Abstract
The medial prefrontal cortex (mPFC), master regulator of higher-order cognitive functions, is the only brain region that matures until late adolescence. During this period, the mPFC is sensitive to stressful events or suboptimal nutrition. For instance, high-fat diet (HFD) feeding during adolescence markedly impairs prefrontal-dependent cognition. It also provokes multiple changes at the cellular and synaptic scales within the mPFC, suggesting that major transcriptional events are elicited by HFD during this maturational period. The nature of this transcriptional reprogramming remains unknown, but may include epigenetic processes, in particular microRNAs, known to directly regulate synaptic functions. We used high–throughput screening in the adolescent mouse mPFC and identified 38 microRNAs differentially regulated by HFD, in particular mir-30e-5p. We used a luciferase assay to confirm the functional effect of mir-30e-5p on a chosen target: Ephrin-A3. Using global pathway analyses of predicted microRNA targets, we identified biological pathways putatively affected by HFD. Axon guidance was the top-1 pathway, validated by identifying gene expression changes of axon guidance molecules following HFD. Our findings delineate major microRNA transcriptional reprogramming within the mPFC induced by adolescent HFD. These results will help understanding the contribution of microRNAs in the emergence of cognitive deficits following early-life environmental events.
Introduction
The medial prefrontal cortex (mPFC) is a complex and highly interconnected brain region critically implicated in the regulation of higher-order cognitive functions such as working memory, attention or behavioral flexibility1. One key feature defining this brain region is its remarkable maturational trajectory; unlike most other cortical areas, maturation in the prefrontal cortex continues until late adolescence, thus being the last brain region to achieve full maturity in humans2–4 and rodents5–7. Therefore, adolescence is a period of extensive remodeling in morphology, functional connectivity and gene expression patterns within the mPFC, and this protracted maturation is thought to confer an extended period of plasticity that supports experience-dependent learning7,8. At the same time, however, such plasticity is also thought to provide a basis for developmental disruption by early-life environmental insults9. Human and rodent studies both have shown that drug use or psychosocial stress associate with a higher risk of developing mental illness or behavioral dysfunctions when the exposure occurs during adolescence10–13. More recent work has shown that unhealthy nutrition during adolescence, in particular consumption of high-fat or high-sugar diets, also associates with deficits in executive functioning, and with a reduction in the volumes of frontal cortical regions in humans14–18. This is alarming as the quality of the human diet has deteriorated in the past few decades, now incorporating increasing levels of processed foods, artificial additives, refined sugars, and unhealthy dietary fats19,20, which may in turn negatively affect the mPFC. Adolescents in particular have a tendency to follow dietary guidelines less closely than adults21–24. Importantly, they are at a vulnerable time point in terms of nutritional training, when they begin making their own decisions about what to eat, yet are still influenced by peer-pressure and media which tend to favor less healthy nutritional options7,23–26.
In previous studies, our groups and others have shown that mice fed high-fat diets (HFD), or high-fat high-sugar diets, develop particularly potent prefrontal-dependent cognitive deficits when the dietary exposure begins during adolescence as compared to adulthood27–31. We also showed that adolescent HFD is associated with multiple and various changes at the cellular and synaptic scales within the mPFC, including modulation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-Methyl-D-aspartate (NMDA) neurotransmission, impairments in synaptic plasticity, or reductions in interneuron-specific protein levels. Notably, some of these neuronal alterations are not observed with a similar dietary exposure during adulthood. Such findings suggest that, at the molecular scale, there may be a number of transcriptional reprogramming events that occur in response to HFD in the mPFC during adolescence, which in turn would modulate expression levels of multiple neuronal proteins and affect mPFC function.
The nature of these transcriptional reprogramming events remains unknown, but could include epigenetic mechanisms that allow modifying gene activity without altering the DNA code. Indeed, accumulating evidence32–35 indicates that epigenetic factors represent a key mechanism linking early-life stress or suboptimal nutrition with changes in brain function36. For instance, early exposure to HFDs was shown to recruit such epigenetic regulatory machineries and to induce the appearance of metabolic abnormalities37,38; such effects could in turn also be valid for the regulation of synaptic and cognitive functions by HFD. The repertoire of epigenetic regulators is large and multi-layered, yet key candidates would include microRNAs (miRNAs), a family of small non-coding RNAs that regulate gene function by inhibiting the expression of their target mRNAs39,40. MiRNAs play important regulatory roles in a variety of cellular and subcellular functions41 and are now recognized as key modulators of dendritic and synaptic maturation and synaptic activity42–44, which in turn will modify cognitive performance45,46. Several studies have indeed characterized the contribution of prefrontal miRNAs to PFC-dependent tasks such as fear extinction and transition to alcohol addiction46,47. For instance, prefrontal mir-128b, whose expression dynamically changes across behavioral training in mice, was shown to regulate expression of several plasticity-related genes within the mPFC and to regulate behavioral performance on a mPFC-dependent task46.
It is thus highly feasible that prefrontal miRNAs could represent a significant link between early adolescent exposure to HFD and prefrontal-dependent cognitive dysfunctions, although such hypothesis has not been examined yet. We thus set out to identify global miRNA mapping within the mPFC in mice exposed to HFD since adolescence using high–throughput screening. We also used global pathway analyses of predicted miRNA targets with the aim of identifying novel biological pathways putatively affected by adolescent HFD. Finally, we validated these analyses using qPCR to confirm the emergence of miRNA-relevant gene expression changes within a top predicted biological pathway, namely Axon Guidance.
Materials and Methods
Animals
C57BL/6 N mice were used throughout the study. C57BL/6 N male and female breeding pairs obtained from Charles River (Sulzfeld, Germany) were maintained in our animal facility to generate a sufficient number of animals for the different experimental series. All animals were kept in groups (2–3 per cage) in a temperature- and humidity-controlled (21 ± 1 °C, 55 ± 5%) vivarium under a reversed light–dark cycle (lights off: 0800 to 2000h). Only male mice were included in all experiments to avoid potential confounds arising from sexual dimorphism. All procedures were approved by the Cantonal Veterinary Office of Zurich and are in agreement with the principles of laboratory animal care in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication No. 86–23, revised 1985).
Chronic HFD and CD feeding
Experimental diets included a HFD consisting of 60% of calories from fat or a control diet (CD) composed of 10% of calories from fat (SSNIFF Diets, Germany). Diets and water were always accessible ad libitum throughout all experiments. Mice had access to HFD or CD starting from postnatal day (PND) 28 for 8 weeks, and body weights were measured throughout. Hence, animals were exposed to HFD or CD throughout adolescent development, covering pre-pubertal and post-pubertal stages of maturation7 (for more detail see Supplementary Information).
Four cohorts of mice were included in the study: experimental series 1 (Cohorts 1 and 2) aimed at assessing behavioral phenotypes of HFD and control mice, while experimental series 2 (Cohort 3) and 3 (Cohort 4) determined changes of miRNA expression, and gene expression, respectively, in naïve animals so as to avoid the possible confounding effects of repeated behavioral testing. Behavioral and molecular analyses were conducted in adulthood.
Spatial working memory in the Y-maze
Working memory is a special short-term memory buffer used to hold relevant information temporarily active in order to guide on-going behavior48. The Y-maze apparatus has been extensively described previously28, and the test procedure is described in the Supplementary Information.
Discrimination reversal learning in the water T-maze
The apparatus and the test procedure are described in detail in the Supplementary Information.
Briefly, during the acquisition training, the animals were required to learn to discriminate the left and right goal arms, with only one of them leading to an escape platform hidden at the far end (right arm for half of the animals; left arm for the other half). A first habituation session was then followed by 6 trials per day sessions, conducted at an intertrial interval of 5 min. Acquisition training continued until an animal had reached criterion performance of 10 correct responses across 2 consecutive days (i.e., 10 correct out of 12 trials). Upon reaching the acquisition criterion, the location of the platform was moved to the other, previously incorrect, arm to assess reversal learning. Reversal training continued until an animal reached criterion performance once again. The percentage of correct arm choices and the errors to criterion were recorded manually and calculated for each animal during acquisition and reversal training.
RNA Purification
Animals from Cohort 2 were sacrificed after an 8-week exposure to HFD or CD. Brains were immediately extracted from the skull and placed dorsal side up on an ice-chilled plate. The medial PFC was dissected as previously established and fully described elsewhere49. Analyses were performed on mPFC tissue (Bregma: +2.3 to +1.3) that included both hemispheres of anterior cingulate, prelimbic, and infralimbic subregions. Brain specimens were collected in 96-well microtiter plates kept on dry ice and allowed to freeze before storage at −80 °C until further use.
Total RNA was isolated using the Qiagen miRNeasy Mini kit (Qiagen, Italy) according to the manufacturer’s instructions, and quantified by spectrophotometric analysis. An aliquot of each sample was then treated with DNase to avoid DNA contamination. The Qiagen miRNeasy Mini kit is optimized for isolating total RNA, including all RNA molecules from 18 nucleotides upwards (miRNAs).
Genome wide microarray analyses of prefrontal miRNAs
Whole genome microRNA analysis (GEO accession number GSE105794) was performed using the Flash Tag Biotin HSR RNA Labeling kit (Affymetrix, Italy), and the miRNA 4.1 array strips (Affymetrix), according to the manufacturer’s protocol. After hybridization, each strip was washed using the Affymetrix Fluidics Station and then scanned in the Affymetrix Imaging station to obtain. CEL files that were then used for further bioinformatics analyses. Affymetrix CEL files were imported into Partek Genomics Suite version 6.6 for data visualization, statistical testing and quality control assessment. All the samples passed the quality criteria for hybridization controls, labeling controls and 3′/5′ Metrics. Background correction was conducted using Robust Multi-strip Average (RMA)50 and normalization was conducted using Quantiles Normalization51. Subsequently, a summarization step was conducted using a linear median polish algorithm52 to integrate probe intensities in order to compute the expression levels for each gene transcript.
Statistical analyses were performed using the Robust MultiChip Average ANOVA statistical test to assess treatment effects. Differential miRNA expression was assessed by applying a p-value filter (for attribute) of p < 0.05 to the ANOVA results. To investigate the effect of HFD, we performed a linear contrast between HFD vs. CD. In this comparison, a maximum filter of p < 0.05 and a minimum absolute fold change cut-off of 1.2 was applied. All pre-miRNAs (i.e. immature forms of miRNAs) were excluded from further analysis. This yielded 38 dysregulated miRNAs.
Bioinformatics analyses
In order to identify the potential gene targets of single miRNAs that were observed to be differentially expressed in HFD vs. CD, two different bioinformatics databases were used jointly: TargetScanMouse (www.targetscan.org) and miRWalk, (www.umm.uni-heidelberg.de/apps/zmf/mirwalk). As the different bioinformatics tools identified different target genes for each miRNA, we performed an overlap of the results obtained by TargetScan and miRwalk for each selected miRNA, which yielded a list of predicted targets that were identified by both TargetScan and miRwalk. MiRNAs that were not annotated in these two databases were excluded from further pathway analysis, yielding a predicted target list for 9 final miRNAs. We then performed single miRNA pathway analysis (using the predicted target gene list identified above by TargetScan and miRWalk) with Ingenuity Pathway Analysis software (Ingenuity Systems, www.ingenuity.com).
In addition, top canonical pathways predicted to be affected by the combinatory of all significantly dysregulated (and annotated) miRNAs (N.B. mir-129b was excluded from this analysis, because it was not annotated in mirPath) were assessed with mirPath software (DIANA TOOLS, 2014)53.
Validation of regulated miRNAs by qRT-PCR
Affymetrix results were validated by performing qRT-PCR on selected candidate miRNAs. cDNA was synthesized by using 10 ng of total RNA with TaqMan-specific RT primers (TaqMan MicroRNA Assay, Applied Biosystems) and a TaqMan miRNA reverse transcription kit (Applied Biosystems) according to the manufacturers’ instructions. Thereafter, quantitative real-time PCR was performed using predesigned assays (TaqMan MicroRNA Assays - Applied Biosystems). PCR reactions were performed as follows: 50 °C for 2 minutes, 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. Relative target gene expression was calculated according to the 2(-Delta Delta C(T)) method, using RnU6 as internal standard. The samples and standard curves were analyzed in triplicate. miRNA fold change data obtained by qRT-PCR was then correlated against miRNA fold change obtained by microarray on a selected 9 miRNAs that included both upregulated and downregulated miRNAs.
Luciferase assay
The reporter plasmid was constructed by inserting Ephrin A3 (EFNA3) mRNA 3′UTR (NM_010108.1) downstream of the Firefly luciferase gene in the pEZX-MT06 vector (GeneCopoeia, Rockville, USA). HEK293T or HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and Pen-Strep 1x (Gibco) and plated 24 h before transfection in 96-well plates at 10e5 cells/well. Cells were transfected with 100 ng of pEZX-MT06/EFNA3-3′UTR or pEZX-MT06 and 10 pmol of mir-30e-5p (#4464066) and negative control (#4464058) miRNAs mimics (mirVana™, Life Technologies, Zug, Switzerland) per well using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions. Two wells of each specific condition were transfected to obtain a mean luciferase activity. All the experiments were performed in triplicate (3 assays for HEK293T and 3 assays for HeLa cells). Luciferase activity was measured 24 h after transfection using the Luc-Pair Duo-Luciferase Assay Kit 2.0 (GeneCopoeia) in a Luminometer (Dynex Magellan Biosciences) according to the manufacturer’s instructions. Normalization of the data included two sequential steps. (1) normalization of Firefly luciferase activity to Renilla luciferase activity (transfection control); (2) normalization against (a) the cellular effects (e.g., RNA binding proteins, endogenous miRNAs, etc.) on luciferase activity and (b) against the exogenous effects of mir-30e-5p on the luciferase coding sequences (see Supplementary Methods for more details and54.
Quantitative Real-Time RT-PCR Analyses
mRNA levels of specific genes from the Axon Guidance pathway were quantified by SYBR Green qRT-PCR (CFX384 real-time system, Bio-Rad Laboratories) using the SsoAdvanced Universal SYBR Green supermix (Bio-Rad Laboratories), following retrotranscription with the iScript cDNA synthesis kit (Bio-Rad Laboratories), as described in the Supplementary Information.
Statistical analyses
All data were analyzed by Student’s t tests or by parametric analysis of variance (ANOVA) followed by Fisher’s least significant difference (LSD) post-hoc comparisons whenever appropriate. Statistical significance was set at p < 0.05. All statistical analyses were performed using the statistical software StatView (version 5.0), unless otherwise specified.
In the first experimental series (cohort 1), percent time in the novel arm was analyzed using an independent Student’s t test (two-tailed). Percent correct trials in the acquisition phase of the T-maze, and in the reversal phase, were analyzed using a 2 × 4 (diet × days) parametric ANOVA and 2 × 6 (diet × days) parametric ANOVA, respectively.
In the second experimental series (cohort 2), fold change in miRNA levels obtained by microarray data was analyzed using an overall ANOVA within the Affymetrix software, as described in the previous paragraph. Fold change in mir-30e-5p levels obtained by qRT-PCR analyses was analyzed using a Student’s t test (two-tailed). Correlative analyses between mean fold change obtained by qRT-PCR and miRNA fold change obtained by microarray were performed using Pearson’s product moment correlations on 9 selected miRNAs. The luciferase gene reporter assay was analyzed using a one-sample t-test to determine whether percent change in luciferase activity due to mir-30e-5p/EFNA3-3′UTR was significantly different from zero.
In the third experimental series (cohort 3), relative gene expression of each target gene was analyzed using a repeated measures 2 × 8 (diet × genes) ANOVA followed by a priori post-hoc comparisons.
Data availability
The datasets pertaining to the miRNA profiling of the prefrontal cortex are available in the GEO repository (accession number GSE105794). The remaining datasets are available from the corresponding author on request.
Results
Chronic HFD throughout adolescence leads to cognitive disturbances
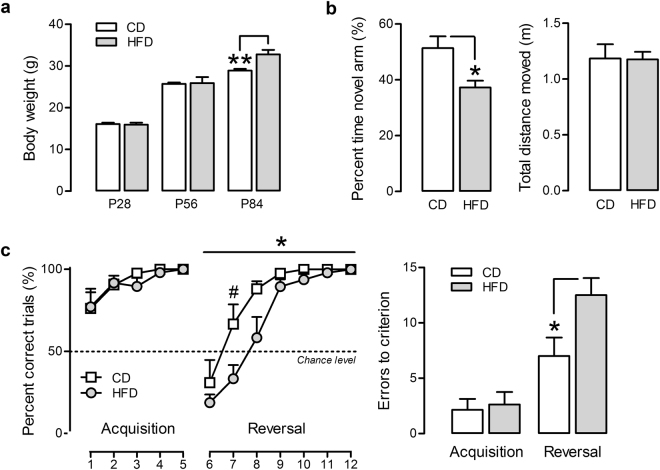
As expected, chronic HFD led to an increase in body weight over time compared to control CD animals, as indicated by a significant effect of diet following 8 weeks on CD or HFD (F(1,28) = 11.33, p < 0.01), following a significant interaction between time and diet (F(2,56) = 6.77, p < 0.01) (Cohorts 3 and 4) (Fig. 1a). We then investigated the effects of HFD on different cognitive paradigms that depend on the integrity of the mPFC. Spatial novelty recognition memory in the Y-maze is a behavioral test that uses the natural tendency of rodents to explore novel over familiar spatial environments and that relies on (dorsal) hippocampal55,56 and prefrontal57–60 circuits. As shown in Fig. 1b, HFD throughout adolescence disrupted novel recognition as indexed by the percent time spent in the novel arm (t(13) = 8.96, p < 0.05). Whilst CD control offspring exhibited a score of ∼51%, the recognition score of HFD-fed mice fell close to chance level (39%). These effects were not confounded by potential changes in locomotor activity, as total distance moved was unchanged by HFD (sample phase) (Fig. 1b).
Figure 1.
Cognitive effects of adolescent high fat diet (HFD). (a) Body weight of HFD and low fat diet (CD) animals at postnatal (P) day 28 (start of HFD), 56 and 84. **p < 0.01 at P84 by post-hoc comparison following significant time x diet interaction (*p < 0.01). n = 15 per diet (b) Left panel: Working memory indexed by the percent time spent in the novel arm during a Y-maze spatial recognition test. *p < 0.05, based on a Student’s t-test analysis. Right panel: Locomotor activity indexed by the total distance moved in Phase 1 of the test. n = 6–8 per diet (c) Cognitive flexibility assessed in a water T-maze left-right discrimination task. Left panel: Number of percent correct trials during the acquisition and reversal phase. *p < 0.05 indicating a main effect of diet in the reversal phase; #p < 0.05 on trial 2 of the reversal phase, based on Fisher’s LSD post-hoc comparisons following the presence of a significant diet × trial interaction (p < 0.05). Right panel: Total errors to criterion in the two phases. *p < 0.05 indicating a main effect of diet in the reversal phase. n = 7–8 per diet. All data are means ± S.E.M.
We then tested the effects of HFD on discrimination reversal learning, a behavioral task that is known to be highly dependent on prefrontal cortex functioning61–63. No changes in performance were detected in the acquisition phase of the test, in which percent correct trials and total errors to criterion were similar in both treatment groups. A significant impairment in discrimination reversal learning was however detected in HFD vs. CD mice, as revealed by a main effect of diet (F(1,13) = 5.83, p < 0.05) as well as a significant diet × trial interaction (F(6,78) = 2.34, p < 0.05). Post-hoc comparisons confirmed that performance of HFD animals was decreased on the second trial of the reversal learning phase (F(1,13) = 5.39, p < 0.05) with a trend on the third trial (F(1,13) = 4.37, p = 0.057). This was further corroborated by the presence of a significant difference in the total errors to criterion between CD and HFD mice during the reversal phase of the test (t(13) = 5.83, p < 0.05) (Fig. 1c).
Cognitive deficits after adolescent HFD associate with changes in the expression of miRNAs in the prefrontal cortex
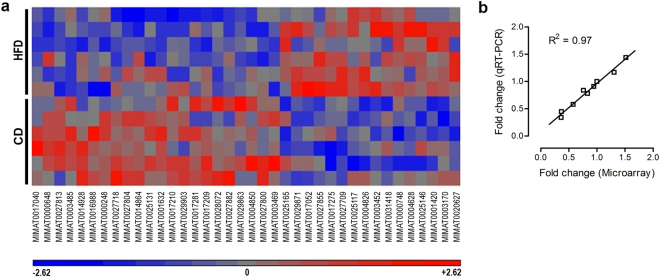
Next, we used genome wide assessment of miRNA expression in order to identify possible alterations in miRNA levels after chronic adolescent HFD. As listed in Table 1, we identified 38 mature miRNAs that were differentially expressed in the mPFC (fold change cutoff: ±1.2%; p < 0.05), including 22 significantly downregulated and 16 upregulated miRNAs. Figure 2a shows hierarchical clustering of miRNA expression changes induced by HFD.
Table 1.
List of significantly affected miRNAs in the PFC induced by adolescent high fat diet (HFD) treatment using a genome wide assessment of miRNA expression by Affymetrix microarray.
| Accession Number | Transcript ID | p-value | Fold-Change (HFD vs. CD) |
|---|---|---|---|
| Significantly upregulated miRNAs - n = 16 | |||
| MIMAT0027855 | mmu-miR-6976–3p | 0.0429968 | 1.20323 |
| MIMAT0025165 | mmu-miR-6412 | 0.0423496 | 1.20987 |
| MIMAT0020627 | mmu-miR-5119 | 0.0296401 | 1.21053 |
| MIMAT0025117 | mmu-miR-6373 | 0.0136909 | 1.21548 |
| MIMAT0003170 | mmu-miR-541-5p | 0.0324367 | 1.21786 |
| MIMAT0000748 | mmu-miR-383-5p | 0.0117387 | 1.22856 |
| MIMAT0004628 | mmu-miR-21a-3p | 0.0281818 | 1.25431 |
| MIMAT0029871 | mmu-miR-7678-3p | 0.0202582 | 1.25933 |
| MIMAT0031418 | mmu-miR-8112 | 0.0389069 | 1.26337 |
| MIMAT0001420 | mmu-miR-433-3p | 0.0145485 | 1.27067 |
| MIMAT0004826 | mmu-miR-146b-3p | 0.0128916 | 1.27534 |
| MIMAT0027709 | mmu-miR-6904-3p | 0.0035105 | 1.30778 |
| MIMAT0025146 | mmu-miR-6395 | 0.0411032 | 1.32443 |
| MIMAT0003452 | mmu-miR-678 | 0.0369783 | 1.32879 |
| MIMAT0017275 | mmu-miR-467c-3p | 0.0260913 | 1.41821 |
| MIMAT0017052 | mmu-miR-210-5p | 0.0311217 | 1.51463 |
| Significantly downregulated miRNAs - n = 22 (1/2) | |||
| MIMAT0001632 | mmu-miR-451a | 0.00716281 | −1.73801 |
| MIMAT0017281 | mmu-miR-511-3p | 0.0232533 | −1.67302 |
| MIMAT0029863 | mmu-miR-129b-3p | 0.032611 | −1.64229 |
| MIMAT0014928 | mmu-miR-344c-3p | 0.0336578 | −1.58691 |
| MIMAT0017209 | mmu-miR-541-3p | 0.0216328 | −1.53285 |
| MIMAT0003469 | mmu-miR-690 | 0.00618111 | −1.52981 |
| MIMAT0027718 | mmu-miR-6909-5p | 0.0147535 | −1.46981 |
| MIMAT0017040 | mmu-miR-350-5p | 0.0245193 | −1.34234 |
| MIMAT0014864 | mmu-miR-3078-5p | 0.0416764 | −1.33395 |
| Significantly downregulated miRNAs (2/2) | |||
| MIMAT0003485 | mmu-miR-455-5p | 0.00250818 | −1.31711 |
| MIMAT0004850 | mmu-miR-883b-5p | 0.0340633 | −1.31545 |
| MIMAT0027804 | mmu-miR-6952-5p | 0.0158051 | −1.30762 |
| MIMAT0027800 | mmu-miR-6950-5p | 0.0299551 | −1.29758 |
| MIMAT0025131 | mmu-miR-6385 | 0.0406697 | −1.283 |
| MIMAT0017210 | mmu-miR-547-5p | 0.0313436 | −1.26247 |
| MIMAT0000648 | mmu-miR-10a-5p | 0.0437868 | −1.25814 |
| MIMAT0016988 | mmu-miR-144-5p | 0.0121198 | −1.25327 |
| MIMAT0027813 | mmu-miR-6956-3p | 0.00668782 | −1.25066 |
| MIMAT0027882 | mmu-miR-6990-5p | 0.0367154 | −1.24339 |
| MIMAT0029903 | mmu-miR-7687-3p | 0.0348489 | −1.23791 |
| MIMAT0028072 | mmu-miR-7083-5p | 0.0130252 | −1.22978 |
| MIMAT0000248 | mmu-miR-30e-5p | 0.0303815 | −1.20456 |
Using a fold change cutoff value of ±1.2% and a p-value cut-off value of p < 0.05, 38 mature miRNAs were identified as being differentially expressed in the PFC. n = 6 per dietary treatment.
Figure 2.
Hierarchical clustering of significantly regulated miRNAs in the PFC in adolescent high fat diet (HFD)- and low fat diet (CD)-treated mice and qRT-PCR validation. (a) Hierarchical clustering diagram revealing significantly regulated miRNAs using a genome wide assessment of miRNA expression by Affymetrix microarray; the analysis comprised all mature miRNAs presented in Table 1, and which were identified using a fold change cut-off value of ±1.2% and a p-value cut-off value of p < 0.05. This analysis included 38 mature miRNAs: 16 significantly upregulated and 22 downregulated miRNAs. (b) Pearson’s product moment correlations revealing a highly significant positive correlation between miRNA fold changes obtained by qRT-PCR and by microarray (r = 0.97, df = 7, p < 0.001) on a selected 9 miRNAs that spanned expression fold change levels from 0.3 to 1.5, thus providing statistical confirmation for the validity of microarray results. n = 6 per diet.
We validated microarray results using qRT-PCR on 9 selected miRNAs that spanned expression fold change levels from 0.3 to 1.5. Pearson’s product-moment correlations revealed a highly significant positive correlation between miRNA fold changes obtained by qRT-PCR and by microarray (r = 0.97, df = 7), n = 9, p < 0.001), thus providing statistical confirmation for the validity of the microarray results (Fig. 2b).
Adolescent HFD reduces the expression of mir-30e-5p, a miRNA with predicted gene targets involved in axon guidance and cognition
In order to identify the potential gene targets of single miRNAs we found to be differentially expressed in HFD vs CD, we performed an overlap of two different bioinformatics databases jointly: TargetScanMouse and miRWalk, that yielded a predicted target list (Supplementary Table 2) derived from the overlap of those gene lists for each miRNA that was annotated in both softwares. We then performed single miRNA Ingenuity Pathway Analysis (IPA) generating a canonical pathway analysis for each miRNA (data not shown).
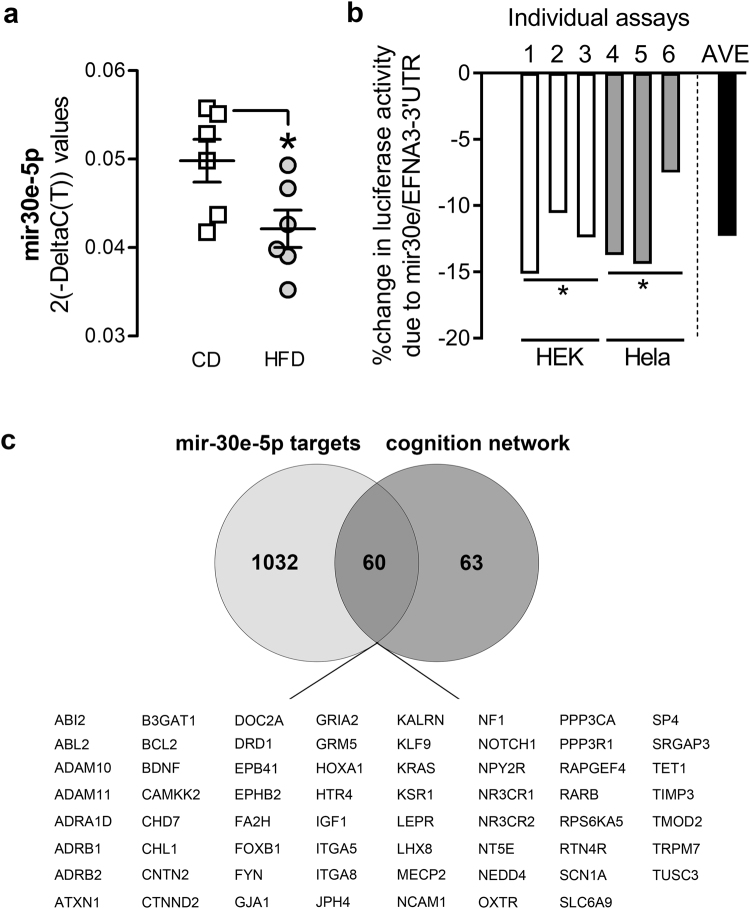
Among the miRNAs that we analyzed, miR-30e-5p was particularly interesting when considering the association between HFD and cognition. Both microarray (Fig. 2a, see MIMAT0000248) and qRT-PCR (Fig. 3a, F(1,10) = 5.75, p < 0.05) analyses revealed that miR-30e-5p was significantly downregulated by adolescent HFD. Among the top-5 significant canonical pathways revealed by IPA analysis, 3 were related to neuronal functions, namely Axon guidance Signaling, Insulin Growth-Factor 1 (IGF 1) Signaling and Nerve Growth-Factor (NGF) Signaling (Table 2). We first aimed at providing a proof-of-principle validation for the predicted targets of miR-30e-5p. We performed a luciferase gene reporter assay so as to evaluate the effects of miR-30e-5p on the expression of Ephrin A3 (EFNA3), an axon guidance molecule. We focused on EFNA3 because it was one of three axon guidance genes (top-1 canonical pathway) that was a predicted target of mir-30e-5p. The EFNA3 molecule itself also has a demonstrated role in cognition and behavior based on previous studies64,65 and regulation of EFNA3 expression is known to depend on microRNA-mediated mechanisms66. We found that mir-30e-5p led to a significant downregulation of the EFNA3 gene found in both HEK cells and HeLa cells (t(2) = 9.55, p < 0.05 and t(2) = 5.42, p < 0.05); Fig. 3b and Supplementary Table 3), thus in line with the predictions of the bioinformatics analyses.
Figure 3.
Targets and expression regulation of mir-30e-5p in the PFC induced by adolescent high fat diet (HFD) (a) Downregulation of mir-30e-5p expression levels, illustrated by the significantly reduced 2(-Delta(C(T)) values and revealed by a Student’s t-test analysis: *p < 0.05. n = 6 per diet. (b) Luciferase reporter gene assay using mir-30e-5p and EFNA3 3′UTR. Mir-30e-5p leads to a downregulation of EFNA3 (average 12% reduction in normalized luciferase activity. *P < 0.05, based on a one-sample t-test in HEK and in HeLa cells assessing difference of the mean from zero. 2 biological replicates/assay (6 assays). (c) Overlap analysis of mir-30e-5p molecular targets and genes belonging to the Ingenuity Pathway Analysis (IPA) ‘cognition’ network, revealing that miR-30e-5p is the first upstream regulator of the IPA ‘cognition’ network (60 genes out of 63).
Table 2.
Ingenuity Pathway Analysis (IPA) for mir-30e-5p, a significantly downregulated miRNA in the PFC induced by adolescent high fat diet (HFD).
| Name | p-Value | # Molecules |
|---|---|---|
| Canonical Pathways | ||
| Axon guidance Signaling | 3.88E-11 | 57/432 |
| IGF-1 Signaling | 1.66E-06 | 18/97 |
| NGF Signaling | 2.73E-05 | 17/107 |
| PKCΘ Signaling in T Lymphocytes | 2.82E-05 | 18/118 |
| Cardiac Hypertrophy Signaling | 7.11E-05 | 26/223 |
| Diseases and Disorders | ||
| Cancer | 1.91E-24−4.60E-04 | 809 |
| Gastrointestinal Disease | 1.61E-17−4.59E-04 | 425 |
| Organismal Injury and Abnormalities | 6.23E-11−4.59E-04 | 419 |
| Reproductive System Disease | 6.23E-11−4.01E-04 | 360 |
| Neurological Disease | 3.32E-09–4.79E-04 | 288 |
| Physiological System Development and Function | ||
| Organismal Survival | 2.77E-21–4.85E-04 | 278 |
| Behavior* | 2.25E-15–3.08E-04 | 141 |
| Nervous System Development and Function | 4.41E-15–4.75E-04 | 277 |
| Tissue Development | 4.41E-15–4.75E-04 | 361 |
| Organismal Development | 7.60E-15–4.75E-04 | 258 |
| *Behavior: Top affected functions | ||
| SubCategories | Diseases or Functions Annotation | p -Value |
| Behavior | Behavior | 2.25E-15 |
| Behavior | Cognition | 2.62E-11 |
| Behavior | Learning | 2.67E-10 |
| Behavior, Nervous System Development and Function | Memory | 5.40E-09 |
IPA analysis: (i) revealed a significant connection between the predicted targets of miR-30e-5p and three neuronal-related pathways, namely Axon guidance Signaling, IGF-1 Signaling and NGF Signaling, (ii) reported ‘behavior’ as the second top biological function affected by the targets of miR-30e-5p, (iii) indicated that targets of miR-30e-5p are implicated in cognition, learning and memory functions and (iv) revealed that miR-30e-5p is the first upstream regulator of the IPA cognition network by which 60 genes (out of 63) are predicted target molecules of miR-30e-5p (targets detailed in Fig. 3).
Furthermore, when further investigating IPA analysis for miR-30e-5p, we found that, although there is no functional evidence in the literature or experimental data regarding the correlation between miR-30e-5p and cognition, IPA reported ‘behavior’ as the second top biological function affected by the targets of miR-30e-5p (Table 2). Further detailed analysis of individual functions belonging to the behavioral network indicated that targets of miR-30e-5p are significantly implicated in cognition and learning and memory functions. Moreover, the IPA software indicated miR-30e-5p as the first upstream regulator of target-genes implicated in cognition (“IPA cognition network”), as 60 genes (out of 63) from the IPA cognition network were predicted target molecules of miR-30e-5p. Predicted targets of miR-30e-5p, which also belong to the cognition network, are reported in Fig. 3c. MiR-30e-5p thus represents a very interesting miRNA that might be implicated in some of the cognitive deficits induced by adolescent HFD.
Global pathways analysis of all HFD-regulated miRNAs identifies biological pathways involved in brain related functions
To further identify biological events putatively affected by global miRNome changes after adolescent HFD, we used mirPath software (DIANA TOOLS) (Vlachos et al.53) on a combinatory of all significantly dysregulated miRNAs. To this purpose, only annotated miRNAs (i.e. miRNAs with known targets) were used and included 23 different miRNAs as listed in Supplementary Table 4. Such analysis allowed the identification of the top 20 canonical pathways predicted to be affected by HFD via miRNAs-related mechanisms, as shown in Table 3. Different disease-related pathways (particularly cancer-related) that are composed of a number of more specific signaling pathways were omitted from the pathway list as performed by others67, to avoid any bias in the interpretation of pathway analyses. Interestingly, Axon guidance (p = 1.3 E-25) yielded the highest score, with 51 genes in the pathway being potential targets of the listed miRNAs. Pathways with important roles for central nervous system (CNS) such as intracellular signaling (mTOR Signaling, PI3K-Akt Signaling, MAPK Signaling, Neurotrophin Signaling, Wnt Signaling), assembly and organization of nervous tissue (e.g., Regulation of Actin Cytoskeleton, Focal Adhesion, Gap Junction) and glutamatergic-related synaptic signaling (Long-Term Depression, Glutamatergic Synapse) resulted as the most significant. Finally, pathway analysis also included immune-related pathways (T-cell Receptor Signaling, TGF-beta Signaling, B-cell Receptor Signaling) and pathways related to protein processing (Ubiquitin Mediated Proteolysis, Protein Processing in Endoplasmic Reticulum) (see Table 3).
Table 3.
DIANA Pathway Analysis of significantly regulated miRNAs in the PFC induced by adolescent high fat diet (HFD).
| # | KEGG pathway | p-value | #genes | #miRNAs |
|---|---|---|---|---|
| 1 | Axon guidance (mmu04360) | 1.29E-25 | 51 | 15 |
| 2 | mTOR signaling pathway (mmu04150) | 7.44E-18 | 27 | 12 |
| 3 | ErbB signaling pathway (mmu04012) | 6.50E-13 | 31 | 11 |
| 4 | T cell receptor signaling pathway (mmu04660) | 2.86E-12 | 34 | 14 |
| 5 | PI3K-Akt signaling pathway (mmu04151) | 2.86E-12 | 79 | 15 |
| 6 | Regulation of actin cytoskeleton (mmu04810) | 1.11E-11 | 56 | 15 |
| 7 | MAPK signaling pathway (mmu04010) | 5.98E-10 | 62 | 17 |
| 8 | Ubiquitin mediated proteolysis (mmu04120) | 3.65E-09 | 39 | 13 |
| 9 | Neurotrophin signaling pathway (mmu04722) | 4.86E-09 | 34 | 14 |
| 10 | Focal adhesion (mmu04510) | 4.93E-09 | 49 | 15 |
| 11 | Gap junction (mmu04540) | 1.17E-07 | 23 | 11 |
| 12 | TGF-beta signaling pathway (mmu04350) | 1.93E-07 | 26 | 9 |
| 13 | Aldosterone-regulated sodium reabsorption(mmu04960) | 2.49E-07 | 14 | 8 |
| 14 | B cell receptor signaling pathway (mmu04662) | 2.49E-07 | 23 | 13 |
| 15 | Long-term depression (mmu04730) | 6.26E-07 | 19 | 9 |
| 16 | Glutamatergic synapse (mmu04724) | 1.22E-06 | 30 | 13 |
| 17 | Wnt signaling pathway (mmu04310) | 2.47E-06 | 38 | 16 |
| 18 | Osteoclast differentiation (mmu04380) | 2.75E-06 | 3 | 12 |
| 19 | Protein processing in endoplasmic reticulum(mmu04141) | 3.01E-06 | 39 | 17 |
| 20 | Insulin signaling pathway (mmu04910) | 3.78E-06 | 33 | 11 |
Analysis was conducted using mirPath software (DIANA TOOLS) (Vlachos et al.53) whereby only annotated miRNAs were used. The 23 annotated miRNAs are listed in Supplementary Table 4. Such analysis allowed identifying the top 20 canonical pathways predicted to be affected by HFD via miRNA mechanisms. Different disease-related pathways (particularly cancer-related) were omitted.
Gene expression analyses confirm downregulation of Axon guidance molecules by adolescent HFD
To provide experimental validation for our bioinformatics analyses, we then decided to evaluate whether the expression levels of genes involved in Axon Guidance signaling, which resulted to be the most significant pathway affected by the modulated miRNAs, were indeed affected by HFD. To do so, we performed qRT-PCR on prefrontal tissue of HFD and CD animals on selected genes involved in this pathway.
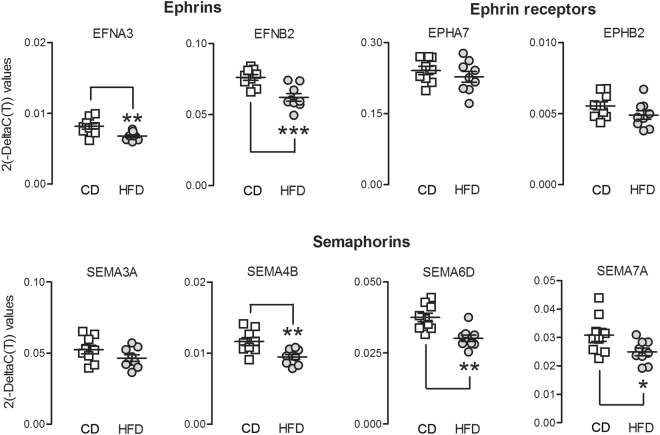
As outlined in Supplementary Table 5, 15 miRNAs altered by HFD had one or several mRNA targets within the Axon guidance pathway, totaling up to 51 genes within this pathway. To this end, we chose to analyze Axon guidance target genes predicted by our miRNA target analyses (DIANA tools) (represented in Supplementary Figure 1). We focused our analyses on Semaphorins and Ephrins (including Ephrin receptors), as these Axon guidance families were both strong targets predicted by the pathway analyses. A repeated-measures ANOVA over all target genes first confirmed a global impact of HFD on Axon guidance gene expression, illustrated by a significant main effect of diet (F(1,16) = 16.56, p < 0.001) but no significant diet × gene interaction. Using a priori post-hoc analyses to identify which genes were specifically affected, we found that Ephrin A3 (F(1,16) = 10.05, p < 0.01), Ephrin B2 (F(1,16) = 16.98, p < 0.001), Semaphorin 4B (F(1,16) = 11.03, p < 0.01), Semaphorin 6D (F(1,16) = 15.35, p < 0.01) and Semaphorin 7A (F(1,16) = 5.26, p < 0.05) were significantly downregulated by HFD as compared to controls by an average 15%, while Ephrin Receptor A7, Ephrin Receptor B2 and Semaphorin 3A remained unaffected by the manipulation (Fig. 4). These analyses thus provide a proof of concept to suggest that miRNAs significantly affected by HFD may have a functional impact on gene expression, through a downregulation of some, but not all, genes within the Axon guidance pathway.
Figure 4.
Gene expression changes of axon guidance molecules induced by high fat diet (HFD) (a) Gene expression levels of Ephrins A3 and B2 indexed by fold change in HFD vs. low fat diet (CD) treatments. **p < 0.01 and ***p < 0.001 by a priori post-hoc comparison (b) Gene expression levels of Ephrin receptors A7 and B2 (c) Gene expression levels of Semaphorins 3A, 4B, 6D and 7A. **p < 0.01 by a priori post-hoc comparison. n = 9 per diet. All data are means ± S.E.M.
Discussion
Despite the well-established effects of HFD on higher-order cognition68, studies had thus far essentially focused on the hippocampus, whereas the effects on the mPFC remained less well understood. Recent work, however, has shown that HFD can indeed affect various forms of cognitive functions that depend on the mPFC27–31. In particular, these studies have shown remarkable effects when HFD begins during adolescence, thus emphasizing that the maturing mPFC is particularly sensitive to such dietary treatments. Interestingly, these investigations have shown that HFD can affect a variety of synaptic and cellular functions within the mPFC, including interneuron function and glutamatergic neurotransmission and plasticity; yet changes at the molecular level within the mPFC remain thus far poorly characterized. In particular, given the functional changes induced by HFD within the mPFC, it would be important to identify potential regulatory mechanisms underlying such changes at the transcriptional level.
The present study first confirmed that chronic HFD consumption throughout adolescent development in mice leads to marked abnormalities in a number of cognitive tasks that are partly dependent on prefrontal functioning such as working memory and discrimination reversal learning. Our study then aimed at assessing potential changes within the prefrontal miRNome induced by HFD feeding, as miRNAs represent important transcriptional regulators39,40, which could potentially link HFD exposure, mPFC gene expression and, eventually, cognitive dysfunctions. In this study, we provide for the first time a global miRNA profiling of the mPFC induced by HFD feeding, showing that HFD leads to an overall remodeling of the prefrontal miRNome. Our study thus substantially extends previous reports by identifying a potential novel mechanism involved in the development of HFD-induced cognitive dysfunctions. MiRNAs are small 22-nucleotide non-coding RNAs that mediate post-transcriptional silencing of gene expression in a sequence specific manner. The capacity of single miRNAs to target different mRNAs makes them essential regulators of a variety of cellular functions41. Importantly, miRNAs represent critical mediators of a number of synaptic events such as dendritic subcellular localization, arborization and synapse formation and maturation42–44 that in turn are essential for information processing during cognitive tasks45,46. MiRNAs have also been identified as key epigenetic molecules that can translate environmental disturbances into lasting changes in cellular expression levels, as has been shown in a number of studies involving adolescent exposure to alcohol or cannabis69,70. Our results are thus in line with the current understanding that miRNAs could represent important cellular nodes that modulate gene expression following early life events to generate anomalies in cognitive functioning.
Although we did not investigate the cognitive implication of miRNA changes after HFD, we identify a number of miRNAs as potential targets for future studies. Of the 38 significantly affected miRNAs (Table 1 and Fig. 2a), mir30e-5p, mir-433-3p and mir-690 are particularly interesting because they are predicted regulators of three biological pathways with essential roles for proper neural functioning, namely Axon guidance, Ephrin Receptor Signaling and Neurotrophin Signaling. In particular, given its strong (predicted) capacity to regulate the expression of numerous genes implicated in cognition (see Fig. 3), mir-30e-5p emerges as one of the most interesting candidate regulators of gene expression and cognitive function after HFD exposure. Interestingly, mir-30e-5p was shown to be similarly down-regulated in an animal model of temporal lobe epilepsy71, whereas mir-30e-5p precursor variants were shown to associate with schizophrenia72,73, supporting the relevance of this miRNA for neurological and neuropsychiatric disease. Examining the functional role of mir-30e-5p in the emergence of HFD-induced cognitive abnormalities would thus represent an interesting extension of the present study and a potential promising avenue for the understanding of cognitive deficits in diseases with prefrontal anomalies.
Furthermore, our study delineates molecular pathways putatively affected by miRNA events, some of which are critically involved in cognitive function. Notably, our results indicate that Axon guidance is the top-1 predicted pathway targeted by miRNAs after HFD. Importantly, we also identified multiple gene expression changes within Axon guidance molecules, in particular ephrins and semaphorins, thus providing proof-of-concept data suggesting that individual miRNA molecules affected by HFD may have functional effects on gene expression. We also provided a direct validation that mir-30e-5p (mentioned above) downregulates one of its predicted targets within the axon guidance pathway: Ephrin A3. Of note when looking at gene expression changes in the mouse mPFC after HFD, we find that although mir-30e-5p expression is reduced (Fig. 3a), EFNA3 expression is also reduced (Fig. 4), even though EFNA3 expression was downregulated by mir-30e-5p in a luciferase cell culture assay. However this seeming contradiction can easily be explained by the fact that multiple miRNAs can regulate the expression of one given mRNA molecule74. Also, it is important to remember that other mechanisms are likely to be involved in gene expression regulation in the in-vivo setting including DNA methylation and histone modifications75,76.
Axon guidance is a tightly regulated process that allows the correct pathfinding of axons during development. The majority of Axon guidance events converge during embryonic and early neonatal life77. Nevertheless, Axon guidance molecules such as ephrins, semaphorins, slits, netrins and a number of extracellular-matrix (ECM) proteins are also found in the adult brain, where they seem to play essential roles in synaptic plasticity, glutamatergic signaling, neuroadaptation78–80 or learning and memory functions81,82. Recent work has also shown that axon guidance molecules contribute to brain maturation during adolescence, in particular within the mPFC5,83,84. Importantly, emerging evidence suggests that axon guidance molecules may also be implicated in the reorganization of the brain after environmental disturbances such as exposure to drugs of abuse, brain injury83,85–87, and now HFD. Interestingly, Axon guidance was selected as one of the top canonical pathways in similar miRNA microarray studies involving animal models of temporal-lobe epilepsy67 or Alzheimer’s disease88. Our study thus extends such reports, indicating that Axon guidance might also play a role in the regulation of prefrontal function by HFD treatments through the involvement of miRNA mechanisms. Future studies are certainly warranted to address such possibilities.
The second top canonical pathway identified by our pathway analyses is the mTOR signaling pathway, a biological pathway involved in the coupling of nutrients and hormones with the regulation of energy-demanding cellular functions89. Interestingly, HFD has been shown previously to modulate hippocampal and cortical mTOR signaling90,91. In the brain, the mTOR pathway is known to play a role in the regulation of synaptic plasticity and cognitive functions89,92. Based on our findings, future studies are therefore warranted to determine a possible link between HFD, miRNAs, and mTOR signaling.
Moreover, our analyses implicate miRNAs in modulating canonical pathways with known roles in neural and cognitive functions. Notably, two related glutamatergic pathways, namely Long-term Depression (LTD) and Glutamatergic Synapse, emerged from such analyses, consistent with previous studies identifying defects in glutamatergic neurotransmission and plasticity in the mPFC of HFD mice28.
Notably, the present results corroborate previous research delineating the effects of HFD on the miRNome of peripheral93 or hypothalamic94 tissues. We identify a number of similarities in the miRNA profiles of hypothalamic and prefrontal tissue after HFD. For example, mir-30e was significantly affected by HFD in both brain regions. Moreover, biological pathways implicated in neurodevelopment such as Axon Guidance and Ephrin Receptor Signaling, as well as the PI3K/Akt and mTOR signaling pathways were commonly recognized as some of the top canonical events putatively affected by miRNAs in these models, suggesting that these cellular pathways might be particularly sensitive to miRNA-dependent signaling in CNS tissue after HFD.
Our present findings are also in line with other studies showing the impact of negative environmental events during adolescence on the prefrontal miRNome. In a study in mice exposed to cocaine during adolescence87, the authors identified similar changes in miRNAs regulating the Axon Guidance and Wnt signaling pathways95. Interestingly, both pathways are known to be involved in the development and maturation of the mPFC. Although speculative, all these studies together suggest that miRNAs, by affecting brain maturational processes, could represent critical contributors to the emergence of behavioral and cognitive deficits following adolescent exposure to environmental disruptors.
Admittedly, our study does not establish a direct causal link between the observed changes in the expression levels of miRNAs and cognitive abnormalities. Nonetheless, the use of available bioinformatic tools provided valuable insights and allowed us to speculate on potential biological networks possibly affected by miRNAs. Also, our luciferase gene reporter assay allowed to provide a proof-of-concept that at least some of the predictions of our bioinformatics analyses are valid; i.e. we showed that EFNA3 is a target for miR-30e-5p. However, more targeted approaches to examine the functional contribution of individual miRNAs are clearly necessary to fully demonstrate the relevance of distinct miRNAs in the emergence of the cognitive deficits described herein. In addition, although beyond the scope of this study, it would be interesting to determine whether changes in miRNA expression and miRNA-regulated gene expression induced by HFD would (partly or fully) recover following a return to a regular diet, as recently suggested by others96.
The present study will be relevant to the clinical appreciation and treatment of cognitive dysfunctions in obesity and in populations that exhibit poor dietary habits, particularly given the dramatic rise in such phenomenon in the past decades97. Notably, our results may have broader implications that reach beyond one’s individual experience of feeding. Indeed, a number of recent studies have also revealed that the metabolic effects of HFD can be transmitted across generations98. Given that epigenetic transmission of behavioral traits are known to partly depend on mechanisms involving germ-line non-coding RNAs such as miRNAs99, the cognitive and miRNA effects of HFD described herein might potentially be transmitted to subsequent generations, although such hypothesis requires investigation.
In conclusion, our results provide the first experimental evidence for the existence of dysregulated prefrontal miRNA expression after HFD exposure and identify target biological pathways putatively affected by miRNAs that include essential neural functions such as Axon guidance. Although further functional studies are required, our findings draw attention to the potential involvement of miRNAs in the development of cognitive deficits, and their implications for obesity and for brain disorders with prefrontal components.
Electronic supplementary material
Acknowledgements
We are grateful to Prof. Urs Meyer, Prof. Marco Riva and Prof. Wolfgang Langhans for infrastructural support and helpful discussions. This work was supported by the University of Zurich (Forschungskredit FK-15–056, Juliet Richetto) and by the Swiss National Science Foundation (Grant 310030_146217, Urs Meyer). Marie Labouesse is recipient of a Swiss National Science Foundation Fellowship (P2EZP3_168841).
Author Contributions
M.A.L. was responsible of the in-vivo experiments, the HFD intervention and the behavioral testing. M.P., A.C. and J.R. performed the molecular analyses related to miRNA profiling and validation. J.R., E.C. and E.M. performed the luciferase assay. M.P., A.C., J.R. and M.A.L. performed the bioinformatic analyses. J.R. and M.A.L. performed the gene expression analyses. F.M. and F.M. assisted in the behavioral and molecular experiments. A.C. assisted in the planning of the miRNA experiments. M.A.L. and J.R. planned the general study design, performed the data analyses and wrote the manuscript, together with input from M.P. and A.C. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26631-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Robbins TW. From arousal to cognition: the integrative position of the prefrontal cortex. Prog Brain Res. 2000;126:469–483. doi: 10.1016/S0079-6123(00)26030-5. [DOI] [PubMed] [Google Scholar]
- 2.Giedd JN, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- 3.Sowell ER, Thompson PM, Holmes CJ, Jernigan TL, Toga AW. In vivo evidence for post-adolescent brain maturation in frontal and striatal regions. Nat Neurosci. 1999;2:859–861. doi: 10.1038/13154. [DOI] [PubMed] [Google Scholar]
- 4.Gogtay N, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci USA. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manitt C, et al. dcc orchestrates the development of the prefrontal cortex during adolescence and is altered in psychiatric patients. Transl Psychiatry. 2013;3:e338. doi: 10.1038/tp.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell P. Adolescent onset of cortical disinhibition in schizophrenia: insights from animal models. Schizophr Bull. 2011;37:484–492. doi: 10.1093/schbul/sbr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spear LP. The adolescent brain and age-related behavioral manifestations. Neurosci Biobehav Rev. 2000;24:417–463. doi: 10.1016/S0149-7634(00)00014-2. [DOI] [PubMed] [Google Scholar]
- 8.Sisk CL, Foster DL. The neural basis of puberty and adolescence. Nat Neurosci. 2004;7:1040–1047. doi: 10.1038/nn1326. [DOI] [PubMed] [Google Scholar]
- 9.Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 10.Cass DK, Thomases DR, Caballero A, Tseng KY. Developmental disruption of gamma-aminobutyric acid function in the medial prefrontal cortex by noncontingent cocaine exposure during early adolescence. Biol Psychiatry. 2013;74:490–501. doi: 10.1016/j.biopsych.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cass DK, et al. CB1 cannabinoid receptor stimulation during adolescence impairs the maturation of GABA function in the adult rat prefrontal cortex. Mol Psychiatry. 2014;19:536–543. doi: 10.1038/mp.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debost JP, et al. Joint Effects of Exposure to Prenatal Infection and Peripubertal Psychological Trauma in Schizophrenia. Schizophr Bull. 2017;43:171–179. doi: 10.1093/schbul/sbw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McEwen BS, Morrison JH. The brain on stress: vulnerability and plasticity of the prefrontal cortex over the life course. Neuron. 2013;79:16–29. doi: 10.1016/j.neuron.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lokken KL, Boeka AG, Austin HM, Gunstad J, Harmon CM. Evidence of executive dysfunction in extremely obese adolescents: a pilot study. Surg Obes Relat Dis. 2009;5:547–552. doi: 10.1016/j.soard.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Maayan L, Hoogendoorn C, Sweat V, Convit A. Disinhibited eating in obese adolescents is associated with orbitofrontal volume reductions and executive dysfunction. Obesity (Silver Spring) 2011;19:1382–1387. doi: 10.1038/oby.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyaradi A, et al. Prospective associations between dietary patterns and cognitive performance during adolescence. J Child Psychol Psychiatry. 2014;55:1017–1024. doi: 10.1111/jcpp.12209. [DOI] [PubMed] [Google Scholar]
- 17.Nyaradi A, Oddy WH, Hickling S, Li J, Foster JK. The Relationship between Nutrition in Infancy and Cognitive Performance during Adolescence. Front Nutr. 2015;2:2. doi: 10.3389/fnut.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overby NC, Ludemann E, Hoigaard R. Self-reported learning difficulties and dietary intake in Norwegian adolescents. Scand J Public Health. 2013;41:754–760. doi: 10.1177/1403494813487449. [DOI] [PubMed] [Google Scholar]
- 19.Haggarty P. Epigenetic consequences of a changing human diet. Proc Nutr Soc. 2013;72:363–371. doi: 10.1017/S0029665113003376. [DOI] [PubMed] [Google Scholar]
- 20.Simopoulos AP. Evolutionary aspects of omega-3 fatty acids in the food supply. Prostaglandins Leukot Essent Fatty Acids. 1999;60:421–429. doi: 10.1016/S0952-3278(99)80023-4. [DOI] [PubMed] [Google Scholar]
- 21.Sluik D, Bezemer R, Sierksma A, Feskens E. Alcoholic Beverage Preference and Dietary Habits: A Systematic Literature Review. Crit Rev Food Sci Nutr. 2016;56:2370–2382. doi: 10.1080/10408398.2013.841118. [DOI] [PubMed] [Google Scholar]
- 22.Sluik D, van Lee L, Engelen AI, Feskens EJ. Total, Free, and Added Sugar Consumption and Adherence to Guidelines: The Dutch National Food Consumption Survey 2007-2010. Nutrients. 2016;8:70. doi: 10.3390/nu8020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Story M, Lytle LA, Birnbaum AS, Perry CL. Peer-led, school-based nutrition education for young adolescents: feasibility and process evaluation of the TEENS study. J Sch Health. 2002;72:121–127. doi: 10.1111/j.1746-1561.2002.tb06529.x. [DOI] [PubMed] [Google Scholar]
- 24.WHO, W. H. O. Who Calls for Stronger Focus on Adolescent Health (2014).
- 25.Deliens T, Clarys P, De Bourdeaudhuij I, Deforche B. Determinants of eating behaviour in university students: a qualitative study using focus group discussions. BMC Public Health. 2014;14:53. doi: 10.1186/1471-2458-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gidding SS, et al. Dietary recommendations for children and adolescents: a guide for practitioners. Pediatrics. 2006;117:544–559. doi: 10.1542/peds.2005-2374. [DOI] [PubMed] [Google Scholar]
- 27.Baker KD, Reichelt AC. Impaired fear extinction retention and increased anxiety-like behaviours induced by limited daily access to a high-fat/high-sugar diet in male rats: Implications for diet-induced prefrontal cortex dysregulation. Neurobiol Learn Mem. 2016;136:127–138. doi: 10.1016/j.nlm.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Labouesse MA, et al. Hypervulnerability of the adolescent prefrontal cortex to nutritional stress via reelin deficiency. Mol Psychiatry. 2017;22:961–971. doi: 10.1038/mp.2016.193. [DOI] [PubMed] [Google Scholar]
- 29.Morin JP, et al. Palatable Hyper-Caloric Foods Impact on Neuronal Plasticity. Front Behav Neurosci. 2017;11:19. doi: 10.3389/fnbeh.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichelt AC. Adolescent Maturational Transitions in the Prefrontal Cortex and Dopamine Signaling as a Risk Factor for the Development of Obesity and High Fat/High Sugar Diet Induced Cognitive Deficits. Front Behav Neurosci. 2016;10:189. doi: 10.3389/fnbeh.2016.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichelt AC, Killcross S, Hambly LD, Morris MJ, Westbrook RF. Impact of adolescent sucrose access on cognitive control, recognition memory, and parvalbumin immunoreactivity. Learn Mem. 2015;22:215–224. doi: 10.1101/lm.038000.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias C, et al. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundakovic M, Champagne FA. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology. 2015;40:141–153. doi: 10.1038/npp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sng J, Meaney MJ. Environmental regulation of the neural epigenome. Epigenomics. 2009;1:131–151. doi: 10.2217/epi.09.21. [DOI] [PubMed] [Google Scholar]
- 35.Szyf M, Bick J. DNA methylation: a mechanism for embedding early life experiences in the genome. Child Dev. 2013;84:49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 37.Langie SA, et al. Maternal folate depletion and high-fat feeding from weaning affects DNA methylation and DNA repair in brain of adult offspring. FASEB J. 2013;27:3323–3334. doi: 10.1096/fj.12-224121. [DOI] [PubMed] [Google Scholar]
- 38.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151:4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 41.O’Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology. 2013;38:39–54. doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischbach S, Kopec AM, Carew TJ. Activity-dependent inhibitory gating in molecular signaling cascades induces a novel form of intermediate-term synaptic facilitation in Aplysia californica. Learn Mem. 2014;21:199–204. doi: 10.1101/lm.033894.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–69. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 44.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Q, et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- 47.Darcq E, et al. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. 2015;20:1261. doi: 10.1038/mp.2014.155. [DOI] [PubMed] [Google Scholar]
- 48.Baddeley A. Working memory: looking back and looking forward. Nat Rev Neurosci. 2003;4:829–839. doi: 10.1038/nrn1201. [DOI] [PubMed] [Google Scholar]
- 49.Bitanihirwe BK, Peleg-Raibstein D, Mouttet F, Feldon J, Meyer U. Late prenatal immune activation in mice leads to behavioral and neurochemical abnormalities relevant to the negative symptoms of schizophrenia. Neuropsychopharmacology. 2010;35:2462–2478. doi: 10.1038/npp.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 52.Tukey JW. Some thoughts on clinical trials, especially problems of multiplicity. Science. 1977;198:679–684. doi: 10.1126/science.333584. [DOI] [PubMed] [Google Scholar]
- 53.Vlachos IS, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campos-Melo D, Droppelmann CA, Volkening K, Strong MJ. Comprehensive luciferase-based reporter gene assay reveals previously masked up-regulatory effects of miRNAs. Int J Mol Sci. 2014;15:15592–15602. doi: 10.3390/ijms150915592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bannerman DM, et al. Double dissociation of function within the hippocampus: a comparison of dorsal, ventral, and complete hippocampal cytotoxic lesions. Behav Neurosci. 1999;113:1170–1188. doi: 10.1037/0735-7044.113.6.1170. [DOI] [PubMed] [Google Scholar]
- 56.Deacon RM, Bannerman DM, Kirby BP, Croucher A, Rawlins JN. Effects of cytotoxic hippocampal lesions in mice on a cognitive test battery. Behav Brain Res. 2002;133:57–68. doi: 10.1016/S0166-4328(01)00451-X. [DOI] [PubMed] [Google Scholar]
- 57.Jones MW. A comparative review of rodent prefrontal cortex and working memory. Curr Mol Med. 2002;2:639–647. doi: 10.2174/1566524023361989. [DOI] [PubMed] [Google Scholar]
- 58.Goldman-Rakic PS. Circuitry of the frontal association cortex and its relevance to dementia. Arch Gerontol Geriatr. 1987;6:299–309. doi: 10.1016/0167-4943(87)90029-X. [DOI] [PubMed] [Google Scholar]
- 59.Kahn JB, et al. Medial prefrontal lesions in mice impair sustained attention but spare maintenance of information in working memory. Learn Mem. 2012;19:513–517. doi: 10.1101/lm.026302.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu D, et al. Medial prefrontal activity during delay period contributes to learning of a working memory task. Science. 2014;346:458–463. doi: 10.1126/science.1256573. [DOI] [PubMed] [Google Scholar]
- 61.Dias R, Aggleton JP. Effects of selective excitotoxic prefrontal lesions on acquisition of nonmatching- and matching-to-place in the T-maze in the rat: differential involvement of the prelimbic-infralimbic and anterior cingulate cortices in providing behavioural flexibility. Eur J Neurosci. 2000;12:4457–4466. doi: 10.1046/j.0953-816X.2000.01323.x. [DOI] [PubMed] [Google Scholar]
- 62.Ragozzino ME. The contribution of the medial prefrontal cortex, orbitofrontal cortex, and dorsomedial striatum to behavioral flexibility. Ann N Y Acad Sci. 2007;1121:355–375. doi: 10.1196/annals.1401.013. [DOI] [PubMed] [Google Scholar]
- 63.Ragozzino ME, Rozman S. The effect of rat anterior cingulate inactivation on cognitive flexibility. Behav Neurosci. 2007;121:698–706. doi: 10.1037/0735-7044.121.4.698. [DOI] [PubMed] [Google Scholar]
- 64.Carmona MA, Murai KK, Wang L, Roberts AJ, Pasquale EB. Glial ephrin-A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proc Natl Acad Sci USA. 2009;106:12524–12529. doi: 10.1073/pnas.0903328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wurzman R, Forcelli PA, Griffey CJ, Kromer LF. Repetitive grooming and sensorimotor abnormalities in an ephrin-A knockout model for Autism Spectrum Disorders. Behav Brain Res. 2015;278:115–128. doi: 10.1016/j.bbr.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fasanaro P, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gorter JA, et al. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol Dis. 2014;62:508–520. doi: 10.1016/j.nbd.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 68.Francis H, Stevenson R. The longer-term impacts of Western diet on human cognition and the brain. Appetite. 2013;63:119–128. doi: 10.1016/j.appet.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 69.Hollins SL, Zavitsanou K, Walker FR, Cairns MJ. Alteration of imprinted Dlk1-Dio3 miRNA cluster expression in the entorhinal cortex induced by maternal immune activation and adolescent cannabinoid exposure. Transl Psychiatry. 2014;4:e452. doi: 10.1038/tp.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prins SA, Przybycien-Szymanska MM, Rao YS, Pak TR. Long-term effects of peripubertal binge EtOH exposure on hippocampal microRNA expression in the rat. PLoS One. 2014;9:e83166. doi: 10.1371/journal.pone.0083166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bot AM, Debski KJ, Lukasiuk K. Alterations in miRNA levels in the dentate gyrus in epileptic rats. PLoS One. 2013;8:e76051. doi: 10.1371/journal.pone.0076051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe Y, et al. Replication in a Japanese population that a MIR30E gene variation is associated with schizophrenia. Schizophr Res. 2013;150:596–597. doi: 10.1016/j.schres.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 73.Xu Y, et al. MicroRNAs and target site screening reveals a pre-microRNA-30e variant associated with schizophrenia. Schizophr Res. 2010;119:219–227. doi: 10.1016/j.schres.2010.02.1070. [DOI] [PubMed] [Google Scholar]
- 74.Wu S, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 75.Tao BB, et al. Evidence for the association of chromatin and microRNA regulation in the human genome. Oncotarget. 2017;8:70958–70966. doi: 10.18632/oncotarget.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poddar S, Kesharwani D, Datta M. Interplay between the miRNome and the epigenetic machinery: Implications in health and disease. J Cell Physiol. 2017;232:2938–2945. doi: 10.1002/jcp.25819. [DOI] [PubMed] [Google Scholar]
- 77.Song H, Poo M. The cell biology of neuronal navigation. Nat Cell Biol. 2001;3:E81–88. doi: 10.1038/35060164. [DOI] [PubMed] [Google Scholar]
- 78.Contractor A, et al. Trans-synaptic Eph receptor-ephrin signaling in hippocampal mossy fiber LTP. Science. 2002;296:1864–1869. doi: 10.1126/science.1069081. [DOI] [PubMed] [Google Scholar]
- 79.Henderson JT, et al. The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron. 2001;32:1041–1056. doi: 10.1016/S0896-6273(01)00553-0. [DOI] [PubMed] [Google Scholar]
- 80.Xiao D, et al. Ephrin/Eph receptor expression in brain of adult nonhuman primates: implications for neuroadaptation. Brain Res. 2006;1067:67–77. doi: 10.1016/j.brainres.2005.10.073. [DOI] [PubMed] [Google Scholar]
- 81.Bayat M, et al. Netrin-1 improves spatial memory and synaptic plasticity impairment following global ischemia in the rat. Brain Res. 2012;1452:185–194. doi: 10.1016/j.brainres.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 82.Dines M, Lamprecht R. EphrinA4 mimetic peptide targeted to EphA binding site impairs the formation of long-term fear memory in lateral amygdala. Transl Psychiatry. 2014;4:e450. doi: 10.1038/tp.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reynolds LM, et al. Amphetamine in adolescence disrupts the development of medial prefrontal cortex dopamine connectivity in a DCC-dependent manner. Neuropsychopharmacology. 2015;40:1101–1112. doi: 10.1038/npp.2014.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reynolds LM, et al. DCC Receptors Drive Prefrontal Cortex Maturation by Determining Dopamine Axon Targeting in Adolescence. Biol Psychiatry. 2017 doi: 10.1016/j.biopsych.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hafner C, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50:490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- 86.Bahi A, Dreyer JL. Cocaine-induced expression changes of axon guidance molecules in the adult rat brain. Mol Cell Neurosci. 2005;28:275–291. doi: 10.1016/j.mcn.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 87.Sillivan SE, et al. Binge cocaine administration in adolescent rats affects amygdalar gene expression patterns and alters anxiety-related behavior in adulthood. Biol Psychiatry. 2011;70:583–592. doi: 10.1016/j.biopsych.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barak B, et al. Opposing actions of environmental enrichment and Alzheimer’s disease on the expression of hippocampal microRNAs in mouse models. Transl Psychiatry. 2013;3:e304. doi: 10.1038/tp.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haissaguerre M, Saucisse N, Cota D. Influence of mTOR in energy and metabolic homeostasis. Mol Cell Endocrinol. 2014;397:67–77. doi: 10.1016/j.mce.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 90.Arnold SE, et al. High fat diet produces brain insulin resistance, synaptodendritic abnormalities and altered behavior in mice. Neurobiol Dis. 2014;67:79–87. doi: 10.1016/j.nbd.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu J, et al. Ursolic acid improves high fat diet-induced cognitive impairments by blocking endoplasmic reticulum stress and IkappaB kinase beta/nuclear factor-kappaB-mediated inflammatory pathways in mice. Brain Behav Immun. 2011;25:1658–1667. doi: 10.1016/j.bbi.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ortega FJ, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59:781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 94.Sangiao-Alvarellos S, Pena-Bello L, Manfredi-Lozano M, Tena-Sempere M, Cordido F. Perturbation of hypothalamic microRNA expression patterns in male rats after metabolic distress: impact of obesity and conditions of negative energy balance. Endocrinology. 2014;155:1838–1850. doi: 10.1210/en.2013-1770. [DOI] [PubMed] [Google Scholar]
- 95.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2014;19:848–852. doi: 10.1038/mp.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boitard C, et al. Switching Adolescent High-Fat Diet to Adult Control Diet Restores Neurocognitive Alterations. Front Behav Neurosci. 2016;10:225. doi: 10.3389/fnbeh.2016.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.WHO, W. H. O. (2016).
- 98.Fullston T, et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013;27:4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- 99.Gapp K, von Ziegler L, Tweedie-Cullen RY, Mansuy IM. Early life epigenetic programming and transmission of stress-induced traits in mammals: how and when can environmental factors influence traits and their transgenerational inheritance? Bioessays. 2014;36:491–502. doi: 10.1002/bies.201300116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets pertaining to the miRNA profiling of the prefrontal cortex are available in the GEO repository (accession number GSE105794). The remaining datasets are available from the corresponding author on request.