Abstract
Colorectal cancer (CRC) genome is unstable and different types of instabilities, such as chromosomal instability (CIN) and microsatellite instability (MSI) are thought to reflect distinct cancer initiating mechanisms. Although 85% of sporadic CRC reveal CIN, 15% reveal mismatch repair (MMR) malfunction and MSI, the hallmarks of Lynch syndrome with inherited heterozygous germline mutations in MMR genes. Our study was designed to comprehensively follow genome-wide expression changes and their implications during colon tumorigenesis. We conducted a long-term feeding experiment in the mouse to address expression changes arising in histologically normal colonic mucosa as putative cancer preceding events, and the effect of inherited predisposition (Mlh1+/−) and Western-style diet (WD) on those. During the 21-month experiment, carcinomas developed mainly in WD-fed mice and were evenly distributed between genotypes. Unexpectedly, the heterozygote (B6.129-Mlh1tm1Rak) mice did not show MSI in their CRCs. Instead, both wildtype and heterozygote CRC mice showed a distinct mRNA expression profile and shortage of several chromosomal segregation gene-specific transcripts (Mlh1, Bub1, Mis18a, Tpx2, Rad9a, Pms2, Cenpe, Ncapd3, Odf2 and Dclre1b) in their colon mucosa, as well as an increased mitotic activity and abundant numbers of unbalanced/atypical mitoses in tumours. Our genome-wide expression profiling experiment demonstrates that cancer preceding changes are already seen in histologically normal colon mucosa and that decreased expressions of Mlh1 and other chromosomal segregation genes may form a field-defect in mucosa, which trigger MMR-proficient, chromosomally unstable CRC.
The genome-wide expression profiling indicates that cancer preceding changes can be detected already in histologically normal mucosa and that decreased expression of specific chromosomal segregation genes are the most initiative cancer preceding markers in colon mucosa.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most common cause of cancer-related deaths worldwide. The incidence rates increase significantly with age and interactions between genetic and environmental factors, including diet, are suggested to play a critical role in its etiology (1,2). Cancer development always includes lack of genomic integrity in cells and different types of genomic instability, such as chromosomal instability (CIN) and microsatellite instability (MIN, also called MSI) are thought to reflect distinct initiating mechanisms in cancer (3). Three different pathways leading to genomic instability in colon cancer have been described. Most CRCs represent CIN, where chromosomes fail to trigger the spindle assembly checkpoint (SAC) leading to aberrant chromosome segregation. In recent years, many new genes have been reported, whose mutations and expression changes disturb chromosomal stability causing aneuploidy and/or comprehensive loss of heterozygosity (LOH) and alterations in chromosome structure (4,5). About 15% of sporadic CRCs and over 95% of CRCs in Lynch syndrome (LS), the most common inherited colon cancer syndrome, represent MSI caused by a defective DNA mismatch repair (MMR) mechanism (6). MMR deficiency causes accumulation of point mutations in the genome and especially in short repeat sequences called microsatellites, and is thought to be the driver defect in MSI carcinomas (7). The third pathway, CpG Island Methylator Phenotype (CIMP), characterized by global genome hypermethylation and tumour suppressor gene silencing, is seen in 20–30% of CRCs (6).
There are different ways to induce MMR malfunction and consequently lead to MSI. In sporadic tumours, the most common mechanism is the MLH1 (mutL homolog 1) promoter hypermethylation often associated with BRAF V600E mutation (8). Also, in CIMP tumours the MLH1 is often inactivated by promoter hypermethylation but usually caused by aging and environmental factors (9). In LS, the first mutation is inherited, mainly in MLH1 (40%), MSH2 (34%) or MSH6 (18%) (10) and generally followed by LOH and somatic inactivation of the MMR gene (8). In LS, the second hit leading to MSI rarely happens through promoter hypermethylation (11). The fact that inactivation of MLH1 is associated with most MSI tumours irrespective of the inactivating mechanism emphasizes MLH1 deficiency as a key defect shared by sporadic and inherited CRC.
Colon cancer research focuses mainly on tumour characteristics, such as genomic instability, which can be utilized in treatment design. Recent findings have revealed that CIN and MSI pathways are not mutually exclusive (4,12), suggesting that also tumours with distinct features and instabilities may share initiative genomic aberrations while different tumour characteristics reflect subsequent alterations during cancer development. Since MLH1 deficiency seems to be a key defect shared by sporadic and inherited microsatellite unstable CRC, our aim was to follow how MLH1 would contribute to colon cancer development. Here, we used a mouse model to study cancer preceding expression changes in colon mucosa, Mlh1 phenotype in tumours and the effect of inherited predisposition (Mlh1+/−) and Western-style diet (WD) on those (13). We conducted a long-term feeding experiment with either a healthy rodent diet AIN-93G (AIN) or WD modified from AIN. WD was used to ensure the development of colon carcinomas, since it has previously been shown to cause CRCs in mice even without any predisposing mutation or carcinogen treatment (14–17). The mouse model provided a valuable tool to study the process of carcinogenesis from the earliest changes in colon mucosa until tumour development and characterization. Moreover, the use of an animal model enabled to distinguish gene expression changes caused by different risk factors, such as age, inherited predisposition and diet and sort out the ones that signal carcinogenesis.
Materials and methods
Mice, experimental study and diets
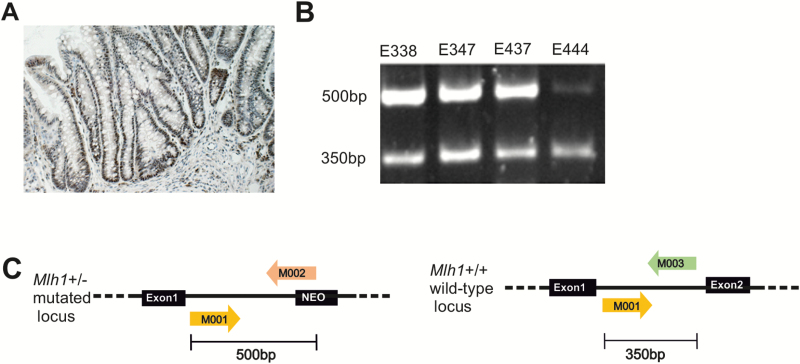
Heterozygote B6.129-Mlh1tm1Rak mice (Mlh1+/−) strain 01XA2 (18) and the C57BL/6 strain were obtained from NCI-MMHCC; National Institutes of Health, Mouse Repository, NCI-Frederick, MD. Altogether 12 animals (equal numbers of sexes), the Mlh1+/− mice and their wild-type C57BL/6 mates, formed six breeder pairs which produced the mouse colony used in our study. Mice were genotyped (Figure 1C) using genomic DNA extracted from earmarks according to the protocol published in our previous work (19). The mice were bred and treated according to the study protocol approved by the National Animal Experiment Board in Finland (ESLH-2008-06502/Ym-23).
Figure 1.
Mlh1 protein expression and loss of heterozygosity analyses. (A) An example of a colon carcinoma showing positive Mlh1 expression analysed by immunohistochemistry (mouse E402, tubular adenocarcinoma). (B) Four CRCs found in heterozygote Mlh1+/− mice showing that the normal Mlh1 allele (350 bp) was still present in tumours. (C) In Mlh1 heterozygote mice, one of the Mlh1 alleles is mutated by replacing the exon 2 with a neomycin cassette. Loss of Mlh1 heterozygosity was analysed using the genotyping primers M001, M002 and M003, which produce two different length fragments, 350 and 500 bp, that separate the normal (M001/M003) and the mutated allele (M001/M002), respectively.
The Mlh1 heterozygote mice and their homozygote wild-type littermates were divided into two dietary groups at the age of 5 weeks. The mice were fed with either healthy rodent control diet AIN-93G (20) or WD modified from AIN (Harlan Teklad, Madison, WI) (19) to resemble, on the nutritional level, the diet consumed in human Western population (high fat and energy content, low amounts of fiber, calcium and vitamin D3) (Supplementary Table 1) (19) Twelve mice per each group (Mlh1+/+ AIN, Mlh1+/− AIN, Mlh1+/+ WD, Mlh1+/− WD) with equal representation of sexes, at time point (tp) 0 (5 weeks of age, Mlh1+/−, Mlh1+/+), tp1 (12 months of age), tp2 (18 months of age) and tp3 (21 months of age), 168 mice in total were sacrificed and sampled.
Collection of tumours and normal colon mucosa samples
All observed colon tumours were collected under dissecting microscope and preserved as FFPE blocks. If a tumour was large enough (3–5 mm in diameter), approximately half of it was embedded in O.C.T compound (VWR, Radnor, Pennsylvania) for cryo sampling. Histological studies, stainings and the grading of neoplasias were carried out at The Finnish Centre for Laboratory Animal Pathology (FCLAP), University of Helsinki, Finland. The neoplasias were graded as hyperplasias, adenomas and carcinomas according to criteria based on consensus rodent intestinal cancer nomenclature (21), (Supplementary Table 2). Longitudinal pieces (excluding the previously harvested tumorous sections), representing approximately one third of the proximal mouse colon were collected for normal mucosa. The mucosa was separated from the underlying submucosa and musculature under a dissecting microscope and samples for RNA extraction were stored in RNAlater (Qiagen, Hilden, Germany) at −80°C (13).
Transcriptome analysis of normal mucosa
Transcriptome analysis was performed using RNA-sequencing method (RNA-seq). Total RNA was prepared from 0, 12 and 18 months old mice (14, 40 and 40 mice, respectively) using the RNeasy Plus Kit (Qiagen) with an extra DNase treatment (Qiagen). The RNA concentration was measured by Qubit 1.0 (Thermo Fisher Scientific, Waltham, MA) and RNA integrity with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only high quality RNA (RNA integrity number RIN ≥ 8) qualified for expression analysis (13).
RNA-seq method followed the single-cell tagged reverse transcription (STRT) (22) protocol with modifications (23). Briefly, 10 ng of total RNA was converted to cDNA and amplified to form an Illumina-compatible library. In total, 25 PCR cycles were used, but as four base-pair unique molecular identifiers were applied, only the absolute number of unique reads were included in the subsequent analysis. The samples were sequenced on a total of six lanes of Illumina HiSeq2000, further processed to fastq files by Casava 1.8.2 (both Illumina, San Diego, CA). Quality control was performed using the STRTprep pipeline (https://github.com/shka/STRTprep) (23). The processed reads were aligned by TopHat2 to the mouse RefSeq mm9 reference genome. STRT captures sequences at the 5′-end of poly(A)+ RNAs and the aligned reads therefore tend to be distributed close to the 5′-end (start site) of genes. STRTprep counts only the aligned reads at the 5′-untranslated region of protein-coding genes, or within the proximal (500 bp) upstream region (13).
Normalizing the RNA-seq data
STRTprep pipeline generated a read count matrix, with genes as rows and samples as columns. Different sample library sizes were normalized using DESeq-style normalization (24). Next shifted log transformation was done to generate more Gaussian like data and the ComBat program was used to filter batch effects. These preprocessing steps and alternative pipelines were evaluated by looking at the hierarchical clustering of samples and by plotting quantiles of expression values for each sample (Supplementary Information, available at Carcinogenesis Online) (13).
Tests for differential gene expression
Since the analyzed data was not any more integer count values after ComBat normalization, we tested three t-test based methods, Voom-Limma, Cyber-T and Shrinkage-T for analysis of differential gene expression. All these methods add a prior to variance estimate. Shrinkage-T is the only method here that allows also testing with unequal variance. This turned out to be important, as the genes with strongest separation between the sample groups had small variance in the analysed subset and medium variance among remaining samples.
The three methods were evaluated by viewing the separation of cancer samples from the remaining samples in the multidimensional Scaling (MDS) plots with top-k genes, which were selected using the evaluated statistic. Parameter k was varied from 25 to few hundreds. Shrink-T showed the best separation in the generated plots across all values of k. Each methods’ ability to find correlations with Gene Ontology classes was also tested. We used t-test scores from each method separately as an input to enrichment analysis tool called GSZ (Gene Set Z-score) (25). Shrink-T again generated strongest results. We therefore applied only Shrink-T in the subsequent analyses (13).
Visualization of sample differences
To detect similarities and differences between the samples, we used MDS that generates a small-dimensional visualisation from the multidimensional data while trying to preserve the pair-wise distances of samples from the multi-dimensional data. PlotMDS distributed in the Limma package was used as a basis of the analysis (26), although modified so that we were able to use any selected score to pick the genes that were used to calculate pair-wise distances (13).
The activity of Mlh1 was visualized with ComBat normalized data. Samples were grouped based on the sample types (genotype, diet and time-point) to highlight the sample differences.
Pathway analysis
To study the biological functions and pathways enriched among the top separating genes, we used QIAGEN’s Ingenuity Pathway Analysis (IPA Software 7.0, Qiagen). Here, we analysed both the top 100 and top 300 genes, which were found to separate the normal mucosa expression patterns in carcinoma mice from the others. The settings for a core analysis were as follows: ingenuity knowledge base (genes and endogenous chemicals) with both direct and indirect relationships, default network interaction settings (include endogenous chemicals, 35 molecules per network and 25 networks per analysis). Data sources were used with stringent confidence (experimentally observed and high predicted) and data obtained in all species was selected with a relaxed filter (13).
MSI and LOH analyses
The MSI status was analysed from all the seven carcinomas (two Mlh1+/+ WD, four Mlh1+/− WD and one Mlh1+/+ AIN mice, Table 1) from which a cryo-preserved sample was available, using four dinucleotide (D18Mit15, D14Mit15, D10Mit2, D7Mit91) and two mononucleotide (JH104, U12235) markers (27). Tumour DNA samples were extracted from the cryo-preserved colon carcinomas using laser micro-dissection for cutting (Zeiss PALM MicroBeam, Carl Zeiss Microscopy GmbH, Jena, Germany) and normal DNA control samples from the tails of the same mice with QIAamp DNA micro Kit, and DNeasy Blood & Tissue Kit (Qiagen), respectively. The genomic DNA was amplified with 6-FAM labeled primers in 11.1× PCR master mix (28) using the following PCR protocol: 1 min at 96°C, 30 cycles of 20 s at 96°C, 30 s at 62°C and 15 s at 70°C, and 7 min at 70°C. The fragments were analysed with ABI3730xl capillary electrophoresis (Thermo Fisher Scientific) and visualized with PeakScanner v1.0 (Thermo Fisher Scientific) (13).
Table 1.
Characteristics of mice and their tumours
| ID | Age | Genotype | Diet | Gender | Tumour characteristics | |||
|---|---|---|---|---|---|---|---|---|
| Histopathology | MSI | LOH | Mlh1 expression | |||||
| E249 | 12 | Mlh1 +/+ | WD | M | Tubular ac | NA | NA | Positive |
| E314 | 18 | Mlh1 +/+ | WD | M | Mucinous ac | NA | NA | Positive |
| E329 | 18 | Mlh1 +/+ | AIN | F | Tubulovillous ac | NA | NA | Positive |
| E333 | 18 | Mlh1 +/+ | WD | F | Tubular ac | NA | NA | Positive |
| E338 | 18 | Mlh1 +/− | WD | F | Tubular ac | MSS | — | Positive |
| E347 | 18 | Mlh1 +/− | WD | F | Serrated ac | MSS | — | Positive |
| E402 | 21 | Mlh1 +/+ | WD | F | Tubular ac | MSS | NA | Positive |
| E409 | 21 | Mlh1 +/+ | WD | M | Mucinous ac | NA | NA | Positive |
| E410 | 21 | Mlh1 +/+ | WD | M | Tubular ac | NA | NA | Positive |
| E421 | 21 | Mlh1 +/+ | AIN | F | Tubular ac | MSS | NA | Positive |
| E437 | 21 | Mlh1 +/− | WD | M | Mucinous ac | MSS | — | Positive |
| E444 | 21 | Mlh1 +/− | WD | F | Mucinous ac | MSS | — | Positive |
| E453 | 21 | Mlh1 +/− | AIN | F | Tubulovillous ac | NA | NA | Positive |
| E214 | 12 | Mlh1 +/+ | WD | F | Adenoma | NA | NA | NA |
| E244 | 12 | Mlh1 +/− | WD | F | Adenoma | NA | NA | NA |
| E246 | 12 | Mlh1 +/− | AIN | F | Adenoma | NA | NA | NA |
| E321 | 18 | Mlh1 +/− | WD | F | Serrated adenoma | NA | NA | NA |
| E337 | 18 | Mlh1 +/+ | WD | F | Adenoma | NA | NA | NA |
| E402 | 21 | Mlh1 +/+ | WD | F | Adenoma | NA | NA | NA |
| E411 | 21 | Mlh1 +/+ | WD | F | Adenoma | NA | NA | NA |
| E416 | 21 | Mlh1 +/+ | WD | M | Adenoma | NA | NA | NA |
| E446 | 21 | Mlh1 +/− | WD | F | Adenoma | NA | NA | NA |
| E448 | 21 | Mlh1 +/− | AIN | F | Adenoma | NA | NA | NA |
| E217 | 12 | Mlh1 +/+ | WD | M | Hyperplasia | NA | NA | NA |
| E307 | 18 | Mlh1 +/− | WD | M | Hyperplasia | NA | NA | NA |
| E328 | 18 | Mlh1 +/− | AIN | F | Hyperplasia | NA | NA | NA |
| E405 | 21 | Mlh1 +/+ | WD | F | Serrated hyperplasia | NA | NA | NA |
| E408 | 21 | Mlh1 +/− | AIN | F | Hyperplasia | NA | NA | NA |
| E409 | 21 | Mlh1 +/+ | WD | M | Hyperplasia | NA | NA | NA |
| E410 | 21 | Mlh1 +/+ | WD | M | Hyperplasia | NA | NA | NA |
| E430 | 21 | Mlh1 +/− | AIN | M | Hyperplasia | NA | NA | NA |
| E433 | 21 | Mlh1 +/+ | AIN | F | Hyperplasia | NA | NA | NA |
| E454 | 21 | Mlh1 +/− | WD | F | Hyperplasia | NA | NA | NA |
ac, adenocarcinoma; AIN, AIN-93G diet; F, female; M, male; NA, not analysed; WD, Western-style diet.
The four colon carcinomas found in the heterozygote Mlh1+/− mice, of which the cryo sample was available, were also studied for loss of Mlh1. Loss of heterozygosity was analysed using the genotyping primers M001 (TGT CAA TAG GCT GCC CTA GG), M002 (TGG AAG GAT TGG AGC TAC GG), and M003 (TTT TCA GTG CAG CCT ATG CTC), which produce two different length fragments, 350 and 500 bp, separating the normal (M001/M003) and the mutated allele (M001/M002), respectively (19) (Figure 1B, C). DNA was amplified with the 11.1× PCR master mix as described above and the fragments were visualized on 1% SB agarose gel.
Immunohistochemical analysis of Mlh1 protein expression in carcinomas
Formalin-fixed, paraffin embedded cancer tissue blocks were studied for Mlh1 expression. The 4 µm thick sections were deparaffinized and rehydrated and heat induced antigen retrieval was performed with 10 mM citrate buffer (pH 6). To detect Mlh1, the slides were incubated overnight at 4°C with the rabbit monoclonal antibody ab92312 (1:1500) (clone EPR3894, Abcam, Cambridge, UK). Stainings were visualized using UltraVision Detection System anti-rabbit HRP/DAB (ThermoFisher Scientific, Waltham, MA) by manufacturer′s instructions. Analysis of staining patterns was conducted at The Finnish Centre for Laboratory Animal Pathology (13).
Analysis of mitoses in carcinomas
A Feulgen with Midori green background stain was used to visualise nuclear material and mitoses in six carcinoma samples (E249, E314, E329, E333, E338 and E347). The samples were deparaffinized and rinsed in 1 M HCl. Mild acid hydrolysis was accomplished by using 60°C 1 M HCl and DNA was stained purple in Schiff′s reagent for 45 min. After several bisulfite washes the samples were counterstained briefly with 1% Midori light green, dehydrated through alcohol series to xylene and mounted with xylene based mounting media. The stained samples were analysed under light microscope (Zeiss Axio Imager.A2, Carl Zeiss Microscopy GmbH) and the mitoses in the malignant areas of carcinomas were compared to mitoses in samples (E305, E311, E322, E323 and E346) from healthy mice (13).
Statistical analysis
Differential expression analysis (DEA) used modified t-tests (Limma, cyber-T and shrinkage-T). With Limma and cyber-T, we used their own P-value estimates. Shrinkage-T does not provide a P-value estimate, which were estimated by re-calculating Shrinkage-T with 1000 permutations for each gene separately. Normal distribution was fitted to the permutations and a one-tailed P-value was obtained from the cumulative distribution. Multiple testing correction was performed using false discovery rate. Importantly, we used DEA mainly to order the genes to most differentially regulated genes. All analysis was performed within the R-environment. Pathway enrichment analysis was done using IPA which uses Fisher’s exact test to analyse over-representation of genes from the analysed gene groups. Here, multiple testing correction was done using the Benjamin–Hochberg method (13).
Results
Carcinomas developed mainly and earlier in WD fed mice
The feeding study was done with offspring produced by crossing two isogenic strains, the heterozygote Mlh1+/− (B6.129-Mlh1tm1Rak) and the wild-type Mlh1+/+ (C57BL/6) mice, and selecting an equal number of both genotypes to the study. Half of the mice fed WD and half the control diet, AIN-93G. In all 168 mice, 24 mice at time point 0 and 48 mice at time points 12, 18 and 21 months, were operated. Approximately 80% of all colon tumours, 10 out of 13 colon adenocarcinomas and 14 out of 20 adenomas and hyperplasias, developed in WD-fed mice (Table 1) (13).
At time points 12, 18 and 21 months, 80, 78 and 64% of all tumours and 100, 80 and 72% of CRCs were found in WD-fed mice, respectively. The overall number of colon tumours increased significantly with time (Supplementary Figure 1), being 5 at 12 months (one adenocarcinoma, two adenomas, one hyperplasia), 9 at 18 months (five adenocarcinomas, two adenomas, two hyperplasias) and 19 at the 21 months time point (seven adenocarcinomas, five adenomas, seven hyperplasias). Tumours were approximately evenly distributed between different genotypes since heterozygote Mlh1+/− mice showed 0, 40 and 43% of carcinomas and 50, 75 and 42% of adenomas and hyperplasias at different time points (Table 1). All the 13 carcinomas were found in the proximal part of colon and the majority of them were either tubular (54%) or mucinous (31%), two were tubulovillous and one carcinoma had serrated histology (Supplementary Table 2, Supplementary Figure 2, available at Carcinogenesis Online).
Mlh1 mutation carriers did not show MSI, LOH and loss of MMR protein in tumours
To check for the typical LS characteristics, seven carcinomas found in 18 and 21 months old mice, four in the Mlh1+/− mice (E338, E347, E437, E444) and three in the Mlh1+/+ mice (E402, E410, E421), were analysed for MSI status and all the 13 carcinomas for Mlh1 expression. Surprisingly, all CRCs showed Mlh1 expression (Figure 1A), indicating that irrespective of the inherited mutation in one Mlh1 allele in the heterozygote mice, the normal allele was still present in the tumours. The presence of the normal allele was further confirmed with LOH study in all the four Mlh1+/− carcinomas (Figure 1B). To study whether the detected Mlh1 protein was functional and MMR proficient, we analysed the stability of six polymorphic microsatellite regions in the mouse genome. The markers and their amplified fragment sizes were as follows: D14Mit15 (148 bp, 150 bp), D18Mit15 (151 bp, 157 bp), D7Mit91 (139 bp, 147 bp), D10Mit2 (117 bp, 122 bp), JH104 (178 bp, 181 bp) and U12235 (79 bp, 83 bp). Altogether six out of seven CRCs were studied (E410 could not be amplified) and shown to be microsatellite stable, since no differences in the fragment lengths were observed between the tumour and corresponding normal DNA (Supplementary Figure 3, available at Carcinogenesis Online) (13).
Mlh1 RNA expression was significantly decreased in normal mucosa of CRC mice
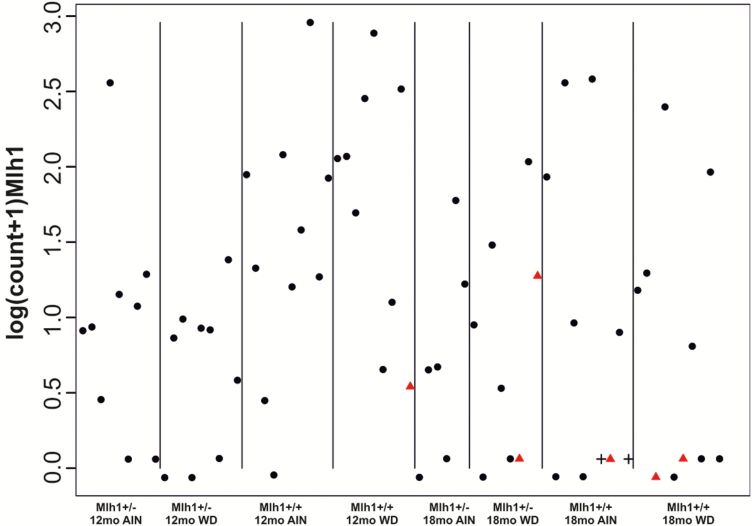
After finding that irrespective of the mouse genotype, functional Mlh1 was still expressed in carcinomas, our interest was to look for potential early drivers of tumorigenesis on a genome-wide scale. Altogether 71 out of 80 normal colon mucosa samples operated from 12- and 18-month old mice were qualified for genome-wide transcriptome analysis. Analysis was done with RNAseq using the single-cell tagged reverse transcription method (STRT) (23,29). The 21-month old mice were left out from the RNAseq study due to many health problems most probably because of their old age. Altogether 12 216 expressed transcripts were identified in the samples. First, we analyzed the Mlh1 gene expression levels from the STRT data (Figure 2). In our previous study (19), we showed that in the beginning of the feeding experiment (at time point 0), the Mlh1 heterozygote mice showed exactly 50% lower Mlh1 expression than the Mlh1+/+ mice. Contrary to varying Mlh1 expression levels in mice in general, 5/6 mice (E249, E314, E329, E333, E338) who developed carcinoma showed remarkably low Mlh1 expression in their normal colon mucosa (P = 0.03) (Figure 2) (13). The mouse E347 whose carcinoma had serrated histology had, however, higher Mlh1 expression level as non-carcinoma mice on average (Table 1; Figure 2).
Figure 2.
The Mlh1 mRNA expression in normal mucosa. The Mlh1 gene expression levels detected in individual mice at different time points, genotypes and diet groups, show that five out of six carcinoma mice (red triangle) show very low Mlh1 expression. Non-carcinoma mice E325 and E332 are shown with black crosses.
Expression profiles in normal mucosa formed a distinct cluster for CRC mice
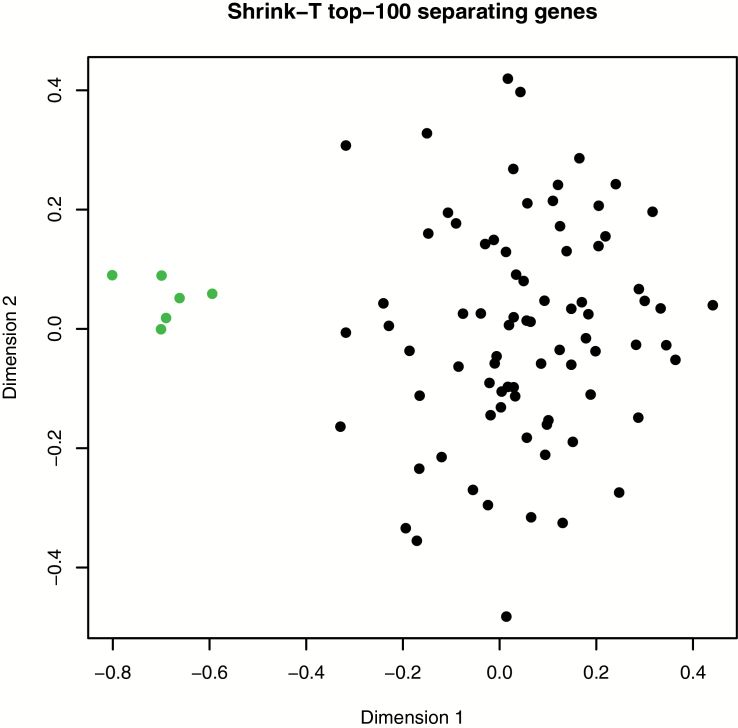
After finding that carcinoma mice had extremely low levels of Mlh1 transcripts in their mucosa, we next compared their genome-wide expression profiles with profiles of all other 12 and 18 months old mice. The normal mucosa expression profiles of the six CRC mice were strikingly different from the profiles of the other mice and formed a distinct cluster as visualized by an MDS plot created with the 100 most altered/differentially regulated genes (Figure 3, Supplementary Table 3, available at Carcinogenesis Online). Altogether 86% of the top 300 differentially regulated genes in CRC mice were down-regulated and 14% were up-regulated (Supplementary Table 3, available at Carcinogenesis Online) (13).
Figure 3.
Genome wide expression profiles in normal colon mucosa. An MDS plot created with the 100 most differentially expressed genes between CRC (green) mice and mice without cancer (black). The expression profiles of the six mice which developed carcinoma up to 18 months of age form a distinct cluster in the plot.
Pathway analysis and shortage of chromosomal segregation gene-specific transcripts suggest problems in cell cycle regulation and mitosis
To further understand the biological functions and pathways enriched among the top separating genes in CRC mice, the expression data were analysed with Ingenuity Pathway Analysis. According to Ingenuity Pathway Analysis, chromosome segregation (P = 2.92 × 10–5), aneuploidy of fibroblasts (P = 5.31 × 10–4), checkpoint control (P = 1.10 × 10–4), DNA replication checkpoint (P = 1.88 × 10–4) and morphology of mitotic spindle (P = 6.45 × 10–5) were among the most affected biological functions. In network analysis, the most affected molecular and cellular functions included cell cycle (P = 9.24 × 10–5), cellular assembly and organization (P = 9.24 × 10–5), DNA replication, recombination and repair (P = 9.24 × 10–5), cell death and survival (P = 7.30 × 10–5) and cellular growth and proliferation (P = 3.07 × 10–3) (Supplementary Table 4, available at Carcinogenesis Online). The analysis was also repeated with different RNA-seq data preprocessing (all mouse samples without ComBat normalization, Supplementary Table 4, available at Carcinogenesis Online). These results confirmed our findings on chromosome segregation (P = 1.03 × 10–5), aneuploidy of fibroblasts (P = 4.57 × 10–4) and checkpoint control (P = 4.29 × 10–4) (13).
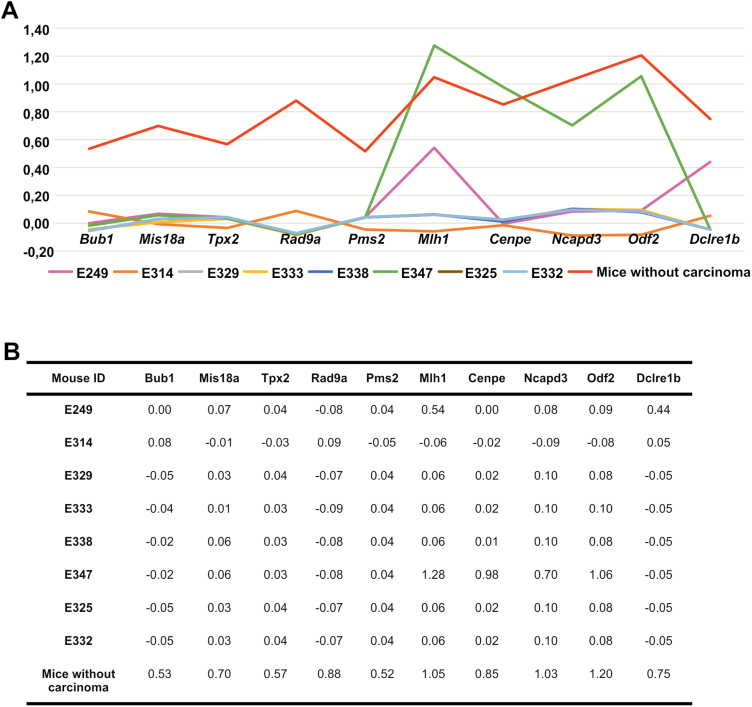
The Ingenuity Pathway Analysis results strongly indicated that there are severe problems in cell cycle regulation and mitosis already in colon mucosa. In the six mice who developed carcinoma up to 18 months, the most altered/differentially expressed genes that pointed to chromosome segregation and SAC were Bub1 (BUB1, mitotic checkpoint serine/threonine kinase), Mis18a (MIS18 kinetochore protein A), Tpx2 (TPX2 microtubule associated), Rad9a (RAD9 checkpoint clamp component A), Pms2 (postmeiotic segregation increased 2), Mlh1 (mutL homolog 1, along with MMR function also triggers checkpoint activation), Cenpe (centromere protein E), Ncapd3 (non-SMC condensing II complex subunit D3), Odf2 (outer dens fiber of sperm tails 2) and Dclre1b (DNA cross-link repair 1B). Five of these 10 genes, Bub1, Mis18a, Tpx2, Rad9a and Pms2, were strongly down regulated in all of the six carcinoma mice (Figure 4). Mlh1, Cenpe, Ncapd3, Odf2 and Dclre1b showed variable level of expression in two CRC mice (E347 and E249). In E249, Mlh1 and Dclr1b showed approximately 50% down regulation when compared to the average expression level in non-carcinoma mice (Figure 4). In E347, whose carcinoma was histologically different from the others and showed typical serrated phenotype, Cenpe, Ncapd3 and Odf2 expression levels were equal and Mlh1 expression level higher than in the non-carcinoma mice. Importantly, among all the 74 mice that did not develop colon carcinoma up to 18 months, only two mice, E325 and E332, showed similar low expression of all the 10 genes (Figure 4). Although, no colonic tumours were found in those mice, E325 had bloody feces and anemia, suggesting undefined mucosal pathology (13).
Figure 4.
The expression levels of 10 chromosomal segregation-specific genes in colon mucosa. The mRNA expression levels (gene expression values after ComBat processing) are described as (A) a line chart and as (B) expression values in the carcinoma mice (E249, E314, E329, E333, E338, E347), two mice with similar expression profiles with CRC mice (E325 and E332), and as the average levels of mice without cancer, including mice E325 and E332.
Abnormal mitoses and CIN in carcinomas
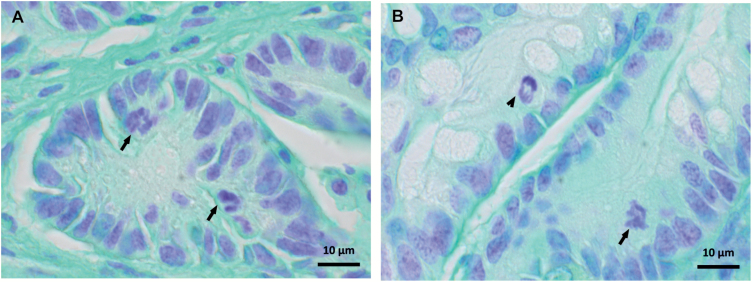
Undisturbed mitosis is a central requirement of the normal cell cycle and division. In cancer cells, mitoses are often aberrant, showing aneuploidy caused by unequal segregation of chromosomes and/or structural changes in chromosomes, both of which lead to CIN. To validate the RNA sequencing results, which suggested impaired cell cycle regulation and mitosis in CRC mice, all the 13 carcinomas were stained with feulgen and analyzed for mitotic aberrations. Although all the carcinomas were well-differentiated early cancers with limited submucosal invasion and relatively lenient cytological changes, they exhibited increased mitotic activity and abundant numbers of unbalanced/atypical mitoses in contrast to normal tissue samples (Figure 5) (13).
Figure 5.
Abnormal mitoses in mouse colon carcinomas. Representative pictures of abnormal mitoses (arrows) in (A) serrated adenocarcinoma (mouse E347) and (B) tubular adenocarcinoma (mouse E333), and a normal mitosis (arrow head).
Discussion
The instability status and other characteristics of colon tumours have been used to define the mechanisms behind colon cancer formation (3). Our present genome-wide expression profiling experiment demonstrates that cancer preceding changes occur and can be detected already in normal colon mucosa. We show that these changes may form a field-defect in histologically normal mucosa and trigger MMR-proficient, chromosomally unstable CRC both in Mlh1+/+ and Mlh1+/− mice.
One common event linked to both sporadic and inherited MSI cancers is the inactivation of MLH1, although the inactivation mechanism varies from genetic mutations in LS to epigenetic silencing in sporadic CRC (7,30). Our mouse study was designed to comprehensively clarify the role of Mlh1 expression during colon tumorigenesis and the effects of genetic predisposition and other risk factors such as WD and aging on it. Mlh1 protein expression was studied in colon tumours and Mlh1 gene expression in histologically normal mucosa. Approximately 70% of all tumours and 80% of colon carcinomas developed in WD-fed mice indicating a strong diet effect on cancer predisposition. 33 % of CRCs and 75% of adenomas and hyperplasias found in mice up to 18 months of age was found in Mlh1+/− mice. Surprisingly, Mlh1 protein was present and there was no MSI in their cancers. Genome-wide expression profiling of histologically normal mucosa however showed that 5/6 mice who developed CRC up to 18 months had significantly decreased mucosal Mlh1 RNA expression. Only in the carcinoma mouse E347 the Mlh1 expression level was higher than the average level of 65 mice without cancer.
Low Mlh1 expression, although a prominent signal, seemed not to be an absolute requirement or sufficient alone to cause colon cancer since several mice without CRC had low Mlh1 expression as well. In order to identify other genes and pathways involved in CRC development, we compared the genome wide expression profiles in the six CRC mice with the profiles of 65 mice without CRC (13). Remarkably, the expression profiles of CRC mice formed a clearly distinct cluster (Figure 3), indicating a field-defect in normal colon mucosa (31,32). By network analysis of top 100 CRC mice separating genes, Mlh1 expression in normal mucosa from CRC mice was found to be low together with significant down regulation of several cancer related genes and pathways (Supplementary Table 3 and Tables 4, available at Carcinogenesis Online) and especially of chromosomal segregation genes, Bub1, Mis18a, Tpx2, Rad9a, Pms2, Cenpe, Ncapd3, Odf2 and Dclre1b. Only two (E325 and E332) of the 65 mice without cancer shared the expression profile of the CRC mice related to chromosomal segregation (Figure 4). Although no colonic tumours were found in those mice, carcinogenesis might been happening in their mucosa. For example, E325 had bloody feces and anemia suggesting pathological problems in mucosa. Differing from the other CRC mice, E347, which did not show decrease in Mlh1 expression, showed decrease only in the expressions of Bub1, Mis18a, Tpx2, Rad9a, Pms2 and Dclre1b, suggesting their remarkable importance in serrated carcinogenesis. Furthermore, in the mouse E249 the Mlh1 and Dclre1b genes showed approximately 50% lower expression than was detected in the non-carcinoma mice on average. Here, the milder decrease may reflect young age of onset.
During cell division the SAC, which is the major target of mitotic alterations, maintains the genome stability by delaying cell division until all chromosomes are accurately aligned in the spindle (33). Aberrant expression of mitotic genes leads to mitotic abnormalities including centrosome defects and improper spindle checkpoint leading to CIN (34) and tumour formation in multiple tissues including colon (35). In this study, the mRNA expression was significantly decreased in five SAC associated genes, Mlh1, Bub1, Rad9a, Dclre1b and Cenpe. Of those, Bub1 is a major player and activator in SAC and its haploinsufficiency (heterozygosity) is known to be responsible for chromosome segregation defects and aneuploidy (35). During mitosis, Bub1 is required for the recruitment of other checkpoint and motor proteins, such as Cenpe, to a kinetochore (36). There is evidence suggesting that inaccurate chromosome segregation with causal implication of Bub1 deficiency drives tumorigenesis through tumour-suppressor gene LOH (37), perfectly in line with our findings that the majority of the carcinoma mice distinguishing genes were tumour suppressor genes, which were down regulated (Supplementary Table 3, available at Carcinogenesis Online). Cenpe, a kinesin-like motor protein which is an efficient stabilizer of microtubule capture at kinetochores and hence essential for metaphase chromosome alignment (38), was strongly down regulated in the mice with carcinoma. While it plays an important role in the movement of chromosomes toward the metaphase plate during mitosis, it is also necessary for the mitotic checkpoint signal at the kinetochore to prevent chromosome loss (39). Dclre1b has a central role in telomere maintenance and protection during S-phase through its 5′–3′ exonuclease activity. Moreover, in case of spindle stress, Dclre1b like Bub1 is involved in prophase checkpoint (40,41). RAD9A, a component of the 9-1-1 cell cycle checkpoint response complex, plays a major role in DNA repair and participates in multiple cell cycle checkpoints and apoptosis and its aberrant expression has been linked to tumorigenesis of multiple tissues (42). Interestingly, Rad9 also physically interacts with the MMR protein MLH1 (43). The MMR mechanism is so essential for normal cell function that it may explain why even a small amount of MLH1 appears to be sufficient for MMR function, whereas its checkpoint activation role seems to require a full complement of the protein (44). It has been argued that the MLH1 heterozygosity/haploinsufficiency may drive the development of cancer by accumulation of insertion/deletion mutations in other gatekeeper genes prior to MSI (45). Indeed, cells with diminished amount of MLH1 protein may still be MMR proficient, although they show defects in DNA damage signalling (44). Consequently, the damaged cells may not activate cell cycle checkpoints and enter apoptosis. Our observation that low mRNA expression of Mlh1 in carcinoma mice together with down regulation of several other genes related to chromosome segregation and checkpoint control supports the proposition that already decreased amount of Mlh1, when MMR is still functional, may have an important role in tumorigenesis (13).
Low expression of Tpx2, Mis18a, Ncapd3 and Odf2 reflects problems in formation of the nuclear spindle and chromosome segregation. Tpx2, Ncpd3 and Odf2, a general scaffolding protein (46), are all involved in microtubules related processes in spindle formation.Tpx2 plays a role in microtubule organization and is involved in centrosome maturation (47). In fact, TPX2-depleted cells fail to form proper mitotic spindles (48). Recent findings suggest that TPX2 also plays an important role in promoting colon tumorigenesis (49). In this study results support a driver role for Tpx2, since it was strongly down regulated in colon mucosa in all carcinoma mice. Ncapd3 functions in the condensin II complex and is needed to establish the chromosomal architecture necessary for proper spindle assembly and chromosome segregation. Chromosome condensation and resolution are compromised when condensin is depleted (50). The MIS18 complex accumulates at the centromere during anaphase to early G1 phase, slightly ahead of the histone H3 variant CENPA and is an absolute requirement for the localization of CENPA at centromeres. Importantly, Mis18a knockout causes severe chromosomal missegregation, lack of CENPA and ultimately cell death (51). Here, along with Mis18a, Cenpa was significantly down regulated in the normal colon mucosa of the CRC mice (Supplementary Table 3, available at Carcinogenesis Online) supporting the finding of improper chromosome segregation.
Our RNA sequencing results suggest that cancer preceding changes occur already in histologically normal mucosa and trigger MMR-proficient chromosomally unstable CRC. Problems in cell cycle regulation and mitosis were confirmed by tumour analysis, which showed increased mitotic activity and abundant numbers of atypical mitoses in carcinomas. Altogether 71 mice were included in genome-wide expression profiling and only two of the 65 mice without cancer shared the expression profile of the CRC mice related to chromosomal segregation, indicating that the aberrant expression of this gene set could be used as signalling carcinogenesis in colon mucosa. In addition, the finding of unexpected tumour phenotypes in Mlh1+/− mice implies that stable microsatellites and normal MLH1 expression in human tumours may not necessarily exclude inherited MLH1 deficiency.
Supplementary material
Supplementary data can be found at Carcinogenesis online.
Funding
This study was supported by grants from Jane and Aatos Erkko Foundation (M.N.), and Helsinki Institute of Life Science (P.T.).
Acknowledgements
The authors wish to thank Laura Sarantaus for help in planning the feeding study and sample collection, Satu Valo and Denis Dermadi-Bebek for help in sample collection, Marja Mutanen for help with the diet planning, Mikael Niku for help with immunohistochemistry, Liisa Kauppi for help in planning the MSI markers and Ingegerd Fransson for help in the sequencing library preparation.
Conflict of Interest Statement: None declared.
Abbreviations
- CRC
colorectal cancer
- CIN
chromosomal instability
- LOH
loss of heterozygosity
- LS
Lynch syndrome
- MDS
multidimensional Scaling
- MSI
microsatellite instability
- MMR
mismatch repair
- RNA-seq
RNA-sequencing method
- SAC
spindle assembly checkpoint
- STRT
single-cell tagged reverse transcription
- WD
Western-style diet
References
- 1. Bingham S., et al. (2004)Diet and cancer–the European prospective investigation into cancer and nutrition. Nat. Rev. Cancer, 4, 206–215. [DOI] [PubMed] [Google Scholar]
- 2. Fearon E.R. (2011)Molecular genetics of colorectal cancer. Annu. Rev. Pathol., 6, 479–507. [DOI] [PubMed] [Google Scholar]
- 3. Abbas T., et al. (2013)Genomic instability in cancer. Cold Spring Harb. Perspect. Biol., 5, a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Sohaily S., et al. (2012)Molecular pathways in colorectal cancer. J. Gastroenterol. Hepatol., 27, 1423–1431. [DOI] [PubMed] [Google Scholar]
- 5. Pino M.S., et al. (2010)The chromosomal instability pathway in colon cancer. Gastroenterology, 138, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colussi D., et al. (2013)Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int. J. Mol. Sci., 14, 16365–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boland C.R., et al. (2010)Microsatellite instability in colorectal cancer. Gastroenterology, 138, 2073–2087.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parsons M.T., et al. (2012)Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J. Med. Genet., 49, 151–157. [DOI] [PubMed] [Google Scholar]
- 9. Ahuja N., et al. (2000)Aging, methylation and cancer. Histol. Histopathol., 15, 835–842. [DOI] [PubMed] [Google Scholar]
- 10. Thompson B.A., et al. (2014)Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet., 46, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valo S., et al. (2015)DNA hypermethylation appears early and shows increased frequency with dysplasia in Lynch syndrome-associated colorectal adenomas and carcinomas. Clin. Epigenetics, 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goel A., et al. (2003)Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res., 63, 1608–1614. [PubMed] [Google Scholar]
- 13. Pussila M. (2017)Cancer-Preceding Gene Expression Changes in Mouse Colon Mucosa. University of Helsinki, Helsinki. [Google Scholar]
- 14. Newmark H.L., et al. (2009)Western-style diet-induced colonic tumors and their modulation by calcium and vitamin D in C57Bl/6 mice: a preclinical model for human sporadic colon cancer. Carcinogenesis, 30, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newmark H.L., et al. (2001)A Western-style diet induces benign and malignant neoplasms in the colon of normal C57Bl/6 mice. Carcinogenesis, 22, 1871–1875. [DOI] [PubMed] [Google Scholar]
- 16. Yang K., et al. (2008)Dietary induction of colonic tumors in a mouse model of sporadic colon cancer. Cancer Res., 68, 7803–7810. [DOI] [PubMed] [Google Scholar]
- 17. Yang K., et al. (2005)Dietary components modify gene expression: implications for carcinogenesis. J. Nutr., 135, 2710–2714. [DOI] [PubMed] [Google Scholar]
- 18. Edelmann W., et al. (1996)Meiotic pachytene arrest in MLH1-deficient mice. Cell, 85, 1125–1134. [DOI] [PubMed] [Google Scholar]
- 19. Pussila M., et al. (2013)Cancer-predicting gene expression changes in colonic mucosa of Western diet fed Mlh1+/- mice. PLoS One, 8, e76865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reeves P.G., et al. (1993)AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr., 123, 1939–1951. [DOI] [PubMed] [Google Scholar]
- 21. Washington M.K., et al. (2013)Pathology of rodent models of intestinal cancer: progress report and recommendations. Gastroenterology, 144, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Islam S., et al. (2012)Highly multiplexed and strand-specific single-cell RNA 5’ end sequencing. Nat. Protoc., 7, 813–828. [DOI] [PubMed] [Google Scholar]
- 23. Krjutškov K., et al. (2016)Single-cell transcriptome analysis of endometrial tissue. Hum. Reprod., 31, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., et al. (2010)Differential expression analysis for sequence count data. Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mishra P., et al. (2014)Gene set analysis: limitations in popular existing methods and proposed improvements. Bioinformatics, 30, 2747–2756. [DOI] [PubMed] [Google Scholar]
- 26. Ritchie M.E., et al. (2015)Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edelmann W., et al. (1999)Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res., 59, 1301–1307. [PubMed] [Google Scholar]
- 28. Kauppi L., et al. (2009)Analysis of meiotic recombination products from human sperm. Methods Mol. Biol., 557, 323–355. [DOI] [PubMed] [Google Scholar]
- 29. Islam S., et al. (2011)Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res., 21, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ollikainen M., et al. (2007)Mechanisms of inactivation of MLH1 in hereditary nonpolyposis colorectal carcinoma: a novel approach. Oncogene, 26, 4541–4549. [DOI] [PubMed] [Google Scholar]
- 31. Park S.K., et al. (2016)Field cancerization in sporadic colon cancer. Gut Liver, 10, 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hawthorn L., et al. (2014)Evidence for field effect cancerization in colorectal cancer. Genomics, 103, 211–221. [DOI] [PubMed] [Google Scholar]
- 33. Lara-Gonzalez P., et al. (2012)The spindle assembly checkpoint. Curr. Biol., 22, R966–R980. [DOI] [PubMed] [Google Scholar]
- 34. Perez de Castro I., et al. (2007)A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis, 28, 899–912. [DOI] [PubMed] [Google Scholar]
- 35. de Voer R.M., et al. (2013)Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are risk factors for colorectal cancer. Gastroenterology, 145, 544–547. [DOI] [PubMed] [Google Scholar]
- 36. Johnson V.L., et al. (2004)Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci., 117(Pt 8), 1577–1589. [DOI] [PubMed] [Google Scholar]
- 37. Sotillo R., et al. (2009)Very CIN-ful: whole chromosome instability promotes tumor suppressor loss of heterozygosity. Cancer Cell, 16, 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Putkey F.R., et al. (2002)Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell, 3, 351–365. [DOI] [PubMed] [Google Scholar]
- 39. Weaver B.A., et al. (2003)Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol., 162, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L., et al. (2009)SNM1B/Apollo interacts with astrin and is required for the prophase cell cycle checkpoint. Cell Cycle, 8, 628–638. [DOI] [PubMed] [Google Scholar]
- 41. Yan Y., et al. (2010)The multifunctional SNM1 gene family: not just nucleases. Future Oncol., 6, 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lieberman H.B., et al. (2011)The role of RAD9 in tumorigenesis. J. Mol. Cell Biol., 3, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He W., et al. (2008)Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res., 36, 6406–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cejka P., et al. (2003)Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J., 22, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L., et al. (2012)Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res., 22, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ishikawa H., et al. (2005)Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol., 7, 517–524. [DOI] [PubMed] [Google Scholar]
- 47. Karsenti E., et al. (2001)The mitotic spindle: a self-made machine. Science, 294, 543–547. [DOI] [PubMed] [Google Scholar]
- 48. De Luca M., et al. (2006)A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle, 5, 296–303. [DOI] [PubMed] [Google Scholar]
- 49. Wei P., et al. (2013)TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J. Transl. Med., 11, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wignall S.M., et al. (2003)The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J. Cell Biol., 161, 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baumann K. (2012)Keeping centromeric identity. Nat. Rev. Mol. Cell Biol., 13, 340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.