Abstract
Here, we report the application of a portable sequencer, MinION, for genotyping the malaria parasite Plasmodium falciparum. In the present study, an amplicon mixture of nine representative genes causing resistance to anti-malaria drugs is diagnosed. First, we developed the procedure for four laboratory strains (3D7, Dd2, 7G8, and K1), and then applied the developed procedure to ten clinical samples. We sequenced and re-sequenced the samples using the obsolete flow cell R7.3 and the most recent flow cell R9.4. Although the average base-call accuracy of the MinION sequencer was 74.3%, performing >50 reads at a given position improves the accuracy of the SNP call, yielding a precision and recall rate of 0.92 and 0.8, respectively, with flow cell R7.3. These numbers increased significantly with flow cell R9.4, in which the precision and recall are 1 and 0.97, respectively. Based on the SNP information, the drug resistance status in ten clinical samples was inferred. We also analyzed K13 gene mutations from 54 additional clinical samples as a proof of concept. We found that a novel amino-acid changing variation is dominant in this area. In addition, we performed a small population-based analysis using 3 and 5 cases (K13) and 10 and 5 cases (PfCRT) from Thailand and Vietnam, respectively. We identified distinct genotypes from the respective regions. This approach will change the standard methodology for the sequencing diagnosis of malaria parasites, especially in developing countries.
Introduction
Malaria is at the forefront of the WHO’s disease eradication programs1. Every year, malaria causes the most fatalities of any parasitic disease, and the most severe symptoms have been reported from Plasmodium falciparum infections2. Spread all over tropical countries across the world, malaria imposes a severe problem in developing countries. While no vaccine is available for the malaria parasite, anti-malaria drugs are readily available. Indeed, medicines against malaria parasites have been known probably since malaria itself was noted approximately 4,000 years ago3. Starting from quinine and progressing to chloroquine, sulfadoxine-pyrimethamine, mefloquine, and the recent artemisinin, these drugs were effective in killing parasites4. However, rapidly increasing cases of drug-resistant malaria are gradually rendering medication difficult. Due to administration of drugs at suboptimal concentrations, partly because of inadequate dosing and the half-life of the medicines themselves, parasite strains that are resistant to these medicines started to appear and are rapidly spreading5. Particularly, P. falciparum has developed resistance to nearly all anti-malarial drugs that are in current use5,6. For example, chloroquine-resistant P. falciparum has been described everywhere in Central and South America, Africa, and Southeast Asia7. Sulfadoxine-pyrimethamine is currently not effective in Africa, Southeast and South Asia, South America, or Oceania8. Notably, the most powerful anti-malaria drug, artemisinin, has been shown to have reduced efficacy in Southeast Asia9. Recently, it has become known that multidrug-resistant malaria is starting to spread from the Mekong region10. To address these concerns, novel anti-malaria drugs are being developed11. However, antimalarial invention takes a long time; artemisinin was found in 1972 but its approval as an antimalarial was in the 1990s12. There is a pressing need to precisely understand the prevalence of drug resistance and decide the proper strategy for how to use the limited repertoire of drugs and their combinations.
Drug resistance is acquired due to mutations in the parasite genes. There have been several parasite genes reported to be associated with drug resistance. For instance, cytochrome B mutations cause resistance to atovaquone13. Mutations in PfCRT and PfMDR1 cause resistance to quinine, chloroquine, amodiaquine, mefloquine, piperaquine, lumefantrine, and primaquine14. Mutations in PfDHFR and PfDHPS cause resistance to sulfadoxine-pyrimethamine8. Likewise, mutations in K13 have been associated with decreased artemisinin susceptibility15. To be more precise, those drug-resistant mutations are mostly realized by single nucleotide polymorphisms (SNPs) and their combinations in these genes. For example, mutations in transmembrane domains 1, 4 and 9 of PfCRT are responsible for the chloroquine-resistant phenotype16. Similarly, four point mutations in PfDHFR and four point mutations in PfDHPS are known to be responsible for hampering the drug effect of sulfadoxine-pyrimethamine8. Five recently described point mutations in the K13 gene are purported to cause artemisinin resistance15. When they occur, K13 mutations neutralize the drug effect of impairing the PI3K signal pathway in the parasite17,18.
To ensure proper use of the drugs, examination of parasites’ genotypes at the early stage of the infections is ideal, preferably before drug administration. For this purpose, sequencing is the most decisive means. Although conclusive, sequencing technologies, including Sanger and massively parallel sequencing, are rarely available in the field hospitals of developing countries, where the immediate diagnoses are needed. A recently available new type of sequencer, MinION, is changing the methodology of sequencing. This technology has a great advantage in addressing the current concerns regarding the use of sequencing technology in developing countries. It does not require a laborious sample processing step by skillful laboratory technicians. Indeed, MinION is a single-use USB-powered sequencer, operated by a PC, requiring no prior instrumental investment19. The application of MinION in the field can greatly assist the identification of pathogens, such as Ebola and Zika viruses20,21. MinION also has been used to assemble bacterial genome de novo22. A pathogen’s relatively small genome size means that whole genome sequence can be easily performed using MinION’s current chemistry. Whole genome sequence of Neisseria gonorrhoae in clinical setting has helped with the antibiotic selection23. Therefore, utilization of MinION is preferable when faced with malaria parasites. The parasite is quick to acquire drug resistance24. Drug resistance against artemisinin is recognized after failure to clear malaria parasites in the third day post-medication based on microscopic examination15. Sequencing of the respective parasite can predict that resistance is expected and therefore a change of drug regimen might be considered. The data is also important for surveillance or further research in finding out the mechanism of resistance.
Here, we report our first use of the MinION sequencer for genotyping P. falciparum in the field.
Results and Discussion
Sequencing of laboratory strains
We selected nine genes in P. falciparum, namely, mitochondrial apocytochrome B (CYTB), sarcoplasmic/endoplasmic reticulum Ca2+-ATPase6 (PfATPase6), multidrug resistance protein 1 (PfMRP1), dihydrofolate reductase-thymidylate synthase (PfDHFR), translationally controlled tumor protein (TCTP), chloroquine resistance transporter (PfCRT), multidrug resistance protein 1 (PfMDR1), dihydropteroate synthase (PfDHPS), and Kelch protein gene (K13), which are associated with resistance to the most representative anti-malaria drugs as shown in Supplementary Table 1. For these genes, we amplified the entire genic regions by PCR and subjected them to MinION sequencing. These genes were not long and had relatively short introns if any. One or two sets of PCR amplicons approximately 1–3 kb long could cover the entire coding regions.
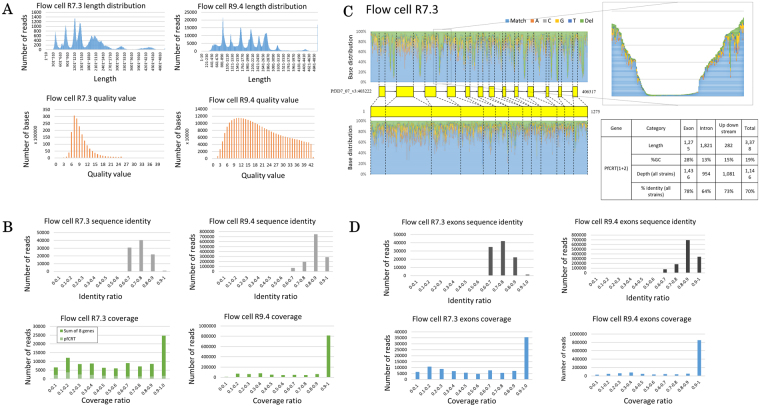
We first sequenced the four commonly used laboratory strains, i.e., 3D7, 7G8, K1, and Dd2. The PCR amplicons of the nine genes were sequenced as a mixture, with one MinION flow cell used for each strain. We sequenced the samples using the obsolete flow cell R7.3 and re-sequenced them with the newest flow cell R9.4. We obtained 179,444 reads for the combined laboratory strains with flow cells R7.3 (Supplementary Table 2) and 1,875,850 with flow cells R9.4 (Table 1). Their average lengths were 1,654 and 2,736 bp for flow cell R7.3 and R9.4, respectively (Fig. 1A, top panel). MinION’s ability to sequence relatively long reads comes with the drawback of low sequence quality is only true in the obsolete flow cells. The average sequencing quality value (QV) with flow cell R7.3 was 9.4, which increased to 20.1 when re-sequenced with the newest chemistry (Fig. 1A, bottom panel). The general low quality of flow cell R7.3, may be partly caused by the fact that P. falciparum is especially difficult organism to sequence. Particularly in the intergenic regions and introns, the AT content exceeded 80%, and there are numerous AT-rich repetitive sequences25. Even in the exonic regions, there are several such AT-rich regions. The MinION principle of deciphering the electric disturbance pattern for every five nucleotides in MinION may have made the base-calling of such regions particularly difficult. On the other hand, the newest chemistry upgrades the nanopores to have better accuracy26. It also employs new base-calling algorithms based on Recurrent Neural Nets (RNN)27. These upgrades might be compatible with the complex malaria parasite genome.
Table 1.
Sequencing and mapping statistics for four laboratory strains of P. falciparum.
| Sample | Total reads (2D) | Reads that are mapped to target (LAST 658, MC Frith et al. 2010 BMC Bioinformatics) | Ratio of mapped reads |
|---|---|---|---|
| 3D7 | 619,172 | 605,491 | 97.79% |
| K1 | 222,110 | 217,971 | 98.13% |
| 7G8 | 797,373 | 754,223 | 94.59% |
| Dd2 | 327,195 | 318,180 | 97.24% |
| Total | 1,875,850 | 1,734,486 | 92.46% |
These results were obtained with flow cell R9.4.
Figure 1.
Sequencing statistics of laboratory strains. Nanopore sequencing covers the whole length of nine genes of the laboratory strains regardless of the flow cell version. The quality value is significantly better for flow cell R9.4 (A). The sequence accuracy is better for flow cell R9.4. Mapped reads are fragmented with flow cell R7.3, especially PfCRT (B). The fragmentation is caused by introns (PfCRT is shown as an example) having higher AT counts than the exons (lower right panel). Upper left panel shows the SNPs distribution of PfCRT, while lower left panel shows the SNPs distribution of PfCRT protein-coding regions only. Light blue color symbolizes the matched sequenced nucleotides to the reference genome, while orange, grey, yellow, dark blue, and green colors symbolize the mutation to adenosine, cytosine, guanine, thymine, and deletion, respectively. Upper right panel shows the magnification of the deleted section of a PfCRT intron. Data for this figure is obtained with flow cell R7.3 (C). When introns are excluded from the analysis, fragmentations decrease with no difference in sequence accuracy on flow cell R7.3, but such problem is not found on flow cell R9.4, even if introns are included in the analysis (D). Note the significant increase in read number on flow cell R9.4 in Figures A, B, and D.
We aligned the obtained reads to the respective genic regions of the reference genome sequence, which is determined based on 3D7 strain. For mapping, we used the alignment program LAST28. We tried various parameters and thresholds as shown in Supplementary Figure 1. Although BWA-MEM outperforms LAST in this test, we have opted to use LAST as the mapping software since we are using a match/mismatch score matrix (“ATMAP”) that is theoretically tuned for the AT-richness of this genome29; in theory this should improve accuracy. Definitively judging which is the best aligner is difficult. But our paper does not depend on this; it requires only that our alignments are good enough to justify our results and conclusions. With the tuned parameters, 57.86% of reads sequenced with flow cell R7.3 were aligned to the target region (Supplementary Table 2) with 73.46% similarity to the reference (Fig. 1B, top left panel). This is comparable to the sequencing of the AT-rich bacterium FSC996, which also yielded an average 79% read accuracy30. Using the new chemistry, however, these numbers greatly increased to 92.46% mapping ratio (Table 1) and 84.56% identity (Fig. 1B, top right panel). During the mapping, we found that the alignments were generally fragmented in the PfCRT gene when sequenced with R7.3 (Fig. 1B, bottom left panel). For this gene, most of the alignment blocks cover only 20% of the target region. A similar observation was also obtained in the other amplicons to varying extents (Supplementary Figure 2A). We examined the cause of the fragmentation. For PfCRT, we found that the sequence depth was especially low in the introns (Fig. 1C). The introns had an extremely low GC content, and the sequences consisted of series of homopolymers, which imposed serious problems for MinION31. When we removed the introns and included exons only for the analysis, the fragmentation problem disappeared (Fig. 1D, bottom left panel). The overall sequencing accuracy also improved to 74.07% (Fig. 1D, top left panel). This problem, however, was not found in flow cell R9.4 as no fragmentations occurred (Fig. 1B, bottom right panel). Excluding introns from the downstream analysis did not generate much difference on the sequence accuracy, either (Fig. 1D, right panel).
We examined the possible cause of the mapping errors and found that the incorrectly amplified PCR amplicons were occasionally sequenced preferentially. The primers that we used to finalize sequencing have been chosen from multiple trial-and-errors. We chose the ones that showed the expected amplicon length. Nevertheless, although we confirmed the presence of single band amplifications before the sequencing, those dubious sequences may have been enriched, perhaps due to preferential sequence efficacy. For example, we found that the earlier peaks observed in the K13 amplicons were derived from incorrectly primed amplicon products (Supplementary Figure 2B). A sequence in the central of the gene has high similarity to the reverse primer, which we postulate can contribute to the mis-priming and give rise to shorter fragment amplification. Furthermore, we do not think this seemingly mis-priming can be solved by alignment alone. When attempts are made to use large sets of sequencing data to reduce the cost per sample, additional care should be taken to further optimize the PCR primers.
Detection of SNPs
Compiling the obtained sequence reads, we attempted to determine the genotype with respect to each parasite gene of interest. For this purpose, we developed a bioinformatics analytical pipeline. To identify SNPs, we first constructed consensus sequences from MinION reads, following the scheme shown in Supplementary Figure 3. Briefly, the consensus base is detected when the depth at a given position is more than a threshold R, where R is depth variable, and when the ratio of a base to the sum of all bases at a given position is larger than another threshold X. For heterozygous sites, a second base was called in a similar way. When the total read count is less than the threshold R or when no base fulfills the condition defined by the threshold X, the position is considered unknown, “n”. A SNP is called if the consensus base is different from the reference.
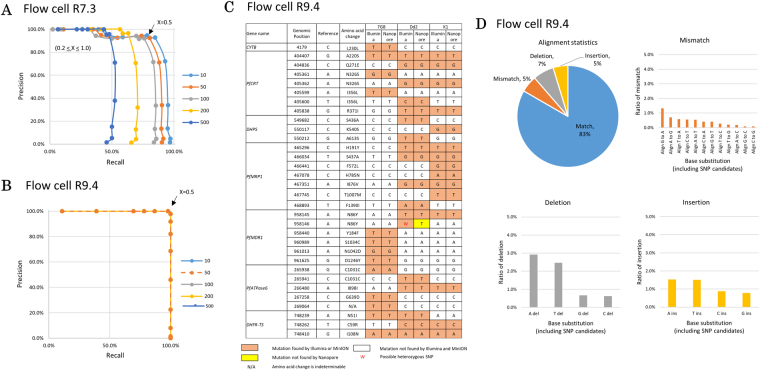
To test the performance of the developed pipeline, we compared the results of the detected SNPs with those from the Illumina sequencing. For Illumina sequencing, the PCR amplicons were subjected to shotgun sequencing. SNPs were called using a standard program, GATK. From the sequencing, we detected 52 SNPs. Using these SNPs as the reference dataset, we evaluated the precision and recall rates of the constructed pipeline at varying parameters of the thresholds R and X. Our analysis revealed that at threshold of R > 50, a reasonable good precision-recall curve was obtained (Fig. 2A). In this curve, threshold of X > 0.5 yielded satisfactorily high precision and recall rates of 0.92 and 0.8, respectively. This was true only for flow cell R7.3, because flow cell R9.4 gave consistent precision and recall with every R value, which are 1 and 0.97, respectively (Fig. 2B). We distinguish SNPs from PCR errors by following the algorithm in Supplementary Figure 3. Since PCR errors are supposed to occur randomly on the elongation of DNA strand, we believe that it would not exceed the threshold X ≥ 0.5 that we employ in our pipeline. An SNP, on the other hand, would exceed the threshold since the SNP should be present in every DNA molecule that is amplified by PCR.
Figure 2.
SNP calling of laboratory strains. The precision and recall of multiple thresholds used for heterozygous SNP calling on data obtained with flow cell R7.3. Precision is defined as the ratio of true positive (i.e., SNPs found by both MinION and Illumina) to the total of true positive and false positive (i.e., SNPs found only by MinION). Recall is defined as the ratio of true positive to the total of true positive and false negative (i.e., SNPs found only by Illumina). The lines show the sequence depth used for analysis (threshold R). The dots on the line show the threshold X. Threshold X ≥ 0.5 with R ≥ 50 sequence depth is used for subsequent analysis because of the reasonable precision and recall values, which are 0.92 and 0.8, respectively (A). On the other hand, flow cell R9.4 gives the same precision and recall for every threshold R with X ≥ 0.5, which is 1 and 0.97, respectively. In this figure, all the lines produced by different threshold R stack up (B). SNPs called with the parameters X ≥ 0.5 and R ≥ 50 on data obtained with flow cell R9.4 show 100% identity to Illumina validation (orange boxes). Exception is made for one position in PfMDR1 (red color font). This position is seemingly heterozygous as called by Illumina. However, MinION could only determine this position as a homozygous SNP (C). The error patterns show that G to A mismatches, A deletions, and A/T insertions are the most frequent polymorphisms in the laboratory strain malaria parasites sequenced with flow cell R9.4 (D).
Sequencing with flow cell R7.3, however, showed some false positive and negative calls (Supplementary Figure 4), assuming the calls by the Illumina sequencing are correct. Of the three “false” positive cases, two cases are in either AT-rich regions or homopolymer tracts, which is a challenge for MinION sequencing (see example in Supplementary Figure 5A). Nine “false” negative cases were derived from the “unconfident” call due to the requirements of X or R values being unsatisfied. One interesting case was found in the PfMDR1 region. In this case, one SNP was heterozygous, which was also confirmed by Sanger sequencing (Supplementary Figure 5B). This heterozygous SNP, which might have been acquired in a minor subpopulation of the parasites before or after the laboratory strain was established, could not be detected due to the unsatisfied requirement of the X parameter. Importantly, all those problems were once common to Illumina sequencing in its early days and have been addressed by specializing protocols for the analysis of P. falciparum later. Supplementary Figure 5C shows another example of validating correct SNPs. As expected, data obtained from flow cell R9.4 showed a very different result. While the mutations found with R7.3 were also found with R9.4, our pipeline performed a lot better with the new flow cell. Previously undetermined positions were confidently called. False positive and negative nucleotides also disappeared. Nevertheless, our pipeline could not call the seemingly heterozygous position of PfMDR1 (Fig. 2C).
For future development, we inspected the causes of incorrect SNP calls. We collected basic information on the sequencing errors in general. We evaluated the error rates occurring in all of the sequence reads sequenced with R9.4 and found that those respective mutation types accounted for 5%, 7% and 5% of the mismatch, deletion, and insertion errors, respectively (Fig. 2D). This is an improvement over flow cell R7.3 (Supplementary Figure 6). Guanine was more likely to be miscalled as adenosine. Deletions were more likely to be accumulated at adenosine or thymine sites. Insertions were mainly adenosine or thymine. Incorrectly called SNPs at the relaxed threshold somewhat represented these general error patterns. Thus, considering the general patterns of the error matrices should give useful information to minimize erroneous SNP calling, especially when relaxed thresholds are used.
To compare the performance of our pipeline, we also employed a variant calling specifically built for MinION, Nanopolish32, that uses hidden Markov model (HMM) signal-level consensus algorithm to call for SNPs. As flow cell R7.3 employs HMM for its base-calling, Nanopolish applies the same algorithm for variant-calling. Using default parameters, we found that Nanopolish variant-calling returned many false positive results for the data obtained with R7.3 (Supplementary Figure 7). Even calling the reference strain yielded false positive nucleotides (Additional File 1). There were instances when Nanopolish found some variants not detected by our pipeline, but the many false positive results indicate that Nanopolish might be too sensitive for variant calling, at least in malaria parasite. On the contrary, our pipeline has been optimized for malaria parasite’s genome to minimize false nucleotides.
Sequencing of clinical samples
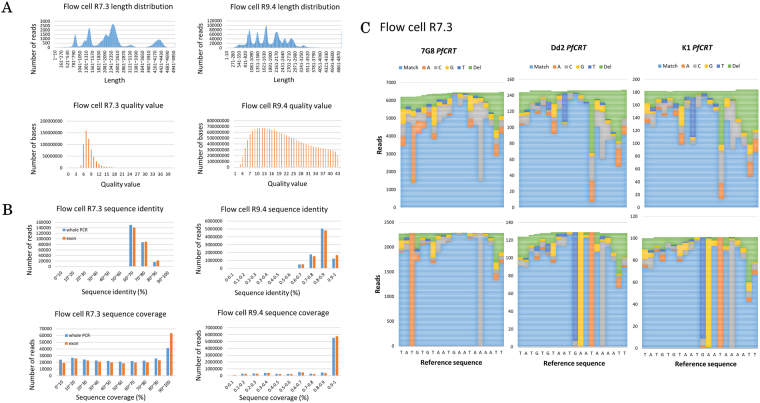
We applied the developed technique to analyze clinical samples. First, ten samples from patients with a positive diagnosis of P. falciparum infection were processed in the same manner as the laboratory strains. A total of 337,104 reads were obtained for the nine genes of ten clinical samples (Table 2). As the case with laboratory strains, the numbers increased greatly with flow cell R9.4 (Supplementary Table 3). Unfortunately, comparison of flow cell R7.3 and R9.4 performance could not be completed with the same clinical samples, because we have used all of the original samples for the analysis with flow cell R7.3. The average read length of 2,334 bp (Fig. 3A, top left panel) and QV of 8.3 was similar to that of the laboratory strains (Fig. 3A, bottom left panel). On the contrary, sequencing with flow cell R9.4 yielded reads with an average read length of 15.9 kB and QV of 20, which was a very significant improvement (Fig. 3A, right panel). Sequencing accuracy and coverage of data obtained with flow cell R9.4 did not change much with the exclusion of introns, while data from flow cell R7.3 gave a significant improvement in coverage with the exclusion of introns (Fig. 3B).
Table 2.
Sequencing and mapping statistics of ten clinical samples.
| Sample | Total reads (2D) | Mapped reads | Best score (≥150) | Ratio of mapped reads |
|---|---|---|---|---|
| 2-#2 | 60,472 | 72,353 | 40,339 | 66.7% |
| 2-#5 | 116,582 | 64,713 | 41,273 | 55.5% |
| 5-#1 | 34,186 | 31,468 | 18,928 | 92.05% |
| 5-#3 | 2,493 | 2,920 | 1,562 | 62.65% |
| 5-#6 | 48,600 | 49,856 | 24,701 | 49.54% |
| 5-#39 | 2,024 | 1,807 | 1,139 | 89.28% |
| 5-#40 | 26,663 | 16,477 | 9,872 | 61.8% |
| 5-#41 | 10,037 | 8,671 | 6,901 | 86.39% |
| 5-#43 | 19,618 | 7,892 | 4,989 | 40.23% |
| 5-#46 | 18,922 | 16,466 | 12,664 | 87.02% |
These results were obtained with flow cell R7.3.
Figure 3.
Sequencing of clinical samples. The read length distributions of the combined clinical specimens show that the sequenced reads are distributed to cover all nine genes with a quality value that is significantly better on flow cell R9.4 (A). The reads obtained with flow cell R7.3 are not fragmented in clinical specimens, although exons are included in the analysis. Nevertheless, removal of introns significantly increased the coverage. On the other hand, the reads obtained with flow cell R9.4 have better accuracy and coverage, regardless of the introns removal (B). Remapping of the sequenced reads (upper panel) to the representative haplotype reference shows confident SNP calling (lower panel). In this process, we removed the reads that have unconfident SNPs called in highly difficult region of amino acid 72–76 of PfCRT. We then remapped the remaining reads to the representative strain reference to increase call confidence (C). Note the significant increase in read number on flow cell R9.4 in Figures A and B. Comparison between data produced by flow cell R7.3 and R9.4 uses different samples due to the exhaustion of the original samples for the flow cell R7.3 analysis.
Using the analytical pipeline described above, we were able to detect SNPs for PfCRT and all other genes using data obtained from flow cell R7.4 (Supplementary Table 4). Some SNPs were shared with the laboratory strains, while novel candidates were also identified uniquely in clinical samples and had never been documented before. There were instances when MinION gave false positive results. As with laboratory strains, these false positive cases were not in the coding region, indicating the difficulty of MinION to sequence the intronic region. Furthermore, the high number of false positives in only one sample raised the suspicion of poor library preparation that caused relatively poor MinION sequencing. Overall, MinION’s precision and recall for this dataset are 0.87 and 0.91, respectively.
In addition to the SNPs occurring at a single base, there is one site in the PfCRT gene where different haplotypes were directly related to chloroquine resistance. The site corresponds to amino acids 72 to 76. There are three main haplotypes reported so far, namely, CVMNK (chloroquine sensitive), CVIET (chloroquine resistant) and SVMNT (chloroquine resistant)33. Consistently, Illumina sequencing detected CVMNK type in 3D7, CVIET in K1 and Dd2 and SVMNT in 7G8. All our clinical samples resembled the 7G8 haplotype (Supplementary Table 5). When we closely looked at the MinION reads, the alignments were hard to determine at this site due to complex patterns of mismatches, deletions and insertions spanning multiple bases (Fig. 3C). For this site, we removed the reads that had unconfident SNP calls. We then remapped the MinION reads for the representative haplotypes. Without any changes to the other parameters, the refined alignment exactly detected the respective haplotypes. Independent procedures may be needed to discriminate different haplotypes, consisting of long and complex patterns of multiple mutations. We also expect that haplotypes spanning longer regions, if there are any, could also be detected by a similar method.
The eventual goal of this study is to predict possible drug resistance for each patient based on the genetic information of the parasites. We categorized the patients with respect to patterns of SNPs that have been associated with drug resistance (Table 3). For chloroquine and sulfadoxine-pyrimethamine, we found that all samples that we processed had mutations for resistance to those drugs. Namely, all the patients had the resistance-causing SNPs at PfCRT, PfMRP1, PfMDR1, DHFR-TS and DHPS. For the resistance to artemisinin, we found inconsistencies with Illumina data for the K13 (see below for further description on K13). However, all the patients had SNPs in the PfATPase6. Nevertheless, this gene has been proved to have no correlation with artemisinin resistance34–36. Another exciting finding in the light of recent research is that our sequencing results suggest that all, except for one, of our clinical samples are sensitive to atovaquone. As a single agent, atovaquone gives a recrudescence rate of approximately 30%, but in combination with proguanil, atovaquone is very effective37, and resistance to it will not spread because atovaquone-resistant parasites cannot develop in vector mosquitoes38.
Table 3.
Summary of drug resistance phenotypes of ten clinical samples, inferred from data obtained with flow cell R7.3.
| Drugs | Patients | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2-#2 | 2-#5 | 5-#1 | 5-#3 | 5-#6 | 5-#39 | 5-#40 | 5-#41 | 5-#43 | 5-#46 | |
| Atovaquone | − | − | − | − | − | − | − | − | − | − |
| Chloroquine/mefloquine | + | + | + | + | + | + | + | + | + | + |
| Sulfadoxine-pyrimethamine | + | + | + | + | + | + | + | + | + | + |
| Artemisinin | − | − | − | − | − | − | − | − | − | − |
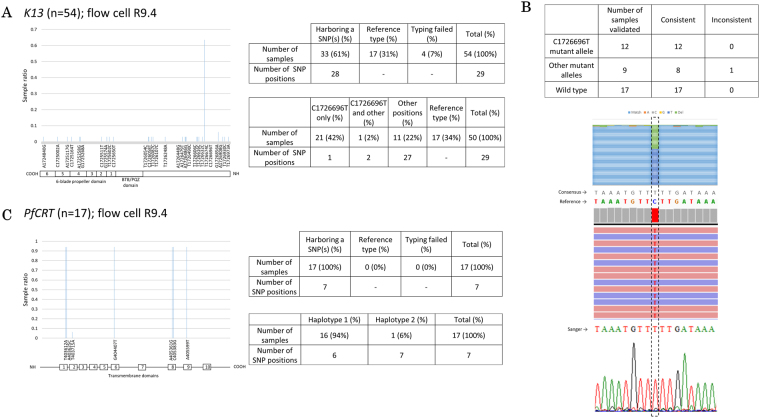
We further intended to expand the dataset for the PfCRT and K13 genes. This extensive analysis also should serve as a model for epidemiological study of a gene. We collected 54 parasite-positive bloods and preserved the DNA using Whatman’s FTA Elute cards, which is a common means for sample collection and storage in the field. We purified the DNA, PCR-amplified K13 for its entire gene region, and MinION-sequenced the amplicons using flow cell R9.4. The genotyping was successful for 50 samples. We identified that 33 samples (61%) had at least one SNP at a total of 28 positions (Fig. 4A, upper right panel). The remaining 17 samples completely matched the genotype of the reference genome. The detected SNPs included a frequent C1726696T SNP which was observed in 22 samples (Fig. 4A, lower right panel; this SNP was also observed by Illumina in 3 of the initially analyzed 10 clinical samples). The other 27 SNPs were mostly observed only in one sample (“singletons”). All these mutations could be compiled into 11 mutation haplotypes (Supplementary Figure 8A). For these SNPs, we validated 20 samples using Sanger and Illumina sequencings. We found that almost all the detected SNPs were validated (Fig. 4B, upper panel). This included the high frequency SNP, which were always validated (example of validation of this SNP is shown in Fig. 4B, lower panel). Lastly, we similarly conducted the genotyping analysis of PfCRT gene for 17 samples (Fig. 4C). All samples had valid mutations at a total of 7 positions, converged to one major and one minor haplotypes (Fig. 4C, lower right panel). The major haplotype was mostly like that of the 7G8 strain (Supplementary Figure 8B). It is intriguing that the distinct mutation patterns between K13 and PfCRT gene should be associated with the fact that the administration of chloroquine has been suspended and substituted with artemisinin for more than ten years due to the wide-spread of the drug-resistant phenotype. This data however cannot tell us whether the detected SNPs in the K13 were caused by normal genetic drift, which is common in malaria parasite39, or functionally relevant SNPs invoked by the selective pressure from frequent use of artemisinin in this area. The low frequency mutation phenomenon in K13 is also common to previous research40. Also, none of our finding is in agreement with the four Asian mutations that have been validated41. Nevertheless, the C1726696T SNP, which changes arginine to lysine at amino acid position 101 (R101K), has now dominated the parasite population in this area. Taking all the results together, we consider that genetic drifts may be emerging world-wide, which may eventually lead to a novel drug-resistant mutation. This is also in agreement with the observation that artemisinin resistance is different from other drugs, in terms that it can sporadically appear42.
Figure 4.
K13 sequencing as a proof-of-concept. We sequenced the whole K13 from 54 samples obtained in Indonesia. There are 61.1% samples with SNPs in 28 positions. A significant number of samples (42%) have SNPs in genomic position 1726696 of chromosome 13, which is in the region of K13. The respective K13 protein domains in regard to genomic positions is shown (A). The polymorphism in genomic position 1726696 is validated consistently with other sequencing means. An example of this validation with Sanger and/or Illumina is shown (B). Sequencing of PfCRT from 17 samples shows SNPs in 7 positions. We further found two haplotypes with most samples are of haplotype 1 (see Supplementary Figure 8B). The respective PfCRT protein domains in regard to genomic positions is also shown (C). All data are obtained from the sequencing of clinical specimens collected with FTA card and sequenced with flow cell R9.4.
These results are consistent with the fact that the parasite population in North Sulawesi is still susceptible to artemisinin, but resistant to chloroquine and sulfadoxine-pyrimethamine in the clinical practices. Nevertheless, one district in East Indonesia is known to have a high degree of resistance to artemisinin combination therapy (ACT), with 52% and 84% response rates for artesunate + amodiaquine and dihydroartemisinin + piperaquine, respectively43. The migration of the parasites from the resistant area to North Sulawesi, if happened, would have significant impact on the treatment strategy in this region as well. A vigilant surveillance is required to monitor the spread of the resistance in the surrounding regions.
Fidelity of the generated dataset
Having collected all the datasets, we carefully re-considered the overall performance of our pipeline, especially because there are already many publications regarding SNPs found in P. falciparum. Therefore, using four public datasets, we compared the SNPs found in this research (from both flow cell R7.3 and R9.4) to the published datasets. PlasmoDB44 is a collection of 1,775,595 SNPs collected from all over the world. P. falciparum Community Project45 curates 681,587 SNPs of 3,488 samples from 23 countries. Another database is Pf3k pilot data release 446, that contains 944,270 SNPs of 2,512 samples from 14 countries. Specifically for K13, we added a comparison with a comprehensive report by Ménard D et al.41. We found that 4 out of a total of 45 positions (excluding K13-propeller) were not overlapping with the datasets, suggesting the possibility of novel SNPs (Table 4). We had validated these SNPs with Illumina (Supplementary Figure 9) and found that these SNPs were not errors found by MinION. In case of K13, only one of our singleton SNPs in the propeller region shared a synonymous mutation with P. falciparum Community Project and Pf3k datasets. Any other mutations in K13 do not intersect with the datasets. While it is true that each of the sequences produced by the MinION is still error-prone, the redundant reads of the sequences may have been able to complement the drawback. We have conducted theoretical validation regarding this issue and found that when the base is covered by >10 sequences with the quality value >10, the sequence error should be less than 0.2% (data not shown). Our novel SNPs met these criteria. We further examined the mutual overlaps of the novel SNPs in our dataset and found that 20% were common to more than one samples.
Table 4.
A summary showing the comparison of SNPs found by this paper’s pipeline to four public databases.
| Database or study name | Total SNPs | Total studies or samples | Total SNPs in database shared with this paper target genes | Total positions shared with the database | Total positions not shared with the database |
|---|---|---|---|---|---|
| PlasmoDB | 1,775,395 | 16 studies | 673 | 34 | 11 |
| P. falciparum Community Project | 681,587 | 3,488 samples from 23 countries | 831 | 39 | 6 |
| Pf3K | 944,270 | 2,512 samples from 14 countries | 884 | 30 | 15 |
| Total positions unique to our sequencing data (excluding K13-propeller) | 4b | ||||
| Ménard D et al.a (K13-propeller only) | 108 | 14,037 samples from 59 countries | 108 | 0 | 10 |
| Total positions unique to our sequencing data (including K13-propeller) | 13c | ||||
Data is obtained with flow cell R7.3 and R9.4.
aMénard, D. et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 374, 2453–2464 (2016). bOne mutation in K13 non-propeller region (R101K) is included here due to its significant presence in the clinical samples. cOne synonymous mutation (627A) in K13-propeller region is excluded because it is found in two other databases.
Also, we tested the consistency of invariant positions with the datasets. As invariant positions are considered neutral, we expect that our sequencing data should also be neutral in these positions. A low consistency would suggest that MinION introduced a lot of errors and therefore low reliability. First, we sought for non-SNP positions in the datasets which intersecting with all three datasets. We found that from a total 21,337 positions in the coding region of the target genes, 20,167 positions were non-SNPs. We then matched these positions to our sequencing data and found that 20,151 (99.9%) positions were intersecting with the non-SNPs positions (Supplementary Table 6). This result tells that MinION can show good consistency when sequencing coding regions. Taking all these results together, we conclude that these novel SNPs should be correctly representing novelty.
Sequencing analysis in other areas
To further enrich our data, we also sequenced PfCRT and K13 of clinical samples from Thailand and Vietnam using flow cell R9.4. As expected, the samples that we acquired from Thai-Cambodia border mostly have the artemisinin-resistance associated SNPs. We found F419L, R539T, and C580Y mutations from Thai samples. On the contrary, the samples from Vietnam contains mutation in the non-propeller region (K189T) (Fig. 5A). These results are in contrast with K13 sequence data acquired in North Sulawesi, Indonesia. Of these mutations, only R539T and C580Y that have been verified by an international consortium as artemisinin-resistance markers15. As for PfCRT, all the samples from Thailand and Vietnam has the classic markers for chloroquine resistance, i.e., K76T and A220S (Fig. 5B).
Figure 5.
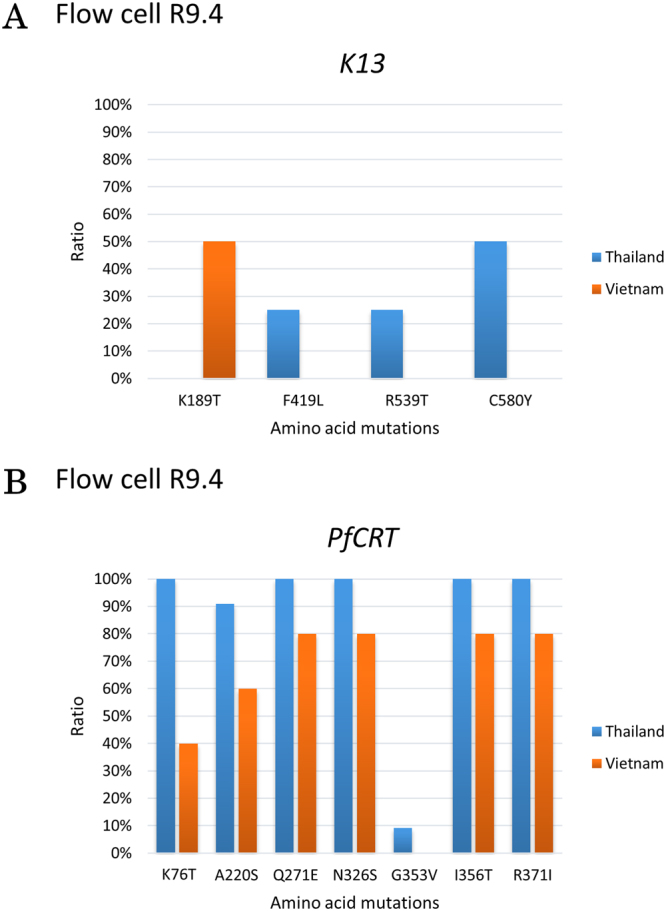
K13 and PfCRT sequencing of Thailand and Vietnam samples. We sequenced 3 and 5 cases (K13) and 10 and 5 cases (PfCRT), from Thailand and Vietnam, respectively. Mutations in K13 from Thai samples show a different pattern compared to Vietnamese samples (A). Nevertheless, mutation pattern of PfCRT is the same between Thai and Vietnamese samples (B). All data are obtained with flow cell R9.4.
We also attempted to address how our approach can detect multiple parasite genomes in a single individual, namely complexity of infection (COI) cases47. Clinically, it can have an effect for the disease outcome. The results of the hitherto described analyses seemed indicating that there is possibility of multiple genome within a sample. At these positions, the base variation could not be explained by simply detecting the presence or absence of a SNP. Since we employed conservative criteria to reduce false positive calls, the called SNPs were homozygous in a given sample (all the sequences represented a variant except for possible sequencing errors). When we closely looked at the seemingly heterozygous SNPs, we detected a total of 13 of such sites. However, closer inspection at the Illumina sequencing data do not support them. Indeed, the areas in Indonesia where we collected the samples do not have malaria cases as high as those areas where mixed infections are indicated. To further address these issues, we examined to what extent multiple strain infections could be detected by our pipeline. We produced a serial mix of 3D7 and Dd2 strains. We amplified PfCRT and PfMRP1 and looked for the known polymorphic sites from these genes as inferred from Fig. 2B. We called for SNPs and counted ratio of SNPs to the wild type nucleotides. As Fig. 6 shows, we theoretically could detect a possible mix infection if the ratio of a SNP is less than 0.5. We could interpolate this result to our pipeline as threshold X < 0.5. By taking into consideration multiple known SNPs and this parameter, we might detect and suspect multiple strain infection. However, at the same time, it is shown that we must be prepared for the increasing rate of the false detections, depending on the mixture rate.
Figure 6.
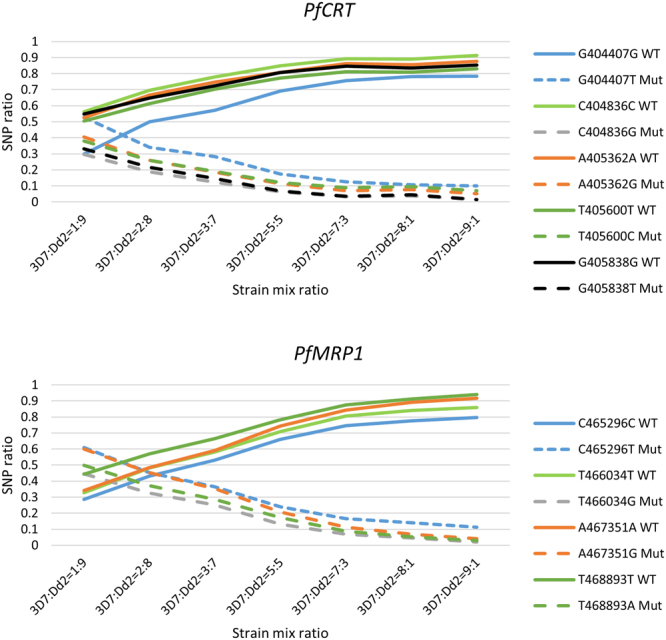
Ratio of wild type and mutated nucleotides in various proportion of a mixture of two P. falciparum strains, 3D7 and Dd2. A mixture of two P. falciparum strains (3D7 and Dd2) with different ratio show that we could infer the possibility of multiple strain infection if the SNP ratio is less than 0.5. Each color in the figure represents the position of the SNPs of PfCRT and PfMRP1 inferred from Fig. 2B. Solid lines show the wild type nucleotides found in 3D7, while the dotted lines show the SNPs found in Dd2. The intersection of the solid and dotted lines is a possible mix infection, which is at the ratio of less than 0.5. Data is obtained with flow cell R9.4.
Conclusions
In this study, we described an application of the MinION sequencer for sequencing of malaria clinical samples. MinION could generate reads with long sequences and acceptable quality. Sequence accuracy was less than 90%, even with the newest flow cell. However, by compiling more than 50 sequences in depth, our in-house-developed bioinformatics pipeline achieved overall precision and recall rates of 1 and 0.97, respectively. Further improvements with new chemistry is proved to significantly increase the precision and recall while reducing the error rate. Additionally, more sophisticated methods, such as Bayesian algorithms or deep learning, could be used to construct a better analytical pipeline. A rapid examination method using portable sequencing technology will be extremely beneficial in many aspects of infectious diseases, particularly in developing countries where modern sequencing instruments are rarely available.
Our method combines PCR amplification and the portability of MinION sequencing. Coupled with PCR, it might seem impractical, but it is still can be performed in the field hospital, especially because there is a portable thermal cycler that is easy to be installed. We agree that Sanger method would be unavailable in the field, but we present Sanger method in this paper as a validation for our results. Therefore, it would not be performed routinely in this method, but as a quality control, we might employ Sanger or Illumina sequencing occasionally.
We believe these extensive analyses have strengthened our claim that the on-site sequencing is the practical approach to genotype malaria parasites, whose acquisition of the drug resistance is a substantial threat to the human health world-wide. Further, the convenient use of the sequencing method will expand our database repertoire of the parasites and will enrich our basic knowledge on its epidemiology regarding their geographical distributions and changes overtime. Towards such goal, we hope this paper paves the first step.
Materials and Methods
P. falciparum laboratory culture and clinical samples
P. falciparum strains 3D7, Dd2, 7G8, and K1 were used in this study. The parasite was incubated at 37 °C, 5% CO2 in a flask containing 10 ml of complete medium (see Supplementary Table 7 for culture recipe) with a starting parasitemia of 0.5% for 4 days. The parasite solution was then collected by 600 × g centrifugation at room temperature for 6 minutes. Clinical samples were obtained from 64 parasite-positive patients visiting a clinic at Sam Ratulangi University in Manado, Sulawesi, Indonesia, with informed consent and approved by the Ethical Committee of Sam Ratulangi University. Ten samples were preserved with PAXGene DNA Blood tubes (PreAnalytix) and 54 samples were preserved with FTA Elute cards (Whatman). Additional 11 and 5 parasite-positive samples were obtained from Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand and National Institute of Hygiene and Epidemiology, Hanoi, Vietnam, respectively. For Complexity of Infection (COI) experiment, we mixed laboratory culture of 3D7 and Dd2 strains with parasitemia ratio of 1:9, 2:8, 3:7, 5:5, 7:3, 8:1, and 9:1. All experiments were performed in accordance with the relevant guidelines and regulations.
DNA extraction
We extracted culture solution and blood specimens using the DNaeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. As for DNA that was preserved with FTA Elute cards, we first eluted the DNA by submerging a part of an FTA card in 100 µl distilled water and boiling at 95 °C for 30 minutes. The subsequent extraction was similar to culture solution and blood specimens. Briefly, we added 100 µl of culture solution, EDTA-treated blood, or FTA card elute to 20 µl proteinase K and adjusted the final volume to 220 µl with PBS. We then added 200 µl of buffer AL to this solution. After incubation at 56 °C for 10 minutes, we added 200 µl of absolute ethanol. Following vortexing, we transferred all volume of the solution to a DNeasy Mini spin column attached to a 2 ml collection tube. Centrifugation at ≥6000 × g was performed for 1 minute. We discarded the flow-through and added 500 µl of Buffer AW1 the spin column. Centrifugation at ≥6000 × g was performed again for 1 minute. After discarding the flow-through, we added 500 µl of Buffer AW2 to the spin column and centrifuged it at 20,000 × g for 3 minutes. Elution was completed with the addition of 200 µl buffer AE to the spin column, incubation for 1 minute at room temperature, and further centrifugation at ≥6000 × g for 1 minute.
Polymerase chain reaction (PCR)
We performed PCR with 1 µl of template using KAPA Hifi (Kapa Biosystems) according to the manufacturer’s protocol. PCR was executed with the following temperatures: 94 °C for 2 minutes; 30 cycles of 94 °C for 15 seconds, 42 °C for 2 minutes, 60 °C for 1 minute; and 60 °C for 2 minutes. Supplementary Figure 10A and B show the part of genes amplified and the primers used to amplify those parts. Supplementary Figure 10C and D show the agarose gel images of the amplicons. DNA was measured and assessed using a BioAnalyzer (Agilent).
MinION library preparation
MinION sequencing libraries were prepared using the Nanopore Sequencing Kit [SQK-MAP005 or SQK-MAP006, Oxford Nanopore Technologies (ONT)] according to manufacturer’s protocol. Four lab strains re-sequencing; 54 additional samples from North Sulawesi, Indonesia; 11 samples from Mahidol University, Bangkok, Thailand; 5 samples from NIHE, Hanoi, Vietnam; and 7 mixtures of 3D7 and Dd2 strains were prepared using the 1D2 Sequencing of Genomic DNA Kit (SQK-LSK308, ONT). Briefly, the protocol for SQK-LSK308 is as follow: a total 1 µg of input DNA (a mixture of PCR amplicons from nine genes) was end-repaired and A-tailed using 1x Ultra II End-prep enzyme (New England Biolabs) incubated in 20 °C for 5 minutes and 65 °C for 5 minutes. Following the reaction, we purified end-prepped PCR amplicons with 1x AMPure XP (Beckman Coulter) and eluted the DNA in 25 µl nuclease-free water. A minimum of 500 ng of DNA was ligated with 1D2 adapter (ONT) using 1x Blunt/TA Ligase (New England Biolabs) in room temperature for 10 minutes. After purification of adapted PCR-amplicons with 1x AMPure XP (DNA was eluted in 46 µl of nuclease-free water), we ligated 400 ng of Barcode Adapter Mix (ONT) to the adapted PCR-amplicons with 1x Blunt/TA Ligase (New England Biolabs) for 10 minutes in room temperature. Purification was achieved with 140 µl of Adapter Binding Buffer (ONT) twice using the magnetic stand and the pre-sequencing mix was eluted with 15 µl of Elution Buffer (ONT). Of this, 12.5 µl of pre-sequencing mix was administered to flow cell version FLO-MIN107 in a mix of 35 µl Running Buffer with Fuel Mix (ONT) and 25.5 µl of Library Loading Buffer (ONT).
MinION sequencing and base-calling through Metrichor
Sequencing was performed for 48 hours using flow cell version FLO-MAP003. After sequencing, base-calling (r7.X 2D Base-calling revision 1.9 or version 1.24, 2D Base-calling for SQK-MAP005 version 1.48, 2D Base-calling SQK-MAP006 version 1.62) was performed via the ONT’s Metrichor cloud-based service. As for sequencing using the new chemistry, we conducted 48 hours sequencing using flow cell version FLO-MIN107. We performed offline base-calling using ONT’s Albacore version 1.2.5 with the command line: full_1dsq_basecaller.exe–flowcell FLO-MIN107–kit SQK-LSK308–output_format fast5,fastq–input [directory of FAST5 file]–save_path [directory of the output FASTQ files] -r -t2.
Alignment of MinION sequencing data
We used Poretools48 version 0.5.1 to convert FAST5 to FASTQ files for experiments base-called with Metrichor. Both “pass” and “fail” reads were used. Two aligners, BWA49 version 0.7.13 and LAST28 version 704, were used with the following parameters: BWA MEM (command line: bwa index [reference genome in FASTA file name] | bwa mem -x ont2d [indexed reference genome file name] [FASTQ file name]), LAST with default parameters (command line: lastdb [reference genome file name] [reference genome in FASTA file name] | lastal -Q1 [reference genome file name] [FASTQ file name]| last-split -m1), LAST with tuned parameters (command line: lastdb [reference genome file name] [reference genome in FASTA file name] | lastal -a12 -A15 -b3 -D1e7 -pATMAP -m100 -Q1 [reference genome file name] [FASTQ file name] | last-split -m1), and LAST-train parameters (command line: lastdb [reference genome file name] [reference genome in FASTA file name] | lastal -a11 -A14 -b3 -B3 -S1 -pATMAP -Q1 [reference genome file name] [FASTQ file name] | last-split -m1). P. falciparum strain 3D7 genome was used as reference genome.
Detection of SNPs
Reads aligned to the 3D7 genome were extracted. The sequencing depth of each base was calculated, and a consensus sequence was constructed. The consensus sequence was compared with the reference sequence, and any difference was considered a SNP. The detected SNVs were validated using Illumina.
Consensus sequences were constructed as shown in Supplementary Figure 3. When the depth of a given position was less than the threshold R, where R is the depth variable, the base was considered “N”. A base (A, T, G, or C) in a given position was called if the ratio for a base to the sum of all other bases was more than the threshold X. When the base could not be determined, the position was called “n”. For heterozygous sites, a second base was called in a similar way. Variant calling using Nanopolish32 version 0.7.1 was performed with default parameters. We conducted these procedures with in-house scripts.
We validated our results with Illumina sequencing. A library was constructed using LT TruSeq DNA according to the manufacturer’s protocol from 1 µg of PCR amplicons. Sanger sequencing was outsourced to FASMAC Japan.
Data access
The MinION sequencing data from the cell lines were deposited in the DNA DataBank of Japan (DDBJ) under the accession number DRA005565.
Electronic supplementary material
Acknowledgements
The authors are grateful to Mr. Hiroyuki Wakaguri, Ms. Kazumi Abe, Ms. Terumi Horiuchi, Ms. Megumi Kombu and Ms. Kiyomi Imamura for their technical support. The authors also gratefully acknowledge support from the Asia-Africa S & T Strategic Cooperation Promotion Program by the Special Coordination Funds for Promoting Science & Technology, Core-to-Core Program, B. Asia-Africa Science Platforms (FY2012-2014) by the Japan Society for the Promotion of Science (JSPS) and a Grant-in-aid for Scientific Research on Innovative Areas in “Genome Science” (221S0002) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author Contributions
Conception and study design: L.R.R., J.S.B.T., A.E.M., W.M., J.Y. and Y.S. Acquisition of data: L.R.R., J.S.B.T., A.E.M., J.Y. and Y.S. Analysis and interpretation of data: L.R.R., W.M., M.C.F., Y.E., R.M., J.Y., and Y.S. L.R.R., W.M., M.C.F., J.Y. and Y.S. wrote the manuscript and all authors assisted with the drafting and revision of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26334-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Global technical strategy for malaria 2016–2030 (2015).
- 2.Hotez, P. & Herricks, J. One million deaths by parasites. at http://blogs.plos.org/speakingofmedicine/2015/01/16/one-million-deaths-parasites/ (2015).
- 3.Kim Y, Schneider KA. Evolution of drug resistance in malaria parasite populations. Nat. Educ. Knowl. 2013;4:6. [Google Scholar]
- 4.Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial drug resistance: Literature review and activities and findings of the ICEMR network. Am. J. Trop. Med. Hyg. 2015;93:57–68. doi: 10.4269/ajtmh.15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White NJ. Antimalarial drug resistance. J. Clin. Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloland, P. B. Drug resistance in malaria. World Health Organization41 (2001).
- 7.Mehlotra, R. K. & Zimmerman, P. A. In Malaria: Genetic and evolutionary aspects (eds Dronamraju, K. R. & Arese, P.) 81–124 (2006).
- 8.Abdul-Ghani R, Farag HF, Allam AF. Sulfadoxine-pyrimethamine resistance in Plasmodium falciparum: A zoomed image at the molecular level within a geographic context. Acta Trop. 2013;125:163–190. doi: 10.1016/j.actatropica.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 9.MalariaGEN Plasmodium falciparum Community Project. Genomic epidemiology of the current wave of artemisinin resistant malaria. bioRxiv 10.1101/019737 (2015).
- 10.Roberts BL. Malaria wars. Science (80-.) 2016;352:398–405. doi: 10.1126/science.352.6284.398. [DOI] [PubMed] [Google Scholar]
- 11.Flannery EL, Chatterjee AK, Winzeler EA. Antimalarial drug discovery - approaches and progress towards new medicines. Nat. Rev. Microbiol. 2013;11:849–62. doi: 10.1038/nrmicro3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White NJ, Hien TT, Nosten FH. A brief history of qinghaosu. Trends Parasitol. 2015;31:607–610. doi: 10.1016/j.pt.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syafruddin D, Siregar JE, Marzuki S. Mutations in the cytochrome b gene of Plasmodium berghei conferring resistance to atovaquone. Mol. Biochem. Parasitol. 1999;104:185–194. doi: 10.1016/S0166-6851(99)00148-6. [DOI] [PubMed] [Google Scholar]
- 14.Petersen I, Eastman R, Lanzer M. Drug-resistant malaria: Molecular mechanisms and implications for public health. FEBS Lett. 2011;585:1551–1562. doi: 10.1016/j.febslet.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. Artemisinin and artemisinin-based combination therapy resistance. at http://apps.who.int/iris/bitstream/10665/208820/1/WHO_HTM_GMP_2016.5_eng.pdf?ua=1 (2016).
- 16.Cooper Ra, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol. Microbiol. 2007;63:270–82. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 17.Straimer J, et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science (80-.) 2014;347:428–31. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbengue A, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oxford Nanopore Technologies. Nanopore Technologies. at https://nanoporetech.com/ (2017).
- 20.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–32. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quick J, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. bioRxiv. 2017;12:098913. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 23.Phillips LT, et al. Hand-held rapid whole genome nanopore sequencing to predict neisseria gonorrhoeae antibiotic susceptibility: steps towards clinic based tailored antimicrobial therapy. Sex Transm Infect. 2017;93:A56–A56. doi: 10.1136/sextrans-2016-052609. [DOI] [Google Scholar]
- 24.The ongoing battle against drug resistant malaria. at https://www.yourgenome.org/stories/the-ongoing-battle-against-drug-resistant-malaria (2016).
- 25.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servick, K. Amid patent lawsuit, genetic sequencing upstart unveils new technology. Science10.1126/science.aaf4165 at http://www.sciencemag.org/news/2016/03/amid-patent-lawsuit-genetic-sequencing-upstart-unveils-new-technology (2016).
- 27.Oxford Nanopore Technologies. New basecaller now performs ‘raw basecalling’, for improved sequencing accuracy. at https://nanoporetech.com/about-us/news/new-basecaller-now-performs-raw-basecalling-improved-sequencing-accuracy (2017).
- 28.Frith MC, Hamada M, Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LAST E-values. at http://last.cbrc.jp/doc/last-evalues.html (2008).
- 30.Karlsson E, Lärkeryd a, Sjödin a, Forsman M, Stenberg P. Scaffolding of a bacterial genome using MinION nanopore sequencing. Sci. Rep. 2015;5:11996. doi: 10.1038/srep11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loman NJ, Watson M. Successful test launch for nanopore sequencing. Nat. Methods. 2015;12:303–304. doi: 10.1038/nmeth.3327. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, J. Nanopolish at, https://github.com/jts/nanopolish.
- 33.Awasthi G, Satya GBK, Das A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem. Inst. Oswaldo Cruz. 2012;107:129–134. doi: 10.1590/S0074-02762012000100018. [DOI] [PubMed] [Google Scholar]
- 34.Zakeri, S. et al. Molecular assessment of atpase6 mutations associated with artemisinin resistance among unexposed and exposed Plasmodium falciparum clinical isolates to artemisinin-based combination therapy. Malar. J. 11, 373 (2012). [DOI] [PMC free article] [PubMed]
- 35.Phompradit, P., Wisedpanichkij, R., Muhamad, P., Chaijaroenkul, W. & Na-Bangchang, K. Molecular analysis of pfatp6 and pfmdr1 polymorphisms and their association with in vitro sensitivity in Plasmodium falciparum isolates from the Thai-Myanmar border. Acta Trop. 120, 130–135 (2011). [DOI] [PubMed]
- 36.Kwansa-Bentum B, et al. Plasmodium falciparum isolates from southern Ghana exhibit polymorphisms in the SERCA-type PfATPase6 though sensitive to artesunate In vitro. Malar. J. 2011;10:187. doi: 10.1186/1475-2875-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaidya AB, Mather MW. Atovaquone resistance in malaria parasites. Drug Resist. Updat. 2000;3:283–287. doi: 10.1054/drup.2000.0157. [DOI] [PubMed] [Google Scholar]
- 38.Goodman CD, et al. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science (80-.) 2016;352:349–353. doi: 10.1126/science.aad9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang HH, et al. Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci USA. 2013;110:20129–20134. doi: 10.1073/pnas.1319857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar. J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ménard D, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts L. Skirmishing over the scope of the threat. Science (80-.) 2016;352:403–403. doi: 10.1126/science.352.6284.403. [DOI] [PubMed] [Google Scholar]
- 43.Rahman MM, Ortega L, Rastogi MM, Thimasarn K. Antimalarial drug resistance. WHO Reg. Heal. Forum. 2011;15:52–56. [Google Scholar]
- 44.Aurrecoechea C, et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MalariaGEN P. falciparum Community Project. at https://www.malariagen.net/projects/p-falciparum-community-project (2016).
- 46.The Pf3K Project: pilot data release 4. at www.malariagen.net/data/pf3k-4 (2015).
- 47.Galinsky K, et al. COIL: a methodology for evaluating malarial complexity of infection using likelihood from single nucleotide polymorphism data. Malar. J. 2015;14:4. doi: 10.1186/1475-2875-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loman NJ, Quinlan AR. Poretools: A toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.