Abstract
The immune system responds vigorously to microbial infection, while permitting life-long colonization by the microbiome. Mechanisms that facilitate the establishment and stability of the gut microbiota remain poorly described. We discovered that a sensor/regulatory system in the prominent human commensal Bacteroides fragilis modulates its surface architecture to invite binding of immunoglobulin A (IgA). Specific immune recognition facilitated bacterial adherence to cultured intestinal epithelial cells and intimate association with the gut mucosal surface in vivo. The IgA response was required for B. fragilis, and other commensal species, to occupy a defined mucosal niche that mediated stable colonization of the gut through exclusion of exogenous competitors. Therefore, in addition to its role in pathogen clearance, we propose that IgA responses can be co-opted by the microbiome to engender robust host-microbial symbiosis.
Main Text
At birth, ecological and evolutionary processes commence to assemble a complex microbial consortium in the animal gut. Community composition of the adult human gut microbiome is remarkably stable during health, despite day-to-day variability in diet and diverse environmental exposures. Instability, or dysbiosis, may be involved in the etiology of a variety of immune, metabolic, and neurologic diseases (1, 2). Longitudinal sequencing studies indicate a majority of bacterial strains persist within an individual for years (3), and for most species there is a single, persistently dominant strain (4) (termed “single-strain stability”). Mucus and components of the innate and adaptive immune systems are thought to influence microbiome stability, independently of diet. For example, immunoglobulin A (IgA), the main antibody isotype secreted in the gut, shapes the composition of the intestinal microbiome via currently unknown mechanisms (5–8). IgA deficiency in mice increases inter-individual variability in the microbiome (9) and decreases diversity (10, 11). The direct effects of IgA on bacteria have largely been studied in the context of enteric infection by pathogens (12). However, early studies of IgA in the healthy gut found that the majority of live bacterial cells in feces are bound by IgA (13), reflecting a steady-state IgA response to persistent indigenous microbes (14). Studies show that IgA promotes adherence of commensal bacteria to tissue-cultured intestinal epithelial cells (15, 16), though the in vivo implications of this observation are unclear. Furthermore, lack of IgA, the most common human immunodeficiency, does not affect lifespan and only modestly increases susceptibility to respiratory and gastrointestinal infections (17), raising the question of why the immune system evolved to invest the considerable energy to produce several grams of IgA daily (18).
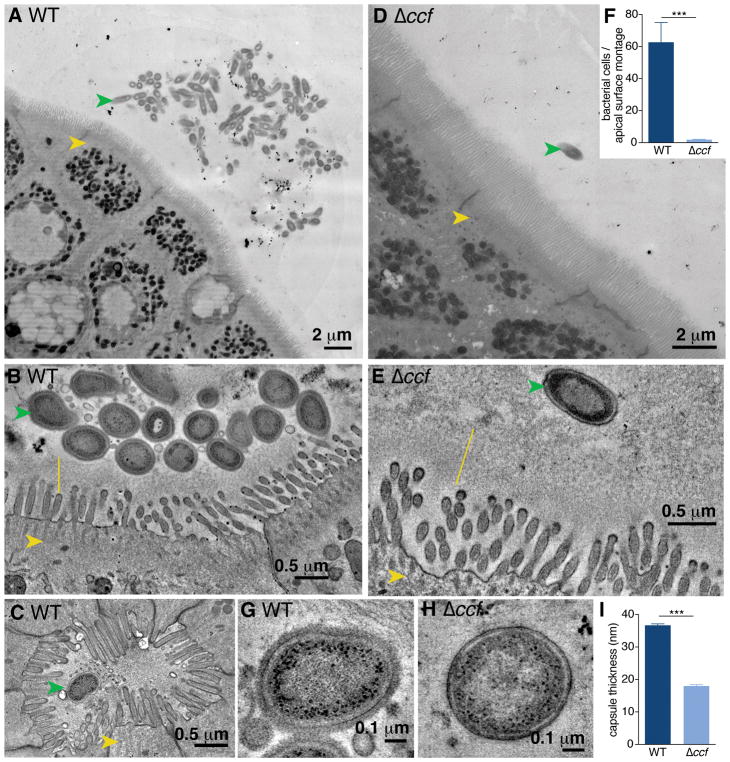
Bacteroides fragilis is an important member of the human gut microbiome, with beneficial properties that ameliorate inflammatory and behavioral symptoms in preclinical animal models (19–22). This commensal exhibits remarkable single-strain stability (23, 24) and enriched colonization of the gut mucosal surface (25). To explore physical features of B. fragilis interaction with the host epithelium, we used transmission electron microscopy (TEM) to visualize colonic tissues of mono-colonized mice. B. fragilis commonly formed discrete aggregates of tightly-packed cells on the apical epithelial surface (Fig. 1A) and penetrated the glycocalyx layer of transmembrane mucins, nearly contacting the microvilli (Fig. 1B and fig. S1A and B). Intact B. fragilis cells were also found nestled in the ducts of the crypts of Lieberkühn (Fig. 1C and S1C). We previously identified a genetic locus in B. fragilis, named the commensal colonization factors (ccfABCDE), which is necessary for colonization of colonic crypts (26). To assess how these genes affect bacterial localization to the mucosal surface, we mono-colonized mice with a ccfCDE (Δccf) mutant. By TEM, B. fragilis Δccf was only found as sparse, individual cells within the epithelial mucosa, excluded from contact with the glycocalyx (Fig. 1D and E), and never observed in aggregates as for wild-type bacteria (Fig. 1F). B. fragilis burden in the colon lumen was identical between strains (fig. S2A), suggesting that the CCF system is required specifically for bacterial aggregation within mucus.
Fig. 1. Bacteroides fragilis resides as aggregates on the colon epithelium in a CCF-dependent manner.
(A) Representative transmission electron microscopy (TEM) projection and (B) high-resolution tomogram of epithelial-associated wild-type B. fragilis in mono-colonized mice. Ascending colons of mice harbored aggregates of B. fragilis (green arrow) under non-pathogenic conditions, that made tight associations with the glycocalyx (yellow line) overlying intestinal epithelial cells (IECs, yellow arrow). (C) Tomogram of wild-type B. fragilis penetrating deep into the duct of a crypt of Lieberkühn. (D) Representative TEM projection image and (E) tomogram of epithelial-associated B. fragilis Δccf. The absence of the CCF system abrogated formation of bacterial aggregates and prevented intimate association with the glycocalyx. (n = 3 mice per group, about 1 mm epithelium scanned per mouse). (F) Quantification of bacterial cells per projection montage (A and D) of epithelial-associated bacteria (unpaired t test, n = 7, 8 images from 4 mice per group). (G and H) Tomogram of the bacterial surface of wild-type B. fragilis (G) in comparison to B. fragilis Δccf (H) revealed a thick fuzzy capsule for wild-type bacteria residing in the colons of mice. (I) Measurement of capsule thickness (unpaired t test, n = 10 cells from 3 mice per group) (*** p < 0.001).
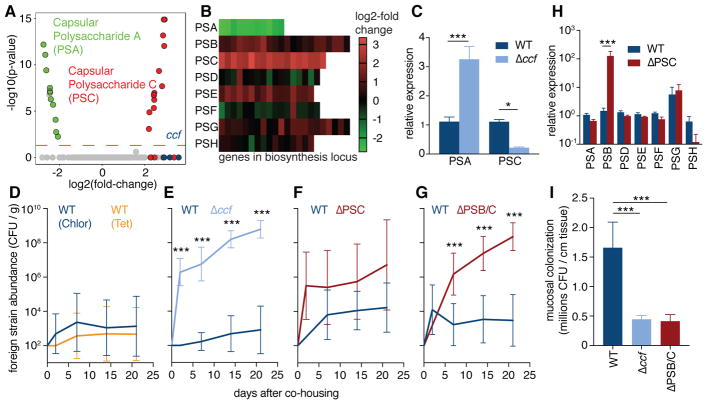
High-resolution tomograms of bacterial cells in vivo revealed the presence of a thick, fuzzy capsule layer covering wild-type B. fragilis (Fig. 1G), which was significantly reduced in B. fragilis Δccf (Fig. 1H and 1I). We sought to investigate the bacterial physiology underlying this ultrastructural change, and potential corresponding effects on colonization. The ccf locus is highly induced during gut colonization (26) and bacterial growth in mucin O-glycans (27), indicating the CCF system may sense a specific host-derived glycan. The ccf genes are homologous to polysaccharide utilization systems in which a sigma factor (ccfA) is activated by extracellular glycan sensing (28), thus we hypothesized that ccfA may activate genes involved in mucosal colonization. We overexpressed ccfA in B. fragilis and assessed global gene expression by RNAseq during in vitro growth (without overexpression ccf is poorly expressed in culture (26)). Of the non-ccf genes regulated by ccfA, 24 out of 25 genes mapped to the biosynthesis loci for capsular polysaccharides A and C (PSA and PSC) (Fig. 2A, 2B and Table S1). Correspondingly, ccf mutation decreased expression of PSC and increased expression of PSA in vivo (Fig. 2C). While phase variation of capsular polysaccharides is known to influence general in vivo fitness of B. fragilis (29, 30), these studies identify a pathway for transcriptional regulation of specific polysaccharides in the context of mucosal colonization.
Fig. 2. Specific capsular polysaccharides, regulated by ccf, are necessary for single-strain stability.
(A) RNAseq gene expression analysis of B. fragilis overexpressing ccfA during laboratory culture growth, relative to empty vector control (n = 3). Green symbols represent PSA genes; red symbols represent PSC genes; blue symbols represent ccf genes. (B) Heat map of expression levels for all capsular polysaccharide loci in B. fragilis following ccfA overexpression during growth in culture. (C) Relative expression using qRT-PCR (ΔΔCt normalized to gyrase) of RNA from colon lumen contents of mice mono-colonized with B. fragilis or B. fragilis Δccf (Sidak 2-way ANOVA, n = 4). (D–G) Abundance of foreign strains exchanged between pairs of co-housed mice each mono-colonized with the indicated strains, in colony forming units (CFU) per gram of feces (Sidak repeated measure 2-way ANOVA on log-transformed data, geometric mean and 95% CI, n = 9–12 pairs per plot). (H) Relative expression levels of capsular polysaccharides analyzed by qRT-PCR (ΔΔCt normalized to gyrase) of RNA from colon lumen contents of mice mono-colonized with B. fragilis or B. fragilis ΔPSC (Sidak 2-way ANOVA, n = 3, 4). (I) Plating of CFU from ascending colon mucus of mice mono-colonized with B. fragilis strains (Tukey ANOVA, n = 8) (* p < 0.05, ** p < 0.01, *** p < 0.001).
We modeled single-strain stability using a horizontal transmission assay, wherein co-housing animals respectively harboring isogenic strains of wild-type B. fragilis resulted in minimal strain transmission from one animal to another (Fig. 2D, S2A). This intra-species colonization resistance is provided through bacterial occupation of a species-specific nutrient or spatial niche (26). However, as previously reported (26), if mice are colonized initially with B. fragilis Δccf, animals were permissive to co-colonization by wild-type B. fragilis after co-housing (Fig. 2E, S2B), indicating a CCF-dependent defect in niche saturation. Mice harboring a mutant in the biosynthesis genes for PSC (ΔPSC) showed highly variable co-colonization by wild-type bacteria (Fig. 2F, S2C). Interestingly, we observed an unexpected increase in expression of the PSB biosynthesis genes in this mutant (Fig. 2H), which may compensate for the loss of PSC. We generated a strain defective in synthesizing both PSB and PSC (ΔPSB/C), and mice mono-associated with the double mutant were consistently unable to maintain colonization resistance (Fig. 2G, S2D–F), though the strain retained ccf expression (fig. S2G). Despite lack of competition in a mono-colonized setting and equal levels of colonization in the colon lumen (fig. S2H), the B. fragilis Δccf and ΔPSB/C strains were defective in colonization of the ascending colon mucus (Fig. 2I), reflecting impaired saturation of the mucosal niche. Accordingly, when we imaged the ΔPSB/C strain in vivo employing TEM, though the capsule was not as thin as in B. fragilis Δccf (fig. S2I and J), the hallmark epithelial aggregation phenotype was abrogated compared to wild-type bacteria (fig. S2K and L). Therefore, we conclude that the CCF system regulates capsule expression to mediate B. fragilis mucosal colonization and single strain stability.
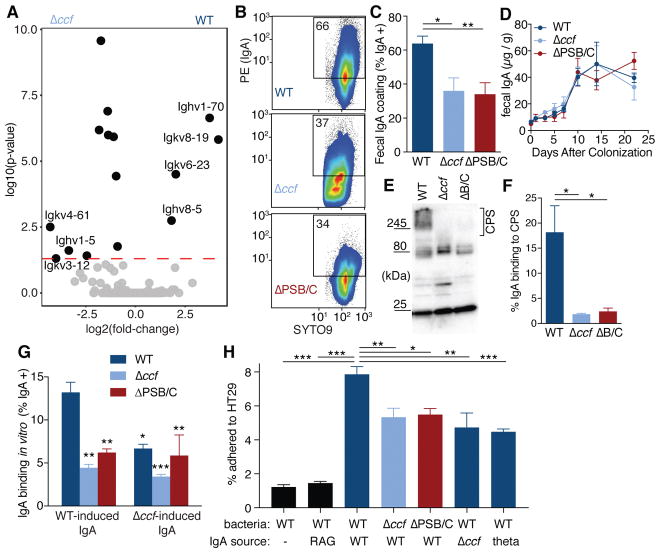
To investigate host responses contributing to mucosal colonization, we defined the transcriptome of the ascending colon during colonization with wild-type B. fragilis or B. fragilis Δccf. Remarkably, 7 of the 14 differentially expressed genes encode immunoglobulin variable chains (Fig. 3A and table S2). We did not observe any elevation of immune responses in Δccf–colonized mice (fig. S3A), indicating that changes in mucosal association are not caused by inflammation. Accordingly, we tested whether capsular polysaccharide regulation by ccf affects IgA recognition of bacteria (31–33). In fecal samples from mono-colonized animals, wild-type B. fragilis was highly coated with IgA, which was significantly diminished in Δccf and ΔPSB/C strains (Fig. 3B, 3C, and S4A). We observed no difference between these strains in the induction of total fecal IgA (Fig. 3D), reflecting equivalent stimulation of nonspecific IgA production (10, 34, 35). To test bacteria-specific responses, IgA extracted from feces of mice mono-colonized with B. fragilis was evaluated for binding to bacteria recovered from mono-colonized Rag1−/− mice (in vivo-adapted, yet IgA-free bacteria). Western blots of bacterial lysates showed that strong IgA reactivity to capsular polysaccharides was abrogated in the Δccf and ΔPSB/C strains (Fig. 3E and F). Although IgA can be polyreactive (10, 34, 35), binding to lysates of Bacteroides was species-specific (fig. S4B) and required induction of IgA following bacterial colonization (fig. S4C and D). Accordingly, in a whole bacteria binding assay, IgA induced by wild-type bacteria maximally coated wild-type B. fragilis compared with the Δccf and ΔPSB/C strains (Fig. 3G). IgA induced by B. fragilis Δccf exhibited reduced binding to wild-type bacteria (Fig. 3G). The addition of IgA to in vivo-adapted, IgA-free bacteria increased adherence of B. fragilis to intestinal epithelial cells in tissue culture (Fig. 3H), yet had no effect on bacterial viability (fig. S4E). Cell lines known to produce more mucus (36) exhibited a greater capacity for IgA-enhanced B. fragilis adherence (fig. S4F), consistent with prior work showing that IgA binds mucus (36–38). Importantly, IgA-enhanced adherence was decreased if targeted bacteria lack ccf or PSB/C, or if the IgA tested was induced by a ccf mutant or Bacteroides thetaiotaomicron (Fig. 3H and S4G). While pathogenic bacteria elaborate capsular polysaccharides for immune evasion, these results suggest B. fragilis deploys specific capsules for immune attraction, potentially enabling stable mucosal colonization.
Fig. 3. B. fragilis induces a specific IgA response, dependent on ccf regulation of surface capsular polysaccharides, which enhances epithelial adherence.
(A) RNAseq gene expression analysis of RNA recovered from whole ascending colon tissue of mice mono-colonized with B. fragilis or B. fragilis Δccf (n = 3). (B) Flow cytometry plots and (C) quantification of IgA coating of B. fragilis from feces of mice mono-colonized with various strains (Tukey ANOVA, n = 11–12). (D) ELISA for total fecal IgA in mono-colonized mice (Sidak repeated measure 2-way ANOVA, not significant, n = 4). (E) Bacterial lysates from feces of mono-colonized Rag1−/− mice probed in Western blots with fecal IgA from B. fragilis mono-colonized mice and (F) quantification of the proportional signal from IgA binding to capsular polysaccharides (CPS) (over 245 kDa) (Tukey ANOVA, n = 3 mice). (G) Binding of fecal IgA extracted from mono-colonized mice to various strains of B. fragilis. Source of IgA is mice colonized with either WT B. fragilis or B. fragilis Δccf. Because ccf is expressed in vivo, IgA-free bacteria from feces of mono-colonized Rag1−/− mice were used as the target for IgA binding (Tukey 2-way ANOVA, *significantly different from WT bacteria with WT IgA, n = 3). (H) In vitro epithelial cell adherence assay using IgA extracted from Swiss Webster mice (or Rag1−/−, second column) mono-colonized with B. fragilis or B. thetaiotaomicron (theta; last column). IgA-free but in vivo-adapted bacteria were isolated from mono-colonized Rag1−/− mice (Tukey ANOVA, n = 4 mice as the source of bacteria) (* p < 0.05, ** p < 0.01, *** p < 0.001).
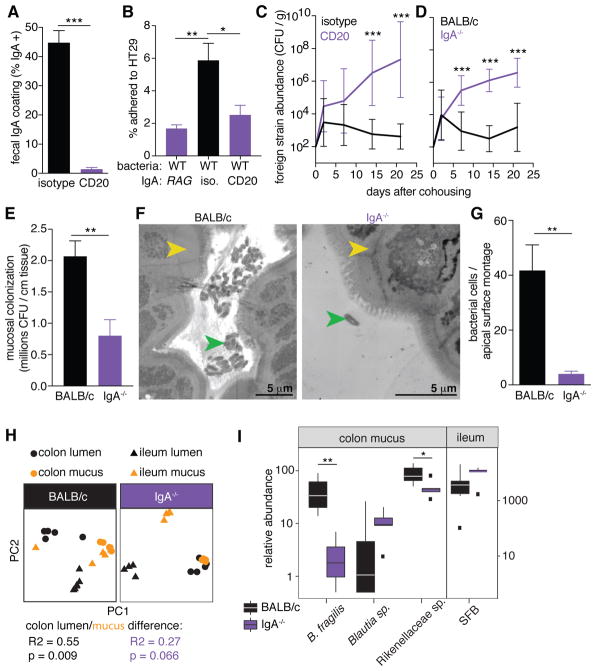
We determined whether IgA coating promotes B. fragilis colonization in mice. Using the horizontal transmission paradigm, Rag1−/− mice colonized with wild-type B. fragilis were readily co-colonized by an isogenic strain from a co-housed animal (fig. S5A and B), showing loss of colonization resistance in the absence of adaptive immunity. We next treated wild-type mice with an anti-CD20 antibody (fig. S5C) (39) to deplete B cells (fig. S5D–F), thus reducing total fecal IgA levels (fig. S5G) and eliminating IgA coating of wild-type B. fragilis during mono-colonization (Fig. 4A). IgA recovered from isotype control treated mice, also mono-colonized with B. fragilis, promoted adherence of wild-type bacteria to epithelial cells in vitro, while IgA from anti-CD20 treated mice had no effect despite being exposed to B. fragilis antigens (Fig. 4B). In the horizontal transmission assay, B cell depleted mice mono-colonized with B. fragilis were readily invaded by wild-type bacteria, while isotype control-injected animals retained colonization resistance (Fig. 4C and S5H). Therefore, active B cell responses to B. fragilis colonization enhance single-strain stability.
Fig. 4. IgA production in vivo is necessary for single-strain stability, mucosal colonization, and epithelial aggregation.
(A) IgA coating of wild-type B. fragilis in feces following injection of anti-CD20 or isotype control antibody (unpaired t test, n = 8). (B) Epithelial cell adherence assay of wild-type B. fragilis incubated with IgA extracted from indicated mono-colonized mice (Tukey ANOVA, n = 4 mice as the source of bacteria). (C) Abundance of foreign strains exchanged between pairs of wild-type B. fragilis mono-colonized mice treated with anti-CD20 or an isotype control (Sidak repeated measure 2-way ANOVA on log-transformed data, n = 10). (D) Foreign strains exchanged between pairs of BALB/c and BALB/c IgA−/− mice mono-colonized with wild-type B. fragilis (Sidak repeated measure 2-way ANOVA on log-transformed data, n = 9). (E) CFU plating of ascending colon mucus of wild-type and IgA−/− mice mono-colonized with wild-type B. fragilis (unpaired t test, n = 9). (F) Representative TEM projections of ascending colon (yellow arrow: epithelial cell) from mice mono-colonized with wild-type B. fragilis (green arrow) (n = 3 mice per group, about 1 mm epithelium scanned per mouse) and (G) quantification of bacterial cells per projection montage (unpaired t test, n = 7, 6 images from 3 mice per group) (H) Principle coordinate analyses of weighed UniFrac distances of 16S community profiles of ex-germ-free BALB/c and BALB/c IgA−/− mice transplanted with a complex mouse microbiota (Adonis test within colon for lumen/mucus difference). (I) Relative abundance of B. fragilis and highly IgA-coated ESVs in ex-germ-free mice (* p < 0.05, ** p < 0.01, *** p < 0.001).
As B cell depletion eliminates all antibody isotypes, germ-free IgA−/− mice (40) were generated and mono-colonized with B. fragilis. We did not observe compensatory coating by IgM (fig. S6A). In a horizontal transmission assay with wild-type (BALB/c) and IgA−/− mice, lack of IgA allowed co-colonization by challenge strains (Fig. 4D, S6B–D), indicating that IgA specifically contributes to single-strain stability. This feature was reproduced in mice with a full microbial community “spiked” with genetically marked B. fragilis strains (fig. S6E and F), revealing that single-strain stability of an individual bacterial species occurs in the context of a complex community. Mono-colonized IgA−/− mice harbored reduced levels of live bacteria in the colon mucus compared to wild-type mice (Fig. 4E), though they had greater numbers of bacteria in the colon lumen (fig. S6G). TEM images of ascending colon tissues reveal that in IgA−/− animals, wild-type B. fragilis failed to aggregate on the epithelial surface (Fig. 4F and 4G), similar to the ccf and PSB/C mutants in wild-type animals. B. fragilis cells also formed aggregates in feces in the presence of IgA (fig. S7), indicating that enhanced mucosal colonization may be due to increased aggregation or growth (41) within mucus. These findings converge to support a model whereby ccf regulates expression of specific capsular polysaccharides to attract IgA binding, allowing for robust mucosal colonization and single-strain stability.
Beyond B. fragilis, we tested whether IgA shapes a complex microbiome following controlled introduction of mouse microbiota to germ-free BALB/c or IgA−/− mice. One month following colonization, despite similar microbiome profiles in feces of both mouse genotypes (fig. S8A), we observed differences for specific taxa (Table S3). We also identified a defect in community stratification between the colonic mucus and lumen of IgA−/− mice (Fig. 4H and S8B), revealing that IgA is required to individualize microbiome profiles between these two anatomic locations. Remarkably, a highly mucus-enriched exact sequence variant (ESV), mapping uniquely to B. fragilis, was significantly decreased in the mucus of IgA−/− mice compared to BALB/c mice (Fig. 4I and S9A), naturally supporting our observations from mono-colonized mice. To extend this analysis to other microbial species, we identified Rikanellaceae, Blautia sp., and segmented filamentous bacteria (SFB) as being highly IgA-coated (fig. S9B) (35), and assessed the abundance of these taxa in the colonic or ileal mucus. Blautia sp. and segmented filamentous bacteria (SFB) displayed increased mucosal association in the absence of IgA (Fig. 4I) (42), demonstrating that IgA can protect the intestinal barrier. However, similar to B. fragilis, Rikanellaceae were highly abundant in colon mucus and significantly depleted in IgA−/− mice (Fig. 4I). We conclude that IgA-enhanced mucosal colonization occurs within complex communities for multiple strains of B. fragilis and other species of the gut microbiome.
Classically viewed, the immune system evolved to prevent microbial colonization. However, not only do animals tolerate a complex microbiome, in the case of B. fragilis provoking an immune response paradoxically enables intimate association with its mammalian host. Related commensal bacteria may also benefit from actively engaging IgA during symbiosis, as Rag2−/− mice devoid of adaptive immunity harbor fewer Bacteroides (43), and both B cell deficient and IgA−/− animals display decreased colonization by the Bacteroidaceae family (44). IgA has been previously shown to increase adherence of Escherichia coli (15), Bifidobacterium lactis, and Lactobacillus ramnosus (16) to tissue-cultured epithelial cells, suggesting that these microorganisms may also benefit from IgA to establish a mucosal bacterial community. Mucosal microbiome instability or loss of immunomodulatory species may underlie the link between IgA deficiency and autoimmune diseases in humans (45). Interestingly, while IgA-coated bacteria from individuals with IBD (46) or nutritional deficiencies (47) exacerbate respective pathologies in mice, IgA-coated bacteria from healthy humans protect mice from disease (47). We propose that during health, IgA fosters mucosal colonization of microbiota with beneficial properties (9), while disease states may induce (or be caused by) IgA responses to pathogens or pathobionts that disrupt healthy microbiome equilibria. Indeed, computational models indicate that IgA can both maintain indigenous mucosal populations and clear invasive pathogens (48). In addition to serving as a defense system, we discover that adaptive immunity evolved to engender intimate association with members of the gut microbiome.
Supplementary Material
Acknowledgments
We thank Drs. Elaine Hsiao, June Round, Hiutung Chu, and members of the Mazmanian laboratory for critical review of this manuscript. The anti-CD20 antibody was provided under an MTA from Genentech. We appreciate technical support from Taren Thron, the Caltech Office of Laboratory Animal Resources, Caltech Genomics Laboratory, and Caltech Flow Cytometry Facility. G.P.D. was supported by an NIH training grant (5T32 GM07616) and NSF Graduate Research Fellowship (DGE-1144469). Support for this research was provided by grants from the National Institutes of Health to the Broad Institute (U19AI110818), the NIH (P50 GM082545 and AI04123) to P.B.J., the NIH (GM099535 and DK083633) to S.K.M., and the Heritage Medical Research Institute to S.K.M. All data and code to understand and assess the conclusions of this research are available in the main text, supplementary materials and via the following repositories: EMBL-EBI accession ERP107727 and NCBI Bioproject accessions PRJNA445716 and PRJNA438372.
References and Notes
- 1.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017 doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 2.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017 doi: 10.1101/gr.216242.116. gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 6.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 7.Macpherson AJ, Köller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 2015;36:460–470. doi: 10.1016/j.it.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kubinak JL, Round JL. Do antibodies select a healthy microbiota? Nat Rev Immunol. 2016;16:767–774. doi: 10.1038/nri.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fransen F, et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunology. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunology. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Mantis NJ, Forbes SJ. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol Invest. 2010;39:383–406. doi: 10.3109/08820131003622635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–3913. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollinger RR, et al. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003;109:580–587. doi: 10.1046/j.1365-2567.2003.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathias A, et al. Potentiation of polarized intestinal Caco-2 cell responsiveness to probiotics complexed with secretory IgA. J Biol Chem. 2010;285:33906–33913. doi: 10.1074/jbc.M110.135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yel L. Selective IgA deficiency. Journal of clinical immunology. 2010 doi: 10.1007/s10875-009-9357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conley ME D. D. A. O. I. medicine. Intravascular and mucosal immunoglobulin A: two separate but related systems of immune defense? Am Coll Physicians. 1987 doi: 10.7326/0003-4819-106-6-892. [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 20.Ochoa-Repáraz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 21.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholz M, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 24.Yassour M, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Science Translational Medicine. 2016;8:343ra81–343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pudlo NA, et al. Symbiotic Human Gut Bacteria with Variable Metabolic Priorities for Host Mucosal Glycans. MBio. 2015;6 doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79:2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moor K, et al. Analysis of bacterial-surface-specific antibodies in body fluids using bacterial flow cytometry. Nat Protoc. 2016;11:1531–1553. doi: 10.1038/nprot.2016.091. [DOI] [PubMed] [Google Scholar]
- 34.Shimoda M, Inoue Y, Azuma N, Kanno C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer’s patches. Immunology. 1999;97:9–17. doi: 10.1046/j.1365-2567.1999.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunker JJ, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358:eaan6619. doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbins HL, Proctor GB, Yakubov GE, Wilson S, Carpenter GH. SIgA binding to mucosal surfaces is mediated by mucin-mucin interactions. PLoS ONE. 2015;10:e0119677. doi: 10.1371/journal.pone.0119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biesbrock AR, Reddy MS, Levine MJ. Interaction of a salivary mucin-secretory immunoglobulin A complex with mucosal pathogens. Infect Immun. 1991;59:3492–3497. doi: 10.1128/iai.59.10.3492-3497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phalipon A, et al. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunology. 2002;17:107–115. doi: 10.1016/s1074-7613(02)00341-2. [DOI] [PubMed] [Google Scholar]
- 39.Sarikonda G, et al. Transient B-cell depletion with anti-CD20 in combination with proinsulin DNA vaccine or oral insulin: immunologic effects and efficacy in NOD mice. PLoS ONE. 2013;8:e54712. doi: 10.1371/journal.pone.0054712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blutt SE, Miller AD, Salmon SL, Metzger DW, Conner ME. IgA is important for clearance and critical for protection from rotavirus infection. Mucosal Immunol. 2012;5:712–719. doi: 10.1038/mi.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moor K, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 2017;103:3–19. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barroso-Batista J, Demengeot J, Gordo I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat Commun. 2015;6:8945. doi: 10.1038/ncomms9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh K, Chang C, Gershwin ME. IgA deficiency and autoimmunity. Autoimmun Rev. 2014;13:163–177. doi: 10.1016/j.autrev.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Science Translational Medicine. 2015;7:276ra24–276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host Selection of Microbiota via Differential Adhesion. Cell Host Microbe. 2016;19:550–559. doi: 10.1016/j.chom.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 49.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 51.Mastronarde DN. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc. 2008;230:212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- 52.McClure R, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson LR, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amir A, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems. 2017;2 doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirarab S, Nguyen N, Warnow T. SEPP: SATé-enabled phylogenetic placement. Pac Symp Biocomput. 2012:247–258. doi: 10.1142/9789814366496_0024. [DOI] [PubMed] [Google Scholar]
- 61.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R. C. Team. R: A language and environment for statistical computing. 2013 [Google Scholar]
- 64.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson MJ. A new method for non−parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 66.Oksanen J, Kindt R, Legendre P, O’Hara B. The Vegan Package—Community Ecology Package. 2007. R package version 2.0-9. [Google Scholar]
- 67.Coyne MJ, Kalka-Moll W, Tzianabos AO, Kasper DL, Comstock LE. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect Immun. 2000;68:6176–6181. doi: 10.1128/iai.68.11.6176-6181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 69.Stevens AM, Shoemaker NB, Salyers AA. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. Journal of Bacteriology. 1990;172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.