Abstract
Background
The aim of this study was to investigate the prognostic relevance of CEP55 in lung cancer (LC).
Material/Methods
LC microarray profile GSE30219 was obtained from the GEO database. The 2-sample t test was performed to clarify the difference in CEP55 expression between LC and normal lung tissue. The chi-square test and logistic regression analysis were preformed to investigate the relationship between CEP55 expression and the clinical features of LC patients. Log-rank test and Cox proportional hazards regression analysis were conducted to evaluate the disease-free survival (DFS) and overall survival (OS) of LC patients. Gene set enrichment analysis was conducted to investigate the related mechanisms.
Results
CEP55 was significantly increased in LC cells relative to normal lung tissues (P<0.0001). Univariate and multivariate correlation analyses demonstrated that CEP55 expression was associated with advanced T and N staging of LC (P<0.0001). Survival analyses indicated that CEP55 expression was an independent risk factor for DFS (HR: 1.515, 95% CI: 1.277–1.797, P<0.0001) and OS (HR: 1.436, 95% CI: 1.278–1.615). CEP55 might affect the proliferation of LC cells through Myc signaling, DNA repair, and G2M checkpoint.
Conclusions
Our results indicated that CEP55 was increased in LC cells and was associated with poor clinical outcomes of LC patients, and could be a prognostic biomarker for LC.
MeSH Keywords: Data Collection, Lung Neoplasms, Prognosis, Survival Analysis
Background
Lung cancer (LC), a malignant lung tumor characterized by uncontrolled cell growth in tissues of the lung, is one of the most common causes of cancer-related death worldwide, affecting both males and females [1,2]. LCs are divided into 2 subtypes according to histological characteristics: non-small cell lung cancer (NSCLC) represents 80–85% of all LCs, and the remaining 15–20% of cases are small cell lung cancers (SCLCs). NSCLCs are usually classified into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Given that most of these NSCLC cases are diagnosed at late stages (stage III/IV), the median survival has been reported to be 18 months in patients with NSCLC. SCLC is another more aggressive form of LC, and the 5-year overall survival of SCLC patients is only about 5% [3–6]. With advances in pathologic and molecular studies of LC, early detection, diagnosis, and treatment are of great significance for the management of LC [7,8]. Hence, identification of molecules that are correlated with the clinical features of LC patients might be helpful in the management of LC.
Centrosomal protein 55 (CEP55), also known as FLJ10540, C10orf3, or URCC6, is an important centrosomal protein for midbody formation in cytokinesis [9]. Previous studies demonstrated that CEP55 is involved in the cell cycle and the PI3K/AKT signaling pathway, and therefore is associated with pathological processes of several human malignances [10,11]. Liu et al. demonstrated that knockdown of CEP55 significantly inhibited proliferation and promoted apoptosis of human LC cells [12], but the correlation between CEP55 expression and clinical outcome of patients with LC remains unclear. Therefore, we conducted correlation analyses between CEP55 expression and clinical features of patients with LC to determine the clinical significance of CEP55 in LC.
Material and Methods
LC microarray
We selected the LC microarray GSE30219 [13], obtained from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database, to conduct our analysis. GSE30219 comprised 293 LC samples and 14 non-tumoral lung samples and was annotated with the Affymetrix Human Genome U133 Plus 2.0 Array. Detailed clinical information including age, sex, histology type, and TNM staging of LC patients were also included.
Statistical analysis
Firstly, mRNA levels of CEP55 (the corresponding probe ID is 218542_at in GSE30219) were assessed between LC patients and normal controls. The 2-sample t test was performed to investigate whether the differences reached statistical significance. LC samples in GSE30219 were divided into 2 subgroups (a CEP55 low-expression group and a CEP55 high-expression group) according to the median of CEP55 expression in this study. Then, we used the chi-square test and logistic regression to investigate the relationship between the expression of CEP55 and clinical features (age, sex, histological type, and TNM staging) of patients with LC. We used the log-rank test to evaluate the disease-free survival (DFS) and overall survival (OS) in these 2 groups, and Kaplan-Meier plots of DFS and OS were generated for LC patients in the CEP55 low-expression group and CEP55 high-expression group. The OS and DFS were calculated as previously reported [10]. Multivariate Cox proportional hazards models were used to determine the associations of various demographic and tumor characteristics with DFS and OS. If the p value was 0.05 or lower, the result was regarded as statistically significant for the above statistical analyses. Furthermore, we conducted gene set enrichment analysis (GSEA) [14,15] based on samples in the CEP55 low-expression group and CEP55 high-expression group to identify relevant molecular mechanisms affecting the progression of LC. The number of permutations was 1000, and h.all.v6.0.symbols were used as a reference gene sets database. Gene sets at Nominal P value less than 0.05 and false discovery rate less than 25% were considered statistically significant.
Results
CEP55 was increased in LC cells
Firstly, the mRNA levels of CEP55 in LC samples and normal lung tissues were analyzed. As shown in Figure 1, CEP55 was significantly upregulated in LC cells (n=293) compared to non-tumoral lung tissues (n=14) (7.458±1.526 vs. 4.538±0.872, P<0.0001). This result indicates that CEP55 might promote the carcinogenesis of LC.
Figure 1.
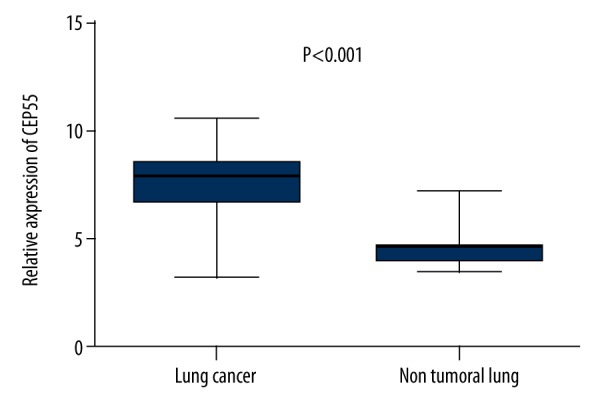
The mRNA level of CEP55 was significantly increased in lung cancer.
CEP55 was associated with worse demographic and tumor characteristics of LC
Secondly, we investigated the associations between CEP55 expression and the age, sex, histological type, and TNM staging of patients with LC. Univariate analysis (chi-square test) suggested that more patients in the CEP55 low-expression group had NSCLC rather than SCLC (P=0.002), and CEP55 expression was demonstrated to be associated with advanced T and N stages of LC (P<0.0001) (Table 1). No significant associations were found between age and sex distribution and CEP55 expression. Owing to a lack of significant correlations between CEP55 expression and the age and sex distribution of LC patients, we included the histological type, T staging, N staging, and M staging in a logistic regression model. The results of logistic regression indicated that patients in the CEP55 low-expression group had better T and N stages (odds ratio (OR) for T staging: 0.51, 95% CI: 0.281–0.926; OR for N staging: 0.351, 95% CI: 0.18–0.685) (Table 1).
Table 1.
Univariate and multivariable analyses of the relation between CEP55 expression and clinical characteristics of patients with lung cancer.
| Clinical characteristics | CEP55 expression | Univariate analysis | Multivariable analysis | ||||
|---|---|---|---|---|---|---|---|
| High (n=146) | Low (n=147) | Chi-square | P value | P value | OR | 95% CI | |
| Age (year) | n~% | ||||||
| ≤60 | 59 (40.4) | 71 (48.3) | 1.997 | 0.158 | – | – | – |
| >60 | 87 (59.6) | 75 (51.0) | |||||
| Gender | n~% | ||||||
| Female | 15 (10.3) | 28 (19.0) | 4.053 | 0.34 | – | – | – |
| Male | 131 (89.7) | 119 (81.0) | |||||
| Histology | n~% | ||||||
| NSCLC | 99 (67.8) | 142 (96.6) | 9.67 | 0.002 | 0.158 | 2.694 | 0.682–10.645 |
| SCLC | 16 (11.0) | 5 (3.4) | |||||
| T staging | n~% | ||||||
| T1 | 63 (43.2) | 103 (70.1) | 23.528 | P<0.0001 | 0.027 | 0.51 | 0.281–0.926 |
| T2–T4 | 81 (55.5) | 40 (27.2) | |||||
| N staging | n~% | ||||||
| N0 | 78 (53.4) | 120 (81.6) | 28.788 | P<0.0001 | 0.002 | 0.351 | 0.18–0.685 |
| N1 | 68 (46.6) | 25 (17.0) | |||||
| M staging | n~% | ||||||
| M0 | 140 (95.9) | 142 (96.6) | 0.514 | 0.473 | 0.39 | 2.4 | 0.326–17.661 |
| M1 | 5 (3.4) | 3 (2.0) | |||||
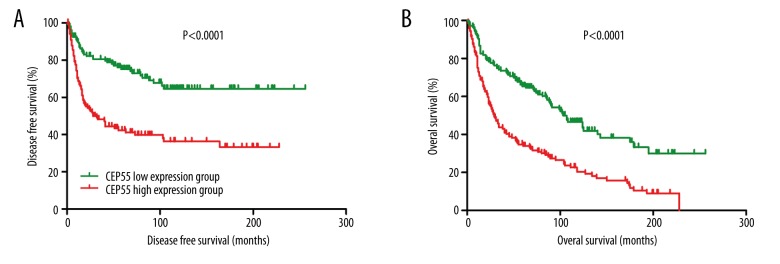
The survival of patients in the CEP55 low-expression group was better than in the CEP55 high-expression group
Thirdly, we investigated the associations between the DFS and OS of LC patients and CEP55 expression. As shown in Figure 2, both DFS and OS were better in patients in the CEP55 low-expression group compared with those in the CEP55 high-expression group (HR for DFS: 0.3666, 95% CI: 0.2515–0.5344, P<0.0001; HR for OS: 2.272, 95% CI: 1.705–3.028, P<0.0001). The results of multivariate Cox proportional hazards analyses indicated that CEP55 expression was an independent risk factor for DFS (HR: 1.515, 95% CI: 1.277–1.797, P<0.0001) and OS (HR: 1.436, 95% CI: 1.278–1.615) (Table 2).
Figure 2.
Disease-free survival (A) and overall survival (B) of lung cancer patients in the CEP55 low-expression group and CEP55 high-expression group.
Table 2.
Multivariable Cox regression analysis on disease free survival (DFS) and overall survival (OS).
| Variables | DFS | OS | ||
|---|---|---|---|---|
| P value | OR (95% CI) | P value | OR (95% CI) | |
| T staging | 0.001 | 0.449 (0.278–0.725) | 0.006 | 0.617 (0.438–0.869) |
| N staging | 0.073 | 0.647 (0.402–1.041) | 0.198 | 0.791 (0.553–1.131) |
| M staging | 0.215 | 0.523 (0.188–1.456) | 0.274 | 0.599 (0.239–1.501) |
| Histological type | 0.431 | 1.316 (0.665–2.607) | 0.142 | 1.525 (0.868–2.680) |
| CEP55 expression | <0.0001 | 1.515 (1.277–1.797) | <0.0001 | 1.436 (1.278–1.615) |
DFS – disease free survival; OS – overall survival.
Gene sets enriched in the CEP55 high-expression group
Finally, we conducted GSEA based on the median of CEP55 expression in GSE3029. As shown in Table 3, LC samples in the CEP55 high-expression group were significantly enriched in Myc signaling (nominal P=0.019, FDR=15.2%), DNA repair (nominal P=0.047, FDR=20.5%), and G2M checkpoint (nominal P=0.034, FDR=19.6%). These results indicate that CEP55 might affect LC cell growth through the above biological processes.
Table 3.
Gene sets enriched in CEP55 high expression group.
| Gene set | ES | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| Myc targets_v1 | −0.58652 | −1.73688 | 0.019342 | 0.152046 |
| Myc targets_v2 | −0.73024 | −1.68969 | 0.009804 | 0.1162 |
| DNA repair | −0.4043 | −1.58924 | 0.046875 | 0.205288 |
| G2M checkpoint | −0.72575 | −1.56384 | 0.033531 | 0.196249 |
ES – enrichment score; NES – normalized enrichment score; NOM p-val – nominal p value; FDR q-val – false discovery rate q value.
Discussion
Because most LC patients were diagnosed at late stages, and the disease becomes metastatic and drug-resistant at the early stage of treatment [16,17], the clinical outcomes of LC remain poor.
We found that CEP55 was increased in LC cells, higher expression of CEP55 was associated with poor T staging and N staging, and LC patients in the CEP55 high-expression group has worse OS and DFS compared with those in the CEP55 low-expression group. GSEA results proved that CEP55 affected the growth of LC cells through Myc signaling, DNA repair, and G2M checkpoint.
Centrosomal protein 55 (CEP55) has been identified as a microtubule-bundling protein required for cytokinesis [18]. The emerging roles of CEP55 in tumorigenesis have been recognized in multiple human cancers [9]. CEP55 was demonstrated to be increased in muscle-invasive bladder cancer relative to that in non-muscle-invasive bladder cancer, making it a diagnostic biomarker for bladder cancer [19]. Waseem et al. suggested that CEP55 accompanied with other molecules (FOXM1 and HELLS) could be used as a biomarker set for early diagnosis of head and neck squamous cell carcinoma [20]. CEP55 was demonstrated to be increased in breast cancer as compared to that in normal breast tissues, and knockdown of CEP55 can induce cycle arrest and apoptosis [21]. Moreover, cell viability assays demonstrated that knockdown of CEP55 inhibited proliferation, induced cell cycle arrest, and promoted apoptosis of human LC cells [22]. Therefore, the increased expression and carcinogenesis property of CEP55 in LC cells are understandable.
TNM staging refers to the invasiveness and metastasis of human cancers (including LC) [23,24]. For LC, patients were often classified into NSCLC and SCLC due to differences in clinical outcomes of LC patients. Patients with higher TNM stages often have poor treatment efficacy and clinical outcomes. Although our univariate and multivariable analyses found no significant correlations between CEP55 expression and histological types of LC, patients in the CEP55 high-expression group had significantly more aggressive T stages and N stages, indicating that CEP55 might promote the invasiveness and migration of LC cells regardless of the histological types. In accordance with these conclusions, log-rank test and Cox proportional hazards model revealed that LC patients in the CEP55 low-expression group had better DFS and OS compared to those in the CEP55 high-expression group. These results indicate that CEP55 might be an independent risk factor and diagnostic marker for LC.
This was a retrospective analysis of gene expression data and clinical information of 293 lung cancer patients and 14 normal controls. Thus, future studies using in vivo and in vitro experiments, as well as prospective clinical trials, are required to verify our conclusions.
Conclusions
Our results indicate that CEP55 is increased in LC cells, is associated with poor clinical outcomes of LC patients, and could be a prognostic biomarker for LC.
Footnotes
Source of support: Departmental sources
References
- 1.Dhanasopon AP, Kim AW. Lung cancer screening and its impact on surgical volume. Surg Clin North Am. 2017;97(4):751–62. doi: 10.1016/j.suc.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman RM, Sanchez R. Lung cancer screening. Med Clin North Am. 2017;101(4):769–85. doi: 10.1016/j.mcna.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inamura K. Lung cancer: Understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193. doi: 10.3389/fonc.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mari-Alexandre J, Diaz-Lagares A, Villalba M, et al. Translating cancer epigenomics into the clinic: Focus on lung cancer. Transl Res. 2017;189:76–92. doi: 10.1016/j.trsl.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Imai H, Kaira K, Minato K. Clinical significance of post-progression survival in lung cancer. Thorac Cancer. 2017;8(5):379–86. doi: 10.1111/1759-7714.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waqar SN, Morgensztern D. Treatment advances in small cell lung cancer (SCLC) Pharmacol Ther. 2017;180:16–23. doi: 10.1016/j.pharmthera.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Chen X, Wei K, et al. Identification of key transcription factors associated with lung squamous cell carcinoma. Med Sci Monit. 2018;24:172–206. doi: 10.12659/MSM.898297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Zhang W, Guo N, et al. Expression of molecular markers in primary sites and metastatic lymph nodes of lung cancer patients. Med Sci Monit. 2018;24:513–20. doi: 10.12659/MSM.898688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffery J, Sinha D, Srihari S, et al. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35(6):683–90. doi: 10.1038/onc.2015.128. [DOI] [PubMed] [Google Scholar]
- 10.Jeffery J, Neyt C, Moore W, et al. Cep55 regulates embryonic growth and development by promoting Akt stability in zebrafish. FASEB J. 2015;29(5):1999–2009. doi: 10.1096/fj.14-265090. [DOI] [PubMed] [Google Scholar]
- 11.Xu ZY, Ma XS, Qi ST, et al. Cep55 regulates spindle organization and cell cycle progression in meiotic oocyte. Sci Rep. 2015;5:16978. doi: 10.1038/srep16978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Mei Q, Zhao J, et al. Suppression of CEP55 reduces cell viability and induces apoptosis in human lung cancer. Oncol Rep. 2016;36(4):1939–45. doi: 10.3892/or.2016.5059. [DOI] [PubMed] [Google Scholar]
- 13.Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis – prone lung cancers. Sci Transl Med. 2013;5(186):186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Chalela R, Curull V, Enríquez C, et al. Lung adenocarcinoma: From molecular basis to genome – guided therapy and immunotherapy. J Thorac Dis. 2017;9(7):2142–58. doi: 10.21037/jtd.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai H, Kaira K, Minato K. Clinical significance of post-progression survival in lung cancer. Thorac Cancer. 2017;8(5):379–86. doi: 10.1111/1759-7714.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Rajendran V, Sethumadhavan R, Purohit R. CEP proteins: The knights of centrosome dynasty. Protoplasma. 2013;250(5):965–83. doi: 10.1007/s00709-013-0488-9. [DOI] [PubMed] [Google Scholar]
- 19.Singh PK, Srivastava AK, Rath SK, et al. Expression and clinical significance of Centrosomal protein 55 (CEP55) in human urinary bladder transitional cell carcinoma. Immunobiology. 2015;220(1):103–8. doi: 10.1016/j.imbio.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Waseem A, Ali M, Odell EW, et al. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46(7):536–42. doi: 10.1016/j.oraloncology.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Jin T, Dai X, Xu J. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends. 2016;10(1):67–73. doi: 10.5582/bst.2016.01010. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Mei Q, Zhao J, et al. Suppression of CEP55 reduces cell viability and induces apoptosis in human lung cancer. Oncol Rep. 2016;36(4):1939–45. doi: 10.3892/or.2016.5059. [DOI] [PubMed] [Google Scholar]
- 23.Kay FU, Kandathil A, Batra K, et al. Revisions to the tumor, node, metastasis staging of lung cancer (8th edition): Rationale, radiologic findings and clinical implications. World J Radiol. 2017;9(6):269–79. doi: 10.4329/wjr.v9.i6.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151(1):193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]