Abstract
In recent years, protocols have been established to differentiate stem and precursor cells into more mature cell types. However, progress in this field has been hampered by difficulties to assess the differentiation status of stem cell-derived cells in an unbiased manner. Here, we present an analysis pipeline based on published data and methods to quantify the degree of differentiation and to identify transcriptional control factors explaining differences from the intended target cells or tissues. The pipeline requires RNA-Seq or gene array data of the stem cell starting population, derived ‘mature’ cells and primary target cells or tissue. It consists of a principal component analysis to represent global expression changes and to identify possible problems of the dataset that require special attention, such as: batch effects; clustering techniques to identify gene groups with similar features; over-representation analysis to characterize biological motifs and transcriptional control factors of the identified gene clusters; and metagenes as well as gene regulatory networks for quantitative cell-type assessment and identification of influential transcription factors. Possibilities and limitations of the analysis pipeline are illustrated using the example of human embryonic stem cell and human induced pluripotent cells to generate ‘hepatocyte-like cells'. The pipeline quantifies the degree of incomplete differentiation as well as remaining stemness and identifies unwanted features, such as colon- and fibroblast-associated gene clusters that are absent in real hepatocytes but typically induced by currently available differentiation protocols. Finally, transcription factors responsible for incomplete and unwanted differentiation are identified. The proposed method is widely applicable and allows an unbiased and quantitative assessment of stem cell-derived cells.
This article is part of the theme issue ‘Designer human tissue: coming to a lab near you’.
Keywords: stem cells, transcriptomics, hepatocytes
1. Introduction: the need to quantify stem cell differentiation
In the past two decades, much progress has been made in establishing protocols for differentiation of stem cells into specific, mature cell types such as cardiomyocytes [1,2], neurons [3,4] and hepatocytes [5,6]. However, further development in stem cell research has been hampered by an overoptimistic interpretation of the differentiation status of stem and precursor-cell-derived types. Often, studies relied on a few selected markers as indicators of mature cell or tissue identity. By way of example, ‘multipotent adult progenitor cells' from bone marrow were reported to differentiate into hepatocyte-like cells (HLCs) with ‘functional characteristics’ of hepatocytes [7]. With more than 800 citations, this article strongly influenced the field of stem cell research and created the impression that the task of generating human hepatocytes from in vitro-expanded non-endodermal progenitor cells can be considered as almost accomplished. Also many further studies using different types of stem and progenitor cells offered similar optimistic views (e.g. [8–12]). However, the far-reaching conclusions of the aforementioned studies were based on a set of selected hepatocyte markers and not confirmed by unbiased genome-wide studies. Although articles on stem cell-derived ‘HLCs' have been published already, some more than 10 years ago [13–20], primary hepatocytes isolated from liver tissue still remain the gold standard.
Some early reports warned of too-optimistic interpretations in stem cell-derived hepatocytes [17,21]. However, it was only upon the application of omics technologies and bioinformatics that the degree of differentiation of these ‘HLCs’ could be objectified [22,23]. These studies compared ‘HLCs' derived from human induced pluripotent cells (hiPSCs) and human embryonic stem cells (hESCs) from multiple centres using different differentiation protocols with freshly isolated and cultivated primary human hepatocytes (PHHs) [22,23]. The ground-breaking result of these studies was that the difference between stem cell-derived ‘HLCs’ and real hepatocytes was so large that the term ‘hepatocyte-like’ rather represents a euphemism. On the one hand, hundreds of genes responsible for differentiated functions of hepatocytes are expressed several orders of magnitude lower compared with primary hepatocytes isolated from human liver or compared with liver tissue. On the other hand, stem cell-derived ‘HLCs' contain ‘unwanted’ features, such as expression of colon or fibroblast genes that are not observed in primary hepatocytes [22,23]. Importantly, this feature of unwanted differentiation seems to be a widespread phenomenon, because it was observed for ‘HLCs' obtained from different centres using different protocols [22,23].
Taken together, it has become clear that in vitro differentiation of stem cells does not represent the clear transition of one defined cell state to another. Rather a continuum seems to exist, in which incomplete differentiation towards a target cell type, further named primary differentiation, coincides with the development of unwanted features, termed secondary differentiation. The advantage of genome-wide characterization of stem cell-derived cells is that not only does it give an unbiased and quantitative measure of primary and secondary differentiation, but it also identifies candidate transcription factors potentially responsible for incomplete or unwanted differentiation. This results in a set of transcriptional regulators with too low and too high activities that may serve as a blueprint for fine-tuning of differentiation protocols. Genome-wide characterization requires RNA-Seq or gene array analysis of RNA isolated from the stem cell-derived cells, which have to be compared with RNA from primary cells or tissue. In the case of human liver, hepatocytes are commercially available from several sources.
In the present article, we describe a bioinformatics pipeline based on publicly available software that allows a quantitative, unbiased assessment of the differentiation status (figure 1). As these methods are cost-efficient and the biostatistics require only few hours for an experienced operator, it is strongly recommended that unbiased genome-wide techniques are used instead of or in addition to selected individual hepatocyte markers to come to an objective assessment. Although the pipeline is described for the example of HLCs, the method is applicable for all cell types of stem or precursor cell-derived cells and tissues.
Figure 1.
Analysis pipeline to characterize the differentiation status of stem cell-derived cells by genome-wide data. Technical descriptions of how to apply the individual analyses are provided in the electronic supplementary material, S1.
2. Analysis pipeline for genome-wide expression data of stem cell-derived cell types
After standard processes, such as normalization, the analysis starts with principal component analysis (PCA), identification of gene groups with similar features by clustering techniques, characterization of gene clusters by over-representation analysis, calculation of metagenes and further characterization by gene regulatory networks (GRNs) (figure 1). Below we describe this standardizable workflow, beginning with definition and principles, illustration by examples and the discussion of limitations. The examples were selected from recently published data [23,24]. An important precondition for application of the pipeline is the availability of high-quality genome-wide transcriptional data based on at least three biological replicates. Our chosen examples are based on three to five biological replicates, which significantly reduces the risk of outlier overestimation.
3. Principal component analysis
(a). Definition and principles
PCA allows a first visualization of global gene expression changes induced by a differentiation protocol; it also gives a first impression of the similarity of stem cell-derived cells and the intended cell type. PCA is a statistical procedure that converts a genome-wide set of several correlated groups of genes into a set of uncorrelated variables named principal components (PCs). The number of PCs is smaller than or theoretically equal to the number of genes, but is, in practice, much smaller because many genes cluster in co-behaving groups. In gene expression analyses, it is usually sufficient to consider up to five PCs. The variance explained by individual PCs may vary from dataset to dataset depending on the influence of confounding variables. The first PC (PC1) accounts for the highest degree of variability, the second PC (PC2) the second highest and so on. Individual PCs can be compared in two-dimensional plots to graphically display their specific influence on data point separation. While the first PCs often provide the major part of variance, it is advisable to consider also combinations of minor PCs, e.g. PC1 versus PC3, PC2 versus PC3 and PC1 versus PC4, to discover overlays and separations of data points not visible in the major PCs alone.
(b). Example of application
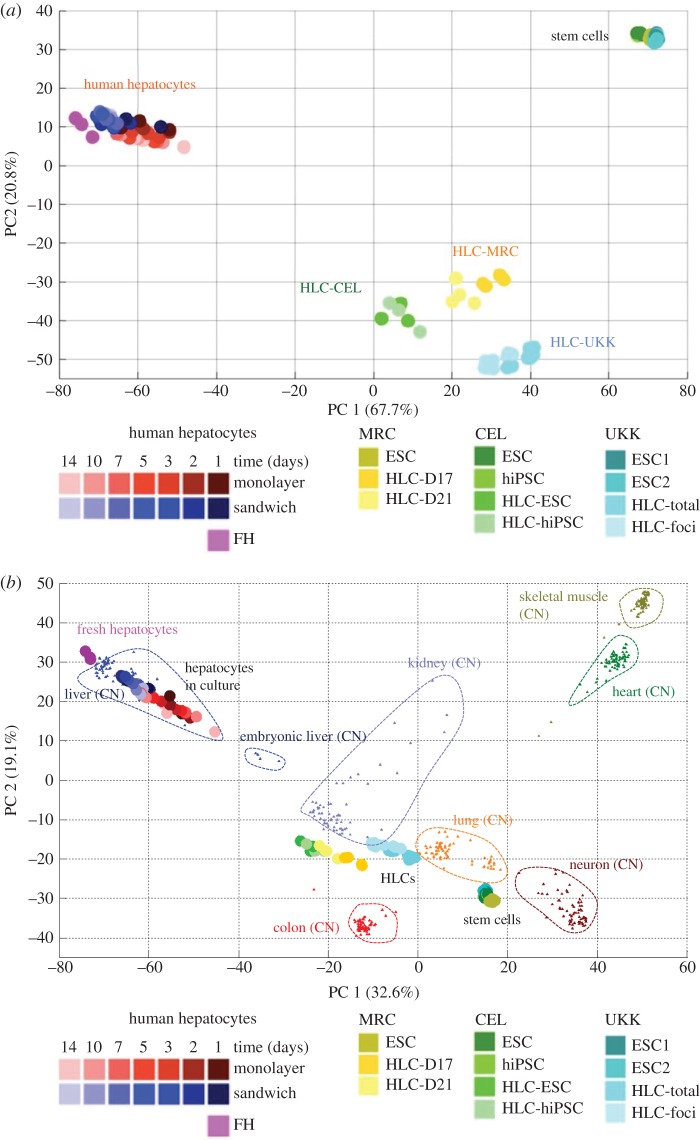
To illustrate the application of PCA, we have chosen an example of transcriptomes of hESCs generated by three different research centres that applied distinct differentiation methods to produce definitive endoderm and ‘HLCs' (figure 2a; from Godoy et al. [23]). The colours yellow, green and blue represent data from three research centres focusing on differentiation of stem cells to HLCs; the circles with identical colour represent three independent experiments. All three centres used hESCs as a starting population. The centre represented by greenish colours additionally included hiPSCs, while the centre represented by yellowish colours additionally compared a 17- and 21-day differentiation period to obtain HLCs. The PCA illustrates the following key features: (i) stem cells before initiating the differentiation process cluster closely together in the upper right corner (figure 2a); there seems to be no major difference between the three involved centres. Also, no major difference seems to exist between hESCs and hiPSCs. After the differentiation process, the generated HLCs shift to the lower middle of the PCA plot. The samples from each centre cluster closely together and the inter-centre influence can be seen, illustrating that the individual protocols and different conditions in the three laboratories have some impact but do not represent the dominant factor of influence. From the PCA presentation, one also learns that there are no major differences between HLCs generated from hESCs or hiPSCs. The 21-day differentiation period leads to clustering at lower values along the PC1 axis compared with the 17-day protocol. Therefore, the results after 21 days are closer to PHHs, but the difference is relatively small. The major goal of genome-wide expression studies is to assess the degree of similarity between HLCs and PHHs. In the PCA plot, PHHs are represented by the purple circles that cluster to the extreme left of PC1 (figure 2a). This illustrates that the stem cells shift along the PC1 in the direction of PHHs, but do not reach their position. Moreover, they shift inversely along PC2, but exceed the position of PHHs. Together, this demonstrates that there are large differences between HLCs and PHHs, despite some promising changes represented by the shift into the inverse orientation of PC1. It is also possible to include further cell types and tissues into the PCA plot, such as human colon, heart, skeletal muscle, neuronal cells, lung, as well as adult and embryonal liver tissue, which can be obtained from public sources (figure 2b). In this plot, human liver tissue from several donors clusters closest to the aforementioned freshly isolated PHHs, which is plausible (figure 2b). It is also understandable that human embryonic liver tissue clusters between the isolated hepatocytes and the stem cell-derived HLCs. However, with respect to quantitative comparisons of the positions of HLCs and the other cell types (lung and kidney), misinterpretations should be avoided as discussed in §3c. A further information obtained from the PCA plot refers to the cultivated PHHs. While the freshly isolated PHHs cluster to the extreme left (purple circles), they time-dependently shift to the bottom right during a 14-day incubation period, independently of whether the hepatocytes are cultivated in monolayer or three-dimensional culture. Interestingly, this cultivation-associated shift brings the PHHs closer to the HLCs. This leads to the hypothesis that owing to the loss of the in vivo environment, PHHs lose the same features that HLCs have not yet or only partially adopted during their incomplete differentiation. The example illustrates that already the explorative PCA may generate hypotheses. Of course, these hypotheses have to be further analysed by more specific methods (as described below) and need to be confirmed by independent experiments.
Figure 2.
Representation of global gene expression changes by PCA. (a) Visualization of stem cells (ESCs and hiPSCs), HLCs and primary human hepatocytes. The HLCs were generated by three different research centres using different protocols, represented by yellowish (centre 1; MRC), greenish (centre 2; CEL) and bluish (centre 3; UKK) colours. Primary human hepatocytes were analysed directly after isolation (FH) or after different periods (1–14 days) in monolayer or sandwich culture. ESC, embryonic stem cells; hiPSC, human induced pluripotent cells; HLC-D17 and HLC-D21, hepatocyte-like cells obtained by a 17-day and 21-day differentiation protocol; HLC-ESC/HLC-hiPSC, hepatocyte-like cells generated from human embryonic stem cells or human induced pluripotent cells, respectively; HLC-total, analysis of the total cell population harvested from the culture dish; HLC-foci, analysis of islets of HLCs manually harvested with the help of a binocular (from Godoy et al. [23]). (b) Similar PCA plot to that shown in (a) with additional cell and tissue types included. All additional expression data were obtained from the CellNet training dataset: adult liver (GEO accession: GSE41804, GSE40873, GSE38941, GSE3526, GSE26627, GSE15239, GSE14668), embryonic liver (GSE15238), colon (GSE37364, GSE8671, GSE9452), lung (GSE14334, GSE21411, GSE31210, GSE33356, GSE37768), kidney (GSE11166, GSE21374, GSE22459), neuron (GSE13564, GSE18696, GSE20589, GSE21935, GSE40438, GSE4757, GSE5281), heart (GSE21610, GSE29819, GSE3526) and skeletal muscle (GSE21496, GSE2328, GSE25462, GSE35661). It should be considered that the inclusion of additional tissues into the PCA plot shifts the relative positions of the original samples (ESCs, HLCs and PHHs) to each other.
(c). Limitations and challenges
PCA plots are often helpful to understand the architecture of larger expression changes. However, PCA can only dissect uncorrelated (orthogonal) differences in gene expression. Other dimension-reduction techniques like independent component analysis [25] or t-distributed stochastic neighbour embedding (t-SNE, see below) [26] can be used to visualize high-dimensional datasets. Graphical user interface applications are available, for example in the ‘Scater’ package (https://doi.org/10.1093/bioinformatics/btw777), that allow the application of t-SNE without advanced programming skills. Additionally, expression changes of a smaller number of genes that are only moderately up- or downregulated would usually not lead to clear shifts in the PCA plot, owing to the variability of the levels of genes whose expression is not altered during differentiation. Therefore, a lack of major shifts in the PCA plot must not be interpreted as the absence of real effects.
A challenge is the choice of the number of genes to be used for generation of the PCA plot. Usually, selection of genes is performed based on the variability of genes in the set of analysed samples. It is advisable to check PCA plots with the 50, 100, 500, 1000 and 5000 genes with highest variability in the set of analysed samples to learn if the observed patterns in a PCA plot are stable. If only few genes are altered during differentiation, it may be useful to include only small numbers, e.g. the 50 genes with highest variability. Large global expression changes as typically observed during stem cell differentiation are usually captured more reliably by including the 1000 or even 5000 genes with highest variability.
A challenge that sometimes becomes obvious at the level of explorative PCA is batch effects. In this case, specific experimental steps are responsible for the separate clustering of some samples. Typically, RNA isolation on different experimental days or hybridization on different chips is among the culprits. Usually, batch effects cause minor changes per gene but affect large numbers of genes, which may result in strong shifts in the PCA plot. Therefore, it may be helpful to reduce the genes used for the PCA to small numbers focusing on the genes with highest variability. Moreover, batch effects can be corrected or ameliorated by specific software. Therefore, it is usually possible to draw conclusions also from datasets with batch effects. Nevertheless, reproduction of the main effects in independent experiments is important.
The PCA plot shown in figure 2b illustrates one further limitation that should be kept in mind when using this technique. It is correct to conclude from this plot that the differentiation protocols caused a systematic shift of the HLCs away from the stem cells. The interpretation that there is still a large difference between HLCs and real hepatocytes (PHHs) is also correct (figure 2b). However, quantitative conclusions can be misleading, e.g. that HLCs are closer to lung or kidney than primary hepatocytes or heart. The PCA technique is definitively not adequate for quantitative analysis of cell-type identity, because it does not weight genes of central importance for a specific cell or tissue type. This challenge can be met by GRN analysis, an approach that weights genes central to a given network more heavily, as discussed in further sections of this review. However, this is not the case for PCA.
The analysis of biological motifs is not routinely included in PCA. However, studies in the field of cancer research have visualized patient cohorts, in a way that each individual patient is represented by a symbol in the PCA cloud [27–29]. Interestingly, genes associated with prognosis, such as proliferation-associated genes or genes indicating immune cell infiltration, showed gradients in the PCA cloud that could be visualized by colour codes. In the case of stem cell research, it can be expected that proliferation-associated genes should decrease during differentiation, while expression of further genes, e.g. drug-metabolizing enzymes in the case of HLCs, should increase. Visualizing genes associated with proliferation or differentiated cell functions should be easily feasible, because the individual genes defining the PCs are known. Although this can be done by programming, a user-friendly software that integrates biological motifs into PCA plots would facilitate explorative data analysis and hypothesis generation.
The aforementioned pipeline with PCA is very well suited for bulk gene expression analysis. However, as discussed below, recent developments in single-cell RNA sequencing technologies generate data with even higher dimensionality and intrinsic noise [30]. Hence, additional algorithms for dimensionality reduction have been implemented for improved data visualization. One of the most widely used is t-distributed stochastic neighbour embedding [26]. This method embeds high-dimensional data points into a space of two or three dimensions. t-SNE tries to preserve local structure of the data points, i.e. low-dimensional neighbourhood should be the same as original neighbourhood. Each data point is assigned to a map point, where the mapping is designed such that similar data points are modelled by nearby map points and dissimilar data points are modelled by distant map points. The resulting map can then be visualized in a scatter plot. The t-SNE algorithm involves two main stages. In the first step, a probability distribution over pairs of the data points is constructed using a Gaussian distribution such that similar points have a high probability of being picked by each other, while dissimilar data points have an extremely low probability of being picked. In the second step, t-SNE defines a similar probability distribution using Student's t-distribution over the map points. The algorithm then fits the locations of the points in the map to minimize the Kullback–Leibler (KL) divergence between the two distributions. Unlike PCA, t-SNE can only be used for data visualization; it is not possible to directly project a new point onto an already computed map. t-SNE has a non-convex objective function. The objective function is minimized using a gradient descent optimization that is initiated randomly. As a result, it is possible that different runs lead to different solutions.
4. Hierarchical clustering and heat map representation
(a). Definition and principles
Like PCA, unsupervised clustering and heat map presentation belong to the first steps of explorative data analysis. Genes and samples with similar expression profiles are automatically grouped together and the expression level of each individual gene is visualized by a colour code.
(b). Example of application
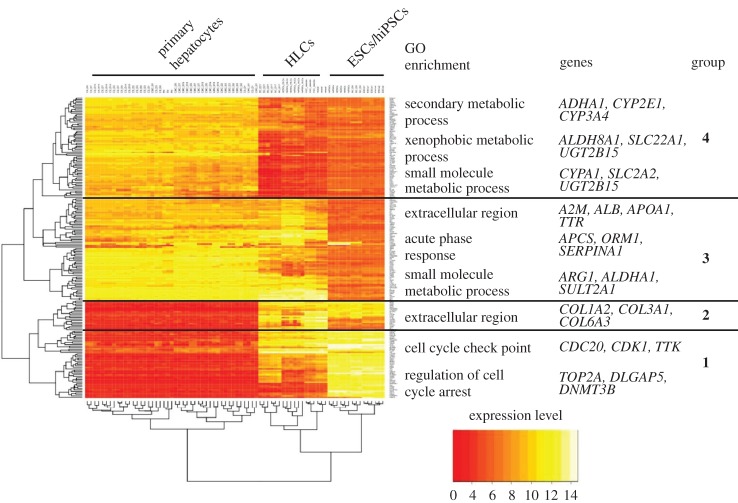
The example of hierarchical clustering and heat map representation in figure 3 includes the same samples already analysed by PCA in the previous paragraph. The 200 genes with highest variance over all samples (hESCs/hiPSCs, HLCs and PHHs) were included. Of course, smaller or larger gene numbers can also be used. In the discussed example, hierarchical clustering correctly groups all samples of PHHs, HLCs and ESCs/hiPSCs together (figure 3). So far, hierarchical clustering offers no additional information to PCA. However, a practical aspect of this representation is that ‘favourable’ and ‘unfavourable’ gene clusters can be differentiated: genes of the topmost cluster in figure 3 show high expression in PHHs and low expression in the stem cells (ESCs/hiPSCs). Unfortunately, expression in HLCs remains similarly low in HLCs and in ESCs. Therefore, genes of this cluster represent an unfortunate situation. The second uppermost gene cluster also shows high expression in primary hepatocytes and low expression in ESCs/hiPSCs (figure 3). Nonetheless, most of the genes increase during the differentiation process, some even close to the levels observed in PHHs, representing a relatively favourable situation.
Figure 3.
Hierarchical clustering and heatmap representation of the top 200 deregulated genes in primary hepatocytes, stem cells and stem cell-derived hepatocytes (from fig. 2a in Godoy et al. [23]; ArrayExpress accession: E-MTAB-4442). Functional groups were established manually based on overrepresentation of Gene Ontology (GO) annotations. Selected genes from the GO annotations are also shown.
(c). Limitations and challenges
Unlike PCA, hierarchical clustering is not helpful if only few genes are differentially expressed in a set of data. The heat map representation after hierarchical clustering is often considered a standard that should be included in a systematic analysis of genome-wide data. However, more complex clustering techniques as described in the next paragraph represent a comprehensive approach for identification of gene groups with similar features.
5. Data mining by clustering: identification of gene groups with similar features
(a). k-means and fuzzy c-means clustering
(i). Definition and principles
Clustering serves to group a dataset, e.g. expression data of genes, into so-called clusters with similar patterns. Among the numerous clustering algorithms available, k-means and fuzzy c-means are two related popular algorithms. In both approaches, the user defines the number of clusters, and both k-means and fuzzy c-means partition the genes into clusters, where genes within clusters are maximally similar. The difference between these algorithms is that fuzzy clustering allows ‘unsharp’ clustering, i.e. each gene can be assigned to multiple clusters, and it assigns those genes that are similar to different clusters into those clusters. In contrast, k-means form clear clusters, where each gene is assigned to only one cluster. In practice, clustering analysis should be typically limited to the most variable genes (e.g. top 1000 genes with highest variance). Clustering may include one cluster to which all genes are assigned that do not fit into any ‘real’ cluster. Fuzzy clustering is particularly helpful when a time course of several differentiation periods is available. However, it is also useful to analyse stem cells and derived differentiated cells generated by different centres in order to identify common features, as described below.
(ii). Example of application
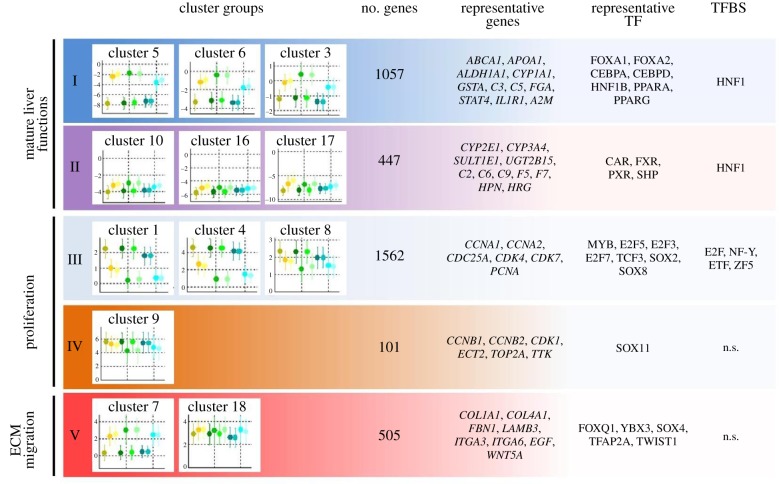
The cluster analysis in figure 4 uses the same data and colour code as already introduced in figure 2a. Fuzzy c-means clustering identified 20 clusters that were assigned to five cluster groups. Only 12 of the 20 clusters are shown, because the remaining clusters showed only very low expression differences compared with PHHs [23]. The first three symbols in yellowish colours in figure 4 represent (i) ESCs, (ii) HLCs at day 17 and (iii) HLCs at day 21, generated in one of the three involved centres (centre 1). For cluster 5 (the uppermost panel in figure 4), a scenario was obtained characterized by low expression in ESCs and a strong increase in HLCs at days 17 and 21. Similar results were generated by the two other centres represented in greenish (centre 2) and bluish (centre 3) colours. The y-axis has a log scale, where zero represents the expression level of PHHs. Therefore, cluster 5 comprises genes that are expressed by hepatocytes and were successfully induced in ESCs during the differentiation process but not completely to the level of PHHs. A similar constellation was obtained for clusters 6 and 3 with the difference that initial expression levels of ESCs were higher. By contrast, clusters 10, 16 and 17, all assigned to cluster group II, show only weak induction of gene expression in ESCs and remain orders of magnitude below the expression level of PHHs (figure 4). Therefore, cluster group II represents less successful scenarios than cluster group I. Moreover, all further clusters show specific patterns that are similar in the ESCs/hiPSCs and HLCs obtained from the three centres.
Figure 4.
Cluster groups obtained by fuzzy c-means clustering. Besides representative genes and transcription factors (TFs), over-represented transcription factor-binding sites (TFBS) are given. The colour code of the symbols is identical to that in figure 2 (from Godoy et al. [23]). Error bars represent standard deviation of the mean. n.s., no significant TFBS enrichment detected in these clusters.
(iii). Limitations and challenges
k-Means and fuzzy c-means clustering are both stochastic algorithms that may result in different clusters every time they are run. Therefore, repeated application is required to ensure stable results. Often one can separate a high number of homogeneous clusters, but to obtain an overview, it may be useful to summarize clusters with similar features into cluster groups. This grouping currently can only be done manually and is part of the interpretation process that will be described below. This process of interpreting clusters can be facilitated by over-representation analysis, but remains a relatively labour-intensive process.
6. Over-representation analysis
(a). Definition and principles
Over-representation or enrichment analysis compares a set of genes annotated to a biological motif or a pathway to a set of genes that results from differential analysis of two phenotypes, e.g. stem cells before and after differentiation. The methods evaluate whether genes annotated to a specific motif are more frequently represented among the phenotype-associated genes than expected by chance. The simplest way to generate sets of phenotype-associated genes is to establish differential gene lists, e.g. between stem cells before and after a differentiation process. However, in many cases, the use of clustering techniques is superior, because the identified gene clusters with similar properties have a higher probability of representing specific biological motifs than lists of differential genes without further processing. Among the most commonly used ontologies of annotated genes are Gene Ontology (GO) terms, which assign genes to molecular functions, cellular components or biological processes [31,32]. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways are pathway maps that represent interactions and relation networks of metabolism, information processing, cellular processes, responses to external stress and disease [33]. Also, commercial knowledge-based analysis platforms, e.g. Ingenuity Pathway Analysis (IPA), have implemented annotations for disease conditions. Moreover, several techniques are available to identify over-represented transcription factor-binding sites in a set of genes [34–36]. In addition, pathway signature databases can be used to explore deregulated (signalling) pathways [37].
(b). Example of application
Clusters of genes with similar features have been identified as described in the previous sections (figures 3 and 4). With the help of over-representation analysis, biological motifs and transcriptional regulators have been identified for these clusters. For example, the uppermost gene cluster (group 4; an ‘unfavourable’ gene cluster) in figure 3 showed a strong over-representation of metabolism-associated GO terms. A well-known phase II-metabolizing gene of this cluster is UGT2B15. Also, genes from group 3, a ‘favourable’ gene cluster, include metabolism-associated genes, such as SULT2A1, another example of a phase II-metabolizing enzyme. Therefore, already the relatively simple technique of unsupervised clustering demonstrates that some genes associated with metabolism of hepatocytes respond to the differentiation protocol (group 3 genes in figure 3), while others remain unresponsive (group 4 genes in figure 3). Cell cycle-associated GO groups are over-represented in cluster group 1 (figure 3). However, this cluster group illustrates one of the limitations of the here applied unsupervised clustering method: the same cluster (group 1) contains cell cycle-associated genes whose expression decreases during the differentiation process (a desired scenario), while further cell cycle-associated genes remain highly expressed in HLCs with levels similar to those in hESCs (representing an undesired scenario illustrated by the uppermost genes in group 1). Such ambiguity can be avoided by fuzzy c-means clustering which can assign ambiguous genes to several clusters, which is often more appropriate (figure 4). Here, cluster groups I and II (figure 4) show a strong overlap of genes presented in groups 3 and 4 of the unsupervised clustering result (figure 3), but also the proliferation-associated cluster groups III and IV (proliferation; figure 4) now have a sufficiently high degree of similarity. These are good conditions for an analysis of transcription factors and over-represented transcription factor-binding sites. The analysis suggests that a lack of transcription factors CAR, FXR and PXR may play a role for the too low levels of some metabolizing genes in cluster group 2 (figure 4). This hypothesis could be experimentally addressed by overexpression of the identified transcription factors. Similarly, the over-representation analysis suggests which transcription factors may be responsible for high expression of cell cycle-associated genes (figure 4) and the unwanted colon as well as fibroblast features of HLCs [23]. These hypotheses could be experimentally validated by knockdown of the identified transcription factors.
(c). Limitations and challenges
Estimation of TFBS over-representation is performed on a defined sequence in promoter/enhancer regions of genes, usually in regions spanning between 1000 and 2000 bp (e.g. −1500 to +500) from transcription start site. Therefore, any transcriptional regulation potentially occurring further up- or downstream of this region will not be detected by this approach. Furthermore, the software assumes that all chromatin is in an open configuration; thereby any TFBS identified in the aforementioned region is assumed to be functional. A more refined estimation of TF activity can be obtained with combined analysis of open chromatin (e.g. ATAC-seq and DNAseI-Seq) and TFBS over-representation [38].
Over-representation analysis based on publicly available annotated genes is an unsupervised, fast and usually helpful first step of gene cluster interpretation. However, it should be considered that over-representation analysis cannot fully replace manual inspection and gene-by-gene interpretation by an expert revisiting the available literature. It has become clear that the human genome contains only approximately 20 000 protein-coding genes. With some training, the human brain will remember these genes, their functions and important aspects of regulation. For comparison, it should be considered that native speakers usually understand and use approximately 40 000 words, and the vocabulary can exceed 200 000 words for individuals speaking foreign languages. After some years of research in the field of gene expression, many scientists become so familiar with genes that they read expression lists like crime novels. It should also be considered that in specific research fields, such as liver physiology and development, already the knowledge of approximately 2000 genes is sufficient for a comprehensive understanding of most functions. Computerized bioinformatics are certainly essential but are ideally used complementarily to the human brain, which intuitively generates hypotheses of possible functional interrelations.
7. Metagenes for quantitative assessment of biological processes
(a). Definition and principles
As soon as gene clusters of highly correlating genes have been characterized by over-representation analysis, they can be used for the calculation of metagenes. A metagene is defined as a pattern of gene expression that associates with a specific biological behaviour, e.g. proliferation or inflammation. Metagenes can be calculated to characterize biological processes in cell or tissue samples of interest. The normalized mean of all genes of the metagene can be used to calculate a score [24]. For example, a metagene of ‘mature liver function’ should include a representative set of genes responsible for liver-specific processes, such as endogenous and xenobiotic metabolism, synthesis of clotting factors and further proteins secreted by the healthy liver. Next, the expression value of each gene of a sample of interest is divided by the mean (or median) of healthy reference livers and the respective ratios of all genes of the list are averaged. Instead of using simple ratios, more elaborate techniques of normalization may be applied that also take into account the variance of the individual genes. If a metagene is established for a diseased state, e.g. liver inflammation, its quality will, of course, depend on the representativeness of the chosen reference tissues. Finally, metagenes can be used to quantitatively compare samples, e.g. hESCs/hiPSCs differentiated by distinct differentiation protocols to understand their degree of differentiation and also unwanted features induced by the protocol.
(b). Example of application
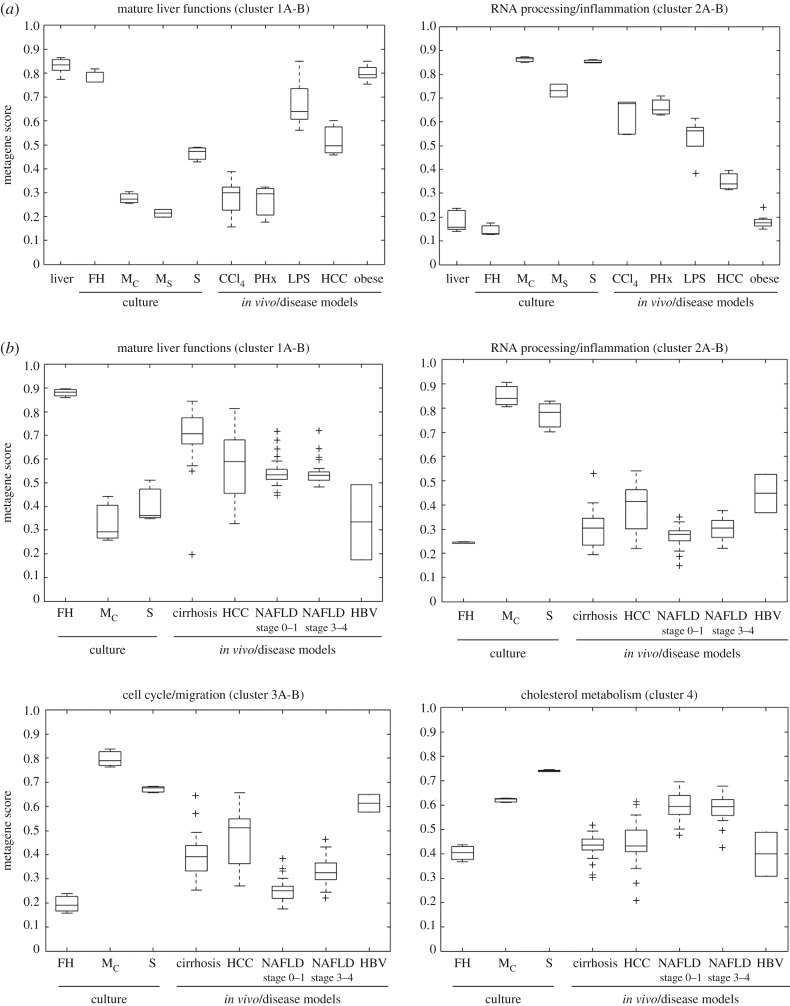
Clusters representative of all ‘mature liver functions', ‘liver inflammation’, ‘cell cycle’ and also more specific processes such as ‘cholesterol metabolism’ have been identified and were used for the calculation of metagenes [24]. This allowed the quantification of the aforementioned biological processes in various cell and disease models. For example, mouse livers after acute intoxication with the hepatotoxic compound CCl4 show a simultaneous response of strongly reduced ‘mature liver function’ and strongly increased ‘inflammation’ metagenes (figure 5a). By contrast, both metagenes remain unaltered in steatotic livers of leptin-deficient obese mice [24]. Interestingly, cultivated hepatocytes from healthy C57BL6/N mice show responses of the ‘mature liver function’ and ‘inflammation’ metagenes similar to those from CCl4-intoxicated mice, demonstrating that hepatocyte cultures represent an in vitro model of the inflamed rather than the healthy liver (figure 5a). Similarly, the metagene approach could be used to quantify, e.g. ‘mature liver functions', ‘inflammation’, ‘cell cycle activity’ and ‘cholesterol metabolism’ in liver tissue of patients (figure 5b).
Figure 5.
Characterization of cells and tissues by metagene scores, where the metagene scores are an artificial variable for all genes within a specific cluster representing a specific biological function. The calculation of the metagene score is based on the median of the scaled gene expression and the resulting value distribution of the metagene score is represented as a boxplot. The bottom and the top of a box represent the first and third quartiles of the distribution and the whiskers mark the most extreme values within 1.5 times of interquartile of the ends of the box. (a) Mouse hepatocytes and liver tissue; (b) human hepatocytes and liver tissue. FH, freshly isolated hepatocytes; Mc, hepatocytes cultivated as monolayers in a confluent state; Ms, hepatocytes cultivated as monolayers in a subconfluent state; S, hepatocytes cultivated as sandwich cultures; CCl4, mouse liver tissue after acute CCl4 intoxication; PHx, mouse livers after two-thirds hepatectomy; LPS, mouse livers after acute intoxication with lipopolysaccharide; HCC, hepatocellular cancer; obese, steatotic livers of leptin-deficient mice; cirrhosis, human cirrhotic liver tissue; NAFLD, human liver tissue of patients with non-alcoholic fatty liver disease; HBV, human liver tissue of patients with hepatitis B virus infection (from Godoy et al. [24]).
(c). Limitations and challenges
Although the metagene approach is an easy-to-use technique that can be adapted to any specific cell or animal model, a limitation remains that all genes selected for a metagene enter the score with identical weight. However, recently, GRN scores have been introduced that give individual genes different weights as described in §8.
8. Cell-type assessment by gene regulatory network-based approaches
(a). Definition and principles
A powerful example for how cell identity can be assessed based on GRNs is the CellNet platform [22,39]. The GRNs in this platform were established using gene expression profiles from over 3000 publicly available studies on diverse cells and tissues, in control (healthy) conditions and also after interventions (e.g. small interfering RNA (siRNA) and transcription factor over-expression), and were processed with the ‘context likelihood of relatedness' algorithm [40]. The GRNs were controlled by comparison with gold standards using ENCODE-generated transcription factor-binding data, expression profiles from stem cells overexpressing specific transcription factors and ChIP–CHIP/ChIP-Seq of transcription factors in stem cells (www.encodeproject.org/). The GRNs were then used to generate a metric of specific cell/tissues based on gene expression profiles, where expressed genes are weighted by their expression levels, their importance to the specific network and to the specific cell/tissue network classifier [39]. This results in a well-refined cell/tissue identity score, because it combines gene expression profiles but also estimates a gene network function based on state-of-the-art knowledge on transcription factor activity. This combined bioinformatics pipeline (transcriptomics/multivariate analysis—PCA/gene clustering/gene set enrichment analysis (GSEA)/CellNet) allows a robust and quantitative characterization of the cell identity obtained by differentiation of stem cells. The CellNet platform is publicly available and easy to use also by scientists without a specific background in bioinformatics.
(b). Example of application
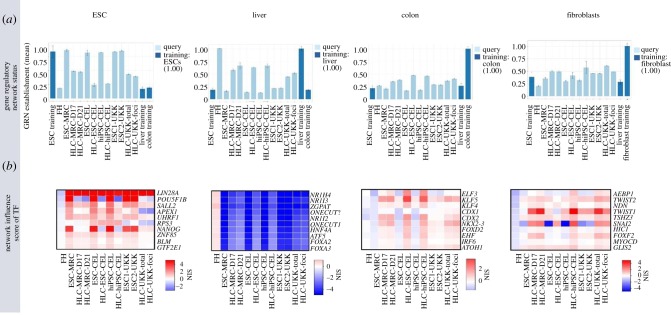
Genome-wide expression data of ESCs/hiPSCs and HLCs as described above (figure 2) were analysed by the CellNet algorithm [22]. The ESC-GRN status was close to 1.0 for the stem cells of each of the three centres, while a low ESC-GRN status of approximately 0.2 was obtained for freshly isolated PHHs and for human liver tissue (figure 6a). An interesting observation was that the ESC-GRN status of HLCs did not decrease to the level of PHHs, with the best result obtained by the protocol of centre 2 (CEL). The GRN status for liver showed an increase from approximately 0.2 (ESCs/hiPSCs) to approximately 0.6 for HLCs from the three centres, suggesting that the HLCs reached a phenotype approximately halfway between stem cells and real hepatocytes. The colon-GRN status indicates an increase for HLCs from all three centres, demonstrating that the protocols also induce an unwanted secondary differentiation. The unwanted colon-GRN status was highest for centre 2, the same protocol that most successfully suppressed the GRN status of ESCs. Possibly, successful suppression of stemness by this protocol was achieved at the expense of unwanted colon differentiation. The fibroblast-GRN status demonstrates a further secondary differentiation for all three protocols/centres.
Figure 6.
Analysis of the same samples shown in figure 2 by GRN approach. (a) GRN status for different cell and tissue types (ESCs, liver, colon and fibroblasts). Error bars represent standard error of the mean value for each tissue identity, for three replicas analysed for each sample. (b) Network influence score (NIS) of transcription factors (TF) for the individual GRNs (from Godoy et al. [23]).
A further possibility offered by the CellNet platform is the calculation of network influence scores of transcription factors (figure 6b). This algorithm identifies the transcription factors with highest influence over individual GRNs. In the colon-GRN, this analysis leads to the suggestion that knockdown of ELF3, KLF5 and KLF4 may suppress the unwanted colon differentiation (figure 6b).
(c). Limitations and challenges
Although CellNet currently is among the most powerful and practical methods to evaluate genome-wide data for similarities with specific tissues, it is important to be aware of its limitations. First, the tissue training datasets used in CellNet were generated with whole-tissue extracts, which may lead to confounding effects due to their multicellular composition. The degree of these confounding effects depends on the individual tissues. In the case of liver, the abundance of hepatocyte mRNA largely overwhelms mRNA from non-parenchymal cells, owing to the fact that hepatocytes represent the largest cell component of the liver (approx. 70%) and that hepatocytes contain approximately four times more mRNA per cell than non-parenchymal cells [13,41]. Hence, using a whole-liver expression profile is a robust reference for assessing hepatocyte differentiation in stem cells. Second, CellNet is limited to the cells/tissues that were included in the training datasets, and the normalization of the output may result in spurious tissue association if the correct tissue was not in the training set. Also, it does not yet include disease or early developmental state datasets in cells and tissues. This can be partially overcome by GO and KEGG enrichment analysis. However, additional knowledge-based curation is required for the identification of disease-related features.
9. Perspectives
(a). Knowledge-based algorithms
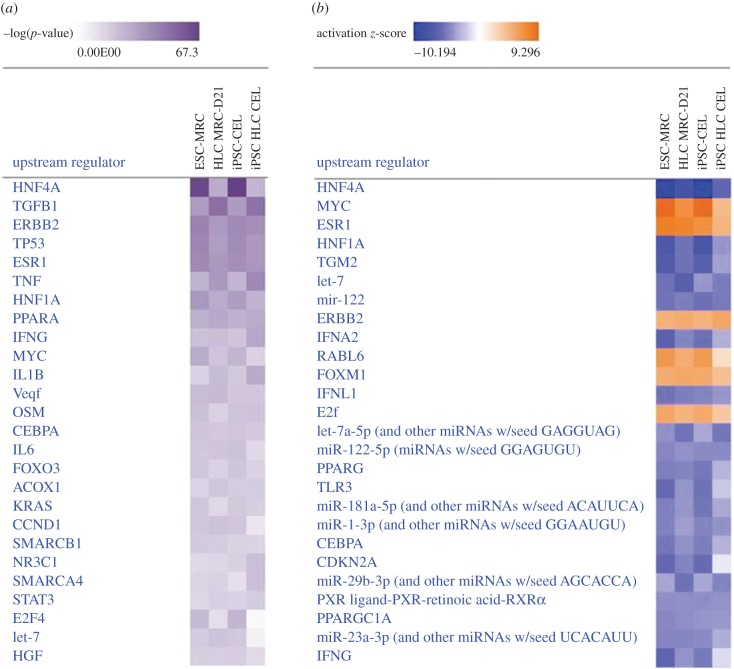
In addition to the aforementioned pipeline, knowledge-based algorithms can be used to generate robust inferences on upstream regulators associated with gene expression profiles. Upstream regulators consist of not only transcription factors, but also molecules such as cytokines, growth factors, microRNA (miRNA) and chemicals that can be associated with alterations in gene expression [42]. These algorithms integrate extensive, manually curated relationships between genes and signalling pathways controlling their expression, based on observations in multiple experimental conditions reported in the scientific literature. Hence, the strength of these algorithms will depend on the breadth of literature and the frequency of updates in the database used for each algorithm. There exists commercial (e.g. IPA, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) and open source software (e.g. DAVID, https://david.ncifcrf.gov/tools.jsp; Cytoscape, http://www.cytoscape.org/; GeneMania, http://genemania.org/). However, only IPA leverages observed cause–effect associations reported in the literature, a feature that usually is not considered in gene set enrichment analysis software. The algorithms used at IPA are supported by a database with more than 5 million findings curated from scientific literature (Ingenuity Knowledge Base) [42]. Upstream regulators can be estimated by determining the overlap of observed and predicted regulated genes for a particular regulator (using Fisher's exact test), and a z-score to assess directionality (predicted up-/downregulation) in genes composing the network related to each upstream regulator [42]. This approach has been applied to iPSC-derived mammary-like organoids and revealed novel transcriptional regulators in embryoid bodies committed to mammary gland differentiation [43]. Here, we provide an example of upstream regulators identified by IPA in two stem cell and stem cell-derived hepatocytes from our previous study [23]. The software identified HNF4 as the top-ranking transcriptional regulator both in embryonic stem cells (ESC-MRC) and in induced pluripotent stem cells (iPSC-CEL), while its score is lower in the corresponding HLCs (figure 7a). The activation z-score indicates that HNF4 ranks as the lowest active regulator in stem cells, while its activity rises in HLCs (figure 7b). This is consistent with the acquisition of hepatocyte GRNs in HLCs caused, in part, by upregulation of HNF4, and it is also consistent with the findings of TFBS enrichment analysis. In addition, IPA identifies regulators not revealed by GSEA, including further hepatocyte-specific regulators with low activity such as PPARG, CEBPA and miRNA species (e.g. miR122 and let-7), and overactive factors such as Myc, TGFb1 and ERBB2 (figure 7a,b). These factors can be added to those identified by GSEA and TFBS enrichment analysis, to generate a more robust assessment of the networks controlling the state of differentiation in HLCs. Furthermore, the relative relevance of each factor can be ranked by the corresponding z-score. However, a weakness of this algorithm is that it is biased towards best-studied transcriptional regulatory networks.
Figure 7.
Example of upstream regulator identification with Ingenuity Pathway Analysis (IPA) software. The analysis was conducted using levels of differentially expressed genes compared with levels in primary human hepatocytes (twofold cut-off, p < 0.05), in embryonic stem cells (ESC-MRC), induced-pluripotent stem cells (iPSC-CEL) and their corresponding HLCs (HLC MRC-D21 and iPSC HLC CEL). (a) The heatmap shows the −log(p-value) of the top 20 most significant upstream regulators. (b) The heatmap shows the activation z-score of the top 20 most significant upstream regulator stem cells and HLCs.
(b). Single-cell gene expression analysis
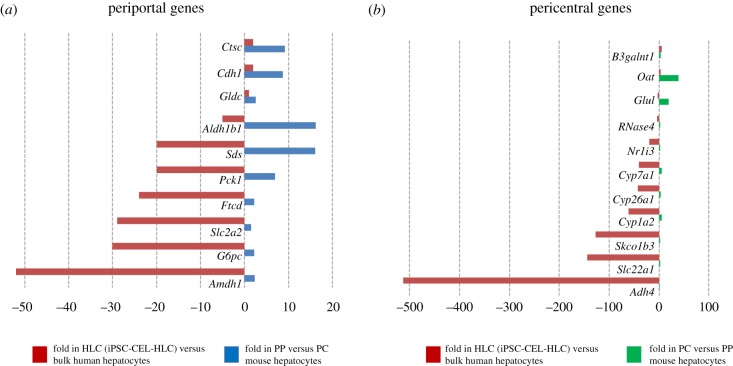
International efforts are underway to catalogue all human cell types using single-cell approaches [44]. Based on these data, more fine-grade analysis of cell-type similarities will be possible, with sets of genes clearly describing a cell type and differentiation state. Hepatocytes are known to show a zonal expression pattern in the liver lobules, where numerous genes are differentially expressed in the pericentral, midzonal and periportal regions [45,46]. Thus, it may be considered as inadequate to expect HLC cultivated under homogeneous conditions to express all zonated hepatocyte genes. Analysis in bulk HLCs of human orthologue genes to periportal (PP)- and pericentral (PC)-enriched mouse genes [45] does not suggest a bias towards a PP or PC profile (figure 8 and electronic supplementary material, table S1). While a few preferentially PP expressed genes (e.g. Ctsc, Cdh1 and Gldc) (figure 8a) and preferentially PC expressed genes (e.g. B3galnt1, Oat, Glul and RNase4) (figure 8b) showed comparable levels in HLCs versus bulk human hepatocytes, many PP (e.g. G6pc, Slc2a2, Pck1 and Sds; Fig 8a) or PC genes (e.g. Adh4, Slc22a1, Slc01b3, Cp7a1 and Nr1i3) (figure 8b) are between tens- and hundreds-fold lower in HLCs compared with bulk freshly isolated hepatocytes (electronic supplementary material, table S1). The analysis of single-cell transcriptomes might reveal small fractions of HLCs achieving a state of differentiation close to that of mature PP or PC hepatocytes, which would not be detectable in bulk RNA-Seq data.
Figure 8.
Comparison of periportal (PP) and pericentral (PC)-enriched genes (in mouse liver) with differentially expressed genes in iPSC-derived HLCs. The graphs in (a) and (b) show the fold gene expression in bulk HLCs compared with bulk freshly isolated human hepatocytes (from Godoy et al. [23]). The graphs also show the fold expression in PC- versus PP-enriched mouse hepatocytes (in (a)) and in PC- versus PP-enriched mouse (in (b)) (from Braeuning et al. [45]).
Recently, a pioneer study on scRNA-Seq analysis of iPSC-derived HLCs [47] from self-generating organoids [48] reported a rather uniform composition of HLCs [47]. Furthermore, the transcriptional profile of HLCs was closer to that of immature hepatocytes (i.e. hepatoblasts) than to mature hepatocytes [47]. This is consistent with the fact that liver zonation is achieved only after birth [49,50]. However, the HLCs in this study represent a unique differentiation procedure during self-assembly into liver bud organoids and transplantation into mouse liver [47,48]. Furthermore, RNA-Seq analysis was conducted in only a few hundred cells. Further studies are required to confidently identify subpopulations of HLCs at different maturation stages, using different methods for HLC differentiation, and higher throughput analysis of thousands of cells.
Supplementary Material
Supplementary Material
Data accessibility
This article has no additional data.
Authors' contributions
P.G., N.B., W.S.-H. and J.G.H. drafted the main text and figures. B.H., J.R., K.K., P.N., D.F. and J.W. drafted the main text.
Competing interests
The authors have no competing interests to declare.
Funding
The study was supported by the BMBF project StemNet (01EK1604A) with some contributions of the projects Liver Simulator (BMBF, 031A355A), DILI (BMBF, 031L0074F), LiSyM (BMBF, 031Loo45), LivSysTransfer (BMBF, 0101-31Q0517), InnoSysTox (BMBF/EU, 031L0021A), WISP1 (DFG, Go1987/2-1), IL-15 (GO1987/3-1), DEEP (BMBF, 01KU1216) and EUToxRisk (EU, no. 681002).
References
- 1.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386. ( 10.1016/j.cell.2010.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudhari U, Nemade H, Sureshkumar P, Vinken M, Ates G, Rogiers V, Hescheler J, Hengstler JG, Sachinidis A. 2018. Functional cardiotoxicity assessment of cosmetic compounds using human-induced pluripotent stem cell-derived cardiomyocytes. Arch. Toxicol. 92, 371–381. ( 10.1007/s00204-017-2065-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. ( 10.1038/nature08797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang ZP, et al. 2011. Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223. ( 10.1038/nature10202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. 2011. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389. ( 10.1038/nature10116) [DOI] [PubMed] [Google Scholar]
- 6.Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Hay DC. 2014. Deriving functional hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 30, 1G51-12 ( 10.1002/9780470151808.sc01g05s30) [DOI] [PubMed] [Google Scholar]
- 7.Schwartz RE, et al. 2002. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J. Clin. Invest. 109, 1291–1302. ( 10.1172/JCI15182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KD, Kuo TK, Whang-Peng J, Chung YF, Lin CT, Chou SH, Chen JR, Chen YP, Lee OK. 2004. In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology 40, 1275–1284. ( 10.1002/hep.20469) [DOI] [PubMed] [Google Scholar]
- 9.Jang YY, Collector MI, Baylin SB, Diehl AM, Sharkis SJ. 2004. Hematopoietic stem cells convert into liver cells within days without fusion. Nat. Cell Biol. 6, 532–539. ( 10.1038/ncb1132) [DOI] [PubMed] [Google Scholar]
- 10.Cai J, et al. 2007. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 45, 1229–1239. ( 10.1002/hep.21582) [DOI] [PubMed] [Google Scholar]
- 11.Banas A, Teratani T, Yamamoto Y, Tokuhara M, Takeshita F, Quinn G, Okochi H, Ochiya T. 2007. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology 46, 219–228. ( 10.1002/hep.21704) [DOI] [PubMed] [Google Scholar]
- 12.Huang P, et al. 2014. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384. ( 10.1016/j.stem.2014.01.003) [DOI] [PubMed] [Google Scholar]
- 13.Godoy P, et al. 2013. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 87, 1315–1530. ( 10.1007/s00204-013-1078-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grinberg M, et al. 2014. Toxicogenomics directory of chemically exposed human hepatocytes. Arch. Toxicol. 88, 2261–2287. ( 10.1007/s00204-014-1400-x) [DOI] [PubMed] [Google Scholar]
- 15.Ghallab A, et al. 2016. Model-guided identification of a therapeutic strategy to reduce hyperammonemia in liver diseases. J. Hepatol. 64, 860–871. ( 10.1016/j.jhep.2015.11.018) [DOI] [PubMed] [Google Scholar]
- 16.Arbo MD, et al. 2016. Hepatotoxicity of piperazine designer drugs: up-regulation of key enzymes of cholesterol and lipid biosynthesis. Arch. Toxicol. 90, 3045–3060. ( 10.1007/s00204-016-1665-3) [DOI] [PubMed] [Google Scholar]
- 17.Hengstler JG, et al. 2000. Cryopreserved primary hepatocytes as a constantly available in vitro model for the evaluation of human and animal drug metabolism and enzyme induction. Drug Metab. Rev. 32, 81–118. ( 10.1081/DMR-100100564) [DOI] [PubMed] [Google Scholar]
- 18.Gebhardt R, et al. 2003. New hepatocyte in vitro systems for drug metabolism: metabolic capacity and recommendations for application in basic research and drug development, standard operation procedures. Drug Metab. Rev. 35, 145–213. ( 10.1081/DMR-120023684) [DOI] [PubMed] [Google Scholar]
- 19.Ehrhardt S, Schmicke M. 2016. Isolation and cultivation of adult primary bovine hepatocytes from abattoir derived liver. EXCLI J. 15, 858–866. ( 10.17179/excli2016-794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhulst S, Best J, van Grunsven LA, Dolle L. 2015. Advances in hepatic stem/progenitor cell biology. EXCLI J. 14, 33–47. ( 10.17179/excli2014-576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brulport M, et al. 2007. Fate of extrahepatic human stem and precursor cells after transplantation into mouse livers. Hepatology 46, 861–870. ( 10.1002/hep.21745) [DOI] [PubMed] [Google Scholar]
- 22.Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. 2014. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158, 889–902. ( 10.1016/j.cell.2014.07.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godoy P, et al. 2015. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 63, 934–942. ( 10.1016/j.jhep.2015.05.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godoy P, et al. 2016. Gene network activity in cultivated primary hepatocytes is highly similar to diseased mammalian liver tissue. Arch. Toxicol. 90, 2513–2529. ( 10.1007/s00204-016-1761-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comon P. 1994. Independent component analysis, a new concept. Signal Process. 36, 287–314. ( 10.1016/0165-1684(94)90029-9) [DOI] [Google Scholar]
- 26.van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605. [Google Scholar]
- 27.Schmidt M, et al. 2008. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 68, 5405–5413. ( 10.1158/0008-5472.CAN-07-5206) [DOI] [PubMed] [Google Scholar]
- 28.Schmidt M, Hengstler JG, von Torne C, Koelbl H, Gehrmann MC. 2009. Coordinates in the universe of node-negative breast cancer revisited. Cancer Res. 69, 2695–2698. ( 10.1158/0008-5472.CAN-08-4013) [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M, et al. 2012. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin. Cancer Res. 18, 2695–2703. ( 10.1158/1078-0432.CCR-11-2210) [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. ( 10.1038/nrg2484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. ( 10.1093/bioinformatics/btl140) [DOI] [PubMed] [Google Scholar]
- 32.Kammers K, Lang M, Hengstler JG, Schmidt M, Rahnenfuhrer J. 2011. Survival models with preclustered gene groups as covariates. BMC Bioinformat. 12, 478 ( 10.1186/1471-2105-12-478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogata H, Goto S, Fujibuchi W, Kanehisa M. 1998. Computation with the KEGG pathway database. Biosystems 47, 119–128. ( 10.1016/S0303-2647(98)00017-3) [DOI] [PubMed] [Google Scholar]
- 34.Zellmer S, et al. 2010. Transcription factors ETF, E2F, and SP-1 are involved in cytokine-independent proliferation of murine hepatocytes. Hepatology 52, 2127–2136. ( 10.1002/hep.23930) [DOI] [PubMed] [Google Scholar]
- 35.Krug AK, et al. 2013. Human embryonic stem cell-derived test systems for developmental neurotoxicity: a transcriptomics approach. Arch. Toxicol. 87, 123–143. ( 10.1007/s00204-012-0967-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldmann T, et al. 2014. Design principles of concentration-dependent transcriptome deviations in drug-exposed differentiating stem cells. Chem. Res. Toxicol. 27, 408–420. ( 10.1021/tx400402j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schubert M, Klinger B, Klunemann M, Sieber A, Uhlitz F, Sauer S, Garnett MJ, Bluthgen N, Saez-Rodriguez J. 2018. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 ( 10.1038/s41467-017-02391-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt F, et al. 2017. Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res. 45, 54–66. ( 10.1093/nar/gkw1061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. 2014. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915. ( 10.1016/j.cell.2014.07.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 ( 10.1371/journal.pbio.0050008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hewitt NJ, et al. 2007. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev. 39, 159–234. ( 10.1080/03602530601093489) [DOI] [PubMed] [Google Scholar]
- 42.Kramer A, Green J, Pollard J, Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. ( 10.1093/bioinformatics/btt703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu Y, Han BC, Gao BW, Bose S, Gong YP, Wawrowsky K, Giuliano AE, Sareen D, Cui XJ. 2017. Differentiation of human induced pluripotent stem cells to mammary-like organoids. Stem Cell Rep. 8, 205–215. ( 10.1016/j.stemcr.2016.12.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozenblatt-Rosen O, Stubbington MJT, Regev A, Teichmann SA. 2017. The human cell atlas: from vision to reality. Nature 550, 451–453. ( 10.1038/550451a) [DOI] [PubMed] [Google Scholar]
- 45.Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M. 2006. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 273, 5051–5061. ( 10.1111/j.1742-4658.2006.05503.x) [DOI] [PubMed] [Google Scholar]
- 46.Halpern KB, et al. 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356. ( 10.1038/nature21065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camp JG, et al. 2017. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538. ( 10.1038/nature22796) [DOI] [PubMed] [Google Scholar]
- 48.Takebe T, et al. 2013. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. ( 10.1038/nature12271) [DOI] [PubMed] [Google Scholar]
- 49.Gordillo M, Evans T, Gouon-Evans V. 2015. Orchestrating liver development. Development 142, 2094–2108. ( 10.1242/dev.114215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke ZD, Reed KR, Yeh SW, Meniel V, Sansom OJ, Clarke AR, Tosh D. 2018. Spatiotemporal regulation of liver development by the Wnt/β-catenin pathway. Sci. Rep. 8, 2735 ( 10.1038/s41598-018-20888-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.