Abstract
Selection of molecular targets based on disease understanding is a dominant paradigm in drug discovery. We argue that a focus on classes of targets with central roles in biology provides a complementary approach that has higher quality outcomes in early discovery efforts.
“In all affairs it’s healthy now and then to hang a question mark on the things you have long taken for granted.”
—Bertrand Russell
Though a disease-centric approach to drug discovery and development has dominated academic and industrial biomedical research, it often fails to leverage hard-won expertise in chemistry, biology and structural biology. Rather, these disciplines thrive in the context of protein-family, target class–focused approaches. Despite the prevalence of target class science in industrial and some academic drug discovery efforts, a set of principles underlying this strategy have yet to be delineated. We provide a brief historical context of successful target class efforts and offer insight into important considerations in its application to a wider breadth of protein families.
The path that a drug discovery program follows is often dictated by selection of a molecular target, which is typically based on disease-focused knowledge. Target selection sets in motion the creation of assays often intended to support a high-throughput screen (HTS) of a diverse chemical library, which, if successful, results in a ‘hit-to-lead/probe’ effort. Only when a high-quality lead/probe is in hand can the pharmacologic relevance of the selected target in cellular or animal models of disease begin to be assessed. Thus, a target-to-hit-to-lead effort can take 2–3 years and consume considerable financial investment before the original hypothesis can be tested pharmacologically. This is a risky endeavor.
Target selection has accordingly been written about at length, with a focus on risk minimization1. Risk can be categorized as ‘validation risk’ (the likelihood that modulation of the target will have a favorable outcome in patients) or ‘technical risk’ (the likelihood that a tolerable molecule that modulates the target in patients can be discovered). A target class strategy is based on the premise that the scientific expertise and physical assets developed within a protein family will greatly diminish the chances for technical failure. In addition to accumulating knowledge, the systematic evaluation of functionally related proteins seeks to reduce technical risk by diversifying the target space of ligandable pockets while also increasing the potential clinical applications, so that failure to validate a single target is not fatal to a program. This approach enables efficient preclinical pharmacologic validity assessment while driving innovation by exploring less well-documented targets with high-quality probes. We believe that a target class approach to drug discovery can complement disease-focused strategies, enhance the role of chemical sciences in driving innovation and address some of the risks inherent in preclinical discovery. The ability of this approach to also prospectively address inevitable ligand selectivity issues within a protein family is another inherent advantage.
Target class selection
Selection of a target class begins with evaluation of a family of structurally and/or functionally related proteins that have been broadly characterized for their biological significance. Ideally, the proteins in the family would be amenable to high-resolution structural studies, and an extant set of structural data for at least some family members would reveal opportunities for small-molecule intervention. For example, when initiating our chemical biology program 9 years ago, we evaluated the methyllysine (Kme) reader proteins as a potential target class2. At that time, chromatin regulation was emerging as an exciting area of biology with therapeutic relevance3. Additionally, Kme readers were known to be a large (>200 members), structurally characterized target class with a conserved functional recognition motif, and yet no ligand discovery efforts had been published. While evidence that some Kme readers had specific disease implications was emerging4, others remained largely unexplored. The major hurdle envisioned for success in this target class was the ability to design or discover high-affinity, selective ligands with sufficient cellular activity to use in target validation studies. As this hurdle was within our skill set to address, Kme readers were chosen as the target class for our program. The success in ligand discovery for the bromodomain target class of acetyllysine readers, which followed soon after, also validated our selection of a protein–protein interaction module to influence chromatin regulation5. Here we use Kme readers, along with the archetypical target classes of kinases6, G-protein-coupled receptors (GPCRs)7 and nuclear receptors (NRs)8, to illustrate the history and proven advantages of this scientific strategy. Though not discussed here, ion channels9 have seen similar successes in the application of target class approaches to ligand discovery.
Though Kme readers fall broadly within the category of epigenetic targets, it is important to specify that ‘epigenetic proteins’ is not a designation for a target class. Specifically, proteins within a target class are interrelated via structural or functional similarities from a mechanistic and chemical viewpoint. For instance, within the writers of the epigenetic code, histone methyltransferases (HMTs) share some similarities in mechanism of action and substrates with histone acetyltransferases (HATs), but HMTs and HATs are not members of the same target class. Crossover between the chemical space of the cofactors (S-adenosylmethionine and acetyl-CoA for HMTs and HATs, respectively) is limited, and the substrate recognition sites are very different, even though they both recognize lysine. Accordingly, the regions of chemical space likely to yield specific HMT ligands should be more enriched for hits versus other HMTs than for HATs.
Building a target class platform
Though different protein classes require domain-specific considerations, a set of defined practices form the basis of a target class approach. Among these, chemical strategies, in vitro assay development and structural studies should be given particular consideration during the early discovery stage (Fig. 1), whereas cellular studies and complex mechanism of action (MOA) considerations become critical as high-quality in vitro ligands emerge.
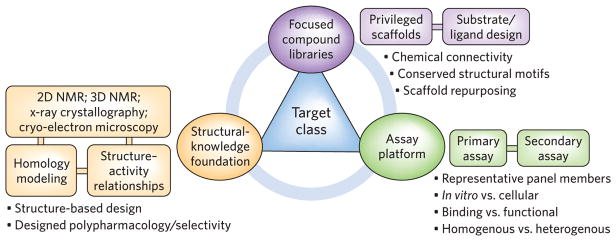
Figure 1.
The three principles of target class discovery. The purposeful design and construction of focused compound libraries based on structural and ligand similarities, often guided by available structural information, can effectively enable target class hit discovery when coupled with robust assay platforms that allow extensive cross-screening against a carefully chosen panel of representative proteins. Ultimately, a target class platform composed of these three interconnected principles will deliver high-quality ligands in a rational, systematic, and productive fashion with diminished chances for technical failure.
Generation of focused libraries
A prerequisite for the optimization of a drug or chemical probe is a synthetically tractable ligand or ‘hit’. Many successful discovery campaigns have relied on HTS of large, diverse, small-molecule libraries for the discovery of such a starting point. This is an expensive endeavor, as the creation, curation and growth of such a screening collection requires staff and resources supported by a long-term funding mechanism10. In contrast, a focused compound screening approach can be more practical, cost efficient and information rich, and is an attractive option for hit discovery within a specific target class. Emerging platforms in chemical biology seek to reduce the investment required for diversity-based hit discovery, and we consider these to be very promising for target class exploitation (Box 1). If resources are available, HTS efforts can generate unanticipated ligands and binding modes that can then enrich focused screening sets.
Box 1. Leveraging target class approaches in chemical biology.
Target class science combines naturally with modern approaches in hit discovery and chemical biology. For example, in our Kme reader work, we recently demonstrated that focused library generation via one-bead-one-compound approaches is amenable to target class ligand discovery, selectivity optimization and screening23, but additional combinatorial approaches including DNA display technologies, other in vitro display platforms (phage and mRNA), and dynamic combinatorial chemistry would benefit from a target class focus. Target class strategies provide a means for rigorous identification of selective hits within these diverse libraries. Additionally, activity-based protein profiling (ABPP) has been applied to target discovery, whereby a reactive probe differentially captures the active site of a class of enzymes in normal and diseased cells24. These differential activities may lead to new strategies for combination therapies (or designed polypharmacology) and identify new molecular targets. Lastly, the emerging power of proteomics combined with thermal profiling presents an unbiased approach to cellular compound profiling to interrogate biological mechanisms both within and outside the target family25.
For emerging target classes, focused library design may begin by considering the overarching chemical connectivity as defined by the structural similarity of the native ligand(s), cofactor(s) or substrate(s) and the concordant recognition elements within each target’s binding sites. For example, in the construction of our Kme reader–biased compound set, we began by making compounds containing secondary, tertiary and quaternary amines attached to peptidic and small-molecule scaffolds to recapitulate the interactions of methylated lysine11. In the case of enzymes, this practice can be expanded to exploit the known geometry and catalytic mechanism of the active site. As conserved ligand recognition motifs among target class members are likely to recognize related chemical features, this increases the potential for discovering chemical entities that interact with several members of the target class. Though the design and creation of such focused libraries requires an initial knowledge base, chemistry resources and a substantial upfront time investment, they serve as an invaluable, enduring asset for target class hit discovery and are meant to evolve and grow over time as knowledge of the druggable features of the proteins and compound binding modes comes to light.
When a target class that has been well studied is being considered, the use of privileged scaffolds has emerged as a means to discover synthetically tractable starting points (Fig. 2). Privileged scaffolds comprise a core structure that has been characterized as having activity against one or more targets within a class. These scaffolds theoretically enable rapid diversification via rational modification to generate libraries that are likely to yield novel inhibitors with potential for improved potency as well as unique selectivity profiles.
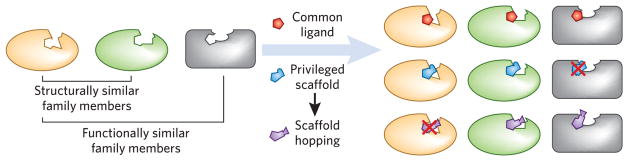
Figure 2.
Overview of target class scaffold hopping and optimization. Evaluating proteins that are connected by their structural similarities or functional redundancies enables diverse ligand hypotheses to be tested against a set of related targets. Such ligand hypotheses can be the result of structure- or ligand-based design efforts. Alternatively, privileged scaffolds, which are known to have activity against one or more targets within the class, may be available and can serve as the basis for creating focused compound libraries that are likely to yield ligands of novel potencies and selectivity profiles. Screening a carefully chosen panel of related targets can also facilitate scaffold hopping when a hit for one target is shown to have affinity for another member of the family.
Taking the kinase target class as an example, early hit-discovery efforts via diversity screening revealed a set of privileged ATP competitive scaffolds that interact with the critical ‘hinge-region’ via 1–2 hydrogen bonds. Subsequently, the different ‘nooks and crannies’ of the ATP binding site, which are not highly conserved among family members, were successfully exploited to improve potency and selectivity12. Over a 20-year period, medicinal chemistry efforts have resulted in more than 30 FDA-approved kinase inhibitors, and advances in structural studies and assay formats has led to both selective covalent and non-ATP competitive inhibitors13. Protein kinases are therefore fully enabled for rapid ligand discovery and target validation efforts, with many unexplored targets remaining. By comparison, the GPCR superfamily recognizes a diverse set of endogenous ligands and might be better described as subfamilies of GPCRs from a ligand recognition perspective. Targeting GPCRs with known ligands can follow a biased library design or privileged scaffold approach, whereas pursuing orphan GPCRs poses a distinct challenge. In this case, library construction and screening must instead focus on chemical diversity and a HTS, which, if successful, can create knowledge and opportunities for focused libraries targeting an unexplored GPCR subfamily.
Assembly of an appropriate assay platform
The selection of a representative protein panel and assay format is essential for a successful target class campaign and is the critical first step in building an assay platform. As many target classes are too large to allow comprehensive screening of all members, the selection of a representative panel of proteins from the target class should consider both highly homologous targets, to explore ligand selectivity at an early stage, and structurally distinct but functionally similar domains with an eye toward inclusion of binding site diversity. The curse of a medicinal chemist is to be ‘almost right’ with a ligand hypothesis and miss a potentially good starting point. By screening multiple targets with a conserved recognition motif, being ‘almost right’, and inactive for one target, can lead to a potent ligand for another member of the class. Thus, diverse ligand hypotheses are assessed versus a set of related targets. Although a screening panel can initially be chosen on the basis of a rational set of criteria, it should be fluid to accommodate new targets as novel biology is revealed and traction is made with specific family members within the target class. As targets in the class are advanced through the hit-to-lead process, new analogs would continue to be cross-screened for selectivity and creation of target class structure–activity relationships (SAR)14.
Following the design of a diverse protein panel, an effective primary in vitro assay should allow extensive cross-screening of focused compound collections to enable hit discovery and SAR development. Ideally, the chosen assay should be robust, adaptable to most proteins within the target class, cost effective, and should generate limited false positive hits. The concurrent use of a counter screen assay, wherein a key component of the assay (such as the target protein) is eliminated, can serve as an effective means to quickly identify compounds interfering with the assay that would otherwise appear as hits. In vitro assays that have been commonly employed for target class screening include binding, competition and enzymatic activity assays, although for some targets (GPCR’s, ion channels, and NRs), a cellular assay is the most appropriate screening method for hit discovery. Orthogonal, secondary assays such as isothermal titration calorimetry (ITC) are essential for confirming preliminary hits before initiating follow-up chemistry. If sufficiently potent, ligands should progress to cellular studies that explore direct target engagement, cell permeability (when applicable) and functional effects of the compound.
Cellular assessment of ligand potency, selectivity and MOA is a rapidly developing area and may become a primary source of hit generation and characterization for some target classes15,16. Deconvoluting on-target from off-target effects in cells requires complementary genetic techniques such as engineered assays, knockouts and inactivating mutations17. Importantly, these techniques often do not yield a clear confirmation that the observed biological effects are the result of disruption of the desired target, and, in these instances, target-specific biology may be distinguished via pharmacology using a structurally distinct ligand with target affinity, but a unique selectivity profile. The cell is the vessel wherein target class strategies and biological knowledge must be integrated to advance a project toward translation, and this can be a challenging area for less explored target classes. In our experience with Kme readers, collaboration with chromatin biology labs in which the appropriate assays and expertise exist for cellular MOA studies is critical.
In the case of GPCRs, for example, numerous factors must be accounted for in the selection of appropriate cellular assays. Though they are efficient, binding assays do not necessarily select compounds that elicit a functional outcome, but functional assays may require substantial upfront investigation and resources to discover the appropriate biological readout for the many orphan receptors of this class18. Additionally, GPCRs signal through multiple pathways and therefore require not only characterization of target selectivity but also functional selectivity19. Fortunately, decades of work within this target class has enabled sophisticated assays that anticipate these issues.
Final characterization of a chemical probe should include extensive profiling against protein target families, both related and unrelated to the primary target, so that compound effects observed in cellular assays can be attributed to the inhibition of the known target(s). A variety of assay systems and technologies have been developed to comprehensively interrogate compounds against various protein families. In our own work in collaboration with the Bedford lab2 targeting Kme readers, protein microarrays have enabled rapid, qualitative profiling of biotin labeled ligands. Over a hundred reader proteins spanning more than six domain types are available for profiling via a chromatin-associated domain array (CADOR) chip20. For many established protein classes such as GPCR’s, kinases, NRs and even bromodomains, there are commercially available assays to enable broad compound profiling.
Construction of a structural-knowledge foundation
Advances in structural biology have substantially expanded our understanding of ligand–protein interactions, and techniques including X-ray crystallography, 2D and 3D NMR and cryoelectron microscopy have revolutionized the strategies that can be employed to discover new ligands for protein targets. Additionally, the use of homology models for proteins that lack structural information provides fodder for ligand hypotheses in the absence of a resolved structure. Structure-based design spurs the creation and expansion of focused compound libraries to elaborate on target class SAR and increase the odds for technical success. Plexxikon’s application of scaffold hopping represents an exceptional example of the crosstalk between biochemical screening and structural biology in target class research, whereby a preliminary hit against one kinase led to the development of a drug for a wholly different kinase (Fig. 2)21. Ultimately, a structural-knowledge base provides invaluable information on routes to designed polypharmacology or, conversely, selectivity that may otherwise be overlooked, especially in the early stages of ligand optimization.
The target class payoff
Tackling an unprecedented target class requires long-term focus and perseverance, as knowledge grows slowly in the beginning. However, physical (compound sets, assay platforms and structural data and systems) and intellectual assets eventually accumulate, making the process far more efficient and increasing the quality of the hypotheses and results. This is demonstrably true for the kinase target class, as over the last 20 years industry and academics have built a well-established platform and knowledge base that enables efficient prosecution of any new kinase target12; however, this effort was initially not purposeful, and the physical assets remain inaccessible to many. With emerging target classes, an intentional effort to create accessible platforms and share learnings and expertise would greatly accelerate biomedical science aimed at new medicines. Our modest success in the Kme reader class required talented and supportive collaborators in the Structural Genomics Consortium and at other academic institutions.
Unfortunately, the single-target, disease-focused paradigm continues to dominate biomedical research. This frequently contributes to the production of low-quality, nonreproducible results, because each target is most often a stand-alone effort without the assay expertise, chemical tools, cellular target engagement knowledge16 and rigor that can emerge from target class–focused science22. Though ‘known unknowns’ and ‘unknown unknowns’ are risks in all areas of drug discovery, they are huge pitfalls in tackling one-off targets: unknown selectivity issues, unknown SAR for homologous targets, unknown MOAs within the cell, among others. Fortunately, unknowns are likely to be exposed over time in a well-explored target class, for example, the capacity for ligands to demonstrate functional selectivity in GPCRs and how to design selective ligands for kinases.
Target class science has led directly to new biologic understanding driven by ligand discovery. For example, in the NR area, GlaxoSmithKline scientists deorphanized a number of targets, and this led to new insights into members of the family that bind with low affinity to highly abundant cellular molecules such as fatty acids and oxysterols8. Likewise, the regulatory function of the NR pregnane X receptor (PXR) was uncovered through realization of the bridge it provides between small molecules and p450 induction. The target class is the best organizing paradigm and intellectual home for experimental design, technology development, data analysis and interpretation in the early stages of drug discovery and needs to be understood, embraced and cultivated by the biomedical community. The synergies between this strategy and disease-oriented research readily emerge once high-quality leads and probes demonstrate compelling outcomes in preclinical models.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Prinz F, Schlange T, Asadullah K. Nat Rev Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 2.James LI, Frye SV. Curr Opin Chem Biol. 2016;33:135–141. doi: 10.1016/j.cbpa.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 4.Ting AH, McGarvey KM, Baylin SB. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 5.Filippakopoulos P, Knapp S. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 6.Kettle JG, Wilson DM. Drug Discov Today. 2016;21:1596–1608. doi: 10.1016/j.drudis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Heilker R, Wolff M, Tautermann CS, Bieler M. Drug Discov Today. 2009;14:231–240. doi: 10.1016/j.drudis.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Willson TM, Moore JT. Mol Endocrinol. 2002;16:1135–1144. doi: 10.1210/mend.16.6.0849. [DOI] [PubMed] [Google Scholar]
- 9.Bagal SK, et al. J Med Chem. 2013;56:593–624. doi: 10.1021/jm3011433. [DOI] [PubMed] [Google Scholar]
- 10.Lane SJ, et al. Drug Discov Today. 2006;11:267–272. doi: 10.1016/S1359-6446(05)03730-X. [DOI] [PubMed] [Google Scholar]
- 11.Herold JM, Ingerman LA, Gao C, Frye SV. Curr Chem Genomics. 2011;5:51–61. doi: 10.2174/1875397301005010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller S, Chaikuad A, Gray NS, Knapp S. Nat Chem Biol. 2015;11:818–821. doi: 10.1038/nchembio.1938. [DOI] [PubMed] [Google Scholar]
- 13.Wu P, Nielsen TE, Clausen MH. Drug Discov Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Frye SV. Chem Biol. 1999;6:R3–R7. doi: 10.1016/S1074-5521(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 15.Patricelli MP, et al. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon GM, Niphakis MJ, Cravatt BF. Nat Chem Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss WA, Taylor SS, Shokat KM. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth BL, Kroeze WK. J Biol Chem. 2015;290:19471–19477. doi: 10.1074/jbc.R115.654764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban JD, et al. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, et al. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bollag G, et al. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 22.Frye SV, et al. Nat Rev Drug Discov. 2015;14:733–734. doi: 10.1038/nrd4737. [DOI] [PubMed] [Google Scholar]
- 23.Barnash KD, et al. ACS Chem Biol. 2016;11:2475–2483. doi: 10.1021/acschembio.6b00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weerapana E, et al. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez Molina D, et al. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]