection is established (10–12). Short-term viral DNA replication occurs in the period between virus infection and latency establishment, which allows the KSHV genome to be amplified from 1 to 50 to 100 copies. This phase of viral DNA replication is termed abortive lytic replication as, unlike lytic replication, it does not produce infectious virions. We and others have found that viral abortive replication is sensitive to phosphonoacetic acid (PAA), a specific inhibitor of herpesvirus DNA polymerase (11, 13), suggesting that the DNA replication in this phase is different from latent replication, which is cellular DNA polymerase dependent but utilizes lytic DNA replication machinery. However, the mechanism that controls abortive replication during de novo infection is elusive.
KSHV DNA replication initiates at an origin (such as ori-Lyt) and requires trans-acting elements, both viral and cellular. Two duplicated copies of lytic DNA replication origin, namely, ori-Lyt (L) and ori-Lyt (R), were identified in the KSHV genome (14, 15). These two ori-Lyts share almost identical 1.1-kb core component sequences and 600-bp GC-rich repeats, which are represented as 20-bp and 30-bp tandem arrays. Each 1.7-kb ori-Lyt sequence is necessary and sufficient as a cis-acting signal for KSHV lytic DNA replication (15). Furthermore, both ori-Lyts carry a transcriptional promoter that directs transcription of a 1.4-kb RNA in ori-Lyt (L) and a 0.7-kb RNA in ori-Lyt (R) (16). The ori-Lyt-associated transcriptions are absolutely required for viral DNA replication, but the functions of the RNAs are unknown (17). Two virus-encoded proteins, namely, replication and transcription activator (RTA) and K8 (K-bZIP), were found to be ori-Lyt binding proteins, and both were essential for KSHV lytic DNA replication in a transient-transfection assay (15, 17, 18). K8 associates with ori-Lyt DNA and recruits DNA replication-relevant proteins, both viral and cellular, to ori-Lyt to form viral core replication machinery complexes (15, 19, 20). Recently, we and others observed that DNA replication still proceeds in K8-null virus during the lytic reactivation phase, suggesting that the function of K8 in viral DNA replication can be substituted for by another protein(s) (13, 21). However, K8 was found to be absolutely essential for viral DNA replication during de novo infection (13).
In addition to its role in viral DNA replication, K8 has been shown to be a global gene expression repressor, inhibiting both viral and cellular gene expression (22, 23). Several studies have uncovered linkages between K8 and epigenetic regulation, such as inhibiting the H3K9me3 demethylase JMJD2A and mediating SUMO-2/3 modification of the KSHV chromosome (24, 25). In addition, K8 was reported to inhibit the activity of CDK2, which plays a pivotal role in cell cycle progression. Inhibition of CDK2 activity by K8 resulted in a prolonged G1 phase with concomitant induction of p21, P27, and C/EBPα (26, 27).
To elucidate the biological functions of K8 in the KSHV life cycle and the mechanisms underlying these actions, we attempted to identify components that interact with K8 in the belief that knowing the K8 binding partners would provide clues to reveal K8 function and its underlying mechanism. In this effort, we found that K8 is a novel RNA binding protein and that its RNA binding ability is essential to its role in viral DNA replication during de novo KSHV infection. Furthermore, a viral RNA, namely, T1.4 or ori-Lyt-associated RNA, was found to mediate the interaction between K8 and ori-Lyt DNA and to be absolutely required for viral DNA replication. Through this investigation, a novel mechanism of viral DNA replication by coordinates actions of K8 protein and viral noncoding RNA was revealed.
RESULTS
K8 predominately interacts with RNA binding proteins.
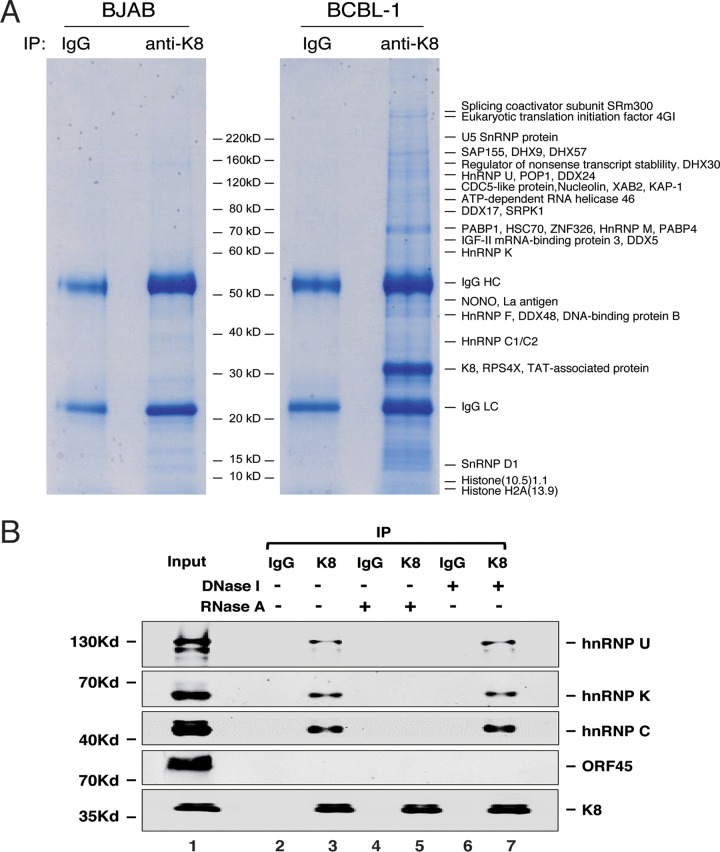
In order to elucidate the mechanisms underlying the K8 actions in viral DNA replication and other events in the viral life cycle, a proteomics approach was undertaken to identify the proteins that interact with K8 in KSHV-infected cells during lytic replication. BCBL-1 cells were induced for reactivation with tetradecanoyl phorbol acetate (TPA) for 48 h and subjected to coimmunoprecipitation (co-IP) with an antibody against K8. The resultant proteins were resolved on SDS-PAGE and revealed by Coomassie blue staining (Fig. 1A). Numerous protein bands were shown in the K8 immunoprecipitated samples but not visually detected in the control samples (mouse immunoglobulin G [IgG] precipitate of induced BCBL-1 nuclear extract and KSHV-free BJAB B lymphoma cells precipitated with anti-K8 antibody). The discrete bands were excised and subjected to mass spectrometry (MS) analysis. This approach led to identification of a series of proteins, including a few viral proteins (RTA, ORF21, and ORF57) (22, 23, 28) and many cellular proteins (Fig. 1A and Table 1). Interestingly, most of the identified proteins were potential RNA binding proteins, including DEA(D/H) box helicases (DDX5, DDX17, DDX24, DDX46, DDX48, DHX9, DHX30, and DHX57), members of the heterogeneous ribonucleoprotein family (hnRNPs U, M, Q, K, F, C1/C2), and RNA-splicing proteins. The finding that K8 predominately interacts with RNA binding proteins suggests that some regulatory function(s) of K8 is involved with RNA.
FIG 1.
Identification of proteins that interact with K8 in BCBL-1 cells. (A) Nuclear extracts of TPA-treated KSHV-positive BCBL-1 cells and KSHV-negative BJAB cells were immunoprecipitated with anti-K8 antibody. Immunoprecipitated proteins were resolved on SDS-PAGE, followed by Coomassie blue staining. The discrete bands were excised and subjected to mass spectrometric analysis. The resultant MS-MS spectra were run against a sequence database with the SEQUEST program. Matched cellular proteins are indicated at the right of the gel. (B) Interaction between K8 and hnRNPs is RNA mediated. Coimmunoprecipitation of TPA-treated BCBL-1 cells was performed with anti-K8 antibody. RNase A or DNase I was added during the washing steps. The precipitates were Western blotted with antibodies against hnRNPs U, K, and C. The Western blot was also probed with an antibody against ORF45, a protein that was known not to interact with K8.
TABLE 1.
K8-associated proteins identified by mass spectrometry
| Accession no.a | Gene | Description | Size (amino acids) | No. of peptidesb | Score |
|---|---|---|---|---|---|
| gi:19923466 | SRRM2 | Splicing coactivator subunit SRm300 | 2,752 | 17 | 15 |
| gi:21655146 | EIF4GI | Eukaryotic translation initiation factor 4 gamma 1 | 1,600 | 10 | 8.62 |
| gi:12667788 | MYH9 | Myosin-9 | 1,960 | 7 | 6.19 |
| gi:28174906 | VIRMA | Vir-like M6A methyltransferase associated | 1,198 | 2 | 1.74 |
| gi:37589132 | EPRS | Glutamyl-prolyl-tRNA synthetase | 869 | 3 | 2.69 |
| gi:100913206 | DHX9 | DExH-box helicase 9 | 1,270 | 22 | 19.6 |
| gi:4033735 | SAP155 | Spliceosome-associated protein 155 | 1,304 | 19 | 16.8 |
| gi:3882183 | LARP1 | La ribonucleoprotein domain family member 1 | 1,096 | 11 | 9.93 |
| gi:39777586 | DEAH 57 | DExH box helicase 57 | 1,386 | 7 | 6.14 |
| gi:15215317 | RRP12 | rRNA processing 12 homolog | 1,297 | 5 | 4.14 |
| gi:27882034 | DHX8 | DEAH box helicase 8 | 1,214 | 3 | 2.69 |
| gi:1575536 | RENT1 | Regulator of nonsense transcript stability | 1,118 | 18 | 15.8 |
| gi:40254861 | UBAP2L | Ubiquitin-associated protein-2 like | 1,087 | 9 | 8.34 |
| gi:20336294 | DHX30 | DExH box helicase 30 | 1,194 | 10 | 8.54 |
| gi:66347698 | C9ORF10 | Chromosome 9 open reading frame 10 | 1,118 | 7 | 6 |
| gi:2498883 | SF3B2 | Splicing factor 3b subunit 2 | 872 | 7 | 6.27 |
| gi:34222504 | FNBP3 | Formin-binding protein 3 | 957 | 2 | 1.9 |
| gi:18071115 | ATXN2L | Ataxin-2-related domain protein | 1,075 | 3 | 2.63 |
| gi:13124451 | POP1 | Ribonucleases P/MRP protein subunit | 1,024 | 2 | 1.86 |
| gi:12803113 | EFTUD2 | Elongation factor Tu GTP binding domain containing 2, U5 SnRNP component | 972 | 26 | 23 |
| gi:10047339 | MOV10 | Mov10 RISC complex RNA helicase | 1,032 | 9 | 8.16 |
| gi:40788339 | MATR3 | Matrin 3 | 853 | 6 | 5.57 |
| gi:4050087 | RBM25 | RNA binding motif protein 25 | 735 | 6 | 4.98 |
| gi:14141161 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 806 | 4 | 3.49 |
| gi:11526793 | DHX36 | DEAH box helicase 36 | 1,008 | 5 | 4.57 |
| gi:14250756 | DDX24 | DEAD box helicase 24 | 859 | 4 | 3.56 |
| gi:11067747 | CDC5L | Cell division cycle 5 like | 802 | 12 | 10.5 |
| gi:128841 | NCL | Nucleolin | 707 | 7 | 6.08 |
| gi:10566459 | XAB2 | XPA binding protein 2 | 855 | 9 | 7.9 |
| gi:1699027 | TRIM28 | Tripartite motif containing 28 | 835 | 8 | 7.01 |
| gi:5032087 | SF3A1 | Splicing factor 3a subunit 1 | 793 | 7 | 5.96 |
| gi:29881667 | SFPQ | Splicing factor proline/glutamine-rich | 707 | 4 | 3.59 |
| gi:27477136 | ZC3HAV1 | Zinc finger CCCH-type antiviral protein 1 isoform 1 | 902 | 5 | 4.61 |
| gi:5762315 | NFAR-2 | Nuclear factor associated with dsRNA 2 | 894 | 6 | 5.44 |
| gi:168776622 | IFI-16 | Interferon, gamma-inducible protein 16 | 736 | 2 | 1.87 |
| gi:10863889 | SART1 | Squamous cell carcinoma antigen recognized by T cells 1 | 800 | 2 | 1.81 |
| gi:21619831 | TFIP11 | Tuftelin-interacting protein 11 | 837 | 2 | 1.81 |
| gi:2696613 | DDX46 | ATP-dependent RNA helicase 46 | 813 | 20 | 18.4 |
| gi:1857944 | SRPK2 | Serine/arginine-rich protein-specific kinase 2 | 686 | 4 | 3.43 |
| gi:2498733 | GP137 | GPI-anchored protein p137 | 709 | 4 | 3.67 |
| gi:14252988 | SRPK1A | Serine/arginine-rich protein-specific kinase 1a | 826 | 7 | 76.45 |
| gi:32880087 | DDX17 | DEAD box helicase 17 | 729 | 8 | 7.25 |
| gi:12653845 | AKAP8L | A kinase (PRKA) anchor protein 8 like | 646 | 2 | 1.86 |
| gi:10863945 | XRCC5 | X-Ray repair cross complementing 5, ATP-dependent DNA helicase II | 732 | 3 | 2.49 |
| gi:37925422 | NUFIP2 | Nuclear fragile X mental retardation-interacting protein 2, proliferation-inducing gene 1 | 695 | 4 | 3.48 |
| gi:1082856 | ILF3 | Interleukin enhancer binding factor 3 isoform a, transcription factor NF-AT 90K chain | 894 | 2 | 1.74 |
| gi:47125447 | ZNF326 | Zinc finger protein 326 | 508 | 14 | 12 |
| gi:73974124 | PABP1 | Polyadenylate binding protein 1 | 690 | 19 | 17.1 |
| gi:55632315 | HSP70 (HSPA5) | 78-kDa glucose-regulated protein | 654 | 11 | 10.3 |
| gi:2832596 | DDX17 | DEAD box helicase 17 | 729 | 13 | 11.5 |
| gi:14141152 | HNRNPM | Heterogeneous nuclear ribonucleoprotein M isoform a | 730 | 15 | 14 |
| gi:5454064 | RBM14 | RNA binding motif protein 14 | 669 | 5 | 4.48 |
| gi:73976763 | PABP4 | Polyadenylate-binding protein 4 | 644 | 13 | 11.6 |
| gi:31542501 | DEF6 | Differentially expressed in FDCP 6 | 631 | 4 | 3.63 |
| gi:30795212 | IGF-II 3 | Insulin-like growth factor 2 mRNA binding protein 3 | 579 | 18 | 16.1 |
| gi:56237027 | IGF-II 1 | Insulin-like growth factor 2 mRNA binding protein 1 | 577 | 8 | 7.07 |
| gi:30584123 | DDX5 | DEAD box helicase 5 | 615 | 10 | 8.51 |
| gi:228008400 | HNRNPQ | Heterogeneous nuclear ribonucleoprotein Q isoform 6 | 562 | 13 | 11.6 |
| gi:34783647 | SNW1 | SNW domain-containing protein 1 | 531 | 6 | 5.33 |
| gi:12751481 | SPATS2 | Spermatogenesis-associated, serine-rich 2 | 545 | 4 | 3.78 |
| gi:15022507 | COAA | Coactivator activator | 669 | 2 | 1.87 |
| gi:14165437 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 464 | 20 | 17.9 |
| gi:2808511 | NONO | Non-POU domain-containing octamer binding protein | 471 | 7 | 6.25 |
| gi:15809586 | HNRNPQ | Heterogeneous nuclear ribonucleoprotein Q isoform 1 | 623 | 2 | 1.84 |
| gi:10835067 | SSB | Sjogren syndrome antigen B, autoantigen LA | 408 | 12 | 10.9 |
| gi:12655035 | RPL4 | Ribosomal protein L4 | 427 | 10 | 8.78 |
| gi:15620927 | PNMA5 | Paraneoplastic Ma antigen family member 5 | 452 | 9 | 7.98 |
| gi:18088719 | TUBB | Tubulin beta polypeptide | 444 | 8 | 7.03 |
| gi:181486 | DBPB | DNA binding protein B | 364 | 8 | 7.53 |
| gi:13177790 | DDX48 | DEAD box polypeptide 48 | 411 | 15 | 13.3 |
| gi:16876910 | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 415 | 8 | 7.23 |
| gi:14250148 | RPL3 | Ribosomal protein L3 | 403 | 9 | 7.84 |
| gi:12655035 | RPL4 | Ribosomal protein L4 | 427 | 5 | 4.27 |
| gi:12654229 | EIF2S2 | Eukaryotic translation initiation factor 2 beta | 333 | 3 | 2.74 |
| gi:1167838 | YBX3 | Y box binding protein 3, DNA binding protein | 372 | 2 | 1.78 |
| gi:2072950 | p40 | LINE retrotransposable element 1 | 338 | 6 | 5.46 |
| gi:532313 | ILF2(NF45) | Interleukin enhancer binding factor 2 | 406 | 6 | 5.45 |
| gi:14249959 | HNRNPC | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | 293 | 10 | 9.04 |
| gi:444021 | PCBP1 | Poly(rC) binding protein 1, Sub2.3 | 299 | 5 | 4.53 |
| gi:18490263 | RPL6 | Ribosomal protein L6 | 288 | 5 | 4.27 |
| gi:14043072 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A isoform B1 | 353 | 2 | 1.8 |
| gi:18089152 | DRG1 | Developmentally regulated GTP binding protein 1 | 367 | 4 | 3.32 |
| gi:12654583 | RPLP0 | Ribosomal protein P0 | 317 | 5 | 4.44 |
| gi:1314308 | RARA | Nucleophosmin-retinoic acid receptor alpha fusion protein NPM-RAR long form | 563 | 3 | 2.82 |
| gi:133252 | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 195 | 3 | 2.74 |
| gi:4377849 | TRA2B | Transformer-2-beta isoform 3 | 188 | 3 | 2.54 |
| gi:48376549 | RPS4X | Ribosomal protein S4, X linked | 243 | 8 | 6.79 |
| gi:1096067 | TAP | Tat-associated protein | 279 | 4 | 3.81 |
| gi:15928608 | SLC25A6 | Solute carrier family 25, member A6 | 298 | 3 | 2.44 |
| gi:14198272 | PGAM5A | Phosphoglycerate mutase family member 5 | 255 | 2 | 1.74 |
| gi:14714596 | H1FX | H1 histone family member X | 213 | 2 | 1.85 |
| gi:21410831 | SAP | Sin3A-associated protein | 153 | 5 | 4.67 |
| gi:32959908 | SNRNPD1 | Small nuclear ribonucleoprotein D1 polypeptide | 119 | 4 | 3.4 |
| gi:1070593 | HIST1H1C | Histone cluster 1 H1 family member C, histone (10.5)1.1 | 213 | 3 | 2.65 |
| gi:122009 | HIST1H2AG | Histone H2A.1 | 129 | 2 | 1.68 |
Accession numbers may be found in the NCBI Protein database (https://www.ncbi.nlm.nih.gov/protein/).
Number of polypeptides detected for the protein by MS.
To confirm the MS results and explore the mode of binding between K8 and these RNA binding proteins, we analyzed the interaction of K8 with several representative RNA binding proteins, such as hnRNPs U, K, and C, in TPA-induced BCBL-1 cells using coimmunoprecipitation, followed by Western blotting. The results showed that K8 indeed interacts with hnRNPs U, K, and C, but not with ORF45, a viral protein known not to interact with K8 (17, 29) and serving as a negative control (Fig. 1B). Since these three hnRNPs are RNA binding proteins, we asked if the interactions of K8 with the hnRNPs are mediated by RNA. To this end, RNase A was added in washing buffer during the coimmunoprecipitation procedure. The results showed that treatment with RNase A eliminated the interaction between K8 and the hnRNPs (Fig. 1B, lane 5). In contrast, treatment with DNase I did not affect these interactions (Fig. 1B, lane 7). Thus, we conclude that the interactions of K8 with hnRNPs are mediated by RNA molecules.
K8 is a novel RNA binding protein.
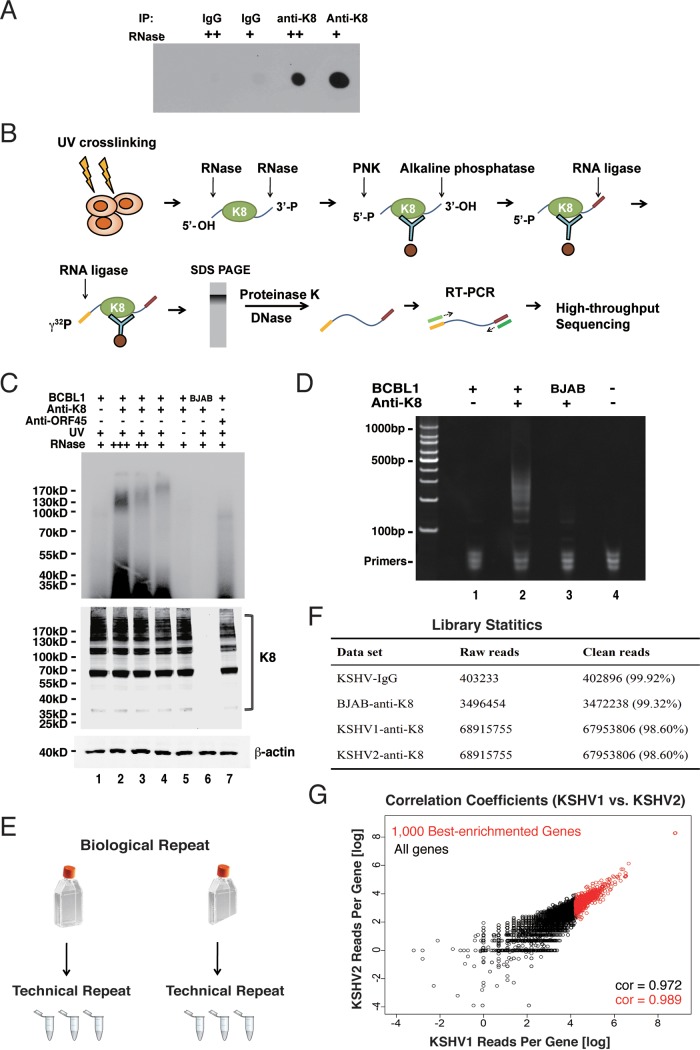
Since K8 predominately interacts with RNA binding proteins and the interactions are mediated by RNA, we asked whether K8 itself is an RNA binding protein. To address this question, we performed UV cross-linking and immunoprecipitation (CLIP) followed by dot blotting. Briefly, TPA-induced BCBL-1 cells were UV irradiated for protein-RNA cross-linking. Then, the cells were lysed and subjected to immunoprecipitation with K8 antibody (mouse IgG was included as a negative control). RNase A at different concentrations was added to the washing buffer. The immunoprecipitated components were labeled with biotin and spotted on a nylon membrane. Biotin-labeled RNA was then detected with a biotin chromogenic detection kit. As shown in Fig. 2A, the K8 immunoprecipitates showed strong RNA-biotin signals, which were not detected in the IgG control. Furthermore, the RNA-biotin signal decreased with increasing concentrations of RNase A in the CLIP experiment, indicating that K8 was associated with RNA.
FIG 2.
Validation of K8 RNA binding property and identification of K8-associated RNAs using a CLIP-seq approach. (A) TPA-induced BCBL-1 cells were UV irradiated and subjected to immunoprecipitation with K8 antibody. Different amounts of RNase A (++, 2 μg/ml; +, 1 μg/ml) were added to the washing buffer during immunoprecipitation. Coprecipitated RNA was labeled with biotin at the 3′ end and spotted onto a nylon membrane. The biotinylated RNA was then analyzed with a chemiluminescent nucleic acid detection module kit. (B) Schematic illustration of the CLIP-seq procedure. TPA-induced BCBL-1 cells were UV irradiated and immunoprecipitated with K8 antibody. Different amounts of RNase A were added to the washing buffer. The coprecipitated RNAs were phosphorylated at the 5′ end with T4 polynucleotide kinase (PNK) and dephosphorylated with alkaline phosphatase at the 3′ end. The RNAs were ligated to the 3′ linker and then to the 5′ linker labeled with γ-32P. The RNA-protein complex was resolved in SDS-PAGE, and RNAs were isolated and amplified by RT-PCR. The PCR products were analyzed by high-throughput sequencing. (C) Autoradiogram of γ-32P-labeled RNA cross-linked to K8. Immunoprecipitation was performed with anti-K8 or anti-ORF45 antibody. (Bottom) Input of K8 and β-actin. (D) RT-PCR products from K8-CLIP. (E) Experimental design of CLIP-seq. For each set of CLIP experiments (IgG, KSHV-negative [BJAB], KSHV1 [BCBL-1], and KSHV2 [BCBL-1]), two biological repeats were carried out. Each biological repeat contained three technical repeats. (F) Library statistics of CLIP-seq. Two biological repeats from K8-CLIP-seq (KSHV1 and KSHV2) contained 67,953,806 and 81,632,254 clean reads, respectively. In contrast, there were only 402,896 and 3,472,238 clean reads in the IgG control and KSHV negative (BJAB) data sets, respectively. (G) Libraries from two biological repeats were highly similar. The correlation coefficient between two biological repeats (KSHV1 and KSHV2) was above 0.97.
Identification of K8 binding RNAs.
To comprehend the biological function and process mediated by K8 and its associated RNAs, we attempted to identify the RNAs that are associated with K8. To this end, cross-linking and immunoprecipitation followed by high-throughput sequencing (CLIP-seq) (30) was employed to determine K8 binding RNAs. The CLIP procedure is schematically illustrated in Fig. 2B. In brief, TPA-induced BCBL-1 cells were UV irradiated to form protein-RNA adducts. Cell lysates were prepared and treated with RNase A to digest the RNAs that were not protected by proteins. Then, the lysates were immunoprecipitated with K8 antibody or mouse IgG. The precipitated RNAs were ligated with linkers and radiolabeled at the 5′ ends with [γ-32P]ATP. In SDS-PAGE, a smeared band around 100 kDa was observed in the UV-cross-linked K8-immunoprecipitated BCBL-1 samples (Fig. 2C, top). Given that the molecular mass of K8 is around 35 kDa, the K8-RNA complexes over 100 kDa apparently resulted from homomultimer K8 (31) associated with RNAs, as evidenced in Western blotting (Fig. 2C, bottom). No band was found in control samples (without UV cross-linking) or BJAB cells (Fig. 2C, lanes 5 and 6). Such bands were also undetectable with the sample from anti-ORF45 immunoprecipitation (lane 7). RNA was purified from the gel and subjected to reverse transcription-PCR (RT-PCR), which yielded a smeared band from 100 to 500 bp long (Fig. 2D, lane 2). Control samples of IgG (lane 1) and BJAB (lane 3) yielded almost no PCR product. The cDNAs in the range of 100 to 200 bp were purified and subjected to high-throughput deep sequencing. Experimental design and library statistics of the RNA sequencing are illustrated in Fig. 2E to G.
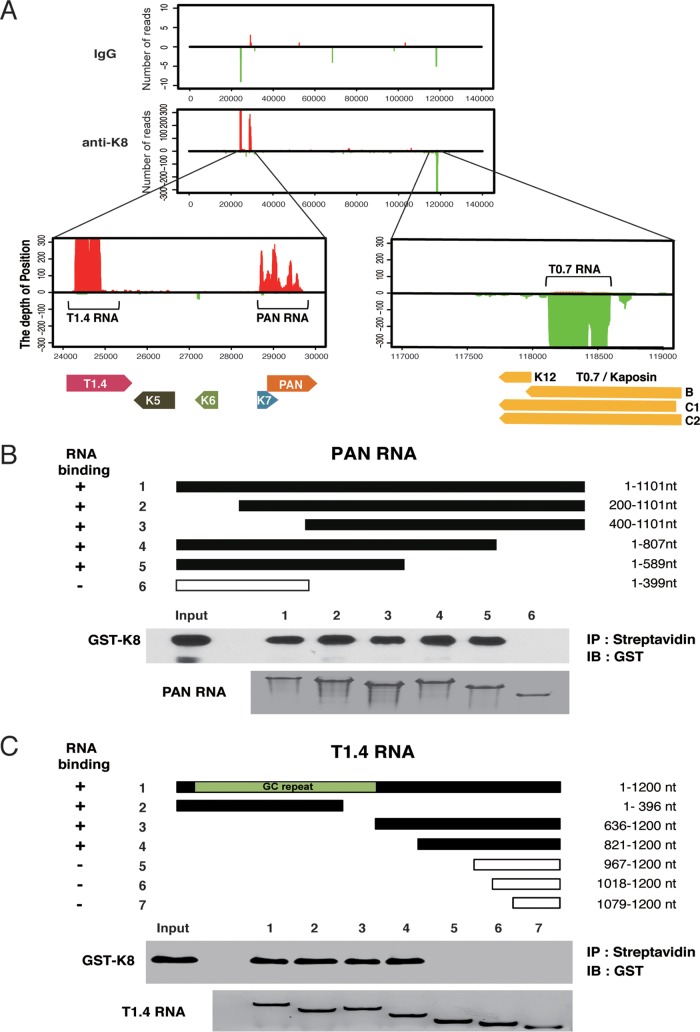
The CLIP-seq reads were first mapped to the KSHV genome (GenBank ID U75698.1). The alignment showed three major peaks in the KSHV genome in the loci of PAN RNA, T1.4 RNA in ori-Lyt (L), and T0.7 RNA in ori-Lyt (R) regions (Fig. 3A). To confirm the CLIP-seq data, PAN RNA and T1.4 RNA were selected for in vitro RNA pulldown assays to verify their interaction with K8 (Fig. 3B and C). Glutathione S-transferase (GST)-K8 protein prepared from Escherichia coli was mixed with biotinylated PAN or T1.4 RNA and subjected to a pulldown procedure with streptavidin beads. Associated protein was analyzed by Western blotting. The results confirmed that K8 directly interacts with PAN RNA and T1.4 RNA in a specific manner. Furthermore, both RNAs were mapped for the regions that are required for K8 binding by using truncation mutants of the two RNAs. It was found that K8 binds to PAN RNA in the region between nucleotides 400 and 589 (Fig. 3B) and interacts with T1.4 RNA in both 5′ GC repeat and 3′ nonrepeat regions (Fig. 3C).
FIG 3.
Three viral noncoding RNAs were identified and confirmed to be directly associated with K8 protein. (A) CLIP-seq reads mapped to the KSHV genome. Enlargement of the K8 cross-linked RNA in the KSHV genomic location showed two peaks (T1.4 RNA and PAN RNA) from the forward strain (left) and one peak (T0.7 RNA) from the reverse strain (right). (B and C) PAN (B) and T1.4 (C) RNAs and their truncation mutants were synthesized in vitro and labeled with biotin. An in vitro RNA pulldown assay was performed to evaluate the abilities of the RNAs and their mutants to be bound by K8. Biotinylated transcripts were mixed with purified GST-K8 protein, and the RNA-protein complexes were precipitated with streptavidin-coupled Dynabeads. The precipitates were analyzed by Western blotting for RNA-bound GST-K8 using anti-GST antibody and by Northern blotting for RNAs using the chemiluminescent nucleic acid detection module.
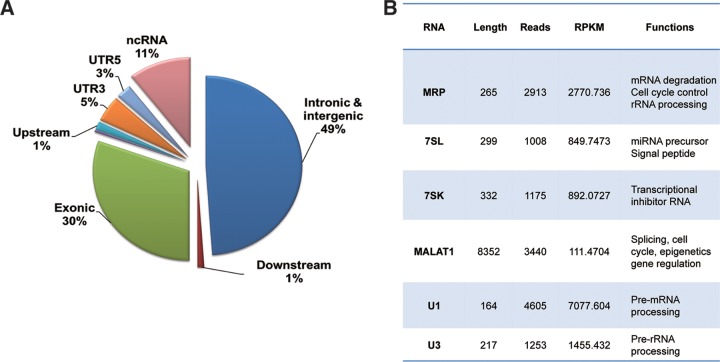
Next, the CLIP-seq reads were mapped to the human genome. About half (49%) of K8 binding RNAs are intronic and intergenic sequence, and 11% are annotated noncoding RNAs, suggesting K8 predominately associates with noncoding RNAs (ncRNAs). Thirty percent of the RNAs were from exons of mRNAs (Fig. 4A). These data imply that ncRNA might play a role in the function of K8. The top six of the abundant cellular ncRNAs that are associated with K8 are MRP, 7SL, 7SK, MALAT1, U1, and U3 RNAs (Fig. 4B). All of them are associated with RNA metabolism and gene regulation, suggesting that K8 might play roles in RNA processing and gene regulation.
FIG 4.
Cellular RNAs identified through K8 CLIP-seq. (A) Percentages of cDNAs from K8–CLIP-seq that mapped to different types of RNAs. UTR, untranslated region. (B) List of K8-associated cellular noncoding RNAs that were identified through K8–CLIP-seq and their known functions. RPKM, reads per kilobase of transcript per million mapped reads.
Mapping of the K8 RNA binding region identified a novel RNA binding motif.
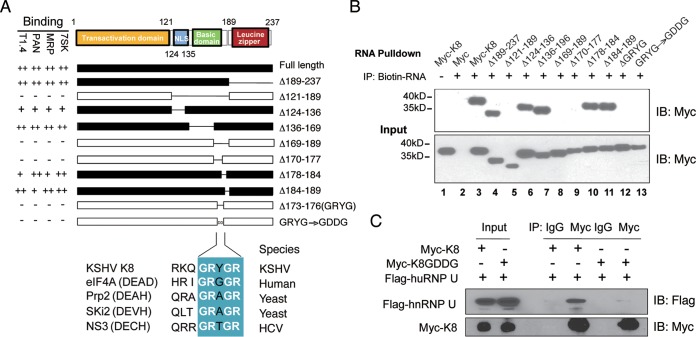
Based on its homology to Epstein-Barr virus (EBV) ZTA protein, K8 protein is divided into three domains: the transactional domain at the N terminus (amino acids [aa] 1 to 121) (32), the basic domain (DNA binding domain; aa 121 to 189) (23), and a leucine zipper domain at its C terminus (aa 189 to 237) (Fig. 5A) (31, 33, 34). To identify the domain(s) in K8 that is required for RNA binding, a series of truncation and deletion mutants of K8 were constructed and examined for RNA binding ability in a biotinylated RNA pulldown assay. 293T cells were transfected with Myc-tagged K8 or its mutants, and whole-cell lysates were prepared. The lysates were incubated with biotinylated transcripts, and the RNA-protein complexes were isolated using streptavidin-coupled Dynabeads (ThermoFisher). RNA-associated proteins were eluted in SDS-PAGE buffer and analyzed by Western blotting with an anti-Myc antibody. Several potential K8 binding RNAs identified in our CLIP-seq were used to map K8 protein for the RNA binding region(s), including the PAN and T1.4 viral RNAs, as well as the 7SK and MRP cellular RNAs. Interestingly, the results with these different RNAs were consistent, as shown in Fig. 5A. A representative Western blot of 7SK pulldown is presented in Fig. 5B. The results showed that the basic domain (aa 121 to 189), but not the leucine zipper domain, was responsible for K8 binding to all four RNAs tested. Further mutagenesis analysis narrowed down the RNA binding motif to 4 amino acids, GRYG (aa 173 to 176), as deletion of these 4 amino acids from K8 led to total loss of RNA binding activity. When GRYG was mutated to GDDG, two negatively charged aspartic acids were introduced into the motif, and K8 completely lost the ability to associate with all the RNAs (Fig. 5B, lane 13). Interestingly, GRYG appears to be part of the RNA binding consensus motif (GRXGR) found in DEAD box family proteins (35–37).
FIG 5.
Mapping of K8 for the RNA binding domain. (A) Schematic presentation of K8 truncation, deletion, or amino acid substitution mutants that were used for mapping the RNA binding domain. The binding properties with T1.4, PAN, MRP, and 7SK RNAs are shown on the left of each construct. NLS, nuclear localization signal. GRXGR is the consensus RNA binding motif of the DEAD box protein. The symbols ++, +, and − represent strong, moderate, and no binding, respectively. (B) Expression vectors of Myc-K8 and mutants were introduced into 293T cells. Cell lysates were prepared and mixed with streptavidin beads coated with biotinylated RNA. The RNA pulldown materials were analyzed by Western blotting with an anti-Myc antibody. Shown are the results from the 7SK RNA pulldown assay. IB, immunoblotting. (C) Effects of GDDG mutation on binding of K8 with hnRNP U determined by immunoprecipitation with anti-Myc antibody and Western blotting using anti-Flag antibody.
Since the interactions of K8 and hnRNPs are RNA mediated (Fig. 1B), we examined whether the GDDG mutation in K8 affects the association between K8 and hnRNPs. Myc-tagged K8 or its GDDG mutant was cotransfected with Flag-tagged hnRNP U into 293T cells. Coimmunoprecipitation was performed with anti-Myc antibody and analyzed by Western blotting using anti-Flag antibody. It was found that hnRNP U interacted with wild-type (WT) K8 but failed to bind the K8 GDDG mutant, indicating that K8 was unable to interact with the hnRNP without the GRYGR-dependent RNA binding activity (Fig. 5C).
The RNA binding ability of K8 is essential for viral DNA replication during primary infection.
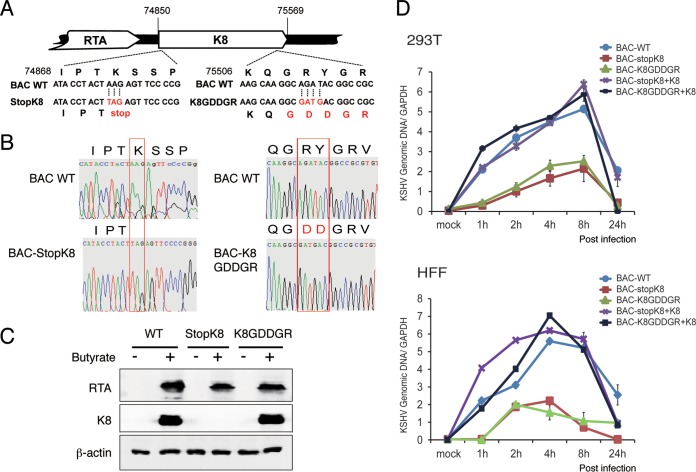
K8 is known to bind on ori-Lyt DNA and plays a role in viral lytic (including abortive lytic) DNA replication. We asked if the function of K8 in viral DNA replication requires RNA binding activity. To address this question in the context of virus, we generated a recombinant virus with the K8 GRYGR motif mutated to GDDGR in the bacterial artificial chromosome (BAC)-cloned KSHV (BAC16) genome (38, 39) using two-step lambda Red-mediated seamless recombination (40), which is designated BAC-K8GDDGR (Fig. 6A). A BAC-stopK8 mutant was also generated as a control (13). Restriction enzyme digestion (data not shown) and sequencing confirmed that the mutants had the desired sequences (Fig. 6B). BAC-cloned viral genomic DNAs were introduced into 293T cells. After validation of the expression of the viral proteins RTA and K8 in 293T cells at 48 h postinduction (Fig. 6C), virions from the cells infected with the BAC-cloned viruses (wild-type BAC16, BAC-stopK8, and BAC-K8GDDGR) were prepared at 5 days postinduction. The wild-type and mutant viruses were used to infect 293T cells and human foreskin fibroblasts (HFF) at a multiplicity of infection (MOI) of 50 (viral genomic DNA copy equivalents). Viral DNA contents of infected 293T cells and HFF with different BAC-cloned viruses were determined at different time points postinfection using quantitative PCR (qPCR) analysis. As expected, BAC-stopK8 virus could not support viral abortive DNA replication in both types of cells, but the lost DNA replication activity could be restored by ectopic expression of K8 (Fig. 6D) (13). Similarly, viral DNA replication of BAC-K8GDDGR decreased significantly in comparison to that of wild-type virus, suggesting that the function of K8 in abortive lytic replication during primary infection is dependent on its RNA binding activity.
FIG 6.
Mutation in the RNA binding motif GRYGR abolishes KSHV DNA replication in the viral context. (A) Schematic diagrams of the structures of K8 in BAC-stopK8 and K8-GDDGR recombinant viruses. The nucleotide sequences refer to GenBank accession number U75698.1. The mutated sequences on the stop codon and the GRYGR motif are indicated in red. (B) Sequence chromatograms of the clones. The mutations from AAG to the stop codon TAG in BAC-stopK8 and from GRYGR to GDDGR in BAC-K8GDDGR are boxed. (C) Expression of RTA and K8 genes in the wild-type and mutant viruses. (D) Viral DNA contents in 293T cells and HFF that were primarily infected by BAC16 (WT), BAC-stopK8, and BAC-GDDGR at an MOI of 50 (viral genomic DNA equivalents). A complementary experiment with ectopic expression of K8 was performed to examine whether the defects caused by K8 mutation could be rescued by wild-type K8. 293T cells and HFF were transfected with K8 expression vector (pCR3.1-K8α). Twenty-four hours posttransfection, the cells were infected with BAC16 (WT), BAC-stopK8, and BAC-GDDGR viruses. At different time points postinfection, total DNA was isolated, and viral genomic DNA was quantified by qPCR with primers specific to ORF73. Viral DNA copy numbers were calculated from a standard graph generated using known concentrations of BAC16 DNA. The error bars indicate standard deviations (SD).
K8 binds to ori-Lyt DNA in an RNA-dependent manner.
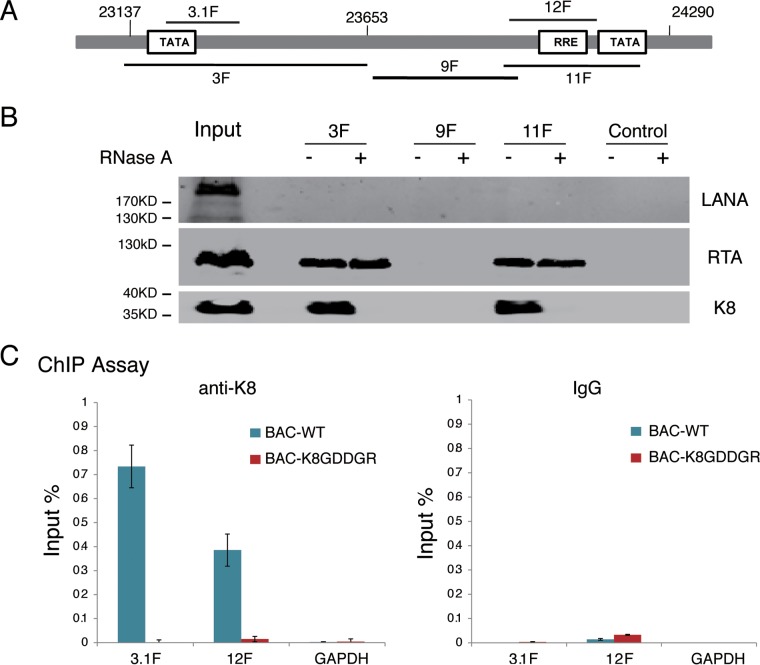
Following the observation that the RNA binding activity of K8 is important for viral DNA replication, we explored the mechanism underlying this K8 function. In KSHV ori-Lyt-dependent DNA replication, K8 and RTA recruit six core replication proteins and other cofactors to ori-Lyt DNA (15, 18, 20). Although K8 binds to the ori-Lyt core region (15–17, 20), this DNA binding was found to be indirect through piggybacking on another component(s) (15, 34). Therefore, we wondered whether RNA contributes to the association of K8 with ori-Lyt DNA. To answer this question, we first adapted a DNA affinity assay (17, 20) to examine the interaction between K8 and the ori-Lyt core domain. Three overlapping DNA fragments representing the core domain of KSHV ori-Lyt labeled with biotin at their 5′ ends were coupled to streptavidin-conjugated magnetic beads and incubated with nuclear extract derived from TPA-induced BCBL-1 cells (Fig. 7A). The affinity-purified materials were then analyzed by Western blotting using antibodies against LANA, RTA, and K8. As expected, both RTA and K8 were able to bind to 3F and 11F fragments (RTA-responsive element [RRE] and TATA box), but not 9F, while LANA did not interact with ori-Lyt DNA at all (20, 41) (Fig. 7B). However, we found that treatment with RNase A during the DNA affinity assay abolished the association of K8 with ori-Lyt but did not affect RTA binding to ori-Lyt DNA (Fig. 8B), suggesting that the interaction between K8 and ori-Lyt DNA is mediated by certain RNAs.
FIG 7.
Association of K8 with ori-Lyt DNA is mediated by RNA. (A) Schematic illustration of the KSHV ori-Lyt core domain and DNA fragments that were used in the DNA affinity assay. (B) Three biotinylated ori-Lyt DNA fragments and an irrelevant DNA fragment from the ORF45 coding region as a control were prepared by PCR, conjugated on magnetic beads, and incubated with TPA-induced BCBL-1 nuclear extract with and without treatment with RNase A. After washing, samples were assayed by Western blotting with antibodies as indicated. (C) Binding of K8 to ori-Lyt DNA was determined in BAC16 (BAC WT) and BAC-K8GDDGR by ChIP assay with anti-K8 antibody. The positions of the amplicons (3.1F and 12F) are shown in panel A. The error bars indicate SD.
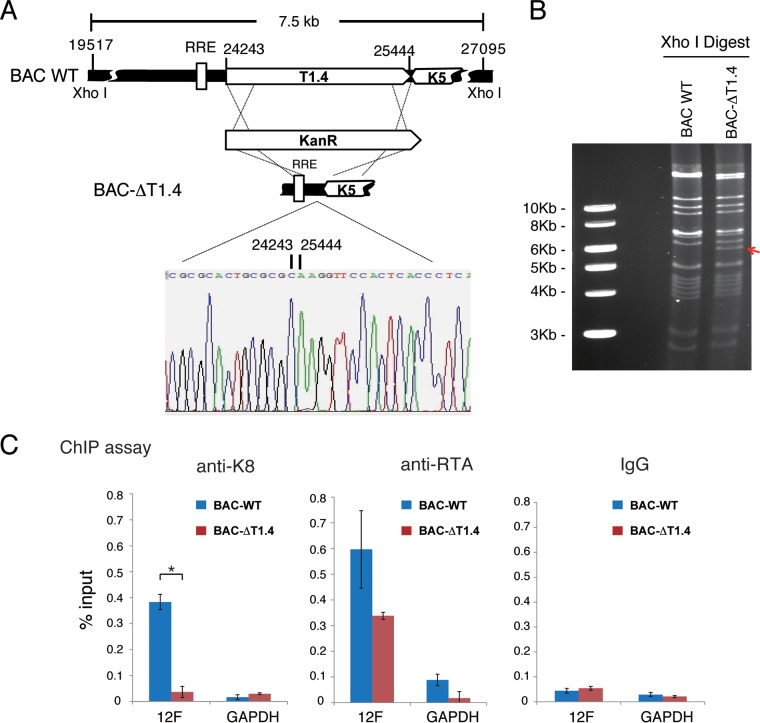
FIG 8.
Binding of K8 to ori-Lyt DNA is mediated by T1.4 RNA. (A) Schematic diagrams of the T1.4 locus in the KSHV genome and strategy for construction of BAC-ΔT1.4. The nucleotide sequences refer to GenBank accession number U75698.1. The sequence chromatogram of the deletion junction in BAC-ΔT1.4 is shown. (B) Electrophoretic analysis of the BAC-ΔT1.4 mutant viral genomes. BAC16 (BAC WT) and BAC-ΔT1.4 DNAs were digested with XhoI, resolved on a 0.8% agarose gel, and stained with ethidium bromide. The red arrow indicates the changes of restriction pattern caused by the deletion of T1.4 in BAC-ΔT1.4. (C) BAC16 and BAC-ΔT1.4 were introduced into 293T cells, and the cells were induced with sodium butyrate for 48 h. A ChIP assay was performed with anti-K8 and anti-RTA antibodies or IgG and detected by PCR with primers for the 12F region or GAPDH. The data are shown as the means ± SD of the results of three separate experiments. *, P < 0.05.
This notion was further tested using mutant K8 with an RNA binding defect (K8GDDGR) in a chromatin-immunoprecipitation (ChIP) assay in the context of virus. 293T cells carrying BAC16 (wild-type K8) and BAC-K8GDDGR mutant viruses were induced for reactivation and treated with formaldehyde for cross-linking at 48 h postinduction. Fragmented chromatin was immunoprecipitated with anti-K8 antibody or IgG, and K8-bound DNAs were quantified by qPCR. The results showed that K8 bound to ori-Lyt DNA (3F and 11F regions), but not to GAPDH DNA, while mutant K8 in BAC-K8GDDGR failed to associate with ori-Lyt DNA (Fig. 7C), suggesting that K8 binding to ori-Lyt DNA is mediated by RNA(s) that associates with K8 through the GRYGR motif.
K8 binding to ori-Lyt DNA is mediated by virus-borne T1.4 RNA.
Among the three virus-borne RNAs identified as encoding K8 binding protein in our CLIP-seq, two, namely, T1.4 and T0.7 RNAs, are ori-Lyt-associated RNAs (16, 17). Previous studies demonstrated that T1.4 RNA is absolutely essential for KSHV ori-Lyt-dependent DNA replication (16, 17) and is expressed at a very high level during KSHV de novo infection (11). Therefore, we asked if this RNA also plays a role in viral DNA replication during de novo infection and, furthermore, whether T1.4 RNA is the RNA that mediates the K8 association with ori-Lyt DNA. To address these questions, a recombinant KSHV with the ori-Lyt (L)-associated T1.4 RNA locus deleted from the BAC16-cloned KSHV genome was constructed and designated BAC-ΔT1.4. The mutant virus was verified by restriction enzyme digestion and sequencing (Fig. 8A and B). The deletion did not affect viral gene expression (LANA, RTA, and K8) in lytic replication, but viral DNA replication and virion production were depleted, consistent with a previous study (data not shown). 293T cells carrying BAC16 wild-type and BAC-ΔT1.4 viruses were used for ChIP assay with anti-K8 and anti-RTA antibodies. The results showed that K8 could not bind to ori-Lyt DNA without T1.4 in BAC-ΔT1.4. In contrast, RTA was able to bind to ori-Lyt regardless of the presence of T1.4 RNA in BAC16 and BAC-ΔT1.4 cells (Fig. 8C), indicating that the interaction between K8 and ori-Lyt DNA is indeed T1.4 RNA mediated.
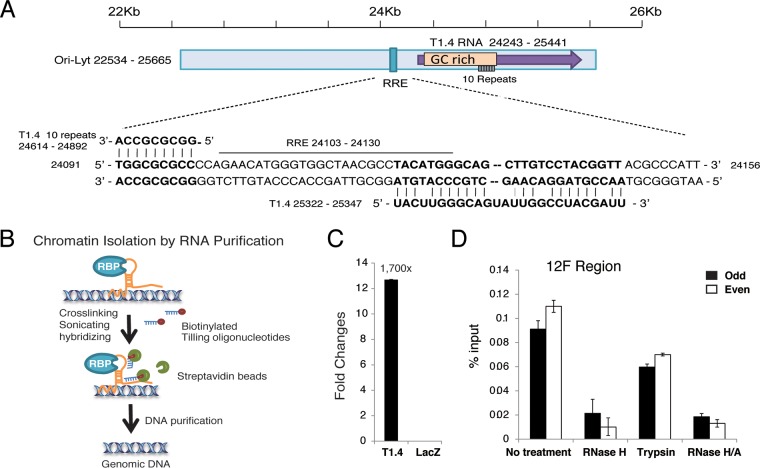
Then, we further investigated how T1.4 mediates K8 binding to ori-Lyt DNA. Sequencing alignment identified potential base pairing between T1.4 RNA and ori-Lyt DNA (Fig. 9A). This led to the hypothesis that T1.4 associates with the viral genome through RNA-DNA base pairing and recruits K8 to ori-Lyt DNA. To test this hypothesis, a chromatin isolation by RNA purification (ChIRP) assay (42) was designed (Fig. 9B). In brief, TPA-induced BCBL-1 cells were cross-linked with 1% glutaraldehyde, and chromatin was sheared by sonication to the size range of 100 to 500 bp. Biotinylated probes complementary to T1.4 and LacZ RNAs (a negative control) were added to the chromatin preparation. T1.4-associated chromatin fragments were purified with streptavidin beads, and T1.4-bound DNA was quantified by qPCR analysis. To validate the T1.4 probe for specific RNA enrichment, a ChIRP assay was performed with T1.4 and LacZ probes. The precipitated RNAs were subjected to RT-qPCR with T1.4 primers, and the result showed the T1.4 RNA level was 1,700-fold higher in the preparation with T1.4 probe than in that with LacZ probe, indicating that the T1.4 probe can specifically enrich T1.4 RNA (Fig. 9C). Then, two sets of T1.4 probes (odd and even) were used for two independent ChIRP experiments. Both sets of probes gave rise to similar results showing that T1.4 associates with ori-Lyt chromatin (Fig. 9D). Furthermore, treatment with RNase H after chromatin pulldown greatly reduced the association between T1.4 RNA and ori-Lyt DNA in the preparation (Fig. 9D). In contrast, treatment with trypsin did not reduce the interaction much. Taken together, these results indicate that T1.4 RNA associates with ori-Lyt DNA through RNA-DNA base pairing rather than mediation by protein(s).
FIG 9.
T1.4 RNA associated with ori-Lyt DNA. (A) Alignment of T1.4 RNA with ori-Lyt DNA sequences in the KSHV genome revealing potential base pairing between them. (B) Schematic illustration of the ChIRP assay procedure. Glutaraldehyde was applied to induce DNA-RNA cross-linking. Chromatin was sonicated and then incubated with biotinylated oligonucleotide probe specifically to the target RNA. Streptavidin beads were added to pull down the biotinylated RNA and bound DNA. qPCR was used to detect the ori-Lyt DNA. (C) Validation of the efficiency of T1.4 probe in pulldown of T1.4 RNA. A ChIRP assay was performed using probes targeting T1.4 or LacZ. Precipitated RNAs were purified and subjected to RT-qPCR with primer against T1.4. (D) T1.4 RNA associates with ori-Lyt DNA. Two sets of probes were used to perform ChIRP experiments. RNase H, RNase A, or trypsin was added in washing buffer during the experiments to digest the RNA or protein. The pulled down DNA was amplified by primers for the 12F region of ori-Lyt. The error bars indicate SD.
T1.4 is essential for KSHV abortive lytic replication following de novo infection.
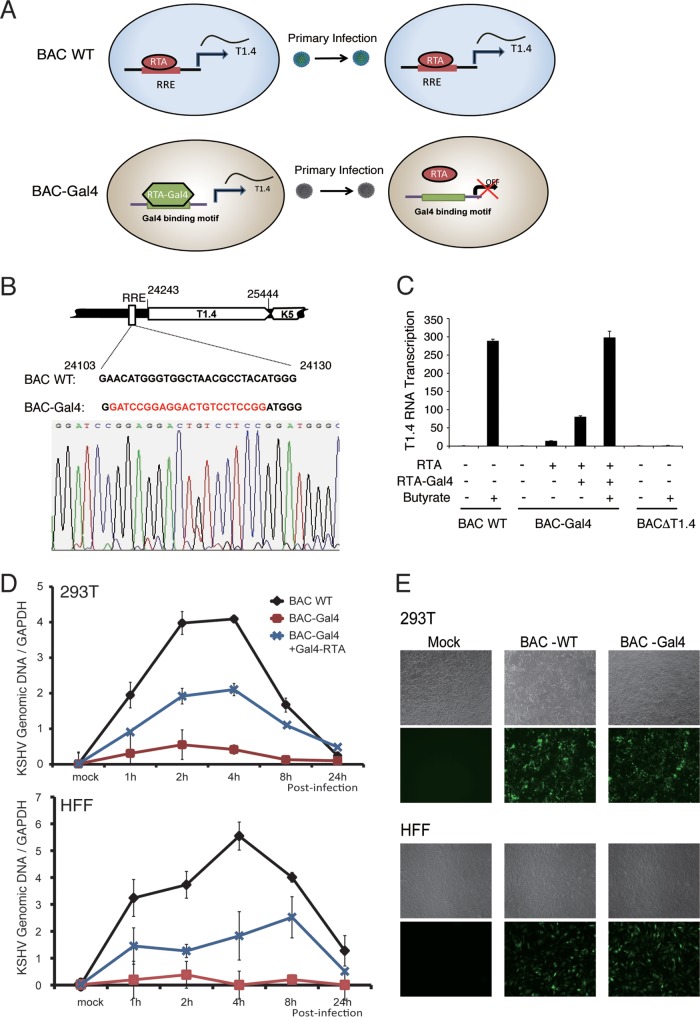
Using the BCBL-1 KSHV reactivation system, we have shown that T1.4 RNA associates with ori-Lyt DNA and facilitates K8 binding to the KSHV genome. However, whether T1.4 functions in viral DNA replication during KSHV de novo infection needs to be confirmed. The BAC-ΔT1.4 mutant virus does not produce infectious virions and therefore cannot be used to study viral de novo infection. Therefore, we generated a T1.4 conditional expression system in which the RRE in the T1.4 promoter was replaced by a Saccharomyces cerevisiae Gal4 binding motif (GATCCGGAGGACTGTCCTCCGG) (Fig. 10B). Thus, RTA could no longer bind to ori-Lyt to transcribe T1.4. However, the T1.4 transcription and ori-Lyt-dependent DNA replication could be restored by ectopic expression of an RTA-Gal4 chimeric protein (Fig. 10A) (17). The recombinant mutant virus, termed BAC-Gal4, was shown to be unable to support T1.4 transcription, and overexpression of RTA was barely able to induce T1.4 transcription. However, ectopic expression of RTA-Gal4 chimeric protein in the cells restored the transcription of T1.4, especially when combined with sodium butyrate treatment (Fig. 10C), as well as virion production (data not shown). BAC16 and BAC-Gal4 virions were prepared and used to infect 293T cells and HFF at an MOI of 50 (viral genomic-DNA equivalents). Viral genomic content in the infected cells at different time points was determined by qPCR. In contrast to BAC16 wild-type virus (in that viral DNA replication continued for 4 h [13]), BAC-Gal4 virus displayed much lower viral DNA copy numbers in both types of infected cells, indicating that T1.4 was essential for viral abortive lytic replication after de novo infection. However, in the presence of RTA-Gal4 protein, viral DNA replication was recovered (Fig. 10D). To rule out the possibility that the lower DNA replication in the mutant virus was caused by the infection rate, we compared infectivity between BAC-WT and BAC-Gal4 recombinant viruses by detection of green fluorescent protein (GFP)-expressng cells with a fluorescence microscope 24 h postinfection. The results showed that the infectivities of these viruses were similar, indicating that T1.4 does not involve virion attachment or entry (Fig. 10E). Taken together, these data showed that T1.4 expression and association with ori-Lyt DNA are critical for KSHV DNA replication; T1.4 RNA mediates the interaction of K8 and the viral genome and, as a result, facilitates the assembly of the viral replication complex and DNA replication during de novo infection.
FIG 10.
T1.4 RNA is essential for KSHV DNA replication following de novo infection. (A) Schematic presentation of the experimental design for conditional expression of T1.4 to demonstrate its role in viral DNA replication using RTA-Gal4 mutant virus. In WT virus, RTA binds the RRE to induce the expression of T1.4 RNA and lytic replication, followed by virion release. During de novo infection, RTA binds to the RRE to induce T1.4 RNA for the initiation of DNA replication. In BAC-Gal4 mutant virus, the RRE in the T1.4 promoter has been replaced with a Gal4 binding sequence. T1.4 expression is induced by ectopic expression of RTA-Gal4 fusion protein, and BAC-Gal4 mutant virus is produced. When BAC-Gal4 virions infect fresh cells, RTA is unable to bind to the Gal4 motif in the T1.4 promoter, so no T1.4 is transcribed. (B) Construction of BAC-Gal4, where the RRE sequence in ori-Lyt has been replaced with a Gal4 binding motif. The mutant virus was confirmed by sequencing. (C) Validation of the expression of T1.4 RNA in BAC16 (BAC WT) and BAC-Gal4. (D) BAC-Gal4 does not support viral DNA replication following de novo infection. 293T cells and HFF were infected by BAC16 (WT) or BAC-Gal4 at an MOI of 50 (viral genomic DNA equivalents). To induce the expression of T1.4 in BAC-Gal4 virus, 293T cells and HFF were transfected with Gal4-RTA expression vector. Twenty-four hours posttransfection, the cells were infected with BAC16 and BAC-Gal4 viruses. The viral DNA content was determined at different time points postinfection by qPCR analysis. The viral genomic DNA copy number was normalized by GAPDH. The error bars indicate SD. (E) The infectivity of BAC-WT and BAC-Gal4 viruses was examined 24 h postinfection by GFP expression (green fluorescence) under a fluorescence microscope at ×5 magnification.
DISCUSSION
In this study, we investigated the functional role of K8 in the KSHV life cycle. The salient features of the outcomes are as follows. (i) The study discovered that K8 is an RNA binding protein and that the majority of K8-associated RNAs are noncoding RNAs, both viral and cellular. This suggests that some of the functions of K8 in the viral life cycle may be carried out in coordination with noncoding RNAs. (ii) A function of K8 in viral DNA replication following KSHV de novo infection (abortive lytic replication) was studied in detail, which led to the conclusion that viral DNA replication following de novo infection initiates at an ori-Lyt and relies on coordinated actions of K8 and the noncoding RNA T1.4.
Unique characteristics and roles of K8 in the KSHV life cycle as an RNA binding protein.
K8 is a structural and positional homolog of EBV ZTA (31, 43). However, unlike ZTA, which is a transcription activator and is capable of initiating an EBV reactivation cascade (44–46), K8 has no transcriptional activity and is not able to initiate viral lytic replication. Instead, K8 is a transcriptional repressor that affects the expression of a subset of viral and cellular genes (22, 23). Unlike EBV ZTA, which binds directly to ZRE DNA within the ZTA promoter (47) and other promoters (45) to regulate gene expression, K8 does not directly bind to viral genomic DNA (15, 34). However, K8 was found to bind to the origin of viral lytic DNA replication (ori-Lyt) of KSHV in an indirect manner, and the binding is absolutely required for viral DNA replication (15, 34). In addition, K8 causes cell cycle arrest at the G1 phase through induction of C/EBPα and p21 (48). Therefore, K8 is a multifunctional protein with its own characteristics. In this study, the unique characteristics of K8 as an RNA binding protein were unveiled, and some of the functions of K8 were found to be executed in coordination with its associated RNAs.
K8-associated RNAs, which include both viral and cellular RNAs, were identified using a CLIP-seq approach. Interestingly, 49% of the K8 binding RNAs are intronic and intergenic sequences, and 11% are annotated noncoding RNAs, suggesting K8 predominately associates with noncoding RNAs. Identification of these noncoding RNAs has provided some clues for us in searching for mechanisms underlying K8 functions in the viral life cycle. For example, K8 is known to function as a global gene expression repressor, but its mechanism remains unknown. Among K8-associated noncoding RNAs, 7SK RNA has also been reported to be a transcriptional repressor. It inhibits gene expression through inactivating positive transcription elongation factor b (P-TEFb) (49). It was shown that P-TEFb–7SK RNA interaction is regulated by hnRNP K, as depletion of hnRNP K increases the binding of 7SK RNA to P-TEFb (50). It can be speculated that K8 may repress global gene expression through modulating the dynamics of the P-TEFb–7SK complex and strengthening the inhibition of P-TEFb. Interaction of K8 with PAN RNA may suggest that K8 represses RTA-mediated transcription of viral delayed-early genes (22, 23), possibly through targeting PAN RNA and blocking its function in recruiting histone demethylases (UTX and JMJD3) to the gene loci (51, 52). These hypotheses warrant future investigation.
The basic region of K8 is an RNA binding domain.
Based on its homology to EBV ZTA, K8 has been proposed to be divided into several functional domains: a transcription activation domain at the N terminus (aa 1 to 121), a DNA binding domain (aa 121 to 189) that contains a basic region (aa 169 to 185), and a leucine zipper domain at the C terminus (aa 191 to 237) (31, 43, 53). The leucine zipper domain has been known to be essential for certain functions of K8, including homodimerization of K8 (31), association with RTA (22, 23), and interaction with histone modification proteins JMJD2A and histone deacetylase (HDAC) (24). In the current study, we demonstrated that the basic region within the previously designated DNA binding domain is actually responsible for RNA binding. Further analysis identified a GRYGR motif (aa 173 to 177) that is required for RNA binding. Interestingly, this motif appears to be a conserved RNA binding motif (motif VI) of the DEAD box protein family (Fig. 5A), and such motifs in other DEAD box family proteins have been shown to be involved in ATPase activity and RNA binding (35–37). When the GRYGR motif in the K8 protein was mutated to GDDGR, the mutant K8 completely lost the ability to bind to the RNAs that we tested (T1.4, PAN, MRP, and 7SK RNAs), as well as viral DNA replication, demonstrating the importance of this motif, as well as the RNA binding property of K8 in the viral life cycle. Thus, this region should be named the RNA binding domain instead of the DNA binding domain, as K8 has never been demonstrated to directly bind to DNA.
The GRYGR motif was found to be responsible for binding of K8 to the diverse noncoding RNAs (T1.4, PAN, MRP, and 7SK RNAs), suggesting that K8 binds to these RNAs through the same RNA recognition mode. A structure analysis of the K8 basic domain with different noncoding RNAs is warranted to reveal the K8 RNA binding mechanism. Since K8 is known as a multifunctional protein, we may further hypothesize that K8 executes different cellular functions through coupling with different RNAs. In the current study, we demonstrated that the function of K8 in viral DNA replication is carried out in coordination with T1.4 RNA. We expect that the functions of K8 in association with PAN and 7SK RNAs will be revealed soon and that it is possibly related to K8-mediated global gene repression. In addition, other functions of K8 have been reported to be associated with the K8 basic domain. It was shown that the basic region is required for cell cycle arrest and cell growth control. Deletion of aa 121 to 189 from the K8 protein resulted in loss of the activity of cell cycle regulation (26). Furthermore, K8 SUMOylation at lysine 158, which was reported to be crucial for the gene repression function of K8, is dependent on the basic domain, as K8 could not be SUMOylated when the basic region was deleted (54). It is worthwhile to discover if the GRYGR mutation affects these events (cell cycle and K8 SUMOylation) and to identify the RNAs that bind to K8 and are responsible for these functions.
KSHV DNA replication following de novo infection initiates at the ori-Lyt and requires coordinated functions of K8 protein and T1.4 RNA.
Besides latent and lytic viral DNA replication, there is a third phase of DNA replication in herpesviruses. After a herpesvirus particle enters a host cell, the virus undergoes DNA replication that results in accumulation of the viral genome to 50 to 100 copies per cell prior to establishment of latency. There is no virion particle produced in this process, so it is called abortive lytic replication. Using K8-null recombinant virus, we previously demonstrated a role of K8 in initial viral DNA replication following de novo KSHV infection. K8-null viruses exhibit much lower viral genome copy numbers than wild-type viruses (13). However, little is known regarding the mode of abortive viral replication and the regulation mechanism. Where does the abortive lytic DNA replication initiate, ori-Lyt, the origin of plasmid replication (ori-P), or another locus in the viral genome? In this study, our data confirmed that KSHV abortive lytic DNA replication initiates at ori-Lyt. In addition, we found that T1.4 noncoding RNA and its mediated K8 binding to ori-Lyt DNA are essential for abortive lytic DNA replication. RTA and its association with ori-Lyt DNA are also critical for DNA replication. These results demonstrate a novel mechanism that regulates DNA replication through coordinated actions of a protein and a noncoding RNA, presenting a new paradigm of eukaryotic DNA replication.
Potential role of T1.4 RNA in KSHV ori-Lyt-dependent DNA replication and a possible mechanism.
T1.4 is highly expressed during de novo infection (11), and its transcription is essential for ori-Lyt-dependent DNA replication (17). In this study, using a Gal4-RTA conditional T1.4 expression system, we demonstrated that KSHV abortive lytic DNA replication is dependent on the expression of T1.4. BAC-Gal4 recombinant virus was unable to initiate DNA replication during de novo viral infection, but the defect could be rescued by ectopic expression of Gal4-RTA, indicating that T1.4 noncoding RNA plays a critical role in ori-Lyt-dependent DNA replication in KSHV primary infection. In addition, our in vitro assay showed that T1.4 mediates K8 binding to ori-Lyt DNA, presenting a new paradigm for noncoding RNA regulating a biological process by mediating protein-DNA interaction.
The mechanism of T1.4 mediation of K8 binding to ori-Lyt DNA has not been fully elucidated. One model is that T1.4 may function as a guide RNA, bringing its associated protein (K8) to DNA by recognizing the DNA sequence through base pairing. Potential base pairing between T1.4 and ori-Lyt DNA was proposed, and an RNase H assay provided supporting evidence for this model (Fig. 9C and D). Another model is that T1.4 may form a G-quadruplex (G4) structure that associates with ori-Lyt DNA and recruits proteins to the DNA. T1.4 RNA is a noncoding RNA with high GC content. QGRS (quadruplex-forming G-rich sequences) Mapper (55) (http://bioinformatics.ramapo.edu/QGRS/index.php) predicts that T1.4 is a potential G-quadruplex RNA. G quadruplexes are noncanonical secondary structures found in guanine-rich regions of DNA and RNA. There are more than 370,000 G-quadruplex-forming sequences in the human genome (56). G-quadruplex structure has also been found in the genomes of many viruses, including HIV-1 (57), KSHV (58), and EBV (59). In EBV latent DNA replication, EBNA1 binds to ori-P and recruits the cellular origin recognition complex (ORC). The interaction between EBNA1 and the ORC is mediated by a noncoding G-rich RNA, which is predicted to possess a G4 structure and which forms a stable complex with EBNA1 and the ORC (60). By treatment with a G-quadruplex-interacting compound, BRACO-19, EBNA1-dependent viral latent DNA replication was inhibited (59). In KSHV, it was also shown that treatment of latently infected cells with a quadruplex-interacting compound, PhenDC3, led to loss of KSHV episomes (58). Whether the function of T1.4 in binding to ori-Lyt and recruiting K8 is dependent on its G-quadruplex structure needs further investigation. If the G-quadruplex hypothesis is proven to be true for T1.4 RNA, it will inform a novel therapeutic strategy to block KSHV primary infection, and possibly lytic replication, as well, using small molecular compounds that interrupt RNA G-quadruplex formation.
MATERIALS AND METHODS
Cell culture.
BCBL-1, a primary effusion lymphoma cell line that carries latently infected KSHV (5), and BJAB, a KSHV-free Burkitt lymphoma B cell line, were maintained in RPMI 1640 medium with 10% heat-inactivated fetal bovine serum (FBS). Human embryonic kidney (HEK) 293T cells, obtained from the ATCC, and HFF (HFF2441), kindly provided by Meenhard Herlyn at the Wistar Institute, were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS. All cultures contained penicillin-streptomycin (50 U/ml) and amphotericin B (1.25 μg/ml).
Antibodies.
Mouse anti-Myc tag, anti-Flag (M2), and anti-β-actin antibodies were purchased from Sigma-Aldrich. Rabbit anti-hnRNP U, anti-hnRNP K, and anti-hnRNP C antibodies were purchased from Abcam. IRDye 680LT/800CW goat anti-rabbit IgG or anti-mouse IgG antibodies were purchased from Li-Cor Biosciences. The mouse monoclonal antibodies against K8 and ORF45 were generated and purified in our laboratory. Mouse monoclonal anti-RTA antibody was provided by Erle Robertson (University of Pennsylvania). Mouse monoclonal anti-LANA antibody was provided by Ke Lan (Wuhan University, China).
Co-IP assay.
TPA-induced BCBL-1 cells (1 × 107) were collected and lysed in 1 ml of immunoprecipitation (IP) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 30 mM sodium fluoride, 1 mM sodium orthovanadate [Na3VO4], 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail) on ice for 30 min. The cell lysates were clarified by centrifugation at 4°C for 10 min. Immunoprecipitation was performed by addition of 2 μl of the antibodies of interest or IgG to cell lysates with gentle agitation at 4°C for 2 h. Then, 30 μl of washed protein G-coated beads was added, and the mixtures were incubated at 4°C for 4 h. The beads were then washed four or five times with cold IP buffer (without protease inhibitor). To eliminate RNA-mediated or DNA-mediated protein interactions, RNase A (100 μg/ml) and DNase I (100 U/ml) were added to washing buffer with 5 mM MgCl2. The precipitates were resuspended in 60 μl of SDS-PAGE loading buffer, boiled for 10 min, and then loaded onto SDS-PAGE gels for Western blot assay.
Western blotting.
Cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 30 mM sodium fluoride,1 mM Na3VO4, 1 mM PMSF, protease inhibitor cocktail [1 tablet in 50 ml lysis buffer]), homogenized for 30 min on ice, and centrifuged at 13,000 rpm for 10 min. The whole-cell extract was resolved by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked in 5% nonfat milk in 1× Tris-buffered saline (TBS) for 1 h and incubated with diluted primary antibodies overnight at 4°C. IRDye 680LT/800CW goat anti-rabbit or anti-mouse antibody (Li-Cor Biosciences) was used as a secondary antibody. The blots were visualized in an Odyssey system (Li-Cor).
Mass spectrometric analysis.
Proteins coimmunoprecipitated with K8 were resolved on 4 to 12% bis-Tris NuPAGE gels (Invitrogen) and stained with a Coomassie G-250 staining kit (Invitrogen). The protein bands were excised and subjected to trypsin digestion. A portion of the peptide digest was injected onto a nanocapillary reverse-phase high-performance liquid chromatograph coupled to a nanoelectrospray ionization source of an ion trap mass spectrometer (ThermoFinnigan LCQ). Mass spectrometry measured peptide masses and then fragmented individual peptides to produce liquid chromatography–tandem-MS (MS-MS) spectra of fragments that reflected the peptide sequence. The MS-MS spectra were run against a nonredundant sequence database using the program SEQUEST. The mass spectrometry was carried out in the protein microchemistry/mass spectrometry facility at the Wistar Institute.
Dot blotting.
RNA was isolated with TRIzol reagent, and the RNA was biotin labeled with the Pierce RNA 3′ end biotinylation kit (Thermo). Briefly, RNA was incubated at 85°C for 3 to 5 min to relax the secondary structure and chilled on ice. Biotinylated cytidine (Bis) phosphate was ligated to the 3′ end of the RNA with T4 RNA ligase at 16°C for 4 h. The biotinylated RNA was purified with chloroform and precipitated with ethanol and glycogen. Biotin signal was detected with a chemiluminescent nucleic acid detection module (Thermo). Two microliters of RNA sample was spotted onto the nitrocellulose membrane. The membrane was dried and blocked with bovine serum albumin (BSA)-TBS with Tween 20 (TBST) and incubated with primary antibody against biotin in BSA-TBST for 30 min at room temperature. The membrane was incubated with secondary antibody conjugated with horseradish peroxidase (HRP) for 30 min at room temperature and then exposed to X-ray film.
CLIP.
CLIP was performed as previously described (30, 61, 62). Briefly, 3 × 107 TPA-induced BCBL-1 cells and BJAB cells were collected by centrifugation, followed by washing with ice-cold phosphate-buffered saline (PBS). The cells were irradiated at 150 mJ/cm2 at 254 nm and then mixed and irradiated again. The cells were spun down and resuspended in 1 ml lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, protease inhibitor cocktail). Two microliters of Turbo DNase (Ambion) was added to the cell lysate. The lysate was incubated on ice for 30 min and spun down at 20,000 × g at 4°C for 10 min to clear the lysate. Two microliters (∼2 μg) of the antibody of interest and 50 μl washed protein A Dynabeads were added per 500 μl lysates. Samples were rotated for 2 h at 4°C. The supernatant was discarded, and the beads were washed twice with 900 μl high-salt buffer (50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate). Then, the beads were washed again twice with 900 μl washing buffer (20 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 0.2% Tween 20) with different amounts of RNase A (high RNase concentration, 2 μg/ml; moderate RNase concentration, 1 μg/ml; and low RNase concentration, 0.2 μg/ml) and an additional 900 μl RNase-free washing buffer twice. The RNAs were dephosphorylated at the 3′ ends. Linker RNA 3′ adapter (Table 2) was ligated to the 3′ ends of the RNAs, and [γ-32P]ATP-labeled linker RNA 5′ adapter (Table 2) was ligated to the 5′ ends of the RNAs. The protein-RNA complexes were resolved on a 4 to 12% NuPAGE Bis-Tris gel and transferred to a nitrocellulose membrane. K8-RNA complexes were cut and subjected to proteinase K digestion. The RNAs were purified with TRIzol and subjected to RT-PCR. Sequencing was performed on an Illumina HiSeq 2500 sequencer at the Beijing Genomics Institute (BGI).
TABLE 2.
Oligonucleotides used in this study
| Purpose | Name of oligonucleotide | Sequence (5′-3′) |
|---|---|---|
| qPCR | KSHV ORF73-LCN | CGCGAATACCGCTATGTACTCA |
| KSHV ORF73-LCC | GGAACGCGCCTCATACGA | |
| GAPDH-F | ACATCATCCCTGCCTCTAC | |
| GAPDH-R | TCAAAGGTGGAGGAGTGG | |
| Construction of plasmids | hnRNP U-F | gcgggatcccaccATGAGTTCCTCGCCTGTTAATGTA |
| hnRNP U-R | gggatccTCAATAATATCCTTGGTGATAATGCTGA | |
| GST-K8 F | cggaattcCGATGCCCAGAATGAAGGACATAC | |
| GST-K8 R | atagtttagcggccgctttTCAACATGGTGGGAGTGGC | |
| ChIP assay | 3.1F | TAGGTGGGACCGTGAGCGACT |
| 3.1R | CCATAATCCTCTGCCCCGC | |
| RRE 12F | ACGGGCCTGGAATCTCGCCTCTGG | |
| RRE 12R | ATGGGCGTAACCGTAGGACAAGCTG | |
| GAPDH-F | ACATCATCCCTGCCTCTAC | |
| GAPDH-R | TCAAAGGTGGAGGAGTGG | |
| RT-PCR of CLIP-seq | RNA 5′ adapter | GUUCAGAGUUCUACAGUCCGACGAUC |
| RNA 3′ adapter | UGGAAUUCUCGGGUGCCAAGG | |
| RT primers for CLIP | GCCTTGGCACCCGAGAATTCCA | |
| CLIP PCR primer | AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA | |
| CLIP PCR primer, index 1a | CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA | |
| BACmid construction | BAC-stopK8-F | GTGTAACCCTGCCAAATGCCCAGAATGAAGGACATACCTACTTAGAGTTCCCCGGGAACGGACATTTAGGGATAACAGGGTAATCGATT |
| BAC-stopK8-R | CAGCTTCATCTTTCTCAGAATTGTCCG TTCCCGGGGAACTCTAAGTAGGTATGTCCTTCATTCTCAACCAATTAACCAATTCTGATTAG | |
| BAC-K8-GDDGR-F | TTAAAGGCCGAAGTATGTGATCAGTCACATTCTCCCACGCGAAAGCAAGGCGATGACGGCCGCGTGTCATCGAAAGCATACAGGATGACGACGATAAGTAGGG | |
| BAC-K8-GDDGR-R | GTCTATACCTGCTGCAGCTGTCTTGTGTATGCTTTCGATGACACGCGGCCGTCATCGCCTTGCTTTCGCGTGGGAGAACAACCAATTAACCAA TTCTGATTAG | |
| BAC-GAL4-F | TAGATGGGGGCCGGGAGGATGGGGCCCCGCCCACCGCTGGCGCGCCCCAGGATCCGGAGGACTGTCCTCCGGAGGATGACGACGATAAGTAGGG | |
| BAC-GAL4-R | AGTTGGTTAACCCGTCTCAAATGGGCGTAACCGTAGGACAAGCTGCCCATCCGGAGGACAGTCCTCCGGATCCTGGGGCGCGCCAGCGGTGGGCGGCAACCAATTAACCAATTCTGATTAG | |
| BACΔT1.4-F | AGAGCAGCCCCAAATGTGCGCGCAACACCCAGCACATGCTCCACATACAGCACCCAGGGCGTCACGTCACATATCTCTGTAGGATGACGACGATAAGTAGGG | |
| BACΔT1.4-R | TTGAACAACCACTTGGGTGCACAGAGATATGTGACGTGACGCCCTGGGTGCTGTATGTGGAGCATGTGCTCAACCAATTAACCAATTCTGATTAG | |
| Probes for ChIRP | T1.4-1 | TAAATCCAAGAGATCCGTC |
| T1.4-2 | GTGTGCTTGTGACTGATACA | |
| T1.4-3 | TTATATGCGCGTGCTTGCGA | |
| T1.4-4 | GCTACCTGTGTAAATTTCGG | |
| T1.4-5 | ACAATGTGAATCCTCTGTGC | |
| T1.4-6 | ATTTCAAGAGCCTATGCTGG |
Index sequence is underlined.
RNA pulldown assay.
To synthesize biotinylated transcripts (7SK, MRP, PAN, and T1.4 RNAs) in vitro, DNA templates were prepared by PCR using forward primers that contained the T7 RNA polymerase promoter sequence. Biotinylated transcripts were synthesized using MEGAscript kits (Thermo Fisher) with biotin-14-CTP (Thermo Fisher). Since T1.4 RNA contains high-GC-repeat sequence, biotinylated T1.4 was generated by adding biotin at the 3′ end of in vitro-transcribed T1.4 RNA with the Pierce biotin 3′ end DNA-labeling kit (Thermo Fisher). The expression vectors for K8 and its mutants were transfected into 293T cells, and cell lysates were prepared 48 h posttransfection in lysis buffer (150 mM KCl, 25 mM Tris-HCl, pH 7.4, 0.5 mM dithiothreitol [DTT], 0.5% NP-40, RNase inhibitor, and protease inhibitor cocktail). Two micrograms of biotin-labeled RNAs and 2 μl yeast tRNA were added to 1 ml cell lysate (or purified GST-K8 protein from E. coli BL21) and incubated with rotation at 4°C for 1 h. Complexes were isolated with streptavidin-coupled Dynabeads, and the RNA-bound proteins were detected by Western blotting. Biotinylated RNAs could be detected by Northern blotting with the chemiluminescent nucleic acid detection module (Thermo Fisher Scientific).
Genetic manipulation of BAC-cloned KSHV genome.
The mutagenesis of BAC16 was performed using a recombineering system as described by Brulois et al. and Tischer et al. (38, 40). In brief, the Kan/I-SceI cassettes were amplified from plasmid pEPKan-S by PCR with oligonucleotides of interest for BAC mutants. The oligonucleotides used for PCR are listed in Table 2. The purified PCR fragments were transformed by electroporation into BAC16-containing GS1783 cells that had been induced at 42°C for 15 min (38). The recombinant clones were selected at 32°C on LB plates containing 34 μg/ml chloramphenicol and 50 μg/ml kanamycin. Positive clones were analyzed by miniassay and restriction enzyme digestion. Confirmed clones were cultured with 1% l-arabinose, induced at 42°C again, and plated on LB plates containing 1% l-arabinose and 34 μg/ml chloramphenicol for secondary recombination. Colonies that survived on the l-arabinose plates were cultured with 34 μg/ml chloramphenicol alone and with both 34 μg/ml chloramphenicol and 50 μg/ml kanamycin. The kanamycin-sensitive clones were analyzed by restriction enzyme digestion, and proper mutations were further confirmed by DNA sequencing.
Quantification of viral genomic DNAs.
Total DNAs were prepared from KSHV-infected cells with a GeneJet genomic DNA purification kit (Fermentas Life Sciences). KSHV genomic DNA was measured by qPCR, and genomic DNA copy numbers were calculated based on external standards of known concentrations of BAC16 DNA. The primers ORF73-LCN and ORF73-LCC were previously described by Krishnan et al. (63). The viral DNA copy numbers were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA.
KSHV virion purification and primary infection.
293T cells carrying BAC16 wild-type or mutant virus were induced by 3 mM sodium butyrate for 4 to 5 days. The culture supernatant was filtered through a 0.45-μm filter and centrifuged at 100,000 × g for 1.5 h at 4°C. The pellet was resuspended in 1/100 volume of 1× PBS or DMEM and stored at −80°C until use. The infection process was previously described (13). Briefly, cells were incubated with concentrated virus in the presence of 4 μg/ml Polybrene and spun at 2,500 rpm for 1 h at room temperature. The cells were washed twice with PBS to remove the unbound or loosely bound virions. At the indicated time points postinfection, cells were collected and total genomic DNA was isolated from infected cells. Viral genomic DNAs were quantified by qPCR with primers specific for KSHV ORF73.
Quantification of genomic and virion DNAs.
DNAs from viral stocks or cells were prepared using the Universal Genomic DNA Extraction Kit 5.0 (TaKaRa). Monolayers of infected cells in 6-well plates were treated with trypsin, washed, and resuspended in 200 μl of PBS. Total DNA was prepared according to the manufacturer's instructions. For virion DNA, 200 μl of virus stocks was pretreated with 3 μl of Turbo DNase I (Ambion) for 1 h at 37°C. The reaction was stopped with 10 μl of 500 mM EDTA, followed by heat inactivation at 70°C. Then, 20 μl of proteinase K solution (Sigma) and 200 μl of buffer AL from the DNeasy kit (Qiagen) were added to the reaction mixture. The mixture was kept at 56°C for 15 min and then extracted with phenol-chloroform. The DNA was precipitated with ethanol and dissolved in Tris-EDTA buffer. DNA was quantitated by qPCR.
Plasmids.
Wild-type K8 was cloned into the pCMV-3tag-2 vector containing 3× Myc tags between the BamHI and EcoRI sites (15). All mutations of K8 were generated using a QuikChange multisite-directed mutagenesis kit (Stratagene). The vector for Flag-hnRNP U was generated from total RNA derived from 293T cells by RT-PCR with primers hnRNP U-F and hnRNP U-R and cloned into the pCMV-3tag-1 vector containing 3× Flag tags at the BamHI site. The vector for GST-K8 was generated from wild-type K8 with primers GST-K8 F and GST-K8 R and cloned into the pGEX-4T-2 vector between the EcoRI and NotI sites.
ChIRP.
The ChIRP protocol was adapted from that of Chu et al. (42). The probes were synthetic DNAs with BiotinTEG at the 3′ end designed to be complementary to T1.4 RNA sequence using the online designer from Biosearch Technologies. The high-GC-repeat region of T1.4 was omitted from the probe. LacZ probes were designed and used as controls. The probes were divided into two pools: the “even” pool containing all probes numbered 2, 4, and 6 and the “odd” pool containing probes numbered 1, 3, and 5. BCBL-1 cells were treated with 20 ng/ml TPA to induce lytic reactivation for 48 h. The cells were treated with 1% glutaraldehyde in PBS for cross-linking for 10 min at room temperature. The cross-linking was quenched with 125 mM glycine in PBS for 5 min. The cells were washed twice and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.0, 10 mM EDTA, 1% SDS, freshly added 1 mM PMSF, complete protease inhibitor [Roche], and 0.1 U/μl Superase-In [Ambion]) for 10 min on ice. The lysates were subjected to sonication to share genomic DNA in the size range of 100 to 500 bp. The samples were centrifuged at 16,000 × g for 10 min, and the supernatants were collected as soluble chromatin. The chromatin solution (100 μl) was mixed with 400 μl hybridization buffer, and 0.5 to 1 μl of 100 μM probes of T1.4 or LacZ was added to the diluted chromatin and rotated at 37°C for 4 h. Streptavidin magnetic beads (Sigma) were washed three times in lysis buffer and blocked with 500 ng/ml yeast RNA (Ambion), 100 mg/ml sheared salmon sperm DNA (Ambion), and 1 mg/ml BSA (Sigma) for 1 h. Then, 50 μl of prewashed and preblocked beads was added per 100 pmol of probes, and the reaction mixture was rotated at 37°C for 30 min. The beads were precipitated and washed 3 times with washing buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% SDS, freshly added 1 mM DTT, and freshly added 1 mM PMSF) and resuspended in buffer with RNase A (100 μg/ml), RNase H (0.1 U/μl), or trypsin. DNA was eluted from the beads with 150 μl elution buffer (50 mM NaHCO3, 1% SDS, 200 mM NaCl) with 100 μg/ml RNase A (Sigma) and 0.1 U/ml RNase H (Ambion). Proteinase K (0.2 U/ml) was added and incubated at 65°C for 45 min. DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with 70% ethanol. The eluted DNA was analyzed by qPCR with primers for the 12F region of ori-Lyt.
DNA affinity purification assay.
The DNA affinity purification assay was performed as previously described (17). Briefly, three biotinylated ori-Lyt DNA fragments (3F, 9F, and 11F) were synthesized using PCR with pOri-A DNA as the template. The biotinylated PCR fragments were coupled to streptavidin-conjugated magnetic beads (Sigma) and then mixed with nuclear extracts prepared from TPA-induced BCBL-1 cells. In each reaction mixture, 2/3 volume of DNA-coupled beads in solution A (20 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.05% NP-40, 15 mM MgCl2, 75 μg/ml salmon sperm DNA) was mixed with 1/3 volume of the nuclear extract in buffer C (20 mM HEPES, pH 7.9, 25% glycerol, 0.2 mM EDTA, 0.42 M NaCl, 1 mM DTT, 0.05% NP-40, 1 mM PMSF, and a protease inhibitor tablet and 100 μg/ml RNase A when needed) and incubated at 25°C for 45 min. The beads were washed four times in D150 buffer (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 150 mM KCl, 1 mM DTT, 0.05% NP-40, 1 mM PMSF, and RNase A was added at 100 μg/ml when needed). The affinity-purified materials were resuspended in SDS-PAGE loading buffer, boiled for 10 min, and resolved on SDS-PAGE.
Accession number(s).
The CLIP-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO accession number GSE104711).
ACKNOWLEDGMENTS
We thank the members of the Yuan laboratory, the Paul Lieberman laboratory (Wistar), and the Erle Robertson laboratory (University of Pennsylvania) for discussions and constructive suggestions. We are grateful for reagents and technical assistance from Erle Robertson, Paul Lieberman, and colleagues.
This work is supported by grants from the NIH (P01CA174439 to Y.Y.), the National Natural Science Foundation of China (81530069 to Y.Y. and 81371793 to Y.W.), and the Guangdong Innovative Research Team Program (no. 2009010058).
REFERENCES
- 1.Antman K, Chang Y. 2000. Kaposi's sarcoma. N Engl J Med 342:1027–1038. doi: 10.1056/NEJM200004063421407. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D. 1996. Human herpesvirus 8 and the biology of Kaposi's sarcoma. Semin Virol 7:325–332. doi: 10.1006/smvy.1996.0040. [DOI] [Google Scholar]
- 3.Schulz TF. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J Gen Virol 79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 4.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol 71:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 6.Grundhoff A, Ganem D. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest 113:124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, Haase AT. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 71:715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol 73:2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W, Wang H, Herndier B, Ganem D. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A 93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jha HC, Lu J, Verma SC, Banerjee S, Mehta D, Robertson ES. 2014. Kaposi's sarcoma-associated herpesvirus genome programming during the early stages of primary infection of peripheral blood mononuclear cells. mBio 5:e02261-14. doi: 10.1128/mBio.02261-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purushothaman P, Thakker S, Verma SC. 2015. Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells. J Virol 89:3093–3111. doi: 10.1128/JVI.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. 2013. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog 9:e1003813. doi: 10.1371/journal.ppat.1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Sathish N, Hollow C, Yuan Y. 2011. Functional characterization of Kaposi's sarcoma-associated herpesvirus open reading frame K8 by bacterial artificial chromosome-based mutagenesis. J Virol 85:1943–1957. doi: 10.1128/JVI.02060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AuCoin DP, Colletti KS, Xu Y, Cei SA, Pari GS. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J Virol 76:7890–7896. doi: 10.1128/JVI.76.15.7890-7896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CL, Li H, Wang Y, Zhu FX, Kudchodkar S, Yuan Y. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J Virol 77:5578–5588. doi: 10.1128/JVI.77.10.5578-5588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Li H, Chan MY, Zhu FX, Lukac DM, Yuan Y. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J Virol 78:8615–8629. doi: 10.1128/JVI.78.16.8615-8629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Tang Q, Maul GG, Yuan Y. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J Virol 80:12171–12186. doi: 10.1128/JVI.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.AuCoin DP, Colletti KS, Cei SA, Papouskova I, Tarrant M, Pari GS. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542–555. doi: 10.1016/j.virol.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Rossetto C, Yamboliev I, Pari GS. 2009. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 K-bZIP modulates latency-associated nuclear protein-mediated suppression of lytic origin-dependent DNA synthesis. J Virol 83:8492–8501. doi: 10.1128/JVI.00922-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Li H, Tang Q, Maul GG, Yuan Y. 2008. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J Virol 82:2867–2882. doi: 10.1128/JVI.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato-Noah T, Xu Y, Rossetto CC, Colletti K, Papouskova I, Pari GS. 2007. Overexpression of the Kaposi's sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion BACmid and yields an enhanced growth phenotype. J Virol 81:13519–13532. doi: 10.1128/JVI.00832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumiya Y, Lin SF, Ellison T, Chen LY, Izumiya C, Luciw P, Kung HJ. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J Virol 77:1441–1451. doi: 10.1128/JVI.77.2.1441-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao W, Tang Y, Lin SF, Kung HJ, Giam CZ. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J Virol 77:3809–3815. doi: 10.1128/JVI.77.6.3809-3815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang PC, Fitzgerald LD, Hsia DA, Izumiya Y, Wu CY, Hsieh WP, Lin SF, Campbell M, Lam KS, Luciw PA, Tepper CG, Kung HJ. 2011. Histone demethylase JMJD2A regulates Kaposi's sarcoma-associated herpesvirus replication and is targeted by a viral transcriptional factor. J Virol 85:3283–3293. doi: 10.1128/JVI.02485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang WS, Hsu HW, Campbell M, Cheng CY, Chang PC. 2015. K-bZIP mediated SUMO-2/3 specific modification on the KSHV genome negatively regulates lytic gene expression and viral reactivation. PLoS Pathog 11:e1005051. doi: 10.1371/journal.ppat.1005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumiya Y, Lin SF, Ellison TJ, Levy AM, Mayeur GL, Izumiya C, Kung HJ. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J Virol 77:9652–9661. doi: 10.1128/JVI.77.17.9652-9661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu FY, Wang SE, Tang QQ, Fujimuro M, Chiou CJ, Zheng Q, Chen H, Hayward SD, Lane MD, Hayward GS. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1). J Virol 77:8893–8914. doi: 10.1128/JVI.77.16.8893-8914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter OV, Sei E, Richardson RB, Conrad NK. 2013. Chromatin immunoprecipitation and microarray analysis suggest functional cooperation between Kaposi's Sarcoma-associated herpesvirus ORF57 and K-bZIP. J Virol 87:4005–4016. doi: 10.1128/JVI.03459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu FX, Li X, Zhou F, Gao SJ, Yuan Y. 2006. Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J Virol 80:12187–12196. doi: 10.1128/JVI.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2011. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp 30:2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin SF, Robinson DR, Miller G, Kung HJ. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol 73:1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Seo T, Hwang S, Lee D, Gwack Y, Choe J. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J Virol 74:11977–11982. doi: 10.1128/JVI.74.24.11977-11982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al Mehairi S, Cerasoli E, Sinclair AJ. 2005. Investigation of the multimerization region of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) protein K-bZIP: the proposed leucine zipper region encodes a multimerization domain with an unusual structure. J Virol 79:7905–7910. doi: 10.1128/JVI.79.12.7905-7910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair AJ. 2003. bZIP proteins of human gammaherpesviruses. J Gen Virol 84:1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- 35.Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 36.de la Cruz J, Kressler D, Linder P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci 24:192–198. doi: 10.1016/S0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 37.Rocak S, Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 38.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira J, O'Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 40.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 41.Lu F, Tsai K, Chen HS, Wikramasinghe P, Davuluri RV, Showe L, Domsic J, Marmorstein R, Lieberman PM. 2012. Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA. J Virol 86:5752–5762. doi: 10.1128/JVI.07216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol 73:5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieberman PM, Berk AJ. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol 64:2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol 64:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor N, Countryman J, Rooney C, Katz D, Miller G. 1989. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J Virol 63:1721–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang YN, Dong DL, Hayward GS, Hayward SD. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol 64:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu FY, Tang QQ, Chen H, ApRhys C, Farrell C, Chen J, Fujimuro M, Lane MD, Hayward GS. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc Natl Acad Sci U S A 99:10683–10688. doi: 10.1073/pnas.162352299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterlin BM, Brogie JE, Price DH. 2012. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA 3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogg JR, Collins K. 2007. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossetto CC, Pari G. 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossetto CC, Pari GS. 2014. PAN′s labyrinth: molecular biology of Kaposi's sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses 6:4212–4226. doi: 10.3390/v6114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang PC, Izumiya Y, Wu CY, Fitzgerald LD, Campbell M, Ellison TJ, Lam KS, Luciw PA, Kung HJ. 2010. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem 285:5266–5273. doi: 10.1074/jbc.M109.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izumiya Y, Ellison TJ, Yeh ET, Jung JU, Luciw PA, Kung HJ. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J Virol 79:9912–9925. doi: 10.1128/JVI.79.15.9912-9925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikin O, D'Antonio L, Bagga PS. 2006. QGRS Mapper: a Web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Todd AK, Johnston M, Neidle S. 2005. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res 33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Nicola B, Lech CJ, Heddi B, Regmi S, Frasson I, Perrone R, Richter SN, Phan AT. 2016. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res 44:6442–6451. doi: 10.1093/nar/gkw432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madireddy A, Purushothaman P, Loosbroock CP, Robertson ES, Schildkraut CL, Verma SC. 2016. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res 44:3675–3694. doi: 10.1093/nar/gkw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norseen J, Johnson FB, Lieberman PM. 2009. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol 83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J 27:3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sei E, Wang T, Hunter OV, Xie Y, Conrad NK. 2015. HITS-CLIP analysis uncovers a link between the Kaposi's sarcoma-associated herpesvirus ORF57 protein and host pre-mRNA metabolism. PLoS Pathog 11:e1004652. doi: 10.1371/journal.ppat.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J Virol 78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]