Summary
Most bacteria break down a significant portion of their cell wall peptidoglycan during each round of growth and cell division. This process generates peptidoglycan fragments of various sizes that can either be imported back into the cytoplasm for recycling or released from the cell. Released fragments have been shown to act as microbe-associated molecular patterns for the initiation of immune responses, as triggers for the initiation of mutualistic host-microbe relationships, and as signals for cell-cell communication in bacteria. Characterizing these released peptidoglycan fragments can, therefore, be considered an important step in understanding how microbes communicate with other organisms in their environments. In this chapter, we describe methods for labeling cell wall peptidoglycan, calculating the rate at which peptidoglycan is turned over, and collecting released peptidoglycan to determine the abundance and species of released fragments. Methods are described for both the separation of peptidoglycan fragments by size-exclusion chromatography and further detailed analysis by HPLC.
Keywords: Peptidoglycan, PG, murein, peptidoglycan fragments, peptidoglycan turnover, pulse-chase, size-exclusion chromatography, HPLC
1. Introduction
Peptidoglycan is a critical structural macromolecule that protects bacterial cells from osmotic rupture and determines cell shape [1]. In most bacteria, peptidoglycan is composed of a lattice made up of strands of repeating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) sugars cross-linked by three-to-five amino acid peptide stems covalently attached to the MurNAc moiety. This dynamic structure is constantly expanded during cell growth, broken during cell division, and remodeled to accommodate the assembly of large membrane-spanning structures [2]. Both Gram-positive and Gram-negative bacteria break down a significant portion of their peptidoglycan in the course of each cell cycle. In E. coli, almost 50% of the cell’s peptidoglycan is turned over each generation [3], with the vast majority being re-imported into the bacterial cytoplasm via the muropeptide permease AmpG and processed for recycling. Gram-positive bacteria lack an AmpG homolog, and most do not recycle peptidoglycan [4], though there are exceptions [5]. Despite the presence of recycling pathways for muropeptides, the assembly and disassembly of cell wall does not represent a closed system, as some fragments of peptidoglycan escape into the environment. These fragments can induce inflammatory responses in hosts, provide protective immune modulatory signals, aid in initiating mutualism, and coordinate bacteria-bacteria interactions (reviewed in [6,7]).
There is a growing appreciation for the ability of bacteria to release soluble peptidoglycan fragments. Among Gram-negative bacteria, the release of peptidoglycan fragments was thought to only be a significant a feature of Bordetella pertussis and Neisseria gonorrhoeae (Fig. 1), which release large amounts of inflammatory peptidoglycan monomers [8,9]. Soluble, released peptidoglycan fragments typically include those generated by the activity of specific peptidoglycanases, including lytic transglycosylases, endopeptidases, and N-acetylmuramyl-L-alanine amidases [10]. Different bacterial species are equipped with varying numbers of peptidoglycan-degrading enzymes with different specificities. Bacteria also vary in the efficiency of peptidoglycan recycling, making it difficult to predict exactly which peptidoglycan fragments are released by a given bacterial species or strain. For this reason, sensitive methods capable of distinguishing released sugar and peptide moieties of peptidoglycan must be utilized in order to determine the composition of peptidoglycan fragments released by bacteria and what role these fragments play in the interaction of bacteria with their environment.
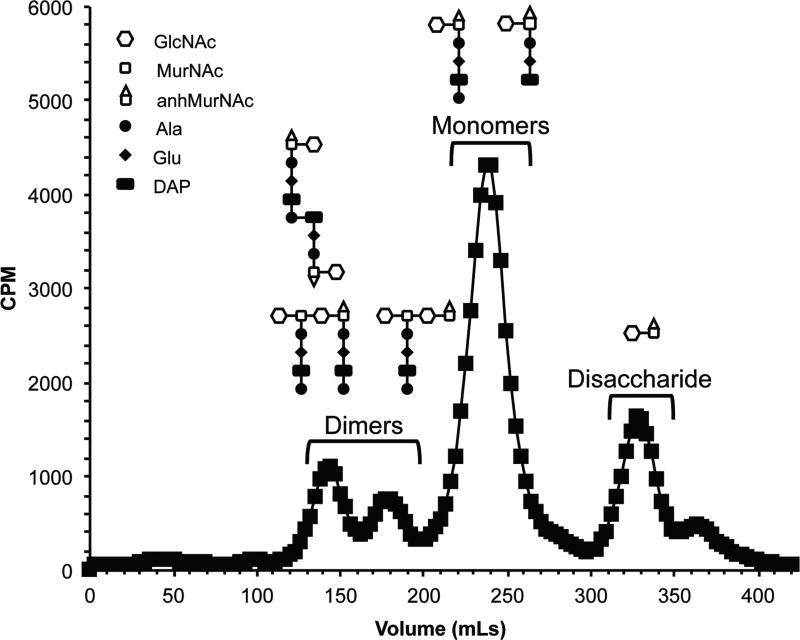
Figure 1.
Fragment release from wild-type Neisseria gonorrhoeae following size-exclusion chromatography using [3H]-glucosamine to label peptidoglycan. The elution of peptidoglycan fragments containing sugars make up clearly defined peaks.
The following protocols describe how to utilize radioactive precursor molecules for labeling peptidoglycan sacculi in order to achieve highly sensitive detection of released fragments and monitor peptidoglycan turnover [11]. Metabolic pulse-labeling using tritiated peptidoglycan precursors, such as D-glucosamine or meso-2,6-diaminopimelic acid allow incorporation of traceable radioactivity into the sugar backbone or peptide chain of peptidoglycan, respectively. Radiolabeling provides a quantifiable way to measure the abundance of particular released fragments within and between bacterial strains. Bacterial species differ in the fragments of peptidoglycan they release (Fig. 2) and can include: free peptides, free GlcNAc-MurNAc disaccharides, monosaccharides, monomers (a single disaccharide and single peptide stem), and/or dimers (two disaccharide subunits and one or two peptide stems). Since all of these molecules exist within a molecular weight range of 200–2000 Daltons, we present a size-exclusion chromatography methodology to differentiate between released fragments within this size range.
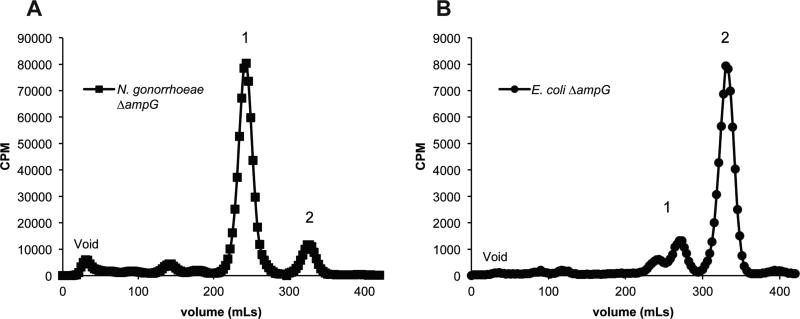
Figure 2.
Peptidoglycan fragments released from N. gonorrhoeae ΔampG (A) and E. coli ΔampG (B). N. gonorrhoeae lacking ampG release mostly peptidoglycan monomer (1), whereas E. coli lacking ampG release mostly disaccharide (2).
More detail is often desired in characterizing released fragments, with HPLC analysis as the standard in the field for the discrimination of chemical differences between similarly-sized muropeptides [12]. Analysis by HPLC can be used to determine such characteristics as the length of peptide chains, the presence of modifications such as acetylation, or the type of bond present on the MurNAc sugar (reducing vs. anhydro), all of which can have implications for the biological function of the released molecules [9,13,14]. For this purpose, we present a typical application of HPLC utilizing a C18 column for separating two related species of peptidoglycan monomer (Fig. 3a) and two configurations of peptidoglycan dimer (Fig. 3b), both of which elute as single peaks from size-exclusion columns. Our goal is to provide these protocols as a framework for the exploration of peptidoglycan fragment release in many species. Determining the peptidoglycan fragments released by various bacteria has the potential to reveal not only novel information about host-bacteria and bacteria-bacteria interactions but also help to better define the basic biology of peptidoglycan metabolism.
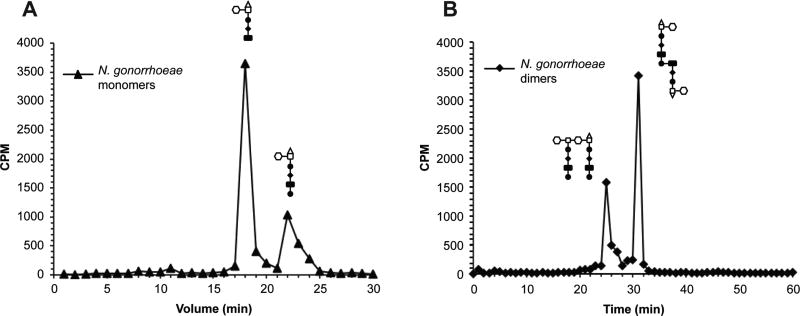
Figure 3.
Reversed-phase HPLC of radiolabelled fragments. Pooled fractions from size-exclusion chromatography of wild-type N. gonorrhoeae supernatant were analyzed by HPLC with a C18 column. (A) Monomer fractions were run using a 4 to 13% gradient of acetonitrile with 0.05% TFA at 1 mL/min for 30 min. Peaks with retention times of 18 minutes and 22 minutes correspond to 1,6-anhydrodissacharide-tripeptide and -tetrapeptide respectively. Quantification of peaks reveals a 3:1 ratio of tripeptide to tetrapeptide peptidoglycan monomers released. (B) Dimer fractions were run over a 5 to 25% gradient of acetonitrile with 0.05% TFA at 1 mL/min for 60 min. Peaks with retention times of 25 minutes and 31 minutes correspond to glycosidically-linked and peptide-linked dimers respectively.
2. Materials
Prepare all media and solutions with deionized water and autoclave or filter sterilize. The media listed below was originally adapted for N. gonorrhoeae (GC), but has also been successfully used for radiolabeling E. coli peptidoglycan. For species with specific nutritional requirements, other growth media can be employed provided a version not containing glucose is used during pulse-labeling.
2.1 [3H]-glucosamine labeling of peptidoglycan sacculi
GC medium base (GCB): 1.5% proteose peptone no. 3, 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl, 0.1% corn starch, and 1.0% agar.
GC medium base broth (GCBL): 1.5% proteose peptone no. 3, 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl, and 0.042% sodium bicarbonate
GC medium base broth with glucose supplements (cGCBL): GCBL with Kellogg supplements I (22.2 mM glucose, 0.68 mM glutamine, 0.45 mM cocarboxylase) and II (1.23 mM Fe(NO3)3).
GCBL with pyruvate supplements. GCBL with Kellogg supplements I (36.35 mM pyruvate, 0.68 mM glutamine, 0.45 mM cocarboxylase) and II (1.23 mM Fe(NO3)3).
GCB plates: GC medium base with Kellogg supplements I and II added.
CO2 incubator.
Petri dishes.
Tube rotator.
Polyester-tipped applicator swabs.
Pipette with disposable tips.
Vortex mixer.
[6-3H]-glucosamine.
Sterile 15 mL conical vials.
Clinical centrifuge.
Microcentrifuge.
Microcentrifuge tubes.
10 mL syringes.
0.2 µm syringe filters.
Scintillation counter.
Scintillation vials.
Scintillation fluid.
Boiling water bath.
50 mM sodium acetate pH = 5.
8% (w/v) sodium dodecyl sulfate solution.
Microcentrifuge cap locks (for boiling steps).
2.2 Size-exclusion chromatography
1L Erlenmeyer flasks for reconstituting and autoclaving beads.
Two 2.5 cm × 75 cm glass columns (e.g., BioRad Econo-Columns).
Column buffer reservoir (500 mL – 1 L capacity).
3.2 mm (internal dimension) silicone tubing.
Stopcocks.
Polyacrylamide beads with low molecular weight exclusion limit (e.g., Bio-Rad Bio-Gel P6, medium size, with 6,000 Da nominal exclusion limit).
Polyacrylamide beads with high molecular weight exclusion limit (e.g., Bio-Rad Bio-Gel P30, medium size, with 40,000 Da nominal exclusion limit).
0.1M Lithium Chloride (LiCl), autoclaved (1–2 L/column run plus 5L if pouring new columns).
Disposable pipettes and pipetting device.
Automated fraction collector.
Disposable borosilicate glass tubes to fit fraction collector.
Scintillation vials with lids.
Scintillation fluid cocktail (i.e. PerkinElmer Ultima-Flo AP)
Repeating scintillation fluid dispenser (recommended).
Vortex mixer.
Scintillation counter capable of measuring tritiated [3H] samples.
2.5 cm × 20 cm (98 mL volume) glass column for desalting.
Speed-vac, lyophilizer, rotary evaporator or other concentrating device.
2.3 Analysis of fragments by reversed-phase HPLC
HPLC, UPLC or other liquid chromatography system with loading loop and trigger-style injector.
C18 analytical column with in-line guard column (protocols described are optimized for a 250 mm × 4.6 mm column with 5 µm pore size).
Water, HPLC-grade, submicron filtered.
Trifluoroacetic acid (TFA).
Acetonitrile (ACN), HPLC grade.
500–1000 mL glass bottles thoroughly cleaned with ultrapure or HPLC-grade water.
Automated fraction collector for HPLC.
Disposable borosilicate glass tubes to fit fraction collector.
3. Methods
3.1 Quantitative metabolic radiolabeling of bacteria and collection of released peptidoglycan fragments
Metabolic radiolabelling of bacteria is accomplished under conditions that promote the rapid incorporation of labeled precursors into peptidoglycan, typically conditions of exponential growth. The conditions listed here have been refined for labeling of Neisseria species, but these conditions also support labeling of E. coli and should be considered adaptable to suit the particular growth requirements of most bacteria (Fig. 2). In general, bacteria should be cultured at a temperature promoting rapid growth (37°C with aeration for E. coli and Neisseria). All media should be prepared and warmed in advance to avoid cold-shock and autolysis. Manipulations of cultures can be done at room temperature, but work should be done quickly.
Streak out frozen stocks of strains to be labeled to single colonies on GCB plates. Incubate plates overnight in a 37°C degree incubator with 5% CO2.
The following day, streak colonies onto GCB plates. Grow plates overnight at 37°C with 5% CO2. Make liquid media: GCBL, GCBL with glucosamine supplements (cGCBL), and GCBL with pyruvate supplements.
Aliquot media needed for the next day’s experiments into 15 mL conical tubes and place at 37°C to warm.
On the following day, begin cultures for radiolabeling. Swab each overnight plate into 3 mL of warm cGCBL, vortex, and measure OD540.
Use swabbed cultures to create 2 × 3 mL cultures at OD540 = 0.25 in cGCBL in 15 mL conical tubes. Work quickly to avoid cold-shock and autolysis.
Grow cultures at 37°C in a roller drum for 3 hours or until late log phase.
After 3 hours, measure the OD540 of cultures to determine the appropriate volume of culture to obtain 2 × 3 mL cultures at OD540 = 0.2 (approximately 1 × 108 cfu/mL) and transfer the culture volume necessary for seeding each culture into a microcentrifuge tube(s).
Centrifuge cells at 15,000 × g for 1 min, discard supernatant, and wash cells with 1 mL of warm GCBL.
Transfer cells to 15 mL conical tubes and bring to a total volume of 3 mL in warm GCBL with pyruvate supplements.
Add 10 µCi/mL of [3H]-glucosamine to each 3 mL culture (2 cultures per strain) and grow as above for 45 min to pulse-label. The pulsing time can be extended to label more peptidoglycan, but longer times will eventually lead to the labeling of other cellular components. If labeling efficiency is determined to be too low, the concentration of [3H]-glucosamine per culture or the total number bacteria labeled can be increased.
At the conclusion of pulse labeling, centrifuge cultures in a clinical centrifuge for 5 min at 3,500 × g. Discard media and use 1 mL warm GCBL to transfer bacteria to a microcentrifuge tube. Centrifuge bacteria for 1 min at 15,000 × g. Remove supernatant and wash pellet with 1 mL warm GCBL, centrifuging again for 1 min at 15,000 × g and discarding media to remove unincorporated label. Suspend each strain in 6 mL warm cGCBL.
For quantitative comparisons between strains, immediately take 60 µL of each 6 mL culture for scintillation counting. Place strains at 37°C while measurement is taking place. Using scintillation measurements, calculate the CPM/mL of each culture and normalize the CPM to match the culture with the lowest reading by removing volume from cultures with higher readings. If only one strain is being labeled or if quantitative comparison is not desired, this step can be skipped.
Split 6 mL volume of each strain into 2 × 3 mL cultures (to provide sufficient aeration) and return cultures to the roller drum at 37°C to continue growth for 2.5 hr.
After the chase period is completed, centrifuge cultures in a clinical centrifuge for 5 min at 3,500 × g. Remove supernatant from each strain and filter through a 0.2 µm syringe filter. If desired, remove 60 µL of filtered supernatants and measure CPM by scintillation counting to determine the percentage of peptidoglycan released relative to the beginning of the chase period.
Store supernatants at −20°C for further analysis by size-exclusion chromatography and/or HPLC.
3.2 Analysis of peptidoglycan turnover
The rate of peptidoglycan turnover is determined from the amount of radiolabelled peptidoglycan remaining in the sacculi during a chase period following pulse-labeling. This method measures the amount of radiolabel in the sacculi as peptidoglycan is removed from the cell wall and either incorporated into other cellular components or released into the environment.
To assess the rate of peptidoglycan turnover, begin by metabolically labeling cultures and normalizing culture volumes to total CPM by completing steps 3.1.1–12.
Return 6 mL cultures to the roller drum at 37°C to continue growth. At desired time points (e.g., 0, 0.5, 1, 2, 4 hrs), remove 1 mL from each culture and transfer to a microcentrifuge tube. Centrifuge for 1 min at 15,000 × g and remove supernatant.
To isolate macromolecular peptidoglycan, suspend bacterial pellets in 165 µL of 50 mM sodium acetate (pH 5) and 165 µL 8% sodium dodecyl sulfate. Boil samples for 30 min using microcentrifuge cap locks.
Add 800 µL unlabeled carrier (see Note 4) and collect insoluble macromolecular peptidoglycan by centrifugation for 30 min at 17,000 × g, 15°C or room temperature (SDS will precipitate at lower temperatures).
Carefully remove the supernatant and suspend the insoluble peptidoglycan pellet in 200 µL of sterile water. Measure CPM from each time point to calculate the amount of labeled peptidoglycan remaining at each time point compared to T=0. The rate of radioactivity loss from the sacculi provides the peptidoglycan turnover rate. The entire peptidoglycan turnover procedure should be repeated for a total of three independent experiments to calculate the significance of observed differences of peptidoglycan turnover.
3.3 Size-exclusion chromatography
Released peptidoglycan fragments in conditioned medium from bacterial culture must be separated from any extraneous labeled sugars, large fragments of lysed sacculi, and components of the culture medium that could impede downstream analysis of fragments. The following method describes the use of size-exclusion chromatography to separate and collect fractions containing commonly observed peptidoglycan fragments. The amount and relative proportions of fragments can be determined by quantifying total CPM of each peak that corresponds to certain released fragments.
3.3.1 Pouring new size-exclusion columns
If pouring a new tandem size-exclusion chromatography column, hydrate and sterilize both the low and high molecular weight exclusion limit beads prior to use. A general rule for packing a column is to use twice the buffer volume as the total column volume for bead hydration. For the Bio-Rad Bio-Gel beads, hydrate 52 g of P30 and 70 g of P6 beads (separately) in 500 mL of 0.1 M LiCl overnight. The next day, autoclave beads on a 30 min liquid cycle and let cool to room temperature.
Once beads have settled, decant as much liquid as possible and add 500 mL of fresh, autoclaved 0.1 M LiCl, swirling to mix. Let settle and repeat the decanting and filling process four times.
Prior to filling, sterilize columns by autoclaving at 121°C for 15 min or by rinsing columns with either 2 N NaOH or 100% ethanol followed by two rinses with 0.1 M LiCl. Securely attach a stopcock or other on/off valve directly to the bottom of each column.
Once the beads have settled, decant ~150 mL of the excess buffer, swirl beads to resuspend, and pour the P6 beads into one column and the P30 beads into the other, making sure to keep the stopcocks open during filling and adding buffer as needed to keep columns hydrated. Save any beads (especially P6) that do not fit initially. Each column will hold ~350mL of hydrated beads. Stop column flow by closing the stopcocks when columns are filled. Allow beads to settle 12–18 hr.
Following settling, pipette off any excess LiCl from the top of the P6 column and fill to the top with any remaining hydrated P6 beads to avoid leaving any air/buffer pocket.
To connect the columns, mount the P30 column above the P6 column in a standing clamp apparatus (a minimum 8 ft of clearance floor-to-ceiling is recommended for proper installation). Remove the stopcock from the column filled with P30 beads and use silicone tubing to attach the bottom of the P30 column to the top of the P6 column. When complete, attach a reservoir to the top of the P30 column. Additional tubing will be needed at the bottom of the P6 column to connect the outflow to an automated fraction collector.
The reservoir at the top of the tandem column apparatus should always be kept with autoclaved 0.1 M LiCl buffer above the level of the beads when the column is not in use. Beads should never be allowed to dry after being hydrated. If a column does dry (typically the upper column), repeat steps 3.3.1.1–7 for the dried column. Used beads can be repoured following rehydration and autoclaving.
3.3.2 Running samples on size-exclusion columns
To introduce sample to prepared columns, carefully remove buffer from the reservoir with a pipette, avoiding disturbing the beads at the top of the column. To completely remove the liquid, excess buffer can be drained off by opening the stopcock.
Once buffer has drained to expose the top of the beads, apply filtered culture supernatant (step 3.1.15) to the top of the column by decanting or pipetting a single labeled sample onto the column. Immediately begin a timer to start tracking the void time (this must be determined empirically but is typically 2–4 hours) (see Note 7). Allow the sample to soak into the column.
Once the sample has entered the column, apply a small amount of 0.1 M LiCl to the top of the column (5–10 mL), and allow this buffer to soak in as above. Adding a small amount of buffer here keeps the column flowing and hydrated, without risking dilution of your sample.
Once a small amount of buffer has flowed into the column, the reservoir can be filled to capacity and covered with a vented lid to avoid contamination. During the column run (12–18 hours) this reservoir should always contain 0.1 M LiCl, which can be achieved through the use of a large reservoir (>1 L) or by establishing a gravity-fed continuous flow or siphon to the column-mounted reservoir from a larger source bottle or tank.
At the conclusion of the void time, initiate collection of fractions, which can be collected based on time or volume. Automated fraction collectors that hold ≥175 collection tubes are recommended to avoid the need to switch collection drums or racks during collection, or collect manually. Automated units that collect volumes typically count drops via an electronic eye and may require some testing to refine the desired volume. For the analysis of peptidoglycan fragments from gonococci, 150–170 × 3 mL fractions (at ~75 drops/fraction) are needed following the void volume, with the exact number based upon the labeling technique (see Notes 1and 2). At the conclusion of fraction collecting, it may be desirable to run the column with buffer for an additional 2–3 hours to assure the voiding of all labeled material prior to the next run. When complete, be sure to stop column flow and cap the column.
To measure radiolabeled peptidoglycan fragments, a portion of each collected fraction should be mixed with scintillation fluid. For [3H]-glucosamine-labeled fragments, a mixture consisting of 0.5 mL of fraction volume and 3 mL scintillation fluid (i.e. Ultima-Flo AP - Perkin Elmer) is sufficient for detection. To increase detection, a mixture of 1 mL of fraction volume and 3 mL of scintillation fluid can be used.
Detection of radiation is performed by scintillation counting and CPM/fraction can be graphed to determine when fragments elute and in what proportions (Fig. 1). Samples labeled together (quantitatively) in a single experiment should be analyzed in successive runs on the same sizing column and can be initiated at the conclusion of the previous run. Graph CPM per fraction, or CPM per column volume, to determine which fractions make up peaks containing peptidoglycan fragments of interest (Fig. 1)
Save the remaining unmeasured portion of each fraction for additional analysis. As peaks of interest are identified from scintillation counting, fractions that make up those peaks can be pooled and stored at −20°C.
3.3.3 Concentration and desalting of collected fractions for HPLC
Analysis of radiolabelled fractions requires only a portion of the total fraction volume, leaving sufficient material for further analysis. Since downstream HPLC analysis is often desired and the injection volume for most HPLC instrumentation is small (10 µL – 2 mL, depending on the column size and injection loop), it is often necessary to concentrate all fractions comprising a single peak into a smaller volume. Concentration of fractions, however, will result in concentration of the LiCl from the column running buffer. It is recommended to remove this salt prior to separation by HPLC, though a shortcut to increase the throughput of analysis is described below (see Note 8).
A 2.5 cm × 20 cm glass column (98 mL volume) should be prepared with hydrated, sterilized P6 beads as above, except that Ultrapure or HPLC-grade water should be substituted for LiCl, since this column will be used for desalting. Column should be fitted with a stopcock on the bottom and reservoir on the top.
Pooled fractions of interest from step 3.3.2.8 should be reduced in volume by dehydration (typically in a speed-vac or lyophilizer apparatus), in equipment approved for radioactive materials.
Once dehydrated and suspended in a smaller volume (<3 mL), sample can be applied to the top of the column (as in steps 3.3.2.2–4), using water to fill the reservoir rather than LiCl. Once sample has been applied, collect the void volume into a graduated cylinder or other container with volume markings. Depending on the size and chemical composition of the fragment of interest (peptidoglycan monomer, peptidoglycan dimer, peptides, etc.), different combinations of void volume and fractions collected may be required and should be determined empirically. Generally for a 2.5 × 20 cm column, collection of a ~20 mL void is followed by manual collection of 15–20 × 1 mL fractions to retrieve peptidoglycan monomers.
Each desalted fraction should be sampled (~100 µL out of 1 mL), mixed with 3 mL scintillation fluid, and measured by scintillation counting to confirm that the radioactive fraction applied to the column was recovered. Peptidoglycan fragments should elute as distinct peaks spread over several fractions.
Aqueous products containing detectable radiation should then be reduced in volume as above and suspended in a small volume (100–500 µL). This final suspension should be measured for levels of radioactivity prior to storage (at −20°C) and is suitable for analysis by HPLC.
3.4 Analysis of fragments by reversed-phase HPLC
Size-exclusion chromatography provides information on the size and quantity of released peptidoglycan fragments, allowing quantitative comparisons of released fragments within a single sample or between various strains, species, or conditions. Details such as the exact peptide stem length among monomers, the linkages present within dimers, and whether fragments have reducing or 1,6-anhydro ends are better explored through additional chromatography techniques. Reversed-phase HPLC using a C18 column is a common approach for analyzing peptidoglycan fragments (and is described here). Other columns including those with different carbon-chain length bonded phases, size-exclusion columns, cation/anion exchange columns, and a variety of length and pore size options could be considered depending on the experimental question.
Prior to separation of peptidoglycan fragments by HPLC, necessary buffers should be made and degassed (under vacuum) in advance. Many effective separations of peptidoglycan monomers, dimers, and peptides on C18 columns can be accomplished with two buffers: A) HPLC-grade water + 0.05% TFA, and B) 25% acetonitrile (in HPLC-grade water) + 0.05% TFA. These buffers should ideally be made fresh or used within one week. Prior to the first use of the day, prime the HPLC lines and clean the column first by flushing with Buffer B then with Buffer A. A blank sample (i.e. buffer only) should be run before each day of use.
While the analysis of radiolabelled fragments will typically involve detection by scintillation counting of fractions, peptidoglycan fragments can also be detected at 206 nm. UV detection should be performed any time fragments are separated by HPLC to assure that the HPLC is operating properly. Turn on the UV lamp and allow to warm for at least 1 hr to stabilize readings.
Create an HPLC program for the samples to be analyzed. For separation of monomers, a gradient of 4–13% acetonitrile over 30 min at 1 mL/min allows separation of 1,6-anhydrodisaccharide-tripeptide monomer from 1,6-anhydrodisaccharide-tetrapeptide monomer (Fig. 3a). Reducing-end fragments have a shorter retention time compared to fragments with anhydro linkages.
Prior to starting the run, prepare the sample from step 3.3.3.5 so that at least 1000 CPM will be injected into the column. Samples can be prepared for loading either via the “partial fill” method (<1/2 loop volume) if sample is limiting, or the “complete fill” method (2–5 loop volumes) for greatest precision.
Immediately following injection, begin fraction collection using an automated fraction collector set to collect fractions using ≤1 min increments over the entire run time.
At the conclusion of the run, combine the entire volume of each fraction with 3 mL scintillation cocktail, vortex thoroughly to mix, and measure by scintillation counting (Fig. 3). Clean column after each run by running 25% acetonitrile (100% of Buffer B) to elute any remaining material from the column, then flushing with Buffer A.
Notes
As an alternative to labeling the peptidoglycan sugar backbone using [3H]-glucosamine, [3H]-rac-2,6-diaminopimelic acid ([3H]-DAP) can be used to label peptide stems. Labeling with [3H]-DAP can be done by substituting GCBL with pyruvate supplements for Dulbecco’s Modified Eagle’s Medium (DMEM) without cysteine. At step 3.1.9, wash and dilute cells into DMEM without cysteine supplemented with 100 µg/mL methionine and 100 µg/mL threonine. Due to less efficient labeling by [3H]-DAP, add 20 µCi/mL [3H]-DAP to each 3 mL culture at step 3.1.10 and proceed as noted above.
Different peptidoglycan precursors and different isotopes are available that can be used to radiolabel peptidoglycan. The best peptidoglycan precursor to use will depend of the species and strain of bacteria to be labeled. For example, meso-DAP is an amino acid specific to peptidoglycan but is only found in Gram-negatives and some Gram-positive rods (Gram-positives typically use L-Lysine in place of meso-DAP). Combinations of precursors and isotopes, such as [14C]-glucosamine in combination with [3H]-DAP, can be used to measure the release of both sugars and peptides simultaneously.
If quantitative labeling is desired (in step 3.1.12) but there is not immediate access to a scintillation counter, collect 60 µL of culture as above and take CPM readings when possible. At the conclusion of the chase period, take another 60 µL sample from the filtered supernatant and measure CPM. It is then possible to normalize the counts from the released fragments to the total counts at the beginning of the chase period. Although this method is not preferred, fractions can be characterized as % of total CPM.
To increase the efficiency of sacculi recovery and make centrifuged material more easily visible, it is common to use unlabeled (cold) carrier when working with small quantities of radiolabelled peptidoglycan. Carriers can be in the form of unlabeled macromolecular peptidoglycan or even whole bacterial cells. One commercially available option is lyophilized preparations of Micrococcus luteus peptidoglycan.
Always monitor a size-exclusion chromatography column for which no large-volume reservoir is providing continuous flow. Never allow a column to run dry. Cracked or caked beads that result from significant drying are signs that the column should be emptied of beads and repoured. Over time, prolonged use will cause the beads in the column to compact, slowing the flow of samples and changing the void time. If unacceptable slowing of runs or broadening of known peaks is observed, the column(s) should be emptied of beads and repoured.
Care should always be taken to avoid spills and overflows of radioactive material. Column void and flow-through material should be collected in a sufficiently sized container placed within secondary containment and surrounded by absorbent material. All fractions and flow-though should be collected, tested, and disposed of in accordance with local and institutional disposal regulations.
When establishing a new size-exclusion chromatography system, or repouring columns, it is recommended to determine the time required for samples to pass entirely through the system (void time), since this will impact the time range for collecting relevant fractions. To determine the void time, prepare a 3–6 mL solution of Blue Dextran (Sigma-Aldrich) in water and apply to the top of the sizing column as in step 3.3.2. Proceed as if running a labeled sample, timing the movement of the blue dextran until it exits the bottom of the columns. The time from addition of blue dextran solution until it exits the column is the void time.
HPLC is routinely used for the desalting of nucleotides and proteins and some columns can handle the introduction of soluble salt without changing retention times. To increase the throughput of fraction analysis from size-exclusion chromatography, it is possible to run a portion of pooled fractions without the lengthy concentration-desalting-concentration protocol. To skip directly from obtaining column fractions to analysis by HPLC, the HPLC injector should be fitted with a larger volume loop (about 2 mL). Pooled fractions from size-exclusion (typically 25 mL from the center of the peak of interest) must also be sufficiently radioactive that >1000 CPM are available in your loaded volume to achieve reliable detection of products. Pooled fractions taken directly from a sizing column (step 3.3.2.8) can then be run as in step 3.4.2, though caution is advised when interpreting comparisons between these runs and any analyses done with primarily aqueous samples loaded from smallervolume loops.
In complex samples analyzed by HPLC, it is possible that multiple products can elute simultaneously. These problems can occasionally be resolved by executing a longer run with a slower ramp or modifying the beginning or ending concentrations of acetonitrile. In some cases it may be necessary to use a different HPLC column for full separation of certain products.
Acknowledgments
This work was supported by the National Institutes of Health through grants AI097157 and AI099539.
References
- 1.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62(1):181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Micro. 2012;10(2):123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodell EW. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163(1):305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reith J, Mayer C. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol. 2011;92(1):1–11. doi: 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- 5.Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, Mayer C. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J Bacteriol. 2010;192(12):3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4(9):710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin J. The medium is the message: interspecies and interkingdom signaling by peptidoglycan and related bacterial glycans. Annu Rev Microbiol. 2014;68:137–154. doi: 10.1146/annurev-micro-091213-112844. [DOI] [PubMed] [Google Scholar]
- 8.Cookson BT, Cho HL, Herwaldt LA, Goldman WE. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun. 1989;57(7):2223–2229. doi: 10.1128/iai.57.7.2223-2229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha RK, Rosenthal RS. Release of soluble peptidoglycan from growing conococci: demonstration of anhydro-muramyl-containing fragments. Infect Immun. 1980;29(3):914–925. doi: 10.1128/iai.29.3.914-925.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32(2):259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 1994;13(19):4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desmarais SM, de Pedro MA, Cava F, Huang KC. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol Microbiol. 2013;89(1):1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magalhaes JG, Philpott DJ, Nahori MA, Jehanno M, Fritz J, Le Bourhis L, Viala J, Hugot JP, Giovannini M, Bertin J, Lepoivre M, Mengin-Lecreulx D, Sansonetti PJ, Girardin SE. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 2005;6(12):1201–1207. doi: 10.1038/sj.embor.7400552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veyrier FJ, Williams AH, Mesnage S, Schmitt C, Taha MK, Boneca IG. De-O-acetylation of peptidoglycan regulates glycan chain extension and affects in vivo survival of Neisseria meningitidis. Mol Microbiol. 2013;87(5):1100–1112. doi: 10.1111/mmi.12153. [DOI] [PubMed] [Google Scholar]