Abstract
Leber congenital amaurosis (LCA) is a genetically and clinically heterogeneous disease, and represents the most severe form of inherited retinal dystrophy (IRD). The present study reports the mutation spectra and frequency of known LCA and IRD-associated genes in 34 Japanese families with LCA (including three families that were previously reported). A total of 74 LCA- and IRD-associated genes were analysed via targeted-next generation sequencing (TS), while recently discovered LCA-associated genes, as well as known variants not able to be screened using this approach, were evaluated via additional Sanger sequencing, long-range polymerase chain reaction, and/or copy number variation analyses. The results of these analyses revealed 30 potential pathogenic variants in 12 (nine LCA-associated and three other IRD-associated) genes among 19 of the 34 analysed families. The most frequently mutated genes were CRB1, NMNAT1, and RPGRIP1. The results also showed the mutation spectra and frequencies identified in the analysed Japanese population to be distinctly different from those previously identified for other ethnic backgrounds. Finally, the present study, which is the first to conduct a NGS-based molecular diagnosis of a large Japanese LCA cohort, achieved a detection rate of approximately 56%, indicating that TS is a valuable method for molecular diagnosis of LCA cases in the Japanese population.
Introduction
Leber congenital amaurosis (LCA; MIM 204000) is the most severe inherited retinal dystrophies (IRDs). Its incidence among the general population is estimated to be only three per 100,000. It is known to affect approximately 20% of children attending schools for the blind, and comprises at least 5% of all IRDs1,2.
LCA is a genetically and clinically heterogeneous disease, whose clinical features include blindness or severe visual impairment within the first year of life, nystagmus, very poor or absent ocular pursuit, and a severely reduced or absent electroretinogram (ERG). The fundus appearance in LCA patients varies, either appearing relatively normal, or exhibiting obvious pigmentary changes similar to those seen in patients with retinitis pigmentosa (RP). Other signs and symptoms which may or may not be present in patients with LCA include photophobia, night blindness, hyperopia, macular/peripheral retinal abnormalities, keratoconus, and cataracts3–5.
LCA is predominantly inherited as an autosomal recessive (ar) trait; however, rare cases of autosomal dominant (ad) LCA have also been reported5. A total of 25 LCA-associated genes have been identified to date (https://sph.uth.edu/retnet/; accessed 14th December 2017). Variants in these genes are estimated to cause approximately 70% of all LCA cases5–8.
Gene therapies represent a promising treatment option for patients with LCA9; however, their application requires the accurate and effective molecular identification of eligible patients. In addition, LCA has many overlapping clinical features to those observed in other IRDs, which can make it difficult to accurately diagnose a patient. Thus, methods for comprehensive variant screening of patients with LCA are urgently required.
In recent years, the development of DNA sequencing technologies such as next generation sequencing (NGS) has enabled the simultaneous detection of multiple known and novel gene variants. The present study thus performed a comprehensive molecular diagnosis of the largest Japanese LCA cohort investigated to date, by using a targeted-NGS (TS) approach to evaluate the mutation spectra and frequency of known LCA-associated genes in Japanese families with a history of the disease.
Results
Patient recruitment
In this study, we newly recruited 33 patients from 31 families who were diagnosed with LCA and fulfilled the three criteria (as described in the Methods section). The results of previously reported 6 patients in 3 families were also included10,11, resulting in a total of 39 patients in 34 families analysed. Among 34 families, 30 families include isolate cases with LCA, three families include two patients with LCA, and one family include three patients with LCA10,11.
Identification of potential pathogenic variants in 19 families
The target-capture panel used in the present study comprised 445,968 bp derived from 1182 target regions of the 74 genes10. The targeted region of each analysed gene included all exons and flanking intronic sequences (i.e. the intronic sequence ±25 bp from each exon boundary), and the coverage rate was 98.98% (details see Supp. Table 1). The analysed target regions achieved an average 223.9 ± 47-fold coverage across the 33 newly recruited samples, and an average of 91.4% and 86.1% of bases in the target regions exhibited 10-fold and 20-fold coverage, respectively. These results indicate that sufficient coverage was achieved for the identification of variants.
The obtained sequence data were analysed using the filtering criteria described in the methods section. Consequently, in 19 of the 34 analysed families, we identified 30 potential pathogenic variants, of which 16 were novel. All potential pathogenic variants identified in this study were confirmed via Sanger sequencing. These included 16 families that harboured variants in nine LCA-associated genes, and three families that exhibited variants in three other IRD-associated genes. The 30 identified variants consisted of 4 nonsense, 9 frameshift11, 3 splice site10,11 and 14 missense variants10 (Tables 1 and 2). The pathogenicity of the novel missense variants was supported by conducted in silico prediction analyses (Supp. Table 2).
Table 1.
Potential pathogenic variants were identified in 19 of the analysed families.
| Family ID | Patient ID | Affected gene | Inherit –ance mode | Identified variant | Zygosity | Origin (as per segregation analysis) | ACMG classifi-cation | Reference |
|---|---|---|---|---|---|---|---|---|
| EYE42 | EYE42 | NMNAT1 | AR | c.1A > G;p.? | Het | Maternal | P | 13 |
| c.709C > T;p.(R237C) | Het | Paternal | P | 12,13 | ||||
| EYE68 | EYE68 | CRB1 | AR | c.668dupT;p.(L223Ffs*4) | Het | Maternal | P | Novel |
| c.733dupG;p.(A245Gfs*16) | Het | Maternal | P | Novel | ||||
| c.1567dupC;p.(L523Pfs*28) | Het | Paternal | P | Novel | ||||
| EYE69 | EYE69 | CEP290 | AR | c.2390delA;p.(K797Sfs*2) | Het | Maternal | P | 43 |
| c.6889A > T;p.(K2297*) | Het | Paternal | P | Novel | ||||
| EYE70 | EYE70 | CRX | AD | c.124G > A;p.(E42K) | Het | de novo | P | 15 |
| EYE115 | EYE115 | CRB1 | AR | c.1334_1740del;p.(C445Yfs*8) | Het | Maternal | P | Novel |
| c.1576C > T;p.(R526*) | Het | Paternal | P | 42 | ||||
| EYE121 | EYE121 | CRB1 | AR | c.2T > C;p.? | Het | NAc | P | Novel |
| c.3068T > G;p.(L1023R) | Het | Maternal | L.P | Novel | ||||
| EYE125 | EYE125 | IMPDH1 | AD | c.590A > C;p.(Q197P) | Het | de novo | L.P | Novel |
| EYE139 | EYE139 | GUCY2D | AR | c.2765A > G;p.(Y922C) | Het | Maternal | L.P | Novel |
| c.2983C > T;p.(R995W) | Het | Paternal | P | 14 | ||||
| EYE149 | EYE149 | RPGRIP1 | AR | c.799C > T;p.(R267*) | Het | Paternal | P | 15 |
| c.1687C > T;p.(R563*) | Het | Maternal | P | Novel | ||||
| EYE156 | EYE156 | PRPH2 | AR | c.730_736delinsCAGCTCCTCCAGACGGGTGCACCAGAC;p.(N244Qfs*19) | Het | Maternal | P | Novel |
| c.748T > G;p.(C250G) | Het | Paternal | L.P | Novel | ||||
| EYE159 | EYE159 | NMNAT1 | AR | c.196C > T;p.(R66W) | Het | Maternal | P | 13 |
| c.709C > T;p.(R237C) | Het | Paternal | P | 12,13 | ||||
| JIKEI-145 | JU1039 | LRAT | AR | c.163C > T;p.(R55W) | Het | Paternal | P | 33 |
| Exon 1–3 deletion | Het | Maternal | P | Novel | ||||
| JU1040 | LRAT | AR | c.163C > T;p.(R55W) | Het | Paternal | P | 33 | |
| Exon 1–3 deletion | Het | Maternal | P | Novel | ||||
| S132 | S132 | NMNAT1 | AR | c.196C > T;p.(R66W) | Het | Paternal | P | 13 |
| c.709C > T;p.(R237C) | Het | Maternal | P | 12,13 | ||||
| EYE50 | EYE50 | RPGR | XL | c.977A > C;p.(K326T) | Hemi | Maternal | L.P | Novel |
| EYE114 | EYE114 | RP2 | XL | c.769–2A > G | Hemi | Maternal | P | Novel |
| EYE187 | EYE187 | BEST1 | AD | c.682G > T;p.(D228Y) | Het | Paternal (mosaic)b | L.P | Novel |
| EYE20a | EYE20 | RPGRIP1 | AR | c.3565_3571delCGAAGGC;p.(R1189Gfs*7) | Hom | Biparental | P | 42 |
| EYE64 | RPGRIP1 | AR | c.3565_3571delCGAAGGC;p.(R1189Gfs*7) | Hom | Biparental | P | 42 | |
| EYE65 | RPGRIP1 | AR | c.3565_3571delCGAAGGC;p.(R1189Gfs*7) | Hom | Biparental | P | 42 | |
| EYE55a | EYE55 | RPGRIP1 | AR | c.1467+1G > T | Het | Paternal | P | 11 |
| Exon 17 deletion (c.2710 + 374_2895 + 74del) | Het | Maternal | P | 11,25 | ||||
| LCA1Ha | Twin 1 | GUCY2D | AR | c.2113 + 2_2113 + 3insT | Het | Paternal | P | 10 |
| c.2714 T > C;p.(L905P) | Het | Maternal | P | 10 | ||||
| Twin 2 | GUCY2D | AR | c.2113 + 2_2113 + 3insT | Het | Paternal | P | 10 | |
| c.2714T > C;p.(L905P) | Het | Maternal | P | 10 |
aMutation analyses for families EYE20, EYE55, and LCA1H were reported previously10,11.
bSee Supp. Figure 2 for sequencing electropherogram data.
cDNA was unavailable for the patient’s father, as he was deceased.
NA, not available; Hom, homozygous; Het, heterozygous; Hemi, hemizygous; P, pathogenic; L.P, likely pathogenic; AD, autosomal dominant; AR, autosomal recessive; XL, X-linked.
Table 2.
Identified potential pathogenic variants in Leber congenital amaurosis (LCA)- and other inherited retinal dystrophies (IRD)-associated genes.
| Disorder | Gene | Accession number | Variant type | Nucleotide change | Location in gene | Patients found to harbour variant | SNP ID | Database listingsa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HGVD | ToMMo | 1000 Genomes | ExAC | ||||||||
| LCA | CRB1 | NM_201253.2 | Insertion | c.668dupT;p.(L223Ffs*4) | Exon 3 | EYE68 | 0 | 0 | 0 | 0 | |
| c.733dupG;p.(A245Gfs*16) | Exon 3 | EYE68 | 0 | 0 | 0 | 0 | |||||
| c.1567dupC;p.(L523Pfs*28) | Exon 6 | EYE68 | 0 | 0 | 0 | 0 | |||||
| Nonsense | c.1576C > T;p.(R526*) | Exon 6 | EYE115 | rs114342808 | 0 | 0.0002 | 0 | 4.94 × 10−5 | |||
| Deletion | c.1334_1740del;p.(C445Yfs*8) | Exon 6 | EYE115 | 0 | 0 | 0 | 0 | ||||
| Missense | c.2T > C;p.? | Exon 1 | EYE121 | 0 | 0 | 0 | 0 | ||||
| c.3068T > G;p.(L1023R) | Exon 9 | EYE121 | 0 | 0.0002 | 0 | 0 | |||||
| NMNAT1 | NM_022787.3 | Missense | c.1A > G;p.? | Exon 2 | EYE42 | 0 | 0 | 0 | 8.26 × 10−6 | ||
| c.196C > T;p.(R66W) | Exon 3 | EYE159, S132 | 0 | 0.0002 | 0 | 0.0001 | |||||
| c.709C > T;p.(R237C) | Exon 5 | EYE42, EYE159, S132 | rs375110174 | 0.0009 | 0.0002 | 0 | 7.42 × 10−5 | ||||
| RPGRIP1 | NM_020366.3 | Deletion | c.3565_3571delCGAAGGC;p.(R1189Gfs*7) | Exon 22 | EYE20, EYE64, EYE65, EYE170 | 0 | 0 | 0 | 1.66 × 10−5 | ||
| Splicing | c.1467 + 1G > T | Intron 11 | EYE55 | 0 | 0 | 0 | 0 | ||||
| Deletion | Exon 17 deletion (c.2710 + 374_2895 + 74del) | Exon 17 | EYE55, EYE16, JU0954, JU0955 | 0 | 0 | 0 | 0 | ||||
| Nonsense | c.799C > T;p.(R267*) | Exon 5 | EYE149 | rs554396590 | 0 | 0 | 0.0002 | 8.36 × 10−6 | |||
| c.1687C > T;p.(R563*) | Exon 13 | EYE149 | 0 | 0 | 0 | 0 | |||||
| GUCY2D | NM_000180.3 | Splicing | c.2113 + 2_2113 + 3insT | Intron 10 | LCA1H (Twin 1 and 2) | 0 | 0 | 0 | 0 | ||
| Missense | c.2714T > C;p.(L905P) | Exon 14 | LCA1H (Twin 1 and 2) | 0 | 0 | 0 | 0 | ||||
| c.2765A > G;p.(Y922C) | Exon 14 | EYE139 | 0 | 0.0003 | 0 | 0 | |||||
| c.2983C > T;p.(R995W) | Exon 16 | EYE139 | rs61750187 | 0 | 0 | 0 | 0 | ||||
| CEP290 | NM_025114.3 | Deletion | c.2390delA;p.(K797Sfs*2) | Exon 23 | EYE69 | rs781670422 | 0 | 0 | 0 | 5.10 × 10−5 | |
| Nonsense | c.6889A > T;p.(K2297*) | Exon 50 | EYE69 | 0 | 0 | 0 | 0 | ||||
| CRX | NM_000554.4 | Missense | c.124G > A;p.(E42K) | Exon 3 | EYE70 | rs863224863 | 0 | 0 | 0 | 0 | |
| IMPDH1 | NM_000883.3 | Missense | c.590A > C;p.(Q197P) | Exon 8 | EYE125 | 0 | 0 | 0 | 0 | ||
| LRAT | NM_004744.4 | Missense | c.163C > T;p.(R55W) | Exon 2 | JU1039, JU1040 | rs527236079 | 0.0018 | 0.0025 | 0 | 0 | |
| Deletion | Exon 1–3 deletion | Exon 1–3 | JU1039, JU1040 | 0 | 0 | 0 | 0 | ||||
| PRPH2 | NM_000322.4 | Deletion/ Insertion | c.730_736delinsCAGCTCCTCCAGACGGGTGCACCAGAC;p.(N244Qfs*19) | Exon 2 | EYE156 | 0 | 0 | 0 | 0 | ||
| Missense | c.748T > G;p.(C250G) | Exon 2 | EYE156 | 0 | 0 | 0 | 0 | ||||
| Other IRD | RP2 b | NM_006915.2 | Splicing | c.769-2A > G | Intron 2 | EYE114 | 0 | 0 | 0 | 0 | |
| RPGR b | NM_000328.2 | Missense | c.977A > C;p.(K326T) | Exon 9 | EYE50 | 0 | 0 | 0 | 0 | ||
| BEST1 c | NM_004183.3 | Missense | c.682G > T;p.(D228Y) | Exon 6 | EYE187 | 0 | 0 | 0 | 0 | ||
aDatabases include: 1000 Genomes database (1000 Genomes; http://www.1000genomes.org/), Exome Aggregation Consortium database (ExAC; http://exac.broadinstitute.org/), Human Genetic Variation Database (HGVD; http://www.genome.med.kyoto-u.ac.jp/SnpDB/), and Tohoku Medical Megabank Organization database (ToMMo; https://ijgvd.megabank.tohoku.ac.jp/).
bRPGR and RP2 mutations have been previously shown to cause X-linked retinitis pigmentosa (RP).
cMutations in BEST1 gene have been shown to cause Best vitelliform macular dystrophy, autosomal recessive bestrophinopathy, adult-onset vitelliform macular dystrophy, autosomal dominant vitreoretinochoroidopathy, and autosomal dominant RP.
AD, autosomal dominant; AR, autosomal recessive; XL, X-linked.
Variants in LCA-associated genes
Of the identified potential pathogenic variants in LCA-associated genes, 25 (in 14 families) were identified to occur in seven known ar LCA-associated genes, while two (in two families) were identified to occur in two known ad LCA-associated genes (Tables 1 and 2). The most frequently mutated genes identified by the present study were CRB1, NMNAT1, and RPGRIP1, which were each found to occur in three of the 19 families (Fig. 1). The three identified NMNAT1 variants (carried by patients EYE42, EYE159, and S132) were missense variants that were previously shown to be pathogenic12,13, while the five identified RPGRIP1 variants, and five of the seven identified CRB1 variants included both previously reported and novel variants that were all shown to induce a loss-of-function (LOF). Of the remaining seven families, five carried CEP290, LRAT, PRPH2, CRX, and IMPDH1 variants, and two were shown to harbour four GUCY2D variants, three of which were missense variants10,14. The conducted segregation analyses of 19 of the identified families confirmed that the pathogenic alleles were on different chromosomes10,11 (Table 1 and Fig. 2). Although TS data suggested that elder (JU1039) and younger (JU1040) affected sisters in family JIKEI-145 had an apparently homozygous missense variant c.163C > T [p.(R55W)] in exon 3 of the LRAT, the conducted segregation analysis revealed that the sisters’ unaffected father had the heterozygous variant and their unaffected mother did not. This finding suggested the possibility that both the sisters had a heterozygous deletion including exon 3 of LRAT. In fact, quantitative real-time polymerase chain reaction (qPCR) analysis revealed that both the sisters and their mother harboured a heterozygous deletion that included LRAT exons 1–3 (Supp. Figure 1), indicating that the pathogenic two alleles were on different chromosomes (Table 1 and Fig. 2). The patients with LCA in families EYE70 and EYE125 carried a known heterozygous missense variant in CRX15 and a novel heterozygous missense variant in IMPDH1, respectively, both of which occurred de novo (Table 1 and Fig. 2), consistent with an ad mode of inheritance.
Figure 1.

Frequency of identified variants in Leber congenital amaurosis (LCA)-associated genes in the 34 analysed Japanese families. In total, 19 of the 34 analysed families were shown to carry potential pathogenic variants, of which 16 were found to harbour variants in nine LCA- (Blue) and three other IRD-associated (Red) genes, respectively. The most frequently mutated genes in the analysed cohort were CRB1, NMNAT1, and RPGRIP1.
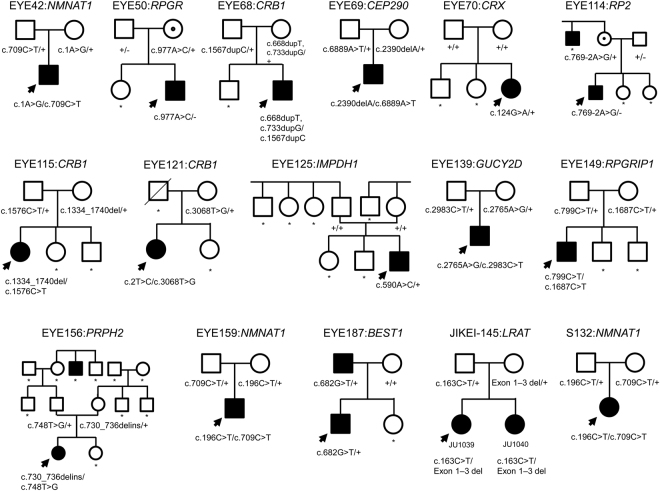
Figure 2.
Pedigrees and segregation analyses for the analysed families. The results of the segregation analyses for families EYE20, EYE55, and LCA1H have been previously reported10,11. Patient EYE68 harboured three variants, of which both p.(L223Ffs*4) and p.(A245Gfs*16) were maternally inherited, and predicted to produce transcripts likely to be targeted for nonsense-mediated mRNA decay, and thus no final protein product. Both variants were thus considered to be likely pathogenic; however, this was not able to be confirmed by the present study. The parents in family EYE156 were normally sighted at the time of their examinations; however, it is possible that this may be a delayed effect of a late-onset form of retinitis pigmentosa (RP) likely to be induced as a result of their identified heterozygous variants in PRPH2, a known autosomal dominant RP-associated gene. The identification code and gene(s) relevant to each family are shown above the pedigree, and the genotype for each family member is shown below their symbol. Probands are indicated by arrows. Square, male; circle, female; black colour, disease-affected; circle with a central black dot, X-linked carrier; slashed symbol, deceased individuals; asterisk, no available DNA sample.
Variants in other IRD-associated genes
Mutations in RPGR and RP2 have been previously shown to cause X-linked RP. We found a novel hemizygous missense variant c.977A > C [p.(K326T)] in RPGR, and a novel hemizygous splice site variant c.769-2A > G in RP2 in families EYE50 and EYE114, respectively, which were maternally transmitted (Table 1 and Fig. 2). BEST1 mutations have been previously shown to cause five clinically distinct retinopathies, including Best vitelliform macular dystrophy, ar bestrophinopathy, adult-onset vitelliform macular dystrophy, ad vitreoretinochoroidopathy, and ad RP16. The present study identified a novel heterozygous BEST1 missense variant c.682G > T [p.(D228Y)] that was carried by the patient with LCA in family EYE187. The patient’s mother was normally sighted, whereas his father had previously been diagnosed with RP. The produced sequencing electropherogram suggested that his father may harbour the variant in a mosaic state (Supp. Figure 2). To evaluate the paternal mosaicism state, it is necessary to perform the segregation analysis using the parental DNA extracted from several tissues. However, the parents did not agree to receive the more elaborate examinations. The p.(D228Y) was a novel missense change at an amino acid residue, where a different missense changes [p.(D228N) and p.(D228H)] were reported to be pathogenic17–19. Xu Y et al. previously described a patient with LCA (patient ID:LH38 in their manuscript) that exhibited a heterozygous BEST1 missense variant p.(D228H)17. Therefore, it is likely that the p.(D228Y) may cause LCA in the patient.
Additional mutation screening
Unsolved families, (i.e. those for which potentially pathogenic variant(s) were not detected using the TS approach) were analysed via additional mutation screening (as described in the Methods section). None of the patients in these families carried the c.2991 + 1655A > G intronic variant in CEP290 that is frequently identified in European, Australian, and Brazilian families with LCA5,7,8. Neither were any of the analysed patients shown to harbour rare variants in five recently discovered LCA-associated genes (CCT2, CLUAP1, DTHD1, GDF6, and IFT140)20–24, nor in exon 15 of RPGR, which is an alternative exon called ORF15 that contains highly repetitive purine-rich sequences (Supp. Tables 3 and 4).
TS data reanalysis and Copy number variation (CNV) analyses
The utilised TS approach identified seven patients (EYE16, EYE47, EYE63, EYE152, EYE170, EYE178, and LCA2H) that carried a single heterozygous rare variant in an ar LCA-associated gene (Supp. Table 3). In addition, the heterozygous known RPGRIP1 exon-17 deletion (c.2710 + 374_2895 + 74del)11,25 was identified in families EYE16 and JIKEI-122 via breakpoint-specific PCR (See the Methods section and Supp. Table 4). These eight families harbour a heterozygous variant in LCA-associated gene; this rate is much higher than the occasional carrier rate of ar variants, suggesting that a second variant is likely located within these genes. Therefore, the generated TS data was used to re-search for splice-site variants within 25 bp of the exon-intron boundaries, and for variants in noncoding exonic regions (see Methods section). The results of these analyses did not reveal any second variants within the analysed genes for these families. Meanwhile, eight of these identified patients, (since patient EYE47 was excluded from further analysis due to a lack of sample DNA), were assessed via a multiplex ligation-dependent probe amplification (MLPA) assay. The results of this assay showed no CNV on the alternate allele in any of the analysed patients (Supp. Table 4).
Patient clinical findings
All enrolled patients met the three criteria (as described in the Methods section), and the 19 patients shown to carry potential pathogenic variants exhibited classical clinical features of LCA. Ophthalmoscopic examinations of the patients revealed various degrees of retinal degeneration, with or without vascular attenuation, macular degeneration, and optic disc pallor. The decimal best-corrected visual acuity (BCVA) in measurable cases ranged from light perception to 0.5.
Among the families assessed by the present study (Fig. 2), the phenotype data collected for family JIKEI-145 (including patients JU1039 and JU1040) were found to be relatively mild (Fig. 3A,B). The proband, JU1039, was a 9-year-old female that exhibited nyctalopia from early childhood. Her BCVA scores at the time of examination were 0.5 and 0.2 (with hyperopia) in the right and left eye, respectively. Her younger sister, JU1040, was a 6-year-old female that also exhibited nyctalopia from early childhood. Her BCVA score at the time of examination were 0.2 and 0.3 (with hyperopia) in the right and left eye, respectively. Fundus examinations revealed almost normal fundi, with only mild diffuse retinal pigment epithelium (RPE) atrophy, and slight retinal vessel narrowing in both patients; however, full-field ERGs were severely decreased and non-recordable for JU1039 and JU1040, respectively. Goldmann perimetry testing showed preserved peripheral visual fields with V-4e isopters, but decreased central sensitivity in both patients. Together these data represent the first report describing the clinical features of Japanese patients with LRAT-associated LCA. Patient EYE139 was found to harbour a GUCY2D mutation, and to exhibit almost normal fundi, with only mild retinal degeneration (Fig. 3C). He was a 9-year-old male with autism, and exhibited nystagmus from birth. His visual acuity (measured using a grating acuity card under binocular conditions), was 0.02 (with hyperopia).
Figure 3.
Clinical findings for six patients with Leber congenital amaurosis-associated mutations. The presented fundus photographs revealed that patients (A) JU1039 (at the age of 9 years), (B) JU1040 (at the age of 6 years), (C) EYE139 (at the age of 22 months), (D) EYE121 (at the age of 8 years), (E) EYE42 (at the age of 18 months), and (F) S132 (at the age of 14 years) all exhibited retinal degeneration, diffuse retinal pigment epithelium (RPE) atrophy, and retinal vessel narrowing; however, the degeneration was milder in patients JU1039, JU1040, and EYE139. Additionally, coloboma-like macular atrophy was observed in patients S132 and EYE42. The accompanying optical coherence tomography (OCT) images presented underneath (A–C), and (F) revealed an unremarkable ellipsoid zone, but a relatively preserved retinal structure and thickness in patients JU1039, JU1040, and EYE139, as well as both severe retinal thinning and a disrupted retinal structure in patient S132.
Conversely, patient EYE121 was found to carry a CRB1 mutation, and to display a more severe phenotype (Fig. 3D). She was a 14-year-old female that exhibited nystagmus from birth. Her BCVA scores at the time of examination were 0.3 and 0.06 (with hyperopia) in the right and left eye, respectively. Fundus examinations revealed marked retinal degeneration with maculopathy, slight retinal vessels narrowing, and pigmentation in the midperiphery. Patients EYE42 and S132 were both shown to carry NMNAT1 mutations, and exhibit very severe phenotypes (Fig. 3E,F). EYE42 was a 16-year-old male with no light perception, while S132 was a 14-year-old female that exhibited only light-perception vision. Both patients exhibited nystagmus from birth, as well as bilateral coloboma-like macular atrophy, diffuse RPE atrophy, and retinal vessel narrowing (identified via the conducted fundus examinations).
Discussion
In this study, we described the mutation screening in a total of 34 Japanese families with LCA (including three previously described families10,11), which we believe is the largest Japanese LCA cohort studied until date. The utilised TS and additional screening methods revealed 30 potential pathogenic variants in 19 of the analysed families. A number of potential pathogenic variants have already been identified in families of various ethnic populations, and several have described variants in Japanese families with LCA25–30. However, this study provides the mutation spectra and frequency of known and novel LCA-causative variants in the analysed Japanese cohorts, which were previously unstudied.
Of the 12 (nine LCA-associated and three other IRD-associated) genes found to contain potential pathogenic variants, the most frequently mutated genes were CRB1 (8.8%, 3/34), NMNAT1 (8.8%, 3/34), and RPGRIP1 (8.8%, 3/34) (Fig. 1). All three of the patients (EYE42, EYE159, and S132) found to harbour a missense variant in NMNAT1 were shown to carry a heterozygous variant c.709C > T [p.(R237C)], and patients EYE159 and S132 were also found to harbour a heterozygous variant c.196C > T [p.(R66W)] (Table 2). While p.(R237C) and p.(R66W) were originally identified as being pathogenic in French, Asian American, and Indian populations12,13, Han J et al. recently reported two unrelated patients from Korean families with LCA that were compound heterozygous for the two variants31. They have since been detected in Japanese families, as well as those of various other ethnicities6–8,12,13,17,31. Conversely, nonsense, frameshift, or canonical splice-site variants in RPGRIP1 were identified in the three of the analysed families (EYE20, EYE55, and EYE149), as was the heterozygous RPGRIP1 exon-17 deletion (see families EYE16, EYE55, and JIKEI-122) (Table 2). A homozygous RPGRIP1 exon-17 deletion variant was previously reported in a Japanese family with LCA25, but not in any other ethnic populations to date. This, together with the results of the present study, suggests that the RPGRIP1 exon-17 deletion variant may be a founder mutation in Japanese LCA.
Other genes found to be mutated in the analysed cohort included GUCY2D (5.9%, 2/34 families), CEP290 (2.9%, 1/34 families), CRX (2.9%, 1/34 families), IMPDH1 (2.9%, 1/34 families), LRAT (2.9%, 1/34 families), PRPH2 (2.9%, 1/34 families), RP2 (2.9%, 1/34 families), RPGR (2.9%, 1/34 families), and BEST1 (2.9%, 1/34 families) (Fig. 1). To the best of our knowledge, this is the first study to report on CEP290, IMPDH1, LRAT, PRPH2, RP2, RPGR, and BEST1 variants in Japanese families with LCA. RP2, RPGR, and BEST1 variants have been previously shown to cause IRDs other than LCA. Only a small number of patients with RPGR- or RP2-associated LCA have been reported in Chinese cohorts6,32; however, there is very limited information regarding patients with BEST1-associated LCA17. Although the BEST1 variant p.(D228Y) identified by the present study was shown to be rare and likely pathogenic based on the conducted in silico analyses, we did not provide experimental evidences supporting the genotype-phenotype correlations. Thus, it remains possible that the variant(s) in other genes not including our designed panel might be pathogenic in patient EYE187. Further analysis with more large-scale cohort study is necessary to better elucidate the genotype-phenotype correlations for patients with LCA that harbour BEST1 variants.
The utilised TS approach was not able to detect mutated genes in 15 of the analysed families (Supp. Table 3). It is likely that these 15 families harbour variants in other IRD-, or novel LCA-associated genes that were not targeted by the utilised TS panel; thus, our research group is planning to re-examine these families using a whole exome sequencing (WES) approach. Likewise, the mutation screening data produced by the present study indicated that eight of the 15 unsolved families carry a single rare heterozygous variant in an ar LCA-associated gene (Supp. Table 3), but no second variants were detected via a reanalysis of the generated TS data or conducted CNV analyses, indicating that additional high-throughput sequencing is required. Therefore, we plan to use a whole genome-sequencing approach to screen for these elusive second variants, since they may be located within gene regulatory regions, deep intronic regions, or unknown exons (e.g. alternative splicing regions).
The present study profiled and compared the mutation spectra in Japanese families with LCA to those previously reported in the literature5–8. The fact that the study detected a large number of novel variants in the analysed cohort suggests that the genetic basis of Japanese families with LCA is unique. The most frequently mutated genes in European, Chinese, Australian, and Brazilian families with LCA are as follows: European, CEP290 (15%), GUCY2D (11.7%), and CRB1 (10%)5; Chinese, CRB1 (16.8%), GUCY2D (10.7%), and RPGRIP1 (7.6%)6; Brazilian, CRB1 (10.7%), CEP290 (10.7%), RPE65 (10.7%)7; Australian, CEP290 (14.7%), GUCY2D (14.7%), and NMNAT1 (8.8%)8. The present study showed that CRB1 was the most frequently mutated gene among the analysed Japanese families and the families of various other ethnic populations, except previously reported Australian families with LCA. Both the present Japanese, and the previously reported Australian families with LCA exhibited a higher proportion (8.8%) of NMNAT1 variants than studied Chinese (2.3%) or Brazilian (3.6%) families with LCA. Conversely, the present Japanese cohort exhibited fewer CEP290 variants (2.9%) than was previously reported to occur in European (15%), Australian (14.7%), and Brazilian (10.7%) families with LCA. Indeed, a large proportion of European, Australian, and Brazilian families with LCA have been shown to carry a founder intronic variant in CEP290 (c.2991 + 1655G > A), that was not detected by the present study (Supp. Table 4). Together, these data strongly suggest that the mutation spectra in Japanese families with LCA are distinctly different to those characteristic of families with LCA of various other ethnic backgrounds.
The present study employed a TS approach that has been demonstrated to capable of accurately and efficiently identifying variants in patients with IRDs10,33,34; however, deep intronic changes and novel genes cannot be identified using this technique. Conversely, the application of WES in genetic diagnosis is aimed for the discovery of novel disease causing genes and WES successfully increases the yield of identification of new causative genes in LCA20–24. Although WES will become a standard method for mutation screening in the near future, the utilised TS approach still has a number of advantages. Firstly, this approach can produce a higher coverage rate for targeted regions. Secondly, it can allow more patients to be analysed in a single assay, which leads to less expensive examinations per patient. Thirdly, output data from TS are relatively small and thus make bioinformatics analyses faster and easier. In addition, it reduces the risk of incidental findings. Thus, the TS approach is currently more practically applicable than WES for screening highly heterogeneous diseases such as IRD.
In conclusion, we reported the results of the first NGS-based molecular diagnosis of the largest Japanese LCA cohort so far to our knowledge. We successfully identified potential pathogenic variants in 19 of the 34 analysed families. The mutation spectra and frequency of LCA-associated genes in the Japanese population appears to be distinctly different from those previously reported for other ethnic populations. Finally, the observed detection rate of approximately 56% indicates that the utilised TS approach is a valuable method for diagnosing LCA, and as such, may facilitate the future application of gene-specific treatments for patients with the disease.
Methods
Ethics statements
This study was approved by the Institutional Review Board for Human Genetic and Genome Research at the Hamamatsu University School of Medicine (permit no. 14-040), the National Centre for Child Health and Development (permit no. 686), the Jikei University School of Medicine (permit no. 24-232 6997), and the University of Occupational and Environmental Health (permit no. H29-03). All study procedures adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained either from the participating individuals or from their legal guardians after all the study procedures were explained in detail.
Patient recruitment
The study participants were recruited from all regions of Japan except the Okinawa islands, and were examined at the Department of Ophthalmology of the National Centre for Child Health and Development Hospital in Tokyo (by NA, SN, and TY), the Department of Ophthalmology at the Jikei University School of Medicine Hospital in Tokyo (by TN, TH, SK, and KM), the Department of Ophthalmology, Hamamatsu University Hospital in Hamamatsu (by YH, MS, AH, KK, and DM), or the University of Occupational and Environmental Health in Fukuoka (by HK). All patients met the following criteria: (1) severe visual impairment (e.g. nyctalopia, nystagmus, very poor or absent ocular pursuit, or oculodigital sign) within the first year after birth; (2) a severely reduced or non-detectable ERG; (3) no systemic abnormality other than neurodevelopmental delay at the time of examination. Clinical diagnoses were based on the results of standard ophthalmic examination techniques including fundus photography, optical coherence tomography, and electroretinography; however, the criteria for classifying LCA varied widely between individuals at the various institutions. Thus, it should be noted that the three listed criteria may not be necessarily sufficient for a standard LCA diagnosis.
DNA extraction
Genomic DNA was extracted from peripheral-blood leukocytes collected from patients and available family members using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
Comprehensive molecular diagnosis of LCA-causative variants
Comprehensive mutation screening was performed via the series of steps summarized in Supp. Figure 3.
Target capture and NGS
The integrity of the TS approach used in this study has been previously evaluated using three families that included six patients with LCA10,11. These previous results were combined with those presented in the current study, which used a target-capture panel targeted to 74 IRD genes to analyse 33 new patients with LCA (from 31 families). The panel was designed, the library prepared, and the target-capture sequencing performed as previously described10.
Bioinformatics analyses
The sequence reads were mapped to the human reference genome sequence (GRCh37/hg19) using Burrows-Wheeler Aligner software (v 0.7.15) after trimming the adapter sequence by cutadapt software (v 1.11) and mapped reads around insertion-deletion polymorphisms (INDELs) were realigned by Genome Analysis Toolkit (GATK; v 3.6)35. Base quality scores were recalibrated by GATK. Variant calls were processed by the GATK HaplotypeCaller, and called single nucleotide variants and INDELs were annotated by ANNOVAR software (v 2016Feb01)36. We focused on nonsynonymous variants and splice site variants which are within 5 bp of the exon-intron boundaries (±5 bp), and excluded synonymous and non-coding exonic variants for the analysis. Common genetic variants (allele frequency, >0.005 for recessive variants or >0.001 for dominant variants) in any of the ethnic subgroups found in the following single nucleotide polymorphism (SNP) databases and synonymous variants were treated as possible non-pathogenic sequence alterations in this study: 1000 Genomes database (http://www.1000genomes.org/), Exome Aggregation Consortium database (http://exac.broadinstitute.org/), Human Genetic Variation Database (HGVD; http://www.genome.med.kyoto-u.ac.jp/SnpDB/) and Tohoku Medical Megabank Organization database (ToMMo; https://ijgvd.megabank.tohoku.ac.jp/). HGVD and ToMMo were used as a reference for Japanese controls. The Human Gene Mutation Database (HGMD; https://portal.biobase-international.com/cgi-bin/portal/login.cgi) was used to identify previously reported variants. A bioinformatics analysis of the analysed region was conducted to exclude false positive variants; however, this may have also filtered out true pathogenic variants. Therefore, for those families in which the utilized TS approach only identified a heterozygous rare variant in ar LCA-associated genes, we expanded the analysed region, and re-examined the our TS data to screen for variants located in intronic regions within 25 bp of the exon-intron boundaries (±25 bp)10, and non-coding exonic regions30.
Prioritization and assessment of identified variants
Identified variants were filtered by applying the following prioritization criteria:
Variants present in the HGMD were considered to be known pathogenic mutations.
Nonsense, frameshift, or canonical splice-site variants were considered to be likely pathogenic.
Missense variants were considered to be potentially pathogenic when at least two in silico computational algorithms, (as applied using either SIFT [http://sift.jcvi.org/www/SIFT_seq_submit2.html], PolyPhen2 [http://genetics.bwh.harvard.edu/pph2/], Mutation Taster [http://www.mutationtaster.org/], or CADD [http://cadd.gs.washington.edu/] software), produced a positive score. Splice-site variants were similarly analysed using Neural Network Splice Site Prediction software (http://www.fruitfly.org/seq_tools/splice.html).
We then screened the remaining variants that matched the patients’ phenotypes and the reported inheritance patterns of the respective genes.
Pathogenicity was considered according to the ACMG guidelines37.
Additional mutation screening
Families for whom no potentially pathogenic variant(s) were identified via the utilised TS method underwent additional mutation screening, as below (Supp. Table 4):
The known CEP290 intronic variant c.2991 + 1655A > G38 was not included in the design of the TS capture panel, so genomic CEP290 fragments encompassing c.2991 + 1655A > G had to be amplified and analysed via Sanger sequencing.
Large deletion and insertion variants were not detectable via the applied TS approach; thus, a long-range PCR assay was used to screen the known RPGRIP1 exon 17 deletion11,25.
The LCA-associated genes CCT2, CLUAP1, DTHD1, GDF6, and IFT14020–24 were identified after the capture panel used in the present study was designed (as reported in RetNet at the time of system design; https://sph.uth.edu/retnet/; accessed 23th January 2014). Thus, all coding exons in GDF6 and IFT140 were screened via Sanger sequencing. Given the rarity of reported LCA-associated variants in CCT2, CLUAP1, and DTHD1, only those exons within these genes that contained previously reported variants were analysed.
The RPGR exon ORF15 is a mutational hotspot for X-linked RP; however, it contains repetitive sequences that cannot be efficiently captured or enriched via conventional targeted capture NGS39. Thus, genomic RPGR fragments encompassing the ORF15 region were amplified and analysed via Sanger sequencing for the unsolved male patients with LCA (i.e. EYE16, EYE103, EYE133, EYE178, EYE182, and JU0955).
CNV analyses
Screening for CNV in identified ar LCA-associated genes was performed for families in which the utilised TS or additional mutation screening approaches revealed only a single heterozygous rare variant (Supp. Table 3). SALASA MLPA probemix reagents (P221 for CRX, and P222 for RPGRIP1, GUCY2D, and CEP290; MRC Holland, Amsterdam, Netherland) were used to conduct an MLPA, as previously described40. A qPCR analysis was performed to confirm the family JIKEI-145 segregation analysis findings, using appropriate primers (available upon request), SYBR Premix EX Taq II (Takara, Japan) and a Thermal Cycler Dice TP800 (Takara, Japan), according to the manufacturer’s instructions. Relative comparative threshold cycle (Ct) values were calculated on the basis of the second derivative maximum method using dedicated software (Thermal Cycler Dice Real Time System software, v 5.11B for TP800, Takara, Japan). The relative copy number (RCN) was determined on the basis of the comparative ddCT method using the unaffected father DNA as a control (where an RCN score of 1.0 represented no copy number change, a score of 0.6–1.4 was considered abnormal, and a score <0.6 and >1.4 represented deletions and duplications, respectively). All reactions were performed in duplicate, and all experiments were repeated in triplicate.
Sanger sequencing and segregation analyses
Potential pathogenic variants detected using the TS approach were validated by performing Sanger sequencing as per the standard protocol41. All utilised primers are available upon request. Sanger sequencing segregation analyses were performed on DNA from family members to investigate the co-segregation of potentially pathogenic variants.
Supplementary data
Supplementary data include three figures and four tables10,13–15,25,26,42–47.
Electronic supplementary material
Acknowledgements
This work was supported by a Psychiatric & neurological disorders (no. 16dk0310018h0003 to N.A.), a Rare/intractable disease project of Japan (no. 17ek0109217h0001 to N.A.), and an Initiative on Rare and Undiagnosed Diseases for Adults grant from the Japan Agency for Medical Research and Development (AMED) (no. 16ek0109151h0002 to Y.H.), a National Centre for Child Health and Development grant (no. 25-7 to N.A.), and two Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (no. 17K111447 to Y.H. and no. 16K11284 to K.H.).
Author Contributions
K.H. designed and performed experiments, analysed data, interpreted results, and authored the manuscript, and obtained funding for the project. S.N. and K.K. performed ophthalmic examinations, provided genetic counselling, and authored the manuscript. T.Y., S.K., D.M., A.H., K.M., T.N., H.K., M.S., and T.H. performed ophthalmic examinations and provided genetic counselling. H.S. and S.M. participated in data interpretation, and reviewed the manuscript. M.F. provided genetic counselling and reviewed samples. N.A. and Y.H. designed the project, interpreted results, reviewed the manuscript, and obtained project funding. All authors approved publication of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26524-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Koenekoop RK. An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv. Ophthalmol. 2004;49:379–398. doi: 10.1016/j.survophthal.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Schappert-Kimmijser J, Henkes HE, Van Den Bosch J. Amaurosis congenita (Leber) A.M.A. Arch. Ophthalmol. 1959;61:211–218. doi: 10.1001/archopht.1959.00940090213003. [DOI] [PubMed] [Google Scholar]
- 3.Hanein S, et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat. 2004;23:306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 4.Hanein S, et al. Leber congenital amaurosis: survey of the genetic heterogeneity, refinement of the clinical definition and phenotype-genotype correlations as a strategy for molecular diagnosis. Clinical and molecular survey in LCA. Adv. Exp. Med. Biol. 2006;572:15–20. doi: 10.1007/0-387-32442-9_3. [DOI] [PubMed] [Google Scholar]
- 5.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, et al. Comprehensive Molecular Diagnosis of a Large Chinese Leber Congenital Amaurosis Cohort. Invest. Ophthal. Vis. Sci. 2015;56:3642–3655. doi: 10.1167/iovs.14-15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porto FBO, et al. Molecular Screening of 43 Brazilian Families Diagnosed with Leber Congenital Amaurosis or Early-Onset Severe Retinal Dystrophy. Genes (Basel). 2017;8:355. doi: 10.3390/genes8120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JA, et al. The genetic profile of Leber congenital amaurosis in an Australian cohort. Mol. Genet. Genomic Med. 2017;5:652–667. doi: 10.1002/mgg3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weleber RG, et al. Results at 2 Years after Gene Therapy for RPE65-Deficient Leber Congenital Amaurosis and Severe Early-Childhood-Onset Retinal Dystrophy. Ophthalmology. 2016;123:1606–1620. doi: 10.1016/j.ophtha.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Hosono, K. et al. Novel GUCY2D Gene Mutations in Japanese Male Twins with Leber Congenital Amaurosis. J. Ophthalmol. 693468, 10.1155/2015/693468 (2015). [DOI] [PMC free article] [PubMed]
- 11.Miyamichi D, et al. Changes in macular structure and retinal function in patients with Leber congenital amaurosis with RPGRIP1 mutations. Invest. Ophthalmol. Vis. Sci. 2017;58:574. [Google Scholar]
- 12.Falk MJ, et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat. Genet. 2012;44:1040–1045. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrault I, et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat. Genet. 2012;44:975–977. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 14.Perrault I, et al. Spectrum of retGC1 mutations in Leber’s congenital amaurosis. Eur. J. Hum. Genet. 2000;8:578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- 15.Li L, et al. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PloS One. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson AA, et al. Disease-causing mutations associated with four bestrophinopathies exhibit disparate effects on the localization, but not the oligomerization, of Bestrophin-1. Exp. Eye Res. 2014;121:74–85. doi: 10.1016/j.exer.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Molecular genetics of Leber congenital amaurosis in Chinese: New data from 66 probands and mutation overview of 159 probands. Exp. Eye Res. 2016;149:93–99. doi: 10.1016/j.exer.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Davidson AE, et al. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am. J. Hum. Genet. 2009;85:581–592. doi: 10.1016/j.ajhg.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Huet RA, et al. The efficacy of microarray screening for autosomal recessive retinitis pigmentosa in routine clinical practice. Mol. Vis. 2015;21:461–476. [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Safieh L, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asai-Coakwell M, et al. Contribution of growth differentiation factor 6-dependent cell survival to early-onset retinal dystrophies. Hum. Mol. Genet. 2013;22:1432–1442. doi: 10.1093/hmg/dds560. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, et al. Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum. Genet. 2015;134:1069–1078. doi: 10.1007/s00439-015-1586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minegishi Y, et al. CCT2 Mutations Evoke Leber Congenital Amaurosis due to Chaperone Complex Instability. Sci. Rep. 2016;6:33742. doi: 10.1038/srep33742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soens ZT, et al. Hypomorphic mutations identified in the candidate Leber congenital amaurosis gene CLUAP1. Genet. Med. 2016;18:1044–1051. doi: 10.1038/gim.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, et al. A novel exon 17 deletion mutation of RPGRIP1 gene in two siblings with Leber congenital amaurosis. Jpn J. Ophthalmol. 2014;58:528–535. doi: 10.1007/s10384-014-0339-z. [DOI] [PubMed] [Google Scholar]
- 26.Kondo H, et al. A homozygosity-based search for mutations in patients with autosomal recessive retinitis pigmentosa, using microsatellite markers. Invest. Ophthalmol. Vis. Sci. 2004;45:4433–4439. doi: 10.1167/iovs.04-0544. [DOI] [PubMed] [Google Scholar]
- 27.Ogino K, et al. [Genotype screening of retinal dystrophies in the Japanese population using a microarray] Nippon Ganka Gakkai Zasshi. 2013;117:12–18. [PubMed] [Google Scholar]
- 28.Katagiri S, et al. RPE65 Mutations in Two Japanese Families with Leber Congenital Amaurosis. Ophthalmic Genet. 2014;37:1–9. doi: 10.3109/13816810.2014.991931. [DOI] [PubMed] [Google Scholar]
- 29.Kuniyoshi K, et al. Longitudinal clinical course of three Japanese patients with Leber congenital amaurosis/early-onset retinal dystrophy with RDH12 mutation. Doc. Ophthalmol. 2014;128:219–228. doi: 10.1007/s10633-014-9436-z. [DOI] [PubMed] [Google Scholar]
- 30.Coppieters F, et al. Hidden Genetic Variation in LCA9-Associated Congenital Blindness Explained by 5′UTR Mutations and Copy-Number Variations of NMNAT1. Hum. Mutat. 2015;36:1188–1196. doi: 10.1002/humu.22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, et al. Diagnostic application of clinical exome sequencing in Leber congenital amaurosis. Mol. Vis. 2017;23:649–659. [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J. Med. Genet. 2013;50:674–688. doi: 10.1136/jmedgenet-2013-101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oishi M, et al. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Invest. Ophthal. Vis. Sci. 2014;55:7369–7375. doi: 10.1167/iovs.14-15458. [DOI] [PubMed] [Google Scholar]
- 34.Oishi M, et al. Next-generation sequencing-based comprehensive molecular analysis of 43 Japanese patients with cone and cone-rod dystrophies. Mol. Vis. 2016;22:150–160. [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Hollander AI, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neveling K, et al. Next-generation genetic testing for retinitis pigmentosa. Hum. Mutat. 2012;33:963–972. doi: 10.1002/humu.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hosono, K., Minoshima, S. & Hotta, Y. Retinitis Pigmentosa in Japanese Population in Advances in Vision Research, Volume I (ed. Prakash, G. & Iwata, T.) 693468 (Springer, Tokyo, 2017).
- 41.Hosono K, et al. Two novel mutations in the EYS gene are possible major causes of autosomal recessive retinitis pigmentosa in the Japanese population. PloS One. 2012;7:e31036. doi: 10.1371/journal.pone.0031036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seong MW, et al. Molecular characterization of Leber congenital amaurosis in Koreans. Mol. Vis. 2008;14:1429–1436. [PMC free article] [PubMed] [Google Scholar]
- 43.Carss KJ, et al. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am. J. Hum. Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerber S, et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur. J. Hum. Genet. 2001;9:561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- 45.Katagiri S, et al. Whole exome analysis identifies frequent CNGA1 mutations in Japanese population with autosomal recessive retinitis pigmentosa. PloS One. 2014;9:e108721. doi: 10.1371/journal.pone.0108721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyagawa M, Naito T, Nishio SY, Kamatani N, Usami S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PloS One. 2013;8:e71381. doi: 10.1371/journal.pone.0071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulz HL, et al. Mutation Spectrum of the ABCA4 Gene in 335 Stargardt Disease Patients From a Multicenter German Cohort-Impact of Selected Deep Intronic Variants and Common SNPs. Invest. Ophthalmol. Vis. Sci. 2017;58:394–403. doi: 10.1167/iovs.16-19936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.