Abstract
Although numbers of genome-wide association studies (GWAS) have been performed for serum lipid levels, limited heritability has been explained. Studies showed that combining data from GWAS and expression quantitative trait loci (eQTLs) signals can both enhance the discovery of trait-associated SNPs and gain a better understanding of the mechanism. We performed an annotation-based, multistage genome-wide screening for serum-lipid-level-associated loci in totally 6863 Han Chinese. A serum high-density lipoprotein cholesterol (HDL-C) associated variant rs1880118 (hg19 chr7:g. 6435220G>C) was replicated (Pcombined = 1.4E-10). rs1880118 was associated with DAGLB (diacylglycerol lipase, beta) expression levels in subcutaneous adipose tissue (P = 5.9E-42) and explained 47.7% of the expression variance. After the replication, an active segment covering variants tagged by rs1880118 near 5′ of DAGLB was annotated using histone modification and transcription factor binding signals. The luciferase report assay revealed that the segment containing the minor alleles showed increased transcriptional activity compared with segment contains the major alleles, which was consistent with the eQTL analyses. The expression-trait association tests indicated the association between the DAGLB and serum HDL-C levels using gene-based approaches called “TWAS” (P = 3.0E-8), “SMR” (P = 1.1E-4), and “Sherlock” (P = 1.6E-6). To summarize, we identified a novel HDL-C-associated variant which explained nearly half of the expression variance of DAGLB. Integrated analyses established a genotype-gene-phenotype three-way association and expanded our knowledge of DAGLB in lipid metabolism.
Introduction
Dyslipidemia, involving abnormal levels of serum triglyceride (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C), is a well-known risk factor for cardiovascular disease (CVDs) [1]. Genetic factors contribute a considerable part (54–66%) to lipid metabolism [2]. A number of loci have been mapped by genome-wide association studies (GWAS). Although vastly expanded sample sizes have been used in combined analyses in European studies, only a limited proportion of heritability has been explained by these loci [3, 4]. Further explorations are needed, especially in East Asians. Similar to other GWAS, only a few of the reported lipid associated tag single nucleotide polymorphisms (SNPs) have been fine-mapped for causal SNP localization [5, 6]. Meanwhile, a limited number of genes mediating the associations between the tagSNPs and lipid phenotypes have been uncovered. The translation and application of the reported variants discovered by classical GWAS strategy are the major challenges currently [7].
According to the results of genotype-phenotype association studies for complex diseases, ~80–90% of these GWAS tag SNPs lie outside of the protein-coding regions, which means that these loci likely manifest their effects via regulating the gene expression or splicing [6, 8–10]. A study performed by Nicolae and his colleagues found that expression quantitative trait loci (eQTLs) are significantly enriched in trait-associated SNPs reported by GWAS for complex traits [11]. Annotation via eQTL can improve the ability to discover true associations and identify the major steps of the regulatory cascade behind the statistical genotype-phenotype associations. A number of projects and databases focused on genotype-expression associations (e.g., MuTHER, SCAN, etc.) launched the eQTL annotation and SNP function prediction after GWAS [12–17]. Recently, the Genotype-Tissue Expression (GTEx) project showed a landscape of eQTL annotation in multiple tissues and accelerated the driver gene prediction in the post-GWAS era [18, 19].
Integrated analyses for GWAS and eQTL in specific tissues have been promising for type 2 diabetes, bipolar disorder, immune-related diseases, etc. [20–24]. We wonder whether such an annotation (eQTL signal in relevant tissues) could be considered as a prior for screening lipid-associated loci in Han Chinese with limited sample size in the discovery stage.
Besides eQTL annotation, chromatin activity regions can be annotated through chromatin immunoprecipitation sequencing (ChIP-seq) or DNase-seq for transcription factors, DNase sensitivity, and histone modification markers such as histone H3 lysine 4 trimethylation (H3K4me3), histone H3 lysine 9 acetylation (H3K9ac), and histone H3 lysine 27 acetylation (H3K27ac) [6, 25–28]. The NIH Roadmap Epigenomics Consortium has published the largest collection of human epigenomes which facilitated the identification of gene regulation in multiple tissues [29].
In the current study, we performed a multistage GWAS for serum lipid levels. First, SNPs associated with gene expression levels in liver and adipose tissue were annotated as candidates for replications. Second, epigenetic modification signals were used for annotating putative regulatory regions. Then, luciferase reporter assay was conducted to verify the segment involving the causal variants for the regulation of the transcription activity. Finally, gene-based expression-trait association analyses were conducted.
Methods
Study subjects
In the discovery stage, 998 participants with metabolic syndrome (according to the criteria of the Metabolic Syndrome Study Cooperation Group of the Chinese Diabetes Society) and 996 healthy controls were recruited from a community-based survey in Hangzhou, Zhejiang Province, China, as described in our previous study [30]. For the replication stage I, six independent community-based cohorts with a total of 3027 subjects were recruited from east China (Dongyang, Liuheng, Zhujiajian, Tongxiang, and Hangzhou) and Northeast China (Shenyang). For replication stage II, 2130 subjects from Shanghai were included [31] (S Table 1). All participants were Han Chinese. Individuals were excluded if they had cancer or serious chronic liver, lung, or kidney disorders. The study protocol was approved by the Research Ethics Committee of the School of Medicine, Zhejiang University. All participants gave informed consent.
Genotyping, imputation, and quality controls
Genomic DNA was isolated from whole blood using a TACO automatic nucleic acid extraction apparatus. The Illumina Omni-Express 760k chip was used for the genome-wide assay of samples in the discovery stage. SNPs with call rates ≥95% and Hardy–Weinberg equilibrium P-values ≥1.0E-6 were included in the discovery stage. Samples were excluded from analyses if they (1) had genotyping call rates ≤95%, (2) were population outliers identified by running EIGENSOFT [32], or (3) had probable relatives (PI_hat>0.25). Then we imputed ungenotyped SNPs via IMPUTE2 [33]. In the replication stage I, genotyping was done using SNPscanTM (Genesky Biotechnologies Inc., Shanghai, China). In the replication stage II, genotyping data were extracted from our previous study using the Affymetrix Genome-Wide Human SNP Array 6.0, the quality control filtering of the genotyping data can be found elsewhere [31]. Genotyping call-rate control (≥95%) and Hardy–Weinberg equilibrium control (P > 1.0E-3) were implemented in the replication stages. After the quality control, 1742, 3027, and 2130 subjects were included in analyses in the discovery, the replication stage I and II, respectively.
Online data acquisition
Tissue-specific eQTL signals were acquired from the Genotype-Tissue Expression (GTEx) project [18], the Multiple Tissue Human Expression Resource (MuTHER) project, GSE26106 (206 and 60 subjects recruited by the University of Chicago and University of Washington) [14], GSE9588 (266 subjects recruited by Merck) [14], and a published meta-analysis in whole-blood samples from 5311 subjects [34]. Subcutaneous adipose-tissue-specific (GTEx and MuTHER) and liver-tissue-specific (GSE26106 and GSE9588) eQTL signals were used for candidate SNP screening. eQTL signals in whole blood, skin tissue, and transformed fibroblasts were used for additional annotations.
Summarized genome-wide lipid-associated signals were downloaded from the Global Lipids Genetics Consortium (GLGC) [3]. For combined analyses, 94,595 European ancestry individuals from 23 studies genotyped with SNP arrays were included.
A cluster of chromatin status was annotated using the data from ENCODE project via the UCSC genome browser. The H3K27ac modification signals were combined from seven cell lines. The DNase I hypersensitivity loci were integrated from 125 cell types. A cluster of 161 transcription factor ChIP-seq data were annotated. Another set of histone modification marks including H3K4me3, H3K9ac, and H3K27ac were used for mapping active regions via ChIP-seq. The ChIP-seq data in primary mononuclear cells (E062) and adipose nuclei (E063) and was acquired from the Roadmap project [29].
Luciferase reporter assay
To evaluate the transcriptional activity of the variant with a potential function, a segment covering the peaks of the transcription activity signals and the variants was amplified by polymerase chain reaction (PCR) from a heterozygote of rs1880118 genomic DNA sample. The recovered fragments were cloned into pGL-3 basic vectors.
Then transient transfections were conducted in the HEK 293T cell line. The cells (1.0E6) were plated into six-well plates and transfected using Lipofectamine 2000. After 48 h of transfection, the fluorescence intensities were evaluated following the protocol of the kit (Promega Dual-Luciferase Reporter Assay System). The pRL-TK vector encoding for the Renilla luciferase was transcribed, which was used as internal controls to normalize firefly luciferase expression. Transfections were done in triplicate and replicated in independent experiments.
Strategies and statistical analyses
First, as shown in Supplementary Fig. 1, linear regressions were performed for the associations between lipid components (including TG, TC, HDL-C, and LDL-C) via PLINK [35]. Second, SNPs associated with at least two lipid components in the discovery stage (P < 0.05 for the association tests) were annotated using eQTL signals either in subcutaneous adipose tissue or in liver tissue. Identical subcutaneous adipose-tissue-specific eQTL signals (cis SNP-gene pairs) from the GTEx (V4) and MuTHER datasets were identified. The liver-tissue-specific SNP-gene pairs (P < 0.05) were combined from three studies. Third, conditional analyses were performed with any two candidate SNPs within ~1 Mb for linkage disequilibrium (LD) pruning. We kept only one of the statistically significant SNPs if the other SNP had a P-value > 0.05 in conditional analyses. Fourth, trait-associated loci with eQTL annotations were considered for replications. Replications I and II with seven independent Han Chinese cohorts were performed. Finally, we combined the effects of replicable SNPs in Han Chinese samples and 94595 individuals of European ancestry reported by GLGC.
For comparison of the transcriptional activities of the vectors, P-value was determined by t-test for independent samples.
Gene-based analyses
After replication for the trait-associated tagSNP, the mediator gene through which these tagSNPs exert their effects on the traits were estimated using gene-based analyses including “TWAS” (transcriptome-wide association study) [36], “SMR”(summary data–based Mendelian randomization) [37], and “Sherlock” [38].
The basic idea of “TWAS” and “SMR” is based on Mendelian Randomization. SNPs are used as instrumental variables for the test between the gene expression and the trait. The method “TWAS” builds a regression model with a cluster of gene expression of associated SNPs through a sample set with genomic and transcriptomic data as a reference. Then the expression levels of the gene were predicted using this model in a much larger cohort with genotype and phenotype data. The expression-trait association was evaluated using the imputed expression data. Similarly, the “SMR” integrates summary-level data from GWAS with data from eQTL studies to identify genes whose expression levels are associated with a complex trait.
The method “Sherlock” identifies potential gene-disease association by matching the genetic signature (GWAS and eQTLs signals) using a Bayesian statistical framework. If a gene’s genetic signature, provided by eQTL data, matches the gene’s signature of GWAS, it suggested that the expression level of the gene was associated with the phenotype of GWAS.
The data used for these analyses were described in Supplementary Methods.
Results
eQTL-based genome wide screening for lipid-associated loci
In the discovery stage, SNPs associated with ≥2 lipid components (including TG, TC, HDL-C, and LDL-C) were annotated with eQTL signals either in subcutaneous adipose tissue or in liver tissue (S Fig. 1). After LD pruning, fourteen SNPs with independent effects were selected for replications (Table 1, S Table 2). In these loci, a reported locus rs1077834 (at LIPC) was removed from the candidate list. Finally, nine SNPs were successfully genotyped in the replication stage I. After the replication stage I, a significant association was found between rs1880118 and HDL-C levels (P = 1.6E-3). The minor allele of rs1880118 (C allele) was associated with increased levels of HDL-C (beta = 0.032, se = 0.010). The effect was consistent with that at the discovery stage (effect allele = C, beta = 0.066, se = 0.019, P = 4.0E-4). Consistent result was also observed in the replication stage II (effect allele = C, beta = 0.038, se = 0.012, P = 1.6E-3). In addition, we checked the effect of rs1880118 in 94,595 individuals of European ancestry and found a significant association between rs1880118 and HDL-C (effect allele = C, beta = 0.022, se = 0.006, P = 1.6E-4). Combining the results of these association studies, the C allele of rs1880118 was associated with increased levels of HDL-C (Han Chinese combined: beta = 0.039, se = 0.007, P = 4.4E-8; all ancestries combined: beta = 0.029, se = 0.005, P = 1.4E-10). The associations between rs1880118 and TC were also shown in Table 2 and S Fig. 2.
Table 1.
Serum lipid-associated loci with eQTL annotations
| Chr | rsid | Major allele and minor allele (minor allele frequency) | Position (hg19) | Associated traits | eGene | eQTL signal (Subcutaneous adipose) | eQTL signal (Liver) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| P_MuTHER | P_GTEx (V4) | P_U_of _C | P_U_of _W | P_Merck | ||||||
| 2 | rs12622106 | C>T (0.44) | 20883561 | HDL-C, LDL-C | C2orf43 | 5.10E-36 | 9.90E-10 | |||
| 2 | rs2834578 | T>C (0.36) | 36059609 | TC, LDL-C | CLIC6 | 2.50E-47 | 8.50E-07 | |||
| 2 | rs2076673 | G>C (0.41) | 36124860 | TC, LDL-C | APOL5 | 2.60E-06 | 2.40E-03 | 4.10E-03 | ||
| 4 | rs1680074 | G>A (0.48) | 1279621 | TG, HDL-C | CTBP1 | 8.60E-06 | 3.00E-02 | 3.50E-02 | ||
| 5 | rs17772583 | A>G (0.43) | 131953510 | TG, TC | RAD50 | <1.0E-16 | 6.50E-06 | 4.60E-03 | ||
| 7 | rs1880118 | G>C (0.19) | 6435220 | TC, HDL-C | DAGLB | 1.00E-04 | 2.20E-12 | |||
| 10 | rs532473 | G>A (0.34) | 27669655 | TC, LDL-C | PTCHD3 | 8.40E-41 | 2.50E-09 | |||
| 10 | rs224045 | T>C (0.42) | 64525547 | TG, TC | ADO | 3.30E-03 | 1.70E-03 | 3.10E-06 | ||
| 12 | rs7296841 | A>C (0.23) | 112444109 | TG, TC | TMEM116 | 8.90E-16 | 1.30E-05 | 3.70E-05 | ||
| 12 | rs11831480 | G>T (0.12) | 132554702 | TG, LDL-C | DDX51 | <1.0E-16 | 2.60E-02 | 5.50E-03 | ||
| 13 | rs2296347 | T>G (0.43) | 52728423 | TG, TC | NEK3 | 6.20E-10 | 3.30E-02 | 1.40E-05 | ||
| 15 | rs1077834 | T>C (0.40) | 58723479 | TG, TC, HDL-C | LIPC | 7.60E-08 | 7.10E-03 | 2.50E-04 | ||
| 17 | rs2286590 | G>A (0.21) | 74683666 | TC, HDL-C | MXRA7 | 1.60E-14 | 2.30E-11 | |||
| 19 | rs3786505 | G>A (0.49) | 45490570 | TC, LDL-C | APOC4 | <1.0E-16 | 9.10E-04 | 1.50E-03 | ||
In the discovery stage, genotype–phenotype associations were performed for SNPs with eQTL annotations either in subcutaneous adipose tissue or in liver tissues. Fourteen eQTLs associated with at least two lipid components are shown in Table 1
The minor allele is considered as the effect allele in analysis
U of C an eQTL study from the University of Chicago, U of W an eQTL study from the University of Washington, Merck an eQTL study from Merck, Chr chromosome, hg19: Human Genome version 19, also known as GRCh37, eGene the gene that has a significant eQTL acting upon it, TC total cholesterol, TG triglyceride, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol
Table 2.
Associations between genotypes of rs1880118 and levels of serum HDL-C and TC in multistage and combined analyses
| Data source | N | Race | BETA | SE | P |
|---|---|---|---|---|---|
| A (trait=HDL-C) | |||||
| Discovery | 1666 | Han Chinese | 0.066 | 0.019 | 4.0E-04 |
| Replication I | 3027 | Han Chinese | 0.032 | 0.010 | 1.6E-03 |
| Replication II | 2094 | Han Chinese | 0.038 | 0.012 | 1.6E-03 |
| Combined (Han Chinese) | 6787 | Han Chinese | 0.039 | 0.007 | 4.4E-08 |
| GLGC | 91033 | European ancestry | 0.022 | 0.006 | 1.6E-04 |
| Combined (All ancestries) | 97820 | All ancestries | 0.029 | 0.005 | 1.4E-10 |
| B (trait=TC) | |||||
| Discovery | 1665 | Han Chinese | 0.087 | 0.040 | 3.2E-02 |
| Replication I | 3025 | Han Chinese | 0.017 | 0.029 | 5.5E-01 |
| Replication II | 2094 | Han Chinese | 0.061 | 0.035 | 8.0E-02 |
| Combined (Han Chinese) | 6784 | Han Chinese | 0.047 | 0.019 | 1.6E-02 |
| GLGC | 91171 | European ancestry | 0.019 | 0.006 | 1.9E-03 |
| Combined (All ancestries) | 97955 | All ancestries | 0.022 | 0.006 | 2.1E-04 |
The minor allele (C allele) was considered as the effective allele in analyses. Beta and SE were calculated using a linear regression model adjusted for age and gender. The effects of rs1880118 were combined using fixed-effect model. No heterogeneities were observed
TC total cholesterol, HDL-C high-density lipoprotein cholesterol, N number of subjects, GLGC Global Lipids Genetics Consortium
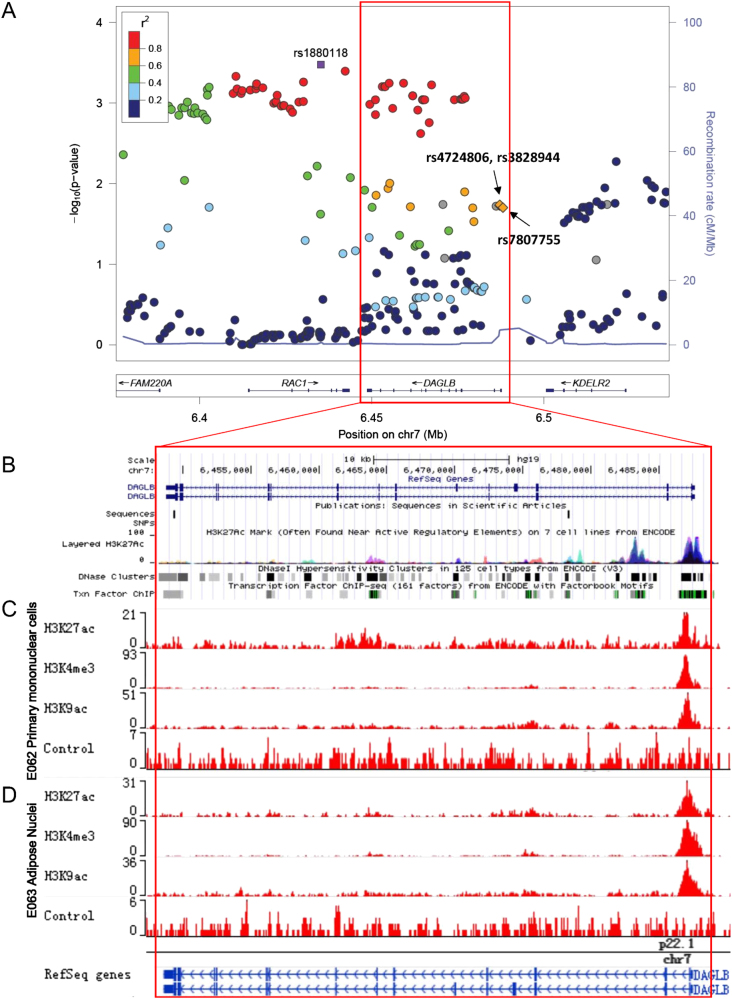
As Fig. 1a shows, the LD block of rs1880118 contains two genes, RAC1 and DAGLB. A variant rs702485, which is located at 3′UTR of DAGLB, has been reported to be associated with serum HDL-C levels in population of European ancestry, so we evaluated the LD between the reported locus and the novel locus of rs1880118. We found that the r2 of LD was only 0.03 in Han Chinese. The conditional analysis was performed by adjusting for rs702485, an independent effect of rs1880118 was observed (S Fig. 3).
Fig. 1.
Regional plot of the replicated SNP and annotations including histone modification, transcription factor binding site, and DNase signals in this region. a Regional plot of the tagSNP rs1880118 plotted using Locuszoom. The tagSNP and another three SNPs located in a potential regulatory region are labeled. b Annotations including DNase and transcription factor binding site ChIP-seq (combined with multiple cell lines) from ENCODE project. c Histone modification signals using primary mononuclear cells from peripheral blood (E062, Roadmap project). d Histone modification signals using adipose nuclei (E063, Roadmap project)
Annotations for the replicated tagSNP rs1880118
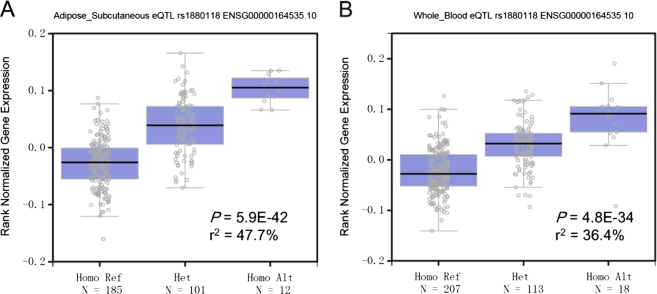
The minor allele of rs1880118 (C allele) was associated with increased mRNA expression levels of DAGLB in subcutaneous adipose tissue. Consistent associations were found in eQTL analyses using data from GTEx (V4, P = 2.2E-12), GTEx (V6, P = 5.9E-42), and MuTHER project (P = 1.0E-4). The associations between rs1880118 and the expression level of DAGLB were also observed in whole blood samples from multiple eQTL data sources (S Table 3). The 47.7% and 36.4% of the DAGLB expression variance could be explained by the variation of rs1880118 in subcutaneous adipose tissue and whole blood samples according to GTEx V6 data (Fig. 2). In addition, the C allele of rs1880118 was associated with the decreased expression level of RAC1 in Transformed fibroblast (GTEx V6, P = 1.9E-14, S Table 3).
Fig. 2.
Boxplots of the DAGLB expression levels classified by the genotypes of rs1880118 in subcutaneous adipose tissue (a) and whole blood (b) samples from GTEx project. The proportions explained (r2, square of the coefficient of determination) by SNP were calculated using summarized data from GTEx
As shown in Fig. 1a, the LD block of rs1880118 captured a region containing genes of DAGLB and RAC1. To identify the putative regulatory region, chromatin state markers including histone modifications and transcription factor binding site were annotated. An active transcription region (hg19, chr7: 6,486,750-6,488,250) near 5′ of DAGLB was uncovered by H3K27ac modification, transcription factor binding site, and DNase I sensitivity signals (Fig. 1b). Histone modification signals using adipose nuclei and primary mononuclear cells from peripheral blood (corresponding to the eQTL analyses) were annotated using data from the Roadmap project (Fig. 1c, d). We considered variants in high LD (r2 > 0.6 in ASN, 1000 genome project phase I) with the proxy SNP rs1880118. Of the tagged variants meeting the LD threshold, three SNPs (rs4724806, rs3828944, and rs7807755, the LD correlation among these variants and the tagSNP rs1880118 is showed in S Fig. 3) were found to be located in this active region. The narrow peaks of the histone modification signals and their relative positions to the three variants are shown in S Table 4 and S Fig. 4. Two loci (rs4724806 and rs3828944) are located in the first intron of DAGLB, and another variant rs7807755 is located in the promoter region of DAGLB.
Allele-specific activities of the active fragment
To evaluate the transcriptional activity of the three candidate loci in the putative regulatory region, 1807-bp segments covering the major alleles (wild-type) and minor alleles (mutant) of three variants (rs4724806, rs3828944, and rs7807755, labeled as “A”, “B”, and “C”, respectively) were cloned into XholI and HindIII sites of pGL-3 basic vectors. Notably, the in silico PCR amplified fragment involving a microsatellite sequence of a poly-T structure including 11 bp repeats of thymine, while the reference sequence contains 40 bp repeats of thymine. In order to make the vectors comparable, we kept the numbers of thymines (11 bp repeats of thymine) in vectors and validated the numbers of thymines using Sanger sequencing.
The activities of the wild-type and the mutant vectors were measured using luciferase reporter assay. As depicted in Fig. 3, we observed that, compared to the wild-type (with major alleles), the mutant (with minor alleles) had significantly higher transcriptional activities (P < 0.001). These findings show that this segment contains three complete LD variants exhibited allele difference in transcription activity and suggested that it acted as a cis-regulatory element of DAGLB. This result is consistent with the eQTL signal in subcutaneous adipose tissue (P = 5.4E-42).
Fig. 3.
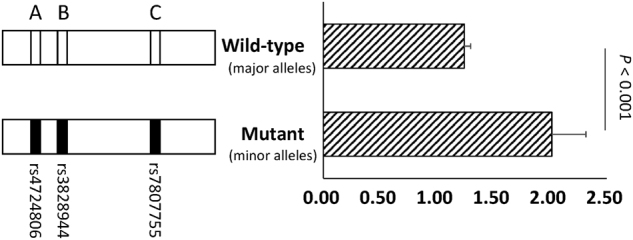
Segment contain the minor alleles of the tagged SNPs increased the transcriptional activity using luciferase reporter assay. The segments of the putative regulatory region containing the three candidate loci were inserted into the PGL-3 basic vector. Then the two vectors were transfected into the 293T cell line. The pRL-TK vector encoding the Renilla luciferase was transcribed, which was used as internal controls to normalize firefly luciferase expression. The error bars denote the SD of the relative activities. Comparisons were performed, and the P-values are labeled in the plot
Expression-trait association analysis
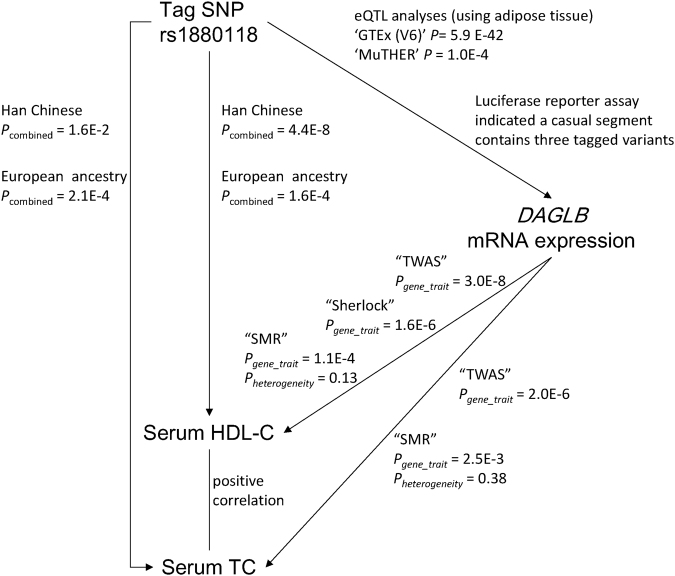
As shown in Fig. 4, replicable evidence has been established between the genotype (rs1880118) to the phenotype (HDL-C) and the genotype (rs1880118) to the expression (DAGLB). The relationship between the expression DAGLB and HDL-C levels remained unknown. We performed the analyses using three novel integrated approaches including TWAS, SMR, and Sherlock for the expression-trait association tests.
Fig. 4.
Integrated plot of three-way association between the tagSNP rs1880118, the mRNA expression levels of DAGLB, and the serum HDL-C levels. Evidence for each line is summarized and marked on the plot. P-values were determined using TagSNP and phenotype: genotype–phenotype linear regression and combined analyses using fixed-effect model; TagSNP and gene expression: eQTL analyses and luciferase reporter assay; gene expression and phenotype: two powerful approaches called “TWAS” and “Sherlock” based on eQTL and GWAS data
Using TWAS, we found that DAGLB levels were significantly associated with serum HDL-C levels (P = 3.0E-8) and TC levels (P = 2.0E-6). Based on SMR, we also observed a significant association between DAGLB and HDL-C (PSMR = 1.1E-4, PHEIDI = 0.13) and TC (PSMR = 2.5E-3, PHEIDI = 0.38). The method “Sherlock” showed that the expression level of DAGLB was associated with serum HDL-C level (logarithm of Bayesian factor = 5.62, P = 1.58E-6).
Discussion
In this study, we identified a novel locus at rs1880118 associated with serum HDL-C levels in Han Chinese. The genotype of rs1880118 explained 47.7% of the variance of the mRNA expression level of DAGLB in subcutaneous adipose tissue. Luciferase reporter assay indicated a causal segment contains three variants of DAGLB transcription regulatory.
As enrichment analysis has shown that trait-associated loci are more likely to be eQTLs [11]. That means transcriptional annotations can enhance discovery of trait-associated SNPs which will raise the possibility that some of the missing heritability would be found. Compared with classical GWAS strategy, eQTL-based screening avoids excessively rigid threshold and reliance on P-value of genotype-phenotype association in discovery stage. Using such strategy, a significant association between the eQTL at rs1880118 and serum HDL-C levels was observed and replicated in Han Chinese. After fine-mapping using epigenetic annotations, transcriptional activity was observed in a segment which contains three candidate causal variants. Reporter assay, Electrophoretic Mobility Shift Assay (EMSA) and ChIP could be performed to verify the regulatory mechanism in the further.
Although we replicated the genotype-phenotype association and found the causal segment, we still could not conclude whether this variant influences the HDL-C level by modulating the expression level of DAGLB because this variant may affect DAGLB and HDL-C independently. Powerful strategies were applied to identify eGene (eQTL gene, the gene that has a significant eQTL acting upon it) whose cis-regulated expression is associated with complex traits. TWAS and SMR have conceptual similarities with the Mendelian randomization (MR) test which aims to identify causal relations using genetic variations as instrument variables [36, 37]. Such strategies could benefit from the MR which will never result in reverse causality [39–41]. Also, only a small number of transcriptome data are required as reference for the analyses [41]. Then we verified the finding using a computational algorithm named Sherlock [38]. Consistent result supporting the association between DAGLB and Serum HDL-C levels was observed.
Similar to DAGLα, a sequence-related enzyme DAGLβ (coded by DAGLB) has a role in the biosynthesis of the endocannabinoid 2-arachidnoyl glycerol (2-AG) [42]. DAGLA is mainly expressed in the nervous system and seems to be the principal regulator of 2-AG [43]. DAGLB is widely expressed in multiple tissues, including adipose tissue, white blood cells, and brain (data from GTEx) [18]. DAGLB has been linked to hypotrichosis and the influence of early life stress [44, 45]. However, the evidence of DAGLB in lipid metabolism is limited. A recent study showed that DAGLβ inactivation lowers 2-AG, arachidonic acid, and eicosanoids in mouse peritoneal macrophages using DAGLB knock-out mice [46]. A corresponding reduction in lipopolysaccharide-induced TNF-α release was observed as well. These findings indicate that DAGLβ plays a role within the lipid network that regulates proinflammatory responses in macrophages [46]. However, it remains unknown whether the expression levels of DAGLB in multiple tissues is responsible for serum HDL-C and TC levels. Gene function studies using cell lines or mouse model with DAGLB knock in or knock down are needed for clarifying the biological role of the gene. In conclusion, we identified a novel serum HDL-C-associated variant rs1880118. The genotype of rs1880118 explained nearly half of the expression variance of DAGLB. After fine-mapping for the tagSNP, a regulatory segment which contributes to the variance of DAGLB was identified. The expression level of DAGLB was associated with serum HDL-C by means of integrated approaches using eQTL and GWAS data. The current study indicates the role of DAGLB and provides novel insight into the lipid mechanism in Han Chinese.
Electronic supplementary material
Acknowledgements
This work was supported by the National Key Technology R & D Program of China (2009BAI80B02, 2012BAI02B03), the 973 Program (2015CB559100), the National Natural Science Foundation of China (81172755, 31325014, 81421061, 81701321), the 111 Project (B13026), the Program for Zhejiang Leading Team of Science and Technology Innovation (2010R50050), Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents, the Program of Shanghai Academic Research Leader (15XD1502200), the National Program for Support of Top-Notch Young Professionals. We would like to acknowledge all the staff and participants in this study for their important contribution.
Compilance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Dan Zhou, Dandan Zhang, Xiaohui Sun, Zhiqiang Li.
These authors jointly supervised this work: Yimin Zhu, Yongyong Shi, Maode Lai.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0108-4) contains supplementary material, which is available to authorized users.
Contributor Information
Yimin Zhu, Email: zhuym@zju.edu.cn.
Yongyong Shi, Email: shiyongyong@gmail.com.
Maode Lai, Email: lmp@zju.edu.cn.
References
- 1.Castelli W. Cholesterol and lipids in the risk of coronary artery disease–the Framingham Heart Study. Can J Cardiol. 1988;4:5A–10A. [PubMed] [Google Scholar]
- 2.Rahman I, Bennet AM, Pedersen NL, De Faire U, Svensson P, Magnusson PK. Genetic dominance influences blood biomarker levels in a sample of 12,000 Swedish elderly twins. Twin Res Human Genet. 2009;12:286–94. doi: 10.1375/twin.12.3.286. [DOI] [PubMed] [Google Scholar]
- 3.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu W, Cheng YC, Chen K, et al. Evidence for several independent genetic variants affecting lipoprotein (a) cholesterol levels. Hum Mol Genet. 2015;24:2390–2400. doi: 10.1093/hmg/ddu731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 6.Edwards S, Beesley J, French J, Dunning A. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolio TA, Fowler DM, Starita LM, et al. Bedside back to bench: building bridges between basic and clinical genomic research. Cell. 2017;169:6–12. doi: 10.1016/j.cell.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Bailey SD, Lupien M. Laying a solid foundation for Manhattan–setting the functional basis for the post-GWAS era. Trends Genet. 2014;30:140–9. doi: 10.1016/j.tig.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YI, Van dGB, Raj A, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–4. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Z, Lupski JR. Non-coding genetic variants in human disease. Hum Mol Genet. 2015;24:R102–10. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamazon ER, Zhang W, Konkashbaev A, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–62. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundberg E, Small KS, Hedman AringK, et al. Mapping cis-and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Innocenti F, Cooper GM, Stanaway IB, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:165–71. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:1020–32. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang L, Morar N, Dixon AL, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–26. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia K, Shabalin AA, Huang S, et al. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012;28:451–2. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaulton KJ, Ferreira T, Lee Y, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters JE, Lyons PA, Lee JC, et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet. 2016;12:e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Luo XJ, Landén M, et al. Impact of a cis-associated gene expression SNP on chromosome 20q11.22 on bipolar disorder susceptibility, hippocampal structure and cognitive performance. Br J Psychiatry. 2015;208:128–37. doi: 10.1192/bjp.bp.114.156976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:568–71. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114–5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degner JF, Pai AA, Piqueregi R, et al. DNaseI sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubert F, Zaugg J, Kasowski M, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015;162:1051–65. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24:R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Zhang D, Zhou D, et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: a multi‐stage genome‐wide association study. J Cell Mol Med. 2017;21:1106–16. doi: 10.1111/jcmm.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Cao L, Li Z, et al. A genome-wide association study identifies a locus on TERT for mean telomere length in Han Chinese. PLoS ONE. 2014;9:e85043. doi: 10.1371/journal.pone.0085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–52. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 38.He X, Fuller C, Song Y, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–80. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 40.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 41.Gamazon ER, Wheeler HE, Shah KP, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wattanamongkhol N, Kumhom P, Chamnongthai K. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163:463–8. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T J, J WM, K M, A S. Diacylglycerol lipaseα (DAGLα) and DAGLβ cooperatively regulate the production of 2-arachidonoyl glycerol in autaptic hippocampal neurons. Mol Pharmacol. 2013;84:296–302. doi: 10.1124/mol.113.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marco EM, Echeverryalzate V, Lópezmoreno JA, Giné E, Peñasco S, Viveros MP. Consequences of early life stress on the expression of endocannabinoid-related genes in the rat brain. Behav Pharmacol. 2014;25:547–56. doi: 10.1097/FBP.0000000000000068. [DOI] [PubMed] [Google Scholar]
- 45.Basit S, Ali G, Wasif N, Ansar M, Ahmad W. Genetic mapping of a novel hypotrichosis locus to chromosome 7p21.3–p22.3 in a Pakistani family and screening of the candidate genes. Hum Genet. 2010;128:213–20. doi: 10.1007/s00439-010-0847-y. [DOI] [PubMed] [Google Scholar]
- 46.Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. DAGLβ inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.