Summary
Ebola virus nucleoprotein (eNP) assembles into higher-ordered structures that form the viral nucleocapsid (NC) and serve as the scaffold for viral RNA synthesis. However, molecular insights into the NC assembly process are lacking. Using a hybrid approach, we characterized the NC-like assembly of eNP, identified novel regulatory elements, and described how these elements impact function. We generated a three-dimensional structure of the eNP NC-like assembly at 5.8 Å using electron cryo-microscopy and identified a new regulatory role for eNP helices α22–α23. Biochemical, biophysical, and mutational analysis revealed inter-eNP contacts within α22–α23 are critical for viral NC-assembly and regulate viral RNA synthesis. These observations suggest that the N-terminus and α22–α23 of eNP function as context dependent regulatory modules (CDRMs). Our current study provides a framework for a structural mechanism for NC-like assembly and a new therapeutic target.
Keywords: Ebola virus, cryoEM, nucleocapsid, nucleoprotein, viral RNA synthesis
Introduction
Ebola virus is a non-segmented negative-sense RNA virus (NNSV), which can cause severe hemorrhagic fever with high case fatality rates during outbreaks as highlighted by the 2013–2016 West African outbreak (Messaoudi et al., 2015; Team et al., 2015). The enveloped Ebola virions contain a single-stranded RNA (ssRNA) genome encoding seven structural proteins known as Ebola nucleoprotein (eNP), viral protein (VP) 35 (eVP35), eVP40, glycoprotein (eGP), eVP30, eVP24 and the large protein (eL) (Messaoudi et al., 2015). This genome is encapsidated by eNP along with other viral proteins, including eVP35, eVP24, eVP30, and eL, to form the nucleocapsid (NC), which protects the ssRNA from degradation and serves as the template for genome replication (Hornung et al., 2006; Ruigrok et al., 2011). Viral genome replication and transcription are carried out on NC-associated RNA by the viral RNA-dependent RNA polymerase (RdRp) eL, whereas eVP35 and eVP30 play critical roles as a polymerase cofactor and transcriptional regulator, respectively (Mühlberger, 2007). eVP24 and eVP35 are also required for NC assembly (Banadyga et al., 2017; Huang et al., 2002). eNP further interacts with the matrix protein eVP40 to allow incorporation of NC into ebolavirus-like particles (Noda et al., 2006). These functions make eNP essential in viral RNA synthesis and virus assembly.
We and others have previously identified a region at the N-terminus of eNP that is conserved among paramyxoviruses and filoviruses, spanning residues 1–450, whereas the C-terminal portion of eNP encompassing residues 451–739 is unique to filoviruses (Leung et al., 2015; Shi et al., 2008; Watanabe et al., 2006) (Figure 1A). Using a combination of biophysical analysis and X-ray crystallographic studies, we determined a structured region in the eNP N-terminus using a truncated N-terminal domain construct (PDB 4YPI)(Leung et al., 2015). We found that the eNP N-terminus (residues 38–383) formed oligomers with a back-to-back double ring configuration that were then stacked in the crystal to form tube-like structures (Leung et al., 2015). Analysis of the ring configuration revealed that the last two helices of each eNP N-terminus molecule formed extensive structural contacts with the last two helices of neighboring eNP molecules (helices α21 and α22), thereby facilitating the formation of oligomeric rings. In contrast, other studies have suggested that these helices are dispensable for oligomerization (Kirchdoerfer et al., 2015). Furthermore, eNP residues 1–24 are also important for oligomerization (Leung et al., 2015). On the other hand, addition of a peptide derived from the N-terminus of eVP35, termed eNP binding peptide (eVP35 NPBP), led to a loss of ring formation and loss of ssRNA binding, suggesting a mechanism for controlling eNP interactions. An X-ray crystal structure of eNP core in the absence of eVP35 NPBP revealed the eVP35 NPBP binding site can be occupied by eNP core helix α20 (Dong et al., 2015). These structural studies highlight the structural plasticity of eNP. However, the functional significance and overall impact of the double ring configuration in the crystal structure, and binding of helix α20 to the eVP35 NPBP binding site observed by Dong et al. on eNP function or assembly are not clear. Given that this region is highly conserved among paramyxoviruses and filoviruses, it is likely that the helical contacts observed are preserved and critical in maintaining eNP NC interactions (Reguera et al., 2014).
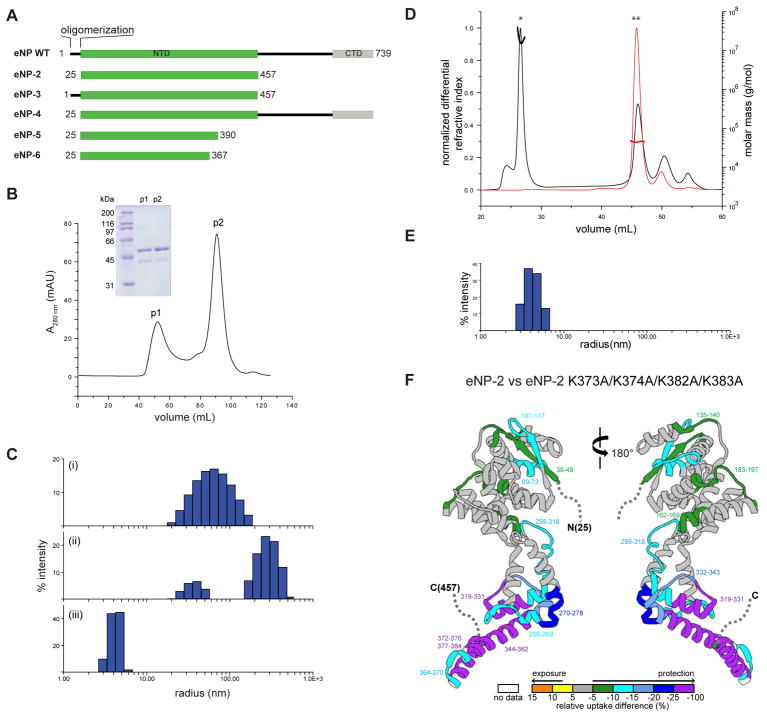
Figure 1. Biochemical characterization of oligomerization in wildtype and mutant eNP proteins.
(A) Domain organization of eNP WT and key constructs used in this study listed below. NTD, N-terminal domain; CTD, C-terminal domain. (B) Elution profile from SEC for eNP-2 in buffer containing 500 mM NaCl. Inset. Coomassie stained SDS-PAGE of corresponding peak fractions from SEC. p1, peak 1; p2, peak 2. (C) DLS data in PBS for: (i) eNP-2 p1 (Rh = 71.6 ± 2.0 nm), eNP-2 p2 (Rh = 33.7 ± 3.6 and 308.8 ± 18 nm), and (iii) eNP-2 p2 + eVP35 NPBP (Rh = 4.2 ± 0.1 nm). Two independent DLS experiments were performed in triplicate for each construct shown. (D) Elution profile from SEC-MALS of eNP-2 WT (black) and eNP-2 K373A/K374A/K382A/K383A (red). Mw = > 104 kDa (*) and 45 ± 1 kDa (**) for the peaks indicated. The theoretical molecular mass for monomeric eNP-2 is 48 kDa. Two independent experiments were performed. (E) DLS data in PBS for eNP-2 K373A/K374A/K382A/K383A (Rh = 4.2 ± 0.2 nm). Two independent DLS experiments were performed in triplicate. (F) Differential HDX-MS highlighted on eNP-2 (PDB: 4YPI); regions not present in the structure are indicated by a dotted line. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-2 K373A/K374A/K382A/K383A from that of eNP-2 at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate.
See also Figure S1.
Here we use a hybrid approach of biochemical, structural, and cell biological assays to better understand how eNP interactions can drive eNP oligomerization, ssRNA binding, and assembly into a NC-like structure. We identify residues in critical structural elements in eNP that are important for mediating eNP intermolecular interactions that also facilitate high affinity binding to ssRNA and viral RNA synthesis. In support of our model, we present a 5.8 Å resolution cryoEM structure of an Ebola virus NC-like tube that explains how individual eNPs are arranged into a helical assembly and how this structure behaves in response to regulatory elements or ssRNA. The impacts of these structural observations were tested in biochemical and functional studies, including ebola virus (EBOV) minigenome (MG) assay that reports on the impact of viral RNA synthesis. Additionally, we compare our structural observations with the recently solved eNP N-terminus (residues 1–450) assembly at 6.6 Å and virus-like particle (VLP) structure of at sub-nanometer resolution by electron cryo-tomography (Wan et al., 2017). Taken together, these studies offer a framework towards understanding Ebola NC architecture and remodeling of the NC-assembly during viral RNA replication.
Results
eNP N-terminus is necessary and sufficient for oligomerization formation
To better define the regions and interactions that drive eNP oligomerization and NC assembly, we generated several eNP constructs (Figure 1A), including eNP, full-length wildtype construct; eNP-2, which we previously used to solve the crystal structure of the N-terminus (Leung et al., 2015); eNP-3, which includes the first 24 residues that are also important for oligomerization; and eNP-4, a near full-length construct that lacks the first 24 residues. Using size exclusion chromatography (SEC), we find that eNP-2 separates into two populations, p1 and p2, in buffer containing 500 mM NaCl (Figure 1B). Fraction p1 elutes in the void volume of the column whereas fraction p2 elutes in the included volume, suggesting that p1 is likely a higher ordered oligomer or aggregate and fraction p2 is a monomer in solution.
We next used dynamic light scattering (DLS) to determine the size distribution of eNP-2 in buffer containing 150 mM NaCl. A subset of the resulting data, shown in Figure 1C, revealed different species for eNP-2. Histogram analysis revealed that p1 of eNP-2 has a relatively broad peak with a measured hydrodynamic radius, Rh, of 71.6 ± 2.0 nm (Figure 1C(i)), suggesting a heterogeneous distribution of eNP oligomers that cannot be resolved. Consistent with this, negative staining electron microscopy (EM) shows that eNP-2 fraction p1 contains rings and tube-like structures of various lengths and diameters (Figure S1A). Fraction p2 of eNP-2 contains two populations with Rh values of 33.7 ± 3.6 nm and 309 ± 18 nm, respectively (Figure 1C(ii)), indicating the presence of two distinct species of rings and tubes that can be visualized by negative stain EM (Figure S1B). Here, the diameter of the rings and the tubes is ~50 nm. We previously showed that addition of the eVP35 NPBP peptide binds and monomerizes eNP-2 (Leung et al., 2015). Addition of eVP35 NPBP to p2 of eNP-2 leads to a single peak in the DLS analysis with an Rh of 4.2 ± 0.1 nm (Figure 1C(iii)). This result corresponds to the loss of eNP-2 oligomerization and formation of a 1:1 heterodimeric complex of eNP-2/eVP35 NPBP, which is consistent with the negative stain EM images showing the loss of rings and tubes and with our previous observations (Figure S1C)(Leung et al., 2015).
Mutations in α-helices α21 and α22 render eNP monomeric
In our previous structure of eNP-2 (PDB 4YPI), the C-terminal α-helices α21 and α22, encompassing residues 345–384, were involved in eNP intermolecular contacts. Examination of the sequence in these helices revealed four conserved basic residues, K373, K374, K382, and K383, that lie outside of the highly basic charged cleft that we previously shown to bind ssRNA (Figure S1D) (Leung et al., 2015). To test the impact of these residues and to validate a role for helix α22 in eNP-eNP oligomerization, we generated alanine mutations of these lysine residues. The resulting eNP-2 K373A/K374A/K382A/K383A mutant protein displayed markedly different profiles in SEC multiangle light scattering (SEC-MALS) experiments compared to eNP-2 WT. The elution volume of eNP-2 K373A/K374A/K382A/K383A mutant corresponds to an experimental molecular weight 45 ± 1 kDa, which is consistent with the theoretical molecular weight for a monomer of 48.4 kDa (Figure 1D). DLS data also shows that the eNP-2 K373A/K374A/K382A/K383A mutant displays a single peak distribution with an Rh of 4.2 ± 0.2 nm (Figure 1E). This finding is similar to that obtained for the eNP-2/eVP35 NPBP complex, suggesting that mutations of these four basic residues in helix α22 also render eNP-2 monomeric (compare Figure 1C with Figure 1E).
Next, we used hydrogen-deuterium exchange mass spectrometry (HDX-MS) to evaluate the effects of eNP-2 K373A/K374A/K382A/K383A mutant on eNP-2. For both proteins, we obtained near 100% peptide coverage (See Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). In comparing the kinetic exchange curves, we observed several regions with notably differential exchange rates, including regions that are protected >25% from exchange (See Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). Raw kinetic curves show that there are several peptides that are highly exposed in the mutant (See Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). In particular, peptides derived from helices α21 and α22 are among the most exposed and undergo rapid deuterium uptake in the eNP-2 K373A/K374A/K382A/K383A mutant compared to eNP-2. We also observe additional areas of protection in the N-terminal lobe of eNP-2 K373A/K374A/K382A/K383A (Figure 1F and Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1)). Together, these results support a model where eNP is monomeric in the context of the eNP-2 K373A/K374A/K382A/K383A mutant with α21 and α22 helices exposed. The observed differential deuterium uptake between eNP-2 and eNP-2 K373A/K374A/K382A/K383A are due to the oligomeric nature of the eNP-2 protein. Consistent with these observations, HDX-MS data comparing eNP-3 and eNP-2 shows a small increase in protection for a limited number of eNP-3 peptides, such as peptide 256–259, as eNP-3 also forms an oligomer (Figure 2A and S2A; see Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). Furthermore, we observed HDX differences in the β-hairpin region involved in eVP35 NPBP binding (peptide 270–278) suggesting that eNP-2 undergoes conformational changes that alter the accessibility of the pocket that overlaps with the eVP35 NPBP binding site. Our HDX-MS data suggests that this conformation depends on the interaction of eNP with other eNP molecules.
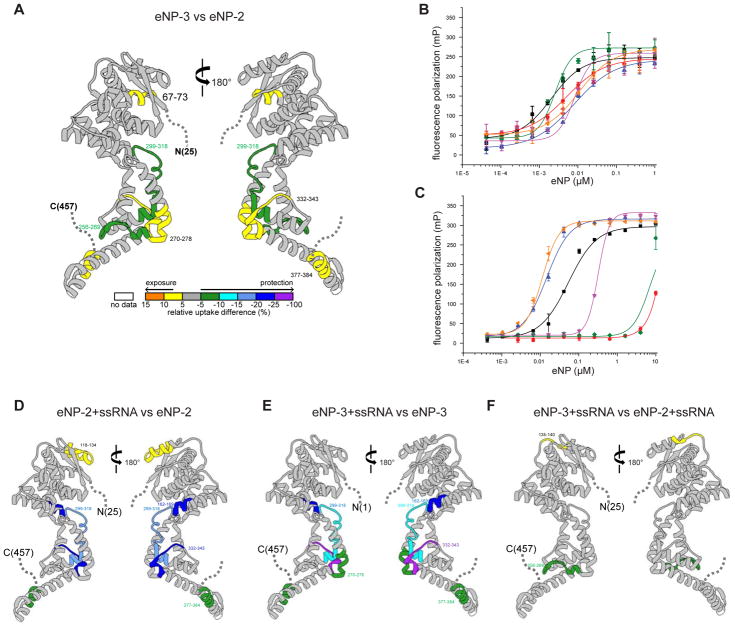
Figure 2. CDRMs modulate eNP function.
(A) Differential HDX-MS between eNP-2 and eNP-3. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-2 from that of eNP-3 at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate.
(B) Fluorescence polarization binding curves of eNP proteins for eVP35 NPBP. The measured KD values are 10.6 ± 2.4 nM (eNP WT,
 ), 1.9 ± 0.2 nM (eNP-2, ■), 5.4 ± 0.3 nM (eNP-2 K373A/K374A/K382A/K383A,
), 1.9 ± 0.2 nM (eNP-2, ■), 5.4 ± 0.3 nM (eNP-2 K373A/K374A/K382A/K383A,
 ), 12.8 ± 6.8 nM (eNP-3,
), 12.8 ± 6.8 nM (eNP-3,
 ), 9.7 ± 2.5 nM (eNP-3 K373A/K374A/K382A/K383A,
), 9.7 ± 2.5 nM (eNP-3 K373A/K374A/K382A/K383A,
 ), and 1.2 ± 1.2 nM (eNP-4,
), and 1.2 ± 1.2 nM (eNP-4,
 ). Each measurement was performed at least twice independently in duplicate.
). Each measurement was performed at least twice independently in duplicate.
(C) Fluorescence polarization binding curves of eNP proteins for 20 nt ssRNA. The measured KD values are 0.013 ± 0.003 μM (eNP WT,
 ), 0.068 ± 0.020 μM (eNP-2, ■), > 10 μM (eNP-2 K373A/K374A/K382A/K383A,
), 0.068 ± 0.020 μM (eNP-2, ■), > 10 μM (eNP-2 K373A/K374A/K382A/K383A,
 ), 0.015 ± 0.005 μM (eNP-3,
), 0.015 ± 0.005 μM (eNP-3,
 ), 0.306 ± 0.006 μM (eNP-3 K373A/K374A/K382A/K383A,
), 0.306 ± 0.006 μM (eNP-3 K373A/K374A/K382A/K383A,
 ), and 3.17 ± 0.28 μM (eNP-4,
), and 3.17 ± 0.28 μM (eNP-4,
 ). Each measurement was performed at least twice independently in duplicate.
). Each measurement was performed at least twice independently in duplicate.
(D) Differential HDX-MS between eNP-2 and eNP-2/ssRNA. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-2 from that of eNP-2/ssRNA complex at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate. (E) Differential HDX-MS between eNP-3 and eNP-3/ssRNA. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-2 complex from that of eNP-3/ssRNA complex at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate. (F) Differential HDX-MS between eNP-2/ssRNA and eNP-3/ssRNA. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-2/ssRNA complex from that of eNP-3/ssRNA complex at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate.
See also Figure S2.
eNP oligomerization mediated through helices α21 and α22 facilitates ssRNA binding
Given that we and others previously suggested that eNP oligomerization and ssRNA binding are correlated through interactions with eVP35 NPBP, we next tested whether the four lysine residues in α22 are also directly involved in ssRNA binding. We used a fluorescence polarization assay (FPA) to test the ability of our eNP constructs (Figure 1A) and the corresponding K373A/K374A/K382A/K383A mutant to bind eVP35 NPBP (Figure 2B) or bind a 20 nt ssRNA (Figure 2C). Consistent with our previous observation that eVP35 NPBP binding to eNP is independent of eNP intermolecular interactions, all eNP proteins tested displayed limited variation in measured affinity for eVP35 NPBP (Figure 2B). Next, we rationalized that if eNP follows the “rule of 6” as some nucleoproteins from other negative RNA viruses have been shown to bind 6 nt (Alayyoubi et al., 2015), then a stretch containing 3 eNPs can bind to a 20 nt ssRNA. We find that eNP-2 binds 20 nt ssRNA with a KD of 0.068 ± 0.02 μM (black curve) (Figure 2C). Addition of the K373A/K374A/K382A/K383A mutation to eNP-2 led to a significant reduction in ssRNA binding with a KD > 10 μM (red curve). To distinguish between the possibility that loss of ssRNA binding is due to loss of oligomerization rather than loss of direct binding of ssRNA to the charged lysine residues, we used the FPA to test the eNP-3 construct, which contains the first 24 residues that further contribute to eNP oligomerization. We find that eNP-3 binds with higher affinity compared to eNP-2, with a KD of 0.015 ± 0.005 μM (blue curve) and that the eNP-3 K373A/K374A/K382A/K383A mutant, which remains oligomeric based on DLS comparison for eNP-3, Rh = 26.3 ± 1.6 nm and eNP-3 K373A/K374A/K382A/K383A Rh = 38.8 ± 3.1 nm (Figure S2A), retains binding to ssRNA albeit with a reduced KD of 0.306 ± 0.006 μM (purple curve). Finally, eNP WT has a similar high affinity for ssRNA (KD = 0.013 ± 0.003 μM, yellow curve) as eNP-3, and deletion of the first 24 residues in the eNP-4 construct results in much lower affinity binding (KD = 3.2 ± 0.03 μM, green curve). Collectively, these results suggest that ssRNA may not bind directly to eNP helices α21 and α22; however, >4-fold difference between eNP-3 and eNP-3 K373A/K374A/K382A/K383A suggests that residues in α21-α22 are important for ssRNA binding. Therefore, ssRNA binding is likely dependent on oligomerization and proper orientation of eNP molecules. Furthermore, the N-terminal eNP 24 residues enhances ssRNA binding while reducing eVP35 NPBP binding by 10-fold simultaneously. In the context of full-length eNP, this potentially corresponds to a striking 1000-fold difference in binding affinities for ssRNA and eVP35 NPBP (see below). Thus, the N-terminal 1–24 residues and residues near α21-α22 helices form an element, which we term context dependent regulatory module (CDRM), which is critical for controlling eNP intermolecular interactions. These observations further strengthen the potential coupling between oligomerization and ssRNA binding that also controls access to RNA template and Ebola viral RNA synthesis.
In order to further define any structural changes in eNP associated with RNA binding, we compared deuterium exchange patterns for eNP-2 and eNP-2/ssRNA complex (Figure 2D and Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). The comparison of eNP-2/ssRNA and eNP-2 revealed that regions that show the most significant HDX changes (>5% change) upon RNA binding are located near the ssRNA binding cleft and in the helices α21–α22 region (boxed region in (See Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). Comparison of eNP-3 with eNP-3 K373A/K374A/K382A/K383A reveals minimal changes, with a few peptides around the eVP35 NPBP binding pocket showing protection from HDX due to eNP-eNP contacts (Figure S2B and Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1). In contrast, we observe a large number of peptides protected in the α22–α23 region when ssRNA binding is considered. Results from these deuterium exchange studies for eNP-2/ssRNA complex (Figure 2D) and eNP-3/ssRNA complex (Figure 2E and Mendeley Data https://data.mendeley.com/datasets/rt3xn85jbv/1) are consistent with the N-terminus having limited impact in the overall interactions with ssRNA. This observation is further confirmed by our comparison of eNP-2/ssRNA with eNP-3/ssRNA, where there are very limited changes in the overall exchange patterns (Figure 2F). The presence of N-terminal residues 1–24 likely confers eNP in a more closed conformation as one major peptide lining the putative ssRNA binding cleft, corresponding to residues 299–318, is more protected in eNP-3 compared to that of eNP-2 in the ssRNA free form (Figure 2A). This suggests that eNP-3 may exist in a more closed conformation in the cleft region that is likely important for high affinity RNA binding observed in Figure 2C.
CryoEM helical reconstruction of eNP-2 NC-like tubes
In order to understand the NC-assembly process and to assess the structural impact of CDRMs in forming and maintaining eNP intermolecular interactions, we obtained cryoEM images of eNP-2 on a JEM 3200FSC electron microscope with a Gatan K2 Summit camera that revealed NC-like tubes (Figure 3A and Table 1). We next performed layer line indexing (Figure S3A and S3B) and used the iterative helical real-space reconstruction (IHRSR) algorithm (Egelman, 2007) to determine the helical parameters of 2.65 Å in rise and −8.53° in twist, which is equivalent to 42.2 asymmetric units per helical turn and a 112 Å helical pitch. Initial 3D refinement using Relion 2 (He and Scheres, 2017; Scheres, 2012) yielded a reconstruction at 7.3 Å resolution, in which free 2D averages exhibited no detailed features (Figure S3C and S3D). This is likely due to the considerable flexibility of the tube caused by the small helical rise and turn, the large number of asymmetric units per helical turn, and 5–6 helical turns in each boxed particle. A rectangular mask was then applied to each raw particle, with a length of ~230 Å along the tube axis that covered two helical turns. 2D class averages of the masked particles showed significantly improved features indicative of increased alignment accuracy (Figure S3E). 3D auto-refinement of the masked particles yielded the final helical reconstruction of the eNP-2 NC-like tubes at 5.8 Å resolution (Figure S3F).
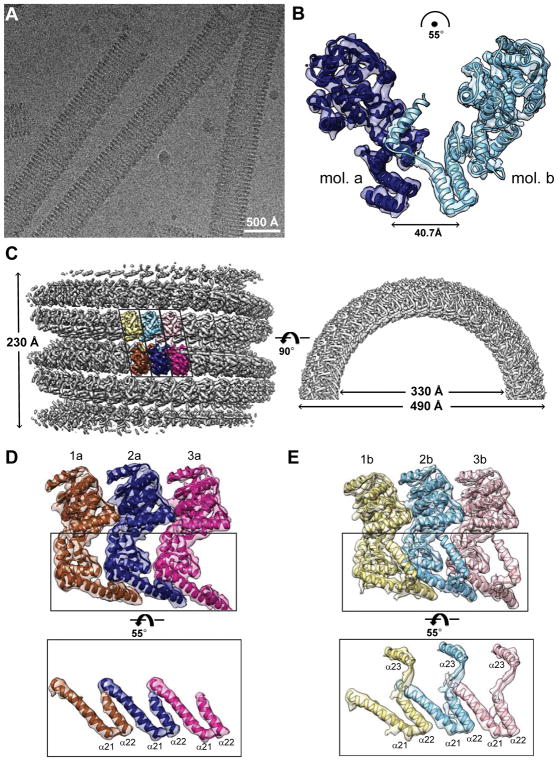
Figure 3. CryoEM helical reconstruction of eNP nucleocapsid-like structure.
(A) Raw cryoEM micrograph. Scale bar represents 500 Å. (B) An asymmetric unit of the tubular structures with two eNP-2 molecules (mol. a, blue; mol. b, cyan) relative to each other without any n-fold symmetry operation. The translation and rotation parameters to superimpose eNP-2 molecules are shown. The extended helix (α23) of eNP-2 molecule b occupies the eVP35 NPBP binding pocket of eNP-2 molecule a. (C) 3D reconstruction of masked eNP structure shows a left-handed hollow tube. Left. Side view. Three asymmetric units colored yellow/brown, cyan/blue, and pink/magenta containing six eNPs are highlighted. Right. Top view. Outer and inner diameters are 490 Å and 330 Å, respectively. (D) Views of three eNPs in the highlighted bottom row in (C), rotated 125° along the same direction of the same rotation axis illustrated in (C). These eNPs lack the density for helix α23 observed in eNPs in (E). (E) Views of three eNPs in the highlighted top row in (C), rotated 70°along the same direction of the same rotation axis illustrated in (C). The coiled-coil motif-like contacts between helices α21 and α22 and position of helix α23 are shown. See also Figure S3–S4 and Movies S1–3.
Table 1.
Electron cryomicroscopy data collection and processing of eNP-2 structure
| Microscope | JEM3200FSC |
| Voltage (kV) | 300 |
| Energy filter slit width (eV) | 30 |
| Detector | Gatan K2 Summit |
| Magnification | 30,000X |
| Pixel size (Å) | 1.2 |
| Defocus range (μm) | 0.5 – 2.0 |
| Electron exposure (e−/Å2) | 24 |
| Micrographs (acquired/used) | 1266 / 1113 |
| Segment step (asymmetric unit) | 5 |
| Number of segments/asymmetric unit | 169,526 / 847,630 |
| Helical twist and rise (°/Å) | −8.53 / 2.65 |
| Map resolution at 0.143 FSC criterion (Å) | 5.8 |
eNP-2 forms helical tubes comprised of heterodimeric eNP subunits
To dissect how eNP assembles, we define an asymmetric unit of our tubular structure made up of two eNPs, called a and b, which are not related by any n-fold symmetry operation (Figure 3B). To demonstrate the molecular interactions between adjacent asymmetric units, we presented three asymmetric units (i.e. six eNPs colored yellow/brown, cyan/blue, pink/magenta, respectively, in Figure 3C). Individual eNPs were segmented with the Segger plugin (Pintilie et al., 2010a; Pintilie et al., 2010b) in UCSF Chimera (Pettersen et al., 2004), using the fitted crystal structure of the eVP35 NPBP bound state of eNP-2 (PDB 4YPI) (Leung et al., 2015) as a guide (Movie S1). The crystal structure of eNP-2 fits well into the density, except for differences at β-strands 14–15 and α-helices 21–22, which lie partially outside the density (Figure S3G). Additionally, we find that molecule b has additional tubular density at the C-terminus extending beyond helix α22, which is absent in both molecule a and the eNP-2 X-ray crystal structure (Figure 3D–E and Figure S3G). Secondary structure element prediction suggests that for residues beyond helix α22 (residues 391–412), there is high probability for forming an additional alpha helix. Thus we call this tubular density in our b molecule as α23, contiguous to α22 by a short extended loop (Figure S3H). The loop and α23 were added to the model with Chimera. A total of 6 rigidly fitted eNP models for three asymmetric units, 3 of them having the added α23, were further relaxed/flexibly fitted into the density using MDFF (Figure S3I (Trabuco et al., 2009)). The cross correlation coefficient (CCC) of the averaged flexibly fitted model to cryoEM map was calculated in UCSF Chimera and improved from 0.79 to 0.87 after flexible fitting. The FSC curve of model to map was calculated in EMAN2 and showed significant improvement (Figure S3J). Uncertainty in the model was calculated using the probabilistic modeling approach (Figure S4A (Pintilie et al., 2016)); an eNP pair from the average model is shown in Figure 3B (See also Figures 3C–E, and Movies S2–S3 for a visualization of all 6 modelled eNPs).
Interestingly, α23 helix of molecule b binds to the β-hairpin region (residues 255–265) of molecule a (Figure 3B and S4A), which has also been shown to bind to eVP35 NPBP (Leung et al., 2015). Therefore, we next tested whether a peptide (residues 370–411 corresponding to α22 and α23) could outcompete for eVP35 NPBP binding using a FPA similar to that described above. Our results show that an unlabeled eVP35 NPBP peptide could compete out FITC-labeled eVP35 NPBP binding to eNP-2 (Figure S4B). However, eNP peptide 370–411 cannot outcompete FITC-labeled eVP35 NPBP binding to eNP-2, even at high concentrations. Isothermal titration calorimetry (ITC) studies also revealed no direct binding of eNP peptide 370–411 to eNP-2 (Figure S4C). These results are similar to that observed for eNP peptide 1–24, which plays a role in eNP oligomerization. Altogether, our results demonstrate that eNP residues 1–24 and 385–411 have unique regulatory roles that are critical for modulating eNP function, but only within the context of the full-length protein as CDRMs and not in the context of isolated peptides. Furthermore, given the eNP arrangement shown in Figure 3C–E and RNA binding groove illustrated in Figure S1D and S4D, our cryoEM structure presents helical RNA binding grooves along the eNP tube on both interior and exterior of the tube. While the exterior RNA binding groove is consistent with the observation by Wan, et al., the presence of an interior RNA binding groove may provide potential functional relevance of the current eNP assembly structure to RNA transcription and replication processes of EBOV (detailed in Discussion).
A key functional role for residues that form eNP α23
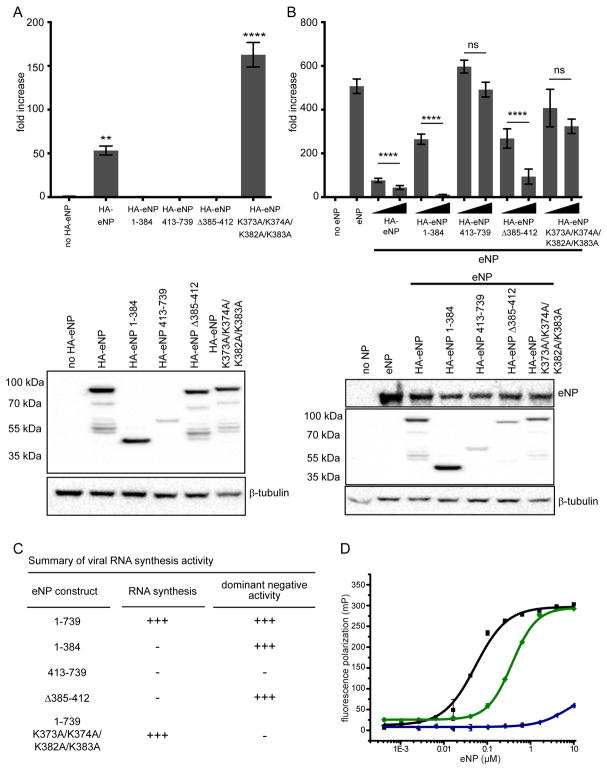
To test whether the additional helix α23 is functionally important as suggested by our cryoEM reconstruction, we used a minigenome (MG) assay to evaluate several eNP truncation mutants (Figure 4A). While viral RNA synthesis is fully functional in the presence of full-length eNP (eNP), truncation mutations lacking either the C-terminus (eNP 1–384), N-terminus (eNP 385–739), or residues 385–412 (eNP Δ385–412) are all nonfunctional. Importantly, eNP K373A/K374A/K382A/K383A was also functional in this assay to levels even higher than eNP alone. On the background of eNP, addition of either eNP 1–384 or eNP Δ385–412, but not eNP 413–739, displayed a dominant-negative phenotype (Figure 4B). However, eNP K373A/K374A/K382A/K383A did not show dominant negative activity in contrast to its activity in the MG assay data. These results suggest that viral RNA synthesis is dependent on eNP oligomerization and residues K373A/K374A/K382A/K383A in α22, but not those in α23, and are important for competing with eNP-eNP oligomers formed by wildtype versus mutant eNP proteins (Figure 4C).
Figure 4. The C-terminal helix α23 is a CDRM that regulates eNP viral RNA synthesis.
(A) A minigenome assay was performed to compare the activities of wild-type eNP or eNP mutants 1–384, 413–739, eNP Δ385–412, and eNP K373A/K374A/K382A/K383A (N-terminus tagged constructs) on EBOV RNA synthesis. (B) The minigenome assay was conducted with eNP +/− HA-eNP wildtype or mutants 1–384, 413–739, eNP Δ385–412, and eNP K373A/K374A/K382A/K383A (N-terminus tagged constructs) to test the dominant negative effect of these proteins on EBOV RNA synthesis. Two concentrations of HA-eNP proteins, 25 ng and 250 ng, were used and the expression was assessed by Western blotting using anti-HA antibody and untagged eNP was assessed by using anti-NP antibody. The luciferase activity was determined 48 h post transfection. The fold increase was determined by normalizing Renilla to firefly luciferase values and setting the value of the “no eNP control” to 1. Statistically significant differences were determined using one-way ANOVA, followed by tukey’s test. *p < 0.001, **** < 0.0001. The error bars denote the standard deviation of three replicates. (C) The summary of RNA synthesis activity and dominant negative activity against wt NP is shown. (D) Fluorescence polarization binding curves of eNP proteins for 20 nt ssRNA. The measured KD values are 0.068 ± 0.020 μM (eNP-2, ■), 0.283 ± 0.129 μM (eNP-5,
 ), and >10 μM (eNP-6,
), and >10 μM (eNP-6,
 ). Each measurement was performed at least twice independently in duplicate.
). Each measurement was performed at least twice independently in duplicate.
Given that residues that encompass α23 also contains a basic residue cluster, we tested several truncation constructs to further define the roles played by α22 basic residues (K373/K374/K382/K383) versus α23 basic residues (R398/K399/R401/R404). To test this, we conducted ssRNA binding assays using eNP-5, which lacks α23, and eNP-6, which lacks α22 and α23. Resulting data shown in Figure 4D reveals that the ssRNA binding difference between eNP-2 and eNP-5 is about 4- to 5-fold, whereas eNP-6, which lacks all residues from α22–α23, results in near complete loss of ssRNA binding. These results indicate that residues within α23 contribute minimally to ssRNA binding.
eNP shows conformational heterogeneity
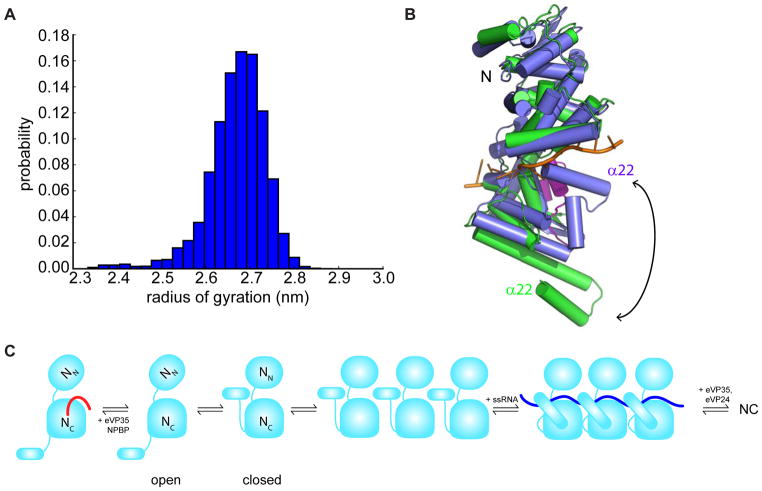
In order to explore the diversity of structures that eNP adopts in solution, we built a Markov state model (MSM). A MSM is essentially a map of a protein’s conformational space, consisting of the dominant structural states the protein adopts and the rates of transitioning between them (Bowman, 2014). In this case, we constructed our MSM from 28 μs of molecular dynamics simulations with explicit solvent. These simulations were initiated from the structure of eNP observed in complex with eVP35 NPBP, but with NPBP peptide removed. Our model reveals extensive conformational heterogeneity, particularly in the helices α21–α22 of eNP. The distribution of Cα RMSDs between the structural states of our MSM and the crystallographic structure is roughly a Gaussian, with a mean of 4.1 Å and a standard deviation of 1.1 Å. Some of the structural states in our model closely resemble the cryoEM structure (Figure S5). This observation suggests that monomeric eNP samples conformations like the oligomeric structure reported in this work, and that this structure is stabilized by the formation of the oligomer.
In order to determine if the helices are capable of curling up to interact with RNA, we queried our model for structural states with particularly low radii of gyration. As shown in Figure 5A, there are a variety of structures with exceptionally low radii of gyration. Visualizing these structures reveals that the helices, including α22 (note that α23 containing structure was not used in these simulations) are indeed curled up towards the RNA-binding cleft (Figure 5B). While the exact conformation of these helices could differ in the presence of RNA and the rest of eNP, this analysis demonstrates that it is plausible for the helices to contribute to RNA binding. Interestingly, there would be steric clashes between the helices in this curled up state and eVP35 NPBP from the holo crystal structure (Figure S5A). This result suggests that eVP35 NPBP peptide may inhibit eNP oligomerization by blocking the helices from folding into a conformation that is permissible for RNA interactions.
Figure 5. Model description for eNP assembly and regulation during viral RNA synthesis.
(A) Population-weighted distribution of the radii of gyration of the structural states from our MSM. (B) Overlay of a representative curled up state from our MSM (blue) with the holo crystal structure (green), highlighting that the helices curl up in an orientation where they interact with RNA. The RNA structure was obtained by aligning both structures to the RNA-bound structure of HMPV NP (PDB 5FVC) and showing the RNA but not the protein from that structure. eVP35 NPBP (magenta) from the holo crystal structure of eNP has steric clashes with the helices of the blue structure from our computational model, suggesting that NPBP binding and curling up of helices are mutually exclusive. (C) A model consistent with the in vitro and computational analyses is shown. See also Figure S5.
Discussion
Using a hybrid structural approach, we have characterized a major Ebola viral NC protein in order to test the functional impact of novel regulatory elements, termed CDRMs, identified by our biochemical studies and cryoEM reconstruction studies. The overall architecture of eNP in the cryoEM structure appears similar to that of the eVP35 NPBP bound eNP X-ray crystal structure. However, experimental data, including equilibrium, kinetic and structural studies show that eNP in NC assembly requires interaction through CDRMs, such as eNP 1–24 and eNP helices α21–α23. Importantly, an additional helix, α23, located at the C-terminus of eNP-2 binds near the eVP35 NPBP binding pocket of the adjacent eNP monomer in the NC-like assembly. Mutation of conserved basic residues in α22–α23 region leads to decreased ssRNA binding without a major impact on the eNP/eVP35 NPBP binding. Interestingly, these same mutants, along with additional mutants, such as eNP-2, eNP-3, and eNP-4, displayed a range of binding affinities to ssRNA, suggesting that both sequence identity and the length of eNP are important for eNP/ssRNA interactions.
A model has emerged that supports the Ebola virus NC assembly through a series of structural rearrangements, both at the N- and C-terminus of eNP-2 (Figure 5C and S5B). The N-terminal lobe (NN) and C-terminal lobe (NC) of eNP-2 likely move relative to each other, where these conformations can be captured either by interactions with other eNP molecules through their CDRMs or with eVP35 NPBP. CDRM interactions, such as those with eNP 1–24 or helix α23, can restrict or facilitate eNP oligomerization, thereby controlling ssRNA binding. Addition of ssRNA and other viral components, such as eVP35 and eVP24, further drives assembly and formation of the viral NC. The interactions identified in this study are important as they likely control the events leading to viral RNA synthesis.
The CDRMs identified in our study, namely the eNP N-terminus and the eNP helices α22 and α23, were functionally validated in a series of biochemical and functional studies. These studies revealed that eNP-eNP interactions leading to eNP oligomerization and NC formation, as well as access to ssRNA (which represents the viral template RNA), may be controlled by CDRMs. Deletion of either CDRM leads to loss of function (Figure 4A and previous studies (Leung et al., 2015)). Expression of the N-terminus of eNP alone, a region previously defined as a region common to NNSVs (as opposed to the C-terminus of eNP, which is defined as filovirus specific)(Reguera et al., 2014; Ruigrok et al., 2011), is not sufficient for viral RNA synthesis (Figure 4A). Similarly, deletion of the CDRM α23 also leads to loss of RNA synthesis in MG assays. As expected, the C-terminus of eNP failed to support viral RNA synthesis in the MG assay. However, eNP K373A/K374A/K382A/K383A (mutations with in α22, a CDRM) is functional in the MG assay, suggesting that this particular mutant is not only able to bind ssRNA, as shown in Figure 2C, but is also able to control viral polymerase access to RNA. This observation is consistent with the ability of this mutant to form oligomers and bind ssRNA with relatively high affinity.
We also assessed the ability of eNP CDRMs to compete with eNP in the MG assay and function in a dominant negative manner. As previously shown, eNP 1–384, which includes CDRMs (N-terminus and α22, but not α23), was able to inhibit the function of eNP. eNP Δ385–411, which lacks α23, was also able to inhibit eNP activity in the MG assay (Figure 4B). In contrast, eNP 413–739 and the eNP C-terminus were unable to inhibit MG activity. However, we note that an eNP construct with mutations in α22 (K373A/K374A/K382A/K383A) was unable to inhibit eNP activity, suggesting that while this construct is able to support viral RNA synthesis, the loss of the α22 CDRM prevents dominant negative activity. One possible explanation for this observation is that the oligomers of eNP formed by wildtype eNP is distinct from other mutant eNP constructs and exhibit higher affinity for the ssRNA template. These results strongly support our model, where oligomerization driven by the CDRMs promote higher affinity toward the RNA template and therefore promote higher activity during viral RNA synthesis. We note that future studies to fully evaluate these observations in the context of additional viral proteins such as eVP35 and eVP24 are required.
Wan and co-workers recently described an eNP helical assembly using a NC-like complex with a construct corresponding to eNP-3 in our study (Wan et al., 2017) (Figure S6). In that structure, all eNPs are arranged in the same orientation to form a helical structure. Helix α23 exists as an extension of helix α22, which may function as a clamp to the ssRNA binding pocket. The N-terminus of eNP-3 forms a helix, under which density attributable to ssRNA is observed. It is likely that the 6.6 Å structure by Wan et al. and the 5.8 Å structure reported here are relevant architectures of the NC assembly process with conformations that represent distinct functional state or metastable structural states d0ue to the absence of RNA and viral factors. In our structure, there are significant differences with respect to asymmetric unit composition and tube diameters. Binding of ssRNA can potentially drive incorporation of helix α23 into α22 helix, forming an extended helix in the previous structure (Wan et al., 2017). Helical extension can then induce additional conformational changes that lead to clamping of the RNA to the binding pocket (Figure 5C and Figure S6). Our HDX-MS data are consistent with these structural rearrangements where the largest changes in solvent exchange, an indirect measure of conformational dynamics, occur near the eVP35 NPBP binding pocket and the α22–α23 region. Together, these studies suggest a highly choreographed process where the conformational flexibility of eNP is critical for regulating the process of NC assembly.
Lower resolution structures were obtained with RNA, where RNA appears to bind to the outer face of the helical NC-like assembly (Wan et al., 2017). This observation would be similar to the NC-like structures of paramxyoviruses showing ssRNA binding to the exterior face (Tawar et al., 2009), which can provide access for the viral polymerase during viral RNA synthesis. Binding of ssRNA to the interior face has also been observed for rhabdoviruses, including rabies and vesicular stomatitis viruses (Albertini et al., 2006; Green et al., 2006), which can provide potential protection of the ssRNA genome from the host environment. Positioning of helix α23 at the eVP35 NPBP binding site, coupled with the presence of basic patches on both sides of the tube, potentially arranges the eNP helical assembly described here in functionally important states during viral RNA synthesis. Future studies, including enzymatic polymerase activity, are likely required to fully dissect NC assembly with respect to its interactions with RNA as well as the impact of such interactions during viral RNA synthesis.
Nucleocapsid-like assemblies from VLPs were generated by Wan et al. in the presence of eVP24 and eVP35, but only eVP24 was seen in the density map (Wan et al., 2017). Although eVP35 could not be docked into the density, eVP24 was assigned to bind eNPs asymmetrically into two distinct sites on eNP. Banadyga, et al. (Banadyga et al., 2017) recently reported crucial eVP24 residues (amino acids 169–173) that make critical contacts with eNP and predicted that these interactions are likely conserved among filoviruses. However, in the Wan et al NC assembly, these eVP24 residues are not directly interacting with eNPs. This discrepancy remains elusive as it is currently unclear what structural state of eNP was used in the study by Banadyga et al z(Banadyga et al., 2017) (Figure S6B). We also note that when interactions in the Wan et al. structure for eNP/eVP24 were mapped on our eNP-2 NC-like assembly, only one set of the asymmetric interactions were compatible with our structure. This may explain the asymmetric binding of eVP24 to eNP nucleocapsid-like assembly as previously described by Bharat et al. (Bharat et al., 2012). Answering these questions will be critical to better understand this highly choreographed NC assembly process.
Insights from our study, from the Wan et al. study, as well as previous works from other negative sense RNA viruses (NSVs) provide an important framework to better understand the highly choreographed process of viral NC assembly. Although nucleoprotein interaction with viral RNA is an important aspect of viral RNA synthesis, additional viral components (such as eVP35, eVP24, and host factors), are likely to impact the structural dynamics observed here as well as RNA synthesis kinetics and ultimately viral assembly. We and others have previously shown that eVP24 may exist in the cytoplasm and in the nucleus (Xu et al., 2014), while similar studies for eNP or eVP30 have yet to be conducted (Biedenkopf et al., 2016). Additionally, eVP30 also interacts with eNP and eVP35 (Biedenkopf et al., 2016). Thus, studies that incorporate additional replication complex components, such as eVP35, eVP30, and eVP24 in filoviruses, as well as those that facilitate membrane association of viral factors are required to obtain a complete picture. These efforts are critical for our understanding and will be part of future effort by the field. We also note that insights gained in our study allow us to target these critical viral interfaces for therapeutic development. For example, binding pockets that are occupied by the CDRMs, which are likely highly populated transient conformations, provide a previously unrecognized opportunity for antiviral development. Targeting the sites occupied by CDRMs using peptidomimetics or small molecule inhibitors can perturb viral RNA synthesis and therefore, these sites should be a major new direction for therapeutic development against filoviruses and NSV family members as CDRMs are likely to be a universal functional element within the NSV family.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gaya K. Amarasinghe (gamarasinghe@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For recombinant protein expression, we used the Escherichia coli strain BL21(DE3) (Agilent). Cells were grown at 37 ˚C as described in the Method Details section.
METHOD DETAILS
Protein Expression and Purification
eNP constructs were expressed as maltose binding protein (MBP) fusion proteins in BL21(DE3) E. coli cells (Novagen). At OD600 of 0.6–0.7, recombinant protein expression was induced with 0.5 mM IPTG for 12–14 h at 18 ˚C. Cells were harvested and resuspended in lysis buffer containing 20 mM Tris pH 7.5, 1 M NaCl, 20 mM imidazole, 5 mM 2-mecaptoethanol (BME). Cells were lysed using an EmulsiFlex-C5 homogenizer (Avestin) and lysates were clarified by centrifugation at 30,000 g at 4 ˚C for 40 min. eNP were purified using a series of affinity and ion exchange chromatographic columns. MBP tag was cleaved using TEV protease prior to final purification with a gel filtration column. After cleavage, three tag-derived residues (GHM) remain at the N-terminus of each construct. Purity of eNP proteins were determined by Coomassie staining of SDS-PAGE.
Negative Staining EM
2 μl of eNP sample at a concentration of 1 mg/ml in PBS was applied to a glow-discharged copper grid (Ted Pella), washed twice with buffer before staining with 2% uranyl acetate for 30 s and air dried. Grids were imaged using a JEOL JEM-1400plus Transmission Electron Microscope operating at 80 kV and recorded with an AMT XR111 high-speed 4k × 2k pixel phosphor-scintillated 12-bit CCD camera.
Dynamic Light Scattering (DLS)
DLS experiments were performed on a DynaPro-PlateReader II (Wyatt Technologies Corporation). Measurements of eNP samples (1 mg/ml) were obtained in 1X PBS buffer at 25 °C and analyzed using Dynamics software (Wyatt).
Multi-angle Light Scattering (MALS)
All experiments were carried out with a Superdex 200 (10/300 GL) connected in-line to a Dawn Heleos II multi-angle light-scattering (MALS) detector (Wyatt Technologies, Santa Barbara, CA). For each experiment, 100 μL of proteins at 2 mg/mL were injected into the column equilibrated in 20 mM Tris, pH 7.5, 150 mM NaCl, and 2 mM tris(2-carboxyethyl)phosphine (TCEP) buffer at room temperature. Molecular weights were determined using Astra software package version 6.1 (Wyatt Technology Corporation, Santa Barbara, CA). All experiments were performed at least in duplicate.
Cryo-EM Data Acquisition
2 μl of eNP-2 tubes were applied onto glow-discharged (40 s) 200-mesh R1.2/1.3 Quantifoil grids. The grids were blotted for 3 s and rapidly frozen in liquid ethane using a Vitrobot Mark IV (FEI). Samples were loaded in a JEM3200FSC cryo-electron microscope (JEOL) operated at 300 kV, condenser lens aperture 70 μm, objective lens aperture 60 μm, spot size 1, equipped with 30 eV in-column energy filter for imaging session. Micrographs were recorded with a K2 Summit direct electron device (Gatan) operating in super resolution mode at a recording rate of 5 raw frames per second and a total exposure time of 8 seconds, yielding 40 frames per stack. Microscope magnification was at 30,000× (corresponding to a calibrated sampling of 1.2 Å per physical pixel), and the total dose was 24 e−/Å2. A total of 1,266 images were manually collected with defocus values below 2 μm.
Single Particle Image Processing and 3D Reconstruction
The raw frames were motion corrected using Motioncorr 2.1. CTFFIND4 (Rohou and Grigorieff, 2015) was used for CTF correction of the whole micrographs. The resulting 1,113 micrographs were subjected to EMAN2.12 (Ludtke, 2016) for evaluation and manual picking of the helical tubes. A total of 200,691 segments (528 pixels × 528 pixels with 5 asymmetric units (ASUs) between segments, which equals to 1,003,455 ASUs) were extracted in Relion 2. A small portion of the entire data set (about 20,000 segments) was initially subjected to selection rule to coarsely determine the helical parameters, followed by 3D reconstructions using EMAN integrated with iterative helical real space reconstruction (IHRSR) to refine the helical parameters to be −8.53° twist and 2.65 Å rise. A 2D rectangular mask (524 pixels (across the helical axis) × 192 pixels (along the helical axis)) was applied to all extracted segments before 2D reference free class averaging. A total of 169,526 segments (847,630 ASUs) from 2D class averages with visible high-resolution features were subjected to auto refinement on helical objects in Relion 2 to yield the final reconstruction at 5.8 Å resolution estimated by the 0.143 criterion of FSC curve. The 5.8 Å Gauss low-pass filter was applied to the final 3D map displayed in UCSF Chimera.
Segmentation and Fitting
The reconstructed density of the helical tube was segmented using the Segger plugin in UCSF Chimera. Smoothing and grouping (5 steps, step size 1) was applied to get an approximate segmentation of each eNP molecule. The crystal structures of several eNP-2 (PDB 4YPI) were then rigidly fitted by alignment to segments corresponding to individual eNP-2. Secondary structure elements in the fitted model matched those visible in the density very well, indicating correct fits.
Flexible Fitting and Modeling
First, the rigidly fitted crystal structure was flexibly fitted to the corresponding extracted density of each eNP-2 in one ASUs using MDFF (Trabuco et al., 2009); this moves helices α21 and α22 and the β-hairpin motif into the density. MDFF was run 10 times starting with the crystal model, which produces 10 slightly different results. The resulting 10 structures were then input into ProMod to create an average model and to calculate uncertainty in residue positions.
For one of the eNP-2, the loop and helix α23 were then added with UCSF Chimera based on the density nearby, using Build Structure functionality extended through several scripts. The new helix α23 was modeled first, orienting it along the clearly tubular density. The loop connecting α22 and the new α23 was then modeled, starting with random φ/ψ angles, further refined by small adjustments to φ/ψ angles to better fit the density and match the two endpoints. The resulting model was flexibly fitted 10 times to the density using MDFF; Promod was then applied to the 10 resulting models to calculate the average model and uncertainties at each residue position.
Fluorescence Polarization Assay (FPA)
FPA experiments were performed on a Cytation5 plate reader (BioTek) operating on Gen5 software. Excitation and emission wavelengths were set to 485 and 528 nm, respectively, with a bandpass of 20 nm. Read height and G factor were set to 8.5 mm and 1.26 using the autogain function. For RNA binding experiments, fluorescein isothiocyanate (FITC) labelled 20 nt ssRNA at a final concentration of 1 nM was loaded on eNP samples (in 20 mM Tris pH 7.5, 150 mM NaCl, 2 mM TCEP) at concentrations ranging from 0.4 nM to 10 μM in a 96-well plate. For eVP35 NPBP binding experiments, FITC labeled eVP35 NPBP at a final concentration of 2 nM were loaded onto eNP samples (in 20 mM Tris pH 7.5, 150 mM NaCl, 2 mM TCEP) at concentrations ranging from 0.04 nM to 1 μM. After 10 min of incubation, fluorescence polarization signals were read. The fluorescence polarization values were then plotted against eNP concentrations to fit the dissociation constant, KD, using ORIGIN software.
Isothermal Titration Calorimetry (ITC)
eNP and peptides samples were dialyzed against 20 mM Tris pH 7.5, 150 mM NaCl, and 2 mM TCEP. Titrations were set up with 100 μM protein in the syringe and 10 μM peptide in the cell using a Microcal VP-ITC microcalorimeter (Malvern). ITC titrations were carried out using a reference power of 5 μcal s−1. All experiments were performed at least in duplicate. The resulting ITC data were processed and fit to a one-site-binding model to determine n (number of binding sites) and KD (dissociation constant) using ORIGIN software.
Hydrogen Deuterium Exchange Mass Spectrometry (HDX-MS)
Each protein solution was buffer exchanged into PBS buffer. Fungal XIII solution in water with 0.1% formic acid was prepared to reach a concentration of 1 mg/mL (25290210)(Li et al., 2014). HDX experiments were initiated by diluting 2 μL of protein solution (50 μM) into 16 μl PBS D2O buffer (final D2O content was 89%, final protein concentration was 5 μM in labeling solution), or H2O buffer for time 0 samples. HDX reactions were measured at various time intervals at 4 °C (10 s, 30 s, 60 s, 360 s, 900 s, and 3600 s). To stop the HDX reaction, ice-cold quench buffer composed of 3 M urea and 1% trifluoreacetic acid was added to the reaction solution. 16 μL of fungal XIII was then added to the mixture and incubated at 4 °C for 2 min before the solution was submitted to on-line pepsin digestion. The resulted peptides were further separated on a reverse-phase C18 column and detected by a Thermo LTQ-FT mass spectrometer, as described previously (Yan et al., 2015). Each measurement was conducted at least in duplicate.
Peptide Identification and HDX-MS Data Analysis
Separate experiments without deuterium labeling were performed to identify peptides generated from the digestion of each protein. Mass spectra of production were collected in a data-dependent mode, in which the six most abundant ions from each scan were selected for MS/MS analysis. The obtained MS/MS files from three parallel runs were then converted to mzXML files using MM File Conversion and submitted to MassMatrix for peptide identification (Xu and Freitas, 2009). Additionally, this search was carried out against a reversed sequence to discard ambiguous identifications. Data analysis of continuous HDX was deconvoluted with HDExaminer (2.0, SierraAnalytics, Inc., Modesto, CA) (Hamuro et al., 2003) and manually inspected. We only show ssRNA free forms of the peptide coverage in the results, but similar coverage was observed for eNP-2, eNP-3 and their mutant forms in the free form (apo) or in complex with ssRNA (holo).
Minigenome Assay (MGA)
The eNP constructs expressing residues 1–384, 413–739, a mutant construct with deletion of residues 384–412 (Δ385–412) and mutant construct (K373A/K374A/382A/383A) were generated in the expression plasmid pCAGGS and include an HA tag at the N-terminus. HEK293T cells were transfected in 96 well format with expression plasmids encoding eL (125 ng), eVP30 (25 ng), eVP35 (31.25 ng), T7 polymerase (50 ng) and firefly luciferase (1 ng), eNP or eNP mutant constructs (62.5ng or as indicated in the figure legend) along with minigenome plasmid construct expressing Renilla luciferase (50ng) using Lipofectamine 2000 (Invitrogen) (Edwards et al., 2015; Xu et al., 2017). Forty-eight hours later the luciferase activity was determined using Dual-Glo luciferase kit (Promega). In parallel, the expression of the eNP constructs was determined by Western blotting using anti-HA or anti-eNP antibody.
Markov State Model (MSM) Construction
We ran 28 simulations, each 1 μs in length, following a previously published protocol (Hart et al., 2016). Each of these simulations was initiated from chain A of PDB ID 4YPI. Simulations were run with the Gromacs software package (Van Der Spoel et al., 2005), using the Amber03 force field (Duan et al., 2003) and TIP3P explicit solvent (Jorgensen et al., 1983). An MSM was constructed using MSMBuilder and the k-hybrids method (Beauchamp et al., 2011; Bowman et al., 2009), ensuring that no two conformations within a single state have a backbone RMSD greater than 2 Å apart. The resulting model had 14,060 states and a lag time of 300 ps.
Data Availability
The hydrogen deuterium exchange mass spectrometry peptide coverage and kinetic plots have been deposited in Mendeley: https://data.mendeley.com/datasets/rt3xn85jbv/1. Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-7343 and Protein Data Bank (PDB; https://www.rcsb.org/): 6C54
Quantification and Statistical Analysis
Statistical parameters including the exact value of n with the description of what n represents, the mean, the SEM and the p value are reported in the Figures and the Figure Legends. Statistical analyses were performed using Prism (GraphPad Software). Statistically significant differences were determined using one-way ANOVA, followed by tukey’s test. In figures, asterisks stand for: *p < 0.001, **** < 0.0001.
Supplementary Material
Figure S1. Negative stain electron microscopy of eNP samples in PBS and electrostatic surface potential of eNP-2.
(A) EM images of eNP-2 p1
(B) EM images of eNP-2 p2
(C) EM images of eNP-2 p2 + eVP35 NPBP
(D) Electrostatic surface potential for eNP-2. Red, white, and blue represent negative, neutral, and positive electrostatic potential (−5 to +5 kBT e−1). Black box indicates RNA binding groove. Orientation is the same as in Figure 1F.
See also Figure 1.
Figure S2. DLS and HDX-MS characterization of eNP proteins.
(A) DLS data in PBS for: (i) eNP-3 (Rh = 26.3 ± 1.6 nm), and (ii) eNP-3 K373A/K374A/K382A/K383A (Rh = 38.8 ± 3.1 nm). Two independent DLS experiments were performed in triplicate for each construct shown.
(B) Differential HDX-MS highlighted on eNP-2 (PDB: 4YPI); regions not present in the structure are indicated by a dotted line. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-3 K373A/K374A/K382A/K383A from that of eNP-3 at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate.
See also Figure 2.
Figure S3. Two dimensional classification and Fourier transform of eNP-2 NC-like tubes.
(A) Fourier transform of the raw micrograph with 3.8 Å water ring indicated.
(B) Layer line indexing of eNP-2 tube.
(C) Two-dimensional class averages before applying the rectangular mask, no detailed features observed.
(D) Fourier shell correlation curve before applying mask. Horizontal dashed lines represent the 0.143 FSC cutoff value.
(E) Two-dimensional class averages after applying the rectangular mask, detailed features improved.
(F) Fourier shell correlation curve after applying mask. Horizontal dashed lines represent the 0.143 FSC cutoff value.
(G) Segmented densities for two eNP conformers within an asymmetric unit with rigidly fitted crystal structure 4YPI for molecule a (left) and molecule b (right).
(H) Secondary structure prediction for eNP sequence from residue 361 to 414 using Jpred.
(I) The model after flexible fitting with MDFF to molecule a (left), and the model after creating an extended C-terminal helix α23 and flexible fitting to the density of molecule b (right).
(J) Model to map FSC curves before (green) and after (blue) flexible fitting.
See also Figure 3.
Figure S4. Analysis and characterization of eNP helix α23.
(A) Probabilistic models for eNP-2 molecules 2a and 2b. Orientation is the same as in Figure 3B. The color of the ribbon representation ranges from green to orange to indicate variance in atomic positions at each residue (green, σ = 0.2 Å; red, σ = 2.7 Å). A majority of the ribbon are colored dark to light green indicating low variance. Other parts with higher variance and uncertainty include loop regions, such as that highlighted in the boxed region.
(B) FP assay of unlabeled eVP35 NPBP peptide (black) or eNP 370–411 peptide (red) competing against FITC-eVP35 NPBP binding to eNP-2. Each measurement was performed two times independently in duplicate.
(C) ITC experiment showing no heat from binding of eNP 370–411 peptide to eNP-2. Each measurement was performed in duplicate.
(D) Electrostatic surface potential for eNP shows eNP oligomerization interfaces have alternating complementary charge distribution. Red, white, and blue represent negative, neutral, and positive electrostatic potential (−5 to +5 kBT e−1). RNA binding grooves locates within the hinge region between N- and C-lobe.
See also Figure 3.
Figure S5. eNP conformational heterogeneity and proposed mechanism for eNP oligomerization and RNA binding.
(A) eNP samples oligomer-like conformations even in the monomeric state. Overlay of a structural state from our MSM (blue) with the holo crystal structure (eNP in green, eVP35 NPBP in magenta) and a monomer from the cryoEM structure (gray).
(B) A proposed mechanism showing that eNP oligomerization is required for RNA binding.
See also Figure 5.
Figure S6. A comparison of the eNP assembly from this study with the eNP assemblies from Wan et al.
(A) Side views of the eNP 1–450 assembly (left) and the eNP-eVP24-eVP35-eVP40 VLP structure (middle) by Wan et al. and the eNP assembly from this study (right). eVP24 subunits from the VLP structure are colored in magenta and yellow (middle). Two eNP-2 molecules in the asymmetric unit from the current study are highlighted in cyan and blue (right).
(B) Six representative eNP molecules from the corresponding assemblies shown in (A) (top). View of the middle two eNP molecules rotated 90° (bottom). The RNA contact surfaces on eNP from the VLP structure are colored in orange and the eVP24 contact surfaces on eNP from the VLP structure are colored in magenta and yellow (middle). This coloring is imposed on the eNP 1–450 assembly (left) and the structure from this study (right). Mapping of the eNP contact surfaces for RNA and eVP24 binding to the eNP-2 assembly from this study suggests potential steric clashes and that the two assemblies are distinct. The red dotted circle on the right panel structure highlights that one set of modeled eVP24 contact interfaces from VLP structure (middle) are not accessible in the structure from the present study.
Movie S2 Probabilistic modeling of eNPs
Movie S3 Illustration of translations and rotations of eNPs
Movie S1 Segmentations and model docking of three asymmetric units
Acknowledgments
We thank Ms. S. Smith for general support and coordination. We also thank the support of National Institutes of Health (P41GM103832 to W.C.; P41GM103422 to M.L.G.; R01GM12400701 to G.R.B; R01AI123926 and U19AI109664 to C.F.B.; P01AI120943 to G.K.A.; R01GM080139 to M.C. via S.J. Ludtke) and by the Department of the Defense, Defense Threat Reduction Agency grant (HDTRA1-16-1-0033 to C.F.B. The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.), and the computing resources provided by the Center for Computational and Integrative Biomedical Research of Baylor College of Medicine and the Texas Advanced Computing Center at the University of Texas at Austin (TGMCB150009). G.R.B. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and a Packard Fellowship for Science and Engineering from the David & Lucile Packard Foundation. C.F.B. is a Georgia Research Alliance Eminent Scholar in Microbial Pathogenesis. We also acknowledge Ms. R. Roth and Drs. R. Zhang and J. Fitzpatrick for support in sample preparation and screening at the Washington University Center for Cellular Imaging supported by Washington University School of Medicine, the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (Grant no: CDI-CORE-2015-505) and the Foundation for Barnes-Jewish Hospital (Grant no: 3770). We also thank E.E.L Amarasinghe and E.O.L. Amarasinghe for comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
G.K.A. and D.W.L. conceived overall project. Z.S., C.W., L.S., P.L., G.D.P., B.J., J.R.P., P.G., M.C., G.L., T.E.F., and J.M.B. performed research. Z.H.Z., G.R.B., C.F.B., M.L.G., D.W.L. W.C., and G.K.A. supervised research. Z.S. C.W. and G.D.P wrote the initial manuscript draft with editing by G.R.B, C.F.B, W.C., D.W.L and G.K.A. All participated in data analysis and assisted in manuscript preparation.
Declaration of Interests:
The authors declare no competing interests.
References
- Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc Natl Acad Sci U S A. 2015;112:E1792–1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Banadyga L, Hoenen T, Ambroggio X, Dunham E, Groseth A, Ebihara H. Ebola virus VP24 interacts with NP to facilitate nucleocapsid assembly and genome packaging. Sci Rep. 2017;7:7698. doi: 10.1038/s41598-017-08167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp KA, Bowman GR, Lane TJ, Maibaum L, Haque IS, Pande VS. MSMBuilder2: Modeling Conformational Dynamics at the Picosecond to Millisecond Scale. Journal of chemical theory and computation. 2011;7:3412–3419. doi: 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci U S A. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkopf N, Lier C, Becker S. Dynamic Phosphorylation of VP30 Is Essential for Ebola Virus Life Cycle. J Virol. 2016;90:4914–4925. doi: 10.1128/JVI.03257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR. An Overview and Practical Guide to Building Markov State Models. In: Bowman GR, Pande VS, Noé F, editors. An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation. Dordrecht: Springer Netherlands; 2014. pp. 7–22. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Huang X, Pande VS. Using generalized ensemble simulations and Markov state models to identify conformational states. Methods. 2009;49:197–201. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Yang P, Li G, Liu B, Wang W, Liu X, Xia B, Yang C, Lou Z, Guo Y, et al. Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution. Protein Cell. 2015;6:351–362. doi: 10.1007/s13238-015-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Edwards MR, Pietzsch C, Vausselin T, Shaw ML, Bukreyev A, Basler CF. High-Throughput Minigenome System for Identifying Small-Molecule Inhibitors of Ebola Virus Replication. ACS infectious diseases. 2015;1:380–387. doi: 10.1021/acsinfecdis.5b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. Journal of Structural Biology. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid Analysis of Protein Structure and Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. Journal of Biomolecular Techniques: JBT. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- Hart KM, Ho CMW, Dutta S, Gross ML, Bowman GR. Modelling proteins’ hidden conformations to predict antibiotic resistance. Nature Communications. 2016;7:12965. doi: 10.1038/ncomms12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Huang Y, Xu L, Sun Y, Nabel GJ. The Assembly of Ebola Virus Nucleocapsid Requires Virion-Associated Proteins 35 and 24 and Posttranslational Modification of Nucleoprotein. Molecular Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- Kirchdoerfer RN, Abelson DM, Li S, Wood MR, Saphire EO. Assembly of the Ebola Virus Nucleoprotein from a Chaperoned VP35 Complex. Cell Rep. 2015;12:140–149. doi: 10.1016/j.celrep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Daisy W, Borek D, Luthra P, Binning Jennifer M, Anantpadma M, Liu G, Harvey Ian B, Su Z, Endlich-Frazier A, Pan J, et al. An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions. Cell Reports. 2015;11:376–389. doi: 10.1016/j.celrep.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rodnin MV, Ladokhin AS, Gross ML. Hydrogen–Deuterium Exchange and Mass Spectrometry Reveal the pH-Dependent Conformational Changes of Diphtheria Toxin T Domain. Biochemistry. 2014;53:6849–6856. doi: 10.1021/bi500893y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ. Single-Particle Refinement and Variability Analysis in EMAN2.1. Methods Enzymol. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Amarasinghe GK, Basler CF. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol. 2015;13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E. Filovirus replication and transcription. Future Virology. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ebihara H, Muramoto Y, Fujii K, Takada A, Sagara H, Kim JH, Kida H, Feldmann H, Kawaoka Y. Assembly and Budding of Ebolavirus. PLOS Pathogens. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie G, Chen DH, Haase-Pettingell Cameron A, King Jonathan A, Chiu W. Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage. Biophysical Journal. 2016;110:827–839. doi: 10.1016/j.bpj.2015.11.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010a;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. Journal of Structural Biology. 2010b;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Cusack S, Kolakofsky D. Segmented negative strand RNA virus nucleoprotein structure. Curr Opin Virol. 2014;5:7–15. doi: 10.1016/j.coviro.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RWH, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Current Opinion in Microbiology. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Huang Y, Sutton-Smith M, Tissot B, Panico M, Morris HR, Dell A, Haslam SM, Boyington J, Graham BS, et al. A Filovirus-Unique Region of Ebola Virus Nucleoprotein Confers Aberrant Migration and Mediates Its Incorporation into Virions. Journal of Virology. 2008;82:6190–6199. doi: 10.1128/JVI.02731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- Team WHOER, Agua-Agum J, Ariyarajah A, Aylward B, Blake IM, Brennan R, Cori A, Donnelly CA, Dorigatti I, Dye C, et al. West African Ebola epidemic after one year--slowing but not yet under control. N Engl J Med. 2015;372:584–587. doi: 10.1056/NEJMc1414992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wan W, Kolesnikova L, Clarke M, Koehler A, Noda T, Becker S, Briggs JAG. Structure and assembly of the Ebola virus nucleocapsid. Nature. 2017;551:394. doi: 10.1038/nature24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Kawaoka YC. Functional mapping of the nucleoprotein of Ebola virus. Journal of Virology. 2006;80:3743–3751. doi: 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. PROTEOMICS. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16:187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Luthra P, Wu C, Batra J, Leung DW, Basler CF, Amarasinghe GK. Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis. Nature Communications. 2017;8:15576. doi: 10.1038/ncomms15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Grant GA, Gross ML. Hydrogen–Deuterium Exchange Mass Spectrometry Reveals Unique Conformational and Chemical Transformations Occurring upon [4Fe-4S] Cluster Binding in the Type 2 l-Serine Dehydratase from Legionella pneumophila. Biochemistry. 2015;54:5322–5328. doi: 10.1021/acs.biochem.5b00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Negative stain electron microscopy of eNP samples in PBS and electrostatic surface potential of eNP-2.
(A) EM images of eNP-2 p1
(B) EM images of eNP-2 p2
(C) EM images of eNP-2 p2 + eVP35 NPBP
(D) Electrostatic surface potential for eNP-2. Red, white, and blue represent negative, neutral, and positive electrostatic potential (−5 to +5 kBT e−1). Black box indicates RNA binding groove. Orientation is the same as in Figure 1F.
See also Figure 1.
Figure S2. DLS and HDX-MS characterization of eNP proteins.
(A) DLS data in PBS for: (i) eNP-3 (Rh = 26.3 ± 1.6 nm), and (ii) eNP-3 K373A/K374A/K382A/K383A (Rh = 38.8 ± 3.1 nm). Two independent DLS experiments were performed in triplicate for each construct shown.
(B) Differential HDX-MS highlighted on eNP-2 (PDB: 4YPI); regions not present in the structure are indicated by a dotted line. The color gradient represented differential HDX shown at the bottom is generated by subtracting the amount of deuterium uptake of eNP-3 K373A/K374A/K382A/K383A from that of eNP-3 at the 60 second time point. Regions colored white are not detected by HDX. HDX-MS measurements were performed in duplicate.
See also Figure 2.
Figure S3. Two dimensional classification and Fourier transform of eNP-2 NC-like tubes.
(A) Fourier transform of the raw micrograph with 3.8 Å water ring indicated.
(B) Layer line indexing of eNP-2 tube.
(C) Two-dimensional class averages before applying the rectangular mask, no detailed features observed.
(D) Fourier shell correlation curve before applying mask. Horizontal dashed lines represent the 0.143 FSC cutoff value.
(E) Two-dimensional class averages after applying the rectangular mask, detailed features improved.
(F) Fourier shell correlation curve after applying mask. Horizontal dashed lines represent the 0.143 FSC cutoff value.
(G) Segmented densities for two eNP conformers within an asymmetric unit with rigidly fitted crystal structure 4YPI for molecule a (left) and molecule b (right).
(H) Secondary structure prediction for eNP sequence from residue 361 to 414 using Jpred.
(I) The model after flexible fitting with MDFF to molecule a (left), and the model after creating an extended C-terminal helix α23 and flexible fitting to the density of molecule b (right).
(J) Model to map FSC curves before (green) and after (blue) flexible fitting.
See also Figure 3.
Figure S4. Analysis and characterization of eNP helix α23.
(A) Probabilistic models for eNP-2 molecules 2a and 2b. Orientation is the same as in Figure 3B. The color of the ribbon representation ranges from green to orange to indicate variance in atomic positions at each residue (green, σ = 0.2 Å; red, σ = 2.7 Å). A majority of the ribbon are colored dark to light green indicating low variance. Other parts with higher variance and uncertainty include loop regions, such as that highlighted in the boxed region.
(B) FP assay of unlabeled eVP35 NPBP peptide (black) or eNP 370–411 peptide (red) competing against FITC-eVP35 NPBP binding to eNP-2. Each measurement was performed two times independently in duplicate.
(C) ITC experiment showing no heat from binding of eNP 370–411 peptide to eNP-2. Each measurement was performed in duplicate.
(D) Electrostatic surface potential for eNP shows eNP oligomerization interfaces have alternating complementary charge distribution. Red, white, and blue represent negative, neutral, and positive electrostatic potential (−5 to +5 kBT e−1). RNA binding grooves locates within the hinge region between N- and C-lobe.
See also Figure 3.
Figure S5. eNP conformational heterogeneity and proposed mechanism for eNP oligomerization and RNA binding.
(A) eNP samples oligomer-like conformations even in the monomeric state. Overlay of a structural state from our MSM (blue) with the holo crystal structure (eNP in green, eVP35 NPBP in magenta) and a monomer from the cryoEM structure (gray).
(B) A proposed mechanism showing that eNP oligomerization is required for RNA binding.
See also Figure 5.
Figure S6. A comparison of the eNP assembly from this study with the eNP assemblies from Wan et al.
(A) Side views of the eNP 1–450 assembly (left) and the eNP-eVP24-eVP35-eVP40 VLP structure (middle) by Wan et al. and the eNP assembly from this study (right). eVP24 subunits from the VLP structure are colored in magenta and yellow (middle). Two eNP-2 molecules in the asymmetric unit from the current study are highlighted in cyan and blue (right).
(B) Six representative eNP molecules from the corresponding assemblies shown in (A) (top). View of the middle two eNP molecules rotated 90° (bottom). The RNA contact surfaces on eNP from the VLP structure are colored in orange and the eVP24 contact surfaces on eNP from the VLP structure are colored in magenta and yellow (middle). This coloring is imposed on the eNP 1–450 assembly (left) and the structure from this study (right). Mapping of the eNP contact surfaces for RNA and eVP24 binding to the eNP-2 assembly from this study suggests potential steric clashes and that the two assemblies are distinct. The red dotted circle on the right panel structure highlights that one set of modeled eVP24 contact interfaces from VLP structure (middle) are not accessible in the structure from the present study.
Movie S2 Probabilistic modeling of eNPs
Movie S3 Illustration of translations and rotations of eNPs
Movie S1 Segmentations and model docking of three asymmetric units
Data Availability Statement
The hydrogen deuterium exchange mass spectrometry peptide coverage and kinetic plots have been deposited in Mendeley: https://data.mendeley.com/datasets/rt3xn85jbv/1. Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-7343 and Protein Data Bank (PDB; https://www.rcsb.org/): 6C54