Abstract
The Y-chromosome is frequently lost in hematopoietic cells, representing the most common somatic mutation in men. However, the mechanisms regulating mosaic loss of chromosome-Y (mLOY), and its clinical relevance, are unknown. Using genotype array intensity data and sequence reads in 85,542 men, we identify 19 genomic regions (P<5x10-8) associated with mLOY. Cumulatively, these loci also predicted X-chromosome loss in women (N=96,123, P=4x10-6). Additional epigenome-wide methylation analyses in whole blood highlighted 36 differentially methylated sites associated with mLOY. Identified genes converge on aspects of cell proliferation and cell-cycle regulation, including DNA synthesis (NPAT), DNA damage response (ATM), mitosis (PMF1-CENPN-MAD1L1) and apoptosis (TP53). We highlight shared genetic architecture between mLOY and cancer susceptibility, in addition to inferring a causal effect of smoking on mLOY. Collectively, our results demonstrate that genotype array intensity data enable a measure of cell-cycle efficiency at population scale, identifying genes implicated in aneuploidy, genome instability and cancer susceptibility.
Introduction
For over a century, errors in cell division have been described which result in too few or too many chromosomes in daughter cells, a cytogenetic feature termed aneuploidy. Although a well-established feature of human cancer cells, it remains unclear whether acquired aneuploidy is a cause or consequence of tumorigenesis. Research into the molecular mechanisms of aneuploidy has focussed largely on the role of mitosis and mitotic checkpoint signalling, primarily in cellular and animal models1,2. Recent human genomic studies have shown that aneuploidy can be estimated using intensity data from standard genotyping arrays; an approach validated by DNA sequencing3–5. These population-based studies demonstrate that mLOY is more frequent than other mosaic chromosomal and structural mutations: indeed, around 1 in 5 men over 80 years of age has detectable Y mosaicism in whole blood-derived DNA4, reflecting the capacity of some cells to survive without this chromosome.
Although a common feature in the general population, it remains unclear whether mLOY is relevant to disease susceptibility, or whether cells in tissues other than peripheral blood undergo similar rates of chromosomal loss. Population studies have identified correlations between mLOY and smoking status, an association which appears transient and reversible after smoking cessation6. Such epidemiological studies have also identified associations with non-hematological cancers4,5 and Alzheimer’s disease7; however, these observations are inconsistent3 and possibly subject to confounding or reverse-causality.
The ability to assay a common measure of aneuploidy in large array-genotyped populations could enable systematic identification of variants/genes involved in cell division errors. This would in turn enable a better understanding of the mechanisms involved, and the potential causal consequences of aneuploidy on cancer risk, inferred using Mendelian randomisation approaches. To date, a single genomic association with mLOY near TCL1A has been reported (N=12,369), suggesting that germline variation influencing mosaic chromosome loss can be detected3. Here, we use data in up to 85,542 men, highlighting widespread genomic, transcriptomic and epigenetic signatures of mosaic Y chromosome loss. We also demonstrate that this approach can successfully identify genes implicated in cell cycle regulation, genome instability and cancer susceptibility.
Results
As a proxy for mLOY, we estimated mean intensity log-R ratio of all array-genotyped Y-chromosome SNPs (mLRR-Y) in a sample of 67,034 male participants from the UK Biobank cohort (UKBB)8. A normal distribution centred around zero was observed (standard deviation = 0.067), with negative values indicating reduced Y chromosome abundance in the clonal blood cell population (Supplementary Figure 1).
Consistent with previous reports3,6, we observed a strong negative correlation between mLRR-Y and age (r=-0.21). A strong association with ‘ever smoking’ status was also observed (P=3.05×10-82), which in combination with age explained 4.74% of the trait variance (age alone = 4.45%). We sought to demonstrate the causal relationship between smoking and mLOY through the principle of Mendelian randomization, using a reported and widely used genetic instrument for smoking frequency9. By modelling genetic variants robustly associated with cigarettes smoked per day at the CHRNA5-CHRNA3-CHRNB4 nicotinic receptor locus, we inferred a causal effect of smoking on decreased mLRR-Y (increased Y loss) (rs1051730 P=0.03 [Pnever-smokers=0.41, Pever-smokers=0.04]). This genetic association was confirmed in independent replication samples (EPIC Norfolk and deCODE combined N=18,508, P=0.009, overall combined P=0.004).
Many autosomal genetic variants are associated with mLOY
To identify novel genetic variants associated with mLOY, we performed a genome-wide association study of mLRR-Y as a quantitative trait in UKB. After stringent quality control (see Methods), the most significantly-associated SNPs were located at the previously reported3 mLOY locus, TCL1A (P=3.6x10-23). In addition, we identified a further 18 novel signals at genome-wide significance (P<5x10-8), with no evidence for significant inflation of test statistics genome-wide (lambda=1.05) (Supplementary Figures 2 and 3). Replication was subsequently performed in an independent set of 9,793 men with array intensity data, in addition to 8,715 men from deCODE with Y loss estimated using sequence reads (see Methods). Both replication datasets provided strong statistical support for the identified loci, with all 19 loci retaining genome-wide significance in a combined model (Table 1). As evaluated in the deCODE data, these loci cumulatively explained 2.7% of the total variance in Y chromosome copy number. We estimated an overall heritability of 34% (25.2-42.4%), suggesting many additional associated variants remain to be discovered.
Table 1. Genome-wide significant associations with Y chromosome loss.
| SNP | Location | Alleles1 | UK Biobank (N=67,034) |
EPIC Norfolk (N=9,793) |
deCODE (N=8,715) |
Replication P | Overall P | Gene4 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect2 | P | Effect2 | P | Effect3 | P | ||||||
| rs17758695 | 18q21.33 | C/T/0.97 | -0.01 | 6.4x10-21 | -0.014 | 3.7x10-04 | -0.020 | 9.1x10-13 | 2.7x10-17 | 1.3x10-33 | BCL2 [NC] |
| rs1122138 | 14q32.13 | C/A/0.84 | -0.005 | 3.6x10-23 | -0.006 | 4.3x10-04 | -0.007 | 1.5x10-04 | 8.0x10-10 | 6.3x10-31 | TCL1A [NEC] |
| rs78378222 | 17p13.1 | G/T/0.01 | -0.013 | 1.3x10-15 | -0.032 | 1.8x10-06 | -0.026 | 3.8x10-10 | 7.3x10-18 | 3.4x10-28 | TP53 [CN] |
| rs59633341 | 3q25.1 | A/AT/0.16 | -0.004 | 2.6x10-18 | -0.009 | 7.5x10-07 | -0.007 | 1.1x10-05 | 8.5x10-14 | 4.1x10-28 | TSC22D2 [N] |
| rs2736609 | 1q22 | T/C/0.36 | -0.003 | 1.9x10-12 | -0.003 | 4.9x10-02 | -0.006 | 2.5x10-07 | 2.4x10-10 | 2.0x10-19 | PMF1 [CFN], SEMA4A [CE] |
| rs13191948 | 6q21 | C/T/0.54 | -0.002 | 1.2x10-11 | -0.006 | 5.4x10-06 | -0.005 | 3.8x10-05 | 4.5x10-12 | 2.2x10-19 | SMPD2 [E], CCDC162P [NE] |
| rs60084722 | 20q11.21 | CT/C/0.79 | -0.003 | 6.6x10-13 | -0.002 | 2.5x10-01 | -0.006 | 9.4x10-05 | 1.5x10-6 | 1.6x10-17 | TPX2 [NEC], BCL2L1 [C], HM13 [E] |
| rs381500 | 6q26 | C/A/0.55 | -0.002 | 5.7x10-11 | -0.002 | 1.9x10-01 | -0.005 | 1.1x10-07 | 1.8x10-7 | 5.0x10-16 | QKI [N] |
| rs56084922 | 5q22.1 | G/A/0.08 | -0.005 | 2.9x10-13 | -0.004 | 1.2x10-01 | -0.005 | 1.6x10-03 | 2.8x10-3 | 3.0x10-15 | NREP [N] |
| rs137952017 | 14q32.2 | C/CT/0.85 | -0.003 | 1.2x10-09 | -0.01 | 1.3x10-07 | -0.004 | 4.0x10-04 | 2.4x10-8 | 4.0x10-15 | DLK1 [N] |
| rs4721217 | 7p22.3 | T/C/0.4 | -0.002 | 6.5x10-10 | -0.005 | 2.8x10-04 | -0.003 | 1.1x10-05 | 1.7x10-6 | 3.5x10-14 | MAD1L1 [NFC] |
| rs35091702 | 8p12 | C/CAAAAAAG/0.74 | -0.002 | 4.2x10-10 | -0.004 | 6.0x10-03 | -0.002 | 3.9x10-02 | 6.5x10-3 | 9.5x10-12 | RBPMS [N] |
| rs4754301 | 11q22.3 | A/G/0.55 | -0.002 | 1.3x10-09 | -0.001 | 5.4x10-01 | -0.002 | 2.8x10-02 | 1.5x10-2 | 6.5x10-11 | NPAT [NF], ATM [C], ACAT1 [E] |
| rs12448368 | 16q23.2 | C/T/0.13 | -0.003 | 9.8x10-10 | -0.002 | 2.5x10-01 | -0.003 | 2.4x10-02 | 2.2x10-2 | 7.1x10-11 | CENPN [NEC], ATMIN [CE] |
| rs11082396 | 18q12.3 | C/T/0.13 | -0.003 | 3.3x10-09 | -0.004 | 6.7x10-02 | -0.003 | 1.2x10-01 | 1.1x10-2 | 1.2x10-10 | SETBP1 [N] |
| rs13088318 | 3q12.3 | G/A/0.34 | -0.002 | 4.1x10-09 | -0.0004 | 7.7x10-01 | -0.003 | 1.7x10-02 | 2.1x10-2 | 2.7x10-10 | SENP7 [E] |
| rs77522818 | 17q21.33 | A/T/0.96 | -0.005 | 1.3x10-09 | -0.004 | 3.0x10-01 | -0.002 | 2.4x10-01 | 1.6x10-1 | 8.8x10-10 | FAM117A (N) |
| rs10687116 | 13q14.11 | AGATG/A/0.8 | -0.002 | 2.6x10-08 | -0.001 | 5.8x10-01 | -0.003 | 5.8x10-02 | 1.0x10-2 | 8.8x10-10 | WBP4 [N] |
| rs115854006 | 3p21.31 | C/T/0.96 | -0.006 | 3.7x10-08 | -0.007 | 5.4x10-02 | 0.002 | 9.3x10-01 | 3.4x10-1 | 4.5x10-08 | TREX1 [C], PLXNB1 [C] |
mLRR-Y lowering allele / increasing allele / lowering allele frequency
Effect estimates in per-allele decreases in raw mean intensity log-R ratio units
Effect estimate per allele for copy number transformed log2(chrY copy-number)
Labelled gene where preceding nomenclature refers to [N] nearest (default), [C] biological candidate, [E] expression mediated by mLRR-Y associated SNPs, [F] non-synonymous variant in gene.
We next used HaploReg10 and sequence data from the deCODE study to functionally annotate identified variants and genes. This highlighted four signals containing highly correlated missense variants, implicating MAD1L1 (rs1801368, r2>0.98), PMF1 (rs1052053, r2=1), NREP (rs11559, r2=0.74) and NPAT (rs2070661, r2=0.97) as potential candidates.
To ascertain whether the identified signals are more likely to reflect gain or loss of Y chromosome material, we performed two analyses comparing the bottom and top 5% of mLRR-Y ranked individuals to the median 25%, as a dichotomous indicator of extreme Y-chromosome loss or gain. All nineteen loci exhibited consistently stronger associations with the bottom 5% of mLRR-Y (greatest mLOY) than with the top 5% (Supplementary Table 1), suggesting their impact was on mosaic Y chromosome loss rather than gain. Analysis of mLRR-Y as a continuous trait across all individuals was, however, the most powerful approach for variant discovery, as only two of the signals reached genome-wide significance in the stratified analysis.
Genome-wide pathway analyses conducted on association results for continuous mLRR-Y highlighted five pre-defined biological pathways enriched for association (study-wise significant FDR<0.05), the most significant of which was ‘Apoptosis’ genes defined per the Kyoto Encyclopaedia of Genes and Genomes (KEGG)11 (Supplementary Table 2). Other significant pathways included sulphur metabolism, susceptibility to colorectal, prostate and thyroid cancers, and progesterone-mediated oocyte maturation.
The impact of mLOY variants on X-chromosome loss in women
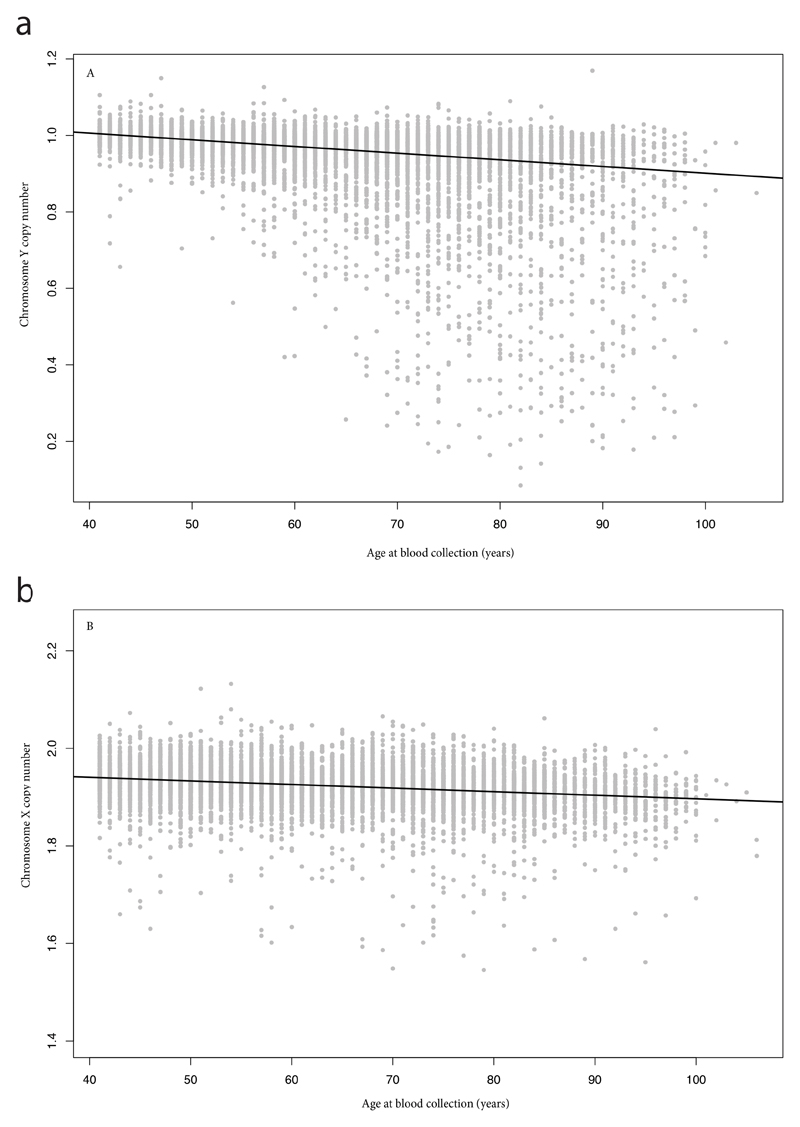
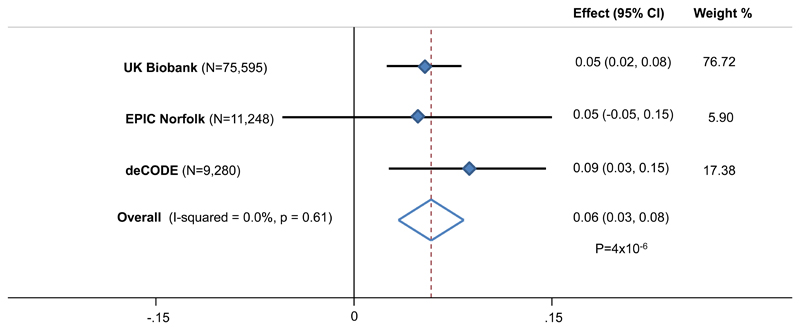
We next sought to understand whether our identified variants acted only on the Y chromosome, or promoted aneuploidy of other chromosomes more generally. Using a combined sample of 96,123 women from three studies, we ascertained X chromosome loss via both array intensity data (N=86,843) and sequence reads (N=9,280, Figure 1). Chromosome X copy number was estimated to have a heritability of 26% (17.4-36.2%) in the deCODE data; comparable to that of Y chromosome loss. Cumulatively, the 19 Y loss SNPs significantly predicted X loss in women, with the expected direction of effect (Figure 2, P=4x10-6).
Figure 1. Estimated X and Y chromosome loss with age in the Icelandic deCODE study.
(A) Y chromosome copy number estimated in 8703 males from whole genome sequencing. (B) X chromosome copy-number for 9280 females. In each case, the black line indicates the line of best fit with age at blood collection as a linear predictor.
Figure 2. Association of 19 SNP mLOY genetic risk score on X loss in women.
The genetic risk score is additive, based on mLRR-Y increasing allele dosage.
Identifying transcriptomic and epigenetic signatures of mLOY
To identify potential functional transcripts mediating Y chromosome loss, we performed summary statistic approaches to infer gene expression associations using three analytical imputation approaches12–14 in independent whole-blood expression datasets (Supplementary Tables 3-5). Across these datasets, eight genes (HM13, SMPD2, TCL1A, SENP7, NPAT, ATM, ACAT1, CENPN) were significantly associated with mLRR-Y, all of which mapped near to one of the 19 associated genetic signals from GWAS.
We additionally identified 36 methylation variable positions (MVPs) correlated with mLRR-Y levels in 569 whole-blood samples from the European Prospective Investigation of Cancer (EPIC)-Norfolk cohort15 (Supplementary Table 6). All significant MVPs were in genomic regions distinct (>500kb) from the 19 mLOY loci, with the exception of four correlated methylation probes within the TP53 gene region. To ascertain if any of the methylation changes represented causal drivers of mLOY, we next identified cis-methylation quantitative trait loci (meQTLs) in publicly available data16 for all associated probes. In total, 20 probes had one or more genetic variants in cis which were associated with methylation levels of the corresponding site (Supplementary Table 7). None of these genetic variants were correlated with the 19 genomic loci; however, one cis-meQTL survived multiple test correction for association with mLRR-Y (rs7208523, cg20116579 methylation P=5.6x10-31, mLRR-Y P=9x10-4). This suggests that genetic variation at the TNK1 locus, a gene with known involvement in tumor growth and survival, may be associated with increased mLOY via an epigenetic mechanism17.
Genetic overlap with cancer susceptibility
Three mLOY signals are correlated with signals previously reported for basal cell carcinoma18, glioma19, neuroblastoma20 (TP53), or testicular cancer21,22 (SEMA4A/PMF1 and MAD1L1). In each case, the mLRR-Y decreasing allele (i.e increased mLOY) was associated with increased cancer susceptibility. We performed a reciprocal lookup of 90 loci previously reported for prostate cancer susceptibility23,24, the most common male non-skin cancer in western populations. There was no obvious enrichment of signal across these loci and no apparent dose-response relationship between the allelic effects on prostate cancer and mLOY (PEGGER-MR = 0.26, Supplementary Table 8, Supplementary Figure 4). Under the hypothesis that susceptibility to many types of cancer may have a common basis in mitotic error, we performed a GWAS in UKB defining men with any diagnosed cancer as a case (N= 7,745 cases, 58,562 controls). This approach was recently used for multiple reproductive cancers, yielding several novel loci25. Applying the 19 mLRR-Y signals as an additive genetic instrument, there was no evidence of a dose-response relationship between genetically-modelled mLOY and cancer risk in men (PEGGER-MR = 0.94, Supplementary Table 9 and Supplementary Figure 5). To test the relationship between cancer risk and mLOY more comprehensively, we estimated the extent of shared genetic architecture across the whole genome using LD score regression26. This revealed an overall significant inverse relationship between mLRR-Y and cancer risk (rg=-0.42, P=0.02), which was not significant when considering only female cancer cases (rg=-0.06, P=0.64).
Discussion
Our findings, together with previous reports, demonstrate that loss of the Y-chromosome in peripheral blood likely represents a proxy trait for the study of aneuploidy in large-scale populations, which can be readily estimated from sequencing reads or array-based genotyping data. The nature of the genes identified by our analyses suggests that genetic determinants of mLOY reflect general mechanisms of aneuploidy, which we speculate most frequently manifest in mLOY due to the higher capacity of cells to tolerate Y-chromosome loss. This hypothesis is supported by the observation that these same SNPs also predicted X chromosome loss in women, the second most frequent large-scale mosaic event27.
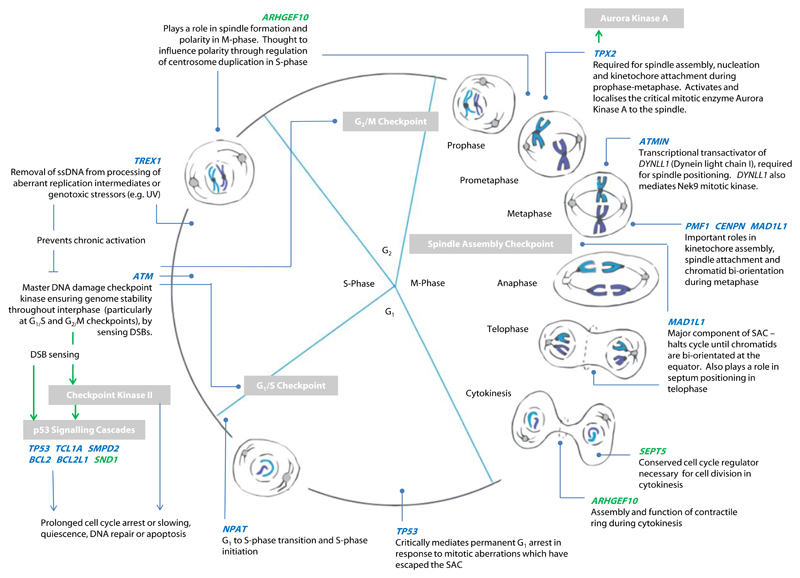
Pathway analyses identified enrichment for cancer and apoptosis pathways associated with mLOY. This is further supported by the many well-established cell cycle regulation genes which we observed either as the closest gene to the association signal, or which were implicated via altered expression or protein coding changes. Major mechanistic aspects of the cell cycle, and key regulators of cell-cycle progression were represented by these findings (Figure 3), including elements of three cell cycle checkpoints, and several genes with complementary functional roles in mitosis. TPX2, CENPN, PMF1 and ATMIN are involved in aspects of chromosome alignment during metaphase, spindle assembly, orientation and attachment to chromatids ahead of segregation28,29. In particular, TPX2 recruits the crucial mitotic enzyme, Aurora Kinase A, to the spindle30, whilst ATMIN regulates expression of a dynein motor component (DYNLL1) which critically mediates spindle positioning31–33 and also modulates Nek9 kinase signalling required for correct spindle formation and function34. Similarly, Rho-GEF 10 (ARHGEF10, for which we observe a nearby methylated signal) regulates centrosome duplication and prevents formation of multipolar spindles35. We identified a missense variant in MAD1L1 (MAD1 mitotic arrest deficient like 1), a major component of the spindle assembly checkpoint (SAC). This represents a key cellular safeguard against chromosome mis-segregation (and subsequent ploidy errors), supressing metaphase-anaphase progression until chromatids are bi-orientated on a bipolar spindle at the metaphase plate1. During cytokinesis, SEPT5 (septin 5, implicated in our methylation analysis) encodes a conserved cell cycle regulator required for effective cell division36, while activation of signalling by Rho-GEF 10 (ARHGEF10) facilitates contractile ring ingression to separate the two daughter cells37.
Figure 3. Overview of identified genes implicated in Y chromosome loss.
Genes falling within GWAS loci are shown in blue, those implicated by methylation analyses in green. Grey boxes highlight specific checkpoints, signalling cascades, or enzymes of note. Green arrows denote activation of a target by phosphorylation, blue arrows a signalling cascade and its ultimate effect.
We also implicated a number of genes with established roles in the replication and stability of nuclear DNA in interphase: replication errors are a key cause of genomic instability and chromosomal fragility38–40. G1 to S-phase transition is dependent on NPAT, at least in part through it promoting histone gene transcription41, while ATM, at least in part in association with ATMIN42, acts as major cell cycle checkpoint kinase dedicated to maintaining genome stability throughout interphase, with particular importance at the G1/S and G2/M checkpoints40. In response to double-stranded DNA breaks (DSBs) indicative of genomic instability, ATM promotes various responses via p53 and other factors to promote DNA repair, arrest cell-cycle progression, or otherwise initiate cell cycle exit strategies including apoptosis and senescence38–40,43. TREX1 encodes 3’ Repair Exonuclease 1, which digests aberrant replication intermediates and single stranded DNA from genotoxic stress to prevent chronic checkpoint activation44. Predicted deleterious missense variants in this gene were recently identified in a mouse GWAS for micronucleus formation, a biomarker of chromosomal breaks, whole chromosome loss and extranuclear DNA45.
At the later stages of the cell lifespan, several genes implicated by our GWAS findings – including TP53, TCL1A, SMPD2, BCL2 and BCL2L1 – functionally impact on apoptotic events46–50. Apoptosis is a prime mechanism by which cells with detected DNA damage or ploidy errors may be eliminated51: indeed, p53 drives multiple cell-cycle exit responses in response to aberrant mitosis, including G1 arrest43,52,53. The TP53 variant associated with mLOY in our analyses is the one previously reported for basal cell carcinoma: for this trait, the risk allele changes the AATAAA polyadenylation signal to AATACA, resulting in impaired 3′-end processing of TP53 mRNA18. Our findings also implicated genes involved in spermatogenesis54,55 (HENMT1 and DAZAP1), and cellular growth and differentiation56 (DLK1).
The genes directly involved in mitotic prophase-metaphase and the SAC have clear roles in averting chromosomal mis-segregation and preventing these from persisting unchecked, however how the broader set of genes we identify here may act to promote mLOY remains less clear. We speculate that either many of these genes act in ways that are not currently recognised, or alternatively that the other highlighted processes outside of cell cycle control and mitosis are important. In particular, as a major mode of cell-cycle exit, our observed enrichment of apoptotic regulatory genes and cascades may play a more passive permissive role in enabling mis-segregated cells to survive with ploidy errors, rather than being directly causative of them.
Although an initial defect during the cell cycle process is required to generate an aneuploid daughter cell, clonal expansion is likely required to drive the lineage to a detectable frequency in the circulating white blood cell population. It is possible that mLOY in haematopoietic precursors confers a proliferative advantage to such cells, leading to a relative enrichment of assayable mLOY progeny. We therefore speculate that some loci may operate through this pathway to further facilitate or promote clonal expansion of these cells. Additional functional experimentation in cellular and animal systems is ultimately required to fully elucidate this issue and the role individual associated genes may play in determining mLOY. We also acknowledge that there are likely other, currently unknown, mechanisms by which our associated loci exert their effects.
We observed a substantial shared genetic architecture between mLOY and cancer susceptibility, suggesting that bivariate analyses of these two traits may help to prioritise novel cancer susceptibility loci and elucidate their functions. We could not, however, find evidence of a dose-response relationship between these two traits. This is perhaps not surprising given that findings from mouse studies in which mitotic checkpoint components are experimentally down-regulated demonstrate an inconsistent relationship between aneuploidy and spontaneous tumorigenesis1. It is possible, therefore, that some of our identified genes may promote benign aneuploidy, whereas others may play a role more generally in genome instability. This makes the use of genetic variants associated with mLOY difficult within a Mendelian randomization framework, as genes with general roles in instability may have different phenotypic consequences to genes that promote aneuploidy in a more stable way. This of course does not preclude identifying causal risk factors for mLOY, exemplified by our positive causal inference for smoking on mLOY, using a genetic instrument for cigarettes per day. More generally, the association between smoking and mLOY suggests that care should be taken to avoid confounding influences such as socioeconomic patterning in epidemiological observations between mLOY and disease. In addition to fully evaluating the broader disease relevance of mLOY, future epidemiological studies should look to assess the differential rates at which mLOY changes in individuals over time, its relevance in other tissue types and further non-genetic modifiable factors which may influence it.
In conclusion, our study highlights that estimation of mLOY using genotype array intensity data may serve as a useful quantitative measure of cell cycle efficiency and genome stability, and may thereby add a new approach to the study of cellular ageing and its associations with disease, particularly cancer.
Data availability statement
The genome-wide discovery data used is from UK Biobank and can be obtained via application from www.ukbiobank.ac.uk. Requests for access to the underlying replication data is limited by participant consent and data sharing agreements; requests should be directed via http://www.srl.cam.ac.uk/epic/) or the corresponding author. Methylation data is available from the same EPIC-Norfolk resource and gene expression datasets are publically available from three resources: MetaXcan (https://github.com/hakyimlab/MetaXcan), SMR (http://cnsgenomics.com/software/smr/) and TWAS (http://gusevlab.org/projects/fusion/).
Online Methods
Estimating Y chromosome mosaicism in UK Biobank
We analysed data from the May 2015 release of imputed genetic data from UK Biobank8, containing ~73M SNPs, short indels and large structural variants in 152,249 individuals. Full details have been published elsewhere57. Briefly, the samples were genotyped on two slightly different arrays - approximately 50,000 on the custom UK BiLEVE study array, and the remainder (~100,000) on the UK Biobank Axiom array (Affymetrix), which was specifically designed to optimize imputation performance in GWAS studies. Removal of SNPs with missing data, multi-allelic SNPs, SNPs with a minor allele frequency (MAF) <1%, and 1,037 sample outliers, resulted in a dataset with 641,018 autosomal SNPs in 152,256 samples for phasing and imputation. Imputation was performed using a reference panel created by merging the UK10K haplotype panel with the 1000 Genomes Phase 3 reference panel.
In addition to the quality control metrics performed centrally by UK Biobank, we defined a subset of “white European” ancestry samples using a K-means clustering approach applied to the first four principle components calculated from genome-wide SNP genotypes. All individuals defined in this group also self-identified by questionnaire as being of white ancestry.
mLOY was estimated by calculating the mean log-R ratio (normalised signal intensity) of SNPs on the male-specific region of the Y chromosome. Signal intensity, genotype call and confidence files from Affymetrix Power Tools software were analysed using the PennCNV-Affy pipeline58 to produce a log-R ratio (LRR) for each SNP. SNPs without LRR calculable on both arrays, or those flagged by UKB as failing QC, were excluded. Whole Y chromosome fluorescence signal intensity was summarised by calculation of mean LRR across all Y chromosome SNPs (mLRR-Y). After omission of monomorphic SNPs, genotyping and QC failures, 253 SNPs were available across all participants for derivation of mLRR-Y.
Association testing and signal selection
Autosomal SNPs were analysed by linear mixed models implemented in BOLT-LMM59 to account for cryptic population structure and relatedness within this group in our genetic association tests. The regression model included age and genotyping array as covariates. SNPs with an imputation quality < 0.4 or MAF < 0.1% were excluded post-analysis. After application of QC criteria, a maximum of 67,034 men were available for analysis with genotype and phenotype data. Samples were subdivided by never (N=32,539] vs ever N=34,329] smoking for the Mendelian Randomization analysis using the CHRNA5-CHRNA3-CHRNB4 rs1051730 locus. Genomic loci were defined on the basis of physical proximity using a 1 Mb window. The following genome-wide significant signals were excluded from further consideration due to concerns of technical artefacts: rs61737590 (Chr1-27Mb), rs115979215 (Chr2-54Mb), rs1857807 (Chr2-115Mb), rs115722056 (Chr2-171Mb), rs73191481 (Chr3-105Mb), rs9289877 (Chr3-152Mb), rs77306208 (Chr3-194Mb), rs9269173 (Chr6-32Mb), rs117810108 (Chr7-130Mb), rs117941885 (Chr12-90Mb), rs118031436 (Chr15-57Mb), rs16961626 (Chr16-84Mb), rs58108384 (Chr20-7Mb), rs73892829 (Chr21-19Mb), rs116446488 (Chr22-24Mb). All were excluded due to fulfilment of 2 or more of the following criteria: a) singletons in regional association plots, b) significantly associated with genotype array status, c) associated with mLRR-Y in women (reflecting technical background intensity).
Replication
Replication was performed in two independent studies using two separate techniques.
The first comprised 9,793 men from the EPIC-Norfolk study15, following the same protocol using GWAS array intensity data as described above.
Secondly, we analyzed whole-blood genome sequences of 8,715 Icelandic males60 (age range 41-105 years, mean 63 years), that had been whole-genome sequenced by Illumina method to a mean depth of 37x.
As an estimate of chromosome Y copy-number we used the average read depth over chromosome Y, using exclusively X-degenerate regions. This was computed by samtools from bam files aligned to hg38 and normalized by genome-wide sequencing coverage for the subject. A total of 12 outlier individuals (copy-number greater than 1.25) were excluded.
Chromosome Y copy-number had a strong negative correlation with age at bleeding (Spearman correlation r=-0.50). For individuals older than 60 years at the time of sample collection, the distribution of chromosome Y copy-number has a heavy left tail with copy-numbers as low as 0.08.
Association analysis was performed using BOLT-LMM59 after inverse normal transformation and adjustment for age at bleeding. To enable comparison with the estimates obtained from GWAS array intensity data, effect sizes for log2(chrY copy-number) were estimated using robust linear regression (rlm from R package MASS).
The fraction of variance explained by a given variant was calculated using the formula 2f(1-f)a^2, in which f denotes the minor allele frequency of the variant and a is the additive effect in standard deviations. Heritability estimates were calculated using the spearman rank correlation of the traits between sibling pairs (max N=1488).
X chromosome loss
Similarly to mLOY, X chromosome loss was estimated using two complementary methods. Firstly, mLRR-X was calculated in UK Biobank (N=75,595) and EPIC Norfolk (N=11,248), using the same methodology described for X loss. Secondly, a similar analysis was performed using whole blood genome sequences of 9,302 Icelandic females (age range 41-106 years, mean 63 years) whole-genome sequenced to a mean depth of 36x. The chromosome X copy-number was estimated from the average read depth over chromosome X, excluding paralogous regions PAR1 and PAR2, the X-transposed region, and the centromere. This estimate was normalized by genome-wide sequencing coverage for the subject and adjusted for the sequencing protocol. A total of 22 outlier individuals (copy-number greater than 2.5 or less than 1.5) were excluded. We observed a Spearman correlation of -0.28 between the chromosome X copy-number and age at bleeding.
Cancer GWAS
To understand the genomic relationship between cancer and mLOY, we defined an ‘any prevalent cancer’ variable in UKB using linked UK cancer registrations. Individuals with a reported age of diagnosis in the cancer registry were coded as a case. Individuals with inconsistent cancer diagnosis (i.e a reported cancer but not age at diagnosis) were set to missing, and controls were defined as any individual with no self-reported or registry-defined cancer. GWAS analysis was performed as described above, including age, sex and genotyping array as covariates.
Genetic correlations (rg) were calculated between mLRR-Y and cancer using LD Score Regression26.
In order to assess the possible causal links between cancer and mLOY we applied Mendelian Randomization methods, which have been described extensively elsewhere61. In order to be as conservative as possible we preferentially report results from the Egger regression method, though inverse weighted, median weighted and penalised median weighted analyses were also calculated.
Gene expression
To identify specific eQTL linked genes, we utilised three complementary approaches – SMR, TWAS and MetaXcan – enabling systematic integration of publicly available gene expression data with our genome-wide dataset.
Summary Mendelian Randomization (SMR) uses summary-level gene expression data to map potentially functional genes to trait-associated SNPs14. We ran this approach against the publicly available whole-blood eQTL dataset published by Westra et al62, providing association statistics for 5,952 transcripts. A conservative significance threshold was set at P<4.9x10-6 reflecting the number of genes tested genome-wide.
MetaXcan, a meta-analysis extension of the PrediXcan method13, was used to infer the association between genetically predicted gene expression (GPGE) and mLRR-Y. PrediXcan is a gene-based data aggregation and integration method which incorporates information from gene-expression data and GWAS data to translate evidence of association with a phenotype from the SNP-level to the gene. Briefly, PrediXcan first imputes gene-expression at an individual level using prediction models trained on measured transcriptome datasets with genome-wide SNP data and then regresses the imputed transcriptome levels with phenotype of interest. MetaXcan extends its application to allow inference of the direction and magnitude of GPGE-phenotype associations with only summary GWAS statistics, which is advantageous when SNP-phenotype associations result from a meta-analysis setting and also when individual level data are not available. As input we utilized GWAS meta-analysis summary statistics for mLRR-Y, LD matrix from the 1000 Genomes project, and as weights, gene-expression regression coefficients for SNPs from models trained with whole-blood transcriptome data from the GTEx Project63. Threshold for statistical significance was estimated using the Bonferroni correction for number of tested genes.
Finally, we used the recently described Transcriptome-wide Association Study (TWAS) approach12 to infer gene expression association using two whole blood datasets (Young Finns Study and Netherlands Twin Registry cohorts). The threshold for significance was set to correct for the number of studies and genes (P<1x10-5). Each of the three approaches described in this section were compared by estimating the correlation (r) of association Z scores across genes present in all three datasets. There was strong concordance between the 2,326 transcripts analysed across the three approaches/datasets; SMR vs. TWAS r=0.72, SMR vs. MetaXcan r=0.54, TWAS vs. MetaXcan r=0.55.
Methylation
DNA methylation in whole blood was measured for 1,378 individuals in the EPIC-Norfolk cohort using the Illumina Human Methylation 450k BeadChip platform. After setting methylation markers with detection p-value ≥ 0.01 to missing, methylation beta values were calculated for each marker. Quantile normalisation of methylation betas was applied separately to different marker groups based on colour channel, probe type and M/U subtypes64. Samples with a sample call rate ≤0.99 were removed (n=77). Methylation beta value distributions of the X, Y and autosomal chromosome markers were analysed separately and a further 11 sample outliers were excluded. Within each sample, markers with a marker call rate ≤ 0.95 were excluded (n=4,423).
All further downstream analyses were restricted to autosomal methylation markers. Signal detection of methylation intensities can be affected by several factors, including SNPs on the probe, repetitive DNA, and cross-reactive probes. We thus calculated the proportion of missing data at each CpG site (marker call rate) and 8,775 CpGs with a call rate ≤ 0.95 were excluded. 3,295 CpGs with multimodal distributions of methylation intensities, identified by the R package ENmix65, which typically arise from technical artefacts were also excluded. A further 18,874 CpG sites which were previously identified as mapping to more than 1 genomic location66 were also excluded. The final cleaned dataset comprised 442,920 autosomal CpG sites. To account for cell composition variability, we estimated counts of T lymphocyte subtypes, natural killer cells, monocytes, granulocytes and B lymphocytes using the minfi R package67,68. These were included as covariates in subsequent epigenome-wide regression models.
To examine the association between methylation markers and mLOY, we performed an epigenome-wide association analysis in all male EPIC-Norfolk methylation samples (n=569). mLRR-Y was regressed separately on each methylation marker, adjusted for type 2 diabetes status, age, current smoking status, estimated cell counts, and sample plate. Bonferroni correction was applied, accounting for the number of markers tested (p=1×10-7). Furthermore, we checked that no significant CpG sites had sequences which also mapped to the Y chromosome.
Association statistics for genetic variants within the probe vicinity and corresponding methylation levels (i.e cis-meQTLs) were available from the BIOS QTL browser (http://www.genenetwork.nl/biosqtlbrowser/)
Pathway analyses
Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA) was used to explore pathway-based associations in the full GWAS dataset. MAGENTA implements a gene set enrichment analysis (GSEA) based approach, as previously described69. Briefly, each gene in the genome is mapped to a single index SNP with the lowest P-value within a 110 kb upstream, 40 kb downstream window. This P-value, representing a gene score, is then corrected for confounding factors such as gene size, SNP density and LD-related properties in a regression model. Genes within the HLA-region were excluded from analysis due to difficulties in accounting for gene density and LD patterns. Each mapped gene in the genome is then ranked by its adjusted gene score. At a given significance threshold (95th and 75th percentiles of all gene scores), the observed number of gene scores in a given pathway, with a ranked score above the specified threshold percentile, is calculated. This observed statistic is then compared to 1,000,000 randomly permuted pathways of identical size. This generates an empirical GSEA P-value for each pathway. Study-wise significance was determined when an individual pathway reached a false discovery rate (FDR) <0.05 in either analysis. In total, 3216 pathways from Gene Ontology, PANTHER, KEGG and Ingenuity were tested for enrichment of multiple modest associations with mLRR-Y.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 9905. This work was supported by the UK Medical Research Council (Unit Programme numbers MC_UU_12015/1 and MC_UU_12015/2). Research in the S. Jackson laboratory is funded by Cancer Research UK (CRUK; programme grant C6/A18796), with Institute core funding provided by CRUK (C6946/A14492) and the Wellcome Trust (WT092096). S. Jackson receives salary from the University of Cambridge, supplemented by CRUK. We thank the MRC Epidemiology genetics group members for useful Friday morning discussions.
Footnotes
Author contributions
All authors reviewed the original and revised manuscripts. Statistical analysis: D.J.W., F.R.D., N.D.K., F.Z., A.C., P.S., R.A.S., J.R.B.P. Individual study sample collection, genotyping and phenotyping: S.S., D.F.G., A.H., N.D.K., A.C., F.Z. Individual study principal investigators: C.L., N.J.W., U.T., K.K.O., K.S., J.R.B.P. Project design and interpretation of results: D.J.W., F.R.D., N.D.K., P.S., D.J.T., J.R.C., S.P.J., C.L., N.J.W., U.T., K.K.O., K.S., J.R.B.P.
Competing financial interests statement
F.Z., P.S., S.S., D.F.G., A.H., U.T. and K.S. are employees of deCODE Genetics/Amgen Inc. (Reykjavik, Iceland). R.A.S. is an employee of GlaxoSmithKline plc.
References
- 1.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou W, et al. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat Genet. 2016;48:563–8. doi: 10.1038/ng.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsberg LA, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–8. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumanski JP, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–3. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumanski JP, et al. Mosaic Loss of Chromosome Y in Blood Is Associated with Alzheimer Disease. Am J Hum Genet. 2016;98:1208–19. doi: 10.1016/j.ajhg.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudlow C, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 15.Day N, et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 16.Bonder MJ, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2016 doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 17.Henderson MC, et al. High-throughput RNAi screening identifies a role for TNK1 in growth and survival of pancreatic cancer cells. Mol Cancer Res. 2011;9:724–32. doi: 10.1158/1541-7786.MCR-10-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey SN, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh KM, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013;37:222–8. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diskin SJ, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106:dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruark E, et al. Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat Genet. 2013;45:686–9. doi: 10.1038/ng.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung CC, et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet. 2013;45:680–5. doi: 10.1038/ng.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Olama AA, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–9. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eeles R, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11:18–31. doi: 10.1038/nrurol.2013.266. [DOI] [PubMed] [Google Scholar]
- 25.Kar SP, et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov. 2016:1–17. doi: 10.1158/2159-8290.CD-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machiela MJ, et al. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat Commun. 2016;7:11843. doi: 10.1038/ncomms11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 29.Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurado S, et al. ATM Substrate Chk2-interacting Zn2+ Finger (ASCIZ) Is a Bi-functional Transcriptional Activator and Feedback Sensor in the Regulation of Dynein Light Chain (DYNLL1) Expression. J Biol Chem. 2012;287:3156–3164. doi: 10.1074/jbc.M111.306019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunsch AK, et al. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198:1039–1054. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaytseva O, et al. The Novel Zinc Finger Protein dASCIZ Regulates Mitosis in Drosophila via an Essential Role in Dynein Light-Chain Expression. Genetics. 2014;196:443–453. doi: 10.1534/genetics.113.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regue L, et al. DYNLL/LC8 Protein Controls Signal Transduction through the Nek9/Nek6 Signaling Module by Regulating Nek6 Binding to Nek9. J Biol Chem. 2011;286:18118–18129. doi: 10.1074/jbc.M110.209080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki T, Ueda S, Kataoka T, Satoh T. Regulation of mitotic spindle formation by the RhoA guanine nucleotide exchange factor ARHGEF10. BMC Cell Biol. 2009;10:56. doi: 10.1186/1471-2121-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–9. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 37.Zuo Y, Oh W, Frost JA. Controlling the switches: Rho GTPase regulation during animal cell mitosis. Cell Signal. 2014;26:2998–3006. doi: 10.1016/j.cellsig.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazouzi A, Velimezi G, Loizou JI. DNA replication stress: Causes, resolution and disease. Exp Cell Res. 2014;329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 39.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–16. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 41.Gao G, et al. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol Cell Biol. 2003;23:2821–33. doi: 10.1128/MCB.23.8.2821-2833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt L, et al. ATMIN is required for the ATM-mediated signaling and recruitment of 53BP1 to DNA damage sites upon replication stress. DNA Repair (Amst) 2014;24:122–130. doi: 10.1016/j.dnarep.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 44.Christmann M, Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: Trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McIntyre RE, et al. A Genome-Wide Association Study for Regulators of Micronucleus Formation in Mice. G3 (Bethesda) 2016;6:2343–54. doi: 10.1534/g3.116.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yabu T, et al. Stress-induced ceramide generation and apoptosis via the phosphorylation and activation of nSMase1 by JNK signaling. Cell Death Differ. 2015;22:258–73. doi: 10.1038/cdd.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laine J, Künstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 49.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 50.Haimovitz-Friedman A, Kolesnick RN, Fuks Z. Ceramide signaling in apoptosis. Br Med Bull. 1997;53:539–53. doi: 10.1093/oxfordjournals.bmb.a011629. [DOI] [PubMed] [Google Scholar]
- 51.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol. 2004;5:752–762. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- 52.Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganem NJ, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim SL, et al. HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse. PLOS Genet. 2015;11:e1005620. doi: 10.1371/journal.pgen.1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu LC-L, et al. DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA. 2008;14:1814–22. doi: 10.1261/rna.1152808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falix FA, Aronson DC, Lamers WH, Gaemers IC. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim Biophys Acta - Mol Basis Dis. 2012;1822:988–995. doi: 10.1016/j.bbadis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank Data: Come and Get It. Sci Transl Med. 2014;6:224ed4–224ed4. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 58.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loh P-R, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–44. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 61.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lehne B, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:e20–e20. doi: 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naeem H, et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics. 2014;15:51. doi: 10.1186/1471-2164-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome-wide discovery data used is from UK Biobank and can be obtained via application from www.ukbiobank.ac.uk. Requests for access to the underlying replication data is limited by participant consent and data sharing agreements; requests should be directed via http://www.srl.cam.ac.uk/epic/) or the corresponding author. Methylation data is available from the same EPIC-Norfolk resource and gene expression datasets are publically available from three resources: MetaXcan (https://github.com/hakyimlab/MetaXcan), SMR (http://cnsgenomics.com/software/smr/) and TWAS (http://gusevlab.org/projects/fusion/).