Abstract
In the sea urchin embryo, gastrulation is characterized by the ingression and directed cell migration of primary mesenchyme cells (PMCs), as well as the primary invagination and convergent extension of the endomesoderm. Like all cell shape changes, individual and collective cell motility is orchestrated by Rho family GTPases and their modulation of the actomyosin cytoskeleton. And while endomesoderm specification has been intensively studied in echinoids, much less is known about the proximate regulators driving cell motility. Toward these ends, we employed anti-sense morpholinos, mutant alleles and pharmacological inhibitors to assess the role of Cdc42 during sea urchin gastrulation. While inhibition of Cdc42 expression or activity had only mild effects on PMC ingression, PMC migration, alignment and skeletogenesis were disrupted in the absence of Cdc42, as well as elongation of the archenteron. PMC migration and patterning of the larval skeleton relies on the extension of filopodia, and Cdc42 was required for filopodia in vivo as well as in cultured PMCs. Lastly, filopodial extension required both Arp2/3 and formin actin-nucleating factors, supporting models of filopodial nucleation observed in other systems. Together, these results suggest that Cdc42 plays essential roles during PMC cell motility and organogenesis.
Keywords: Sea urchin, Gastrulation, Cdc42, Filopodia, Morphogenesis
1. Introduction
During morphogenesis, cells undergo a series of genetically orchestrated changes in cell shape, adhesion and migration that lead to the formation of tissues and organs. These highly stereotypical movements rely on intrinsic and extrinsic signals that must ultimately converge on the proximate regulators of the actomyosin cytoskeleton to facilitate cell shape change. And while comprehensive gene regulatory networks (GRNs) that drive specification for a growing list of organisms (Cheatle Jarvela and Hinman, 2015; Ettensohn, 2013; Peter and Davidson, 2011) have been described, how these gene regulatory cascades ultimately drive changes in cellular behavior remains an area of active investigation.
The sea urchin embryo has been and remains a powerful model for studying collective and individual cell migration during development, due in part to the simplicity of the embryo, ease of manipulation and comprehensive GRNs described for the endomesoderm (Davidson et al., 2002; Lyons et al., 2012; Martik et al., 2016). In the sea urchin embryo, morphogenesis begins with alterations in cell adhesion at the vegetal plate resulting in an Epithelial-Mesenchymal Transition (EMT) of the Primary Mesenchyme Cells (PMCs) (Burdsal et al., 1991; Fink and McClay, 1985; McClay and Fink, 1982). Following ingression into the blastocoel, PMCs migrate in a pattern defined by the ectoderm, eventually fusing to form a common syncytium into which PMCs deposit skeletogenic material, serving as a template for the larval skeleton (Lyons et al., 2014). PMCs initially form a ring around the center of the vegetal plate, and as migration proceeds, PMCs organize into two ventro-lateral clusters (VLCs) (Peterson and McClay, 2003) located where the dorsal-ventral margin of the embryo intersects with the border ectoderm (McIntyre et al., 2014). Establishment of the VLCs and the PMC organization into a ring is patterned by interactions between PMCs and the blastocoel wall through VEGF signaling (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007). PMC motility and detection of chemotactic cues is mediated by actin-based filopodia that contact the blastocoelar wall and search three-dimensional space (Gustafson and Wolpert, 1967; Malinda and Ettensohn, 1994; Malinda et al., 1995; McClay, 1999; Miller et al., 1995). And while other non-skeletogenic mesenchymal cells elaborate filopodia in the sea urchin embryo, PMCs are particularly notable for their requirement of filopodia for both their patterning and motility. How PMCs regulate actin polymerization during filopodial-based motility, however, is not well understood.
The Rho family of small GTPases (Rho, Rac and Cdc42) act as the proximate regulators of the actomyosin cytoskeleton, integrating extracellular signals to mediate cell shape change (Nobes and Hall, 1995a). Early studies using bacterial toxins and activity-modulating mutants suggested that these three proteins affect distinct cellular behaviors (Nobes and Hall, 1995b; Paterson et al., 1990; Ridley and Hall, 1992; Ridley et al., 1992), and it was soon appreciated that these molecules played critical roles in morphogenesis (Barrett et al., 1997; Chen et al., 1996; Harden et al., 1995; Settleman, 2001). In Drosophila, Rho, Rac and Cdc42 are necessary for proper dorsal closure (Glise et al., 1995; Glise and Noselli, 1997; Harden et al., 1995; Noselli, 1998). In Xenopus, both Rho and Rac regulate distinct and complementary pathways involved in cell intercalation during convergent extension of the axial mesoderm (Tahinci and Symes, 2003) while Cdc42 influences convergent extension of the dorsal mesoderm and posterior neuroectoderm downstream of non-canonical Wnt signaling (Choi and Han, 2002). Ascribing general roles for the Rho GTPases during animal development is complicated by the large diversity of regulatory proteins (exchange factors and GTPase activating proteins), the expression of which is subject to spatiotemporal regulation during embryogenesis (Denk-Lobnig and Martin, 2017). Further, the participation of a particular G protein in a given developmental process may also be influenced by synergistic and antagonistic relationships amongst the other GTPases and their respective regulators (Guilluy et al., 2011). Thus, while the developmental roles of small GTPases have been studied in a wide array of model organisms, there are multiple parameters that determine the participation of a G protein in a particular morphogenetic event.
Filopodia are comprised of unbranched, bundled actin filaments that are typically thought to be nucleated by the formin family of actin nucleation factors (Mellor, 2010), although models have been proposed whereby branched actin nucleated by the Arp2/3 complex also contribute to filopodia formation (Svitkina et al., 2003; Young et al., 2015). Both formins and the Arp2/3 family of actin nucleation factors act downstream of Rho family GTPases (Kühn and Geyer, 2014; Rotty et al., 2013), but to date, there have been few reports examining the role of Rho family GTPases in sea urchin development, and none addressing filopodia formation in PMCs. RhoA has been shown to be required for primary invagination, but appears dispensable for PMC ingression and motility (Beane et al., 2006). In this report, we examined the role of Cdc42 in sea urchin morphogenesis. Here, we demonstrate that while Cdc42 was dispensable for PMC ingression and primary invagination of the gut, Cdc42 activity was required for proper PMC migration and patterning as well as elongation of the archenteron. Inhibition of Cdc42 activity blocked filopodia formation by PMCs, resulting in disorganized PMC migration, formation and maintenance of the PMC syncytium and skeletogenesis. Lastly, filopodia formation in PMCs required the action of both formin- and Arp2/3-based actin nucleation, consistent with the convergent elongation model whereby both nucleation factors contribute to filopodial extension (Svitkina et al., 2003). Together these results identify Cdc42 as a key modulator of PMC motility and organogenesis through its modulation of actin networks.
2. Results
2.1. Cdc42 is required for early cleavages and PMC organization
In the Strongylocentrotus purpuratus embryo, Cdc42 transcripts rise approximately six-fold between the egg and mesenchyme blastula stages (Tu et al., 2014), and Cdc42 and its downstream effectors (WASP and the Arp2/3 complex) are enriched in PMCs as demonstrated by in situ hybridization (Rafiq et al., 2012). As a first estimation as to whether Cdc42 plays a functional role in the early morphogenetic events of sea urchin development, fertilized S. purpuratus eggs were injected with mRNA encoding wild-type (WT) or dominant-negative (DN, T17N) Cdc42, and phenotypes of injected embryos were examined 24 h post-fertilization, when embryos have normally undergone EMT (Fig. 1). While embryos expressing WT-Cdc42 underwent PMC ingression normally (Fig. 1A and F, blue), expression of dominant-negative Cdc42 had profound effects on early embryonic development. The percentage of morphologically normal mesenchyme blastulae decreased dramatically between the two concentrations of injected mRNA, with an increase in embryos where PMCs were randomly distributed throughout the blastocoel (Fig. 1C and F, pink). The fraction of blastulae exhibiting no PMC ingression (Fig. 1B and F, green) was very low, suggesting that PMCs were not blocked from ingressing into the blastocoel. Interestingly, embryos with defects in early cell divisions were observed in both DN concentrations (Fig. 1E and F, yellow), where development was halted before reaching the blastula stage. Embryos in this group arrested in the early cleavage stages, frequently with multiple nuclei, suggesting a possible role for Cdc42 in coordinating cytokinesis in the early embryonic stages.
Fig. 1.
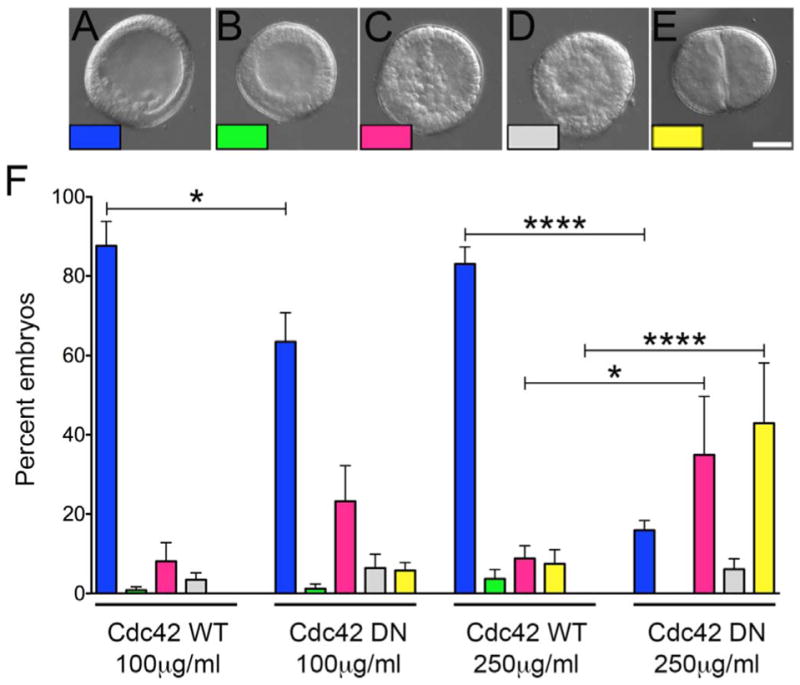
Cdc42 is required for early divisions and PMC organization in the mesenchyme blastula. A–E) S. purpuratus eggs were injected with mRNAs encoding WT or DN-Cdc42, and embryos were scored for developmental progression 24 h post-fertilization when embryos normally reach the mesenchyme blastula stage (A–E; Bar, 50 μm). Embryos were scored as mesenchyme blastula (1A, blue); blastula (1B, green); embryos where PMCs had ingressed but were scattered within the blastocoel (1C, pink); embryos with abnormal ectodermal epithelium (1D, gray); or embryos with cytokinetic defects (1E, yellow). F) Expression of DN-Cdc42 resulted in an increase in cell division defects (1E and F, yellow bars) as well as defects in PMC retention at the vegetal pole, with disorganized cells distributed throughout the blastocoel (1C; F, pink bars). Each experimental condition represents six experimental replicates, with a minimum of 145 embryos scored per condition. * p < 0.05; *** * p < 0.0001.
2.2. PMC migration and archenteron elongation require Cdc42 activity
Expression of DN-Cdc42 resulted in a number of developmental defects, with up to half of embryos failing to progress beyond the cleavage stage (Fig. 1F, yellow). As an alternative approach, morpholino anti-sense oligonucleotides (MASO) were employed to inhibit new translation from maternal or zygotic transcripts, while allowing maternal protein stores to support Cdc42 function during early divisions. A translation-blocking morpholino was designed to specifically target the first 25 nucleotides of the S. purpuratus Cdc42 open reading frame (Fig. S1A). Examination of embryos over a range of MASO concentrations revealed that at the mesenchyme blastula stage, depletion of Cdc42 failed to reveal a significant effect on development, suggesting that maternally loaded Cdc42 was capable of sustaining cell divisions through the blastula stage (Fig. S1C). However, dramatic effects were observed when morphants were examined at 48 h post-fertilization, when embryos are typically at the gastrula stage, with PMCs organized around the posterior and the archenteron having extended to nearly its full length (Fig. 2A). At 500 μM Cdc42 MASO, only 22% of Cdc42 morphants reached the gastrula or mid-gastrula stages in comparison to over 70% of controls (Fig. 2A and B; F, blue and green). The most prominent phenotype displayed by Cdc42 morphants was a lack of PMC organization, with PMCs scattered throughout the blastocoel (Fig. 2D; F, pink). Additionally, these embryos lacked an archenteron. And while there was variation between experimental replicates, both the delay in gastrulation and the increase in scattered PMCs in the morphants were statistically significant (Fig. 2F, p < 0.001). PMC organization and gut elongation could be rescued by the expression of human Cdc42, which shares 90% identity and 95% similarity with S. purpuratus Cdc42 at the amino acid level, but is resistant to the SpCdc42 MASO (Figs. S1A–B, 2F). Injections of concentrations lower than 500 μM failed to demonstrate a statistically significant phenotype, whereas higher concentrations (1 mM) generated phenotypes that could not be rescued by HsCdc42, suggesting that the observed phenotypes at the highest concentrations of morpholino were not specific. Together, these experiments suggested that Cdc42 was playing a role in sea urchin morphogenesis, and that once maternal stocks of the GTPase were depleted, new translation was required for further development.
Fig. 2.
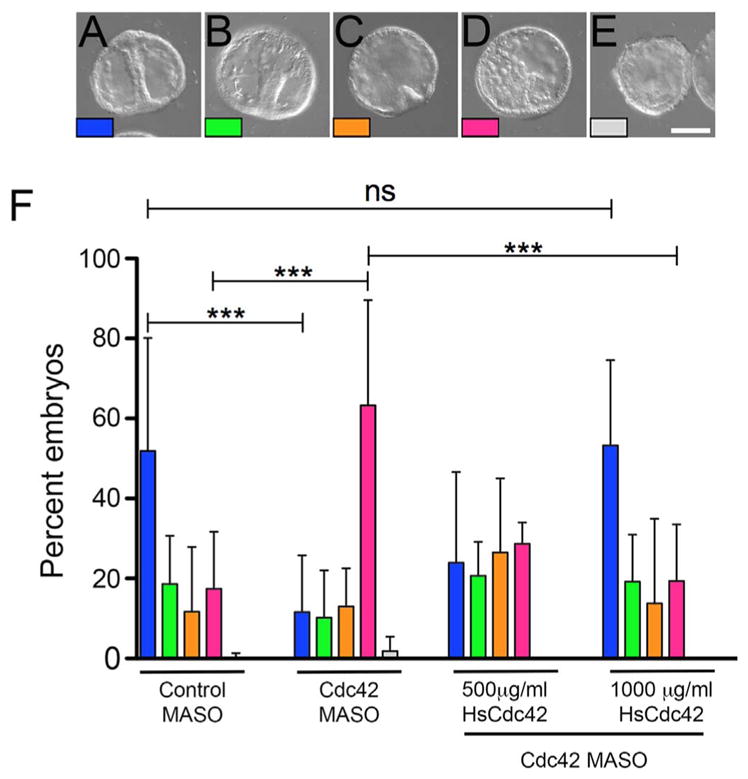
Archenteron elongation and PMC organization are disrupted upon depletion of Cdc42. A–E) S. purpuratus eggs were injected with untargeting control or Cdc42 morpholinos, and embryos were scored for developmental progression 48 h post-fertilization, when embryos normally reach the gastrula stage (Bar, 50 μm). Embryos were scored as gastrulae (with archenterons extended at least 75% of total length and spicules present, 2A, blue); mid-gastrulae, with elongating archenteron and spicules (2B, green); embryos with a primary invagination and a lack of organized PMCs (2C, orange); embryos containing disorganized PMCs and lacking an archenteron (2D, pink); and embryos displaying both disrupted mesenchymal cells and epithelia (2E, gray). F) Quantification of phenotypes in embryos injected with control or Cdc42 antisense morpholinos at 48 h post-fertilization. While control embryos were in the gastrula or mid-gastrula stages, in embryos injected with 500 μM MASO showed defects in PMC organization and archenteron elongation (2H, pink bars). These defects were rescued with co-injection of human Cdc42. Each experimental condition represents at least three experimental replicates, with a minimum of 100 embryos per condition. *** p < 0.001; ns: p = 0.9765.
2.3. Cdc42 is required for PMC alignment, fusion and skeletogenesis
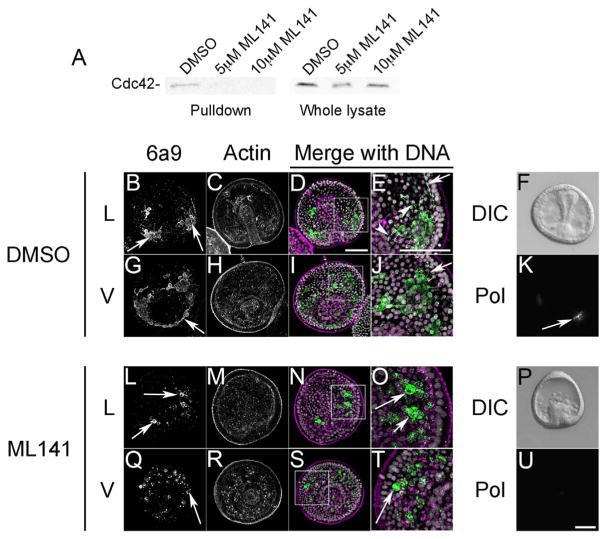
Abrogation of Cdc42 activity using a dominant-negative mutant or antisense morpholinos implicated a role for Cdc42 not only for the early divisions and proper blastula formation but also in morphogenetic movements of the endoderm and skeletogenic mesoderm (Figs. 1 and 2). To exert finer temporal control over Cdc42 function, we employed the Cdc42 inhibitor, ML141, which specifically inhibits Cdc42 nucleotide dissociation and activation (Hong et al., 2013), and reduced active Cdc42 levels in sea urchin embryos as measured with a Cdc42 pull-down assay (Fig. 3A). L variegatus embryos were treated at the early blastula with DMSO or ML141, and were examined by immunolocalization or imaged live using DIC and polarizing microscopy when controls reached the gastrula stage (Fig. 3). Lateral (L) or ventral (V) views of control embryos revealed PMCs organized into ventro-lateral clusters (Fig. 3B, arrows) and organized into a ring around the vegetal pole (Fig. 3G, arrow). In these gastrula-stage embryos, PMCs were forming a syncytium and extending filopodia along the blastocoel wall (Fig. 3E and J, arrows). In contrast, PMCs in ML141-treated embryos could be found near the site of the ventrolateral clusters (Fig. 3L–O, arrows), but did not fuse or extend filopodia (Fig. 3O and T, arrows). Moreover, the 6a9 marker in many PMCs appeared to be internalized (Fig. 3O, arrows). Polarizing microscopy, which highlights the birefringent spicule matrix, revealed the presence of tri-radiate spicules in controls (Fig. 3K, arrow) that were not detectable in ML141-treated embryos (Fig. 3U).
Fig. 3.
Cdc42 activity is required for correct PMC migration, filopodia formation and initiation of skeletogenesis. A) Cdc42 activity assay. Lysates from gastrulae treated with carrier control (0.1%) DMSO or the Cdc42 inhibitor ML141 were incubated with PAK beads, and Cdc42 bound to the beads or present in unfractionated lysates was detected by Western blotting. B–U) L. variegatus embryos were treated with 5 μM ML141 at the blastula stage, and then fixed and processed for immunolabeling with PMC-specific (6a9, green) and anti-actin (magenta) antibodies or analyzed live by polarization microscopy. Maximum intensity projections of Lateral (L) or Vegetal (V) views of control or ML141-treated embryos revealed differences in PMC morphology and migration. Control embryos exhibited PMCs organized in ventrolateral clusters (VLCs) (3B, arrows) and in a ring around the vegetal pole (3G, arrow), with extended filopodia contacting the ectoderm (3E and J, arrows). Inhibition of Cdc42 resulted in PMCs organized around VLCs and the vegetal ring, but lacked filopodia (3O and T, arrows), and the tight organization of controls (3L and Q). Imaging by polarization microscopy revealed the presence of birefringent, tri-radiate spicules in controls (3K) but not detectable in ML141-treated embryos (3U). Bar, 50 μm.
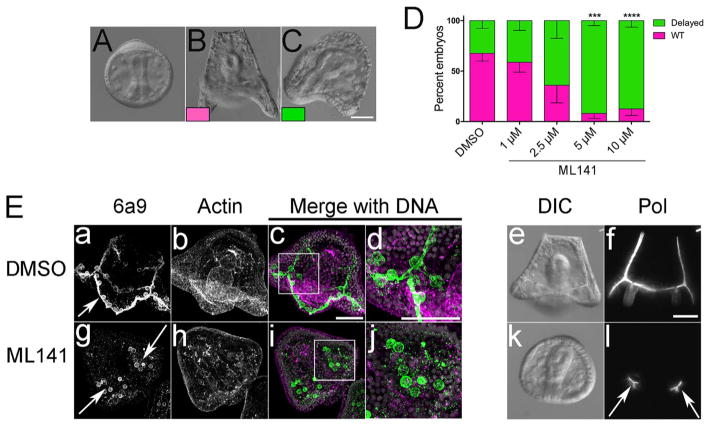
To further examine whether Cdc42 was required for skeletogenesis after PMC alignment and fusion, L. variegatus gastrulae were treated for 24 h with DMSO or ML141 when PMCs had aligned along the dorsal-ventral margin and begun forming tri-radiate spicules (Fig. 4A). ML141 treatment resulted in dose-dependent effects on developmental progression (Fig. 4D). Whereas control cultures were primarily in the prism or early larval stages (Fig. 4B, D, pink), embryos treated with ML141 were delayed in skeletal elongation and extension of the larval arms (Fig. 4C, D, green). Immunolabeling revealed that at this stage, control embryos were characterized by syncytial PMCs whose cell bodies were distributed along the skeletal arms and rods (Fig. 4E, panels a–d), which were also evident by polarizing microscopy (Fig. 4E, panel f). In ML141-treated embryos, PMCs were loosely organized around ventrolateral clusters (Fig. 4E, panel g, arrow), but there was no evidence of fusion or filopodial extension (Fig. 4E, panel j). Moreover, these embryos appeared to have lost the organization and morphology of PMCs at the time of treatment (gastrula-stage embryos, Fig. 3B and E). Similarly, examination of ML141-treated embryos by polarizing microscopy revealed that the birefringent spicules had not elongated beyond what was observed in gastrula-stage embryos at the time of treatment initiation (Fig. 4E, panel l, arrows). Together, these results suggest that Cdc42 was not only required for PMC migration and patterning, but also for skeletogenesis.
Fig. 4.
PMC syncytium formation and skeletogenesis requires Cdc42 activity. A–D) L. variegatus embryos were treated with ML141 at the gastula stage (A), and embryos were scored for developmental progression when controls reached the prism/early larval stage. D) Quantification of phenotypes represented in B and C for three experimental replicates, with an average of 250 embryos scored per condition per experiment. ***, p < 0.0003; *** *, p < 0.0001. E) L. variegatus embryos with treated with 5 μM ML141 at the gastrula stage, and then fixed and processed for immunolabeling with PMC-specific (6a9, green) and anti-actin (magenta) antibodies or analyzed live by polarization microscopy. Control embryos presented a well-formed common synctytium (panels a–d) and larval skeleton (panel f), whereas PMCs in embryos incubated with ML141 failed to form a common syncytium (panels g–j). Deposition of skeletal material was limited to what was generated at the time of treatment (panel l). Bar, 50 μm.
Disruption of Cdc42 activity using three different approaches had dramatic effects on PMC morphology and function. However, the morphology of cells within the blastocoel (Figs. 1C, 2D) as well as the accumulation of 6a9-positive cell fragments (Figs. 3Q, 4E, panel g) raised the possibility that Cdc42 inhibition compromised PMC viability. To test if the observed phenotypes resulted from a loss of skeletogenic gene expression, L. variegatus embryos were treated with either DMSO or ML141 beginning at the blastula stage, and mRNA was harvested from gastrula (24 h) and early larval stages (48 h). Quantitative PCR analysis revealed that there were no changes in the transcript levels of small GTPases (Cdc42 and Rac), PMC-specific markers (C-lectin, SM-32 and MSP130-5) or ectodermal markers (FGFa and NK2.1) equal to- or greater than 3 fold, suggesting that Cdc42 inhibition had no effect on transcriptional regulation of genes related to skeletogenesis (Fig. S2). To further assess possible toxic effects of Cdc42 inhibition on PMCs, we probed control or treated embryos with a marker for apoptosis (Fig. S3). Control embryos treated at the blastula stage (Fig. S3A, panels a–d) exhibited patterned PMCs with few detectable apoptotic cells (Fig. S3B), none of which were PMCs (Fig. S3C). ML141-treated embryos displayed the characteristically scattered PMCs, and apoptotic cells could be detected in the blastocoel (Fig. S3A, panels e–h, S3B). Instances of 6a9-positive apoptotic cells could be detected (Fig. S3A, panels g and h, arrows), but these averaged only 15% of PMCs in the embryo (Fig. S3C). ML141 treatment at the gastrula stage also resulted in more apoptotic cells relative to controls (Fig. S3A, panels i–p, S3B) but only 3% of PMCs were apoptotic (Fig. S3C). Further, the 6a9-positive cell fragments that were prominent in ML141-treated embryos (S3A, panel p, green) were not positive for cleaved caspase, suggesting that these were not apoptotic cell remnants. Thus, while ML141 treatment did result in an increase in apoptotic cells within the embryo, overall PMC viability did not appear to be selectively sensitive to Cdc42 inhibition.
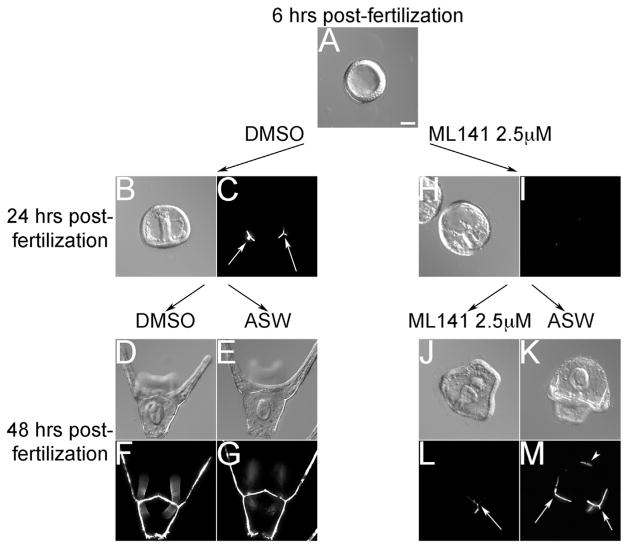
The ability of PMCs to recover from Cdc42 inhibition after ML141 washout was tested. L. variegatus blastulae (Fig. 5A), were treated with DMSO or ML141, and at 24 h, embryos from both treatments were either released into seawater or placed back into treatment for an additional 24 h. At 24 h post-fertilization, control embryos were at the gastrula stage with fully elongated guts (Fig. 5B) and tri-radiate spicules as visualized with polarization microscopy (Fig. 5C). By 48 h, both DMSO and mock washout embryos were in the early larval stage with fully elongated skeletal arms (Fig. 5D–G). At 24 h, ML141-treated embryos displayed the characteristic phenotype of an arrested gut extension and minimally visible spicules (Fig. 5H and I). If embryos were kept in the presence of the inhibitor, there was only slight detectable evidence of skeletogenesis (Fig. 5L). However, if embryos were released into seawater lacking ML141, they were able to resume deposition of skeletal matrix as evidenced by the presence of elongating skeletal rods (Fig. 5M, arrows). Skeletal material deposition did not always follow the normal pattern, and skeletogenic material could be found at irregular locations (Fig. 5M, arrowhead), possibly a reflection of the scattering of PMCs observed in other experiments (Fig. 4E, panel g). Thus, PMCs were able to recover their physiological function and resume skeletogenesis after drug removal, indicating that the effect of Cdc42 inhibition did not compromise PMC viability.
Fig. 5.
PMCs resume skeletogenesis upon reversal of Cdc42 inhibition. Viability of PMCs following ML141 treatment was confirmed by treating embryos at the blastula stage with 2.5 μM ML141 and then releasing embryos from Cdc42 inhibition. As viewed by polarization microscopy, ML141-treated embryos that failed to initiate spiculogenesis by 24 h were able to reinitiate skeletogenesis upon removal of ML141 (5 M, arrows). Arrowhead denotes the presence of an ectopic spicule. Bar, 50 μm.
2.4. Cdc42 inhibition affects filopodia formation and dynamics in PMCs
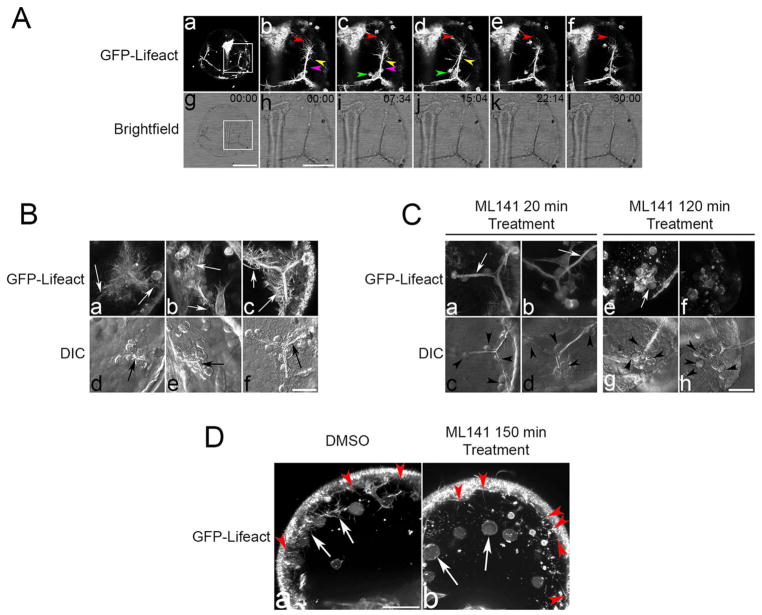
One defining characteristic of PMCs is the use of filopodia to sense spatial cues, establish cell contacts and move within the blastocoel (Malinda et al., 1995; Miller et al., 1995). Cdc42 inhibition resulted in a reduction of filopodial extension and PMC patterning (Fig. 3). To further characterize the influence of Cdc42 on filopodia in PMCs, gastrulae expressing the actin probe GFP-Lifeact (Riedl et al., 2008) were imaged live by confocal microscopy (Fig. 6). In gastrula stage embryos, GFP-Lifeact labeled all cells in the embryo, but was particularly evident in the PMC syncytial cytoplasm and filopodia along the triradiate spicules (Fig. 6A, panels b–f). All along the length of the skeletal rod, filopodia could be observed extending and retracting on a minute time scale (Fig. 6A, panels b–d, pink and yellow arrowheads). Abundant, more stable filopodia could be observed extending from the anterior tip (Fig. 6A, panels b–f, red arrow) and non-skeletogenic mesenchymal cells could be observed making frequent contacts with filopodia all along the structure (Fig. 6A, panels c and d, green arrow).
Fig. 6.
Live-cell imaging of filopodia in the presence or absence of Cdc42 activity. L. pictus gastrulae expressing GFP-Lifeact were imaged by brightfield and confocal microscopy. A) Control embryos form dynamic filopodia along long the length of the spicule (pink and yellow arrowheads), at the distal tips (red arrowhead), and in contacts with non-skeletogenic mesenchymal cells (green arrowhead). Bar, 50 μm. B and C) Observation of ventro-lateral clusters (VLCs) at early maturation stages showed PMCs associated with small tri-radiate spicules (6B, panels d and e, black arrows) and later to the growing spicules (6B, panels c and f). In all stages abundant filopodia extended from the PMCs and spicule rods (6B, panels a–c, white arrows). Embryos incubated with 10 μ ML141 for 20 min exhibited PMCs associated with skeletal rods but with decreased filopodia (6 C, panels a–d) compared to controls (6B, panels a–c). Longer treatments resulted in stronger effects on PMCs organization and filopodia processes, and loss of syncytial cytoplasm associated with the spicule rods (6C, panels e and f). D) Control S. purpuratus gastrulae expressing GFP-Lifeact exhibited dynamic filopodia processes elaborated from PMCs migrating along the ectoderm (6D, panel a, arrows) as well as from the basal membrane of ectodermal cells (6D, panel a, red arrowheads). Treatment with ML141 for 2.5 h inhibited filopodia formation in PMCs (6D, panel b, arrows), whereas ectodermal filopodia were unaffected by Cdc42 inhibition (6D, panel b, red arrowheads). Bar, 25 μm.
Examination of PMCs in ventro-lateral clusters (VLCs) revealed that in control cultures, abundant filopodia could be observed extending outward, some of which made contacts with the ectoderm (Fig. 6B, panels a, b, d and e, arrows). In more mature VLCs (Fig. 6B, panels c and f), which contained more PMCs and longer spicules, filopodia could be observed extending from the cytoplasmic cables enveloping the growing rods (Fig. 6B, panel c, arrows). Examination of VLCs exposed to brief ML141 treatment exhibited diminished filopodia (Fig. 6C, panels a and b, arrows), with PMCs positioned primarily along the tips of the rods (Fig. 6C, panels c and d, arrowheads). However, the PMC cytoplasm enveloping the growing spicule remained intact (Fig. 6C, panels a and b, arrows). Longer treatments resulted in a more dramatic phenotype, with PMCs lacking filopodia (Fig. 6C, panel e, arrow) but remaining associated with the spicules (Fig. 6C, panels g and h, arrowheads). Moreover, the cytoplasmic sheath enveloping the spicules was lost, and cellular fragments could be observed in the blastocoel (Fig. 6C, panels e and f). These results closely resembled the morphology of PMCs treated with ML141 at the gastrula stage (Fig. 4E), and suggested that Cdc42 is not only required for filopodia formation, but also for the maintenance of the syncytial cytoplasm that envelopes the growing spicule.
PMC patterning is a directed by ectodermal signaling (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007) and it has been suggested that filopodia elaborated by ectodermal epithelia participates in the communication of migrational cues to the PMCs (Karp and Solursh, 1985). Ectodermal cells exhibit dynamic extension and retraction of short filopodia from the basal membrane into the blastocoel (Miller et al., 1995), and to determine whether Cdc42 inhibition affected one or both filopodia populations, we imaged ectodermal filopodia in live gastrulae expressing GFP-Lifeact (Fig. 6D). To better distinguish filopodia from the two cell types, cells distal to the VLCs were imaged, where migrating PMCs in control gastrulae contacted each other through active filopodia (Fig. 6D, panel a, arrows), while dynamic ectodermal filopodia could be observed extending to the blastocoel (Fig. 6D, panel a, red arrowheads). Treatment with ML141 resulted in a loss of PMC filopodia (Fig. 6D, panel b, arrows), as observed for PMCs in the ventrolateral clusters (Fig. 6C, panels e and f). Interestingly, ectodermal filopodia was not diminished following Cdc42 inhibition (Fig. 6D, panel b, red arrowheads), suggesting that basal filopodia are likely regulated by a different upstream actin modulator.
To further assess the specificity of Cdc42 in PMC filopodia formation and eliminate the possibility of non-specific effects on actin-based motility, we examined Cdc42 inhibition on pigment cells, a population of non-skeletogenic mesenchymal cells that play a role in innate immune responses and display pseudopodial motility (Ch Ho et al., 2016). Pigment cells in control embryos were found subjacent to the ectodermal layer with multiple cellular protrusions (Fig. S4A–E), and ML141-treated embryos were indistinguishable from controls (Fig. S4F–J). However, inhibition of Arp2/3, an actin-nucleating factor required for lamellapodial motility (Bailly et al., 2001), completely blocked protrusion formation, resulting in a distinctive rounded phenotype (Fig. S4K–O). Thus, the effects of Cdc42 on mesenchymal motility in sea urchin embryos appeared to be selective for filopodia formation.
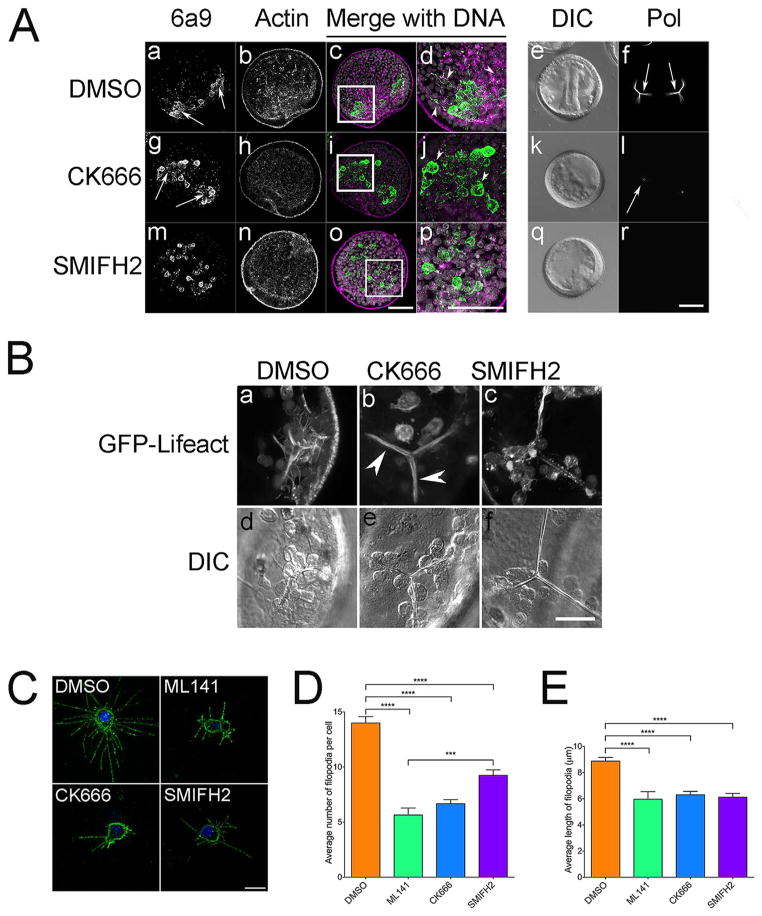
Actin-based motility downstream of Cdc42 is driven by the nucleation of unbranched and branched actin networks nucleated by formins and the Arp2/3 complex, respectively (Campellone and Welch, 2010). To examine the contributions of Cdc42 and the different actin nucleating factors in promoting filopodial formation, embryos were treated at the blastula stage with DMSO or inhibitors for the Arp2/3 complex (CK666) or formins (SMIFH2) (Fig. 7A), which have been shown to inhibit branched and linear actin networks in sea urchin coelomocytes (Henson et al., 2014, 2015). Whereas control embryos at the gastrula stage exhibited organized VLCs, filopodia and growing spicules (Fig. 7A, panel a, arrows; panel d, arrowheads; panel f, arrows), PMCs in CK666-treated embryos failed to form organized VLCs, extend filopodia or grow spicules (Fig. 7A, panel g arrows; panel j, arrowheads; and panels l, arrow). SMIFH2 inhibition of formins resulted in a similar phenotype, with disorganized PMCs lacking filopodia and no visible spicules (Fig. 7A, panels m–r). Treatment of GFP-Lifeact-expressing gastrulae largely mirrored these results, with the only notable difference being that the cytoplasmic sheath was maintained in CK666-treated embryos (Fig. 7B, panel b), whereas it was lost in formin-inhibited embryos (Fig. 7B). To better quantify filopodia formation under these conditions, PMCs were isolated and the number and length of filopodia were measured following treatment with Cdc42, Arp2/3 and formin inhibitors (Fig. 7C–E). ML141 caused a significant decrease in the number of filopodia as well as filopodial length, as did inhibition of either formins or Arp2/3, (Fig. 7C–E). SMIFH2 inhibits formin function by blocking the Formin Homology 2 (FH2) domain (Rizvi et al., 2009), and formin inhibition had a milder but still significant effect on filopodial nucleation in comparison to ML141 and CK666. CK666, which blocks Arp2/3-mediated actin nucleation (Hetrick et al., 2013) had a strong effect on both filopodia number and length. Overall, this quantitation indicated Cdc42 activation is required for formation and elongation of filopodia in PMCs, possibly through the activation of both Arp2/3 and formins.
Fig. 7.
Contribution of Cdc42, formins and Arp2/3 in sea urchin PMC filopodia formation. A) L. pictus embryos were treated at the blastula stage with either 0.1% DMSO, 100 μM Arp2/3 inhibitor (CK666, ) or 10 μM formin inhibitor (SMIFH2) for 24 h before being fixed and processed for immunolabeling with PMC-specific (6a9, green) and anti-actin (magenta) antibodies or analyzed live by polarization microscopy. Bar, 50 μm. B) L. pictus gastrulae expressing GFP-Lifeact were treated with 0.1% DMSO, 100 μM CK666 or 50 μM SMIFH2 and imaged by confocal microscopy. Live-cell imaging of PMCs in ventrolateral clusters from L. pictus gastrulae revealed that embryos treated with Arp2/3 and formin inhibitors did not exhibit the dynamic filopodia observed in controls. Bar, 25 μm. C–E) Isolated PMCs from L. variegatus embryos were treated with Cdc42 (10 μM ML141), Arp2/3 (100 μM CK666) and formin (5 μM SMIFH2) inhibitors, and cultures were fixed, probed with 6a9 and the number and length of filopodia were quantified for 30 cells/experiment, 3 experimental replicates. *** p = 0.0001; *** * p < 0.0001. Bar, 10 μm.
3. Discussion
In the sea urchin embryo, epithelial-mesenchymal transitions, guided cell migration, apical constriction and convergent extension all occur under the direction of gene regulatory networks that specify the endomesoderm and initiate morphogenesis. The goal of this study was to begin identifying the proximate regulators of the actin cytoskeleton that drive these cellular behaviors, that with the exception of one study (Beane et al., 2006) remain largely unknown. We specifically analyzed the input of the small GTPase Cdc42 in driving morphogenetic cell movements, and found that Cdc42 was particularly critical for PMC motility, patterning and skeletogenesis. Filopodial-based PMC motility required not only Cdc42, but also the actin nucleating activities of both formins and Arp2/3, suggesting that PMCs nucleate these cell processes through a process known as convergent elongation (Yang and Svitkina, 2011). And while the upstream signals that regulate Cdc42 activity were not directly addressed, this study begins the process of connecting the patterning signals that dictate PMC behavior to the actin-based cell shape changes required for morphogenesis and organogenesis.
3.1. Cdc42 is necessary for filopodia formation in PMCs
Cdc42 inhibition, either by dominant-negative mutant, MASO or small molecule inhibitor resulted in delays in morphogenesis (Figs. 1–4). The archenteron frequently failed to elongate past the primary invagination stage (Fig. 2), although treatments with the Cdc42 inhibitor later in development (i.e. during late gastrulation) did not have as strong an effect on archenteron differentiation (Fig. 4C). Cdc42 has been implicated in regulating convergent extension movements in Xenopus and zebrafish embryos (Choi and Han, 2002; Penzo-Mendèz et al., 2003; Yeh et al., 2011), and thus a more careful analysis of the role of Cdc42 in archenteron elongation is clearly warranted. However, the most penetrant phenotype for all manipulations was the requirement of Cdc42 for PMC motility, organization and skeletogenesis. While PMCs were able to ingress and were sometimes positioned at the vegetal plate near the site of VLC formation, they lacked the tight organization of controls and were frequently scattered throughout the blastocoel (Fig. 3L and Q; and 4E, panel g). Treatment with ML-141 at the blastula through gastrula stages (Figs. 3 and 4) demonstrated that once inside the blastocoel, PMCs required Cdc42 to carry out directed migration and patterning. Cdc42 has been classically described as a main driver of filopodia formation in fibroblasts and other cell types (Kozma et al., 1995; Miki et al., 1998; Nobes and Hall, 1995b). PMC filopodia elongation and retraction dynamics are likely dictated by ligand-receptor contacts with the cellular environment (Miller et al., 1995), and abrogation of Cdc42 function blocked filopodia formation as assessed by immunolabeling of fixed cells as well as by live cell imaging (Figs. 3 and 6).
Traditionally, PMC filopodia have been classified as thick (1 μm diameter or greater) and thin filopodia (0.2–0.4 μm) (Gustafson and Wolpert, 1961; Miller et al., 1995), but both are thought be involved in the signaling and mechanical properties of PMCs. In this study, we did not distinguish between these populations, but observed a generalized effect on all filopodial processes elaborated by PMCs. Interestingly, Cdc42 inhibition did not suppress ectodermal filopodia (Fig. 6D). Ectodermal epithelial cells extend short, dynamic filopodia from their basal membrane, and it is thought that these may play a direct role in guiding PMC migration (Miller et al., 1995). However, simultaneous imaging of both PMCs and the overlying ectoderm revealed that while the network of PMC filopodia broke down following ML141 treatment, ectodermal filopodia were unperturbed (Fig. 6D, panel b). What regulates the formation of these processes from the basolateral membrane is unknown, but given that several other small GTPases such as Rif and Ral have been shown to regulate filopodia formation (Ohta et al., 1999; Pellegrin and Mellor, 2005), suggesting that there are likely multiple possible mechanisms for Cdc42-independent filopodia formation. Identifying the distinct mechanisms by which PMC and ectodermal filopodia are regulated would significantly contribute to our understanding of how these cell-cell interactions pattern the skeletogenic mesoderm in these embryos.
Cdc42 can promote both linear actin arrays through formins, as well as branched actin networks through the Arp2/3 complex (Kühn and Geyer, 2014; Rohatgi et al., 1999). To gain a better mechanistic understanding of how Cdc42 directed filopodia formation, we blocked the activity of Cdc42, formins and Arp2/3 in PMCs both in vivo and in vitro, and found that all three factors contributed to filopodia (Fig. 7). There are currently two models for filopodia formation: the tip nucleation model where formins are the sole factors that nucleate and elongate unbranched actin filaments during filopodia formation (Gupton and Gertler, 2007); and the convergent elongation model, where dendritic actin networks formed by Arp2/3 serve as a substrate for filopodia elongation by formins (Gundersen and Barrett, 1980; Mattila and Lappalainen, 2008; Svitkina et al., 2003; Zheng et al., 1996). Whether a cell uses a particular strategy may depend on the cell type, with flattened, adherent cells using tip nucleation while less adherent cells employing convergent elongation (Young et al., 2015). The rounded morphology of PMCs, as well as the dependence on both formins and Arp2/3 (Fig. 7) support a model whereby Cdc42, acting downstream of positional cues, regulates filopodia formation through convergent elongation of linear actin arrays from a scaffold of Arp2/3-nucleated actin. Further analyses of filopodial dynamics in PMCs in the presence or absence of these nucleators, as well as identification of the specific formins involved will greatly increase our understanding of how this cell type carries out morphogenesis in response to extracellular cues.
3.2. Cdc42 is essential for proper skeletogenesis
Cdc42 inhibition in embryos after the initiation of spiculogenesis resulted in a reversible arrest in skeletal rod elongation (Fig. 4E, panel l, Fig. 5), and imaging of fixed and live embryos suggested that PMCs were not only unable to elaborate filopodia, but were also unable to maintain the cytoplasmic cables that envelop the growing skeletal rods (Figs. 4E, panel g; and 6C, panels e and f). Cytoplasmic fragments could be seen in the blastocoel in both live and fixed cells (Fig. 4E, panels g and j; Fig. 6C, panels e and f; 6D, panel b), and while it is unclear whether these fragments are remnants of the cytoplasmic cables, active caspase 3 labeling suggested that these were not fragments from dead PMCs (Fig. S3). How might Cdc42 contribute to the integrity of the PMC syncytium? In addition to its demonstrated roles in actin-based motility, Cdc42 has documented functions in the regulation of cell polarity (Etienne-Manneville, 2004; Johnson, 1999; Mack and Georgiou, 2014) and membrane trafficking (Adamo et al., 2001; Bretou et al., 2014; Estravís et al., 2011; Harris and Tepass, 2010; Mohammadi and Isberg, 2013), and it is entirely possible that it may play a role in these events as PMCs undergo polarized secretion of skeletal matrix. However, given that the entire cytoplasmic sheath collapses upon the inhibition of Cdc42 or formins (Figs. 6 and 7), we propose that Cdc42-mediated polymerization of linear actin arrays is required for the structural integrity of these cellular processes. Interestingly, inhibition of Arp2/3 did not have the same effect (Fig. 7B, panel b), suggesting that while Arp2/3 was important for filopodia formation, branched actin networks were less essential once the syncytium had been established. Whether Arp2/3 participates in skeletogenesis through its well-established roles in membrane trafficking remains to be determined (Gasman et al., 2004; Koseoglu et al., 2015; Tran et al., 2015).
In summary, our data point toward a model whereby spatial cues from the ectoderm stimulate Cdc42 to elaborate filopodia through the combined actions of two actin modulators that nucleate these thin processes through convergent elongation of linear actin filaments. Once PMCs fuse and initiate spiculogenesis, Cdc42 helps maintain the integrity of the cytoplasmic sheath that envelopes the growing skeletal arms. How Cdc42 activation is tied to the signaling pathways guiding PMC motility was not addressed in this study, although Cdc42 may be activated by exchange factors shown to act downstream of the VEGF receptor (Abraham et al., 2015; Garrett et al., 2007), raising the possibility that Cdc42 promotes PMC motility downstream of VEGF. Further experimentation will determine whether this is, indeed, the case.
4. Materials and methods
4.1. Embryo culture and isolation of Primary Mesenchyme Cells (PMCs)
Strongylocentrotus purpuratus were obtained from Marinus Scientific, Point Loma or used on site at the Friday Harbor Laboratories, University of Washington. Lytechinus variegatus (Reeftopia) and Lytechinus pictus (Marinus) were also used for some experiments. Gametes were obtained by coelomic injection with 0.5M KCl and fertilized in artificial (ASW) or filtered sea water (FSW). To remove fertilization envelopes, S. purpuratus eggs were fertilized in the presence of 1mM 3-amino-triazole, and envelopes were stripped by passage through 80 μm nitex mesh. To strip fertilization envelopes from Lytechinus sp., eggs were resuspended in calcium free sea water (CaFSW) immediately after fertilization, and passed through 150 μm nitex mesh. Embryo were cultured at 14 °C for S. purpuratus and L. pictus, and at 20 °C for L. variegatus.
To examine filopodia in isolated primary mesenchyme cells (PMCs), L. variegatus embryos, were cultured in ASW to mesenchyme blastula stage. Embryos were washed three times in CaFSW and resuspended vigorously during the third wash to dissociate blastomeres. The cell suspension was passed through a 66 μm nylon mesh and plated for 1 h at 4 °C on coverslips previously coated with 1 mg/ml wheat germ agglutinin (WGA, Sigma) and blocked with 3% BSA. Following incubation, coverslips were washed two times with sterile ASW containing 50 μg/ml gentamycin (Ettensohn and McClay, 1987; Wilt and Benson, 2004). PMCs were cultured in the presence of carrier control or inhibitors for 24 h in sterile ASW containing gentamycin (50 μg/μl) and 4% horse serum and then fixed and prepared for immunolocalization as described below.
4.2. Pharmacological treatments
All lyophilized inhibitors were resuspended in Dimethyl Sulfoxide (Sigma), aliquoted and stored at −80 °C. To block Cdc42 activity, embryos were cultured in the presence of the Cdc42 small molecule inhibitor ML141 (Tocris). To block Arp2/3 and formin-mediated actin polymerization, embryos were cultured in the presence of CK666 and SMIFH2 (Tocris), respectively (Henson et al., 2014, 2015). DMSO alone (0.1%) was used as a carrier control for all pharmacological treatments.
The inhibitory activity of ML141 was confirmed using a Cdc42 Pulldown Activation Assay (Cytoskeleton, BK034). Briefly S. purpuratus gastrulae were treated with 0.1% DMSO, 5 or 10 μM ML141 for two hours. Control and ML141 treated embryos were lysed for 10 min on ice with 10 volumes of lysis buffer (50mM Tris-HCl, 10mM MgCl2, O.5M NaCl and 2% IGEPAL, pH 7.5) supplemented with 0.1 volumes of protease inhibitor cocktail (Amresco). Lysates were clarified by centrifugation and snap frozen in liquid nitrogen and stored at −80 °C. Thawed lysates were incubated with 10 μl of PAK-GST beads at 4 °C for 1 h. Beads were washed once with 25mM Tris-HCl pH 7.5, 30mM MgCl2, 40mM NaCl and bound protein was resolved on 4–15% gradient SDS-polyacrylamide SDS gels (Bio-Rad) and transferred to Immobilon membranes (Millipore). Cdc42 was detected with a rabbit polyclonal anti-Cdc42 antibody (Abcam, ab64533). Bound primary antibodies were detected with peroxidase-conjugated anti-rabbit IgG (GE Healthcare, NA934V) and Immuno-Star chemiluminescent kit (Bio-Rad).
4.3. Immunolocalization
L. pictus and L. variegatus embryos were fixed in 3.7% formaldehyde in ASW for 30 min at room temperature. Fixed embryos were permeabilized with three washes in Phosphate-Buffered Saline with 0.1% Triton X-100 (PBST, 10mM Na2HPO4, 137mM NaCl, 2.7mM KCl, 1.8mM KH2PO4) and blocked for 3 h in 3% BSA in PBST. Embryos were probed with mouse monoclonal antibody 6a9 to detect primary mesenchyme cells (1:500; Charles Ettensohn, Carnegie-Mellon University), rabbit anti-cleaved caspase-3 (Cell Signaling) to identify apoptotic cells and rabbit anti-nonmuscle actin (Sigma). To examine pigment cell morphology, S. purpuratus embryos were fixed in cold methanol containing 5mM EGTA at −20 °C for 30 min. Fixed larvae were then rehydrated and permeabilized in PBST and blocked in 3% BSA-PBST. Embryos were probed with mouse Sp1 monoclonal antibody to detect pigment cells (Developmental Studies Hybridoma Bank) and counterstained with rabbit anti-septin 2 (Abcam). After incubation with Alexa Fluor-labeled secondary antibodies (Life Technologies) and Hoescht 33342 (to label nuclei), all embryos were mounted in 90% glycerol in PBS and imaged as described below.
4.4. Quantitative PCR analysis of gene expression
Beginning at the early blastula stage (4.5 h post-fertilization), L. variegatus embryos were cultured in either 0.1% DMSO or 2.5 μM ML141 (three biological replicates per experiment, three experiments). At 24 h, post-fertilization, half of each culture was harvested and the remaining embryos were transferred to fresh solutions of the inhibitor and control (DMSO) and incubated up to 48 h post-fertilization. Total RNA was extracted from each time point and control, as well as from unfertilized eggs and pre-treatment blastulae. Embryos were washed with RNA Wash Buffer (0.01M Tris pH 8.0, 0.1M EDTA, 7.3% glycerol), followed by RNA isolation with TriReagent (Molecular Research Center). Total RNA was purified using RNeasy columns (Qiagen) and DNA contamination removed by DNase treatment (Life Technologies). cDNA synthesis was performed using Super Script III Reverse Transcriptase (Life Technologies) in a reaction that included RNaseOUT Recombinant Ribonuclease Inhibitor (Life Technologies).
Primers for 7 transcripts were designed using Primer3 Software (http://primer3.ut.ee) or PrimerQuest (https://www.idtdna.com/primerquest/Home/Index) (Table 1). Transcript levels were quantified using qPCR, with each sample performed in triplicate for the three different experimental trials using IQ SYBR Green Supermix (Bio-Rad,). The reactions were run using ICycler IQ multicolored real-time PCR machine (Bio-Rad). Non-template reactions using nuclease-free water were used as negative control. The 2−ΔΔCt equation was used to normalize data to a housekeeping gene (ubiquitin) and to calculate changes in fold change compared to the unfertilized egg.
Table 1.
L. variegatus qPCR Primers
| Target | Sequence |
|---|---|
| Ubiquitin forward | CCAAGGTGGTGTGTTTTTCC |
| Ubiquitin reverse | CCATTGCTGTTGATGTTTGG |
| Cdc42 forward | CTCTCAGCTATCCCCAGACG |
| Cdc42 reverse | TGACAATGGTGAGTGATCTCG |
| Rac forward | AGTGGCATCCAGAAGTTAGC |
| Rac reverse | ATGTGATGGGTCTGAGTTTCC |
| Fgfa forward | GTCGACGACGACGTTATCAAA |
| Fgfa reverse | CTCTCACCTTGCTCTGTTCTATT |
| C-lectin forward | GCCACTAGAGCTCCCAAAGC |
| C-lectin reverse | TGGCACAGGGGTAGTAGTCC |
| SM-32 forward | AAGCGTCAGCACGTAACC |
| SM-32 reverse | GCTACCTCACAGACGAAAGC |
| MSP130-5 forward | GTCCAGAATGTGAATCCCTAGC |
| MSP130-5 reverse | GAAGCTATCGAACTCTGGTATCG |
| nk2-1 forward | CGACTCCATTGCTTCCGATAA |
| nk2-1 reverse | TAGTTCGTATGTCTGGGCTTTG |
4.5. Generation and in vitro transcription of expression constructs
All PCR primers and Gblocks were synthesized by Integrated DNA Technologies. Actin morphology was highlighted using the actin probe, EGFP-Lifeact cloned into pSP64T (gift of Mamiko Yajima, Brown University)(Riedl et al., 2008). For morpholino rescue experiments, the open reading frame of human Cdc42 was synthesized as a Gblock (IDT) and subcloned into pLAGFP2A (Table 3), a pCS2P+ derivative that contains a viral 2A peptide sequence and EcoRV cloning site added to the 3′ end of EGFP-Lifeact, allowing for bicistronic expression from a single mRNA (Trichas et al., 2008). To mark morpholino-injected embryos, PH domain of PLCδ fused to EGFP was amplified from pGFP-C1-PLC-delta-PH (a gift from Tobias Meyer, Addgene, Cambridge, MA, 21179), was subcloned into StuI-digested pCS2P+ using HD In-fusion cloning kit (Takara) according to manufacturer’s specifications.
TABLE 3.
G-Block sequences
| Fragment | Sequence |
|---|---|
| HsCdc42 (Subcloned into pCS2P+) |
ATGCAGACAATTAAGTGTGTTGTTGTGGGCGATGGTGCTGTTGGTA AAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGA ATATGTACCGACTGTTTTTGACAACTATGCAGTCACAGTTATGATT GGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGGCAAG AGGATTATGACAGATTACGACCGCTGAGTTATCCACAAACAGATGT ATTTCTAGTCTGTTTTTCAGTGGTCTCTCCATCTTCATTTGAAAAC GTGAAAGAAAAGTGGGTGCCTGAGATAACTCACCACTGTCCAAAGA CTCCTTTCTTGCTTGTTGGGACTCAAATTGATCTCAGAGATGACCC CTCTACTATTGAGAAACTTGCCAAGAACAAACAGAAGCCTATCACT CCAGAGACTGCTGAAAAGCTGGCCCGTGACCTGAAGGCTGTCAAGT ATGTGGAGTGTTCTGCACTTACACAGAAAGGCCTAAAGAATGTATT TGACGAAGCAATATTGGCTGCCCTGGAGCCTCCAGAACCGAAGAAG AGCCGCAGGTGTGTGCTGCTATGA |
| GFP-Lifeact-2A (Subcloned into the pCS2P+) |
TTCAAGGCCTCTCGAACCATGGGGCCCATGGGTGTCGCAGATTTGA TCAAGAAATTCGAAAGCATCTCAAAGGAAGAAGGGGATCCACCGG TCGCCACCATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGT GCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTC AGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGA CCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCC ACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTA CCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCC GAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCA ACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGT GAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAAC ATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCT ATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAA GATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCAC TACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGA CAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAAC GAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCG GGATCACTCTCGGCATGGACGAGCTGTACAAGAAAGGGTCATCATC ACCATCACCATTGAACGCGAGTGGAGAGGGCAGAGGAAGTCTGCTA ACATGCGGTGACGTCGAGGAGAATCCTGGCCCAGATATCTAGATGA TCGAGCCTCTAGATT |
PCR Primers for S. purpuratus Cdc42 (SpCdc42) were designed from predicted genes (SPU_019494) in Echinobase (http://echinobase.org/), and amplified from random-primed egg cDNA, which was in turn subcloned into XhoI-linearized pCS2P+(a gift from Mark Kirschner, Addgene 17095) by In-Fusion cloning (Takara). Dominant negative (T17N) and constitutively active (Q61L) mutants for Cdc42 were generated using QuickChange II XL site-directed mutagenesis kit (Agilent Technologies).
Capped and polyadenylated mRNAs were synthesized from linearized plasmid templates using the SP6 mMESSAGE mMACHINE and Poly(A) Tailing kits (Life Technologies) according to manufacturer’s specifications. In vitro transcribed mRNA was precipitated with lithium chloride, resuspended in nuclease-free water, quantified by spectrophotometry and stored at −80 °C.
4.6. Microinjection
All microinjections were performed within the first 20 min of development using a Picospritzer II pressure injection system (Parker Hannifen) and micromanipulators mounted on either a Zeiss Axiovert 200M microscope or a Nikon TS100 inverted microscope, equipped with a temperature-controlled microscope stage (Brook Industries). Embryos were injected with either non-targeting control or translation-blocking morpholino antisense oligonucleotides (Gene-tools) directed against SpCdc42 (Table 2) over a range of concentrations, and GFP-Lifeact, GFP-PH mRNA or Rhodamine Dextran (Life Technologies) were co-injected to mark injected embryos. To verify the specificity of the morphant phenotypes, eggs were co-injected with EGFP-Lifeact-2A-HsCdc42. Morpholinos were injected at 250 μM, 500 μM and 1mM concentrations, and the 500 μM dose was found to be the optimal dose where phenotypes could be observed that could be still rescued by co-injection of HsCdc42. Similarly, S. purpuratus embryos were injected with mRNA for dominant-negative (T17N), constitutively-active (Q61L), or wild type SpCdc42 over a range of concentrations, and GFP-Lifeact or GFP-PH mRNA were co-injected to identify injected embryos. Fluorescent embryos were imaged live at 24 and 48hrs post-fertilization using DIC and fluorescent microscopy and phenotypes of injected embryos were scored for each condition.
Table 2.
Morpholino sequences
| Morpholino | Sequence |
|---|---|
| S. purpuratus Cdc42 antisense morpholino | CTACTACACATTTTATCGTCTGCAT |
| Standard control oligo | CCTCTTACCTCAGTTACAATTTATA |
4.7. Image Acquisition and analysis
Embryos were imaged live using a Zeiss Axiovert 200M inverted microscope equipped for DIC and wide-field epifluorescence microscopy. Images were acquired using a Zeiss MrM 12 bit CCD camera, driven by Axiovision 4.8 software. DIC and polarization images were acquired on a Zeiss Axiovert 200M equipped with QImaging 12 bit Retiga camera driven by Micro-manager software (version 1.4.14). Embryos were imaged by confocal microscopy using either a Leica TCS SP5 resonant-scanning confocal microscope driven by Leica Application Suite Software, or an Andor Dragonfly spinning disc confocal microscope driven by Fusion Software. Projection images were then prepared using ImageJ Software (NIH, Ver. 1.6.0-65) or Bitplane Imaris (version8.1) software. Figures were prepared using Adobe Photoshop software.
4.8. Statistical analysis
Statistical significance was determined using one or two way Analysis of Variance (ANOVA) test followed by Tukey-Kramer post hoc test using Graphpad Prism 6 with a 95% confidence interval. For frequency data, data were arcsin-square root transformed followed by two way ANOVA and a Tukey-Kramer post-hoc test.
Supplementary Material
Acknowledgments
The authors would like to thank Anne Meyer-Miner and Richard de la Rosa for their assistance in subcloning sea urchin Cdc42, as well as Charles Ettensohn (Carnegie Mellon University) for generously providing the 6A9 antibody. This work was supported by collaborative research grants to J.H.H. (MCB-1412688) and C.B.S. (MCB-1412734) and National Institutes of Health R15HD080533 to C.B.S. S.S. and L.T. were supported by Charles Lambert fellowships from the Friday Harbor Laboratories and grants from the National Council of Science and Technology in Mexico (CONACyT).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2018.03.015.
References
- Abraham S, Scarcia M, Bagshaw RD, McMahon K, Grant G, Harvey T, Yeo M, Esteves FO, Thygesen HH, Jones PF, Speirs V, Hanby AM, Selby PJ, Lorger M, Dear TN, Pawson T, Marshall CJ, Mavria G. A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis. Nat Commun. 2015;6:7286. doi: 10.1038/ncomms8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo JE, Moskow JJ, Gladfelter AS, Viterbo D, Lew DJ, Brennwald PJ. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA. Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development. 2013;140:4214–4225. doi: 10.1242/dev.100479. [DOI] [PubMed] [Google Scholar]
- Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11:620–625. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Beane WS, Gross JM, McClay DR. RhoA regulates initiation of invagination, but not convergent extension, during sea urchin gastrulation. Dev Biol. 2006;292:213–225. doi: 10.1016/j.ydbio.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Bretou M, Jouannot O, Fanget I, Pierobon P, Larochette N, Gestraud P, Guillon M, Emiliani V, Gasman S, Desnos C, Lennon-Duménil AM, Darchen F. Cdc42 controls the dilation of the exocytotic fusion pore by regulating membrane tension. Mol Biol Cell. 2014;25:3195–3209. doi: 10.1091/mbc.E14-07-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdsal CA, Alliegro MC, McClay DR. Tissue-specific, temporal changes in cell adhesion to echinonectin in the sea urchin embryo. Dev Biol. 1991;144:327–334. doi: 10.1016/0012-1606(91)90425-3. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch Ho E, Buckley KM, Schrankel CS, Schuh NW, Hibino T, Solek CM, Bae K, Wang G, Rast JP. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol Cell Biol. 2016;94:861–874. doi: 10.1038/icb.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Hinman VF. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015;6:3. doi: 10.1186/2041-9139-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chen S, Yap SF, Lim L. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J Biol Chem. 1996;271:26362–26368. doi: 10.1074/jbc.271.42.26362. [DOI] [PubMed] [Google Scholar]
- Choi SC, Han JK. Xenopus Cdc42 regulates convergent extension movements during gastrulation through Wnt/Ca2+ signaling pathway. Dev Biol. 2002;244:342–357. doi: 10.1006/dbio.2002.0602. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Denk-Lobnig M, Martin AC. Modular regulation of Rho family GTPases in development. Small GTPases. 2017:1–8. doi: 10.1080/21541248.2017.1294234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134:2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- Estravís M, Rincón SA, Santos B, Pérez P. Cdc42 regulates multiple membrane traffic events in fission yeast. Traffic. 2011;12:1744–1758. doi: 10.1111/j.1600-0854.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42–the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA. Encoding anatomy: developmental gene regulatory networks and morphogenesis. Genesis. 2013;51:383–409. doi: 10.1002/dvg.22380. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, McClay DR. A new method for isolating primary mesenchyme cells of the sea urchin embryo. Panning on wheat germ agglutinin-coated dishes. Exp Cell Res. 1987;168:431–438. doi: 10.1016/0014-4827(87)90015-2. [DOI] [PubMed] [Google Scholar]
- Fink RD, McClay DR. Three cell recognition changes accompany the ingression of sea urchin primary mesenchyme cells. Dev Biol. 1985;107:66–74. doi: 10.1016/0012-1606(85)90376-8. [DOI] [PubMed] [Google Scholar]
- Garrett TA, Van Buul JD, Burridge K. VEGF-induced Rac1 activation in endothelial cells is regulated by the guanine nucleotide exchange factor Vav2. Exp Cell Res. 2007;313:3285–3297. doi: 10.1016/j.yexcr.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasman S, Chasserot-Golaz S, Malacombe M, Way M, Bader MF. Regulated exocytosis in neuroendocrine cells: a role for subplasmalemmal Cdc42/N-WASP-induced actin filaments. Mol Biol Cell. 2004;15:520–531. doi: 10.1091/mbc.E03-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glise B, Bourbon H, Noselli S. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- Glise B, Noselli S. Coupling of Jun amino-terminal kinase and Decapentaplegic signaling pathways in Drosophila morphogenesis. Genes Dev. 1997;11:1738–1747. doi: 10.1101/gad.11.13.1738. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network? Trends Cell Biol. 2011;21:718–726. doi: 10.1016/j.tcb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen RW, Barrett JN. Characterization of the turning response of dorsal root neurites toward nerve growth factor. J Cell Biol. 1980;87:546–554. doi: 10.1083/jcb.87.3.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Gustafson T, Wolpert L. Studies on the cellular basis of morphogenesis in the sea urchin embryo. Directed movements of primary mesenchvme cells in normal and vegetalized larvae. Exp Cell Res. 1961;24:64–79. doi: 10.1016/0014-4827(61)90248-8. [DOI] [PubMed] [Google Scholar]
- Gustafson T, Wolpert L. Cellular movement and contact in sea urchin morphogenesis. Biol Rev Camb Philos Soc. 1967;42:442–498. doi: 10.1111/j.1469-185x.1967.tb01482.x. [DOI] [PubMed] [Google Scholar]
- Harden N, Loh HY, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding protein Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. Development. 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- Harris KP, Tepass U. Cdc42 and vesicle trafficking in polarized cells. Traffic. 2010;11:1272–1279. doi: 10.1111/j.1600-0854.2010.01102.x. [DOI] [PubMed] [Google Scholar]
- Henson JH, Gianakas AD, Henson LH, Lakin CL, Voss MK, Bewersdorf J, Oldenbourg R, Morris RL. Broadening the spectrum of actin-based protrusive activity mediated by Arp2/3 complex-facilitated polymerization: motility of cytoplasmic ridges and tubular projections. Cytoskeleton. 2014;71:484–500. doi: 10.1002/cm.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Yeterian M, Weeks RM, Medrano AE, Brown BL, Geist HL, Pais MD, Oldenbourg R, Shuster CB. Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol Biol Cell. 2015;26:887–900. doi: 10.1091/mbc.E14-07-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol. 2013;20:701–712. doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Kenney SR, Phillips GK, Simpson D, Schroeder CE, Nöth J, Romero E, Swanson S, Waller A, Strouse JJ, Carter M, Chigaev A, Ursu O, Oprea T, Hjelle B, Golden JE, Aubé J, Hudson LG, Buranda T, Sklar LA, Wandinger-Ness A. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J Biol Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp GC, Solursh M. Dynamic activity of the filopodia of sea urchin embryonic cells and their role in directed migration of the primary mesenchyme in vitro. Dev Biol. 1985;112:276–283. doi: 10.1016/0012-1606(85)90398-7. [DOI] [PubMed] [Google Scholar]
- Koseoglu S, Peters CG, Fitch-Tewfik JL, Aisiku O, Danglot L, Galli T, Flaumenhaft R. VAMP-7 links granule exocytosis to actin reorganization during platelet activation. Blood. 2015;126:651–660. doi: 10.1182/blood-2014-12-618744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S, Geyer M. Formins as effector proteins of Rho GTPases. Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DC, Kaltenbach SL, McClay DR. Morphogenesis in sea urchin embryos: linking cellular events to gene regulatory network states. Wiley Interdiscip Rev Dev Biol. 2012;1:231–252. doi: 10.1002/wdev.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DC, Martik ML, Saunders LR, McClay DR. Specification to biomineralization: following a single cell type as it constructs a skeleton. Integr Comp Biol. 2014;54:723–733. doi: 10.1093/icb/icu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack NA, Georgiou M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 2014;5:10. doi: 10.4161/21541248.2014.973768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinda KM, Ettensohn CA. Primary mesenchyme cell migration in the sea urchin embryo: distribution of directional cues. Dev Biol. 1994;164:562–578. doi: 10.1006/dbio.1994.1224. [DOI] [PubMed] [Google Scholar]
- Malinda KM, Fisher GW, Ettensohn CA. Four-dimensional microscopic analysis of the filopodial behavior of primary mesenchyme cells during gastrulation in the sea urchin embryo. Dev Biol. 1995;172:552–566. doi: 10.1006/dbio.1995.8044. [DOI] [PubMed] [Google Scholar]
- Martik ML, Lyons DC, McClay DR. Developmental gene regulatory networks in sea urchins and what we can learn from them. F1000Research. 2016:5. doi: 10.12688/f1000research.7381.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- McClay DR. The role of thin filopodia in motility and morphogenesis. Exp Cell Res. 1999;253:296–301. doi: 10.1006/excr.1999.4723. [DOI] [PubMed] [Google Scholar]
- McClay DR, Fink RD. Sea urchin hyalin: appearance and function in development. Dev Biol. 1982;92:285–293. doi: 10.1016/0012-1606(82)90175-0. [DOI] [PubMed] [Google Scholar]
- McIntyre DC, Lyons DC, Martik M, McClay DR. Branching out: origins of the sea urchin larval skeleton in development and evolution. Genesis. 2014;52:173–185. doi: 10.1002/dvg.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miller J, Fraser SE, McClay D. Dynamics of thin filopodia during sea urchin gastrulation. Development. 1995;121:2501–2511. doi: 10.1242/dev.121.8.2501. [DOI] [PubMed] [Google Scholar]
- Mohammadi S, Isberg RR. Cdc42 interacts with the exocyst complex to promote phagocytosis. J Cell Biol. 2013;200:81–93. doi: 10.1083/jcb.201204090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995a;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995b;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Noselli S. JNK signaling and morphogenesis in Drosophila. Trends Genet. 1998;14:33–38. doi: 10.1016/S0168-9525(97)01320-6. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol. 1990;111:1001–1007. doi: 10.1083/jcb.111.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Penzo-Mendèz A, Umbhauer M, Djiane A, Boucaut JC, Riou JF. Activation of Gbetagamma signaling downstream of Wnt-11/Xfz7 regulates Cdc42 activity during Xenopus gastrulation. Dev Biol. 2003;257:302–314. doi: 10.1016/s0012-1606(03)00067-8. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, McClay DR. Primary mesenchyme cell patterning during the early stages following ingression. Dev Biol. 2003;254:68–78. doi: 10.1016/s0012-1606(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Rafiq K, Cheers MS, Ettensohn CA. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development. 2012;139:579–590. doi: 10.1242/dev.073049. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Settleman J. Rac ‘n Rho: the music that shapes a developing embryo. Dev Cell. 2001;1:321–331. doi: 10.1016/s1534-5807(01)00053-3. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahinci E, Symes K. Distinct functions of Rho and Rac are required for convergent extension during Xenopus gastrulation. Dev Biol. 2003;259:318–335. doi: 10.1016/s0012-1606(03)00206-9. [DOI] [PubMed] [Google Scholar]
- Tran DT, Masedunskas A, Weigert R, Ten Hagen KG. Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat Commun. 2015;6:10098. doi: 10.1038/ncomms10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Davidson EH. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2014;385:160–167. doi: 10.1016/j.ydbio.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt FH, Benson SC. Isolation and culture of micromeres and primary mesenchyme cells. Methods Cell Biol. 2004;74:273–285. doi: 10.1016/s0091-679x(04)74012-1. [DOI] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adhes Migr. 2011;5:402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CM, Liu YC, Chang CJ, Lai SL, Hsiao CD, Lee SJ. Ptenb mediates gastrulation cell movements via Cdc42/AKT1 in zebrafish. PLoS One. 2011;6:e18702. doi: 10.1371/journal.pone.0018702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Heimsath EG, Higgs HN. Cell type-dependent mechanisms for formin-mediated assembly of filopodia. Mol Biol Cell. 2015;26:4646–4659. doi: 10.1091/mbc.E15-09-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Wan JJ, Poo MM. Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient. J Neurosci. 1996;16:1140–1149. doi: 10.1523/JNEUROSCI.16-03-01140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.