ABSTRACT
Recently, two new influenza A-like viruses have been discovered in bats, A/little yellow-shouldered bat/Guatemala/060/2010 (HL17NL10) and A/flat-faced bat/Peru/033/2010 (HL18NL11). The hemagglutinin (HA)-like (HL) and neuraminidase (NA)-like (NL) proteins of these viruses lack hemagglutination and neuraminidase activities, despite their sequence and structural homologies with the HA and NA proteins of conventional influenza A viruses. We have now investigated whether the NS1 proteins of the HL17NL10 and HL18NL11 viruses can functionally replace the NS1 protein of a conventional influenza A virus. For this purpose, we generated recombinant influenza A/Puerto Rico/8/1934 (PR8) H1N1 viruses containing the NS1 protein of the PR8 wild-type, HL17NL10, and HL18NL11 viruses. These viruses (r/NS1PR8, r/NS1HL17, and r/NS1HL18, respectively) were tested for replication in bat and nonbat mammalian cells and in mice. Our results demonstrate that the r/NS1HL17 and r/NS1HL18 viruses are attenuated in vitro and in vivo. However, the bat NS1 recombinant viruses showed a phenotype similar to that of the r/NS1PR8 virus in STAT1−/− human A549 cells and mice, both in vitro and in vivo systems being unable to respond to interferon (IFN). Interestingly, multiple mouse passages of the r/NS1HL17 and r/NS1HL18 viruses resulted in selection of mutant viruses containing single amino acid mutations in the viral PB2 protein. In contrast to the parental viruses, virulence and IFN antagonism were restored in the selected PB2 mutants. Our results indicate that the NS1 protein of bat influenza A-like viruses is less efficient than the NS1 protein of its conventional influenza A virus NS1 counterpart in antagonizing the IFN response and that this deficiency can be overcome by the influenza virus PB2 protein.
IMPORTANCE Significant gaps in our understanding of the basic features of the recently discovered bat influenza A-like viruses HL17NL10 and HL18NL11 remain. The basic biology of these unique viruses displays both similarities to and differences from the basic biology of conventional influenza A viruses. Here, we show that recombinant influenza A viruses containing the NS1 protein from HL17NL10 and HL18NL11 are attenuated. This attenuation was mediated by their inability to antagonize the type I IFN response. However, this deficiency could be compensated for by single amino acid replacements in the PB2 gene. Our results unravel a functional divergence between the NS1 proteins of bat influenza A-like and conventional influenza A viruses and demonstrate an interplay between the viral PB2 and NS1 proteins to antagonize IFN.
KEYWORDS: influenza viruses, NS1 protein, bat, interferons
INTRODUCTION
Influenza A viruses (IAVs) maintain a large reservoir of genetic diversity in animals, including swine, horses, and, in particular, birds, which represent the largest reservoir of IAVs (1). Recently, two new IAVs have been discovered in bats by metagenomic approaches, A/little yellow-shouldered bat/Guatemala/060/2010 (HL17NL10) and A/flat-faced bat/Peru/033/2010 (HL18NL11) (2, 3), changing our understanding of the influenza virus ecology. Serological studies have demonstrated a high prevalence of bat influenza A-like viruses in Peruvian bats (3). Moreover, cross-reactive antibodies against avian H9 IAVs have been detected in 30% of sampled African bats, indicating that infection of bats with IAV is not limited to the HL17 and HL18 viruses (4). Nevertheless, the HL17NL10 and HL18NL11 viruses have proved to be very particular, since preliminary attempts at isolation were unsuccessful (5, 6), suggesting that they have replication requirements distinct from those of other conventional IAVs. Moreover, several of the known canonical functions of the hemagglutinin (HA) and neuraminidase (NA) proteins of IAVs are missing in the corresponding hemagglutinin-like (HL) and neuraminidase-like (NL) proteins of influenza A-like strains (7–12). Recently, infectious bat influenza A-like viruses have been generated by reverse genetics (12). These viruses demonstrate a unique cell type tropism not restricted to bat cells, highlighting the importance of further research to understand the biology of such viruses and their potential ability to infect nonbat mammalian hosts.
IAV nonstructural protein 1 (NS1) is a multifunctional virulence factor that performs a plethora of protein-protein and protein-RNA interactions during IAV infection. Nonetheless, the major ascribed NS1 function is as an antagonist of type I interferon (IFN) (13). At the pretranscriptional level, NS1 is able to bind to double-stranded RNA (dsRNA) species (14) and to the host proteins RIG-I, TRIM25, and PKR, blocking virus sensing, IFN induction, and host antiviral activities (15–17). At the co- and posttranscriptional levels, NS1 interferes with cellular pre-mRNA processing, shutting down host gene expression, including that of IFN-related genes (13). More interestingly, these interactions have been proposed to be a species-specific signature for several influenza A virus strains (18–21). In particular, experiments in which the NS1 protein from HL17NL10 and HL18NL11 (denoted NS1HL17 and NS1HL18 proteins, respectively) was overexpressed demonstrated the effective specific inhibition of IFN induction by NS1 but did not indicate that NS1 blocked general host gene expression (22, 23). However, other authors showed that truncations of NS1HL17 and NS1HL18 did not dramatically affect viral lung replication in mice in the context of chimeric viruses containing the glycoproteins HA and NA from influenza A/Puerto Rico/8/1934 (PR8) H1N1 viruses and all the internal proteins from the HL17NL10 and HL18NL11 viruses (6). Some of the main functional and structural similarities (and differences) between the NS1 protein from the bat influenza A-like viruses and NS1 from the PR8 virus have been previously described (22, 24).
In the study described here, we generated bat NS1 recombinant HL17NL10 and HL18NL11 viruses in a conventional PR8 IAV backbone and characterized these viruses (r/NS1HL17 and r/NS1HL18, respectively) in bat and notbat mammalian cells and in mice. We show that although these recombinant viruses were able to replicate in cell culture and in mice, there was an evident attenuation imposed by the NS1 protein of the bat influenza A-like viruses. In contrast, the bat recombinant viruses r/NS1HL17 and r/NS1HL18 showed a phenotype more similar to that of the recombinant PR8 virus containing the NS1 protein of the PR8 wild type (r/NS1PR8) in STAT1−/− (IFN signaling-deficient) systems. After multiple mouse passages, bat NS1 viruses acquired single point mutations in the PB2 viral polymerase that reverted their phenotype in mice. In contrast to the parental r/NS1HL17 and r/NS1HL18 viruses, the viruses containing the compensatory mutations in PB2 (r/NS1HL17 P5 and r/NS1HL18 P7, respectively) were able to inhibit IFN production by hindering interferon regulatory factor 3 (IRF3) phosphorylation. Our results demonstrate an interplay between the NS1 and the PB2 viral proteins to counteract the host antiviral response.
RESULTS
Bat NS1 recombinant PR8 viruses are attenuated in bat and nonbat mammalian cells and in vivo.
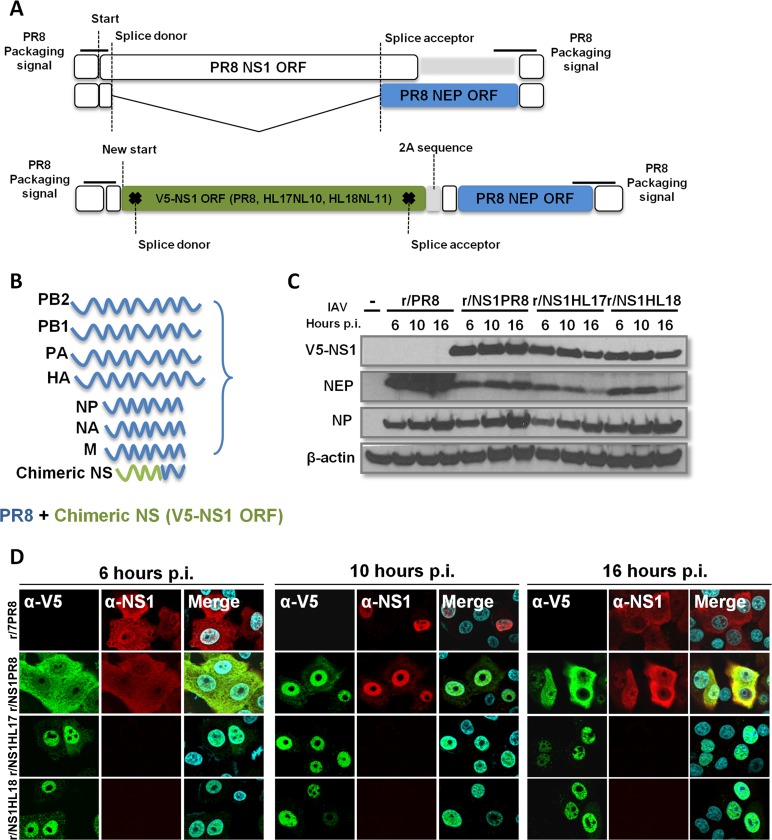
It has been demonstrated that the NS1 proteins of bat influenza A-like viruses (NS1HL17 and NS1HL18 proteins, respectively) are able to prevent IFN induction and that this is strictly dependent on their RNA binding domains (22). To assess if the NS1HL17 and NS1HL18 proteins from bat influenza A-like viruses could functionally and efficiently replace the NS1 protein of a conventional influenza A virus, we used reverse genetics to generate (25) in a PR8 backbone recombinant viruses containing the NS1 open reading frame (ORF) of the PR8 wild type (WT) (for comparison purposes), the NS1 ORF of HL17NL10, and the NS1 ORF of HL18NL11 (generating the r/NS1PR8, r/NS1HL17, and r/NS1HL18 viruses, respectively) (Fig. 1A and B). To prevent modifications in the overlapping nuclear export protein (NEP) ORF of the IAV NS gene, the NS1-coding regions were inserted in a modified NS segment that expresses the NS1 and NEP proteins as a contiguous ORF separated by a 2A picornaviral sequence that allows expression of the NS1 and NEP proteins as individual polypeptides (26). The NS1 proteins were also ligated in frame at their N termini with a V5 tag, a small epitope derived from the P and V proteins of the paramyxovirus simian virus 5 (SV5) (protein sequence, GKPIPNPLLGLDST). In order to verify NS1 expression from these viruses, we performed infections in human lung epithelial (A549) cells at a multiplicity of infection (MOI) of 2 over time for indirect immunofluorescence and Western blotting (Fig. 1C and D). The r/NS1HL17 and r/NS1HL18 viruses expressed the NS1 and NEP proteins. Although the NS1 protein expression levels from the bat recombinant viruses were slightly lower than the NS1 protein expression level from PR8, as measured by Western blotting, no striking differences in NEP expression level from that in the counterpart r/NS1PR8 virus were found, suggesting that increased NS1 nuclear localization might be responsible for decreased NS1 extraction prior to SDS-PAGE and Western blotting, giving the impression of lower expression levels. However, unlike the NS1 protein from PR8 (denoted NS1PR8 protein), which showed a varying nuclear and cytoplasmic distribution depending on the time postinfection (p.i.), the intracellular localization of the bat influenza A-like virus NS1 proteins was mainly nuclear at all times tested, suggesting possible functional differences between NS1 from bat influenza A-like viruses and the NS1PR8 protein.
FIG 1.
Generation of bat NS1 recombinant viruses. (A) Representation of the WT PR8 NS segment and mutated bat influenza A-like NS segment. (B) Color-coded illustration of genes included in recombinant NS1 viruses. Blue, genes from PR8; green, chimeric NS segment indicating the shifted NS1 ORF for PR8, HL17NL10, and HL18NL11. (C) Determination by Western blotting of NS1 expression levels in A549 cells infected at an MOI of 2 (or mock infected [−]) at the indicated times p.i. Lysates were prepared at 6, 10, and 16 h p.i. NS1, the V5 tag, and NP were detected using specific antibodies. β-Actin served as a loading control. (D) Indirect immunofluorescence of NS1 protein localization in A549 cells infected at an MOI of 2 at the indicated times p.i. Costaining was performed using a polyclonal anti-NS1 serum (NS1-155; green) and anti-V5 tag (red), together with 4′,6′-diamidino-2-phenylindole (DAPI) to stain the nuclei (blue).
Next, we analyzed the growth properties of the bat NS1 recombinant viruses by comparing their multicycle growth curves at an MOI of 0.01 in different mammalian cell lines of bat and nonbat origin. The r/NS1HL17 and r/NS1HL18 viruses showed an attenuation in growth in cells of canine (Madin-Darby canine kidney [MDCK]) and human (A549) origin compared to the r/NS1PR8 control virus, with more than 2-log differences in viral titers being detected at 24 and 36 h postinfection in both cell lines, respectively (P < 0.001) (Fig. 2A). To test whether this reduction in viral titers was related to an intrinsic host feature present in cells of bat origin, we assessed the replication ability of these viruses in primary kidney cell lines from Sturnira lilium and Artibeus lituratus bats, the two South American species of microbats in which these viruses were first discovered in 2012 and 2013 (2, 3). The primary kidney bat cell lines supported replication of all the tested viruses (Fig. 2B); however, the recombinant viruses containing a bat virus NS1 were still significantly attenuated compared to the corresponding r/NS1PR8 control.
FIG 2.
Characterization of bat NS1 recombinant viruses in vitro. (A and B) Viral growth of r/NS1HL17, r/NS1HL18, and r/NS1PR8 in MDCK (canine), A549 (human), and A549 STAT1−/− (human) cells and in primary kidney bat cell lines from Sturnira lilium and Artibeus lituratus. Cells were infected at an MOI of 0.01, and the viral supernatant was titrated as the number of PFU per milliliter at the indicated time points postinfection (p.i.). Shown are the means and SDs from three biological replicates. ANOVA for multiple comparisons was used, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (***, P < 0.001; **, P < 0.01). (C and D) Generation of a STAT1 knockout A549 cell line using the CRISPR-Cas9 method. (C) qRT-PCR analyses of representative interferon-stimulated gene (ISG) induction in vitro. WT and STAT1 KO cells were treated with 1,000 units of universal IFN for 12 h. Cells were harvested, and total RNA was isolated and subjected to RT-PCR to measure the transcriptional induction of the indicated interferon-induced genes. (D) The WT and STAT1 KO cell lines were tested for STAT1 expression by Western blotting using STAT1- and actin-specific antibodies. Rel., relative; -/-, STAT1−/− cells.
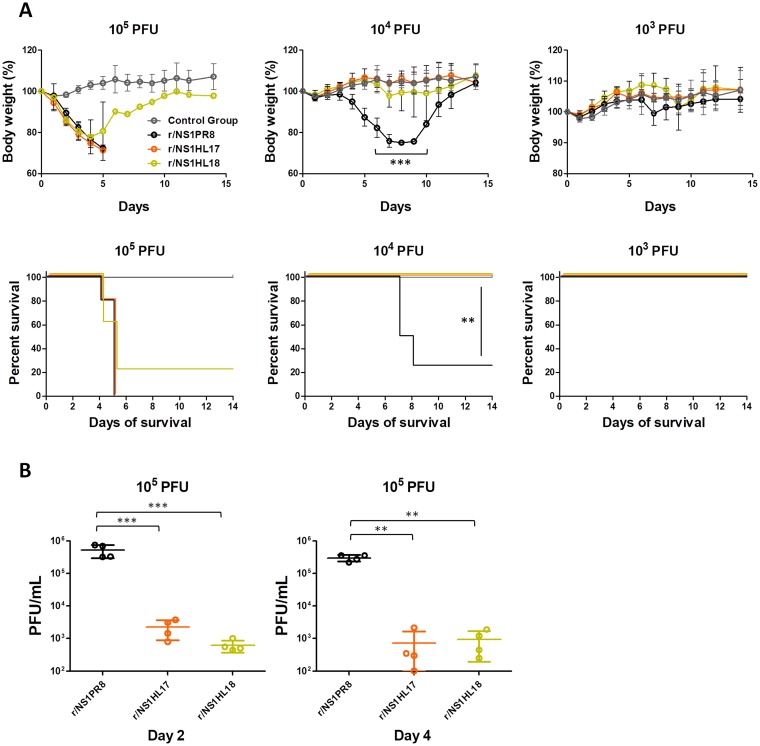
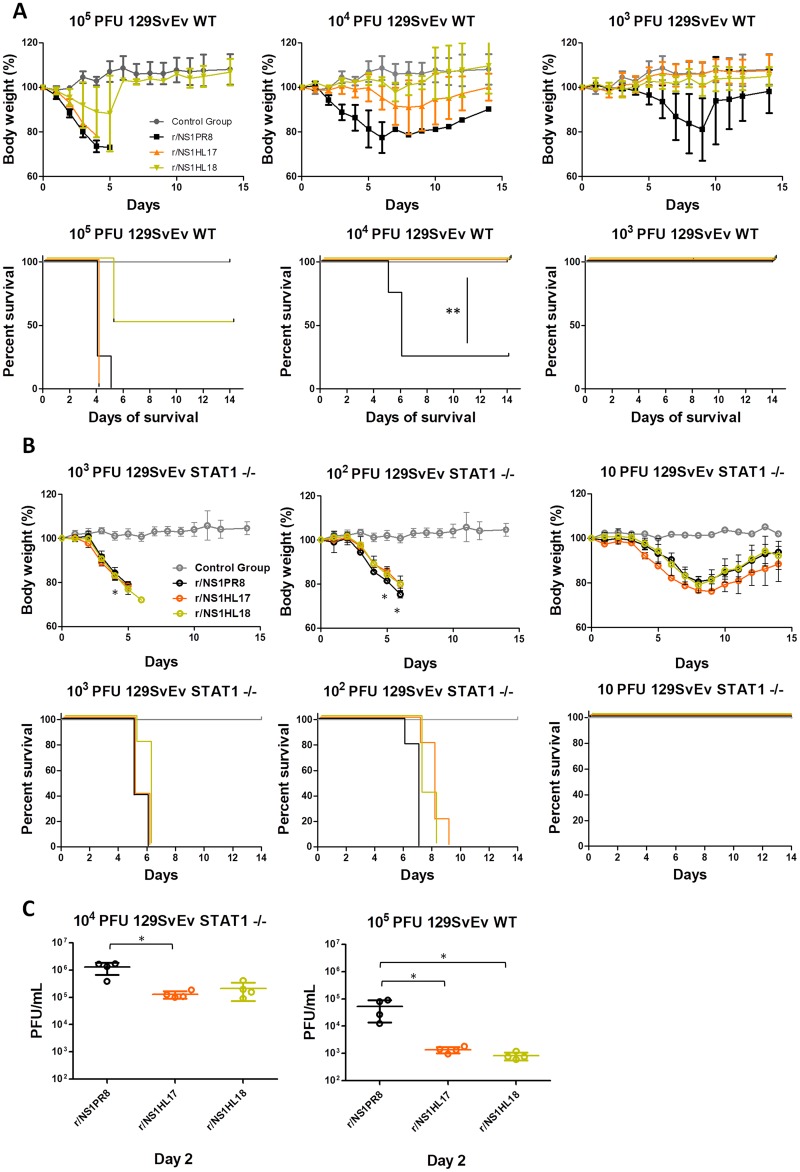
In order to assess the impact of the substitution of the NS1PR8 protein for the NS1HL17 and NS1HL18 proteins on virus replication and pathogenesis in vivo, we infected C57BL/6 mice intranasally with 105, 104, and 103 PFU of the recombinant viruses and determined their body weights postinfection as an indication of disease severity as well as mortality. In addition, lungs from mice infected with 105 PFU were used for the assessment of virus replication at days 2 and 4 postinfection. Similar to their behavior in tissue culture, the r/NS1HL17 and r/NS1HL18 viruses were attenuated in mice compared to the r/NS1PR8 control virus. While the r/NS1PR8 virus induced 75% mortality at an infectious dose of 104 PFU, none of the mice died or showed significant signs of morbidity after infection with 104 PFU of the r/NS1HL17 and r/NS1HL18 viruses (P < 0.01) (Fig. 3A). Although these viruses were lethal at 105 PFU, they demonstrated reduced viral replication (almost 3 logs) compared to that of the r/NS1PR8 virus in lung homogenates at days 2 (P < 0.001) and 4 (P < 0.01) postinfection (Fig. 3B).
FIG 3.
Characterization of bat NS1 recombinant viruses in vivo. (A) Six- to 8-week-old C57BL/6 mice were infected intranasally with the indicated amounts of viruses, and PBS was used for mock infection (gray). Mean body weights and Kaplan-Meier curves are shown (n = 5 mice/group). Error bars represent SDs. ANOVA for multiple comparisons was used to compare the body weights at each time point, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (***, P < 0.001 for r/NS1PR8 versus r/NS1HL17 and r/NS1HL18). The Mantel-Cox test was used to compare survival (**, P < 0.01). (B) Viral lung titers (in numbers of PFU per milliliter per lung; n = 4 mice/group) at a 105-PFU total infectious dose at days 2 and 4 postinfection. Horizontal lines, mean values and SDs. ANOVA for multiple comparisons was used to compare the viral lung titers at each time point, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (***, P < 0.001; **, P < 0.01).
Bat NS1 recombinant PR8 viruses regain their replication ability and virulence in IFN signaling-deficient systems.
It is well-known that the NS1 protein of influenza viruses has evolved mechanisms to evade the antiviral host innate immune responses (27). Thus, we investigated whether the attenuation of bat NS1 recombinant viruses was due to reduced IFN antagonism. For that, we determined viral replication in engineered clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 A549 cells unable to respond to type I IFN (A549 STAT1−/− cells). To generate the STAT1-deficient A549 cell line, we followed a previously described protocol (28) targeting exon 7 of the human STAT1 locus by guide RNA (gRNA). After transfection of the Cas9 expression plasmid and the gRNA, cloned cells were screened for the loss of expression of STAT1 by Western blotting and the loss of IFN signaling by quantitative reverse transcription-PCR (qRT-PCR) of mRNAs derived from IFN-stimulated genes (ISGs) after IFN stimulation (Fig. 2C and D). We performed multicycle growth curves of the NS1 recombinant viruses in STAT1-deficient A549 cells after infection at an MOI of 0.01. Although there was a slight decrease in virus titers for the bat NS1 recombinant viruses compared to those for r/NS1PR8 at late times postinfection, these differences were not statistically significant (Fig. 2A). This is in contrast to the results for wild-type A549 cells. These data suggest that NS1 from bat influenza A-like viruses antagonizes the type I IFN pathway to a lesser extent than the NS1 protein from the PR8 virus in the context of IAV infection in vitro.
We next compared the sensitivity of WT control (strain 129S6/SvEv) and STAT1−/− mice to infection by bat NS1 recombinant viruses (Fig. 4A and B). Mice of both genotypes were intranasally infected with 105, 104, and 103 PFU of the r/NS1HL17, r/NS1HL18, and r/NS1PR8 viruses, and morbidity and mortality were recorded. Interestingly, the r/NS1HL17 and r/NS1HL18 viruses induced similar morbidity and displayed 50% mouse lethal doses (MLD50) similar to the MLD50 of the r/NS1PR8 virus (Fig. 4B). Moreover, when we assessed viral replication in WT and STAT1−/− mouse lungs after 2 days postinfection with 105- and 104-PFU infectious doses, respectively, we found that the r/NS1HL17 and r/NS1HL18 virus titers were closer to the r/NS1PR8 virus titers in STAT1−/− mice than WT mice (Fig. 4C). The results indicate that most of the attenuation of the r/NS1HL17 and r/NS1HL18 viruses compared to the activity of the r/NS1PR8 virus was lost in IFN signaling-deficient (STAT1−/−) mice.
FIG 4.
Characterization of bat NS1 recombinant viruses in mice unable to respond to type I IFN because of disruption of the STAT1 gene. (A) Six- to 8-week-old 129S6/SvEv WT mice were infected intranasally with the indicated amount of viruses, and PBS was used for mock infection. Mean body weights and Kaplan-Meier curves are shown (n = 5 mice/group). Error bars represent SDs. The Mantel-Cox test was used to compare survival (**, P < 0.01). (B) Six- to 8-week-old 129S6/SvEv STAT1−/− mice were infected intranasally with the indicated amounts of viruses, and PBS was used for mock infection (gray). Mean body weights and Kaplan-Meier curves are shown (n = 5 mice/group). Error bars represent SDs. ANOVA for multiple comparisons was used to compare the body weights at each time point, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (*, P < 0.05 for r/NS1PR8 versus r/NS1HL17 and r/NS1HL18). (C) Viral lung titers (in numbers of PFU per milliliter per lung; n = 4 mice/group) at 104- and 105-PFU total infectious doses for 129S6/SvEv STAT1−/− and 129S6/SvEv WT mice, respectively. Horizontal lines, mean values and SDs at day 2 p.i. ANOVA for multiple comparisons was used to compare viral lung titers, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (*, P < 0.05).
Serial passage of bat NS1 recombinant viruses in mice resulted in phenotypic reversion due to mutations in the C-terminal domain of PB2.
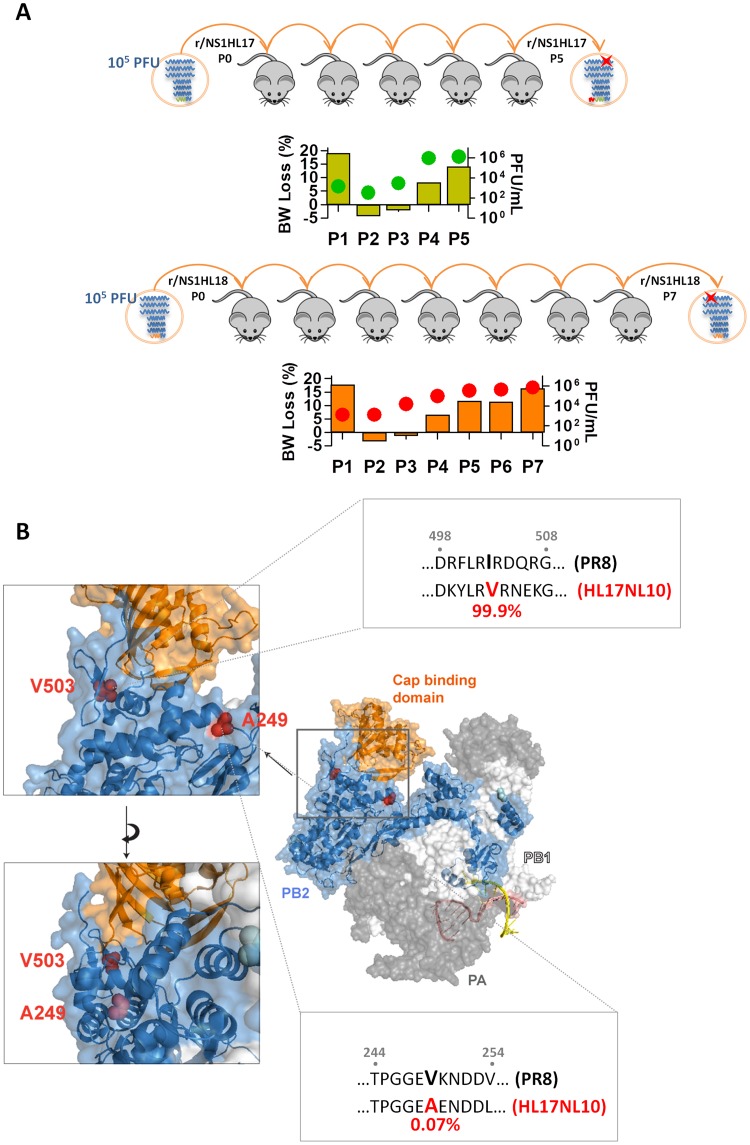
Next, we serially passaged the bat NS1 recombinant viruses in mice in order to investigate whether their attenuation could be overcome by the selection of compensatory mutations. Two mice per virus were infected with 105 PFU, and after 3 days the mice were euthanized and the lungs were excised. After lung homogenization, subsequent mice were infected with 50 μl of the resulting supernatant. Viral lung titers and body weight loss were monitored after every passage, with significant increases being detected at passages 5 and 7 for viruses expressing the NS1 proteins of HL17NL10 and HL18NL11 (r/NS1HL17 P5 and r/NS1HL18 P7, respectively) (Fig. 5A). RNA isolated from lung homogenate supernatants was subjected to Sanger sequencing. The resulting viral genome sequence analysis revealed two different amino acid substitutions in the PB2 gene of each virus provided by the PR8 IAV H1N1 backbone: I503V (A1510G; ATC → GTC) in r/NS1HL17 P5 and V249A (T749C; GTG → GCG) in r/NS1HL18 P7. Also, in the r/NS1HL17 P5 virus, a new start codon resulted in the insertion of 7 amino acids (aa) upstream of the V5 tag in the NS segment. One silent mutation was also identified in the PA gene of the r/NS1HL17 P5 virus, P653P (A1959G; CCA → CCG), and one was identified in the PB2 gene of the r/NS1HL18 P7 virus, G222G (T666A; GGT → GGA). Remarkably, no other nonsynonymous mutations were present in the mouse-passaged viruses at the level of resolution of Sanger sequencing.
FIG 5.
Multiple mouse passages of bat NS1 recombinant viruses resulted in compensatory mutations in PB2. (A) Six- to 8-week-old C57BL/6 mice were infected intranasally with the indicated amount of viruses. Lungs were excised on day 3 postinfection. The resulting supernatants were used to determine lung viral titers and for further inoculations in mice. Illustrations also show body weight (BW) loss (in percent; bars, left axis) and viral lung titers (in numbers of PFU per milliliter; dots, right axis) of mice in every passage for the r/NS1HL17 and r/NS1HL18 viruses. (B) Superimposition of the crystal structure of the HL17NL10 polymerase complex (29). The space-filling representation is color coded according to the different subunits (gray, PA; white, PB1; blue, PB2), with the cap-binding subdomain being highlighted in orange. The selected mutations after mouse passages are shown in red (V249A and I503V). Highlighted squares represent the region of PR8 and HL17NL10 from aa 244 to 254 and aa 498 to 508 after multiple-sequence alignment of viruses from multiple hosts and years and of different subtypes. PB2 sequences (n = 12,996 unique PB2 sequences) were obtained from the Influenza Research Database (https://www.fludb.org/brc/home.spg?decorator=influenza). Consensus sequences in the vicinity of the selected mutations and the percentages of amino acids of interest are shown for the representative viruses. Black, A/Puerto Rico/8/1934 (GenBank accession number AF389115.1); red, A/little yellow shouldered bat/Guatemala/153/2009 (GenBank accession number CY103873.1).
According to the published crystal structure of the HL17NL10 PB2 protein (29), both the I503V and V249A mutations are located relatively close together, in the linker regions of the protein without known functional properties and outside the well-characterized cap-binding domain (Fig. 5B). After alignment of 12,996 unique PB2 sequences obtained from the Influenza Research Database (http://www.fludb.org) representing IAV strains from different years and hosts, including strains of swine, avian, human, and bat origin (Fig. 5B), we noticed that the I503V substitution in r/NS1HL17 was a change to the most common amino acid residue at that position in the PB2 protein of influenza viruses (including also the HL17NL10 and HL18NL11 viruses). However, the original valine in the r/NS1HL18 parental virus at position 249 was conserved in the vast majority of strains aligned (99.9%), while its replacement by the alanine present in r/NS1HL18 P7 was found in only a very few strains, including in the PB2 of both bat influenza A-like viruses, HL17NL10 and HL18NL11.
Compensatory mutations found in mouse-passaged viruses restore virulence and IFN antagonism.
It has previously been described that IAV polymerase complex proteins contribute to counteracting antiviral host responses (30–37). Thus, we investigated whether the selected PB2 protein single mutations compensated for the loss of fitness of the original bat NS1 recombinant viruses through an enhancement of IFN antagonism. For that, we plaque purified the r/NS1HL17 P5 and r/NS1HL18 P7 viruses in MDCK cells and amplified them in eggs once in order to generate a working stock for later characterization. First, we determined the phenotype of the mouse-passaged viruses compared to that of the parental ones in vitro. For that, multicycle growth curves were performed at an MOI of 0.01 in MDCK and A549 cells. As shown in Fig. 6A, after acquiring the compensatory mutations, the recombinant viruses showed an advantage in replication compared to the parental viruses, which was particularly significant for the r/NS1HL17 P5 virus. Interestingly, the V249A and I503V PB2 mutations did not result in increased polymerase activity, as measured by a minigenome luciferase reporter assay, suggesting that the minigenome polymerase activity assay may not recapitulate completely the replication properties of the r/NS1HL17 P5 and r/NS1HL18 P7 viruses within the context of infection. Nonetheless, the attenuated replication of the r/NS1HL18 P7 virus in MDCK and A549 cells was consistent with a reduced polymerase activity associated with the V249A PB2 amino acid substitution, as measured by reconstitution of the RNA-dependent RNA polymerase together with the nucleoprotein (NP) in a minigenome luciferase reporter assay (Fig. 6B).
FIG 6.
Enhancement of replication properties but not polymerase activity of the recombinant viruses after selection of the compensatory mutations. (A) Viral growth properties of r/PR8, r/NS1PR8, r/NS1HL17, r/NS1HL17 P5, r/NS1HL18, and r/NS1HL18 P7 in MDCK (canine) and A549 (human) cells. Cells were infected at an MOI of 0.01, and the viral supernatant was titrated as the number of PFU per milliliter at the indicated time points postinfection (p.i.). Shown are the means and SDs from three biological replicates. ANOVA for multiple comparisons was used, and those that were statistically significant (P ≤ 0.05) were compared in groups two by two and the Bonferroni correction was applied (***, P < 0.001). (B) Minigenome polymerase activity assay of PB2 from the PR8 strain and mutated PB2 V249A and I503V in HEK293T cells (human). The firefly total luciferase level was measured at 24 h posttransfection. The values were averaged and normalized to the values for Renilla luciferase. Mean luminescence levels relative to those for mock-transfected cells are shown.
Next, we measured the expression of beta IFN (IFN-β) mRNA in human A549 cells by quantitative PCR (qPCR) after infection (MOI, 10). After 6 h p.i., total RNA was extracted and reverse transcription was performed using oligo(dT). Subsequently, quantification of mRNA was performed using specific primers against IFN-β. The values were normalized against those for mock-infected samples. As shown in Fig. 7A, the r/NS1HL17 and r/NS1HL18 viruses induced more IFN-β mRNA than the control recombinant PR8 (r/PR8) virus, demonstrating a defect of the NS1 from bat influenza A-like viruses in inhibiting IFN induction in the context of IAV infections. Interestingly, the V249A and I503V PB2 mutations did not result in increased inhibition of IFN induction when overexpressed (Fig. 7B). However, the viruses containing the PB2 compensatory mutations regained the IFN antagonism ability, as indicated by their lower levels of IFN-β mRNA induction in virus-infected human A549 cells. Remarkably, both viruses containing the compensatory mutations, r/NS1HL17 P5 and r/NS1HL18 P7, blocked the activation of IRF3 in human A549 cells (MOI, 5) after 6 h p.i., as determined by measurement of reduced levels of IRF3 phosphorylation and translocation into the nucleus in virus-infected cells (Fig. 7C and D) compared with the levels in cells infected with the parental r/NS1HL17 and r/NS1HL18 viruses. No differences in NS1 intracellular localization were found between the parental and the mouse-passaged viruses by immunofluorescence. Although higher levels of V5-NS1 were detected by Western blotting (Fig. 7D) for the mouse-adapted viruses than for the parental ones, no differences in the expression levels of NEP were found (data not shown).
FIG 7.
Compensatory mutations found in mouse-passaged viruses restore IFN antagonism. (A) qRT-PCR analyses of IFN-β induction in vitro. A549 cells were infected with the indicated amount of virus. Cells were harvested after 6 h postinfection (p.i.). Total RNA was extracted, and the levels of IFN-β mRNA were quantified in triplicate by qPCR. The values were averaged and normalized to those for 18S mRNA. Mean induction levels relative to the mRNA levels in mock-infected cells are shown. (B) IFN-β promoter assay in human HEK293T cells of PB2 from the PR8 strain and mutated PB2 V249A and I503V. Cells were cotransfected with expression plasmids expressing the indicated proteins for 24 h. NS1 from PR8 was used as a control. After infection with a DI-rich SeV preparation for a further 16 h, the amount of firefly total luciferase was determined. Values were normalized to the value for the empty vector plus SeV (for which the value was set to 100%). (C) Indirect immunofluorescence of IRF3 localization in A549 cells infected at an MOI of 5 at 6 h p.i. Costaining was performed using rabbit anti-IRF3 (green), goat anti-V5 tag (light blue), and mouse anti-NP (red) together with 4′,6′-diamidino-2-phenylindole (DAPI) to stain the nuclei (blue). (D) Determination by Western blotting of protein expression levels in A549 cells infected at an MOI of 5 (or mock infected [−]). Lysates were prepared for the indicated times p.i. Phospho-IRF3 (pIRF3), IRF3, the V5 tag, NP, and PB2 were detected using specific antibodies. β-Actin served as a loading control.
Next, we determined the phenotype of the r/NS1HL17 P5 and r/NS1HL18 P7 viruses compared to that of their parental ones in vivo. For that, C57BL/6 mice were subjected to infection with the r/NS1HL17 and r/NS1HL18 viruses and the viruses containing the compensatory mutations at 104, 103, and 10 total PFU, with the WT r/PR8 virus (and r/NS1PR8; data not shown) being used as a positive control. As shown in Fig. 8A, in the body weight loss and Kaplan-Meier curves, the adapted viruses showed an advantage over the parental ones, as the bat NS1-containing viruses regained virulence in mice to levels similar to but not higher than those in r/PR8. This phenotype was also confirmed according to the mouse viral lung titers, with higher levels of replication being found for the r/NS1HL17 P5 and r/NS1HL18 P7 viruses at day 2 p.i. (Fig. 8B). Moreover, when we measured the level of IFN-β mRNA in the mouse lungs after infection at the same infectious dose (Fig. 8C), both viruses with the selected compensatory mutations, r/NS1HL17 P5 and r/NS1HL18 P7, revealed a clear enhancement of IFN antagonism. To confirm that the phenotypic reversion was due to the acquired PB2 mutations, we also rescued recombinant viruses not containing mutations in any other genes but containing the PB2 single point mutations, r/NS1HL17 I503V and r/NS1HL18 V249A, by reverse genetics. Then we infected mice with these viruses at 105 PFU and excised the lungs at day 2 to assess viral replication. The experiments revealed that the recombinant viruses containing single PB2 mutations still showed enhanced replication properties compared to those of r/NS1HL17 and r/NS1HL18 (Fig. 8D), demonstrating that the acquisition of the compensatory mutations in the PB2 protein (I503V and V249A) is responsible for the regained phenotype observed in the previous experiments with the r/NS1HL17 P5 and r/NS1HL18 P7 viruses in vitro and in vivo.
FIG 8.
Antagonism of type I IFN confers virulence to bat influenza A-like viruses containing the compensatory mutations. (A) Six- to 8-week-old C57BL/6 mice were infected intranasally with the indicated amount of viruses; PBS was used for mock infection (gray), and r/PR8 was used as a control. Mean body weights and Kaplan-Meier curves are shown (n = 4 mice/group) for mice infected with the NS1 parental recombinant viruses compared to those for mice infected with the mouse-adapted viruses. Horizontal lines, mean values and SDs. The Mantel-Cox test was used to compare survival (***, P < 0.001). (B) Viral lung titers (in numbers of PFU per milliliter per lung; n = 4 mice/group) at a 105-PFU total infectious dose at days 2 and 4 postinfection. Horizontal lines, mean values and SDs. (C) qRT-PCR analyses of IFN-β induction in vivo. Six- to 8-week-old C57BL/6 mice (2 mice per group) were infected intranasally with a 105-PFU total infectious dose. Lungs were harvested after 6 h postinfection (p.i.). Total RNA was extracted, and the levels of IFN-β mRNA were quantified in triplicate by qPCR. Values were averaged and normalized to those for 18S mRNA. Mean induction levels relative to the mRNA levels in mock-infected mice are shown. (D) Six- to 8-week-old C57BL/6 mice were infected intranasally with the indicated amount of viruses using r/PR8 as a control. Viral lung titers (in numbers of PFU per milliliter per lung; n = 4 mice/group) at a 105-PFU total infectious dose at day 2 postinfection are shown. Horizontal lines, mean values and SDs.
DISCUSSION
In this study, we generated recombinant influenza viruses expressing the NS1 proteins of the bat influenza A-like HL17NL10 and HL18NL11 viruses in a PR8 backbone and tested the specific impact of these NS1 proteins in the context of an infection both in mammalian cells and in mice. Our results demonstrate that the bat r/NS1HL17 and r/NS1HL18 viruses have impaired IFN antagonism compared to the r/NS1PR8 control virus. Specifically, the r/NS1HL17 and r/NS1HL18 viruses were attenuated for replication in MDCK and A549 cells and mouse lungs and displayed less virulence in mice than the control r/NS1PR8 virus. However, their attenuation properties were significantly reverted in IFN signaling-deficient (STAT1−/−) cells and mice. Thus, this indicates that an impaired IFN antagonism conferred by the substitution of the WT IAV NS1 protein for the NS1 proteins of bat influenza A-like viruses is likely responsible for their attenuation. Indeed, infection with bat NS1-expressing IAVs resulted in higher levels of IRF3 activation and IFN induction in cells and mice, consistent with an impaired ability of the NS1 from bat influenza A-like viruses to block the RIG-I-sensing pathway and IFN induction in virus-infected cells. Thus, despite the observed inhibition of IFN induction in the context of overexpression (22, 23), the NS1 proteins of bat influenza A-like viruses are less capable of inhibiting IFN induction than the NS1 protein from a conventional IAV in the context of infection, illustrating that overexpression of a gene product may lead to biologically misleading conclusions compared to those drawn after productive viral replication in mammalian systems.
Bat NS1-mediated attenuation of viral replication was not restricted to nonbat cells. IAVs expressing the NS1 from bat influenza A-like viruses were also attenuated in cells derived from the bat species in which these viruses were discovered. Nonetheless, it is still possible that some specific features of the bat immune system are not totally present in the in vitro model studied here; thus, further studies in bats are necessary in order to dissect the specific virus-host interactions of these unconventional influenza viruses.
After serial passaging in mice, compensatory mutations in the viral polymerase subunit PB2 were acquired by both r/NS1HL17 and r/NS1HL18, demonstrating the versatility of influenza viruses in overcoming host restriction barriers. These mutations translated into a more effective inhibition of IFN-mediated antiviral responses and in the restoration of viral replication and lethality in mice. Although one of the passaged viruses also contained a small amino acid insertion in the NS1 gene (7 aa), this appeared to be an irrelevant passenger mutation. As shown in Fig. 8D, phenotypic reversion mapped to the PB2 mutation and not to the NS1 mutation, as determined by the rescue of recombinant viruses containing single mutations by reverse genetics. However, the lung viral titers of r/NS1HL17 I503V and r/NS1HL18 V249A showed a slight decrease compared to those of the mouse-passaged viruses. Thus, the possibility that the synonymous replacements also identified in the mouse-adapted bat NS1-containing viruses made a specific contribution cannot be ruled out.
Our results therefore indicate an interplay between the viral polymerase and the NS1 protein in viral evasion of the host innate immune system. It is plausible that in bat influenza A-like viruses the PB2 proteins also compensate for the weak IFN antagonism by their NS1 proteins. Consistent with this, the PB2 proteins of both the HL17 and HL18 viruses have a V249A substitution. Further experimentation is required to identify the mechanisms by which the PB2 mutations increase IFN antagonism of the bat NS1-containing viruses. Interestingly, the PB2 V249A and I503V mutations did not result in increased polymerase activity (Fig. 6B) or in increased inhibition of IFN induction when overexpressed (Fig. 7B). Further research will be needed to identify the mechanism responsible for the epistatic interactions between the NS1 and PB2 proteins in preventing the induction of IFN.
It is still intriguing how bats can harbor a large diversity of emerging viruses, such as rabies virus or other paramyxoviruses, often asymptomatically, since no major differences between the immunology of bats and the immunology of other mammals have been found. Remarkably, some of the main players of the innate immune response against viruses, such as pattern Toll-like receptors and RIG-I-like helicases, have previously been identified in some species of bats. Moreover, IFN and ISGs are upregulated upon cellular recognition of viral infection in bat cells (38–44). Although no human infections with the bat influenza A-like viruses have been reported so far, there is still the possibility that HL17NL10 and HL18NL11 may overcome host restriction barriers, as has happened with other influenza viruses in the past (e.g., the pandemic [H1N1] 2009 virus) (45). Unraveling the complex mechanisms of the virus-host interaction of HL17NL10 and HL18NL11 will help to clarify the zoonotic potential of these divergent influenza viruses.
MATERIALS AND METHODS
Plasmid constructions.
Modified NS segments containing the NS1-coding sequence from influenza A/Puerto Rico/8/1934 (PR8), A/little yellow-shouldered bat/Guatemala/060/2010 (HL17NL10), or A/flat-faced bat/Peru/033/2010 (HL18NL11) (2, 3) and the NEP-coding sequence from PR8 were cloned into the ambisense pDZ rescue vector (46). The NS segments were modified to prevent the splicing mechanism and translate the NS1 and NEP proteins independently, as previously described (26). In addition, the packaging signal, consisting of the first 109 nucleotides from the PR8 NS segment with a mutated start codon, was duplicated at the beginning of the segment, to ensure efficient packaging into the viral particle. The V5 tag sequence was fused to the N terminus of the NS1 ORFs to facilitate immune detection. The modified V5-NS1 ORFs from PR8, HL17NL10, and HL18NL11 were followed by the porcine teschovirus 1 (PTV-1) 2A autoproteolytic cleavage site (ATNFSLLKQAGDVEENPGP) and the entire sequence of the NEP ORF from PR8.
Virus rescue.
Virus rescues were performed as previously described by the eight-plasmid reverse genetics system in PR8 (5, 25). These viruses, r/NS1PR8 (for comparison purposes), r/NS1HL17, and r/NS1HL18 or r/NS1HL17 I503V and r/NS1HL18 V249A, were plaque purified in MDCK cells and passaged in eggs once in order to generate a working stock. Rescued recombinant NS1 viruses were confirmed by sequencing, and viral titers were determined by plaque assay in MDCK cells.
Cell lines.
Human embryonic kidney 293T (HEK293T) cells, Madin-Darby canine kidney (MDCK) cells, and human lung epithelial (A549) cells were originally purchased from the American Type Culture Collection (ATCC). A master cell bank was created for each cell line after purchase, and early-passage cells were thawed in every experimental step. Once in culture, cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U penicillin per ml, and 100 mg streptomycin per ml for not longer than 2 months to guarantee genotypic stability. The cells were also monitored by microscopy. For preparation of bat cell lines, bats were sampled between 2014 and 2015 in Costa Rica and captured using mist nets that were set at a height of 2 m in open areas near drinking or feeding sites to maximize capture success. Tissue samples from different organs were obtained to establish primary cell cultures. Briefly, tissue fragments were incubated for 30 to 60 min at 37°C in a solution of 0.2 Wūnsch units/ml of Liberase (Sigma-Aldrich) in DMEM–nutrient mixture F-12 (DMEM–F-12) (Thermo Fisher) and 1% penicillin-streptomycin (Thermo Fisher). Digestion was stopped by adding warm DMEM–F-12 completed with 15% FBS (HyClone) and 1% penicillin-streptomycin. Tissue was mechanically dislodged by pipetting, followed by several centrifugations and washing steps. The pellet was resuspended and plated in a cell culture-ready flask and incubated at 37°C in 5% CO2 for 5 to 7 days to allow cell migration from within the tissue fragments and attachment to the flask surface. Primary kidney cells from Sturnira lilium and Artibeus lituratus bats were immortalized by expressing human telomerase reverse transcriptase (hTERT) through lentiviral transduction and maintained in DMEM–F-12 supplemented with 20% bovine calf serum (BCS), 100 U penicillin per ml, and 100 mg streptomycin per ml. For generation of the STAT1−/− A549 cell line, we followed the CRISPR-Cas9 protocol described before (28). Specifically, exon 7 of the human STAT1 locus (sequence, CCTGTATTGGGGGGCCGCCCAAT) was targeted by designing gRNA against it and using the open-source CRISPR design tool (http://crispr.mit.edu/). The gRNA was cloned into the pSpCas9(BB)-2A-GFP (PX458) plasmid vector and transfected into A549 cells. At 48 h posttransfection, green fluorescent protein (GFP)-positive cells were sorted by fluorescence-activated cell sorting and plated for colony formation. Single-cell-derived colonies were picked and screened for STAT1 knockout (KO) by Western blotting. Cell clones negative for STAT1 protein expression were expanded and used for the experiments (Fig. 2C and D).
Infection of cell cultures.
MDCK, A549, STAT1−/− A549, and primary kidney bat cell monolayers in 6-well plates were infected with the virus suspension at an MOI of 0.01 in DMEM containing 0.2% bovine serum albumin (BSA). After 1 h, the infection medium was removed and the cells were incubated with 2 ml of DMEM–0.2% BSA and supplemented with 0.5 to 1 μg of TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin/ml, according to the specific requirement of each cell line, to allow the production of fusion-competent viruses. Supernatants were collected at 12, 24, 36, and 48 postinfection, and viral titers were determined by plaque assay in MDCK cells.
Infection of mice.
Mouse experiments were performed using 6- to 8-week-old female mice. Specific-pathogen-free C57BL/6 and 129S6/SvEv mice were purchased from The Jackson Laboratory and Taconic Biosciences, respectively, while homozygous STAT1 KO mice on a 129/Sv mouse background (129S6/SvEv-Stat1tm1Rds) were initially purchased from Taconic Biosciences (27). The latter STAT1−/− mice were then bred locally. Animals had free access to food and water and were kept on a 12-h light and 12-h dark cycle. All animal experiments were designed to address the principles of the 3 Rs (reduction [minimal number of mice required for assessment of statistically significant differences in pulmonary virus titers and body weights], refinement [all procedures performed were designed to cause minimal pain or distress, resulting in better animal well-being during the duration of the experiment], and replacement [the mouse is the smallest mammalian animal model that supports influenza virus replication]), and all efforts were made to minimize suffering. For virus inoculation, each mouse was anesthetized by inhalation of 4% isoflurane. The mice were monitored daily and euthanized if they lost more than 25% of their body weight after virus inoculations. Euthanasia of the mice was conducted by inhalation of 2% CO2 followed by cervical dislocation. The lungs were excised on day 2 or 4 postinfection. Following homogenization and centrifugation (10,000 × g, 20 min, 4°C), the resulting supernatants were used to determine the viral titer to assess replication. Bat recombinant NS1-containing viruses were serially passaged in mice to allow the selection of compensatory mutations. For this, mice were inoculated at 105 PFU (50 μl) at passage 0 (P0). The animals were euthanized at 3 days postinfection, and the lungs were excised. Lung supernatants were used for further inoculations in mice. The experimental endpoint was established as a significant increase in the viral lung titers and a body weight loss of greater than 10 to 15% compared to the titers and body weights of mice inoculated with the original NS1 bat recombinant viruses. These experiments were not expected to generate viruses more virulent than the virus with the parental WT PR8 backbone.
Immunofluorescence.
A549 cells were seeded in glass-bottom plates and infected following standard procedures. After incubation, the cells were fixed with 4% paraformaldehyde and permeabilized with a solution of phosphate-buffered saline (PBS) containing 0.2% Triton X-100 for 10 min (Sigma). Then, the cells were washed and stained for 1 h with primary antibodies, followed by staining with fluorescence secondary antibodies in a 5% bovine serum albumin (BSA; Thermo Scientific) solution in PBS. Primary antibodies against the nucleoprotein (NP; 71372; Santa Cruz), V5 tag (9137; Abcam), and interferon regulatory factor 3 (IRF3; 11904; Cell Signaling) were purchased from the indicated suppliers. The antibody against the NS1 protein (NS1-155) was kindly provided by Peter Palese (47).
Western blotting.
Lysates were prepared under the conditions indicated above in each experiment and subjected to SDS-PAGE, followed by transfer to polyvinylidene difluoride membranes. Next, the membranes were incubated overnight with primary antibodies followed by 1 h of incubation with secondary antibodies in a 5% bovine serum albumin (BSA; Thermo Scientific) solution in Tris-buffered saline and 0.1% Tween 20. Primary antibodies against the nucleoprotein (NP; A01506-40; GenScript), V5 tag (46-0805; Invitrogen), polymerase 2 (PB2; GTX125926; Genetex), NS1 protein (NS1-155) (47), nuclear export protein (NEP; A01499-100; GenScript), M1 protein (127356; Genetex), M2 protein (125951; Genetex), interferon regulatory factor 3 (IRF3; 11904; Cell Signaling), phospho-interferon regulatory factor 3 (pIRF3; 4947; Cell Signaling), and signal transducer and activator of transcription (STAT1; 3987; Abcam) were purchased from the indicated suppliers. β-Actin served as a loading control.
qRT-PCR.
Total RNA from the infection experiments or total RNA stimulated with the type I IFN protein (R&D Systems) was isolated using an EZNA HP total RNA isolation kit (Omega) or a Direct-zol RNA miniprep kit (Zymo Research) for in vitro and in vivo approaches, respectively. Reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) and specific primers, oligo(dT), or random primers in every case. Real-time qPCR was performed in 384-well plates for three individual biological samples, and the RNA in each of the samples was measured in triplicate using SYBR green I master mix (Roche) in a Roche LightCycler 480 system. Relative mRNA values were calculated using the ΔΔCT threshold cycle (CT) method (48) and 18S rRNA as an internal control for human cells and mouse tissues, and the results are plotted as relative values or fold change by normalization to the values for mock-infected control samples. The primer sequences were as follows: for influenza virus M1 protein, forward (Fw) primer 5′-AGATGAGTCTTCTAACCGAGTCG-3′ and reverse (Rv) primer 5′-TGCAAAAACATCTTCAAGTCTCT-3′; for 18S rRNA, Fw primer 5′-GTAACCCGTTGAACCCCATT-3′ and Rv primer 5′-CCATCCAATCGGTAGTAGCG-3′; for human IFN-β, Fw primer 5′-TCTGGCACAACAGGTAGTAGGC-3′ and Rv primer 5′-GAGAAGCACAACAGGAGAGCAA-3′; for mouse IFN-β, Fw primer 5′-CAGCTCCAAGAAAGGACGAAC-3′ and Rv primer 5′-GGCAGTGTAACTCTTCTGCAT-3′; for human IFIT1, Fw primer 5′-AGTGTGGGAATACACAACCTACT-3′ and Rv primer 5′-GGTCACCAGACTCCTCACATTT-3′; for human ISG15, Fw primer 5′-TCCTGGTGAGGAATAACAAGGG-3′ and Rv primer 5′-GTCAGCCAGAACAGGTCGTC-3′; and for human IFIT2, Fw primer 5′-GGAGGGAGAAAACTCCTTGGA-3′ and Rv primer 5′-GGCCAGTAGGTTGCACATTGT-3′.
Multiple-sequence alignment and protein model.
The protein sequences of the PB2 proteins of influenza viruses were obtained from the NIAID Influenza Research Database through the website at http://www.fludb.org. All sequences were grouped, resulting in a data set of 12,996 unique PB2 protein sequences from different hosts, including viruses of swine, avian, human, and bat origin. The sequences within the data sets were aligned using the MUSCLE (multiple-sequence comparison by log expectation) algorithm. The sequences obtained from the alignments were compared using the Geneious tool (49), calculating mean pairwise identities. Molecular graphics of the crystal structure of the HL17NL10 polymerase complex were prepared by superimposition (29) using the PyMOL molecular graphics system (version 1.8; Schrödinger, LLC).
Minigenome polymerase activity assay.
For reconstitution of the polymerase complex in HEK293T cells, plasmids (A/Puerto Rico/8/1934) expressing the PB2 WT, the PB2 I503V and V249A mutants, PB1, PA (each 50 ng), and NP (200 ng) were cotransfected with the firefly luciferase (FF-Luc)-expressing viral minigenome construct pPoI-FFLuc-RT (200 ng) and a plasmid (pRL-TK; 50 ng) expressing the Renilla luciferase. Expression of the minigenome was driven by the human polymerase promoter, as previously described (5). Firefly and Renilla luciferase activities were measured using a dual-luciferase reporter assay at 24 h posttransfection (1910; Promega).
IFN-β promoter assay.
HEK293T cells in 24-well plates were cotransfected with plasmids (25 ng) expressing PR8 NS1 as a control, the PB2 WT, and the PB2 V249A and I503V mutants, together with an IFN-β promoter-dependent firefly luciferase expression plasmid (50 ng) and a constitutively active Renilla luciferase expression plasmid (pRL-TK; 25 ng), which was used for normalization. The total amount of plasmid DNA was kept constant with an empty expression vector. At 24 h posttransfection, the cells were infected with a defective interfering (DI) particle-rich stock of Sendai virus (SeV) for 16 h to stimulate the IFN-β promoter. Firefly and Renilla luciferase activities were measured using a dual-luciferase reporter assay at 24 h posttransfection (1910; Promega).
Statistical analysis.
Variables were expressed as the mean ± standard deviation (SD) if the values were adjusted to a normal distribution, and the results were evaluated by the Shapiro-Wilk or Kolmogorov-Smirnov test when appropriate. For bivariate analysis of quantitative variables, the Mann-Whitney test or Student's t test was used on the basis of their distributions. If the variance was not homogeneous (Levene test), the Welch test (analysis of variance [ANOVA]) was applied. Two-way ANOVA with Bonferroni's posttest was used for multiple comparisons. Statistical significance was established at a P value of <0.05. All reported P values are based on two-tailed tests.
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (50). All mouse procedures were approved by Institutional Animal Care and Use Committee (IACUC) of the Icahn School of Medicine at Mount Sinai and performed in accordance with the IACUC guidelines (protocol number IACUC-2013-1408, Center for Research on Influenza Pathogenesis). All bat specimens were collected and processed after approval of the Institutional Committee of Care and Use of Animals (IACUC) of the University of Costa Rica (permit number CICUA-36-13) according to national guidelines for animal care described in the Costa Rica National Law for Animal Welfare 7451.
ACKNOWLEDGMENTS
We thank Megan Shaw for the firefly luciferase-expressing viral minigenome construct pPoI-FFLuc-RT and Peter Palese for the anti-NS1-155 antibody. We also thank Jaime Tome for technical assistance. Microscopy was performed at the Microscopy CoRE at the Icahn School of Medicine at Mount Sinai.
The study was supported by the Center for Research on Influenza Pathogenesis (CRIP), an NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS contract number HHSN272201400008C to A.G.-S), by the Deutsche Forschungsgemeinschaft (SCHW632/15-1 to M.S.), and by FEES-CONARE VI-803-B4-656 and the European Union's Horizon 2020 to E.C.-A. (ZIKAlliance grant agreement no. 734548).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freidl GS, Binger T, Muller MA, de Bruin E, van Beek J, Corman VM, Rasche A, Drexler JF, Sylverken A, Oppong SK, Adu-Sarkodie Y, Tschapka M, Cottontail VM, Drosten C, Koopmans M. 2015. Serological evidence of influenza A viruses in frugivorous bats from Africa. PLoS One 10:e0127035. doi: 10.1371/journal.pone.0127035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juozapaitis M, Aguiar Moreira E, Mena I, Giese S, Riegger D, Pohlmann A, Hoper D, Zimmer G, Beer M, Garcia-Sastre A, Schwemmle M. 2014. An infectious bat-derived chimeric influenza virus harbouring the entry machinery of an influenza A virus. Nat Commun 5:4448. doi: 10.1038/ncomms5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou B, Ma J, Liu Q, Bawa B, Wang W, Shabman RS, Duff M, Lee J, Lang Y, Cao N, Nagy A, Lin X, Stockwell TB, Richt JA, Wentworth DE, Ma W. 2014. Characterization of uncultivable bat influenza virus using a replicative synthetic virus. PLoS Pathog 10:e1004420. doi: 10.1371/journal.ppat.1004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Sastre A. 2012. The neuraminidase of bat influenza viruses is not a neuraminidase. Proc Natl Acad Sci U S A 109:18635–18636. doi: 10.1073/pnas.1215857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Sun X, Li Z, Liu Y, Vavricka CJ, Qi J, Gao GF. 2012. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc Natl Acad Sci U S A 109:18897–18902. doi: 10.1073/pnas.1211037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, Stevens J, Wilson IA. 2012. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci U S A 109:18903–18908. doi: 10.1073/pnas.1212579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Shi Y, Lu X, He J, Gao F, Yan J, Qi J, Gao GF. 2013. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep 3:769–778. doi: 10.1016/j.celrep.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, Garcia-Sastre A, Schwemmle M. 2015. Expected and unexpected features of the newly discovered bat influenza A-like viruses. PLoS Pathog 11:e1004819. doi: 10.1371/journal.ppat.1004819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreira EA, Locher S, Kolesnikova L, Bolte H, Aydillo T, Garcia-Sastre A, Schwemmle M, Zimmer G. 24 October 2016. Synthetically derived bat influenza A-like viruses reveal a cell type- but not species-specific tropism. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1608821113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayllon J, Garcia-Sastre A. 2015. The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- 14.Qian XY, Chien CY, Lu Y, Montelione GT, Krug RM. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA 1:948–956. [PMC free article] [PubMed] [Google Scholar]
- 15.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min JY, Li S, Sen GC, Krug RM. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 18.Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. 2014. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 88:12146–12151. doi: 10.1128/JVI.01567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayllon J, Hale BG, Garcia-Sastre A. 2012. Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence. J Virol 86:5366–5370. doi: 10.1128/JVI.06722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo RL, Krug RM. 2009. Influenza A virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J Virol 83:1611–1616. doi: 10.1128/JVI.01491-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turkington HL, Juozapaitis M, Kerry PS, Aydillo T, Ayllon J, Garcia-Sastre A, Schwemmle M, Hale BG. 2015. Novel bat influenza virus NS1 proteins bind double-stranded RNA and antagonize host innate immunity. J Virol 89:10696–10701. doi: 10.1128/JVI.01430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Tefsen B, Li Y, Qi J, Lu G, Shi Y, Yan J, Xiao H, Gao GF. 2016. The NS1 gene from bat-derived influenza-like virus H17N10 can be rescued in influenza A PR8 backbone. J Gen Virol 97:1797–1806. doi: 10.1099/jgv.0.000509. [DOI] [PubMed] [Google Scholar]
- 24.Turkington HL, Juozapaitis M, Tsolakos N, Corrales-Aguilar E, Schwemmle M, Hale BG. 13 December 2017. Unexpected functional divergence of bat influenza virus NS1 proteins. J Virol doi: 10.1128/JVI.02097-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. 1999. Rescue of influenza A virus from recombinant DNA. J Virol 73:9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 28.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, Liu CH, Zhou L, Krug RM. 2014. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe 15:484–493. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog 7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortigoza MB, Dibben O, Maamary J, Martinez-Gil L, Leyva-Grado VH, Abreu P Jr, Ayllon J, Palese P, Shaw ML. 2012. A novel small molecule inhibitor of influenza A viruses that targets polymerase function and indirectly induces interferon. PLoS Pathog 8:e1002668. doi: 10.1371/journal.ppat.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Chen H, Sutton T, Obadan A, Perez DR. 2014. Interactions between the influenza A virus RNA polymerase components and retinoic acid-inducible gene I. J Virol 88:10432–10447. doi: 10.1128/JVI.01383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graef KM, Vreede FT, Lau YF, McCall AW, Carr SM, Subbarao K, Fodor E. 2010. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J Virol 84:8433–8445. doi: 10.1128/JVI.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwai A, Shiozaki T, Kawai T, Akira S, Kawaoka Y, Takada A, Kida H, Miyazaki T. 2010. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon beta promoter stimulator 1. J Biol Chem 285:32064–32074. doi: 10.1074/jbc.M110.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamayoshi S, Watanabe M, Goto H, Kawaoka Y. 2015. Identification of a novel viral protein expressed from the PB2 segment of influenza A virus. J Virol 90:444–456. doi: 10.1128/JVI.02175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowled C, Baker M, Tachedjian M, Zhou P, Bulach D, Wang LF. 2011. Molecular characterisation of Toll-like receptors in the black flying fox Pteropus alecto. Dev Comp Immunol 35:7–18. doi: 10.1016/j.dci.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowled C, Baker ML, Zhou P, Tachedjian M, Wang LF. 2012. Molecular characterisation of RIG-I-like helicases in the black flying fox, Pteropus alecto. Dev Comp Immunol 36:657–664. doi: 10.1016/j.dci.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iha K, Omatsu T, Watanabe S, Ueda N, Taniguchi S, Fujii H, Ishii Y, Kyuwa S, Akashi H, Yoshikawa Y. 2010. Molecular cloning and expression analysis of bat Toll-like receptors 3, 7 and 9. J Vet Med Sci 72:217–220. doi: 10.1292/jvms.09-0050. [DOI] [PubMed] [Google Scholar]
- 41.Kepler TB, Sample C, Hudak K, Roach J, Haines A, Walsh A, Ramsburg EA. 2010. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics 11:444. doi: 10.1186/1471-2164-11-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virtue ER, Marsh GA, Baker ML, Wang LF. 2011. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS One 6:e22488. doi: 10.1371/journal.pone.0022488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou P, Cowled C, Todd S, Crameri G, Virtue ER, Marsh GA, Klein R, Shi Z, Wang LF, Baker ML. 2011. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J Immunol 186:3138–3147. doi: 10.4049/jimmunol.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Cowled C, Wang LF, Baker ML. 2013. Bat Mx1 and Oas1, but not Pkr are highly induced by bat interferon and viral infection. Dev Comp Immunol 40:240–247. doi: 10.1016/j.dci.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez-Gatell H, Alpuche-Aranda CM, Chapela IB, Zavala EP, Guevara DM, Checchi F, Garcia E, Hugonnet S, Roth C, WHO Rapid Pandemic Assessment Collaboration. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol 79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. 2011. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]