ABSTRACT
CCCTC-binding factor (CTCF) is a conserved, essential regulator of chromatin architecture containing a unique array of 11 zinc fingers (ZFs). Gene duplication and sequence divergence during early amniote evolution generated the CTCF paralog Brother Of the Regulator of Imprinted Sites (BORIS), which has a DNA binding specificity identical to that of CTCF but divergent N- and C-termini. While healthy somatic tissues express only CTCF, CTCF and BORIS are normally co-expressed in meiotic and post-meiotic germ cells, and aberrant activation of BORIS occurs in tumors and some cancer cell lines. This has led to a model in which CTCF and BORIS compete for binding to some but not all genomic target sites; however, regulation of CTCF and BORIS genomic co-occupancy is not well understood. We recently addressed this issue, finding evidence for two major classes of CTCF target sequences, some of which contain single CTCF target sites (1xCTSes) and others containing two adjacent CTCF motifs (2xCTSes). The functional and chromatin structural features of 2xCTSes are distinct from those of 1xCTS-containing regions bound by a CTCF monomer. We suggest that these previously overlooked classes of CTCF binding regions may have different roles in regulating diverse chromatin-based phenomena, and may impact our understanding of heritable epigenetic regulation in cancer cells and normal germ cells.
KEYWORDS: CTCF, CTCFL, BORIS, 1xCTS, 2xCTS, chromatin, ChIP-seq
CTCF and BORIS
CTCF is an essential, ubiquitously expressed DNA-binding factor conserved from Drosophila to human.1 CTCF is a master regulator of chromatin architecture and participates in transcriptional activation and repression,2–5 imprinting,6,7 chromatin insulation,8,9 formation of higher-order chromatin structures,10–12 and X-chromosome inactivation13 and inactivation escape,14 among many other chromatin-based phenomena. In addition to CTCF, vertebrates express a paralogous gene termed Brother Of the Regulator of Imprinted Sites (BORIS, also known as CTCFL).15 BORIS arose from a gene duplication early in the evolution of amniotes16 and subsequent divergence from the ancestral CTCF sequence during vertebrate evolution. The regulation of CTCF and BORIS in humans and in other placental mammals has diverged to such an extent that they are co-expressed only during gametogenesis, while all normal embryonic and adult somatic tissues are BORIS-negative due to methylation of the BORIS promoters17 and express only CTCF. The CTCF gene evolved in chromosomal contexts homologous to the human 16q22 region, recognized for recurrent LOH in tumors,18 while its paralog BORIS has co-evolved in different chromosomal bands homologous to 20q13, known for amplification during immortalization in culture and cancerous transformation in vivo.19 In line with its germline-restricted expression, BORIS-null male mice have notable infertility due to meiotic defects.20,21 BORIS is also activated in a variety of human cancers and so is classified as a cancer-testis antigen (CTA)22 shown to be suitable for immunotherapy in animal models.23
The central 11-ZF DNA binding domains of CTCF and BORIS are highly conserved in terms of genomic architecture and amino acid sequence15 (Fig. 1A-B) and have identical DNA binding specificities. The existence of at least two cellular settings (male germ cells and cancer cells) in which CTCF and BORIS are co-expressed raises the question of whether they associate cooperatively or competitively with a finite number of recognition sequences in the genome. A favored model is that of zero-sum competition, in which one protein completely replaces the other at a given target site. This model has been proposed to be supported by ChIP-seq analysis of CTCF and BORIS, showing single peaks of both paralogous 11-ZF proteins overlapping at thousands of genomic loci.21 However, this model does not account for a number of binding regions that contain two closely spaced CTCF motifs simultaneously bound in EMSA experiments by not one but two 11 ZF DBDs of both paralogs,24,25 which apparently cannot be resolved by ChIP-seq experiments with CTCF-specific antibodies alone.
Figure 1.
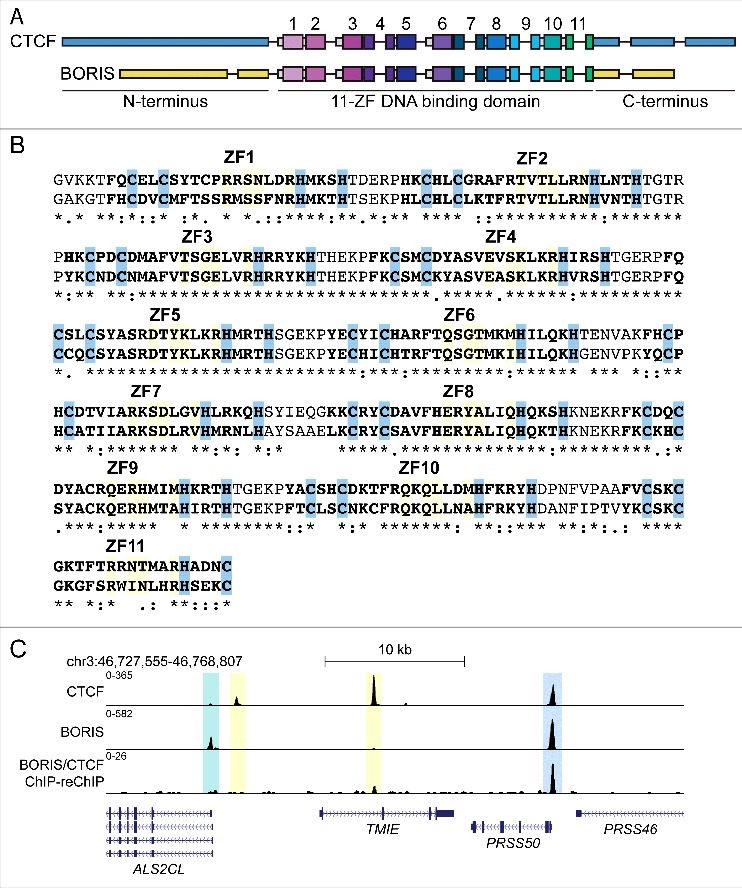
The DNA binding domains of CTCF and BORIS are highly conserved and bind to shared and distinct sites in the human genome. (A) Schematic representations of the genomic architecture of the CTCF and BORIS genes. C- and N-terminal exons are represented by thin blue (CTCF) or yellow (BORIS) lines. Conserved ZF-encoding exons are represented by thick colored boxes, with exons corresponding to each ZF colored differently and the exon(s) numbers according to which ZF they encode. ZFs 4, 7, 9, and 11 are split between two exons; in these cases, the ZF number is written above the intervening intron. Portions of exons corresponding to ZF linkers are represented by thin grey boxes. Introns are represented by black lines. (B) Alignment of the 11-ZF DNA binding domains of CTCF and BORIS. Zinc fingers are in bold and numbered. Zinc-coordinating residues are highlighted in blue and DNA-contacting residues are highlighted in gold. The asterisk (*) indicates amino acid conservation, the colon (:) represents a strongly conservative amino acid substitution, and the period (.) represents a weakly conservative amino acid substitution. The alignment was generated with Clustal Omega. (C) Human K562 cell ChIP-seq and ChIP-reChIP-seq data demonstrating the presence of two major classes of CTCF and BORIS binding regions. A robust CTCF&BORIS-bound 2xCTS element at the PRSS50 (TSP50) gene promoter has been characterized in detail in an earlier study24 and verified by analysis of DNase I footprinting data26 as well as ChIP-reChIP-seq (BORIS ChIP with antibodies verified for lack of CTCF crossreactivity followed by CTCF reChIP-seq) is highlighted in blue. Two CTCF-only peaks are highlighted in yellow, and a BORIS-only peak is highlighted in green.
Genome-wide characterization of CTCF and BORIS binding
To understand the global patterns of selective DNA occupancy by co-expressed CTCF and BORIS, we performed comprehensive ChIP-seq and ChIP-reChIP-seq analyses of both proteins in several human somatic cancer cell lines as well as mouse post-meiotic round spermatids.26 This revealed tens of thousands of specific peaks for both proteins in the BORIS-positive K562, OVCAR8, and Delta47 cancer cell lines. CTCF but not BORIS peaks were also observed in normal human dermal fibroblasts, used as a positive control for CTCF ChIP-seq and a negative control for BORIS ChIP-seq. Strikingly, 29–38% of all detected CTCF peaks overlapped with BORIS peaks. ChIP-reChIP-seq analysis in myeloid K562 and lymphoid Delta47 cells confirmed simultaneous occupancy of CTCF and BORIS at these overlapping binding regions. A similar result was observed in purified post-meiotic mouse round spermatids, where 25% of CTCF peak regions were also occupied by BORIS. Regions co-bound by CTCF and BORIS contained at least two closely-spaced, robust CTCF binding motifs, while CTCF-only peaks contained one or no match to an established CTCF motif.27 Due to their motif content, we termed CTCF-bound ChIP-seq peak regions also displaying BORIS recruitment as 2xCTSes and those bound by CTCF alone 1xCTSes. We also observed a number of 2xCTSes bound by homodimeric BORIS, though these were fewer in number than the 2xCTS elements co-bound by CTCF and BORIS together in BORIS-positive cells. A representative genome browser view of ChIP-seq and ChIP-reChIP-seq data from K562 cells showing distinct CTCF and BORIS binding features of 1xCTS and 2xCTS elements is displayed in Fig. 1C.
To directly elucidate the binding potential of 1x and 2xCTS elements, we performed EMSA analyses using the in vitro synthesized central 11 ZF of CTCF. A characteristic double shift was observed with DNA probes derived from 2xCTSes but not 1xCTSes, indicating that two binding events of similar affinity occur within all analyzed 2xCTS-containing DNA sequences. This result also suggests that 2xCTSes are occupied by CTCF homodimers in BORIS-negative cells. Consistent with this, analysis of DNase I footprinting data revealed two closely spaced footprints within 2xCTSes, regardless of whether the cell type analyzed did or did not express BORIS, and single footprints within 1xCTSes. DNase I footprints coincided with regions of conservation, likely representative of conserved consensus motifs. Examples of DNase I footprints and conservation at representative 1xCTS and 2xCTS regions are shown in Fig. 2A, and heatmaps showing aggregated DNase I footprinting data at 1xCTS and 2xCTS regions are shown in Fig. 2B. We conclude that 2xCTSes are indicative of cooperative protein binding events between one molecule of CTCF and one molecule of BORIS or two molecules of BORIS in BORIS-positive cells and two molecules of CTCF in BORIS-negative cells. Such cooperative interactions may be induced by DNA-dependent spatial constraints, perhaps due to molecular crowding-dependent phase separation.28
Figure 2.
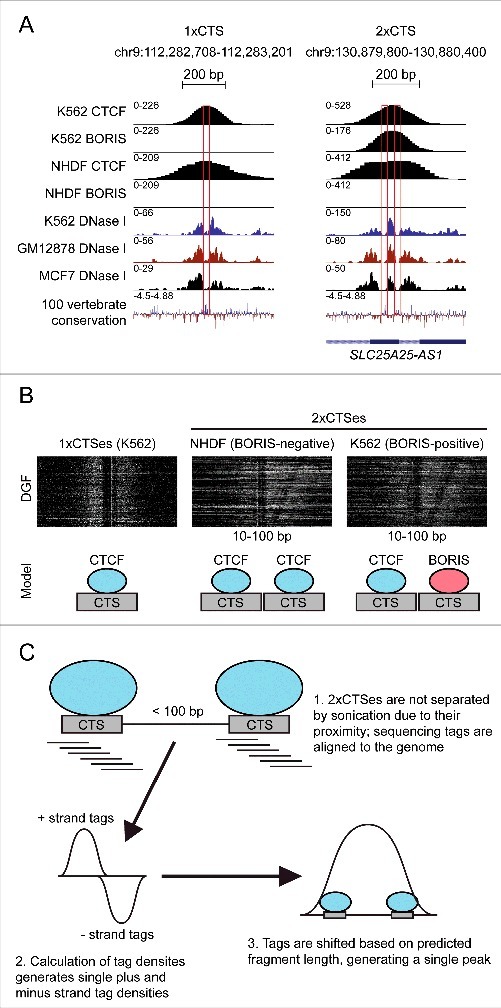
2xCTSes are resolved by nuclease footprinting. (A) UCSC Genome Browser comparison of ChIP-seq and DNase I footprinting data at a selected 1xCTS and 2xCTS regions. K562 CTCF and BORIS ChIP-seq and NHDF BORIS ChIP-seq data are from Pugacheva et al.26 NHDF CTCF ChIP-seq and DNase I overlap signal data are from the ENCODE consortium. DNase I footprints are boxed in red. (B) Heatmaps showing digital genomic footprinting (DGF) data at 1xCTSes in BORIS-positive K562 cells and 2xCTSes in BORIS-negative NHDF cells and K562 cells. A model of the organization of CTSes and bound molecules within each class of region is shown. (C) Schematic of a possible explanation for the failure of standard ChIP-seq to resolve individual binding sites within 2xCTSes.
Notably, ChIP-seq analysis of overexpressed BORIS in the BORIS-negative MCF7 cancer cell line recapitulated the specific profile of BORIS binding observed in K562 cells. This result suggests that the primary DNA sequence context within and in the vicinity of 2xCTS-bearing peaks serves as a major determinant of genomic DNA occupancy by CTCF homodimers in normal BORIS-negative somatic cells and also targets replacement of CTCF homodimers BORIS-positive germ or cancer cells with CTCF&BORIS heterodimers and, less often, with BORIS-only homodimers.
Analysis of CTCF and BORIS binding to repetitive elements also revealed striking distinctions between repeat-contained CTCF-only, CTCF&BORIS, and BORIS-only sites.29 CTCF-only sites were enriched in evolutionarily ancient and inactive types of repeats, while CTCF&BORIS sites were mainly located in uncharacterized tandem repeats. In contrast, BORIS-only sites were found primarily within the evolutionarily young SVA class of repeats. SVA elements are primate-specific, active retrotransposons30 and so their uncontrolled activity presents a threat to the stability of the germline.31 Analysis of repeat expression by RNA-seq following BORIS knockdown revealed a widespread upregulation of SVA expression, suggesting that BORIS acts as a repressor of SVA transcription.29 Given that SVA repeats are primate-specific, these observations suggest that germline-restricted BORIS continued to evolve after the divergence of the primate lineage, acquiring a specific function in germline defense.
The regulatory potential of 2xCTS-containing CTCF regions
Aside from their occupancy by BORIS in BORIS-positive cells and the presence of clustered CTCF motifs, 2xCTSes are distinguishable from 1xCTSes by chromatin features and binding of additional proteins. First, 2xCTSes display a histone modification profile consistent with active transcription: they are enriched for H3K4me3, associated with promoter activity, and H3K27ac, linked to active promoters and enhancers. Moreover, 2xCTSes display robust enrichment of the histone variant H2A.Z, which is associated with active transcription, as well as increased chromatin accessibility as measured by DNase I hypersensitivity. Regions containing 2xCTSes were also selectively enriched for transcription factors including ZNF143, C-MYC, and YY1 as well as histone modifying enzymes such as P300 and SET1B.
As in human cancer cells, 2xCTSes in mouse round spermatids are enriched for H3K4me3 and H3K27ac, as well as RNA polymerase II (RNAPII).26 Intriguingly, 2xCTSes in round spermatids are also associated with regions that retain histones in mature sperm. We recently extended these observations using ChIP-seq datasets for transcriptional regulators with known roles in male germ cell development and found that, like their counterparts in human cancer cells, mouse round spermatid 2xCTSes are often enriched for transcriptional regulators relative to 1xCTSes.32 Consistent with a function in positive transcriptional regulation, genes associated with 2xCTSes bound by transcriptional regulators were expressed more highly than those associated with non-transcriptional regulator-occupied 2xCTSes or 1xCTSes.
Previous studies identified what are now recognized as 2xCTSes in a number of important regulatory regions. For example, two closely-spaced CTCF/BORIS binding sites were found to be required for the activity of the promoter of the TSP50 gene, encoding a testis-specific protease aberrantly expressed in cancer.24 Other studies identified clustered CTCF motifs in alternative BORIS promoters,17 the promoter of the NY-ESO-1 gene,33 mouse Igh enhancers,34 the mouse KvDMR1 imprinting control region,7 and the BAX promoter.25
Genomic 2xCTS sequences and genome architecture
A notable exception to the higher enrichment of transcription factors and chromatin modifiers at 2xCTSes compared to 1xCTSes is the absence of the cohesins RAD21 and SMC3 from BORIS-only regions, suggesting that BORIS alone is insufficient to recruit the cohesin complex. CTCF is well known to cooperate with cohesin in regulating genome architecture,35 and increased expression of BORIS in previously BORIS-negative cancer cells could thus rewire genome architecture by replacing one or two CTCF molecules at some of its target regions. Another non-mutually exclusive possibility is that BORIS interacts with an alternative, cancer-testis specific set of architectural factors which are normally co-expressed together with CTCF and BORIS only during gametogenesis. Since BORIS is present in male germ cells during and after meiosis, and so an attractive hypothesis is that BORIS at 2xCTS elements may interact with at least one of three meiosis-specific subunits of cohesin complexes to contribute to a stage-by-stage re-establishment of genome architecture in haploid post-meiotic round spermatids.
Previous work has shown that N-terminal fusion of EGFP to the 11-ZF DNA binding domain of CTCF is sufficient to disrupt the intra-chromosomal loop between the maternal imprinting control region (ICR) and the maternally imprinted IGF2 gene,36 suggesting that the EGFP/11-ZF chimera functions as a dominant-negative decoy that associates with DNA but cannot interact with the appropriate factors to form this loop. As the N- and C- termini of BORIS are highly diverged from those of CTCF, it stands to reason that BORIS might similarly serve as a decoy by both disrupting CTCF-mediated loops and establishing new BORIS-dependent loops.
A specific way in which BORIS overexpression could alter genome conformation is through the disruption of topologically associating domains (TADs). TADs are megabase-scale segments of the genome displaying high levels of self-interaction that are dependent on CTCF for proper folding.37 Disruption of TADs is associated with activation of proto-oncogenes in cancers including T-cell acute lymphoblastic leukemia,38 glioma,39 and colorectal carcinoma.40 Thus, even transient overexpression of BORIS, potentially in combination with mutation or loss of genomic CTSes,41 dysfunction of CTCF, and/or malfunction of BORIS and/or CTCF-interacting proteins, could be a powerful driving force in carcinogenesis via reorganization of chromatin architecture and concomitant activation of proto-oncogenes.
CTCF is also proposed to mediate the formation of chromatin loops much smaller than TADs.42 Such loops are thought to result from pairs of distal CTCF motifs (‘loop anchors') in a convergent orientation. Loss of a CTCF molecule from one loop anchor could thus impair formation of the loop, leading to inappropriate transcriptional consequences.43 Notably, our previous work has shown that dual CTCF&BORIS-bound regions are enriched at the RNAPII-bound anchors of chromatin loops specific to K562 cells.26 Furthermore, the same regions were occupied by CTCF and RNAPII in BORIS-negative MCF7 cells, but the associated chromatin loops were different.
Conclusions and future directions
Despite having potentially fundamental implications for understanding the regulatory DNA lexicon responsible for transmission of epigenetic memories by mitotic cancer cells through an aberrantly immortal growth in tumors in vivo and in tissue culture in vitro and by post-meiotic germ cells in fertile males throughout continuous rounds of normal germ cell development, the majority of distinct single and bipartite 11 ZF binding regions have been overlooked in chromatin studies based on mapping mouse and human CTCF binding sites by ChIP-seq, even in the K562 cell line, which expresses high levels of BORIS and has been a robust model for functional analysis of BORIS.26,29,44
Moving forward, it will be of interest to understand the architecture of 2xCTSes and how they might induce protein-protein interactions (PPIs) between molecules of CTCF and/or BORIS. A rather strict limitation on the total length of 2xCTSes (<100 bp between adjacent motifs) suggests that this relatively short spacing may be required for the proximity-induced dimerization shown to occur in vitro.26 Additionally, a restricted spacer length might be important for exclusion of a single nucleosome between CTCF motifs, which could interfere with DNA-dependent proximity-induced CTCF/BORIS PPIs. To test this, alteration of the spacing between adjacent CTCF motifs in known 2xCTSes followed by EMSA analysis could be employed, followed by in vivo editing of 2xCTSes using CRISPR. CRISPR editing of 2xCTSes in cell lines would also be a useful way in which to understand the potential roles of 2xCTSes in the regulation of transcription and chromatin architecture. We also note that CTCF multimerization could be mediated by interactions with RNA, as has been shown for the Wrap53 antisense transcript of p53.45 Further study of the structure of 2xCTSes in vivo will also require a way to deconvolve closely-spaced binding sites not resolved by standard ChIP-seq. We presume that 2xCTSes are convolved into single peaks because the spacer DNA between CTSes is not sheared during sonication, leading to the accumulation of sequencing tags from two closely-spaced but distinct binding sites into single pileups, leading to assignment of those fragments into single peaks (Fig. 2C). A corollary of this idea is that 2xCTSes would not be resolved by ChIP-exo46 or ChIP-nexus,47 as CTCF and/or BORIS crosslinked to DNA would act again as a chemically stable exonuclease barrier, preventing digestion of the short intervening stretch of DNA that provides spatial proximity to adjacent CTCF motifs and may contribute to obligatory hetero-dimerization of CTCF and/or BORIS at these regions. However, it is possible that micrococcal nuclease may be able to resolve 2xCTSes on native chromatin, given that it has endo- and exonuclease activities. Resolution of 2xCTSes is unlikely to be achieved by computational methods such as BRACIL48 designed for deconvolution of two spatially constrained CTCF binding events, because this and similar mathematical attempts to enhance bipartite binding site resolution would still require an input of short reads that could not be generated by DNA ends absent inside ultra-sonicated but chemically crosslinked 2xCTS-containing chromatin fragments. In contrast, Fig. 2A-B illustrates that DNase I footprinting provides a promising alternative for resolving dual CTCF footprints without fixation of 2xCTS-based chromatin complexes.
It will also be of interest to understand why natural duplication of the highly conserved 11-ZF coding exons of ancestral CTCF gene occurred in spite of potential competition between two 11-ZF DBDs for the same target sequences, as well as to determine how divergence of BORIS, the “second CTCF gene” in mammals, would result in strictly testis-specific expression of the human BORIS gene (on chr20q13) so that it could allow ubiquitously-expressed human CTCF (on chr16q22) to avoid functional interference during continuous rounds of human development and reproduction.
Finally, it remains to be determined whether BORIS associates with additional factors involved in the regulation of genome architecture, particularly with meiotic cohesins in normal meiotic and post-meiotic germ cells, and whether these BORIS partners are aberrantly co-activated with BORIS in immortalized cancer stem cells responsible for tumor initiation in BORIS-negative somatic tissues. Co-upregulation of meiotic cohesins with BORIS in cancer could influence genome-wide organization of chromatin and orchestration of transcriptional regulation through the two markedly distinct types of regulatory regions described here. Cataloging of the tens of thousands of CTCF-associated DNA sequences in dozens of human cell lines detected by the ENCODE Consortium (http://www.factorbook.org/human/chipseq/tf/CTCF) will be a valuable step toward understanding “binary CTCF code” emerging in the regulatory DNA language of all heritable epigenomes from combinatorial diversity of two or more individual DNA motifs within CTCF target regions bound equally well by the 11-ZF regions of CTCF and BORIS.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Elena Pugacheva for assistance with manuscript preparation.
Funding
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health [to VVL] and Indiana University startup funds [to GEZ].
References
- 1.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, et al.. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab‐8 insulator. EMBO Rep. 2005;6:165–70. https://doi.org/ 10.1038/sj.embor.7400334. PMID:15678159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, et al.. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–24. https://doi.org/ 10.1128/MCB.13.12.7612. PMID:8246978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, et al.. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–13. https://doi.org/ 10.1093/nar/28.8.1707. PMID:10734189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850–60. https://doi.org/ 10.1093/nar/gki989. PMID:16326864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernukhin I, Shamsuddin S, Kang SY, Bergström R, Kwon Y-W, Yu W, et al.. CTCF Interacts with and Recruits the Largest Subunit of RNA Polymerase II to CTCF Target Sites Genome-Wide. Mol Cell Biol. 2007;27:1631–48. https://doi.org/ 10.1128/MCB.01993-06. PMID:17210645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, et al.. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. https://doi.org/ 10.1073/pnas.0600326103. PMID:16815976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzpatrick GV, Pugacheva EM, Shin J-Y, Abdullaev Z, Yang Y, Khatod K, et al.. Allele-Specific Binding of CTCF to the Multipartite Imprinting Control Region KvDMR1. Mol Cell Biol. 2007;27:2636–47. https://doi.org/ 10.1128/MCB.02036-06. PMID:17242189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell AC, West AG, Felsenfeld G. The Protein CTCF Is Required for the Enhancer Blocking Activity of Vertebrate Insulators. Cell. 1999;98:387–96. https://doi.org/ 10.1016/S0092-8674(00)81967-4. PMID:10458613. [DOI] [PubMed] [Google Scholar]
- 9.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, et al.. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–43. https://doi.org/ 10.1038/ng570. PMID:11479593. [DOI] [PubMed] [Google Scholar]
- 10.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al.. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–8. https://doi.org/ 10.1038/ng.857. PMID:21685913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin David U, et al.. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–10. https://doi.org/ 10.1016/j.cell.2015.07.038. PMID:26276636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, et al.. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E65. https://doi.org/ 10.1073/pnas.1518552112. PMID:26499245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a Candidate Trans-Acting Factor for X-Inactivation Choice. Science. 2002;295:345–7. https://doi.org/ 10.1126/science.1065982. PMID:11743158. [DOI] [PubMed] [Google Scholar]
- 14.Filippova GN, Cheng MK, Moore JM, Truong J-P, Hu YJ, Di Kim N, et al.. Boundaries between Chromosomal Domains of X Inactivation and Escape Bind CTCF and Lack CpG Methylation during Early Development. Dev Cell. 2005;8:31–42. https://doi.org/ 10.1016/j.devcel.2004.10.018. PMID:15669143. [DOI] [PubMed] [Google Scholar]
- 15.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, et al.. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci USA. 2002;99:6806–11. https://doi.org/ 10.1073/pnas.092123699. PMID:12011441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hore TA, Deakin JE, Marshall Graves JA. The Evolution of Epigenetic Regulators CTCF and BORIS/CTCFL in Amniotes. PLoS Genet. 2008;4:e1000169. https://doi.org/ 10.1371/journal.pgen.1000169. PMID:18769711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renaud S, Pugacheva EM, Delgado MD, Braunschweig R, Abdullaev Z, Loukinov D, et al.. Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Res. 2007;35:7372–88. https://doi.org/ 10.1093/nar/gkm896. PMID:17962299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, et al.. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. https://doi.org/ 10.1002/(SICI)1098-2264(199805)22:1%3c26::AID-GCC4%3e3.0.CO;2-9. PMID:9591631. [DOI] [PubMed] [Google Scholar]
- 19.Klenova EM, Morse Iii HC, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. https://doi.org/ 10.1016/S1044-579X(02)00060-3. PMID:12191639. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Kosaka-Suzuki N, Pack S, Shin DM, Yoon J, Abdullaev Z, et al.. Expression of a testis-specific form of Gal3st1 (CST), a gene essential for spermatogenesis, is regulated by the CTCF paralogous gene BORIS. Mol Cell Biol. 2010;30:2473–84. https://doi.org/ 10.1128/MCB.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sleutels F, Soochit W, Bartkuhn M, Heath H, Dienstbach S, Bergmaier P, et al.. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics & Chromatin. 2012;5:1–21. https://doi.org/ 10.1186/1756-8935-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall AD, Bailey CG, Rasko JEJ. CTCF and BORIS in genome regulation and cancer. Curr Opin Genet Dev. 2014;24:8–15. https://doi.org/ 10.1016/j.gde.2013.10.011. PMID:24657531. [DOI] [PubMed] [Google Scholar]
- 23.Mkrtichyan M, Ghochikyan A, Davtyan H, Movsesyan N, Loukinov D, Lobanenkov V, et al.. Cancer-testis antigen, BORIS based vaccine delivered by dendritic cells is extremely effective against a very aggressive and highly metastatic mouse mammary carcinoma. Cell Immunol. 2011;270:188–97. https://doi.org/ 10.1016/j.cellimm.2011.05.007. PMID:21641588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosaka-Suzuki N, Suzuki T, Pugacheva EM, Vostrov AA, Morse HC, Loukinov D, et al.. Transcription factor BORIS (Brother of the Regulator of Imprinted Sites) directly induces expression of a cancer-testis antigen, TSP50, through regulated binding of BORIS to the promoter. J Biol Chem. 2011;286:27378–88. https://doi.org/ 10.1074/jbc.M111.243576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez-Catalá CF, Gretton S, Vostrov A, Pugacheva E, Farrar D, Ito Y, et al.. A Novel Mechanism for CTCF in the Epigenetic Regulation of Bax in Breast Cancer Cells. Neoplasia (New York, NY). 2013;15:898–912. https://doi.org/ 10.1593/neo.121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Méndez-Catalá CF, Kang S, Suzuki T, et al.. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015;16:1–24. https://doi.org/ 10.1186/s13059-015-0736-8. PMID:25583448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al.. Analysis of the vertebrate insulator protein CTCF- binding sites in the human genome. Cell. 2007;128:1231–45. https://doi.org/ 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. https://doi.org/ 10.1016/j.cell.2017.02.007. PMID:28340338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugacheva EM, Teplyakov E, Wu Q, Li J, Chen C, Meng C, et al.. The cancer-associated CTCFL/BORIS protein targets multiple classes of genomic repeats, with a distinct binding and functional preference for humanoid-specific SVA transposable elements. Epigenetics & Chromatin. 2016;9:35. https://doi.org/ 10.1186/s13072-016-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. https://doi.org/ 10.1038/nrg2640. PMID:19763152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodier JL. Restricting retrotransposons: a review. Mobile DNA. 2016;7:16. https://doi.org/ 10.1186/s13100-016-0070-z. PMID:27525044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivero-Hinojosa S, Kang S, Lobanenkov VV, Zentner GE. Testis-specific transcriptional regulators selectively occupy BORIS-bound CTCF target regions in mouse male germ cells. Scientific Reports. 2017;7:41279. https://doi.org/ 10.1038/srep41279. PMID:28145452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, et al.. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–74. https://doi.org/ 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 34.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, et al.. Chromatin Architecture near a Potential 3′ End of the Igh Locus Involves Modular Regulation of Histone Modifications during B-Cell Development and In Vivo Occupancy at CTCF Sites. Mol Cell Biol. 2005;25:1511–25. https://doi.org/ 10.1128/MCB.25.4.1511-1525.2005. PMID:15684400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annual Review of Genomics and Human Genetics. 2016;17:17–43. https://doi.org/ 10.1146/annurev-genom-083115-022339. PMID:27089971. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Niu B, Hu J-F, Ge S, Wang H, Li T, et al.. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. The Journal of Cell Biology. 2011;193:475–87. https://doi.org/ 10.1083/jcb.201101021. PMID:21536749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, et al.. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–44. https://doi.org/ 10.1016/j.cell.2017.05.004. PMID:28525758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, et al.. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016:1454–1458. https://doi.org/ 10.1126/science.aad9024. PMID:26940867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al.. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–4. https://doi.org/ 10.1038/nature16490. PMID:26700815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stutz AM, et al.. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2017;49:65–74. https://doi.org/ 10.1038/ng.3722. PMID:27869826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, et al.. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–21. https://doi.org/ 10.1038/ng.3335. PMID:26053496. [DOI] [PubMed] [Google Scholar]
- 42.Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. https://doi.org/ 10.1038/nrg3663. PMID:24614316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norton HK, Phillips-Cremins JE. Crossed wires: 3D genome misfolding in human disease. The Journal of Cell Biology. 2017:3441–3452. https://doi.org/ 10.1083/jcb.201611001. PMID:28855250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teplyakov E, Wu Q, Liu J, Pugacheva EM, Loukinov D, Boukaba A, et al.. The downregulation of putative anticancer target BORIS/CTCFL in an addicted myeloid cancer cell line modulates the expression of multiple protein coding and ncRNA genes. Oncotarget. 2017;8:73448–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saldaña-Meyer R, González-Buendía E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, et al.. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–34. https://doi.org/ 10.1101/gad.236869.113. PMID:24696455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee HS, Pugh BF. Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell. 2011;147:1408–19. https://doi.org/ 10.1016/j.cell.2011.11.013. PMID:22153082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotech. 2015;33:395–401. https://doi.org/ 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes ALC, Abeel T, Peterson M, Azizi E, Lyubetskaya A, Carvalho L, et al.. Decoding ChIP-seq with a double-binding signal refines binding peaks to single-nucleotides and predicts cooperative interaction. Genome Res. 2014;24:1686–97. https://doi.org/ 10.1101/gr.161711.113. PMID:25024162. [DOI] [PMC free article] [PubMed] [Google Scholar]


