Abstract
Here, we transcriptionally and phenotypically characterized the clpB gene from Enterococcus faecalis. Northern blot analysis identified a monocistronic mRNA strongly induced at 48 and 50 °C. In silico analysis identified that the clpB gene encodes a protein of 868 aa with a predicted molecular mass of approximately 98 kDa, presenting two conserved ATP-binding domains. Sequence analysis also identified a CtsR-binding box upstream of the putative −10 sequence, and inactivation of the ctsR gene resulted in an approximately 2-log increase in clpB mRNA expression, confirming ClpB as a member of the CtsR regulon. While expression of clpB was induced by heat stress, a ΔclpB strain grew relatively well under many different stressful conditions, including elevated temperatures. However, expression of ClpB appears to play a major role in induced thermotolerance and in pathogenesis, as assessed by using the Galleria mellonella virulence model.
INTRODUCTION
Enterococcus faecalis inhabits the gastrointestinal tract of humans and animals and can also colonize the oral cavity, genito-urinary tract and skin. This bacterium is also an important opportunistic pathogen, particularly as an aetiological agent of nosocomial infections accounting for approximately 12 % of the hospital-acquired infections in the USA (Murray, 1990; Fisher & Phillips, 2009). E. faecalis can cause a variety of diseases including bacteraemia, endocarditis, intra-abdominal, urinary tract and soft tissue infections (Murray, 1990). The strong association of this organism with infections in hospital settings has been attributed to its inherent capacity to withstand harsh environmental conditions, and to its innate and acquired resistance to multiple antibiotics. The ability of E. faecalis to survive under adverse conditions is particularly important, and many of its stress-resistant characteristics have been used to distinguish enterococci from other Gram-positive cocci (Murray, 1990). For example, E. faecalis can grow at temperatures ranging from 5 to 50 °C and in a wide pH range (4.6–9.9), and can tolerate and grow in the presence of 6.5 % NaCl or 40 % bile salts (Murray, 1990).
The stress responses of E. faecalis have been the subject of extensive research. It has been demonstrated that pre-exposure to sublethal stresses, oligotrophic conditions or glucose starvation leads to an overall increase in stress resistance (Flahaut et al., 1996a, b, 1997; Giard et al., 1997; Hartke et al., 1998). A correlation has been made between the capacity to adapt to stress and the cross-protection phenomenon with the increased synthesis of many proteins, but, with a few exceptions such as the general stress protein Gls24 and the molecular chaperones GroEL and DnaK, the identity of the majority of these proteins remains unknown (Flahaut et al., 1996a, b, 1997; Hartke et al., 1998; Giard et al., 2000; Laport et al., 2004, 2006). In many bacteria, Clp ATPases and ClpP proteolytic complexes are protein machines that play crucial roles in promoting folding, assembly or degradation of proteins during normal growth and, in particular, under stress-inducing conditions (Houry, 2001; Frees et al., 2004). Two classes of Clp ATPases have been identified based on the presence of one or two ATP-binding domains and the occurrence of specific signature sequences (Wawrzynow et al., 1996). Class 1 consists of relatively large proteins, such as ClpA, ClpB, ClpC, ClpE and ClpL, with two distinct ATP-binding domains, whereas class 2 proteins, such as ClpX and ClpY, display a single ATP-binding domain (Schirmer et al., 1996). In low-GC Gram-positive bacteria, transcription of clpP and selected clp ATPase genes is negatively regulated by CtsR (for class three stress repressor), which recognizes a repeated heptanucleotide (A/GGTCAAANANA/GGTCAA) that usually overlaps with the transcription initiation site or the –35 and –10 sequences of the promoter of the corresponding gene (Derré et al., 1999). In addition to repression by CtsR, additional mechanisms involved in control of clp expression have been identified in low-GC Gram-positive bacteria, including dual repression of clpP by CtsR and HrcA in Streptococcus salivarius (Chastanet & Msadek, 2003).
Given the variety of environmental stresses that E. faecalis withstands, it is likely that the Clp proteins may play a role in stress survival and virulence. The genome of E. faecalis encodes a homologue of the gene encoding the ClpP peptidase, three orthologues of class 1 Clp ATPases (ClpB, ClpC and ClpE), and one class 2 Clp ATPase (ClpX) (Paulsen et al., 2003). Among the class 1 Clp ATPases found in E. faecalis, ClpB is distinguished from the other large Clp ATPases by a relatively longer spacer region between the two ATP-binding domains (Houry, 2001), and by the absence of the ClpP recognition tripeptide. In Escherichia coli, ClpB has been shown to act synergistically with the DnaK system to remodel or dissolve protein aggregates that arise from heat stress (Doyle et al., 2007), but it is not known whether ClpB–DnaK interactions also occur in Gram-positive bacteria. In both Gram-positive and Gram-negative bacteria, inactivation of clpB normally results in reduced thermotolerance (Chastanet et al., 2004; Frees et al., 2004; Yuan et al., 2007). ClpB has also been associated with multiplication or survival of Staphylococcus aureus and Porphyromonas gingivalis within mammalian cells (Frees et al., 2004; Capestany et al., 2008). Moreover, animal models have been used to demonstrate that ClpB is important for virulence of bacterial pathogens, including Listeria monocytogenes and P. gingivalis (Chastanet et al., 2004; Yuan et al., 2007).
In this study, we report the genetic and physiological characterization of the clpB gene in E. faecalis. Transcriptional analysis revealed that clpB expression is induced by a variety of stress conditions and negatively regulated by CtsR. Although inactivation of clpB had no major impact on the ability of E. faecalis to grow under different stress conditions, induced thermotolerance and virulence in the Galleria mellonella insect model were significantly reduced in the absence of ClpB.
METHODS
Bacterial strains and general culture conditions.
Bacterial strains used in this study are listed in Table 1. Escherichia coli strains were routinely grown in Luria–Bertani medium and used for plasmid construction and propagation. E. faecalis strains and its derivatives were grown in brain heart infusion (BHI) medium at 37 °C. When required for selective growth of strains, erythromycin (10 μg ml−1 for E. faecalis, 300 μg ml−1 for Escherichia coli), spectinomycin (1000 μg ml−1), rifampicin (200 μg ml−1) or fusidic acid (25 μg ml−1) was added to the growth medium.
Table 1.
Bacterial strains used in this study
Fus, Fusidic acid; Rif, rifampicin; Sp, spectinomycin.
| Strains | Relevant characteristic | Source |
|---|---|---|
| E. faecalis | ||
| OG1RF | Laboratory strain; Rifr Fusr | Lab stock |
| CK111 | OG1Sp upp4 : : P23repA4 | Kristich et al. (2007) |
| EF-clpB | clpB deletion mutant of OG1RF, ΔclpB | This study |
| EF-ctsR | ctsR deletion mutant of OG1RF, ΔctsR | This study |
| cΔclpB | EF-clpB harbouring pTG55 (pMSP3535 expressing clpB) | This study |
| Escherichia coli | ||
| DH10B | Cloning host | Grant et al. (1990) |
| EC1000 | Host for cloning RepA-dependent plasmids | Leenhouts et al. (1996) |
Construction of E. faecalis ΔclpB or ΔctsR strains.
A markerless genetic exchange system was used for construction of the clpB and ctsR deletion mutants in E. faecalis OG1RF (Kristich et al., 2007). To create each strain, two PCR fragments (each approximately 1 kb in size) flanking the target gene were obtained with the primers listed in Table 2. After digestion with the appropriate restriction enzymes, the two PCR products flanking the gene of interest were simultaneously cloned into pGEM7 (Promega) to give plasmids pGclpB and pGctsR in Escherichia. coli DH10B. The 2 kb fragments containing the clpB (pGclpB) and ctsR (pGctsR) up–down fragments were subcloned into pCJK47 using Escherichia coli EC1000 as the host strain. The resulting plasmids, pCJK-clpB and pCJK-ctsR, were electroporated into competent E. faecalis CK111 (donor strain). The E. faecalis CK111 strains containing the pCJK-clpB and pCJK-ctsR plasmids were conjugated with wild-type E. faecalis OG1RF and transformants were selected on BHI agar medium containing rifampicin and erythromycin. Single colonies were subjected to the PheS* negative counter-selection system to isolate double crossover deletions as described elsewhere (Kristich et al., 2007). The clpB and ctsR gene deletions were confirmed by PCR sequencing of the insertion site and flanking sequences.
Table 2.
Oligonucleotide primers and probes used in this study
The underlined bases correspond to restriction sites included to aid in the subsequent cloning of the PCR products.
| Primer name | Sequence (5′–3′) | Application |
|---|---|---|
| FW clpB UP | GGAGGATGG AATTCTCGCTTTTCT | clpB deletion |
| RV clpB UP | TGAGCTTCACCCGGGGCCTCTTGA | clpB deletion |
| FW clpB DOWN | GAAGGTGTGCCCGGGGAAGGAAC | clpB deletion |
| RV clpB DOWN | AACCCTTGGGCATGCGACAGACAA | clpB deletion |
| FW ctsR UP | CTTTGCAAAAGGATCCGCG | ctsR deletion |
| RV ctsR UP | AGCCTCAAGAATTCCTGACGTATT | ctsR deletion |
| FW ctsR DOWN | GGAAGGCAAGAATTCGCTAGCTG | ctsR deletion |
| RV ctsR DOWN | CCAACTACTGGATCCAAAC | ctsR deletion |
| clpB forw 3 | CAAAGGATCCAAGAGTTGGTCAAAC | ΔclpB complementation |
| clpB rev 3 | GAATTCCGTCTCGAGTTACACTTC | ΔclpB complementation |
| clpB forw 2 | GATGCTGGTTTAGATGTTGACG | qRT-PCR |
| clpB rev 2 | CGAAGTGAATCAGCTTCTTGC | qRT-PCR |
| 16S RNA-F | CGCTAGTAATCGTGGATCAGAATG | qRT-PCR |
| 16S RNA-R | TGTGACGGGCGGTGTGTA | qRT-PCR |
| clpB forw 1 | ATCATTGGTCGTGACGAA | clpB probe |
| clpB rev 1 | ATTCACTCATGTCAATCC | clpB probe |
Construction of complemented clpB strain.
To express the clpB gene in trans, the full-length clpB gene including the ribosome-binding site was amplified by PCR with primers containing BamHI restriction sites (Table 2) and ligated into pMSP3535 (Bryan et al., 2000), which had been digested with BamHI and XhoI. The ligation mixture containing the pMSP3535 expressing clpB was electroporated onto CK111 and subsequently conjugated with E. faecalis ΔclpB. The resulting plasmid (pTG55) contains the intact clpB gene under the control of the lactococcal nisin promoter. Expression of clpB from pTG55 was induced by adding 5 ng nisin ml−1 to exponentially grown cultures.
Stress challenges.
To investigate the role of the E. faecalis ClpB in stress responses, cells of the wild-type OG1RF and its isogenic clpB mutant were subjected to a variety of stress challenges. For Northern blot analysis, cells were grown in BHI to mid-exponential phase (OD600∼0.5) and subjected to heat shock at 42, 45, 48 or 50 °C. To evaluate the capacity of E. faecalis strains to grow under stress conditions (5 % NaCl, pH 5, pH 9 and 2 mM H2O2), mid-exponential phase cultures grown in BHI were diluted 1 : 100 into fresh BHI medium adjusted to each specific condition and growth was monitored using a Bioscreen C growth monitor (Oy Growth Curves Ab). Disc inhibition assays were used to assess the sensitivity of the strains to H2O2, HCl or NaOCl. Briefly, a uniform layer of exponentially grown cells was spread onto BHI agar plates using a sterile swab, and filter paper discs (1 mm diameter) saturated with each solution (5 % H2O2, 3 M HCl or 10 % NaOCl) were placed onto the agar. All plates were incubated at 37 °C for 48 h after which the halo of inhibition was recorded. To test the involvement of ClpB in induced thermotolerance, cultures were grown in BHI or BHI containing erythromycin for the complemented mutant. Briefly, cultures were grown exponentially at 37 °C to an OD600∼0.4 and split in two aliquots; one aliquot remained at 37 °C while the other was incubated at the sublethal temperature of 50 °C. After 30 min incubation at 37 or 50 °C, both cultures were incubated at a lethal temperature of 60 °C for 30 and 60 min. ClpB expression in the complemented ΔclpB strain (cΔclpB) was induced by adding nisin (5 ng ml−1) to exponentially grown cultures (OD600∼0.1). Cell survival was evaluated by plating serially diluted aliquots on BHI plates that were incubated at 37 °C for 48 h prior to counting c.f.u. The capacity of the ΔclpB mutant to tolerate oxidative stress was further investigated in an H2O2 killing assay. Briefly, exponentially grown cultures were incubated at 37 °C in the presence of 30 mM H2O2. Every 30 min for up to 2 h, aliquots were taken and serially diluted, plated on BHI plates and incubated at 37 °C for 48 h before colonies were counted.
Biofilm assay.
Biofilm formation on polystyrene microtitre plates was quantified essentially as described previously (Loo et al., 2000). Briefly, biofilms were grown in a semi-defined biofilm medium (BM) containing glucose as the carbohydrate source for 24 h at 37 °C before adherent bacteria were stained with 0.1 % crystal violet. The bound dye was extracted from the stained cells with 33 % acetic acid solution, and the biofilms were quantified by measuring OD575.
Metabolic labelling of proteins and Western blotting.
Overnight cultures were diluted in BHI to 107 c.f.u. ml−1 and incubated for 30 min at 37 °C. For continuous labelling at different temperatures, cells were concentrated by centrifugation to 108 c.f.u. ml−1 in a methionine-free essential medium (MEM; Difco) containing 200 μCi ml−1 (7.4 MBq ml−1) [32S]methionine. Control cells were labelled at 37 °C, whereas heat-shocked cells were labelled at 42, 45, 48, 50 or 52 °C. After 30 min incubation at the different temperatures, protein lysates from each set of samples obtained by lysozyme treatment were separated by 7.5 % SDS-PAGE. After separation, the gel was stained with Coomassie blue, destained, dried and exposed to an X-ray film for detection of radiolabelled proteins. Alternatively, a duplicate gel was transferred to a nitrocellulose membrane for Western blot analysis using a polyclonal antibody raised against the Synechococcus spp. ClpB (a generous gift from Dr Adrian K. Clarke) (Eriksson & Clarke, 1996). Immune reactivity was visualized using the Western blotting luminol reagent according to the manufacturer's recommendations (Santa Cruz Biotechnology).
RNA methods.
For each growth condition, cell pellets were collected by centrifugation and immediately treated with the RNA protect reagent (Qiagen). Total RNA was isolated by the hot acid–phenol method as described previously (Abranches et al., 2006). The crude RNA was treated with DNase I (Ambion) and then further purified by using the RNeasy mini kit (Qiagen), including an on-column DNase I digestion (Qiagen). For Northern analysis, RNA was separated on a 1.2 % agarose–formaldehyde denaturing gel and blotted to a nitrocellulose membrane as described elsewhere (Sambrook & Russell, 2001). The membrane was probed with a fragment of the E. faecalis clpB gene labelled with [α-32P]dCTP and the ready-to-go DNA labelling beads system according to the manufacturer's recommendations (Amersham Pharmacia Biotech). To confirm the integrity and the amount of RNA analysed in the Northern blot, a replica gel was stained with ethidium bromide. For quantitative real-time reverse-transcriptase PCR (qRT-PCR) analysis, cDNA was generated from three independent RNA samples using the SuperScript first-strand synthesis system kit (Invitrogen) and gene-specific primers. The primers used for qRT-PCR were designed using the Beacon designer 2.0 software (Premier Biosoft International). qRT-PCR was carried out by following the protocols described elsewhere (Ahn et al., 2005). A Student's t test was performed to verify significance of the qRT-PCR results.
Galleria mellonella infection.
For the G. mellonella killing assays, insects in the final larval stage were purchased from Vanderhorst Inc., stored at 4 °C in the dark and used within 5 days of shipment. Groups of 20 larvae, ranging from 200 to 300 mg in weight and with no signs of melanization, were randomly chosen and used for subsequent infection. A 10 μl Hamilton syringe was used to inject 5 μl aliquots of bacterial inoculum (5×105 c.f.u. ml−1) into the haemocoel of each larva via the last left proleg. Bacterial colony counts on BHI plates were used to confirm the initial inoculum. Groups injected with saline solution or heat-inactivated (20 min at 75 °C) cells were used as controls in each experiment. After injection, larvae were incubated at 37 °C, and survival was recorded at selected intervals. Larvae were scored as dead when they displayed no movement in response to touch. Kaplan–Meier killing curves were plotted and estimation of differences in survival were compared by using the log-rank test. A P-value ≤0.001 was considered significant. All data were analysed with GraphPad Prism 4.0 software. Experiments were performed independently three times with similar results.
RESULTS AND DISCUSSION
In silico analysis of E. faecalis clpB
Analysis of the E. faecalis V583 genome sequence (Paulsen et al., 2003) identified the gene encoding the ClpB ATPase. The clpB gene encodes a protein of 868 aa with a predicted molecular mass of approximately 98 kDa and an estimated pI of 4.78. The protein presents two conserved nucleotide-binding domains (NBD) separated by a relatively long spacer (154 aa). The NBDs present the consensus sequences Walker-A and Walker-B (Fig. 1a). Analysis of the amino acid sequence confirmed the presence of the N-terminal region and the two C-terminal signature sequences characteristic of ClpB (Fig. 1b). The predicted start codon is preceded by a putative σA-type promoter (TTGACC-N17-TATAAT) (Fig. 2), and a putative transcription terminator [ΔG=−45.3 kcal mol−1 (189.5 kJ mol−1)] was identified downstream of the predicted stop codon. In addition, a CtsR-binding sequence (GGTCAAA-N3-GGTCAAT) was identified starting 30 bp upstream of the −10 sequence. A putative ribosome-binding site (GGGAGG) was found 8 bp upstream of the ATG start codon. Of note, sequence analysis identified a second initiation codon (GTG) and a putative ribosome-binding site (AGGAGG) downstream of the ATG start codon (Fig. 2). This potential internal translation initiation site suggests that a smaller form of ClpB, with a theoretical molecular mass of approximately 80 kDa, may be produced. Of note, sequence analysis of the clpB gene from several other E. faecalis strains with complete genomes available at the Broad Institute (http://www.broadinstitute.org/annotation/genome/enterococcus_faecalis/MultiHome.html) indicated that the second putative translational initiation region inside clpB is fully conserved in other E. faecalis strains.
Fig. 1.

Structural features of the ClpB proteins of E. faecalis. (a) Amino acid sequence deduced from the E. faecalis clpB gene indicating the consensus sequences. (b) Consensus motifs of the ClpB proteins. ATP-1 and ATP-2 represent the Walker-type nucleotide-binding sites: Walker-A is a glycine-containing segment and Walker-B represents hydrophobic segment motifs. I, N-terminal; II and III, middle; and IV and V, C-terminal consensus sequences.
Fig. 2.
Nucleotide sequence of the promoter region and the 5′ coding region of the E. faecalis clpB gene. Regulatory sequences, in bold type and underlined, are as follows: one putative CtsR element (consensus sequence GGTCAAANANGGTCAAA) indicated with an arrow below, the putative −35 and −10 regions, the initiation codon ATG and its ribosome-binding site (RBS), and a second putative initiation codon GTG and its related RBS (boxed).
Induction of clpB mRNA and ClpB in response to heat shock
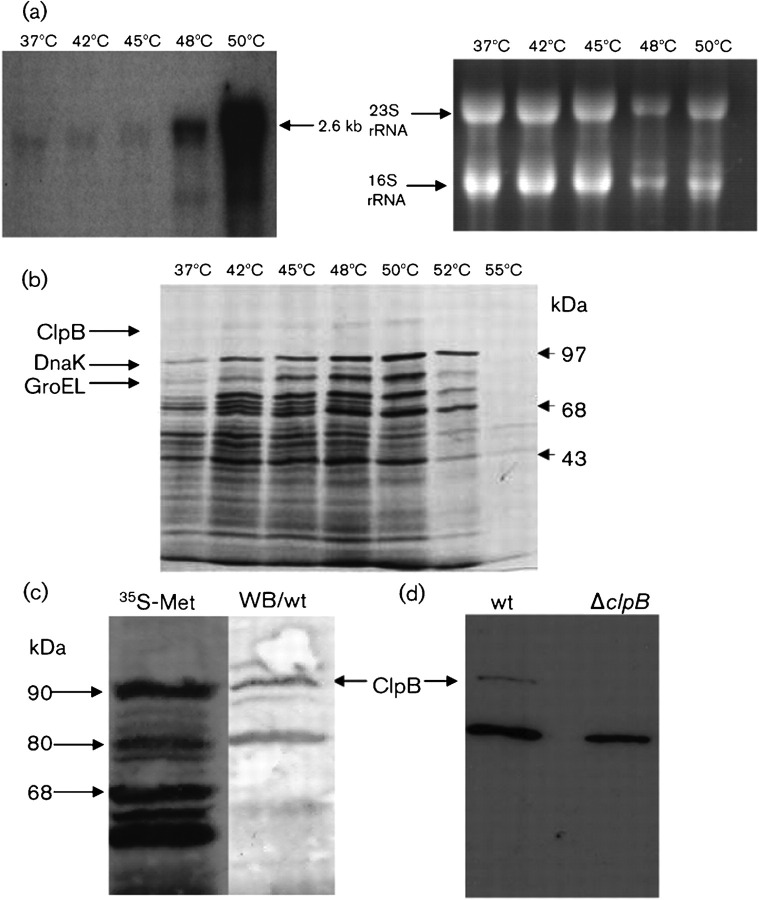
To determine whether ClpB expression is modulated by heat stress, Northern blot, 35S-pulse protein labelling and Western blot analyses were performed. Northern analysis revealed a single stress-inducible transcript of 2.6 kb, compatible with a clpB monocistronic transcript (Fig. 3a). No induction was observed when the temperature was shifted from 37 to up to 45 °C. However, at 48 and 50 °C, a significant increase in induction of 10- and 35-fold, respectively, was observed (Fig. 3a). The result of the autoradiogram of 35S-labelled cells and Western blotting further confirmed ClpB as a heat-shock protein. Autoradiograms of polypeptides labelled at temperatures ranging from 37 to 55 °C revealed two heat-inducible bands of approximately 95 and 80 kDa (Fig. 3b), as well as the induction of the molecular chaperones DnaK and GroEL that have been identified previously (Laport et al., 2001). All heat-inducible bands reached a maximum induction at temperatures between 48 and 50 °C. Western blotting using a polyclonal antibody against ClpB identified two bands of approximately 95 and 80 kDa, suggesting that ClpB may be present in two isoforms (Fig. 3c). The predicted molecular mass of ClpB is 98 kDa, whereas in silico analysis suggested the presence of a putative internal translational initiation site that could potentially yield a smaller 80 kDa protein. Although we cannot rule out that the smaller band is a result of enzymic cleavage of the 98 kDa ClpB, it is tempting to speculate that the 80 kDa protein is a product from a second and internal translational initiation site. Of note, similar observations have been reported in Escherichia coli and in the unicellular cyanobacterium Synechococcus sp., in which clpB was shown to contain two initiation sites for translation, resulting in the synthesis of ClpB polypeptides of ∼95 and ∼80 kDa (Park et al., 1993; Clarke & Eriksson, 2000). Interestingly, in the cyanobacterium Synechococcus sp., the smaller ClpB displayed the same capacity as the larger ClpB for the acquisition of thermotolerance (Eriksson & Clarke, 2000). Moreover, a potential internal translation initiation site preceded by a typical ribosome binding site has also been identified in L. monocytogenes (Chastanet et al., 2004), suggesting that the existence of two different-sized polypeptides may be a common feature of ClpB proteins. Finally, Western blot analysis with the ΔclpB strain confirmed that the 98 kDa polypeptide was ClpB. The 80 kDa band was still observed in the ΔclpB strain albeit not as strongly as it was detected in the parent strain (Fig. 3d). Initially, this finding indicates that the 80 kDa band is not the predicted ClpB internal product but another heat-inducible protein that cross-reacts with the ClpB antibody. However, given the precedents from the ClpB literature and our own findings, we believe that ClpB is in fact produced in two different isoforms and that another protein, which co-migrates with the smaller ClpB, cross-reacted with the ClpB antibody. Considering that Clp ATPases are relatively conserved, it is possible that the cross-reacting protein is one of the two other large class 1 Clp ATPases, ClpC and ClpE, encoded in the E. faecalis genome. Of note, ClpC has a predicted molecular mass of 92 kDa and shares 61 % identity with the E. faecalis ClpB, whereas ClpE has a predicted molecular mass of 82.9 kDa and shares 55 % identity with ClpB. Work is underway to identify the 80 kDa heat-inducible protein(s).
Fig. 3.
Induction of clpB mRNA and ClpB in response to heat shock. (a) Northern blot analysis of the E. faecalis clpB gene. In order to stimulate the production of the heat-shock mRNA, the cells were either cultivated at 37 °C (control cells) or stressed by upshift to 42, 45, 48 or 50 °C for 10 min. Total RNA was separated under denaturing conditions, transferred to a nitrocellulose membrane, and hybridized to a clpB-specific probe (left). A parallel gel was stained with ethidium bromide to show equal loading of the slots (right). (b) Influence of supra-optimum temperatures on protein synthesis. Wild-type cells were either incubated at 37 °C (control) or treated at 42, 45, 48, 50, 52 or 55 °C in the presence of [35S]methionine. Labelled proteins were separated by SDS-PAGE and analysed by autoradiography. The ClpB protein is indicated. DnaK and GroEL, which were previously identified, are also indicated (Laport et al., 2001). (c) Identification of the ClpB protein. Wild-type (wt) cells were subjected to heat shock at 45 °C in the presence of [35S]methionine. Labelled proteins were separated by SDS-PAGE and analysed by either autoradiogram (35S-Met) or Western blotting using polyclonal antibodies raised against Synechococcus spp. ClpB diluted 1 : 1000. (d) Western blotting of wild-type (wt) or ΔclpB cells subjected to heat shock at 45 °C showing the cross-reaction of the ClpB polyclonal antibody.
clpB is a member of the CtsR regulon
Due to the identification of a CtsR consensus-binding motif in the promoter region of the clpB gene, a ΔctsR strain was constructed to investigate the role of CtsR in controlling clpB expression. qRT-PCR analysis revealed that, under non-stressful conditions, CtsR is indeed responsible for clpB repression since an approximate 2-log increase in clpB expression was observed in the ΔctsR strain compared with the wild-type strain (Table 3). Expression of the transcript of the housekeeping gene 16S rDNA was used as an internal standard and no difference was observed between samples (data not shown). Interestingly, induction after exposure to 48 °C was also observed in the ΔctsR strain, suggesting that the clpB gene may have an additional layer of regulation. In Gram-positive bacteria, HrcA and CtsR are two major transcriptional repressors that regulate expression of distinct heat-shock genes (Narberhaus, 1999). The HrcA repressor recognizes a DNA element called CIRCE (for controlling inverted repeat of chaperone expression) and is mainly involved in the transcriptional regulation of the groE and dnaK operons. In some cases, however, heat-shock genes are under dual negative control by the HrcA and CtsR repressors. This is the case for the dnaK operon in Staphylococcus aureus, clpP in Streptococcus salivarius and the groE operon in several streptococci (Chastanet et al., 2001, 2003; Lemos & Burne, 2002; Chastanet & Msadek, 2003). Sequence analysis of the promoter regions of the clpB gene did not indicate the presence of a CIRCE element, indicating that HrcA does not play a role in the regulation of clpB in E. faecalis.
Table 3.
qRT-PCR-based expression profiles of clpB
Total RNA was isolated from mid-exponential phase cultures that were kept at 37 °C (control) or incubated at 48 °C for 30 min. The results were obtained using qRT-PCR as described in Methods. Data are means±sd of three independent experiments performed with RNA isolated from three independent cultures. All samples were assayed in triplicate. nd, Not done.
| Strain | Relative clpB mRNA (copies μl−1) |
|---|---|
| OG1RF (37 °C) | 8.53×104±1.33×104 |
| OG1RF (48 °C) | 1.14×106±4.41×105* |
| ΔctsR (37 °C) | 4.37×106±1.99×106 |
| ΔctsR (48 °C) | 1.00×107±nd* |
*Significant difference compared with the control (P≤0.05).
Growth characteristics, stress tolerance of and biofilm formation by the ΔclpB strain
Analysis of the growth rates and doubling time of the ΔclpB strain under normal growth conditions (i.e. aerobic conditions at 37 °C) indicated that the mutant grew slightly slower than the parent strain (Table 3; P≤0.05). The capacity of the ΔclpB strain to grow under a variety of stress conditions was also examined. No significant differences in growth between the mutant and parent strains were observed when cells were grown at different temperatures (45 and 48 °C), at pH 5 or in the presence of NaCl or H2O2. However, the slower growth of ΔclpB at pH 9 compared with its parent was statistically significant (Table 4; P≤0.05). We also tested the susceptibility of the mutant strain to HCl, H2O2 and NaOCl using disc diffusion assays. Based on the diameter of the zone of inhibition for each compound tested, no significant differences were observed between strains. Finally, the capacity of the mutant to form biofilms on polystyrene microtitre plates was also assessed. Quantitative analysis of biofilms formed by the ΔclpB mutant did not differ from biofilms formed by the parent OG1RF strain (data not shown).
Table 4.
Growth characteristics of the OG1RF and ΔclpB strains grown under different conditions
Doubling time data are means±sd of three independent experiments.
| Strain | Doubling time (min) when grown in/at: | ||||||
|---|---|---|---|---|---|---|---|
| pH 7 | pH 5 | pH 9 | 5 % NaCl | 2 mM H2O2 | 45 °C | 48 °C | |
| OG1RF | 71±6.1 | 209.3±17.4 | 60.1±3.6 | 144.3±4.7 | 82.3±9.6 | 227±49 | 458.3±58.9 |
| ΔclpB | 80.8±4.6 | 234.8±13.1 | 72.2±2.4* | 150±5.2 | 80.7±2.6 | 203±20 | 334.8±31.6 |
*Significant difference compared with OG1RF (P≤0.05).
ClpB contributes to thermotolerance
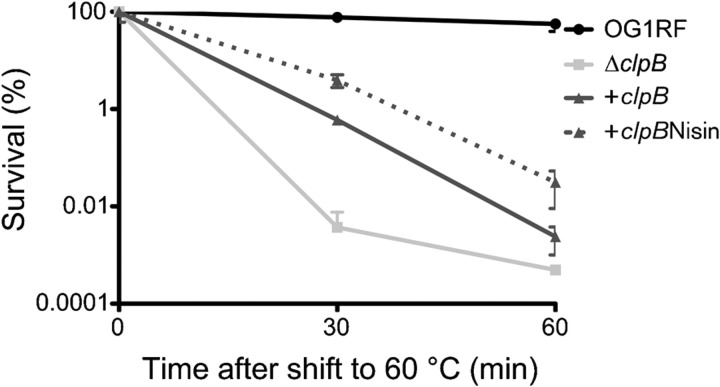
While expression of E. faecalis clpB was highly induced upon heat shock, the ΔclpB strain did not show any inherent growth defect at elevated temperatures. Here, we tested whether ClpB is necessary for the acquisition of thermotolerance in E. faecalis (Fig. 4). In the wild-type strain, pre-incubation of the cells at 50 °C for 30 min prior to exposure to a lethal temperature (60 °C) resulted in full protection for up to 60 min (Fig. 4). In contrast with the wild-type strain, pre-incubation of ΔclpB at 50 °C resulted in only an intermediate level of protection against lethal 60 °C treatment. The introduction of pTG55 (clpB plasmid) partially restored thermotolerance in ΔclpB (cΔclpB strain). Notably, after 60 min at 60 °C, the cells grown in the presence of nisin (induced) showed an approximately 15-fold increased heat tolerance compared with the ΔclpB strain, whereas cΔclpB grown in the absence of nisin displayed fivefold increased survival (Fig. 4). This result showed that ClpB plays an important role in the induction of thermotolerance and this fact might contribute to the persistence of E. faecalis in different environments. Notably, ClpB has been implicated in the acquisition of thermotolerance in Escherichia coli and in other Gram-positive pathogens, such as L. monocytogenes and Staphylococcus aureus (Thomas & Baneyx, 1998; Chastanet et al., 2004; Frees et al., 2004). In Escherichia coli, the ClpB requirement for induction of thermotolerance is related to its ability to act as a molecular chaperone rescuing protein aggregates formed during heat-shock treatment (Thomas & Baneyx, 1998; Doyle et al., 2007). This aggregation-reversing activity of ClpB requires cooperation with the DnaK chaperone machinery (Doyle et al., 2007) but evidence of ClpB–DnaK interactions in Gram-positive bacteria are yet to be demonstrated.
Fig. 4.
ClpB is important for development of thermotolerance. Cultures of wild-type OG1RF, ΔclpB, the complemented cΔclpB and nisin-induced cΔclpB strains were grown exponentially at 37 °C to OD600∼0.4. At this point, cultures were pre-incubated at 50 °C for 30 min prior to exposure to 60 °C (a lethal temperature) for 30 and 60 min. Cell survival was evaluated by plating diluted samples on BHI plates. The result presented is a representative of five independent experiments; error bars, sd.
Virulence of the ΔclpB strain is attenuated in the G. mellonella model
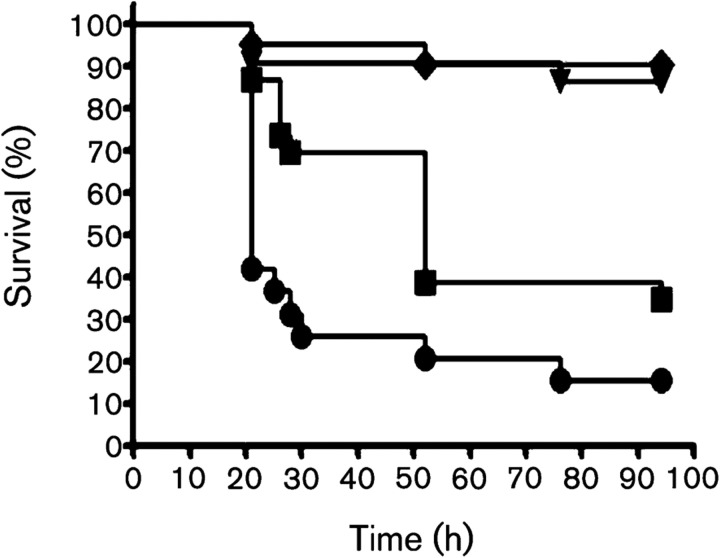
Killing of the larvae of G. mellonella has been routinely used as a virulence model for several pathogenic bacteria, including E. faecalis (Park et al., 2007; Lebreton et al., 2009). Therefore, G. mellonella was tested as a model to verify whether E. faecalis ClpB is involved in virulence. As shown in Fig. 5, the rates of killing were significantly lower in larvae infected with ΔclpB than in those infected with the wild-type strain (P≤0.001). To confirm that killing was due to an infectious process, we also assessed the survival of larvae injected with heat-killed E. faecalis or saline with minimal killing observed within the two groups during the course of the experiment. Of note, the ClpB protein has been implicated in the expression of virulence by other Gram-positive pathogens. In Staphylococcus aureus and P. gingivalis, ClpB was associated with intracellular survival and multiplication (Frees et al., 2004; Capestany et al., 2008), and virulence of L. monocytogenes and P. gingivalis lacking clpB was significantly attenuated in murine models of infection (Chastanet et al., 2004; Yuan et al., 2007).
Fig. 5.
Killing of G. mellonella larvae infected with E. faecalis OG1RF and ΔclpB at 37 °C. Survival (Kaplan–Meyer) plots of larvae injected with OG1RF (•) and ΔclpB (▪) strains. Larvae injected with saline solution (▾) or heat-killed OG1RF (⧫) were used as controls with minimum killing. The experiments were repeated three times, and the results are representative of a typical experiment. Compared with the wild-type OG1RF strain, ΔclpB showed attenuated virulence (P≤0.001).
In G. mellonella, the production of superoxide and other reactive oxygen species (ROS) via a respiratory burst by the insect specialized phagocytic cells, known as haemocytes, is an important line of defence against microbial infections (Bergin et al., 2005). Thus, we hypothesized that the reduced virulence of the ΔclpB strain in G. mellonella could be due to increased sensitivity to ROS. To verify this possibility, we tested the ability of the wild-type and mutant strains to survive a lethal treatment with H2O2. Our results indicated that both strains exhibited similar sensitivity to H2O2 killing (data not shown), suggesting that other host-derived factors may be responsible for the attenuated virulence of ΔclpB in this particular model.
In conclusion, we report the identification and characterization of the ClpB ATPase. We demonstrate that ClpB is a member of the CtsR regulon and, even though it is induced by heat stress, it is not required for growth at high temperatures. However, expression of ClpB appears to play a major role in induced thermotolerance and in pathogenesis, as assessed using the G. mellonella virulence model. Future efforts will be devoted to the identification of proteins that interact with ClpB, which will help to disclose the underlying mechanisms by which ClpB affects virulence traits in E. faecalis.
Acknowledgments
The authors express their thanks to Dr Celia A. Soares, for encouragement, and to Dr Adrian K. Clarke, for providing anti-ClpB. This study was supported by Brazilian grants [Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) and Pronex/FAPERJ] to M. G.-deM. N. E. M. O. was a recipient of a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brazil and a Travel scholarship from CNPq.
Footnotes
Abbreviation: qRT-PCR, quantitative real-time reverse-transcriptase PCR
Edited by: K. E. Weaver
References
- Abranches, J., Candella, M. M., Wen, Z. T., Baker, H. V. & Burne, R. A. (2006). Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans J Bacteriol 188, 3748–3756. 10.1128/JB.00169-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J., Lemos, J. A. & Burne, R. A. (2005). Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol 187, 3028–3038. 10.1128/JB.187.9.3028-3038.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergin, D., Reeves, E. P., Renwick, J., Wientjes, F. B. & Kavanagh, K. (2005). Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun 73, 4161–4170. 10.1128/IAI.73.7.4161-4170.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, E. M., Bae, T., Kleerebezem, M. & Dunny, G. M. (2000). Improved vectors for nisin-controlled expression in Gram-positive bacteria. Plasmid 44, 183–190. 10.1006/plas.2000.1484 [DOI] [PubMed] [Google Scholar]
- Capestany, C. A., Tribble, G. D., Maeda, K., Demuth, D. R. & Lamont, R. J. (2008). Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis J Bacteriol 190, 1436–1446. 10.1128/JB.01632-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet, A. & Msadek, T. (2003). clpP of Streptococcus salivarius is a novel member of the dually regulated class of stress response genes in Gram-positive bacteria. J Bacteriol 185, 683–687. 10.1128/JB.185.2.683-687.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet, A., Prudhomme, M., Claverys, J. P. & Msadek, T. (2001). Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J Bacteriol 183, 7295–7307. 10.1128/JB.183.24.7295-7307.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet, A., Fert, J. & Msadek, T. (2003). Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol Microbiol 47, 1061–1073. 10.1046/j.1365-2958.2003.03355.x [DOI] [PubMed] [Google Scholar]
- Chastanet, A., Derre, I., Nair, S. & Msadek, T. (2004). clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J Bacteriol 186, 1165–1174. 10.1128/JB.186.4.1165-1174.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. K. & Eriksson, M. J. (2000). The truncated form of the bacterial heat shock protein ClpB/HSP100 contributes to development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol 182, 7092–7096. 10.1128/JB.182.24.7092-7096.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derré, I., Rapoport, G. & Msadek, T. (1999). CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol Microbiol 31, 117–131. 10.1046/j.1365-2958.1999.01152.x [DOI] [PubMed] [Google Scholar]
- Doyle, S. M., Hoskins, J. R. & Wickner, S. (2007). Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci U S A 104, 11138–11144. 10.1073/pnas.0703980104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M. J. & Clarke, A. K. (1996). The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol 178, 4839–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M. J. & Clarke, A. K. (2000). The Escherichia coli heat shock protein ClpB restores acquired thermotolerance to a cyanobacterial clpB deletion mutant. Cell Stress Chaperones 5, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, K. & Phillips, C. (2009). In vitro inhibition of vancomycin-susceptible and vancomycin-resistant Enterococcus faecium and E. faecalis in the presence of citrus essential oils. Br J Biomed Sci 66, 180–185. [DOI] [PubMed] [Google Scholar]
- Flahaut, S., Benachour, A., Giard, J. C., Boutibonnes, P. & Auffray, Y. (1996a). Defense against lethal treatments and protein synthesis induced by NaCl in Enterococcus faecalis ATCC 19433. Arch Microbiol 165, 317–324. 10.1007/s002030050333 [DOI] [PubMed] [Google Scholar]
- Flahaut, S., Hartke, A., Giard, J. C., Benachour, A., Boutibonnes, P. & Auffray, Y. (1996b). Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis FEMS Microbiol Lett 138, 49–54. 10.1111/j.1574-6968.1996.tb08133.x [DOI] [PubMed] [Google Scholar]
- Flahaut, S., Hartke, A., Giard, J. C. & Auffray, Y. (1997). Alkaline stress response in Enterococcus faecalis: adaptation, cross-protection, and changes in protein synthesis. Appl Environ Microbiol 63, 812–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frees, D., Chastanet, A., Qazi, S., Sorensen, K., Hill, P., Msadek, T. & Ingmer, H. (2004). Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus Mol Microbiol 54, 1445–1462. 10.1111/j.1365-2958.2004.04368.x [DOI] [PubMed] [Google Scholar]
- Giard, J. C., Hartke, A., Flahaut, S., Benachour, A., Boutibonnes, P. & Auffray, Y. (1997). Glucose starvation response in Enterococcus faecalis JH2-2, survival and proteins analysis. Res Microbiol 148, 27–35. 10.1016/S0923-2508(97)81897-9 [DOI] [PubMed] [Google Scholar]
- Giard, J. C., Rince, A., Capiaux, H., Auffray, Y. & Hartke, A. (2000). Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotropic effect on cell morphology, stress sensitivity and gene expression in Enterococcus faecalis J Bacteriol 182, 4512–4520. 10.1128/JB.182.16.4512-4520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, S. G., Jessee, J., Bloom, F. R. & Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87, 4645–4649. 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartke, A., Giard, J. C., Laplace, J. M. & Auffray, Y. (1998). Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl Environ Microbiol 64, 4238–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry, W. A. (2001). Chaperone-assisted protein folding in the cell cytoplasm. Curr Protein Pept Sci 2, 227–244. 10.2174/1389203013381134 [DOI] [PubMed] [Google Scholar]
- Kristich, C. J., Chandler, J. R. & Dunny, G. M. (2007). Development of a host-genotype-independent counterselectable marker and high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis Plasmid 57, 131–144. 10.1016/j.plasmid.2006.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laport, M. S., Castro, A. C., Villardo, A., Lemos, J. A., Bastos, M. C. F. & Giambiagi-deMarval, M. (2001). Expression of the major heat shock proteins DnaK and GroEL in Streptococcus pyogenes: a comparison to Enterococcus faecalis and Staphylococcus aureus Curr Microbiol 42, 264–268. [DOI] [PubMed] [Google Scholar]
- Laport, M. S., Lemos, J. A., Bastos, M. C. F., Burne, R. A. & Giambiagi-deMarval, M. (2004). Transcriptional analysis of the groE and dnaK heat-shock operons of Enterococcus faecalis Res Microbiol 155, 252–258. 10.1016/j.resmic.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Laport, M. S., Santos, L. L., Lemos, J. A. C., Bastos, M. C. F., Burne, R. A. & Giambiagi-deMarval, M. (2006). Organization of the heat-shock dnaK and groE operons of the nosocomial pathogen Enterococcus faecium Res Microbiol 157, 162–168. 10.1016/j.resmic.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Lebreton, F., Riboulet-Bisson, E., Serror, P., Sanguinetti, M., Posteraro, B., Torelli, R., Hartke, A., Auffray, Y. & Giard, J.-C. (2009). ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immun 77, 2832–2839. 10.1128/IAI.01218-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts, K., Buist, G., Bolhuis, A., ten Berge, A., Kiel, J., Mierau, I., Dabrowska, M., Venema, G. & Kok, J. (1996). A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253, 217–224. 10.1007/s004380050315 [DOI] [PubMed] [Google Scholar]
- Lemos, J. A. C. & Burne, R. A. (2002). Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans J Bacteriol 184, 6357–6366. 10.1128/JB.184.22.6357-6366.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, C. Y., Corliss, D. A. & Ganeshkumar, N. (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182, 1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, B. E. (1990). The life and times of the Enterococcus Clin Microbiol Rev 3, 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus, F. (1999). Negative regulation of bacterial heat shock genes. Mol Microbiol 31, 1–8. 10.1046/j.1365-2958.1999.01166.x [DOI] [PubMed] [Google Scholar]
- Park, S. K., Kim, K. I., Woo, K. M., Seol, J. H., Tanaka, K., Ichihara, A., Ha, D. B. & Chung, C. H. (1993). Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem 268, 20170–20174. [PubMed] [Google Scholar]
- Park, S. Y., Kim, K. M., Lee, J. H., Seo, S. J. & Lee, I. H. (2007). Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun 75, 1861–1869. 10.1128/IAI.01473-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, I. T., Banerjei, L., Myers, G. S., Nelson, K. E., Seshadri, R., Read, T. D., Fouts, D. E., Eisen, J. A., Gill, S. R. & other authors (2003). Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis Science 299, 2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Schirmer, E. C., Glover, J. R., Singer, M. A. & Lindquist, S. (1996). HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci 21, 289–296. 10.1016/0968-0004(96)10038-4 [DOI] [PubMed] [Google Scholar]
- Thomas, J. G. & Baneyx, F. (1998). Roles of the Escherichia coli small heat shock proteins ibpA and ibpB in thermal stress management: comparison with ClpA, ClpB and HtpG in vivo J Bacteriol 180, 5165–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynow, A., Banecki, B. & Zylicz, M. (1996). The Clp ATPases define a novel class of molecular chaperones. Mol Microbiol 21, 895–899. 10.1046/j.1365-2958.1996.421404.x [DOI] [PubMed] [Google Scholar]
- Yuan, L., Rodrigues, P. H., Belanger, M., Dunn, J. R. W. & Progulske-Fox, A. (2007). The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo FEMS Immunol Med Microbiol 51, 388–398. 10.1111/j.1574-695X.2007.00326.x [DOI] [PubMed] [Google Scholar]