Abstract
Mycobacterium ulcerans is the causative agent of Buruli ulcer (BU), a WHO-defined neglected tropical disease. All Japanese BU causative isolates have shown distinct differences from the prototype and are categorized as M. ulcerans subspecies shinshuense. During repeated sub-culture, we found that some M. shinshuense colonies were non-pigmented whereas others were pigmented. Whole genome sequence analysis revealed that non-pigmented colonies did not harbor a giant plasmid, which encodes elements needed for mycolactone toxin biosynthesis. Moreover, mycolactone was not detected in sterile filtrates of non-pigmented colonies. Mice inoculated with suspensions of pigmented colonies died within 5 weeks whereas those infected with suspensions of non-pigmented colonies had significantly prolonged survival (>8 weeks). This study suggests that mycolactone is a critical M. shinshuense virulence factor and that the lack of a mycolactone-producing giant plasmid makes the strain non-pathogenic. We made an avirulent mycolactone-deletion mutant strain directly from the virulent original.
Introduction
Buruli ulcer (BU) is one of twenty neglected tropical diseases (NTD) as defined by the World Health Organization (WHO)1. After leprosy and tuberculosis, BU is now the third most common mycobacterial disease worldwide1. BU is characterized as a chronic, indolent, necrotizing disease of skin and soft tissue that manifests as massive ulcers that result in permanent, disabling scars2.
The first reported case of BU in Japan occurred in a young woman who had never traveled abroad3,4. Based on differences in growth and biochemical properties from prototypic M. ulcerans, the causative agent for this patient’s BU was determined to be a M. ulcerans subspecies, M. ulcerans subsp. shinshuense (hereafter “M. shinshuense”)3–5. The growth rate of “M. shinshuense” was more rapid than that of M. ulcerans and “M. shinshuense” was positive for urease activity3. The incidence of “M. shinshuense” in Japan has gradually increased; as of December 2017, a total of 67 cases have been reported and 35 clinical strains have been isolated in our laboratory6,7.
Although “M. shinshuense” is considered to be an M. ulcerans subspecies, a recent whole genome sequence (WGS) analysis, which included a giant plasmid from the reference strain, revealed significant genetic differences between them8. Nonetheless, both M. ulcerans and “M. shinshuense” carry the giant plasmid (pMUM001) that harbors genes encoding the polyketide synthases required for mycolactone synthesis9,10. The macrolide mycolactone is a lipid-like exotoxin that has a structure similar to immune-suppressive agents such as cyclosporine, rapamycin, and tacrolimus11,12. Production of this toxin is critical for M. ulcerans pathogenesis.
We have noticed pigmented and non-pigmented colonies in the culture of “M. shinshuense”, which led us to explore the possible difference of the nature of these two phenotypes.
Results
Biochemical characteristics
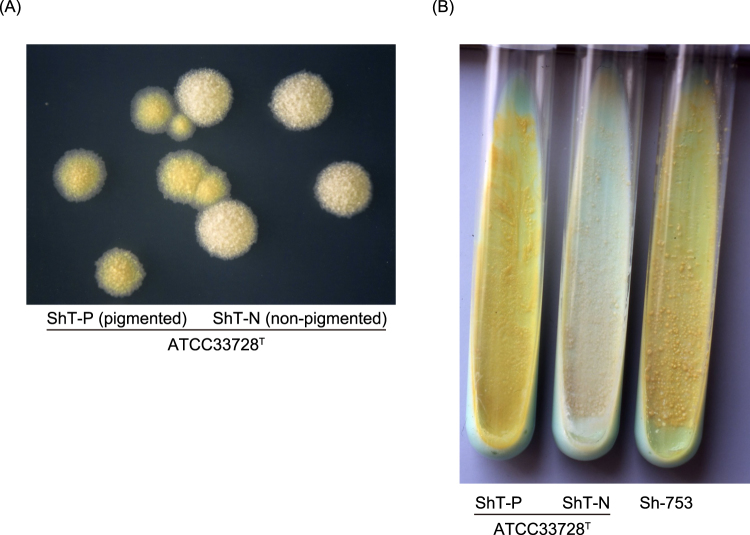
We found ivory (non-pigmented) colonies among yellow (pigmented) “M. shinshuense” reference strain ATCC 33728 colonies during routine culture work (Fig. 1A). Each type of colony (labeled as ShT-P (pigmented) or ShT-N (non-pigmented)) was sub-cloned and purified by continuous three generation passage on 2% Ogawa egg medium slants (Fig. 1B).
Figure 1.
Characteristics of ShT-N and ShT-P colonies. (A) A mixture of pigmented (ShT-P) and non-pigmented (ShT-N) colonies of ATCC 33728T on 7H11 solid medium. (B) Subcloned colonies of ATCC 33728T. ShT-P and ShT-N were isolated and propagated. Only pigmented, yellow colonies were produced by ShT-P, whereas for ShT-N only non-pigmented, ivory colonies reproduced. The mycolactone-producing “M. shinshuense” strain Sh-753 was used as a positive control.
Both ShT-P and ShT-N formed rough colonies that were visible within 14 days of inoculation. ShT-P retained the yellow pigment (and even when glowed in the dark), whereas ShT-N was not pigmented. Both formed visible colonies at 25 °C and 32 °C, but not at 37 °C or 42 °C. Neither grew on a medium supplemented with 500 μg/ml p-nitrobenzoic acid or 5% NaCl. Both isolates were negative for Tween 80 hydrolysis, nitrate reduction, arylsulfatase (3 days), pyrazinamidase, iron uptake, and MPB64 production, but were positive for semiquantitative catalase, 68 °C catalase and urease activities. The biochemical and growth characteristics of both “M. shinshuense” isolates, a different clinical isolate of “M. shinshuense” (Sh-753), and several M. ulcerans strains are summarized in Table 1. The results were in accordance with those of a previous report on M. ulcerans, except for the positive urease activity of the “M. shinshuense” isolates2.
Table 1.
Characteristics of bacteria tested in this study.
| “M. shinshuense” | M. ulcerans | ||||
|---|---|---|---|---|---|
| ATCC 33728 | MU-4 | MU-8 | |||
| ShT-P (pigmented) | ShT-N (non-pigmented) | Sh-753 | |||
| Mycolactone production | + | − | + | + | − |
| Colony: | |||||
| Color (pigmentation) | + | − | + | + | − |
| Morphology | R | R | R | R | R |
| Growth on 2% Ogawa medium slant | |||||
| 14 days | + | + | + | − | − |
| 30 days | + | + | + | + | + |
| Growth at: | |||||
| 25 °C | + | + | + | + | + |
| 32 °C | + | + | + | + | + |
| 37 °C | − | − | − | − | − |
| 42 °C | − | − | − | − | − |
| Growth on 500 μg/ml p-nitrobenzoic acid | − | − | − | − | − |
| Growth on NaCl medium (5%) | − | − | − | − | − |
| Tween 80 hydrolysis | − | − | − | − | − |
| Nitrate reduction | − | − | − | − | − |
| Arylsulfatase | − | − | − | − | − |
| Pyrazinamidase | − | − | − | − | − |
| Iron uptake | − | − | − | − | − |
| MPB64 Ag production | − | − | − | − | − |
| Semi−quantitative catalase | + | + | + | + | + |
| 68 °C catalase | + | + | + | + | + |
| Urease | + | + | + | − | − |
R: rough. See Fig. 2 for visualization of mycolactone production by TLC.
Genetic characteristics of M. shinshuense chromosomes
All multi-locus sequence typing (MLST) showed identical sequences between ShT-P and ShT-N. The 1,475–base pair (bp) 16S rRNA gene sequences of ShT-P and ShT-N were also identical, but both partially differed from those of related species such as M. ulcerans, M. marinum, and M. pseudoshottsii (Supplemental Table 1). The internal transcribed spacer (ITS) (272-bp), rpoB (315-bp), and hsp65 (401-bp) sequences of ShT-P and ShT-N were also identical (data not shown).
A direct comparison of WGS data for ShT-P and ShT-N showed that the chromosomes of both isolates were almost identical except for 8 single nucleotide polymorphisms (SNPs) and 20 small insertions or deletions (Indels). Four discordant SNPs located in coding regions or intergenic regions, all of which could be aligned with WGS data and validated by Sanger sequencing. There was a non-synonymous mutation in three coding regions, whereas the other one SNP was located in intergenic region. Among 20 small insertions of ShT-N, most of them were on hypothetical proteins (n = 2), pseudogenes (n = 5), or intergenic regions (n = 10) (Supplemental Table 2).
Existence of mycolactone biosynthesis proteins produced by a giant plasmid
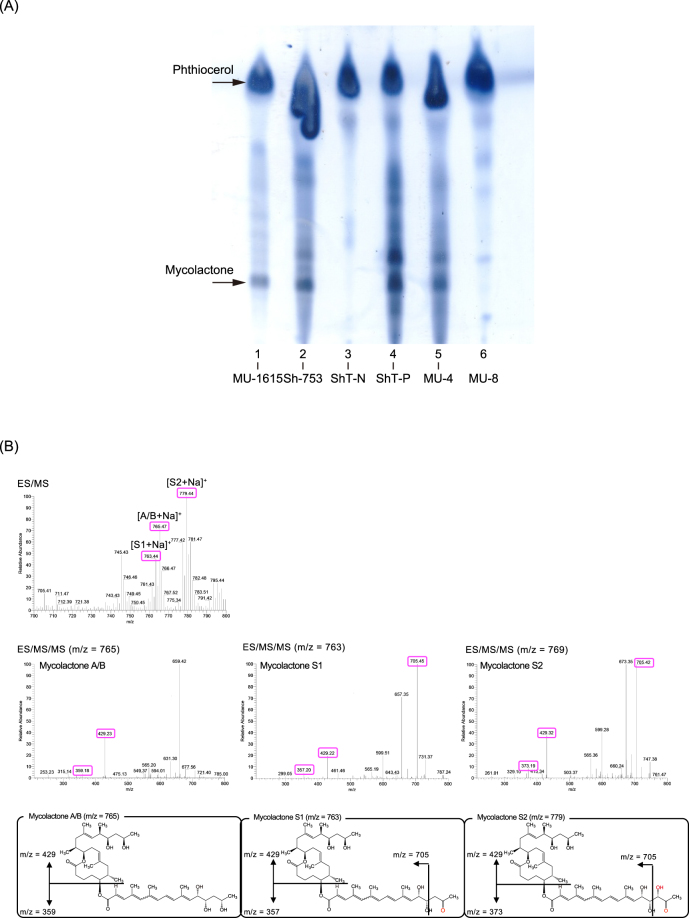
During the course of our experiments we noticed that the avirulent MU-8 strain was also non-pigmented. Although this strain carries the giant plasmid, several deletions (see below) caused some open reading frames (ORFs) to become dysfunctional12,13. Therefore, we hypothesized that non-pigmented ShT-N would be similar to MU-8 in that it does not produce functional mycolactone. To test this hypothesis, we first isolated mycolactone from culture filtrates by thin-layer chromatography (TLC). Mycolactone was not detected in the ShT-N and MU-8 samples, whereas a control lipid, phthiocerol, was present. In contrast, both mycolactone and phthiocerol were present in culture filtrates from ShT-P, MU-1615, and MU-4, which are all virulent strains (Fig. 2A). In detail, we analyzed the precise structures of mycolactone from M. shinshuense ShT-P by electrospray ionization mass spectrometry (ESI/MS) and MS/MS14. The ESI/MS spectrum of acetone soluble lipids (ASLs) from the strain ShT-P detected the peaks of m/z 765.4, 763.4, 779.4 as [M + Na]+. In each MS/MS spectrum, the distinct ions were found m/z 359 and 429 for mycolactone A/B; m/z 357, 429 and 705 for mycolactone S1; m/z 373, 429 and 705 for mycolactone S2, respectively (Fig. 2B). The strain ShT-P produces mycolactone A/B, S1, and S2 as previously described15.
Figure 2.
Silica thin-layer chromatography and ESI/MS/MS analyses of acetone soluble lipids (ASLs) from M. ulcerans or “M. shinshuense” isolates. (A) ASLs extracted from the bacterial cell mass (CM). Samples were loaded as follows: lane 1, MU-1615 (M. ulcerans Malaysian strain, virulent); lane 2, Sh-753 (“M. shinshuense”, virulent); lane 3, ShT-N (ATCC 33728, non-pigmented colony); lane 4, ShT-P (ATCC 33728, pigmented colony); lane 5, MU-4 (M. ulcerans 97–107 African strain, virulent); lane 6, MU-8; (M. ulcerans 5143 Mexican strain, avirulent). Arrows show the position of mycolactone and phthiocerol (control lipid). (B) The ESI/MS spectrum of ASLs from the strain ShT-P detected the peaks of m/z 765.4, 763.4, 779.4 as [M + Na]+. In each MS/MS spectrum, the distinct ions were found m/z 359 and 429 for mycolactone A/B; m/z 357, 429 and 705 for mycolactone S1; m/z 373, 429 and 705 for mycolactone S2, respectively. These molecular and fragment ions were fixed the proposed structures of each mycolactone.
We confirmed the deletion of the giant plasmid in ShT-N from ShT-P by three methods: PCR, pulse field gel electrophoresis (PFGE), and WGS. We used regular PCR to check for the presence of eight ORFs that are associated with mycolactone expression and are expected to be in the giant plasmid (Table 2 and Supplemental Fig. 1)9,16. The ShT-P isolate produced all expected products, as did the virulent reference strains MU-4 and MU-1615. We did not detect any of the ORFs in ShT-N samples, and in the avirulent MU-8 strain only the MUP011, MUP038, and mlsAT(II) ORFs were absent. In PFGE experiments, we detected a <194 kbp band corresponding to the size of the giant plasmid (estimated ~167 kbp8) in the ShT-P isolate; however, the ShT-N isolate did not produce this band (Fig. 3 and Supplemental Fig. 2)17. Results for WGS analysis were consistent with the previous experiments in that sequences from the giant plasmid were absent from the ShT-N isolate, but were present in the ShT-P isolate (data not shown).
Table 2.
PCR detection of eight giant plasmid-associated genes in M. ulcerans or “M. shinshuense” strains.
| pMUM001 marker genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| repA | parA | MUP011 (STPK) | mls | mlsAT | MUP038 (TEII) | MUP045 (KSIII) | MUP053 (p450) | |
| ShT-P | + | + | − | + | + | + | + | + |
| ShT-N | − | − | − | − | − | − | − | − |
| Sh-753 | + | + | − | + | + | + | + | + |
| MU-4 | + | + | + | + | + | + | + | + |
| MU-1615 | + | + | + | + | + | + | + | + |
| MU-8 | + | + | − | + | − | − | + | + |
ShT-P: ATCC 33728, pigmented strain; ShT-N: ATCC 33728, non-pigmented strain; Sh-753: “M. shinshuense”, virulent; MU-4: M. ulcerans 97–107 African strain, virulent; MU-1615: M. ulcerans Malaysian strain, virulent; MU-8: M. ulcerans 5143 Mexican strain, avirulent; STPK, serine/threonine protein kinase gene; mlsAT, acyltransferase domain of mls; TEII, type II thioesterase gene; KSIII, type III ketosynthase gene; p450, p450 hydroxylase gene.
Figure 3.
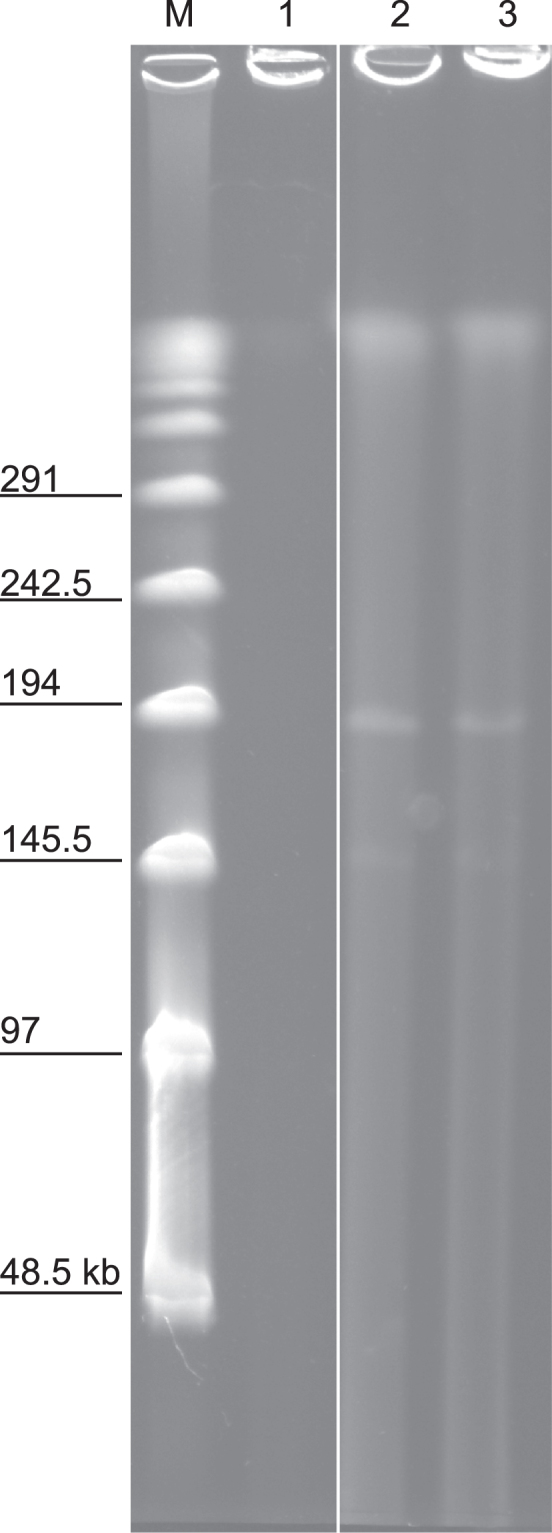
Pulse field gel electrophoresis of “M. shinshuense” or M. ulcerans. Samples were loaded as follows: lane 1, ShT-N (ATCC 33728, non-pigmented colony); lane 2, ShT-P (ATCC 33728, pigmented colony); lane 3, Sh-753 (“M. shinshuense”, virulent); M, lambda PFG DNA size ladder. Arrow indicates the size of a giant plasmid, approximately 167 kbps.
Cytotoxicity of “M. shinshuense” ShT-P and ShT-N isolates
Previous work demonstrated that mycolactone can induce apoptosis and eventually cell death in various primary cells or cell lines, including murine L929 fibroblast cells10,18. To evaluate the cytotoxicity of the ShT-P and ShT-N strains, we incubated L929 cells for 24 hours in the presence of filter-sterile bacterial culture supernatant (SF). L929 cells rounded up and detached from the bottom of the culture dish when exposed to ShT-P SF, Sh-753 SF, or MU-4 SF, suggesting that mycolactone induced cellular damage. In contrast, cells retained a normal morphology when incubated with ShT-N SF or MU-8 SF (Supplemental Fig. 3).
Lethality of “M. shinshuense” ShT-P and ShT-N isolates in mice
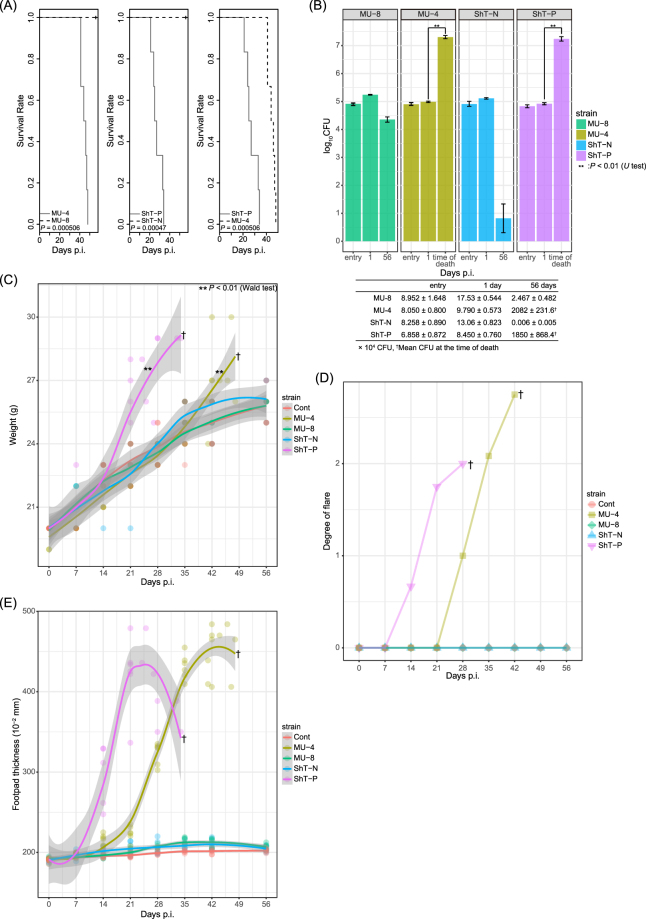
Several reports showed that M. ulcerans mycolactone causes apoptosis in vitro and/or ulcers in vivo12,18–21. To verify that the lack of mycolactone can prevent disease in vivo, we inoculated live bacilli from ShT-P or ShT-N isolates into the footpads of BALB/c mice. All mice given ShT-P were dead within 5 weeks of inoculation, which was significantly faster than those given MU-4, a control virulent strain (p < 0.00051, Fig. 4A right panel). However, all mice inoculated with ShT-N or MU-8 survived for the full course of the experiment (>8 weeks). The number of inoculated bacteria and the infectivity measured as colony-forming units (CFU), 24 hours after inoculation did not have a statistically significant (Fig. 4B, entry and 1 day). For mice given ShT-P, local swelling and erythematous lesions and/or ulcerative and necrotic lesions were observed. Two weeks after the inoculation, these mice showed weight, erythema and swelling of the hind footpads that increased over time (Fig. 4C–E), whereas necrotic skin lesions were observed after three weeks (data not shown). At the time of death or 8 weeks post-inoculation, significant swelling and/or erythema were observed in mice inoculated with ShT-P, but not ShT-N (Supplemental Fig. 4).
Figure 4.
(A) Survival curve of infected mice after inoculation with whole, live bacteria. Similar bacterial loads of “M. shinshuense” (ShT-P or ShT-N) or M. ulcerans (MU-4 or MU-8) were inoculated into the footpads of BALB/c mice (n = 6, each group). The mice inoculated with ShT-P died within 5 weeks of inoculation (middle, right panels) whereas those inoculated with ShT-N were all alive even after 8 weeks (middle panel). (B) Bacterial load in hind footpads of mice inoculated with “M. shinshuense” or M. ulcerans at entry, 1 day and 8 weeks after inoculation or at the time of death. Bacterial load is shown as log10 CFU/foot pad. ShT-N CFUs were absent after 8 weeks. (C) Body weight of infected mice or control mice after inoculation with bacteria. Body weight of infected ShT-P and MU-4 was significantly increased relative to that of control animals (p < 0.01). (D) Degree of erythema in the hind footpads of the inoculated mice. 1+ : slight, 2+ : moderate, 3+ : severe. (E) Measurement of hind footpad swelling induced by “M. shinshuense” or M. ulcerans.
Histology and bacterial burden of local lesions
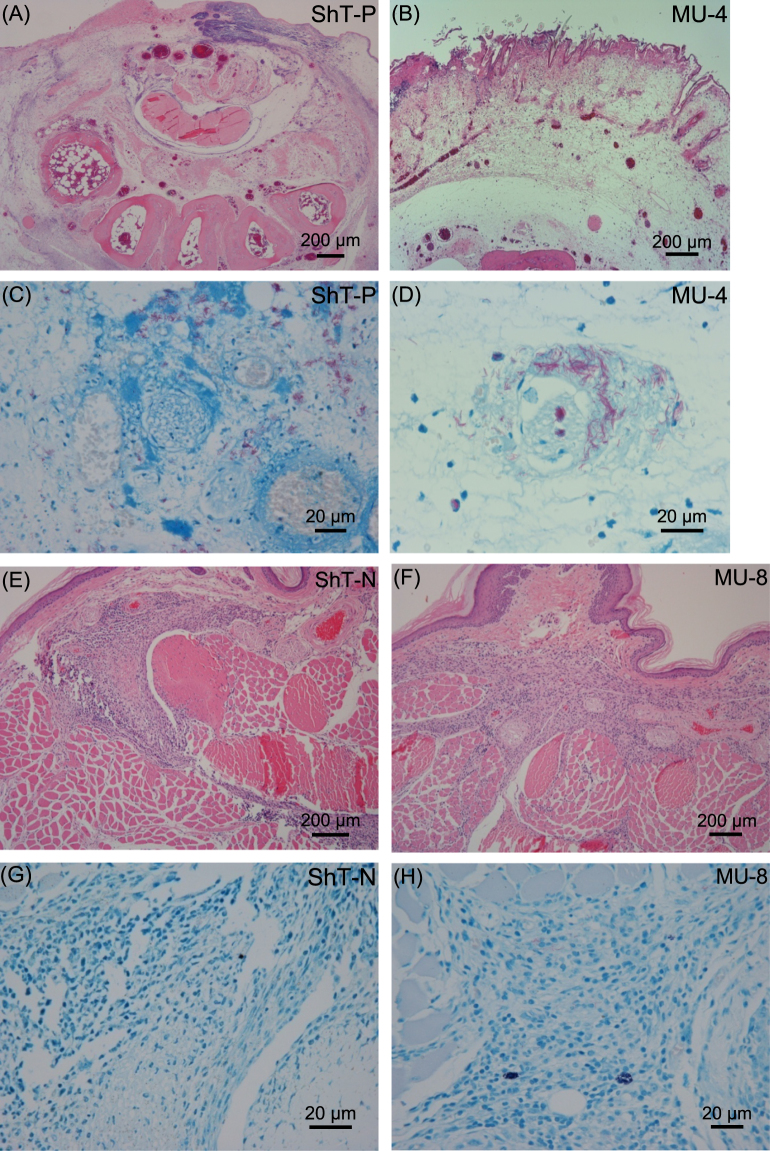
At the time of death, mice inoculated with ShT-P or MU-4 displayed remarkable, deep skin ulcers and extensive subcutaneous edema that was associated with prominent vascular dilatation and occasional thickening of the vascular wall (Fig. 5A). The dermis was edematous with little inflammatory infiltration and mild fibrinous exudation (Fig. 5B). Large numbers of acid-fast bacilli formed clusters mainly in the edematous stroma and focally in the monocytes (Fig. 5C,D). In control mice, some cell infiltration and mild granulomatous change occurred at the inoculation site, but neither necrotic change nor ulceration was seen (Fig. 5E–H). The number of bacilli purified from the hind footpads was significantly higher in ShT-P- or MU-4-inoculated mice compared to ShT-N- or MU-8 inoculated mice (Fig. 4B).
Figure 5.
Representative images of skin lesion tissue sections stained with H&E (A,B,E,F) or Fite-Faraco (C,D,G,H) at the time of death (ShT-P and MU-4) or 8 weeks after inoculation (ShT-N and MU-8). M. ulcerans MU-4 and MU-8 were used as pathogenic positive and negative controls, respectively.
Discussion
During routine culture, we isolated a mycolactone-deficient strain of M. ulcerans subsp. shinshuense from a mycolactone-producing reference strain and confirmed the toxicity of mycolactone. The ORFs required for synthesis of this polyketide toxin (mls) are carried on a giant plasmid found in many strains of M. ulcerans22. We showed that ShT-N lacks this giant plasmid by several ways including WGS analysis and that this plasmid is essential for M. ulcerans and “M. shinshuense” pathogenicity in vitro and in vivo.
Differential diagnosis among nontuberculosis mycobacteria is important since pathogenesis and drug susceptibility can differ significantly among similar species or even subspecies23. Genetic analysis techniques, such as MLST, and conventional biochemical analyses are routinely performed to obtain a differential diagnosis23,24. More recently, WGS has been used to compare species and/or subspecies23. In this study, we demonstrated that WGS analysis can be used to directly compare differences in nucleotides between the ShT-P genome and that of ShT-N by analyzing large numbers of nucleotides to show that the genomes of these two strains were almost identical except for non-synonymous alterations in three ORFs and for frameshifts in five ORFs. These SNPs or Indels could be used in the future to induce deletion of the giant plasmid from “M. shinshuense” strains or reduce “M. shinshuense” pathogenicity. Especially, recent several studies suggest that GntR family transcriptional regulator might be associated with bacterial virulence or pathogenicity25–27.
Mycolactone is a powerful toxin in mammals11. In a previous study, subcutaneous injection of purified toxin into guinea pigs produced severe skin lesions that are similar to human ulcers12. Moreover, when M. ulcerans MU-4 was inoculated in the footpads of mice, necrotic changes without apparent inflammatory infiltration were observed and the affected mice died21. Our study showed that loss of the giant plasmid and mycolactone production from “M. shinshuense” led to a complete loss of bacterial pathogenicity and completely prevented death of inoculated mice. This result supports the idea that mycolactone is critical for virulence of M. ulcerans strains.
Heterogeneity of mycolactone production by various clinical isolates of M. ulcerans has been reported by others13,16,22,28,29. For the MU-8 strain, the giant plasmid is present, but its lack of several key elements for mycolactone synthesis inhibits production of a functional toxin (Fig. 2A). However, studies that compared mycolactone-producing isolates to non-producing isolates from different sources13 were unclear on how much change in virulence occurred. We observed a color change of bacterial colonies during culture of the original giant-plasmid-harboring strain of “M. shinshuense” on Ogawa egg medium slants. We also noted that mlsAT(II) and/or MUP038 might be critical for virulence because both avirulent strains ShT-N and MU-8 were deficient in these genes. On the other hand, the absence of MUP011 from the virulent strains ShT-P and Sh-753 suggests that this gene might not be critical for pathogenesis.
A comparison of the survival curves for mice given ShT-P (“M. shinshuense”) and MU-4 (M. ulcerans) indicated that the pathogenesis of “M. shinshuense” in mice might be stronger than that of M. ulcerans (Fig. 4A). The described below is several speculations to explain the difference. (1) Because both strains possess mycolactone, this difference may be due chromosomal changes. We are currently performing genomic comparison analyses between “M. shinshuense” and M. ulcerans to explore this possibility. (2) Besides with the difference of chromosome, we found the quantitiy of containing plasmids were different, roughly four plasmids harbored in one chromosome in ShT-P, whereas approximately two plasmids in one chromosome in Agy99, typical strain of M. ulcerans30. (3) The quality of mycolactone might be different. M. ulcerans pocesss mycolactone A/B whereas “M. shinshuense” mycolactone A/B, S1, and S2. (4) The in vivo growth rate of bacteria might be different, which may lead the difference of mycolactone production.
We observed that the number of CFU increased between day1 and time of death for mycolactone producing mycobacteria (ShT-P and MU-4) and not for mycolactone deficient mycobacteria (ShT-N and MU-8) (Fig. 4B). This result suggests that the persistence of mycobacteria in mice tissue is due to the presence of mycolactone. In addition, the clearance of CFU is complete for strain ShT-N but not for the non-mycolactone-producing strain MU-8 (Fig. 4B). As neither strains produce functional mycolactone, ShT-N lost a plasmid but MU-8 still has a mutated plasmid, which may lead the difference of the clearance.
In conclusion, we made an avirulent mycolactone-deletion mutant strain directly from the virulent prototypic strain. Our assessment of this strain indicates that “M. shinshuense” pathogenicity was mainly due to mycolactone production. Surprisingly, “M. shinshuense” pathogenesis was stronger than that of M. ulcerans. Because the genomes of these two strains are nearly identical and the most significant differences are whether the giant plasmid is present, this pair would be beneficial for future pathogenicity comparison analyses of the M. ulcerans group.
Materials and Methods
Bacterial strains
The first clinical isolate of “M. shinshuense” was deposited in the ATCC (ATCC 33728) and is considered as the reference strain9. All bacteria were cultured as previously described9. We isolated ShT-P (pigmented) and ShT-N (non-pigmented) strains from “M. shinshuense” strain ATCC 33728s, which was received in December 2004 from The Research Institute of Tuberculosis, Japan (Fig. 1). The virulent “M. shinshuense” clinical isolate JATA 753 (Sh-753)31, virulent M. ulcerans 1615 Malaysian strain (MU-1615), virulent M. ulcerans 97–107 African strain (MU-4), and avirulent M. ulcerans 5143 Mexican strain (MU-8) were used in all experiments. MU-1615, MU-4, and MU-8 were generously provided by Dr. Françoise Portaels (Antwerp, Belgium)9,13. M. marinum ATCC 927T and M. pseudoshottsii JCM 15466T, provided from the Riken Japan Collection of Microorganisms, were used for comparison in some genomic assays, such as MLST6.
Examination of phenotypic properties
Bacterial morphology and acid–alcohol fastness were determined by Ziehl-Neelsen staining as previously described32. Colony morphology, pigmentation, and the ability to grow at various temperatures (25 °C, 32 °C, 37 °C, and 42 °C) were observed on Middlebrook 7H11-OADC agar (Nippon Becton Dickinson, Tokyo, Japan) and 2% Ogawa egg medium slants (Kyokuto Pharmaceutical Industrial, Tokyo, Japan). The following biochemical tests were performed as previously described6,33: Tween 80 hydrolysis, nitrate reductase, semiquantitative catalase, heat-stable catalase (68 °C), arylsulfatase activity (3-day), urease activity, pyrazinamidase, iron uptake, and MPB64 Ag production.
Thin-layer chromatography (TLC)
Mycolactone analyses were performed using silica gel thin-layer chromatography (TLC) as described previously with some modifications34. ASLs were prepared from culture filtrates and analyzed by silica TLC developing with the solvent; chloroform-methanol-water (90:10:1, vol/vol/vol). Lipids were visualized by a phosphomolybdic acid ethanol solution13. The presence of phthiocerol and mycolactone was determined by mass spectroscopy15,35. In briefly, the ASLs dissolved in ethanol were directly perfused into an electrospray ionization source on Thermo Finnigan LCQ Advantage Max LC/MS/MS ion trap mass spectrometer (Thermo Scientific) by using syringe pump YSP-101 (YMC CO. LDT.). The ESI/MS conditions were optimized to the synthetic mycolactone A/B provided by Yoshito Kishi (Harvard University)14. The conditions were as follows, positive mode; infusion rate, 1.0 µl/min; dry temperature, 320 °C. The mycolactones were determined by the molecular ions of ESI/MS and their characteristic fragment ions of ESI/MS/MS.
Pulse field gel electrophoresis (PFGE)
PFGE was performed as previously described36,37. In brief, plugs containing undigested DNA were loaded onto 1% agarose gel and run in Tris-borate-EDTA buffer (0.025 M Tris, 0.5 mM EDTA, 0.0254 M boric acid). PFGE was carried out using a CHEF-DR II system (Bio-Rad Laboratories, Richmond, CA) at 14 °C and 200 V for 20 h. The lambda PFG ladder (Bio-Rad) was used as a molecular size marker. After staining with ethidium bromide, the gel was photographed using a UV transilluminator.
DNA extraction, PCR, and multilocus sequence typing (MLST)
DNA was extracted as previously described35,38–40. In brief, a loop of bacilli was suspended in 400 μl sterilized phosphate buffered saline supplemented with 0.05% Tween 80, and then stored at −80 °C. The frozen sample was crushed with zirconia beads (2 mm diameter) in a bead beating instrument (MagNA Lyser, Roche Diagnostics). Total genomic DNA was purified from the crushed suspension using a High Pure PCR Template Preparation Kit (Roche Diagnostics) according to the manufacturer’s instructions and stored at −20 °C.
PCR products from eight regions of pMUM001 that code for mycolactone-producing enzymes (plasmid replication protein A (repA) (413 bp), plasmid maintenance protein A (parA) (501 bp), M. ulcerans plasmid (MUP) 011 (serine/threonine protein kinase, 479 bp), M. ulcerans type I polyketide synthases loading domain (mls(load)) (560 bp), M. ulcerans type I polyketide synthases acyltransferase domain (mlsAT(II)) (504 bp), MUP038 (type II thioesterase, 500 bp), MUP045 (type III ketosynthase, 496 bp), and MUP053 (p450, 500 bp)) were compared between ShT-P, ShT-N, Sh-753, MU-4, MU-1615, and MU-816. The primers were previously reported and are listed in Supplemental Table 316.
The sequences of the 16S rRNA, the internal transcribed spacer between the 16S and 23S rRNA genes (ITS region), rpoB, and hsp65 genes were analyzed with the primers listed in Supplemental Table 16,41. Amplified PCR products (sizes shown in Supplemental Table 1) were directly sequenced using the ABI Prism 310 PCR genetic analyzer (Applied Biosystems, Foster City, CA)9,35,38–40. ShT-P and ShT-N strains were compared to six related reference strains: “M. shinshuense” Sh-753, M. ulcerans MU-4, M. ulcerans MU-1615, M. ulcerans MU-810, M. marinum ATCC 927T, and Mycobacterium pseudoshottsii JCM 15466T. A similarity search was also performed with ShT-P, ShT-N, and other mycobacterial reference strains using the DNA Data Bank of Japan (DDBJ)6.
Short-read DNA sequencing
WGS analysis of the ShT-P strain is described elsewhere8. The DDBJ/ENA/GenBank accession numbers of the ShT-P chromosome and giant plasmid are AP017624 and AP017625, respectively, whereas that for the ShT-N chromosome is AP017635. The draft genome sequence of the ShT-N strain was determined using the 454 GS FLX Titanium system (Roche Diagnostics). A total of 469,267 single-end and 109,625 paired-end (8 kb insert) reads were assembled with the GS Assembler software version 2.6 (Roche) into one scaffold containing 249 gaps. Illumina paired-end reads (150 × 2; 1,369,886 reads) obtained by a MiSeq sequencer (Illumina, San Diego, USA) were used for sequence-error correction8,42–46.
SNP calling
To detect SNPs between ShT-P and ShT-N strain, we performed the best practice method presented by the authors of the Genome Analysis Toolkit (GATK)47. Briefly, raw sequence reads (FASTQ format) of ShT-N were mapped against the genome sequence of ShT-P, and SNPs and insertions/deletions (indels) were detected from the resulting mapping. Then, the BWA program48. was used to map reads against the genome of ShT-P, resulting in a file of aligned reads in sequence alignment/map (SAM) format49. Using utilities in the SAM tools package49, the SAM file was converted into the binary alignment/map (BAM) format for subsequent processing. Duplicate reads were detected and marked using the “MarkDuplicates” feature of Picard (http://broadinstitute.github.io/picard/). The “RealignerTargetCreator” and “IndelRealigner” functions of GATK50,51 were used to improve the alignment of the reads. The GATK function “HaplotypeCaller” was used to generate a variant call format (VCF) file52, which counts detected SNPs and indels. The “VariantFiltration”, “BaseRecalibrator” and “PrintReads” functions of GATK were used to remove false positives by calibrating the base-quality scores. The Integrative Genomics Viewer (IGV)53,54 was used to visualize the detected SNPs and indels.
Cytotoxicity assay
An assay to determine the toxicity of the different bacterial strains to a murine cell line was performed as previously described12,18,20. Briefly, murine L929 cells were plated at 5 × 104/well in 24-well plates, and sterile filtrate from mycobacterial cultures was added to the wells. At 24 hours post-treatment, the L929 cells were inspected microscopically for cell rounding and detachment.
In vivo infection of mouse footpads
The culture of bacteria in 7H9 broth was allowed to sit for 20 min for the large bacterial clumps to settle down. The resident bacterial suspension was diluted with 7H9 and its optical density was adjusted at 540 nm and then used as inoculum for infection. A suspension of PBS (25 μL) containing various loads of “M. shinshuense” (ShT-P or ShT-N) or M. ulcerans (MU-4 or MU-8) was inoculated into the footpads of 5-week-old female BALB/c mice (n = 6, each group) as previously described21,55. Bacterial load was measured at 24 hours and 8 weeks after inoculation or at the time of death. Hind footpads of mice were disinfected and homogenized in PBS (−) by using a glass homogenizer. The resultant homogenate was subjected to serial 10-fold dilutions with normal saline and 0.1 mL was inoculated onto 7H11 medium. The number of CFU was counted 8 weeks of incubation at 32 °C. Care and treatment of animals followed the regulations of the Animal Care and Use Committee of the National Institute of Infectious Diseases, Japan.
Histopathological examination
Histopathological examination was performed as previously described19 8 weeks after inoculation or at the time of death. After induction of deep anesthesia, mice were fixed by perfusion with 10% formalin. Hind limbs and general organs were then embedded in paraffin, cut into 4 μm sections, and histopathological examination was performed after hematoxylin and eosin (H&E) and Fite-Faraco acid-fast staining. Ulcerative lesion(s), granuloma formation(s), and acid-fast bacilli were evaluated.
Statistical analysis
All statistical analyses were performed with R software (www.r-project.org). The survival curves were plotted according to the Kaplan-Meier method, and statistical significance between infected strains in the assay was assessed by log-lank test. To statistically assess CFUs at each time point, the Mann-Whitney U test was used. To statistically assess the weight of the infected mice, a GLM, which is an extension of the normal linear model, was used. The ANCOVA model, a blend of ANOVA and regression in a multiple linear model, was applied to test the effect of infection on the weight of the mice in the manner of the GLM. The statistical model was defined as η(w) = β0 + β1t · s, where η is the link function, w is a response variable (weight of mice), t is the time post-infection, s is the infected strains, β0 is the intercept, β1 is the fixed effect of interaction between time post-infection and infected strains, of which the explanatory variable t · s of the interaction term was defined as a product of t and s in the linear model. The P values calculated by the Wald test for β1 of the maximum likelihood estimation in each infected strain was used to assess whether the strain affected mouse weight.
Electronic supplementary material
Acknowledgements
We are grateful to Makoto Nakaya and Aiko Shojo (Soai University) for controlling ESI/MS/MS and providing technical assistance and appreciate Yoshito Kishi (Harvard Univeristy) for the donation of synthetic mycolactone A/B. This work was in part supported by a grant from the Japan Agency for Medical Research and Development/Japan International Cooperation Agency (AMED) to Y. Hoshino (jp18fk0108043, jp18fk0108064, jp18fk0108075 and jp18jm0510004), by Grant-in-Aids for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS) to Y. Hoshino (jp18K08312), K. Nakanaga (23591664), and M Goto (jp15K08466) by Grant-in-Aid for Young Scientists (B) to M. Yoshida (jp17K16066) and by Grant-in-Aids for Early-Career Scientists to H. Fukano (jp18K15966). Additional support was provided in part by a JSPS Grant-in-Aid for Scientific Research (S) for Innovative Areas in ‘Genome Science’ (221S0002 and jp16H06279).
Author Contributions
K.N. principally collected, analyzed, and interpreted data. Y.O., A.T., M.Y., and T.O. were responsible for analyzing genome sequences and interpreted data. H.F., N.F., Y.M., N.N., Y.K., S.Mae., M.G., K.T. and N.I. collected and interpreted data. S.Mit., K.S., M.A. and T.H. contributed to interpretation of data. Y.H. was responsible for study design, interpreted data, drafted the manuscript, and revised the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Kazue Nakanaga and Yoshitoshi Ogura contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26425-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.W. H. O. Neglected tropical diseases, http://www.who.int/neglected_diseases/en/ (2016).
- 2.W. H. O. Buruli ulcer, http://www.who.int/mediacentre/factsheets/fs199/en/ (2016).
- 3.Mikoshiba H, Shindo Y, Matsumoto H, Mochizuki M, Tsukamura M. A case of typical mycobacteriosis due to Mycobacterium ulcerans-like organism (author’s transl) Nihon Hifuka Gakkai Zasshi. 1982;92:557–565. [PubMed] [Google Scholar]
- 4.Tsukamura M, Mikoshiba H. A new Mycobacterium which caused skin infection. Microbiol Immunol. 1982;26:951–955. doi: 10.1111/j.1348-0421.1982.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsukamura M, Kaneda K, Imaeda T, Mikoshiba H. A taxonomic study on a mycobacterium which caused a skin ulcer in a Japanese girl and resembled Mycobacterium ulcerans. Kekkaku. 1989;64:691–697. [PubMed] [Google Scholar]
- 6.Nakanaga K, Hoshino Y, Yotsu RR, Makino M, Ishii N. Nineteen cases of Buruli ulcer diagnosed in Japan from 1980 to 2010. J Clin Microbiol. 2011;49:3829–3836. doi: 10.1128/JCM.00783-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanaga K, et al. Buruli ulcer and mycolactone-producing mycobacteria. Jpn J Infect Dis. 2013;66:83–88. doi: 10.7883/yoken.66.83. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida, M. et al. Complete Genome Sequence of Mycobacterium ulcerans subsp. shinshuense. Genome Announc4, 10.1128/genomeA.01050-16 (2016). [DOI] [PMC free article] [PubMed]
- 9.Nakanaga K, et al. “Mycobacterium ulcerans subsp. shinshuense” isolated from a skin ulcer lesion: identification based on 16S rRNA gene sequencing. J Clin Microbiol. 2007;45:3840–3843. doi: 10.1128/JCM.01041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stinear TP, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci USA. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demangel C, Stinear TP, Cole ST. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat Rev Microbiol. 2009;7:50–60. doi: 10.1038/nrmicro2077. [DOI] [PubMed] [Google Scholar]
- 12.George KM, et al. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 13.Mve-Obiang A, Lee RE, Portaels F, Small PL. Heterogeneity of mycolactones produced by clinical isolates of Mycobacterium ulcerans: implications for virulence. Infect Immun. 2003;71:774–783. doi: 10.1128/IAI.71.2.774-783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishi Y. Chemistry of mycolactones, the causative toxins of Buruli ulcer. Proc Natl Acad Sci USA. 2011;108:6703–6708. doi: 10.1073/pnas.1015252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hande SM, et al. Synthesis and structure of two new mycolactones isolated from M. ulcerans subsp. shinshuense. Org Lett. 2012;14:4618–4621. doi: 10.1021/ol302072b. [DOI] [PubMed] [Google Scholar]
- 16.Stinear TP, et al. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J Bacteriol. 2005;187:1668–1676. doi: 10.1128/JB.187.5.1668-1676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip MJ, et al. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol. 2007;189:2021–2029. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George KM, Pascopella L, Welty DM, Small PL. A Mycobacterium ulcerans toxin, mycolactone, causes apoptosis in guinea pig ulcers and tissue culture cells. Infect Immun. 2000;68:877–883. doi: 10.1128/IAI.68.2.877-883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.En J, et al. Mycolactone is responsible for the painlessness of Mycobacterium ulcerans infection (buruli ulcer) in a murine study. Infect Immun. 2008;76:2002–2007. doi: 10.1128/IAI.01588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George KM, Barker LP, Welty DM, Small PL. Partial purification and characterization of biological effects of a lipid toxin produced by Mycobacterium ulcerans. Infect Immun. 1998;66:587–593. doi: 10.1128/iai.66.2.587-593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto M, et al. Nerve damage in Mycobacterium ulcerans-infected mice: probable cause of painlessness in buruli ulcer. Am J Pathol. 2006;168:805–811. doi: 10.2353/ajpath.2006.050375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doig KD, et al. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics. 2012;13:258. doi: 10.1186/1471-2164-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino Y, Suzuki K. Differential diagnostic assays for discriminating mycobacteria, especially for nontuberculous mycobacteria: what does the future hold? Future Microbiol. 2015;10:205–216. doi: 10.2217/fmb.14.120. [DOI] [PubMed] [Google Scholar]
- 24.Stinear TP, Jenkin GA, Johnson PD, Davies JK. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J Bacteriol. 2000;182:6322–6330. doi: 10.1128/JB.182.22.6322-6330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su HZ, et al. Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep. 2016;6:19862. doi: 10.1038/srep19862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu K, et al. CpsR, a GntR family regulator, transcriptionally regulates capsular polysaccharide biosynthesis and governs bacterial virulence in Streptococcus pneumoniae. Sci Rep. 2016;6:29255. doi: 10.1038/srep29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Yan Q, Wang N. Deciphering the regulon of a GntR family regulator via transcriptome and ChIP-exo analyses and its contribution to virulence in Xanthomonas citri. Mol Plant Pathol. 2017;18:249–262. doi: 10.1111/mpp.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi W, Kaser M, Roltgen K, Yeboah-Manu D, Pluschke G. Genomic diversity and evolution of Mycobacterium ulcerans revealed by next-generation sequencing. PLoS Pathog. 2009;5:e1000580. doi: 10.1371/journal.ppat.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira MS, et al. Infection with Mycobacterium ulcerans induces persistent inflammatory responses in mice. Infect Immun. 2005;73:6299–6310. doi: 10.1128/IAI.73.10.6299-6310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stinear TP, Pryor MJ, Porter JL, Cole ST. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology. 2005;151:683–692. doi: 10.1099/mic.0.27674-0. [DOI] [PubMed] [Google Scholar]
- 31.Kazumi Y, et al. Mycobacterium shinshuense isolated from cutaneous ulcer lesion of right lower extremity in a 37-year-old woman. Kekkaku. 2004;79:437–441. [PubMed] [Google Scholar]
- 32.Chapin, K. C. Manual of Clinical Microbiology. 9th edn, 182–191 (American Society for Microbiology, 2007).
- 33.Kent, P. T. & Kubica, G. P. Public health mycobacteriology: a guide for the level III laboratory. (1985).
- 34.Parez JJ, Fauville-Dufaux M, Dossogne JL, de Hoffmann E, Pouthier F. Faster identification of mycobacteria using gas liquid and thin layer chromatography. Eur J Clin Microbiol Infect Dis. 1994;13:717–725. doi: 10.1007/BF02276054. [DOI] [PubMed] [Google Scholar]
- 35.Nakanaga K, et al. Multiple cases of cutaneous Mycobacterium massiliense infection in a “hot spa” in Japan. J Clin Microbiol. 2011;49:613–617. doi: 10.1128/JCM.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekizuka T, et al. Complete genome sequence and comparative genomic analysis of Mycobacterium massiliense JCM 15300 in the Mycobacterium abscessus group reveal a conserved genomic island MmGI-1 related to putative lipid metabolism. PLoS One. 2014;9:e114848. doi: 10.1371/journal.pone.0114848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace RJ, Jr., et al. DNA large restriction fragment patterns of sporadic and epidemic nosocomial strains of Mycobacterium chelonae and Mycobacterium abscessus. J Clin Microbiol. 1993;31:2697–2701. doi: 10.1128/jcm.31.10.2697-2701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanaga K, et al. Mycobacterium shigaense sp. nov., a novel slowly growing scotochromogenic mycobacterium that produced nodules in an erythroderma patient with severe cellular immunodeficiency and a history of Hodgkin’s disease. J Dermatol. 2012;39:389–396. doi: 10.1111/j.1346-8138.2011.01355.x. [DOI] [PubMed] [Google Scholar]
- 39.Nakanaga K, Hoshino Y, Yotsu RR, Makino M, Ishii N. Laboratory procedures for the detection and identification of cutaneous non-tuberculous mycobacterial infections. J Dermatol. 2013;40:151–159. doi: 10.1111/1346-8138.12047. [DOI] [PubMed] [Google Scholar]
- 40.Nakanaga K, et al. Discrimination of Mycobacterium abscessus subsp. massiliense from Mycobacterium abscessus subsp. abscessus in clinical isolates by multiplex PCR. J Clin Microbiol. 2014;52:251–259. doi: 10.1128/JCM.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portaels F, et al. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J Clin Microbiol. 1996;34:962–965. doi: 10.1128/jcm.34.4.962-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukano, H. et al. Complete Genome Sequence of Mycobacterium stephanolepidis. Genome Announc510.1128/genomeA.00810-17 (2017). [DOI] [PMC free article] [PubMed]
- 43.Yoshida, M. et al. Complete Genome Sequence of a Type Strain of Mycobacterium abscessus subsp. bolletii, a Member of the Mycobacterium abscessus Complex. Genome Announc6, 10.1128/genomeA.01530-17 (2018). [DOI] [PMC free article] [PubMed]
- 44.Yoshida, M. et al. Draft Genome Sequence of Mycobacterium sp. Strain shizuoka-1, a Novel Mycobacterium Isolated from Groundwater of a Bathing Facility in Shizuoka, Japan. Genome Announc5, 10.1128/genomeA.01309-17 (2017). [DOI] [PMC free article] [PubMed]
- 45.Yoshida, M., Miyamoto, Y., Ogura, Y., Hayashi, T. & Hoshino, Y. Complete Chromosome Sequence of a Mycolactone-Producing Mycobacterium, Mycobacterium pseudoshottsii. Genome Announc5, 10.1128/genomeA.01363-17 (2017). [DOI] [PMC free article] [PubMed]
- 46.Yoshida, M. et al. The Complete Genome Sequence of Mycobacterium ulcerans subspecies shinshuense. Genome Annoucements (2016). [DOI] [PMC free article] [PubMed]
- 47.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakanaga K, Saito H, Ishii N, Goto M. Comparison of inhibitory effect of rifalazil and rifampicin against Mycobacterium ulcerans infection induced in mice. Kekkaku. 2004;79:333–339. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.