Abstract
Aggregation of misfolded protein and resultant intracellular inclusion body formation are common hallmarks of mutant superoxide dismutase (mSOD1)-linked familial amyotrophic lateral sclerosis (FALS) and have been associated with the selective neuronal death. Protein disulfide isomerase (PDI) represents a family of enzymatic chaperones that can fold nascent and aberrant proteins in the endoplasmic reticulum (ER) lumen. Recently, our group found that S-nitrosylated PDI could contribute to protein misfolding and subsequent neuronal cell death. However, the exact role of PDI in the pathogenesis of ALS remains unclear. In this study, we propose that PDI attenuates aggregation of mutant/misfolded SOD1 and resultant neurotoxicity associated with ER stress. ER stress resulting in PDI dysfunction therefore provides a mechanistic link between deficits in molecular chaperones, accumulation of misfolded proteins, and neuronal death in neurodegenerative diseases. In contrast, S-nitrosylation of PDI inhibits its activity, increases mSOD1 aggregation, and increases neuronal cell death. Specifically, our data show that S-nitrosylation abrogates PDI-mediated attenuation of neuronal cell death triggered by thapsigargin. Biotin switch assays demonstrate S-nitrosylated PDI both in the spinal cords of SOD1 (G93A) mice and human patients with sporadic ALS. Therefore, denitrosylation of PDI may have therapeutic implications. Taken together, our results suggest a novel strategy involving PDI as a therapy to prevent mSOD1 aggregation and neuronal degeneration. Moreover, the data demonstrate that inactivation of PDI by S-nitrosylation occurs in both mSOD1-linked and sporadic forms of ALS in humans as well as mice.
Keywords: ALS, ER stress, Protein disulfide isomerase, S-nitrosylation, SOD1
Introduction
Amyotrophic lateral sclerosis (ALS), the most common adult-onset motor neuron disease, is a fatal neurodegenerative condition characterized pathologically by progressive loss of upper and lower motor neurons from the brainstem motor nuclei, anterior horn of the spinal cord, and cerebral cortex [1–4]. Oxidative injury, misfolded protein aggregates, altered axonal transport, impaired energy and calcium metabolism, excess glutamate activity, and malfunctioning mitochondria have all been implicated in motor neuron death in ALS; nonetheless, the pathogenic mechanism of and satisfactory treatment options for ALS remain unclear [1–4]. Familial ALS (FALS), which accounts for 5–10 % of ALS cases, has provided a number of potentially useful experimental models of the pathogenic mechanism of this disease. Recently, TDP-43 was described in SALS and in FALS with SOD-1 (Cu/Zn superoxide dismutase) mutations, potentially linking two pathologically distinct pathways of motor neuron degeneration. The neuropathology of ALS is characterized by the abnormal accumulation of insoluble proteins in the cytoplasm of degenerating motor neurons. A number of studies have confirmed these neuronal cytoplasmic inclusions (NCIs) to be a highly sensitive and specific marker for ALS [5]. A recent study showed the TAR DNA binding protein, TDP-43 to be a major component of NCIs in SALS, as well as in the most common pathological subtype of frontotemporal dementia [6]. The most common genes currently known to be associated with FALS include SOD1, TDP-43 [7], FUS [8], and the more recently discovered C9ORF72 [9] and UBQLN2 [10]. Among these, mutations in SOD1 have been linked to motor neuron death in up to 20 % of patients with FALS. Although the mechanism of motor neuron death associated with SOD1 mutation is still unclear, it may be related to a toxic function of the SOD1 mutant protein. SOD1 is an intracellular homodimeric metalloprotein that forms a stable intrasubunit disulfide bond. Wild-type SOD1 is normally cytoplasmic, with smaller amounts found in the nucleus, mitochondrial outer membrane, intermembrane space, and matrix. Although SOD1 is not traditionally associated with the ER, morphological ER abnormalities and SOD1 aggregates that colocalize with ER markers have been reported in patients with ALS and in transgenic mouse models. Mutations in SOD1 are known to be involved in motor neuron death in some forms of FALS [4, 11, 12]. Biochemical evidence suggests that the disulfide-reduced monomer of mutant superoxide dismutase (mSOD1) forms inclusion bodies, and aggregates of misfolded mSOD1 are commonly associated with motor neuron cell death. The most prevalent change observed is destabilization of the SOD1 protein, resulting in high levels of protein aggregation. The instability of mutant (m) SOD1 protein has been attributed to loss of metal ions and reduction of the intramolecular disulfide bond [13]. Misfolded protein aggregation and resultant intracellular inclusion body formation are common hallmarks of the selective neuronal cell death in mSOD1 FALS and sporadic ALS (SALS); in fact, similar phenomena are observed in almost all neurodegenerative diseases [14].
Recent reports have suggested that protein misfolding and interrelated endoplasmic reticulum (ER) stress play a critical role in the pathogenesis of many neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, ALS, Huntington’s disease, and prion diseases [15–21]. In mSOD1 mouse models and human ALS patients, SOD1 misfolding in ER lumen can activate signal transduction pathways known collectively as the unfolded protein response (UPR) [22, 23]. The UPR attenuates ER stress by inhibiting protein translation, increasing production of chaperone proteins, and enhancing protein degradation. One important family of UPR-induced chaperones is protein disulfide isomerase (PDI), which may protect against misfolded protein aggregation and is known to be increased in human brain during neurodegenerative conditions in an apparently compensatory manner [24].
Wild-type SOD1 forms a highly conserved intramolecular disulfide bond. A thiol–disulfide group in SOD1 plays a regulatory role in determining its folding/misfolding pathways [25–27]. The enzymatic activity of SOD1 catalyzes the removal of superoxide anion radicals at the bound copper ion, and the formation of an intramolecular disulfide bond is required for the stabilization of mature SOD1 [28]. However, mutant SOD1 can form monomers or insoluble high-molecular-weight multimers within the ER [29]. PDI catalyzes thiol–disulfide exchange, and can thus facilitate disulfide bond formation and inducing correct protein folding [30–33]. Considering its chaperone function that decreases ER stress as well as its disulfide isomerase function that refolds proteins, PDI is thought to represent an important regulator of protein aggregation in neurodegenerative diseases [24, 34–37]. Atkin and colleagues [22] showed that pharmacological inhibition of PDI enzymatic activity increased the presence of mSOD1 inclusions. Recently, PDI overexpression was reported to decrease mSOD1A4Vaggregation and toxicity in NSC-34 cells, but the exact mechanism by which PDI might affect ALS pathogenesis remains unclear [33].
Overproduction of reactive nitrogen species (RNS), mainly nitric oxide (NO), induces cytotoxicity and is considered a mediator of neuronal death in various neurodegenerative diseases. Among the various roles of NO, S-nitrosylation, defined as the covalent addition of NO to a critical cysteine thiol/sulfhydryl to form an S-nitrosothiol derivative (R-SNO), has been shown to modulate the function of many proteins [38, 39]. A number of proteins related to neurodegenerative conditions have been shown to be aberrantly S-nitrosylated [40–46]. For example, S-nitrosylation of PDI decreases both its chaperone function and its disulfide isomerase activity and has been shown to increase protein misfolding and neuronal cell death in models of both Alzheimer’s disease and Parkinson’s disease [37]. More recently, S-nitrosylated (SNO)–PDI has been implicated in mSOD1 aggregation. In cultured cell lines, blocking NO generation with the NO synthase (NOS) inhibitor N-nitro-L-arginine (NNA) significantly attenuated the formation of S-nitrosylated PDI and mSOD1 aggregates [47]. Importantly, in the present study, we detected an increase in the levels of SNO–PDI in spinal cord tissues obtained from human patients with sporadic ALS. Additionally, in cell culture models, we found that overexpression of PDI attenuated not only SOD1 aggregation but also neurotoxicity associated with ER stress in an NO-dependent fashion. Since S-nitrosylation of PDI inhibits its activity, our findings are consistent with the notion that denitrosylation of PDI might provide therapeutic benefit in ALS.
Materials and Methods
Cell Culture and ER Stress
HEK293A cells and NSC-34 cells (CELLutions Biosystems) stably expressing SOD1WT or SOD1G93A were grown in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Salt Lake City, UT, USA) containing 10 % heat-inactivated fetal bovine serum (Hyclone), 2 % penicillin/streptomycin, and 2 mM glutamine in a humidified incubator at 37 °C under 5 % CO2. Primary rat cerebrocortical cultures were prepared as described [48, 49], exposed to NMDA (50 μM plus 5 μM glycine for 20 min) in Earle’s balanced salt solution and then rinsed and placed in the original conditioned medium. Neurons were subsequently assessed for survival. All animal experimental procedures were performed with institutional approval and in accordance with the Guidelines of the National Institutes of Health (NIH). In neurons transfected with SOD1and PDI, SOD1 protein aggregation was examined by Western blot and filter trap assay. NSC-34 cells and mSOD1 (G93A) cells were transfected with PDI and exposed to thapsigargin (a Ca2+-ATPase inhibitor causing ER stress) or S-nitrocysteine (SNOC, an NO donor) to induce cell death. For these experiments, cell death was quantified by PI staining.
Constructs and Transfection
Wild-type (WT) or mutant (G93A) human Cu/Zn SOD1 cDNA (Dr. Lawrence J. Hayward, University of Massachusetts) was cloned into the XhoI and EcoRI site of pcDNA3.1/myc-His A (Invitrogen), the HindIII and EcoRI sites of pcDNA3.1/V5-His A (Invitrogen) or pEGFP-N1 (CLONTECH Laboratories). For stable transfection, NSC-34 cells (1×106 cells/60-mm dish, Falcon) were transfected using Superfect (Qiagen, Valencia, CA), sorted, and then selected with geneticin (G418 sulfate, Gibco, Grand Island, NY). In brief, 24 h after transfection, cells were selected in a medium that contained geneticin (400 μg/ml). This selection was continued for 14 days after which the cells were maintained as a population, and 14 days later single or pooled colonies were used for experiments after determining human SOD1 expression by Western blot and immunocytochemical staining. Cells expressing human SOD1 were cultured in medium containing geneticin (200 μg/ml). Transient transfection was also used for experiments by transfecting HEK293A or NSC-34 cells with vectors using Superfect. Cells were cultured for 3 days prior to the experiment. Wild-type human (GeneCopoeia) and mutant PDI (C36S, C39S, C383S, and C386S) were constructed in myc-tagged PDI; myc-tag was inserted between amino acids E497 and D498 (upstream of the KDEL sequence) in the backbone of pCR3.1 vector.
Western Blot Analysis and Filter Trap Assay
Western blots were performed as previously described [50]. Cell lysates were prepared, subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blotted with primary antibodies. The filter trap assay was performed using a Hybri-Dot manifold (Bio-Rad) and cellulose acetate membrane filter with pore size of 0.2 μm. The cell lysates were prepared as for Western blot analysis. The same amount of protein from each experimental condition was diluted to 100 μl in PBS with 2 % SDS and applied onto the membrane. Soluble proteins were removed by vacuum suction, whereas the SDS-resistant aggregates remained “trapped” on the filter. Wells were washed three times with 2 % SDS/PBS and suction was maintained for 20 min to allow complete and tight trapping of SDS-insoluble material. Membranes were subsequently blocked with 5 % skim milk and immunoblot was performed.
Immunoprecipitation Analysis
Cell pellets were suspended in a lysis buffer containing protease inhibitors, and lysates were centrifuged at 13,000 rpm for 10 min. Equal amounts of protein were precipitated with SOD1 and PDI. The complexes formed were immunoprecipitated using protein A-sepharose. The sepharose beads were boiled in SDS-PAGE sample buffer, and the samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Western blot analysis was performed using anti-PDI or anti-SOD1 antibody.
Quantitative Real-Time (qRT-)PCR
Total RNA was extracted from a motor neuron cell line by TRIzol reagent (MRC, Cincinnati, OH, USA). RNA (50 ng) were used as a template for qRT-PCR amplification, using SYBR green real-time PCR master mix (Toyobo, Osaka, Japan). Primers were standardized in the linear range of the cycle before the onset of the plateau. GAPDH was used as an internal control. Two-step PCR thermal cycling for DNA amplification and real-time data acquisition were performed with an ABI 7500 real-time PCR system.
Cell Death/Cell Viability Assays
In some experiments, PI (Invitrogen) was used to evaluate cell death. PI binds to DNA and RNA and, in nonfixed cells, can only penetrate the membrane of dead cells. As an additional measure of cell viability, we used Cell Counting Kit-8 (CCK-8). CCK-8 allows facile assessment of cell viability using WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H-tetrazolium, monosodium salt], which produces a water-soluble formazan dye upon bioreduction in the presence of an electron carrier, 1-Methoxy PMS. NSC-34 cells were plated on 96-well plates at 2×104 cells/well and incubated for 2 h in a humidified incubator at 37 °C under 5 % CO2. Then, the 96-well plates were measured at 450 nm using an ELISA plate reader. To obtain an index of apoptotic cell death, neuronal nuclei were labeled with Hoechst 33342 dye, as described below.
Biotin Switch Assay for Detecting S-Nitrosylated Proteins
Cell lysates and brain tissue extracts were prepared in HENC or HENT buffers (250 mM Hepes pH 7.5, 1 mM EDTA, 0.1 mM neocuproine, 0.4 % CHAPS, or 1 % Triton X-100). A range of protein concentrations was tested in these assays, but typically, 1 mg of cell lysate and up to 2 mg of tissue extract were used. Blocking buffer [2.5 % SDS, 20 mM methyl methane thiosulfonate (MMTS) in HEN buffer] was mixed with the samples and incubated for 30 min at 50 °C to block free thiol groups. After removing excess MMTS by acetone precipitation, nitrosothiols were then reduced to thiols with 1 mM ascorbate. Ascorbate was omitted as a control. The newly formed thiols were then linked with the sulfhydryl-specific biotinylating reagent N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (Biotin-HPDP). We then pulled down the biotinylated proteins with Streptavidin-agarose beads and performed Western blot analysis to detect the amount of protein remaining in the samples [29]. The input for these blots was typically 2 μg of cell lysate or 100 μg of tissue extract loaded in each lane.
Immunocytochemistry and Immunofluorescence Studies
NSC-34 cells were transfected with SOD1 and/or PDI (1 μg each) using Lipofectamine 2000 reagent in Opti-MEM. The cells were placed in fresh medium (DMEM) containing fetal calf serum approximately 5 h after transfection, exposed to 100 μM SNOC or control solution, and then incubated for another 19 h. Cells were fixed with 4 % paraformaldehyde (PFA) for 10 min at room temperature and then washed three times in PBS, permeabilized with 0.5 % Triton X-100 for 5 min, and washed three times. Nonspecific antibody binding was minimized by incubation in blocking solution (10 % goat serum in PBS) for 1 h at 37 °C. Cells were incubated with antibodies to NeuN (Millipore) for 12 h at 4 °C. After washing, the cells were incubated with anti-mouse antibody conjugated with Alexa 594 for 1 h at 37 °C and then incubated in Hoechst 33342 dye to fluorescently stain nuclei and allow assessment of nuclear morphology to determine apoptosis. Rat cortical neuronal cells were transfected with SOD1 and PDI according to the manufacturer’s protocol. Cells were washed with phosphate buffered saline (PBS) 72 h post-transfection and fixed with 4 % PFA in PBS for 10 min. Cells were exposed to Hoechst due prior to mounting in fluorescent mounting medium. Apoptotic nuclei were defined as condensed or fragmented (as evidenced by multiple, condensed Hoechst-positive bodies in one cell), and counted as the percentage of apoptotic to total cells for at least 100 cells expressing both transfected constructs per treatment in three independent experiments. Cells expressing only one transfected plasmid or undergoing cell division were excluded from the analysis.
Animals
Transgenic TgN (SOD1-G93A) 1GUR mice, which express a transgenic human mSOD1 (G93A) allele containing the Gly 93 to Ala (G93A) substitution, were obtained from Jackson Laboratory (Bar Harbor, ME, USA). SOD1 (G93A) transgenic mice were bred with B6SJLF1/J hybrid females. The human mSOD1 (G93A) transgene was confirmed by PCR assay on samples obtained from tail tissue of transgenic offspring. All mice were housed under a 12-h light/dark cycle. The animal studies were carried out in accordance with the institutional guidelines and approval.
Human Subjects
Supplementary Table 1 lists all human subjects used in the current study and includes subject age, postmortem interval, and gender. Human samples were analyzed with Institutional permission under California and NIH guidelines. Informed consent was obtained according to procedures approved by UCSD and Burnham Institute Review Boards.
Statistical Analysis
Data are presented as mean + SEM or SD. Statistical evaluation was performed by analysis of variance (ANOVA) followed by a post hoc Schéffe test for multiple comparisons. Each experiment was performed in triplicate and three or more separate times.
Results
PDI Decreases SOD1 Monomers as well as Multimers
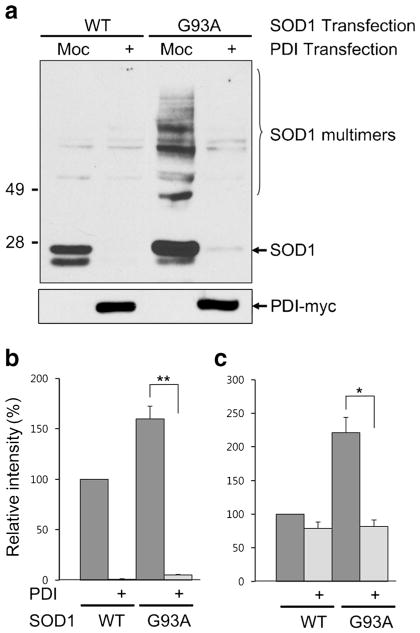
To assess cellular defenses against mSOD1 misfolding and aggregation via PDI expression, we transfected HEK293A cells with SOD1 and PDI, and changes in SOD1 protein were examined by Western blotting under nonreducing conditions (Fig. 1a, b). Overexpression of PDI resulted in decreased G93A mSOD1 protein, with reductions observed in both monomers (Fig. 1a) and multimers (Fig. 1c). Interestingly, the PDI-induced decrease in SOD1 was accentuated in wild-type and mSOD1 monomers over multimers (Fig. 1b). In order to confirm this finding, the experiment was repeated under reducing conditions. When SOD1 multimers were converted to monomers by reducing the disulfide bond between SOD1 molecules, we more clearly observed a PDI-induced decrease in total SOD1 (Fig. 2a, b). It should be noted that mSOD1 is prone to aggregation, and a large amount of mSOD1 protein is insoluble even with detergent treatment. Thus, we next examined whether PDI had an effect on the detergent-insoluble fraction of SOD1 in the same manner. Indeed, we observed that PDI decreased the mSOD1 protein in this fraction as well (Supplementary Fig. 1). For these expirements, we verified a similar degree of transfection efficiency using a green fluorescent protein (GFP) reporter plasmid. The number of GFP-positive cells was determined using flow cytometry. Co-transfected NSC-34 cells with pcDNA empty vector, wild-type SOD1 or mSOD1, wild-type PDI or mutant PDI, displayed similar amounts of GFP. We thus confirmed that the transfection efficiency was consistent among all experimental groups (Supplementary Fig. 2a). As a further control, to check the stability of SOD1 protein, we performed cycloheximide (CHX)-based protein-chase experiments in the absence or presence of wild-type PDI in wild-type SOD1-transfected NSC-34 cells, followed by Western analysis. When CHX (10 μg/ml) was chased for the indicated period of time, the stability of SOD1 protein was not affected by PDI overexpression(Supplementary Fig. 2b). Thus, it seems likely that PDI overexpression does not modulate SOD1 protein stability. We next examined the possibility that PDI overexpression appears to be the reduction of total SOD1 level due to the changes in the transcription level. SOD1 messenger RNA (mRNA) level in the absence or presence of wild-type PDI or mutant PDI in wild-type SOD1–transfected NSC-34 cells were determined by quantitative real-time polymerase chain reaction (qRT-PCR). We found that wild-type PDI or mutant PDI overexpression did not decreased SOD1 levels. These results suggest that SOD1 mRNA level is not regulated by PDI overexpression (Supplementary Fig. 2c).
Fig. 1.
PDI decreases SOD1 monomers as well as multimers. a Lysates from HEK293A cells transfected with SOD1 and PDI were examined by Western blotting under nonreducing conditions. b, c PDI decreased the relative level of mSOD1 (G93A) protein (shown on Y-axis), and the decrease was observed for both monomers (b) and multimers (c). However, the PDI-induced decrease in SOD1 levels was accentuated for SOD1 monomers of both the wild-type and mutant (SOD1: WT wild-type, G93A G93A mutant, Moc mock-transfected). Values are mean + SEM, n =4; *P <0.01, **P <0.001
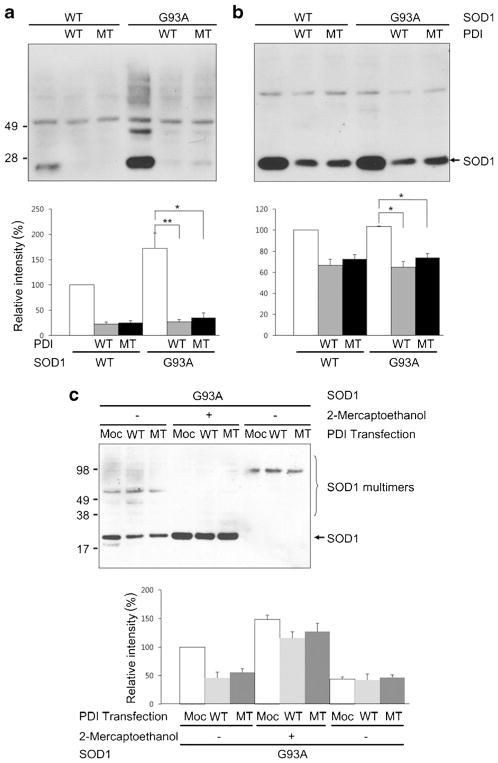
Fig. 2.
Overexpression of PDI decreases SOD1 monomers as well as multimers. a, b HEK293A cells were transfected with SOD1 and wild-type (WT) PDI or mutant (MT) PDI, and the change in SOD1 protein was examined by immunoblotting under nonreducing (a) and reducing (b) conditions. In mutant PDI, the four cysteine residues were mutated to serine (C36S, C39S, C383S, and C386S). Expression of PDI decreased total SOD1 protein. c Media of NSC-34 cells expressing SOD1 and/or PDI was monitored for secreted SOD1. SOD1 in the media was seen in the multimer form and was not significantly different in regardless of the presence of PDI. Values are mean + SEM, n =4; *P <0.01, **P <0.001
Effect of Mutant PDI on Decreasing SOD1
Among the various functions of PDI, chaperone and isomerase functions are well known. PDI has two thioredoxin-like domains containing four cysteine residues as a Cys-Gly-His-Cys sequence in the N- and C-terminal motifs. These domains are thought to represent the active site for isomerase function, and the four cysteine residues are key sites for thiol–disulfide exchange with substrate proteins, thus facilitating disulfide bond formation [51]. In order to clarify whether the PDI-induced decrease in SOD1 levels is mediated by its isomerase function, we mutated these four cysteine residues to serine (C36S, C39S, C383S, and C386S) and then performed immunoblotting to monitor SOD1 protein levels. Although wild-type was somewhat more effective, expression of mutant PDI also decreased total SOD1 (Fig. 2a) and SOD1 monomers (Fig. 2b). The PDI-induced decrease in SOD1 was more evident for SOD1 monomers.
Since PDI overexpression has been reported to increase protein secretion from cells [52], this suggested the possibility that PDI enhances SOD1 secretion, resulting in an intracellular decrease in SOD1 [53]. To exclude this possibility, we collected media from NSC-34 cells expressing SOD1 and/or PDI and measured the secreted levels of SOD1. We found that the levels of SOD1 in the media (observed as multimers) were not significantly different in the presence or absence of PDI expression (Fig. 2c).
PDI But Not SNO–PDI Decreases mSOD1 Aggregation
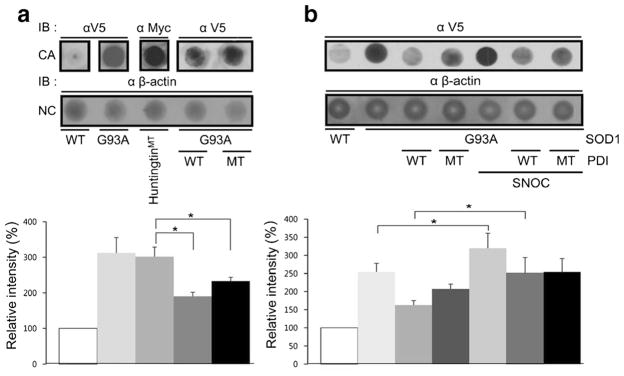
Protein oligomers or aggregates are thought to play a role in the pathogenicity of many neurodegenerative diseases, and protein aggregation in the form of inclusion bodies is a hallmark of neurodegenerative disorders including ALS. SOD1 has a strong propensity to aggregate when it is mutated and can cause FALS. Thus, we wanted to examine whether PDI specifically decreases mSOD1 aggregation. For this purpose, we used a filter trap assay. We found that mSOD1 was trapped on a cellulose acetate membrane filter (and hence aggregated to the point that it could not pass through the filter) to a far greater degree than wild-type SOD1, as previously seen for mutant huntingtin protein [54]. β-Actin served as a control to show that total cell protein remained the same. Interestingly, overexpression of either wild-type or mutant PDI decreased the quantity of trapped mSOD1 (Fig. 3a). This finding is consistent with prior reports showing the protein chaperone function of PDI can be independent of these active cysteine residues, whereas the isomerase activity of the enzyme is linked to the integrity of the cysteine amino acid residues (which were mutated here). These results suggest that PDI attenuates aggregation of SOD1-G93A via its chaperone activity but not isomerase activity since cysteine-mutant PDI blocked aggregation to the same degree as wild-type PDI [55].
Fig. 3.
Filter trap assay. a Overexpression of PDI or mutant PDI decreased the quantity of mSOD1 accumulating on the filter. b SNOC significantly attenuated the ability of PDI to decrease filter-trapped SOD1. Quantification from n =4 experiments shown below sample filter sets. Values are mean + SEM; *P <0.01
Overproduction of RNS is a common feature in neurodegenerative disorders, with NO often generated via activation of neuronal NOS due to Ca2+ influx mediated by stimulation of N-methyl-D-aspartate (NMDA)-type glutamate receptors. NO can modulate protein function and tertiary structure by S-nitrosylation, a reaction transferring an NO group to a critical cysteine thiol [38, 39]. In fact, prior work has shown that PDI can be S-nitrosylated, resulting in inhibition of its enzymatic chaperone as well as its isomerase activity [37]. For example, S-nitrosylation of PDI leads to increased aggregation of proteins such as synphilin 1 and abrogates PDI-mediated attenuation of ER stress [37]. One possible explanation for this finding in light of the foregoing experiments with cysteine mutants is that S-nitrosylation may cause a conformational change that inhibits chaperons activity, whereas mutations in the critical cysteine residues to prevent nitrosylation does not induce this conformation change. Both nitrosylation and mutation, however, would abrogate the interaction of substrate with these critical cysteine residues, hence blocking isomerase activity. Hence, we wanted to examine here if S-nitrosylation of PDI would inhibit its ability to decrease SOD1 aggregation by disrupting PDI chaperone activity. To assess this possibility, we added the physiological NO donor S-nitrosocysteine (SNOC) to PDI prior to its addition to the filter trap assay. Under these conditions, SNOC significantly attenuated the ability of PDI to decrease the amount of filter-trapped mSOD1 (Fig. 3b).
PDI Colocalizes in Part with SOD1
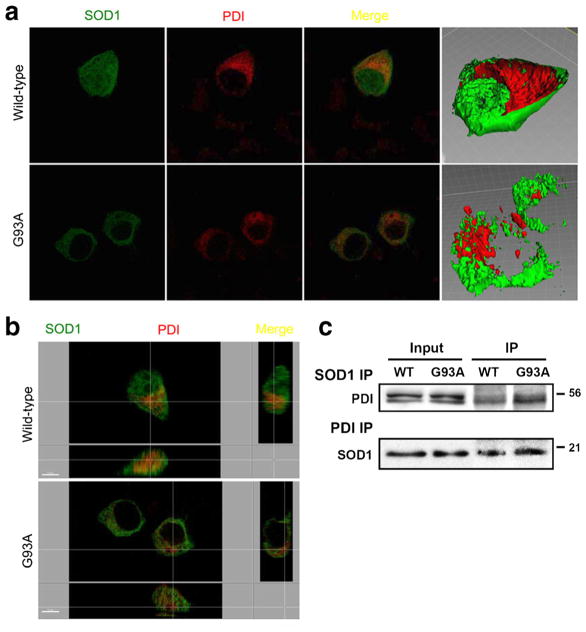
PDI and SOD1 are known to be located in the ER and cytosol, respectively. In order to begin to decipher the mechanism whereby PDI decreases SOD1 aggregation, we transfected NSC-34 cells with GFP-labeled SOD1 and myc-tagged PDI constructs. Wild-type SOD1 stained diffusely throughout the cell, consistent with its previously reported distribution in the cytosol, while mSOD1 appeared to be excluded from nucleus (Fig. 4a, b). The anti-SOD1 antibody co-precipitated PDI, and the anti-PDI antibody co-precipitated SOD1 in NSC-34 cells transfected with wild-type SOD1 or m SOD1. Western blot analysis revealed that the SODI antibody co-precipitated PDI (Fig. 4c, upper panel), indicating a physical interaction between SOD1 and PDI in NSC-34 cells transfected with wild-type SOD1 or m SOD1 under our conditions. The reverse experiment was also performed to confirm the interaction observed binding between PDI and SOD1 in this system. Cell lysates from NSC-34 transfectants were immunoprecipitated using the PDI antibody (Fig. 4c, lower panel). In both cases, a band was observed (Fig. 4c, lower panel), consistent with the presence of a PDI-SOD1 association in NSC-34 cells transfected with wild-type SOD1 or m SOD1. We next investigated whether mSOD1 (G93A) would differentially increase cell death in HEK293A cells vs. NSC-34 cells. We found significantly greater cell death in mSOD1-transfected motor neuronal cell lines than in mSOD1-transfected HEK294A cell lines (Supplementary Figs. 3 and 4). Moreover, transfection with mSOD1 led to significantly increased cell death compared to transfection with wild-type SOD1 in NSC-34 cells (Supplementary Fig. 4). In contrast, transfection of HEK293A cells with wild-type SOD1 or mSOD1 did not induce cell death (Supplementary Fig. 3). These findings indicate that mSOD1 renders motor neurons more susceptible to death than other cell types.
Fig. 4.
Co-localization of overexpressed wild-type SOD1, SOD1 (G93A) mutant, and PDI in NSC-34 cells. Cells receiving the PDI construct were stained immunocytochemically 48 h after transfection. Images of wild-type SOD1 (green), SOD1 (G93A) mutant (green), and PDI (red) and merge were monitored under confocal microscopy. Wild-type SOD1 stained diffusely inside of cells, consistent with its previously reported cytosolic distribution (a), while mSOD1 did not stain the nucleus, indicating its nuclear-sparing pattern of distribution (b). Wild-type and m SOD1 coprecipitate with PDI and induce PDI expression in NSC-34 cells. The anti-SOD1 antibody coprecipitated PDI, and the anti-PDI antibody coprecipitated SOD1 in NSC-34 cells transfected with wild-type SOD1 or m SOD1. SOD1 thus interacts with PDI in NSC-34 cells transfected with wild-type SOD1 or m SOD1 (c)
Effect of PDI on Thapsigargin- and SNOC-Induced Cell Death
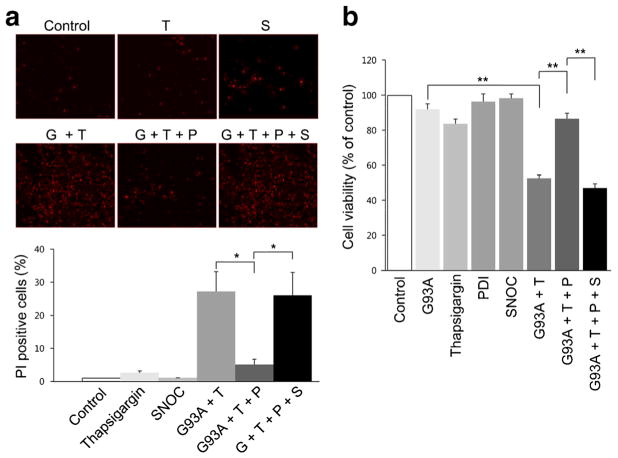
Thapsigargin increases calcium accumulation and cell apoptosis in NSC-34 cells [56]. Thapsigargin acts by inhibiting Ca2+-ATPase in the ER, thus blocking sequestration of calcium into the ER and causing an increase in the intracellular concentration of calcium. Thapsigargin also causes accumulation of unfolded or misfolded proteins, upregulation of GRP78, and activation of caspase-3-mediated apoptosis. Here, we found that 10 μM thapsigargin greatly increased cell death in NSC-34 cells after they were transfected with mSOD1 (G93A) (Fig. 5a). In contrast, transfection with PDI decreased thapsigargin-induced cell death as well as mSOD1 aggregation.
Fig. 5.
Influence of overexpression of PDI on thapsigargin-induced cell death of mSOD1 (G93A)-transfected NSC-34 cells. a Transfection with PDI reduced cell death in the presence of thapsigargin in mSOD1 (G93A)-transfected NSC-34 cells. In contrast, SNOC prevented the beneficial effect of PDI. b Cell viability monitored by CCK-8 assay kit. Treatments: G93A or G, mSOD1 (G93A). T thapsigargin, S SNOC, P PDI. Values are mean + SEM, n =4 for each group; *P <0.01, **P < 0.001
To determine whether the NO-donor SNOC also triggers cell death in NSC-34 cells in an SOD1-dependent manner, we exposed cells to SNOC after transfection with either wild-type or mSOD1. The results showed that cells transfected with mSOD1 were more susceptible to cell death when exposed to SNOC, and this occurred in a dose-dependent manner with 200 μM SNOC but not 100 μM leading to neuronal death (Supplementary Fig. 5). In contrast, even 200 μM SNOC did not induce death in cells transfected with wild-type SOD1.
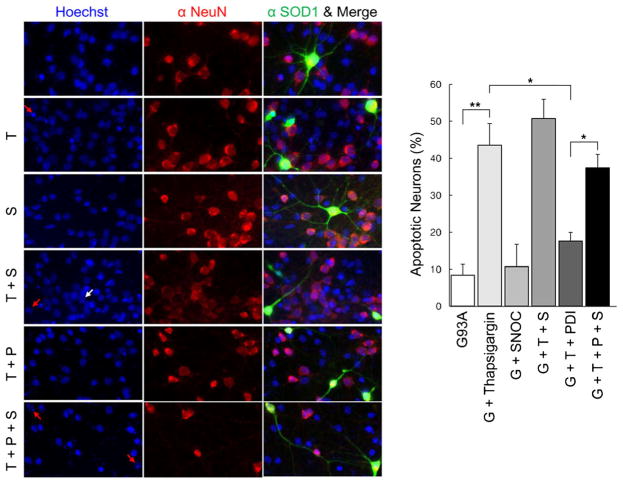
Additionally in NSC-34 cells, SNOC abrogated the protective effect of PDI in the face of thapsigargin insult in the presence of mSOD1 (G93A) (Fig. 5b). Under similar conditions, we found that primary cerebrocortical neurons displayed similar results with regard to protection by PDI and reversal of this protective effect after exposure to SNOC (Fig. 6). These results are consistent with the notion that PDI could reduce mSOD1 aggregation and ER stress, and that NO donors could inhibit this effect of PDI.
Fig. 6.
NO donor prevents PDI-mediated protection of primary neuronal cells from thapsigargin insult after transfection with mSOD1 (G93A). mSOD1 (G93A) (green), NeuN (red), and merged images were monitored under confocal microscopy. Hoechst stain shows nuclei (blue). Expression of PDI significantly decreased apoptotic cell death in rat primary cortical neurons induced by thapsigargin, whereas SNOC significantly prevented this beneficial effect of PDI. Treatments: G93A or G, mSOD1 (G93A). T thapsigargin, S SNOC, P PDI. Values are mean + SEM, n =4. *P <0.01, **P < 0.001
SNOC-Induced S-Nitrosylation of PDI
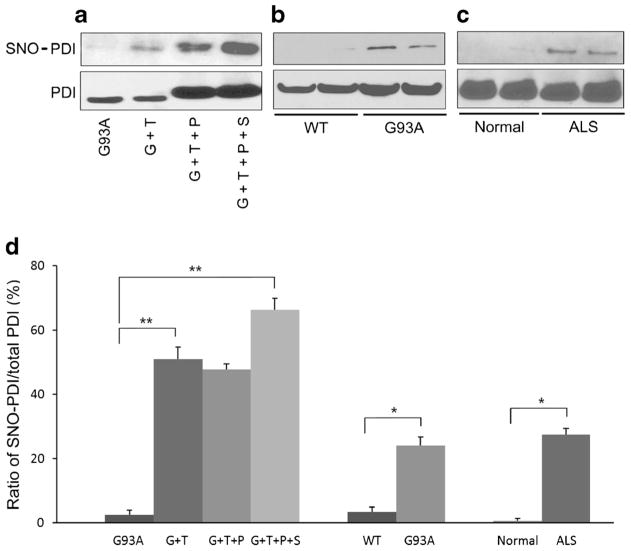
Under our conditions, we found that 100 μM SNOC by itself did not have an effect on cell viability. However, 100 μM SNOC induced S-nitrosylation of PDI in SOD1 (G93A) mutant-transfected neurons, and the degree of this S-nitrosylation was substantially greater after exposure to SNOC than to 10 μM thapsigargin by biotin switch assay (Fig. 7a, d). This result is consistent with the notion that exposure to 100 μM SNOC abolished the protective effect of PDI on thapsigargin-induced cell death via increased S-nitrosylation of PDI. We next investigated whether aberrant S-nitrosylation of PDI occurs in animal models and in human cases of ALS (Fig. 7b–d). Our group had previously shown that PDI can be S-nitrosylated during ER stress, which is involved in the pathogenesis of several neurodegenerative diseases [37]. We found increased levels of SNO–PDI in spinal cord samples from SOD1 (G93A) mice and in human patients with sporadic ALS. Of note, 90–95% of sporadic ALS cases manifest a TDP-43 proteinopathy. Immunoblotting of input samples showed that total PDI was also increased in patients with ALS and transgenic SOD1 (G93A) mice, confirming previous observations. Importantly, however, the ratio of SNO–PDI to total PDI was increased in both the ALS mouse model and human cases of ALS. In fact, we found similarly increased ratios in our cell-based models, which led to cell death linked to the formation of SNO–PDI [37], suggesting that pathophysiologically relevant levels of SNO–PDI were present in the mouse and human ALS spinal cord.
Fig 7.
S-Nitrosylation of PDI (forming SNO–PDI) in vitro and in vivo. Biotin switch shows SNO–PDI and standard immunoblot, total PDI. a Levels of total PDI and SNO–PDI increased in mSOD1 (G93A)-transfected NSC-34 cells in the presence of thapsigargin or SNOC. b, c SNO–PDI was found in the spinal cords of SOD1 (G93A) mice and human patients with sporadic ALS but not in normal controls by biotin switch assay. Immunoblotting of input samples showed that total PDI was also increased in transgenic mSOD1 (G93A) mice (b) and ALS patients (c). d Increased ratio of SNO–PDI (by biotin switch) to total PDI (by immunoblot) in the in vitro conditions shown in (a), in mSOD1 (G93A) mice, and in sporadic ALS patient spinal cord. Values are mean + SEM, n =4, n =2 (normal control). *P <0.01, **P <0.001
Discussion
PDI is a major component of ER stress pathway and catalyzes the formation and dissolution of disulfide bonds, thereby facilitating correct disulfide bond formation [25–28, 57]. In addition, PDI acts as a molecular chaperone, assisting in the folding of polypeptides [25–28, 57]. In the present study, we determined that PDI attenuates aggregation of mSOD1 misfolded proteins as well as motor neuron cell death associated with ER stress. Recently, PDI has been implicated in the pathophysiology of ALS and appears to be particularly correlated with SOD1 misfolding [58–60]. Recent studies have also shown that inhibition of PDI activity can increase aggregation of mSOD1 in neuronal cells, whereas overexpression of PDI decreases mSOD1 aggregation and mSOD1-induced neuronal cell death [25–28]. Taken together, these findings suggest that ER stress may contribute to the pathophysiology of ALS and that increased PDI activity may reduce mSOD1 aggregation and promote neuronal survival. Moreover, increased levels of S-nitrosylated PDI have also been reported in human ALS, suggesting that SNO–PDI may contribute to the pathogenesis of ALS [20, 37, 43–46, 61]. However, it has remained uncertain whether SNO–PDI is involved in protein aggregation and motor neuron injury in sporadic ALS in the absence of SOD1 mutation.
The major finding of the present study concerns the effect of PDI on mSOD1 aggregation in ALS. We show that PDI may serve an important role in decreasing mSOD1 aggregation and SOD1-mediated neurodegeneration. Furthermore, S-nitro-sylation of PDI correlates with a diminished beneficial effect, consistent with the known ability of S-nitrosylation to inhibit PDI activity [37]. Since we also show that SNO–PDI is present in sporadic cases of human ALS, S-nitrosylation may play a pathogenic role in the disease. As mentioned above, 90–95 % of sporadic ALS cases display abnormal aggregates of TDP-43. A recent report showed that PDI colocalized with TDP-43 and SOD1 in neuronal cytoplasmic inclusions in patients with sporadic ALS. This report went on to suggest that NO inhibited PDI and led to the accumulation of unfolded proteins in ALS [62], as we had previously reported in other neurodegenerative conditions [37]. Our new results with PDI transfection suggest that PDI attenuates the formation of SOD1 aggregates in HEK293A cells and in NSC-34 motor neuron cells. Collectively, these findings point to the potential importance of SOD1–PDI interactions in preventing mSOD1 aggregation, and hence, upregulation of PDI may have therapeutic implications. Since recent reports suggest that SNO–PDI disrupts normal protein folding in various neurodegenerative diseases [37], it is likely that formation of SNO–PDI may also contribute to the pathogenesis of ALS.
Our study also shows that SNO–PDI may be a pathological hallmark and possible biomarker for ALS because of the presence of an increased ratio of SNO–PDI to total PDI in the central nervous system of ALS patients. Furthermore, our work is consistent with the notion that increased SNO–PDI leads to incorrect disulfide cross-linking of the immature, misfolded mSOD1 proteins, which may contribute to their aggregation. Hence, PDI may have potential as a novel therapeutic agent in ALS, either by further upregulating the levels of total PDI or by denitrosylating existing PDI in order to defend against protein misfolding and motor neuron apoptotic cell death.
Nevertheless, several issues remain for further study. While PDI may exert a protective action in mSOD1 models of ALS, as yet the mechanistic relationship between PDI and mSOD1 in alleviating ER stress remains unclear. Interestingly, overexpression of either wild-type or mutant PDI decreased the quantity of aggregated (filter-trapped) mSOD1 (Fig. 3). These findings suggest that NO-induced aggregation of SOD1 (G93A) is dependent on PDI chaperone activity but not isomerase activity since the nonnitrosylatable mutant PDI blocked aggregation to approximately the same degree as wild-type PDI [54].
In summary, we used a novel strategy to explore a potential therapeutic role for PDI in suppressing mSOD1-induced neuronal degeneration in models of ALS. Our findings suggest that the presence of increased PDI, as found in motor neurons of SOD1 (G93A) transgenic mice and patients with ALS, may represent a cellular defense against misfolding of mSOD1 and possibly other proteins, and their subsequent aggregation. Moreover, our results demonstrate that S-nitrosylation, and thus presumed inhibition, of PDI is a feature of both mSOD1-linked and sporadic forms of ALS.
Supplementary Material
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010–0011008). Additional support came in part from NIH grants P01 HD29587, P01 ES016738, and P30 NS076411 (to SAL).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12035-013-8562-z) contains supplementary material, which is available to authorized users.
Contributor Information
Gye Sun Jeon, Department of Neurology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Republic of Korea.
Tomohiro Nakamura, Del E. Webb Center for Neuroscience, Aging, and Stem Cell Research, Sanford-Burnham Medical Research Institute, La Jolla, CA 92037, USA.
Jeong-Seon Lee, Department of Neurology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Republic of Korea.
Won-Jun Choi, Department of Neurology, Hanyang University Hospital, Hanyang University College of Medicine, Seoul, Republic of Korea.
Suk-Won Ahn, Department of Neurology, Chung-Ang University Hospital, Chung-Ang University College of Medicine, Seoul Republic of Korea.
Kwang-Woo Lee, Department of Neurology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Republic of Korea.
Jung-Joon Sung, Department of Neurology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Republic of Korea.
Stuart A. Lipton, Del E. Webb Center for Neuroscience, Aging, and Stem Cell Research, Sanford-Burnham Medical Research Institute, La Jolla, CA 92037, USA
References
- 1.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto A, Okada Y, Nakamichi M, Nakamura M, Toyama Y, Sobue G, Nagai M, Aoki M, Itoyama Y, Okano H. Disease progression of human SOD1 (G93A) transgenic ALS model rats. J Neurosci Res. 2006;83:119–133. doi: 10.1002/jnr.20708. [DOI] [PubMed] [Google Scholar]
- 3.Tripathi VB, Al-Chalabi A. Molecular insights and therapeutic targets in amyotrophic lateral sclerosis. CNS Neurol Disord Drug Targets. 2008;7:11–19. doi: 10.2174/187152708783885110. [DOI] [PubMed] [Google Scholar]
- 4.Muyderman H, Hutson PG, Matusica D, Rogers ML, Rush RA. The human G93A-superoxide dismutase-1 mutation, mitochondrial glutathione and apoptotic cell death. Neurochem Res. 2009;34:1847–1856. doi: 10.1007/s11064-009-9974-z. [DOI] [PubMed] [Google Scholar]
- 5.Lowe J. New pathological findings in amyotrophic lateral sclerosis. J Neurol Sci. 1994;124(Suppl):38–51. doi: 10.1016/0022-510x(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 7.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith BN, Newhouse S, Shatunov A, et al. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. Eur J Hum Genet. 2013;21:102–108. doi: 10.1038/ejhg.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, Chen W, Zhai P, Sufit RL, Siddique T. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 12.Ferraiuolo L, Heath PR, Holden H, Kasher P, Kirby J, Shaw PJ. Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J Neurosci. 2007;27:9201–9219. doi: 10.1523/JNEUROSCI.1470-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart PJ. Pathogenic superoxide dismutase structure, folding, aggregation and turnover. Curr Opin Chem Biol. 2006;10:131–138. doi: 10.1016/j.cbpa.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 15.Kim HT, Russell RL, Raina AK, Harris PL, Siedlak SL, Zhu X, Petersen RB, Shimohama S, Smith MA, Perry G. Protein disulfide isomerase in Alzheimer disease. Antioxid Redox Signal. 2000;2:485–489. doi: 10.1089/15230860050192260. [DOI] [PubMed] [Google Scholar]
- 16.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura T, Lipton SA. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 2009;14:455–468. doi: 10.1007/s10495-008-0301-y. [DOI] [PubMed] [Google Scholar]
- 18.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honjo Y, Ito H, Horibe T, Takahashi R, Kawakami K. Protein disulfide isomerase-immunopositive inclusions in patients with Alzheimer disease. Brain Res. 2010;1349:90–96. doi: 10.1016/j.brainres.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Cieplak P, Cho DH, Godzik A, Lipton SA. S-nitrosylation of Drp1 links excessive mitochondrial fission to neuronal injury in neurodegeneration. Mitochondrion. 2010;10:573–578. doi: 10.1016/j.mito.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SB, Shi Q, Xu Y, Xie WL, Zhang J, Tian C, Guo Y, Wang K, Zhang BY, Chen C, Gao C, Dong XP. Protein disulfide isomerase regulates endoplasmic reticulum stress and the apoptotic process during prion infection and PrP mutant-induced cytotoxicity. PLoS One. 2012;7:e38221. doi: 10.1371/journal.pone.0038221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, Horne MK. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- 23.Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–407. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Conn KJ, Gao W, McKee A, Lan MS, Ullman MD, Eisenhauer PB, Fine RE, Wells JM. Identification of the protein disulfide isomerase family member PDIp in experimental Parkinson’s disease and Lewy body pathology. Brain Res. 2004;1022:164–172. doi: 10.1016/j.brainres.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa Y, O’Halloran TV. Posttranslational modifications in Cu, Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid Redox Signal. 2006;8:847–867. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chattopadhyay M, Valentine JS. Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antioxid Redox Signal. 2009;11:1603–1614. doi: 10.1089/ars.2009.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seetharaman SV, Prudencio M, Karch C, Holloway SP, Borchelt DR, Hart PJ. Immature copper-zinc superoxide dismutase and familial amyotrophic lateral sclerosis. Exp Biol Med (Maywood) 2009;234:1140–1154. doi: 10.3181/0903-MR-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa Y, Torres AS, O’Halloran TV. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi H, Almer G, Yamashita S, Guegan C, Nagai M, Xu Z, Sosunov AA, McKhann GM, 2nd, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci U S A. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YS, Harel NY, Strittmatter SM. Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J Neurosci. 2009;29:13850–13859. doi: 10.1523/JNEUROSCI.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD. Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis. Brain. 2010;133:105–116. doi: 10.1093/brain/awp267. [DOI] [PubMed] [Google Scholar]
- 34.Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- 35.Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16:653–662. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hetz C, Russelakis-Carneiro M, Walchli S, Carboni S, Vial-Knecht E, Maundrell K, Castilla J, Soto C. The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J Neurosci. 2005;25:2793–2802. doi: 10.1523/JNEUROSCI.4090-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 38.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura T, Lipton SA. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011;18:1478–1486. doi: 10.1038/cdd.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metallo-proteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 41.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises Parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 42.Yao D, Gu Z, Nakamura T, Shi Z-Q, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates it E3 ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benhar M, Forrester MT, Stamler JS. Nitrosative stress in the ER: a new role for S-nitrosylation in neurodegenerative diseases. ACS Chem Biol. 2006;1:355–358. doi: 10.1021/cb600244c. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura T, Lipton SA. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxid Redox Signal. 2008;10:87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 45.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu Z, Nakamura T, Lipton SA. Redox reactions induced by nitrosative stress mediate protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Mol Neurobiol. 2010;41:55–72. doi: 10.1007/s12035-010-8113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Zhang X, Li C, Guan T, Shang H, Cui L, Li XM, Kong J. S-nitrosylated protein disulfide isomerase contributes to mutant SOD1 aggregates in amyotrophic lateral sclerosis. J Neurochem. 2013;124:45–58. doi: 10.1111/jnc.12046. [DOI] [PubMed] [Google Scholar]
- 48.Budd SL, Tenneti L, Lishnak T, Lipton SA. Mitochondrial and extramitochondrial apoptotic signaling pathways in cerebrocortical neurons. Proc Natl Acad Sci U S A. 2000;97:6161–6166. doi: 10.1073/pnas.100121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto S, Li Z, Ju C, Scholzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn SW, Kim JE, Park KS, Choi WJ, Hong YH, Kim SM, Kim SH, Lee KW, Sung JJ. The neuroprotective effect of the GSK-3β inhibitor and influence on the extrinsic apoptosis in the ALS transgenic mice. J Neurol Sci. 2012;320:1–5. doi: 10.1016/j.jns.2012.05.038. [DOI] [PubMed] [Google Scholar]
- 51.Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulfide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- 52.Inan M, Aryasomayajula D, Sinha J, Meagher MM. Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol Bioeng. 2006;93:771–778. doi: 10.1002/bit.20762. [DOI] [PubMed] [Google Scholar]
- 53.Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP. Chromogranin-mediated secretion of mutant superoxide dis-mutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 54.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, Nukina N. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 55.Quan H, Fan G, Wang CC. Independence of the chaperone activity of protein disulfide isomerase from its thioredoxin-like active site. J Biol Chem. 1995;270:17078–17080. doi: 10.1074/jbc.270.29.17078. [DOI] [PubMed] [Google Scholar]
- 56.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 57.Karala AR, Ruddock LW. Bacitracin is not a specific inhibitor of protein disulfide isomerase. FEBS J. 2010;277:2454–2462. doi: 10.1111/j.1742-4658.2010.07660.x. [DOI] [PubMed] [Google Scholar]
- 58.Nagata T, Ilieva H, Murakami T, Shiote M, Narai H, Ohta Y, Hayashi T, Shoji M, Abe K. Increased ER stress during motor neuron degeneration in a transgenic mouse model of amyotrophic lateral sclerosis. Neurol Res. 2007;29:767–771. doi: 10.1179/016164107X229803. [DOI] [PubMed] [Google Scholar]
- 59.Kanekura K, Suzuki H, Aiso S, Matsuoka M. ER stress and un-folded protein response in amyotrophic lateral sclerosis. Mol Neurobiol. 2009;39:81–89. doi: 10.1007/s12035-009-8054-3. [DOI] [PubMed] [Google Scholar]
- 60.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 61.Faccenda A, Bonham CA, Vacratsis PO, Zhang X, Mutus B. Gold nanoparticle enrichment method for identifying S-nitrosylation and S-glutathionylation sites in proteins. J Am Chem Soc. 2010;132:11392–11394. doi: 10.1021/ja103591v. [DOI] [PubMed] [Google Scholar]
- 62.Honjo Y, Kaneko S, Ito H, Horibe T, Nagashima M, Nakamura M, Fujita K, Takahashi R, Kusaka H, Kawakami K. Protein disulfide isomerase-immunopositive inclusions in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:444–450. doi: 10.3109/17482968.2011.594055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.